Abstract
We show that positively supercoiled [(+) SC] DNA is the preferred substrate for Escherichia coli topoisomerase IV (topo IV). We measured topo IV relaxation of (−) and (+) supercoils in real time on single, tethered DNA molecules to complement ensemble experiments. We find that the preference for (+) SC DNA is complete at low enzyme concentration. Otherwise, topo IV relaxed (+) supercoils at a 20-fold faster rate than (−) supercoils, due primarily to about a 10-fold increase in processivity with (+) SC DNA. The preferential cleavage of (+) SC DNA in a competition experiment showed that substrate discrimination can take place prior to strand passage in the presence or absence of ATP. We propose that topo IV discriminates between (−) and (+) supercoiled DNA by recognition of the geometry of (+) SC DNA. Our results explain how topo IV can rapidly remove (+) supercoils to support DNA replication without relaxing the essential (−) supercoils of the chromosome. They also show that the rate of supercoil relaxation by topo IV is several orders of magnitude faster than hitherto appreciated, so that a single enzyme may suffice at each replication fork.
Keywords: Topoisomerase IV, positively supercoiled DNA, single-molecule biophysics, type II topoisomerases, DNA replication
Topoisomerases have the daunting task of regulating the topology of megabases of DNA. In prokaryotic cells, these essential enzymes maintain the (−) supercoiling of the chromosome required for DNA replication, the transcription of many operons, and several types of recombination (for review, see Kanaar and Cozzarelli 1992; Wang, 1996, 1998). In addition, topoisomerases must remove all of the links between the complementary strands of DNA to ensure segregation of one copy of each chromosome to the daughter cells at division (for review, see Ullsperger et al. 1995).
Topoisomerases can add or remove supercoil, catenane, or knot links by catalyzing the passage of one DNA segment through a transient break in another. Type II enzymes introduce a double-strand break in one DNA duplex, termed the gate (G) segment. ATP binding allows passage of a second duplex, the transport (T) segment, through the break (for review, see Wang 1998; Baird et al. 1999). According to the two-gate model, the T segment is then released from the complex through the other end of the enzyme (Roca and Wang 1994). Hydrolysis of ATP resets the enzyme for another catalytic cycle.
Bacteria have two type II topoisomerases, DNA gyrase and topoisomerase IV (topo IV), which have specialized functions in vivo (for review, see Levine et al. 1998). Gyrase is unique in its ability to introduce (−) supercoils into DNA. It plays a critical role in maintaining the (−) supercoiling of the chromosome and acts ahead of the replication fork to remove the (+) supercoils generated by the separation of the strands of the DNA duplex. The signature activity of topo IV, on the other hand, is decatenation. In vitro, topo IV decatenates DNA ∼30-fold faster than it relaxes (−) supercoils (Ullsperger and Cozzarelli 1996). In vivo, mutants in topo IV replicate DNA at normal rates, but the replicated chromosomes and plasmids fail to separate (Khodursky et al. 1995; Zechiedrich and Cozzarelli 1995; Hiasa and Marians 1996).
There is, however, clear evidence that the role of topo IV is not limited to disentanglement and decatenation. In vivo, topo IV can remove the (+) supercoils generated during transcription ahead of the moving polymerase complex (Khodursky et al. 2000). Topo IV can also support replication elongation fully in vitro (Hiasa and Marians 1994) and partially in vivo (Khodursky et al. 2000) in the absence of gyrase. To do so, topo IV must take over gyrase' s role of removing (+) supercoils generated by replication. Although topo IV relaxes both (−) and (+) supercoils (Kato et al. 1992; Hiasa and Marians 1996), its rate of removal of (−) supercoils is only 1/20 the rate at which gyrase removes (+) supercoils (Ullsperger and Cozzarelli 1996) and nearly four orders of magnitude lower than the in vivo rate of link removal. If topo IV relaxes (+) supercoils equally poorly, it could not fill in for gyrase ahead of the replication fork. The (+) supercoils ahead of the replication fork equilibrate with (+) precatenane links behind the fork in vitro (Peter et al. 1998) and with stalled forks in vivo (Sogo et al. 1999). Therefore, topo IV could, in principle, support replication elongation by removing the precatenane links behind the fork. It is not known, though, whether this equilibration occurs with active replication forks in vivo (Postow et al. 1999).
We tested directly whether topo IV could remove (+) supercoils at a rate comparable to that of gyrase. The availability of methods to readily produce highly (+) supercoiled DNA (Musgrave et al. 1991; Strick et al. 1996) allowed us to compare the activity of topo IV with (−) and (+) supercoiled substrates in vitro. We measured topo IV relaxation in real time on tethered, single DNA molecules to complement conventional ensemble measurements. The former method revealed a pulsative relaxation by single enzyme molecules and detected individual relaxation events. We found a marked preference of topo IV for (+) supercoils. Only (+) supercoils were removed at low enzyme concentration, the enzyme was strikingly more processive on (+) SC DNA, and DNA cleavage promoted by quinolone antibiotics was much more pronounced with (+) SC DNA.
This strong preference for (+) SC DNA helps to resolve two paradoxes about topo IV activity. It has been difficult to understand why (−) supercoil relaxation, an intramolecular event, was so much slower than the intermolecular decatenation reaction (Ullsperger and Cozzarelli 1996). Equally puzzling was how the cell could have an enzyme active enough to remove (+) links fast enough to support replication and transcription and yet retain the essential (−) supercoiling of the chromosome. Our results indicate that (+) supercoil removal and decatenation are the highly efficient in vivo functions of topo IV. The single-molecule measurements in particular showed that the activity of topo IV has been vastly underestimated, so that only one or a few enzyme molecules would be sufficient at each replication fork. In contrast, (−) supercoiled DNA is a poor substrate for topo IV. We propose this is because of an intrinsic chirality in the orientation of the T and G segments on the enzyme that is disfavored by (−) supercoiling.
Results
Topoisomerase IV preferentially relaxes (+) supercoiled DNA
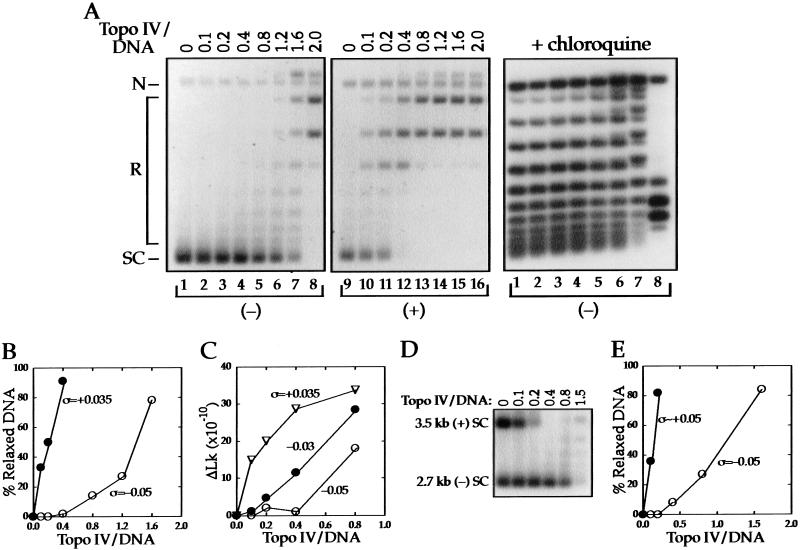
We compared the amount of topo IV needed to relax (+) and (−) SC 2.7-kb DNAs in an enzyme titration experiment. Increasing amounts of topo IV were incubated separately with the two substrates and the reactions analyzed by agarose gel electrophoresis (Fig. 1A). With (−) SC DNA (ς = −0.05; lanes 1–8), there was little relaxation at stoichiometries much less than one. In contrast, topo IV relaxation of the (+) SC DNA (ς = +0.035; lanes 9–16) began at the lowest stoichiometry tested. We also analyzed the reactions with (−) SC DNA on a gel containing chloroquine (right), which resolves the DNA sufficiently to detect relaxation of individual topoisomers. However, no relaxation was detected in the range of stoichiometries at which the (+) SC DNA was completely relaxed (lanes 1–4).
