Abstract
Certain haploid strains of Saccharomyces cerevisiae can undergo meiosis, but meiotic prophase progression and subsequent nuclear division are delayed if these haploids carry an extra chromosome (i.e., are disomic). Observations indicate that interactions between homologous chromosomes cause a delay in meiotic prophase, perhaps to allow time for interhomolog interactions to be completed. Analysis of meiotic mutants demonstrates that the relevant aspect of homolog recognition is independent of meiotic recombination and synaptonemal complex formation. A disome in which the extra chromosome is circular sporulates without a delay, indicating that telomeres are important for homolog recognition. Consistent with this hypothesis, fluorescent in situ hybridization demonstrates that a circular chromosome has a reduced capacity to pair with its homolog, and a telomere-associated meiotic protein (Ndj1) is required to delay sporulation in disomes. A circular dimer containing two copies of the same chromosome delays meiosis to the same extent as two linear homologs, implying that physical proximity bypasses the requirement for telomeres in homolog pairing. Analysis of a disome carrying two linear permuted chromosomes suggests that even nonhomologous chromosome ends can promote homolog pairing to a limited extent. We speculate that telomere-mediated chromosome movement and/or telomere clustering promote homolog pairing.
Keywords: Telomeres, meiosis, homolog pairing, chromosome synapsis
At the first division of meiosis, homologous chromosomes move to opposite poles of the spindle apparatus. This coordination in chromosome behavior depends on complex processes and elaborate structures that bring homologs together in meiotic prophase and hold them together until anaphase of meiosis I (for review, see Roeder 1997).
Understanding how a meiotic chromosome finds and recognizes its homolog is one of the most intriguing problems in the study of meiosis. Cytological studies suggest that chromosome pairing during meiotic prophase can be divided into two distinct substages (for review, see Roeder 1997). At the first stage, called presynaptic alignment, homologs line up side-by-side, but are separated by a variable and often considerable distance (∼300 nm); physical connections between homologs are not yet evident. At the second stage, referred to as synapsis, homologs become intimately associated through the formation of the synaptonemal complex (SC) (for review, see Roeder 1997). Within the SC, the proteinaceous cores of paired chromosomes are separated by a uniform distance of ∼100 nm and held together by the transverse filaments of the central region of the SC. A number of observations indicate that homolog alignment not only precedes, but is separable from synapsis. First, synapsis (i.e., SC formation) can take place between nonhomologous chromosomes or chromosome segments. Second, in certain meiotic mutants, chromosomes become homologously aligned, but they fail to synapse.
Although the molecular mechanisms responsible for presynaptic alignment remain elusive, a number of studies suggest a role for telomeres. In many organisms, including plants and vertebrates, meiotic chromosomes form a bouquet in which the ends of chromosomes are attached to a small region of the nuclear envelope that is juxtaposed to the microtubule-organizing center (for review, see Dernburg et al. 1995). At least in some organisms, bouquet formation is a two-step process in which telomeres first attach to the nuclear envelope and then move along the envelope to a common location (Scherthan et al. 1996). Bouquet formation precedes the initiation of chromosome synapsis and is approximately coincident with presynaptic alignment (Scherthan et al. 1996; Bass et al. 1997). The tight clustering of telomeres may facilitate homolog alignment by bringing homologous sequences near chromosome ends into close proximity.
An important role for telomeres has been demonstrated by studies of meiosis in Schizosaccharomyces pombe (Chikashige et al. 1994, 1997; Scherthan et al. 1994). Time-lapse images of living cells have demonstrated that the nucleus is dragged back and forth over the length of the cell several times during meiotic prophase, producing elongated “horse tail” nuclei (Chikashige et al. 1994). At this stage, all of the telomeres are attached to the spindle pole body, which is present at the leading edge of the travelling nucleus. Mutations that impair telomere clustering (Shimanuki et al. 1997; Cooper et al. 1998; Nimmo et al. 1998) or prevent nuclear movement (A. Yamamoto, R.R. West, J.R. McIntosh, and Y. Hiraoka, pers. comm.) substantially reduce meiotic recombination, suggesting that these processes facilitate homolog alignment in fission yeast.
The results presented below demonstrate that telomeres also play an important role in meiosis in Saccharomyces cerevisiae. Our data indicate that homolog recognition causes cells to delay in meiotic prophase, perhaps to allow time for interhomolog interactions to be completed. The delay in meiosis is evident when disomic strains (i.e., haploids carrying an extra copy of one chromosome) are compared to simple haploids. A circular chromosome and its linear homolog are unable to delay meiosis, demonstrating that the pertinent aspect of homolog recognition requires both chromosomes to have ends.