Figure 1.
Relaxation of (−) and (+) supercoiled DNA by topo IV. (A) Negatively (ς = −0.05) supercoiled (lanes 1–8) and (+) (ς = +0.035) supercoiled (lanes 9–16) 2.7-kb DNAs were incubated with the indicated molar ratios (top) of topo IV to DNA. The reacted DNA was analyzed by agarose gel electrophoresis, and autoradiograms of Southern blots of the gels are shown. The positions of nicked (N), relaxed (R), and supercoiled (SC) DNA for the first two panels are indicated. In the panel at right, reactions shown in lanes 1–8 were analyzed on a gel containing 1 μg/mL chloroquine to resolve the topoisomers. Relaxation, which results in a shift in the distribution of topoisomers and the appearance of new bands, is first detected in lane 5. (B) Quantification of the data from the left and middle panels in A. The percent of relaxed DNA was calculated from the disappearance of DNA from the supercoiled band. (C) Three DNA substrates of different ς values were reacted with topo IV and relaxation evaluated by agarose gel electrophoresis in the absence (ς = +0.035) or presence (ς = −0.03 and −0.05) of 1 μg/mL chloroquine. The ΔLk was calculated as described in Materials and Methods. (D) Negatively supercoiled 2.7-kb (ς = −0.05) and (+) SC 3.5-kb (ς ∼ +0.05) DNAs were mixed and reacted with increasing amounts of topo IV. An autoradiogram of a Southern blot of the portion of the agarose gel containing the supercoiled substrates is shown. (E) Quantification of the data from D measured by the disappearance of DNA from the supercoiled bands.
The relaxation activity of topo IV was quantified by measuring the disappearance of the supercoiled substrates as a function of stoichiometry (Fig. 1B). The relaxation of (+) SC DNA is linear with enzyme concentration, but the curve is sigmoidal with (−) SC DNA, in which we detected no activity at low enzyme concentration. Approximately 10-fold more topo IV was needed to relax the same amount of (−) SC DNA as the (+) SC DNA relaxed by the lowest enzyme concentration tested.
Because the (+) and (−) SC DNAs used in this experiment had different absolute values of ς, it was possible that the preferential relaxation of the (+) SC DNA was due to the lower number of supercoils in that substrate. To address this, we carried out a topo IV titration using two (−) SC DNAs whose |ς| values bracketed that of the (+) SC DNA. To compare topo IV activity on the three substrates, we quantified the number of supercoils removed from the substrate at each enzyme concentration rather than just the fraction of DNA that was relaxed (see Materials and Methods). The results given in Figure 1C show that many more supercoils are removed from the (+) SC DNA than from either of the (−) SC DNAs at all stoichiometries tested. Thus, preferential relaxation of the (+) SC DNA at low stoichiometry is not due to its lower |ς| value compared with (−) SC DNA.
Interestingly, more enzyme was needed to relax the final (+) supercoils, as shown by the plateau in the curve with (+) SC DNA after removal of only 80% of the substrate links. This experiment also showed that the ς = −0.05 DNA was a poorer substrate than the ς = −0.03 DNA; the threshold level of enzyme for relaxation was higher.
We next determined whether topo IV could discriminate between (−) and (+) SC substrates present in the same reaction. We carried out an enzyme titration with (−) SC 2.7-kb DNA (ς = −0.05) and (+) SC 3.5-kb DNA (ς ∼ +0.05). The size difference in the substrates allowed us to analyze the DNAs simultaneously by agarose gel electrophoresis (Fig. 1D). The (+) SC substrate was completely relaxed before any disappearance of the (−) SC DNA band was detected. Quantification of the data (Fig. 1E) gives a result very similar to that shown in Figure 1B, in which the DNAs were reacted separately. Relaxation was linear with enzyme concentration for (+) SC DNA but required a threshold amount of topo IV before relaxation of the (−) SC DNA was detected. Approximately 10-fold more topo IV was needed with (−) SC DNA to match (+) SC DNA relaxation at the lowest enzyme concentration. Comparable results were obtained in the reciprocal mixing experiment, in which the 2.7-kb DNA was (+) SC and the 3.5-kb DNA was (−) SC (data not shown).
To verify that the difference in relaxation of (−) and (+) SC DNA was not limited to our preparation of topo IV, we performed an enzyme titration with topo IV provided by H. Hiasa (University of Minnesota, Minneapolis). Relaxation of the (−) SC substrate again showed a threshold and required 10-fold more enzyme to relax the same percent of the substrate (data not shown). The specific activity of this topo IV was even higher than that of our enzyme, so that both substrates were completely relaxed by substoichiometric amounts of topo IV.
Topo IV relaxes (+) supercoiled DNA at a 20-fold faster rate than (−) supercoiled DNA
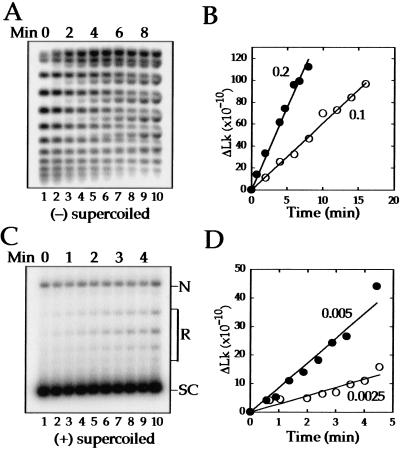
Next, we turned to kinetic experiments to measure the relaxation rates of (−) and (+) supercoils as well as the processivity of the reactions. In these experiments, we used a fivefold higher concentration of DNA, which we determined was a saturating amount of substrate for both (+) and (−) SC DNA. Under these reaction conditions, we measured maximal rates and observed relaxation at lower stoichiometries than in the titration experiments above. Topo IV was added to (−) SC 2.7-kb DNA (ς = −0.05) at stoichiometries of 0.1 or 0.2 and the DNA analyzed by agarose gel electrophoresis in the presence of chloroquine (Fig. 2A). The chloroquine relaxes the substrate sufficiently to resolve the topoisomers (lane 1). As supercoils are removed, the topoisomers become (+) SC and migrate between the substrate topoisomers. The quantification in Figure 2B shows that (−) SC DNA is relaxed linearly with time over the course of the experiment. The rate obtained at 30°C of one strand passage/enzyme/min at both stoichiometries is slightly faster than the rate of 0.3 strand passages/enzyme/min at 23°C reported previously (Ullsperger and Cozzarelli 1996).
Figure 2.
Kinetics of relaxation of (−) and (+) supercoiled DNA by topo IV. (A) Negatively supercoiled 2.7-kb DNA (ς = −0.05) was reacted at a molar ratio of 0.2 topo IV/DNA, and samples were removed at one-min intervals. Relaxation was analyzed by electrophoresis through an agarose gel containing 1 μg/mL chloroquine. An autoradiograph of a Southern blot of the gel is shown. (B) Quantification of relaxation of the (−) SC substrate at molar ratios of 0.1 (○) and 0.2 (●) topo IV/DNA. The rates of relaxation were one strand passage/enzyme/min at both stoichiometries. Negative supercoils were removed at a lower stoichiometry than in Fig. 1, because the DNA concentration was fivefold higher. (C) Positively SC 2.7-kb DNA (ς ∼ +0.05) was reacted with topo IV at a stoichiometry of 0.0025 and samples were removed at 30-sec intervals. Relaxation was analyzed by agarose gel electrophoresis. An autoradiograph of a Southern blot of the gel is shown. The positions of nicked DNA (N), relaxed topoisomers (R), and (+) supercoiled DNA (SC) are indicated. (D) Quantification of the data for relaxation of the (+) supercoiled substrate at stoichiometries of 0.0025 (○) and 0.005 (●). The rates of relaxation were 19 and 29 strand passages/enzyme/min at stoichiometries of 0.0025 and 0.005, respectively.
The processivity, or the number of strand passages per formation of a Michaelis complex, is low with a (−) SC substrate. The concurrent change in Lks for all the topoisomers (Fig. 2A) demonstrates the distributive nature of the reaction, in which the enzyme relaxes only one or a few supercoils before releasing from the substrate, as reported previously (Ullsperger and Cozzarelli 1996). To estimate the processivity, we assume for simplicity that the processive removal of subsequent links is fast compared with the rate of removal of the first link (R1). Therefore, the processivity (P), equals RT/R1, in which RT is the overall rate. To avoid the complication of a topoisomer being both a substrate and a product, we measured R1 from the disappearance of the two bands with the highest |ς|, as they cannot be generated from any other topoisomer. We obtained an average value for R1 of 0.7 strand passages/enzyme/min. From the measured value of RT of 1, we calculate that P = 1.4. Thus, the enzyme is almost perfectly distributive (P = 1).
To measure the topo IV relaxation rate with (+) SC DNA (ς ∼ +0.05), we needed much lower stoichiometries; 0.0025 and 0.005 (Fig. 2C). There is a striking increase in the processivity of the enzyme with the (+) SC substrate. Partially relaxed molecules were not observed even when the reactions were analyzed on a gel containing netropsin, which overwinds the DNA and increases resolution of the (+) topoisomers (data not shown). Moreover, fully relaxed DNA appears at the earliest time point when most of the substrate is unreacted.
Quantification of the data (Fig. 2D) gives relaxation rates of 19 and 29 strand passages/enzyme/min at stoichiometries of 0.0025 and 0.005, respectively. Thus, on average, the overall rate (RT) is 24-fold faster with (+) SC than with (−) SC DNA. We were unable to measure R1 directly because we cannot resolve all of the (+) topoisomers. However, to remove all 12 of the supercoils in this substrate processively, P must be at least six strand passages. R1 is then about four strand passages/enzyme/min for this (+) SC DNA. Thus, topo IV works faster on a (+) SC DNA and carries out more strand passages once it binds.
Topo IV preferentially cleaves (+) supercoiled DNA in a competition experiment
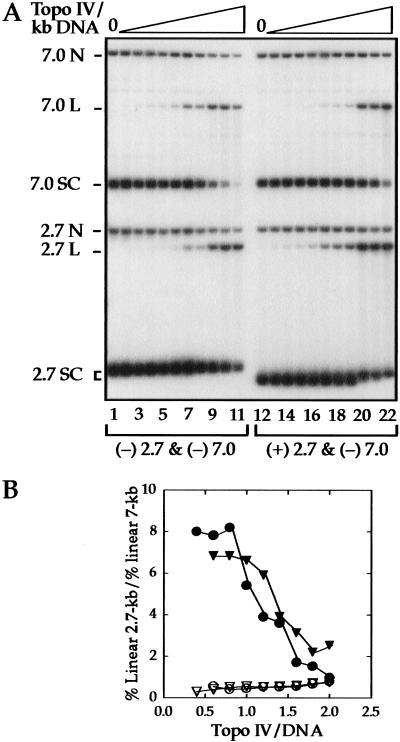
To determine whether topo IV can distinguish (+) SC DNA prior to strand passage, we measured DNA cleavage by topo IV. In the presence of the drug norfloxacin, the cleaved substrates are trapped as complexes in which the double-strand break in the DNA is bridged by the enzyme. Addition of SDS denatures the protein and releases DNA covalently attached to the ParC subunit of topo IV. There are several advantages to a cleavage assay. First, cleavage is an early step in the reaction, occurring before strand transport, DNA religation, and enzyme turnover. Second, processivity is irrelevant, because cleavage is stoichiometric with enzyme. Third, cleavage is simply and accurately quantified.
We used a competition assay in which both a 7-kb (−) SC (ς ∼ −0.05) and a 2.7-kb (+) SC DNA (ς = +0.035) were present in the same reaction. A control contained the same (−) SC 7-kb DNA, but a 2.7-kb DNA that was (−) SC (ς = −0.05) instead of (+) SC DNA. The reactions were analyzed by agarose gel electrophoresis (Fig. 3A), and cleavage was evaluated as the percent of linear DNA molecules.
Figure 3.
Competition between (−) and (+) supercoiled DNAs for cleavage by topo IV. (A) Equal amounts of 2.7- and 7-kb plasmids were incubated with ATP and increasing amounts of topo IV in the presence of norfloxacin to trap enzyme–DNA cleavage complexes. After addition of SDS and deproteinization, the reacted DNA was analyzed by agarose gel electrophoresis in buffer containing ethidium bromide. An autoradiograph of a Southern blot of the gel is shown. Cleavage was measured by the percent of the DNA in linear form. In lanes 1–11, both the 2.7-kb (ς = −0.05) and the 7-kb DNAs were (−) supercoiled. The percent linear DNA in lane 6, for example, was 1.8 for the 2.7-kb DNA and 4.0 for the 7-kb DNA. In lanes 12–22, the 2.7-kb DNA was (+) supercoiled (ς = +0.035) and the 7-kb DNA was (−) supercoiled. The percent linear DNA in lane 17 was 8.7 for the 2.7-kb and 2.4 for the 7-kb DNA. The position of nicked (N), linear (L), and supercoiled (SC) DNA for each substrate is indicated. The number of molecules of topo IV per kilobase of DNA ranged from ∼0.1–0.8. (B) Quantification of the competition cleavage data for reactions carried out in the presence or absence of ATP, expressed as the ratio of cleaved 2.7-kb to cleaved 7-kb substrate. For comparison with other experiments, the topo IV/DNA ratios are based on all of the DNA being 2.7 kb. (+) SC 2.7-kb DNA and (−) SC 7-kb DNA with (●) and without (▾) ATP; (−) SC 2.7-kb and (−) SC 7-kb DNA with (○) and without (▿) ATP.
The ratio of cleaved 2.7-kb DNA to cleaved 7-kb DNA for both sets of reactions is plotted in Figure 3B. In the control, in which both DNAs were (−) SC, the DNAs were cleaved at a frequency proportional to their lengths, 0.4, as expected. However, when the 2.7-kb DNA was (+) SC, it was cleaved at an eightfold higher frequency than the (−) SC 7-kb DNA at low enzyme to DNA ratios. After correction for DNA length, this corresponds to 20-fold higher cleavage of the (+) SC DNA. As the enzyme concentration was increased, the difference between (+) and (−) SC substrates progressively diminished.