Results
Meiosis is delayed in disomic strains compared to haploids
Haploid cells of S. cerevisiae can undergo meiosis and sporulation if they express information from both mating types and carry a mutation in the SPO13 gene (Klapholz and Esposito 1980; Wagstaff et al. 1982). These haploids undergo a single round of equational chromosome segregation to produce dyads containing two viable haploid spores. There are three ways in which expression of both mating types can be achieved in a haploid. (1) A haploid (MATa or MATα) strain can carry the opposite type of mating information on an integrated or replicating plasmid. (2) The haploid may carry a sir mutation, which allows expression of the otherwise silent cassettes of mating type information (HMLα and HMRa) (for review, see Palladino and Gasser 1994). (3) The haploid can carry an extra copy of chromosome III, with MATα on one copy of the chromosome and MATa on the homolog. Unexpectedly, we found that strains disomic for chromosome III undergo meiotic nuclear division 2–3 hr later than isogenic haploids constructed by either of the first two methods.
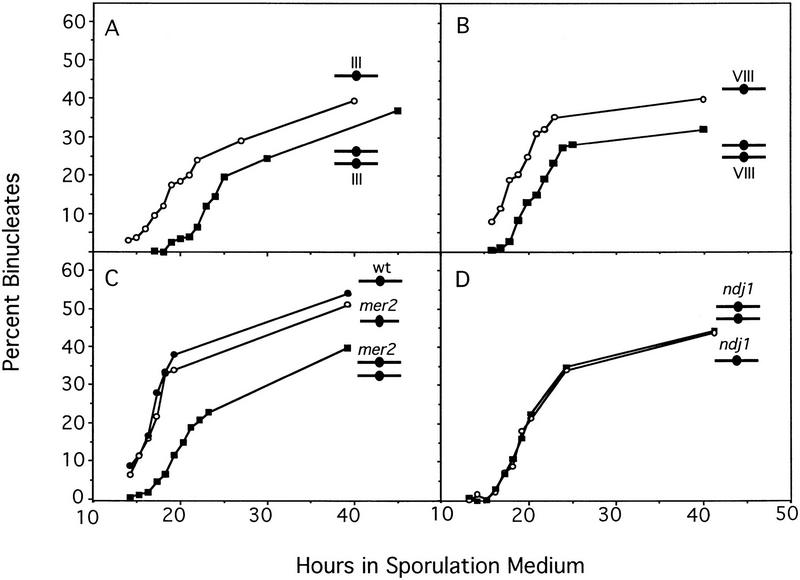
To examine the kinetics of meiotic nuclear division, cells harvested at various time points after the introduction into sporulation medium were stained with the DNA-binding dye, 4′,6-diamidino-2-phenyl-indole (DAPI), and examined by fluorescence microscopy. Cells that have undergone nuclear division contain two nuclei (referred to as binucleates). Figure 1A shows the kinetics of nuclear division in a haploid strain disomic for chromosome III and in an isogenic MATα haploid strain in which MATa has been integrated at an ectopic location. Nuclear division occurs ∼3 hr later in the disome than in the haploid. There is a corresponding delay in spore formation (data not shown).
Figure 1.
Kinetics of meiotic nuclear division in wild-type and mutant haploids and disomes yeast strains were sporulated and the percentage of cells that had undergone nuclear division was determined as described in Materials and Methods. (A) Binucleate formation in a haploid (BR3090, ○) and an isogenic strain disomic for chromosome III (BR3091, ▪). The time at which 50% of the cells that eventually sporulate had sporulated (T50) was 20.9 hr for the haploid and 24.6 hr for the disome. (B) Binucleate formation in a haploid (BR3001, ○, T50 = 18.8 hr) and an isogenic strain disomic for chromosome VIII (BR3002, ▪, T50 = 21.2 hr). (C) Binucleate formation in a mer2 haploid (BR3092, ○, T50 = 18.1 hr), an isogenic mer2 strain disomic for chromosome III (BR3093, ▪, T50 = 20.6 hr), and a wild-type haploid (BR3090; •, T50 = 17.8 hr). (D) Binucleate formation in an ndj1 haploid (BR3231, ○, T50 = 21.1 hr) and an isogenic ndj1 strain disomic for chromosome III (BR3200-12D, ○, T50 = 20.9 hr).