We conclude that the preferential action of topo IV on (+) SC DNA can be prior to strand passage. This discrimination does not require ATP, as the same preferential cleavage of (+) SC DNA was observed in the absence of nucleotide (Fig. 3B). Thus, the recognition of substrate chirality by topo IV is distinguished from the ability of type II topoisomerases to remove DNA topology below equilibrium values (Rybenkov et al. 1997), because that activity requires ATP. The ability of topo IV to discriminate between (+) and (−) supercoils must involve a different aspect of the topo IV reaction mechanism.
Topo IV relaxation of (+) and (−) supercoils in single DNA molecules
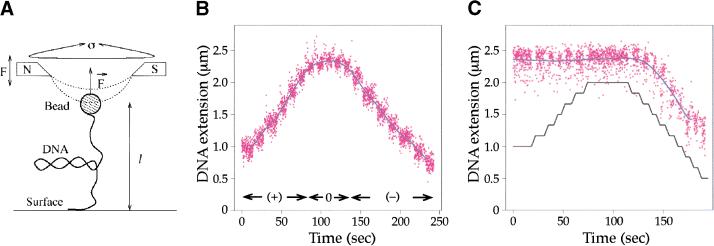
The recent development of techniques to observe enzymes acting on single DNA molecules offered us the opportunity to measure topo IV relaxation of (+) and (−) supercoils on the same substrate molecule (Strick et al. 1996, 1998). In this system, we used a DNA molecule tethered at one end to the floor of a capillary tube. The free end of the DNA is linked to a superparamagnetic bead that can be rotated by external magnets to introduce (−) or (+) plectonemic (interwound) supercoils in the DNA (Fig. 4A). Thus, at a given concentration of topo IV, we could compare the relaxation of (−) and (+) supercoils alternately on the same DNA molecule.
Figure 4.
Relaxation of a single supercoiled DNA molecule by topo IV. (A) Schematic of the experimental system. Here the tethered DNA is stretched by a weak force (F = 0.2 pN) and plectonemic supercoils are generated by rotating the magnets (designated N and S) above the bead. Enzyme-mediated relaxation of supercoils is monitored by measuring the change in extension, l, of the DNA. (B) DNA extension as a function of added supercoils. When the magnets are rotated counterclockwise or clockwise, (+) or (−) supercoils, respectively, are added to the DNA molecule, and its extension changes. The raw data (distance of the bead from the surface in successive video frames) are given in magenta and the filtered data as a blue line. After 40 counterclockwise turns of the magnets, the DNA molecule is (+) supercoiled at time 0. As the magnets are rotated clockwise, (+) supercoils are removed and the extension of the DNA increases. Then, as (−) supercoils are added, the DNA shortens. For the force and ionic conditions used here, a change in extension of 0.12 μm corresponds to two supercoils added or removed. (C) Positive supercoils but not (−) supercoils are relaxed at low enzyme concentration, a Sisyphean effect. At time zero, the reaction mixture contained 50 ng/mL topo IV, 1 mM ATP, and relaxed DNA. When the magnets were rotated counterclockwise (0 to ∼70 sec) to introduce (+) supercoils, the extension of the molecule did not change, indicating that topo IV was relaxing the supercoils as fast as the magnets could add them. When the magnets were rotated clockwise (−) supercoils accumulated (120–190 sec), and the DNA extension decreased, as if no enzyme were present. The black lines indicate steps of five turns.
The number of supercoils in the molecule is known precisely from the extension of the DNA. Because the conformation of SC DNA is force dependent, we carried out our experiments at low force (0.2 pN), in which the molecules should have a plectonemic conformation as found with circular molecules in solution. The molecule is fully extended for the given force when ς = 0 and shortens as (+) or (−) supercoils are introduced. This is shown in Figure 4B, in which the DNA starts out (+) SC, but the counterrotation of the magnets first relaxes and then (−) supercoils the DNA. A data point (magenta) is taken every 40 msec, and the blue line is a filtered average of the data. In our first test of topo IV chirality using this setup, we added topo IV to a relaxed DNA molecule. When the magnets were turned counterclockwise at 0.8 turns/sec to introduce (+) supercoils, there was no change in the extension of the DNA, because relaxation was faster than supercoil addition (Fig. 4C). In contrast, when the direction of rotation of the magnets was reversed to introduce (−) supercoils, the extension of the DNA decreased as though no enzyme were present, because the activity of the enzyme was insufficient to remove supercoils at the rate at which they were added (Fig. 4C). If the direction of rotation of the magnets was again reversed, (+) supercoils again did not accumulate (data not shown), indicating that the enzyme was still active. Thus, at a low concentration of topo IV (50 ng/mL), the preferential relaxation of (+) supercoils was complete.
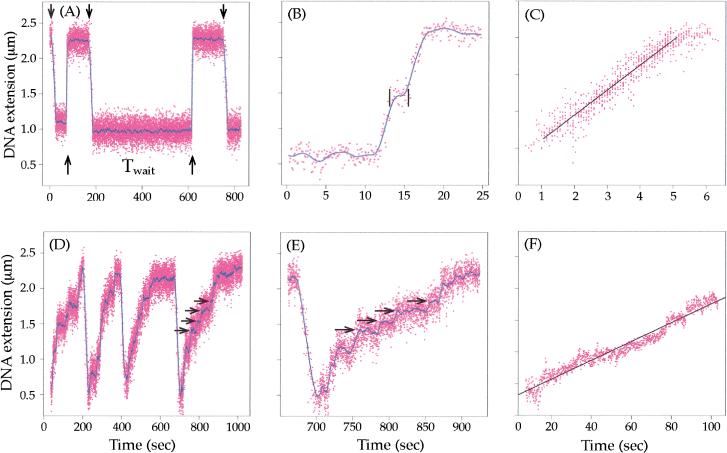
To measure accurately the rate of removal of (+) supercoils, we used a concentration of enzyme that was low enough (10 ng/mL) that the waiting time before the initiation of relaxation was long relative to the time for the enzyme to fully relax the DNA (Fig. 5A). This should ensure that there is only one active topoisomerase per DNA. In these conditions, we saw a rapid and processive removal of (+) supercoils (Fig 5A). Out of a total of 15 experiments, all of the (+) supercoils were processively removed in 10. In the remaining five, the supercoils were removed in two bursts, separated by a small pause (Fig. 5B). As the maximal number of supercoils added in these experiments was 30, we conclude that the processivity of topo IV on (+) SC DNA is on the order of 15 strand passages. The average relaxation from these 15 experiments, after editing out waiting times and pauses much longer than the relaxation time, is shown in Figure 5C. This curve describes the enzyme when it is acting completely processively. Relaxation is linear with time at a rate of 180 strand passages/min. Note that the rate is independent of ς from the initial value of +0.03 to near-complete relaxation. When the rate was slowed down by reducing the ATP concentration to 10 μM, the characteristic steps-of-two change in Lk were observed (data not shown).
Figure 5.