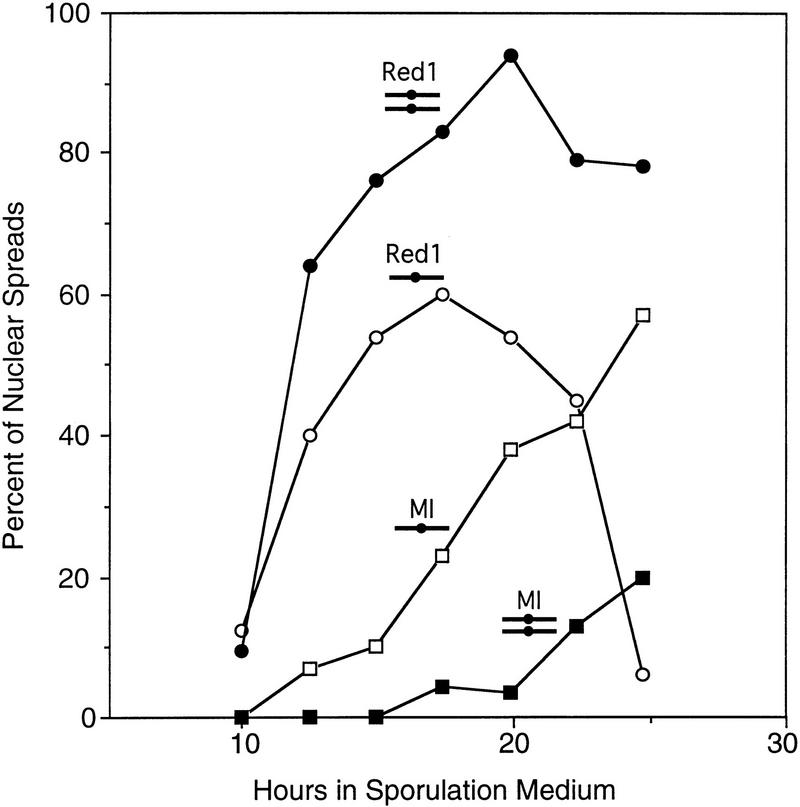
To determine the stage in meiosis at which disomic strains are delayed, meiotic nuclei from cells harvested at different time points in sporulation medium were surface spread and stained with antibodies to tubulin (to assess spindle formation) and with antibodies to the Red1 protein. Red1 is a component of the proteinaceous cores of meiotic chromosomes; the protein localizes to chromosomes during the leptotene, zygotene, and pachytene stages of meiotic prophase (Smith and Roeder 1997). The data presented in Figure 2 demonstrate that Red1 assembles onto chromosomes at the same time in haploid and disomic strains. However, in the disome, Red1 staining persists longer, and spindles are formed later. Thus, disomic strains are delayed in early or mid prophase of meiosis I.
Figure 2.
Kinetics of meiotic prophase events in haploid and disomic strains. Anti-Red1 antibody staining (circles) and spindle formation (squares) were scored in meiotic nuclear spreads of a haploid (BR3133, ○ and ) and a strain disomic for chromosome III (BR3108, • and ▪).
The delay in meiotic nuclear division is not specific to strains disomic for chromosome III. As shown in Figure 1B, nuclear division in a sir2 strain that is disomic for chromosome VIII is delayed compared to an isogenic sir2 haploid. These data demonstrate that an extra chromosome inhibits meiotic cell-cycle progression in haploids.
The delay in meiosis in disomes is due to the presence of homologs
The delay in meiotic progression in disomic strains could result from an increase in the absolute number of chromosomes (from 16 to 17). Alternatively, the delay might be due to the presence of a pair of homologous chromosomes in an otherwise haploid genome. To distinguish between these possibilities, we constructed and analyzed a haploid strain that carries an extra chromosome, but does not contain homologs. This was accomplished by fragmenting chromosome III into two smaller chromosomes, through the introduction a plasmid that contains a centromere and a telomere-seeding cassette (Fig. 3A).
Figure 3.