Single-molecule measurements of the rate of (+) and (−) supercoil relaxation by topo IV. For all panels, the raw data are given in magenta and the filtered data (A,B,D,E) in blue. The raw data were filtered at 0.2 Hz in A, D, and E, and at 0.8 Hz in B. The best fit lines to the data in C and E are in black. (A) Relaxation of (+) supercoils by 10 ng/mL topo IV was monitored by the change in the DNA extension. Arrows pointing down indicate the start of mechanical overwinding of the DNA to ς = +0.03 (30 turns), and arrows pointing up show the initiation of relaxation. A long waiting time (Twait) between the introduction of supercoils and the onset of relaxation is indicated. (B) Relaxation of (+) supercoiling in two bursts, separated by a small pause located between the vertical black lines. Such small pauses were edited out from the data shown in C. (C) Composite data of 15 bursts of (+) supercoil relaxation, totaling about 140 enzymatic cycles, are plotted. The best linear fit to the data yields the rate of supercoil removal of 180 strand passages/min. In this analysis, waiting times and pauses longer than the relaxation time were removed (Strick et al. 2000). (D) Relaxation of (−) supercoils by topo IV. The DNA at time zero was negatively supercoiled to ς = −0.035, and relaxation by 200 ng/mL topo IV was monitored as in A. Four cycles of (−) supercoiling followed by relaxation are shown. The horizontal arrows show the change in extension corresponding to a ΔLk of +2, a single enzyme turnover. (E) Blow-up of the last relaxation event in D. (F) Composite data of ten bursts of relaxation of (−) supercoils, totaling ∼80 enzymatic cycles, are shown. The slope of the best fit line yields the rate of supercoil removal of eight strand passages/min. In this analysis, waiting times and pauses were not removed as they are not clearly differentiated from the slow relaxation events.
When (−) supercoils were introduced into the DNA at the same enzyme concentration (10 ng/mL), no relaxation events were observed. By increasing the concentration to 200 ng/mL, we were able to observe slow relaxation of (−) supercoils (Fig. 5D). At this high concentration of enzyme, the waiting time before relaxation was always shorter than the relatively slow relaxation times. Thus, we cannot rule out the possibility that under these conditions more than one enzyme is acting on the (−) SC DNA. Nevertheless, individual relaxation events are clearly seen throughout relaxation (horizontal arrows in Fig. 5E), in agreement with the distributive mechanism observed in the ensemble kinetics. Average data from 10 experiments are given in Figure 5F, yielding a rate of eight strand passages/min, 23-fold slower than the rate of (+) supercoil relaxation. Because the waiting times were less than the relaxation time, waiting times were not edited out. If more than one enzyme is acting on each DNA, then relaxation of (−) SC DNA would be even poorer.
There was no concentration of topo IV at which we could measure the relaxation rate of both (−) and (+) supercoils. With sufficient topo IV to measure (−) supercoil relaxation, (+) supercoils were relaxed as fast as they were added. When the topo IV concentration was low enough to measure relaxation of (+) supercoils by a single enzyme, no relaxation of (−) supercoils was seen.
Discussion
Using single-molecule and ensemble enzymology, we find that Escherichia coli topo IV greatly prefers (+) SC over (−) SC DNA. This preference was exhibited in four types of measurements that highlight different aspects of the reaction. First, topo IV cleaved (+) SC DNA more frequently than (−) SC DNA in a competition assay. Second, topo IV relaxed only (+) supercoils at low enzyme concentrations. Third, the enzyme removed (+) supercoils an order of magnitude more processively than (−) supercoils. Fourth, topo IV relaxed (+) supercoils at a 20-fold faster rate than (−) supercoils, largely due to the increased processivity with (+) SC DNA.
We begin by discussing the complementary advantages of the single-molecule and ensemble methods we used. Next, we present a model to explain our findings and discuss our results in the context of the model. Finally, we consider the physiological implications of our findings.
Complementarity of single-molecule and ensemble enzymology
Single-molecule measures of topoisomerase function provided a number of advantages over conventional kinetic measures. First, the same DNA molecule can be (+) or (−) supercoiled, so that the experiments are precisely controlled. The substrate is a single topoisomer, whose ς value is known exactly. Second, rates are measured only with active enzyme molecules. Third, the range of rates that can be measured is very large and extends to individual strand passages by a single enzyme. Fourth, the processivity of the enzyme can be measured directly and separately from the overall reaction rate. Fifth, and most remarkably, the enzyme is revealed to act in bursts, separated by variable periods of waiting or pausing. In ensemble measurements, the periods of activity and inactivity are averaged out.
Traditional population measures have a complementary advantage in being averages over a large number of enzyme molecules. They are also more amenable to varying the concentration of enzyme and substrate. Ensemble methods allowed us to measure DNA cleavage and the competition between substrates.
The agreement of our topo IV results with the distinctive single-molecule and ensemble methods adds substantial credence to our conclusions.
Model for the preferential relaxation of (+) supercoils by topo IV
Our results show that a feature of (+) SC DNA allows selective cleavage and processive relaxation of a (+) SC substrate by topo IV. Gyrase, too, is far more active on (+) SC DNA than on relaxed or (−) SC DNA (Westerhoff et al. 1988; Kozyavkin et al. 1990). However, substrate discrimination by gyrase is due to a chiral wrapping of 135 bp of DNA around the enzyme that is favored by (+) supercoiling (Morrison et al. 1980; Kirkegaard and Wang 1981). The footprint of topo IV on DNA, in contrast, is only 34 bp (Peng and Marians 1995), far too small for a stable wrap. Therefore, topo IV must use a different mechanism to discriminate between (−) and (+) SC substrates.
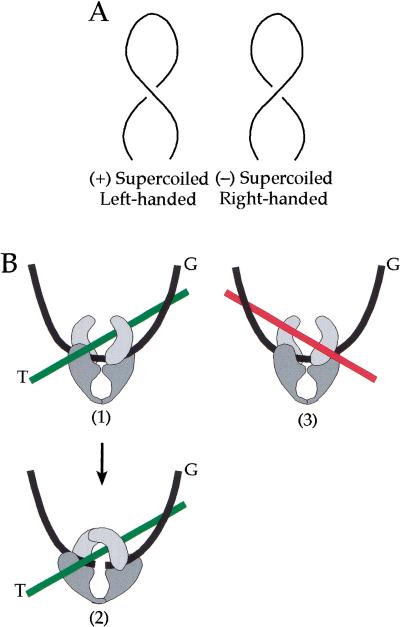
Supercoiled DNA forms a plectonemic superhelix with sharp apical bends (Fig. 6A) (Vologodskii and Cozzarelli 1994). The ascending and descending helical paths are antiparallel in both (+) and (−) SC DNA, but differ in the handedness of the superhelix; left for (+) SC DNA and right for (−) SC DNA. We propose that it is a local geometric arrangement of the DNA segments in (+) SC DNA that is the basis for the recognition of substrate chirality by topo IV. We do not believe the enzyme detects directly the topological sign of supercoiling, because it is a global property of the DNA molecule, dependent on the connectivity of crossing segments as well as the local geometry.
Figure 6.
Model for the differential binding of topo IV to (+) and (−) supercoiled DNA. (A) Both positively (left) and negatively (right) plectonemic supercoiled DNAs have sharply bent apical regions and antiparallel crossings of the DNA but different local DNA geometry. The superhelix of (+) SC DNA is left-handed, whereas that of (−) SC DNA is right-handed. (B) Topo IV is depicted as a clamp-like structure by analogy to yeast topo II (Berger et al. 1996). The black line represents the G segment, which is bent as a consequence of topo IV binding. (1) When the DNA is (+) SC, a second DNA segment binds properly as the T segment (green line) in a left-handed relationship to the G segment. (2) The proper binding of the G and T segments allows cleavage of the G segment to take place. (3) When the DNA is (−) SC, the second segment (red line) is at the wrong angle relative to the G segment to bind properly as the T segment. However, there is an interaction between the second segment and the enzyme that stabilizes the complex, but in a nonproductive way.
To recognize the geometry of the superhelix, the enzyme may bind to two DNA segments. These could be the segments at a crossing of the upward and downward paths or the juxtaposition of the apex of the superhelix with another segment. Type II topoisomerases have been visualized by electron microscopy preferentially bound at both locations in (−) SC DNA; Drosophila and calf thymus topo II at interior crossings in earlier studies (Zechiedrich and Osheroff 1990) and topo IV and yeast topo II at apices in more recent investigations (A. Vologodskii, W. Zhang, V. Rybenkov, A. Podtelezhnikov, D. Subramanian, J. Griffith, and N. Cozzarelli, in prep.). The latter two enzymes were also shown to bend the DNA upon binding, presumably to the G segment (A. Vologodskii, W. Zhang, V. Rybenkov, A. Podtelezhnikov, D. Subramanian, J. Griffith, and N. Cozzarelli, in prep.). Because the bend is roughly planar, the enzyme should bind the G segment about equally well with (−) and (+) SC DNA, barring a large change of twist upon binding. Therefore, G-segment binding is unlikely to be the basis for discrimination between (−) and (+) SC substrates. We propose instead that it is the proper binding of the T segment relative to the G segment that is favored with (+) SC DNA.
For topoisomerase action, the G and T segments need to be bound in a particular geometry, much like the assembly of a synaptic complex for site-specific recombination (Craigie and Mizuuchi 1986; Boocock et al. 1987). Moreover, there is evidence with Drosophila topo II that binding of both segments is required even for cleavage to occur (Corbett et al. 1992). A similar conclusion was reached for DNA gyrase, which is structurally homologous to topo IV (Kampranis et al. 1999). In Figure 6B, we show how the local geometry of (+) SC DNA could favor T-segment binding. In panel 1, after one DNA segment has bound to the enzyme as the G segment, the T segment can then be readily bound in a juxtaposition to the G segment that we postulate is favored by (+) supercoiling. After proper binding of the T segment, the G segment could be cleaved (panel 2). Because cleavage does not require ATP, topology discrimination can be made in the absence of ATP, as we observed in our cleavage experiment. The addition of ATP results in transport of the T segment through the cleaved G segment and release of the T segment from the enzyme (for review, see Wang 1998).
In (−) SC DNA, after binding of the G segment, the crossing DNA segment would then be at the wrong angle to bind correctly as the T segment (panel 3). However, because topo IV binds preferentially to (−) SC DNA compared with relaxed DNA (Peng and Marians 1995), it seems reasonable to postulate that there is an interaction of a second segment with the enzyme, but in a nonproductive manner (panel 3). This off-pathway binding must be rearranged to form the productive G/T binding, or the T segment must be recruited from a distant site. This nonproductive binding may be more frequent when the substrate has more (−) supercoils, in agreement with our observation that ς = −0.05 DNA was a poorer substrate than −0.03 DNA (Fig. 1C).
There is direct evidence with yeast topo II that the enzyme prefers to bind the G segment in a (−) SC DNA but cannot then readily bind the T segment. With a catenated substrate containing one (−) SC and one nicked ring, yeast topo II preferentially binds and cleaves the (−) supercoiled ring but decatenates rather than relaxes (Roca and Wang 1994). Therefore, the G segment came from the supercoiled ring, but the T segment was from the relaxed ring. There are parallel results with topo IV. When topo IV acts on a (−) SC catenane, it decatenates the struc-ture before relaxing it (Ullsperger and Cozzarelli 1996).
We emphasize that this is a working model and more experiments are needed to test and refine it. However, it is consistent with our major experimental results. The preferential cleavage of (+) SC DNA (Fig. 3) requires that discrimination between (+) and (−) supercoils by topo IV occurs prior to strand passage. In terms of our model, the facilitation of productive T-segment binding with (+) SC DNA results in enhanced cleavage. The favored geometry with (+) SC DNA can also explain the 20- to 25-fold faster rate of strand passage by topo IV when the substrate was (+) SC (Figs. 2 and 5). Both the single-cycle rate constant and processivity were higher with (+) SC DNA. Thus, the formation of an active enzyme–substrate complex was faster with (+) SC DNA, and the enzyme remained bound to the (+) SC DNA after the first reaction cycle. The enhanced processivity can be explained by the preferential productive binding of topo IV to the (+) SC DNA. After a strand passage, the enzyme can quickly capture another T segment before it dissociates. With (−) SC DNA, the enzyme may need to dissociate from one or both DNA segments to rebind two segments in a productive manner, resulting in the distributive relaxation we observed.
An earlier study found the rate of (+) supercoil relaxation by topo IV to be even slower than the rate we measured with (−) SC DNA (Hiasa and Marians 1996). A method for making highly (+) SC DNA was not available at that time, and the slow rate was likely due to the low ς of the (+) SC DNA used (∼+0.01). We found that topo IV is not efficient in removing the final few supercoils from a substrate.
We do not know why the preference for (+) SC DNA is exclusive at low enzyme concentrations in both ensemble and single-molecule experiments or the basis for the sigmoidal dependence of (−) supercoil relaxation on enzyme concentration, but several possible explanations for these results are consistent with our model. One possibility is that after binding of the G segment of a (−) SC DNA, a nonproductive interaction with a second segment is favored. Therefore, more enzyme molecules may be required for one of them to successfully form a productive complex. Alternatively, the enzyme concentration dependency may reflect the way the active form of the enzyme is initially assembled on the DNA. The active tetrameric topo IV may be in equilibrium with a form that lacks one or more subunits. Although the active yeast topo II dimer is stable at all enzyme concentrations tested (Tennyson and Lindsley 1997), it is not known how stable the topo IV tetramer is. The assembly of the active form of topo IV may be highly cooperative with a (+) SC substrate. Once assembled, the enzyme fully relaxes the DNA before disassembly. However, with (−) SC DNA, assembly of the active enzyme may not be cooperative and must be assisted by a high protein concentration. At high enzyme concentration, (+) SC DNA loses its advantage, as seen in the competition cleavage experiment (Fig. 3).
Physiological implications
In prokaryotes, (−) supercoiling is essential for promotion of DNA double helix opening and chromosome compaction. The physiological importance of (+) supercoiling, however, is often overlooked because (+) supercoiling has been studied far less and because of its transient nature compared with the longevity of (−) supercoiling. However, (+) supercoiling is rapidly generated by transcription (Liu and Wang 1987) and by helicase action at a DNA replication fork (for review, see Ullsperger et al. 1995; Wang 1996). There are two avenues for release of this (+) ΔLk during replication: relaxation of (+) supercoils ahead of the fork and removal of (+) precatenane links behind the fork (Hiasa and Marians 1996; Peter et al. 1998). It is clear that both gyrase and topo IV can promote replication fork movement (Hiasa and Marians 1994, 1996; Khodursky et al. 2000). Previously, removal of (+) precatenanes behind the fork appeared to be the only mechanism for topo IV to rapidly reduce Lk, because (−) supercoil relaxation, and by inference (+) supercoil relaxation, was so weak. Our experiments show that topo IV can also act in front of the fork to remove (+) supercoils.
There is direct evidence for strong and preferential (+) supercoil relaxation in vivo by topo IV. Transcription generates (+) supercoils ahead of and (−) supercoils behind the polymerase, via the twin domain effect (Liu and Wang 1987). The activity of topo IV completely prevents the appearance of (+) supercoils when gyrase is absent, but does not effectively remove (−) supercoils from the same plasmid (Khodursky et al. 2000; Zechiedrich et al. 2000).