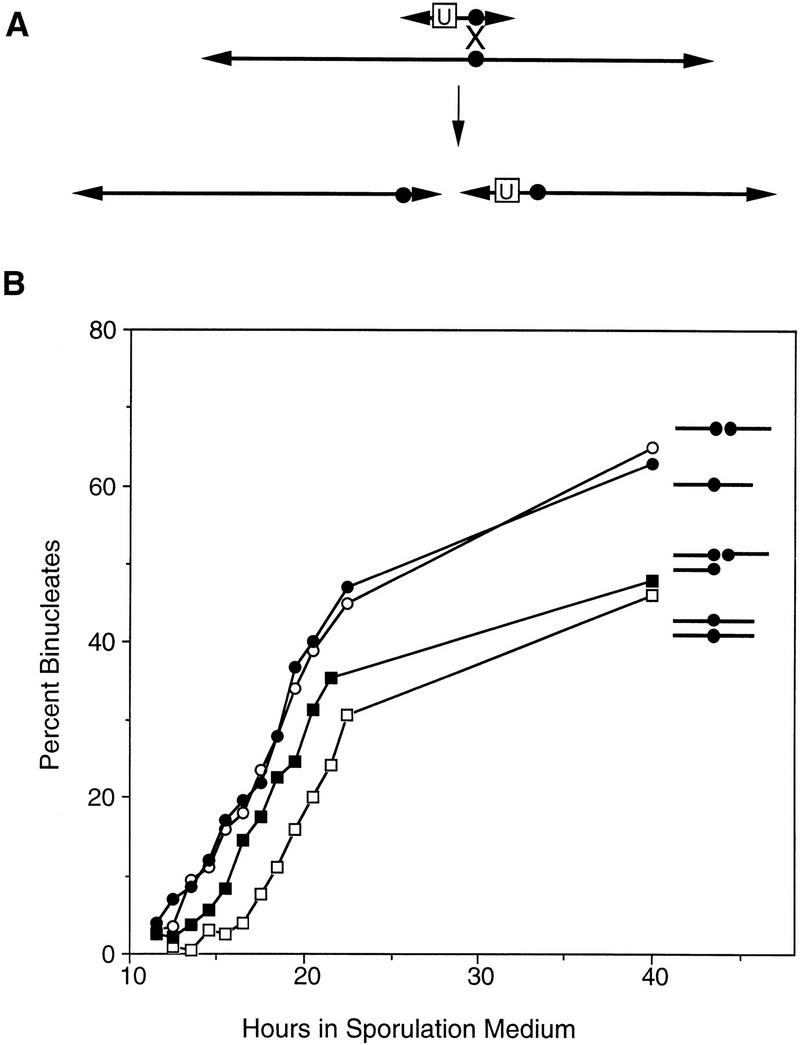
Construction and analysis of a haploid with a broken chromosome III. (A) Diagram of the bisection of chromosome III (see Materials and Methods). (Arrowheads) Telomeres; (•) centromere. The box labeled “U” indicates the URA3 gene and the X indicates the position of a crossover. (B) Binucleate formation was assayed in a haploid (BR3133, ○, T50 = 19.0 hr), a strain disomic for chromosome III (BR3108, , T50 = 21.2 hr), a haploid strain in which chromosome III had been bisected (BR3110, •, T50 = 18.7 hr), and a strain carrying two copies of the chromosome III fragment corresponding to the left arm and one copy of the fragment derived from the right arm (BR3111, ▪, T50 = 19.6 hr). All strains are isogenic.
A haploid strain carrying the two chromosome III fragments undergoes meiotic nuclear division with the same timing as an isogenic haploid (Fig. 3B), indicating that increasing the number of chromosomes does not necessarily delay meiosis. A strain that carries two copies of the chromosome III fragment corresponding to the left arm (IIIL), and a single copy of the fragment derived from the right arm (IIIR), displays a delay in meiotic progression (Fig. 3B). These data indicate that it is the presence of homologous chromosomes, not the increase in chromosome number, that is responsible for the delay in meiosis in disomic yeast.
The timing of meiotic nuclear division in the strain disomic for chromosome IIIL is intermediate between that of a haploid and that of a strain disomic for the intact chromosome (Fig. 3B). This observation suggests that homolog recognition is less efficient when the amount of homology is decreased. Cells from the chromosome IIIL disome may be heterogeneous in behavior, with some cells delayed in meiosis (because of homolog pairing) and others preceding through meiosis on time (because of a failure of homolog recognition). Alternatively, homolog recognition may be unaffected by the decrease in chromosome size, and the intermediate effect may be due to a subset of cells that lost the chromosome IIIL fragment during vegetative growth (see Materials and Methods). Such segregants would sporulate rapidly because of the absence of homologs.