The rate and processivity of (+) supercoil removal by topo IV measured in single-molecule experiments are nearly the same as those for Drosophila topo II relaxation of both (−) and (+) supercoils (Strick et al. 2000). Thus, the slow and distributive removal of (−) supercoils by topo IV both in vitro (Ullsperger and Cozzarelli 1996) and in vivo (Zechiedrich et al. 2000) underscores a fundamental difference between the two enzymes that may be a consequence of the critical role of free (−) supercoils in prokaryotes. Negative supercoiling in bacteria has many important roles, so that even a small drop in (−) supercoiling is lethal. Moreover, relaxation of (−) supercoils actively opposes replication because it increases Lk. Thus, the preferential relaxation of (+) supercoils by topo IV and their conversion to (−) supercoils by gyrase allows bacteria to remove unwanted (+) supercoiling without compromising essential (−) supercoiling.
Our findings provide a resolution to a quantitative paradox about topoisomerases in E. coli replication. The maximum rate of fork movement at 37°C requires ∼100 links to be removed every second, or 3000 strand passages/min, at each fork (Bird and Lark 1970). The rate of relaxation of (−) SC plasmids by topo IV in vivo at 37°C is only on the order of one strand passage/min (Zechiedrich et al. 2000), the same rate we measured at 30°C. Topo IV can act in place of gyrase in fork movement by the relaxation of supercoils. One resolution of this difficulty is that there are thousands of topo IV molecules to make up the rate deficit. However, there are not enough enzyme molecules at the fork in the cell or enough DNA to accommodate this scenario (Huang et al. 1998).
Our data, particularly the single-molecule results, show instead that only one or a few molecules of topo IV could suffice at each fork to support replication elongation. The rate of (+) supercoil relaxation by a single topo IV is 180 strand passages/min at 25°C. Given that topo IV supports chain growth in vivo at about one-third of the normal rate (∼1000 strand passages/min) (Khodursky et al. 2000), then the in vivo rate is only about five times the in vitro single-molecule rate. Raising the temperature from 25° to 37°C increases the rate twofold (data not shown), thus halving the discrepancy. We believe the remaining difference is not significant, particularly if topo IV also removes (+) precatenanes behind the replication fork.
The single-molecule measures of relaxation are about eightfold higher than ensemble measurements for both (+) and (−) SC DNA. Whereas ensemble measures average out the activity of functioning enzymes with those that are paused on the DNA, not bound to substrate, or completely inactive, the single-molecule experiments measure only active enzyme. We believe that the single-molecule data are a more accurate reflection of the in vivo activity of topo IV. These data show, not just for topoisomerases but also for polymerases (Davenport et al. 2000; Maier et al. 2000; Wuite et al. 2000) and ion channels (Sakmann 1992), that activity occurs in bursts. In Figure 5, we see long pauses between intense relaxation activity. These pauses may reflect the disassembly of the active enzyme–DNA complex. We presume that factors in the cell deliver topo IV to the replication fork and keep it from a pulsative behavior, which would cause intermittent replication. Analogous factors have been shown to aid RNA polymerase in proceeding through pause sites in both prokaryotes (greA and greB) (Borukhov et al. 1993) and eukaryotes (TFIIS) (for review, see Wind and Reines 2000).
We suggest that (+) supercoiling is the physiological substrate for prokaryotic type II topoisomerases acting in replication. In addition to our data here on topo IV, there are a number of experiments that show that DNA gyrase is much more active on (+) than (−) supercoils. Its activity is reduced by 10-fold at ς = −0.06 compared with relaxed DNA (Westerhoff et al. 1988; Kozyavkin et al. 1990). Given the essential role of (−) supercoiling, we suppose that there is a patch of (+) supercoiling around the relication fork that is maintained by topological domain barriers. There is a pleasing biological unity to this view, as the DNA in eukarya and archaea must be (+) supercoiled around the replication fork, because the only mechanism open to them to reduce Lk in the absence of gyrase is by relaxation of a (+) ΔLk.
Materials and methods
Proteins
Topo IV (Ullsperger and Cozzarelli 1996), wheat germ topo I (Dynan et al. 1981), and the archaeal histone HMfB (Sandman et al. 1994) were prepared as described. Another preparation of topo IV was obtained from H. Hiasa (University of Minnesota).
Negatively supercoiled DNA substrates
Negatively SC substrates used were pUC18 (2.7 kb), pLR3.5i (3.5 kb) (Crisona 1999), and pAB4 (7.0 kb) (Benjamin and Cozzarelli 1990). pUC18 DNA of ς = −0.03 was prepared by treatment of ς = −0.05 DNA with topo I in the presence of ethidium bromide (EtBr). The ς values of pUC18 and pLR3.5I, were determined by band counting on gels containing chloroquine and are relative to DNA completely relaxed by topo I under the topo IV reaction conditions.
Positively supercoiled DNA substrates for ensemble experiments
We prepared (+) SC DNA by incubating plasmid DNA with the archaeal histone HMfB in the presence of wheat germ topo I (Musgrave et al. 1991). The (+) SC pUC18 DNA used in all experiments was from two large-scale reactions containing 100 μg of DNA. Reactions containing 50 mM Tris-HCl (pH 8), 2.5 mM EDTA (pH 8), 50 mM NaCl, 2 mM potassium phosphate (pH 7), 50 μg/mL (−) supercoiled DNA, and HMfB were incubated for 15 min at 25°C. Topo I was added and incubation continued for 30 min. The reactions were treated with Proteinase K, extracted with phenol–chloroform, precipitated with ethanol, and analyzed by agarose gel electrophoresis in TAE buffer (40 mM Tris-acetate, 1 mM EDTA) containing 5 μg/mL of chloroquine. The (−) SC DNA is relaxed, whereas the (+) SC DNA migrates as a tight band of high mobility.
The ς of the (+) SC DNAs was measured by electrophoretic band counting in TAE buffer containing a saturating amount (10 μM) of netropsin (Boehringer Mannheim). Netropsin overwinds the DNA, resulting in a ΔLk of −4. This allowed the determination of the center of the (+) topoisomer distribution for DNAs of ς ≤ +0.04. For more (+) SC DNA, the ς is an estimate, because the entire topoisomer distributions were not resolved. The ς values of the two preparations were +0.035 and ∼+0.05.
Reactions
Topo IV titration reactions contained 25 mM Tris-HCl (pH 7.6), 100 mM potassium glutamate, 10 mM MgCl2, 0.5 mM dithiothreitol (DTT), 50 μg/mL BSA, 1 mM ATP, 5% glycerol, and 150 ng (85 fmole) of DNA in a 15 μL volume. Reactions were incubated at 30°C for 30 min and terminated by the addition of EDTA. The DNA was analyzed by agarose gel electrophoresis in TAE buffer. Quantification was performed from Southern blots of the gels with a Fuji PhosphorImager.
Relaxation kinetics reactions contained a saturating concentration of DNA (50 μg/mL) to ensure that the rates were maximal. Samples were removed at time points and added to EDTA on ice to terminate the reaction. Negatively SC samples were run on a 1.2 % agarose gel containing 1 μg/mL of chloroquine. To quantify relaxation, we assigned a ΔLk to each topoisomer and calculated the total number of links in the DNA at each time point on the basis of percent of each topoisomer present. Positively SC samples were run on a 1% agarose gel without chloroquine. The total number of links at each time point was calculated from the average number of links per substrate molecule, the ΔLks of the relaxed topoisomers, and the percent of the DNA in each species.
Topo IV competition cleavage reactions contained 75 ng each of the 2.7-kb pUC18 DNA and a 7-kb DNA (pAB4) and 20 μg/mL norfloxacin. After a 30-min incubation at 30°C, SDS was added to 1% and incubation continued for 15 min. EDTA was added, and the reactions were treated with Proteinase K and analyzed on a 1% agarose gel containing 0.5 μg/mL of EtBr.
Single-molecule experiments
Relaxation of (−) and (+) supercoils by topo IV was measured on tethered 11-kb linear DNA molecules in buffer lacking glycerol (Strick et al. 1996, 1998). The DNA was multiply labeled with biotin at one end and digoxigenin at the other, bound to streptavidin-coated magnetic beads, and tethered to the bottom of a square glass capillary tube coated with antidigoxigenin. The capillary tube was attached to a buffer flow system and placed above a 60× oil-immersion lens on an inverted light microscope. Magnets above the capillary can pull on and rotate the magnetic bead: clockwise (counterclockwise) rotation generates negative (positive) supercoiling in the DNA. The stretching force can be increased (decreased) by lowering (raising) the magnets. From a force versus extension curve, a persistence length of the DNA is calculated for each molecule used. The experiment is continued only if the value of the persistence length indicates that a single DNA molecule is attached to the bead (Strick et al. 1998). The extension of the DNA is a function of the force and the number of plectonemic supercoils and is monitored by video tracking (at 12.5 Hz) the height of the bead relative to the surface. The DNA is fully extended at a given force when ς = 0. Experiments were done at the low force of 0.2 pN, which causes no denaturation of the DNA. The raw data (dots) indicate the bead height for each video frame, and the continuous curve represents the same data filtered with an integration time of 5 sec. Because the noise is Gaussian, the filtered line is a moving average of the raw data.
Acknowledgments
We thank Drs. A. Bates, J. Berger, and A. Vologodskii for helpful discussions. This work was supported by grants from the NIGMS, NSF, and ARC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ncozzare@socrates.berkeley.edu; FAX (510) 643-1079.
Article and publication are at www.genesdev.org/cgi/doif/10.1101/gad.838900
References
- Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by Hydrolyzing one ATP. Proc Natl Acad Sci. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin HW, Cozzarelli NR. Geometric arrangements of Tn3 resolvase sites. J Biol Chem. 1990;265:6441–6447. [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- Bird R, Lark K. Chromosome replication in Escherichia coli 15T– at different growth rates: Rate of replication of the chromosome and the rate of formation of small pieces. J Mol Biol. 1970;49:343–366. doi: 10.1016/0022-2836(70)90249-4. [DOI] [PubMed] [Google Scholar]
- Boocock MR, Brown JL, Sherratt DJ. Topological specificity in Tn3 resolvase catalysis. In: McMacken R, Kelly TJ, editors. DNA replication and recombination. Vol. 47. New York: Alan R. Liss; 1987. pp. 703–718. [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Zechiedrich EL, Osheroff N. A role for the passage helix in the DNA cleavage reaction of eukaryotic topoisomerase II. A two-site model for enzyme-mediated DNA cleavage. J Biol Chem. 1992;267:683–686. [PubMed] [Google Scholar]
- Craigie R, Mizuuchi K. Role of DNA topology in Mu transposition: Mechanism of sensing the relative orientation of two DNA segments. Cell. 1986;45:793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- Crisona NJ, Weinberg RL, Peter BJ, Sumners D, Cozzaretti NR. The topological mechanism of phage λ integrase. J Mol Biol. 1999;289:747–775. doi: 10.1006/jmbi.1999.2771. [DOI] [PubMed] [Google Scholar]
- Davenport RJ, Wuite GJL, Landick R, Bustamonte C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Jendrisak JJ, Hager DA, Burgess RR. Purification and characterization of wheat germ DNA topoisomerase I (nicking-closing enzyme) J Biol Chem. 1981;256:5860–5865. [PubMed] [Google Scholar]
- Hiasa H, Marians KJ. Topoisomerase IV can support oriC DNA replication in vitro. J Biol Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- ————— Two distinct modes of strand unlinking during theta-type DNA replication. J Biol Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- Huang W, Libbey J, van der Hoeven P, Yu S. Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. Proc Natl Acad Sci. 1998;95:4652–4657. doi: 10.1073/pnas.95.8.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci. 1999;96:8414–8419. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Cozzarelli NR. Roles of supercoiled DNA structure in DNA transactions. Curr Opin Struct Biol. 1992;2:369–379. [Google Scholar]
- Kato J-i, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- Khodursky AB, Zechiedrich EL, Cozzarelli NR. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NC. Analysis of topoisomerase function in bacterial replication fork movement: Use of microarrays. Proc Natl Acad Sci. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K, Wang JC. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981;23:721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Kozyavkin SA, Slesarev AI, Malkhosyan SR, Panyutin IG. DNA linking potential generated by gyrase. Eur J Biochem. 1990;191:105–113. doi: 10.1111/j.1432-1033.1990.tb19099.x. [DOI] [PubMed] [Google Scholar]
- Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: Biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochem Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Higgins NP, Cozzarelli NR. Interaction between DNA gyrase and its cleavage site on DNA. J Biol Chem. 1980;255:2211–2219. [PubMed] [Google Scholar]
- Musgrave D, Sandman K, Reeve J. DNA binding by the archael histone HMf results in positive supercoiling. Proc Natl Acad Sci. 1991;88:10397–10401. doi: 10.1073/pnas.88.23.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Marians K. The interaction of topoisomerase IV with DNA. J Biol Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- Postow L, Peter BJ, Cozzarelli NR. Knot what we thought before: The twisted story of replication. BioEssays. 1999;21:805–808. doi: 10.1002/(SICI)1521-1878(199910)21:10<805::AID-BIES1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Roca J, Wang J. DNA transport by a type II DNA topoisomerase—evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Ullsperger CU, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- Sakmann B. Elementary steps in synaptic transmission revealed by currents through single ion channels. Science. 1992;256:503–512. doi: 10.1126/science.1373907. [DOI] [PubMed] [Google Scholar]
- Sandman K, Grayling RA, Dobrinski B, Lurz R, Reeve JN. Growth-phase-dependent synthesis of histones in the archaeon Methanothermus fervidus. Proc Natl Acad Sci. 1994;91:12624–12628. doi: 10.1073/pnas.91.26.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Stasiak AM, Martinez-Robles ML, Krimer DB, Hernandez P, Schvartzman JB. Formation of knots in partially replicated DNA molecules. J Mol Biol. 1999;286:637–643. doi: 10.1006/jmbi.1998.2510. [DOI] [PubMed] [Google Scholar]
- Strick TR, Allemand J-F, Bensimon D, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- ————— Behavior of supercoiled DNA. Biophys J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- Tennyson RB, Lindsley JE. Type II topoisomerase from Saccharomyces cerevisiae is a stable dimer. Biochemistry. 1997;36:6107–6114. doi: 10.1021/bi970152f. [DOI] [PubMed] [Google Scholar]
- Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J Biol Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- Ullsperger CJ, Vologodskii AA, Cozzarelli NR. Unlinking of DNA by topoisomerases during DNA replication. In: Lilley DMJ, Eckstein F, editors. Nucleic acids and molecular biology. Vol. 9. Berlin, Germany: Springer-Verlag; 1995. pp. 115–142. [Google Scholar]
- Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- ————— Moving one double helix through another by a type II DNA topoisomerase: The story of a simple molecular machine. Q Rev Biophys. 1998;31:104–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- Westerhoff HV, O'Dea MH, Maxwell A, Gellert M. DNA supercoiling by DNA gyrase. A static head analysis. Cell Biophys. 1988;12:157–181. doi: 10.1007/BF02918357. [DOI] [PubMed] [Google Scholar]
- Wind M, Reines D. Transcription elongation factor SII. BioEssays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Cozzarelli NR. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes & Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DMJ, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8102–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]