Homolog recognition is independent of recombination and synapsis
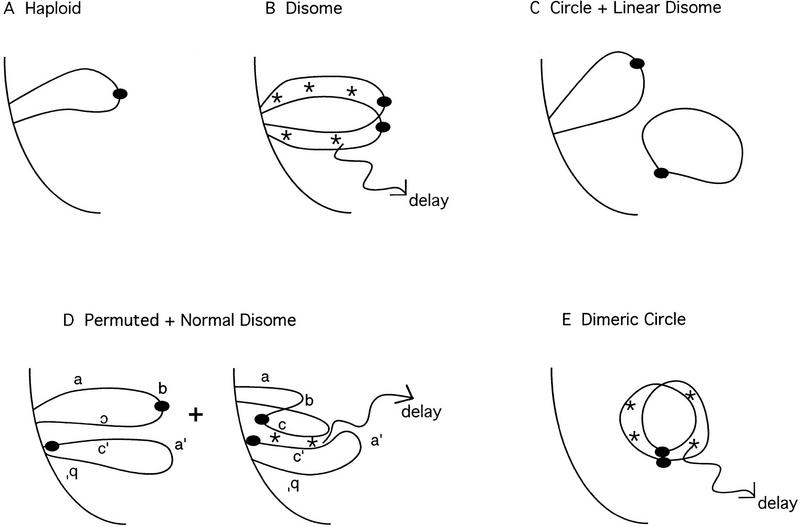
The delay in meiosis in disomic strains might reflect the cell’s ability to assess its ploidy. In a disome or diploid, interactions between homologous chromosomes could allow the cell to recognize that homologs are present. Under these circumstances, mechanisms might operate to delay meiotic chromosome segregation to allow time for all chromosomes to pair (Fig. 4B).
Figure 4.
Cartoon of the relationship between homolog pairing and meiotic progression. Shown is a diagrammatic representation of chromosome pairing in haploids and disomes carrying normal and rearranged copies of chromosome III. The telomeres of linear chromosomes are anchored in the nuclear membrane (curved line on the left of each panel). (*) Homolog recognition. (A) In a haploid, homologs are not present, and meiosis is not delayed. (B) In a disome containing two normal copies of chromosome III, the homologs recognize each other, and this recognition elicits a delay. (C) In a disome containing one linear and one circular chromosome, the circular chromosome is not attached to the nuclear membrane, and therefore not close enough to its homolog to be recognized. (D) In a disome containing permuted linear chromosomes (a, b, c, and b′, c′, a′), the anchoring of telomeres to the nuclear membrane brings homologs into proximity. However, homology is recognized only when one chromosome becomes twisted such that homologous sequences (e.g., c and c′) are aligned. The subset of cells in which homologs are recognized experience a delay in meiotic progression. (E) In a strain carrying a dimeric circular chromosome, the two homologs are physically attached, allowing for efficient homolog recognition in the absence of telomeres and attachment to the nuclear membrane.
What aspect of the interaction between homologous chromosomes allows the cell to assess its ploidy? To address this question, we analyzed strains carrying mutations in genes involved in various aspects of meiotic recombination and/or chromosome synapsis, including red1, mer1, dmc1, rec104, mer2, mek1, and spo11 (Fig. 1C; data not shown) (for review of mutant phenotypes, see Roeder 1997). In every case, meiosis in a mutant disome was delayed compared to the isogenic mutant haploid. Three of the mutations tested (mer2, rec104, and spo11) abolish the meiosis-specific double-strand breaks (DSBs) that initiate meiotic recombination (Cao et al. 1990; Rockmill et al. 1995; Bullard et al. 1996), and at least two of these (mer2 and spo11) prevent the formation of SC (Giroux et al. 1989; Loidl et al. 1994; Rockmill et al. 1995). Thus, these data indicate that homolog recognition is independent of both recombination and synapsis.
Chromosome ends are important for homolog recognition
To determine whether telomeres are important for homolog recognition, we constructed strains carrying a circular version of chromosome III (Fig. 5A). A haploid strain containing the circular chromosome, and a disomic strain carrying one linear and one circular chromosome III, were examined in meiotic time courses. Both strains undergo nuclear division with the same timing as a haploid carrying a single linear version of the chromosome, and earlier than a disome containing two normal linear chromosomes (Fig. 5B). These data imply that the linear and circular chromosomes III do not undergo the aspect of homolog recognition that delays sporulation. Thus, homolog recognition, as assessed in this assay, requires both chromosomes to have ends (Fig. 4C).
Figure 5.
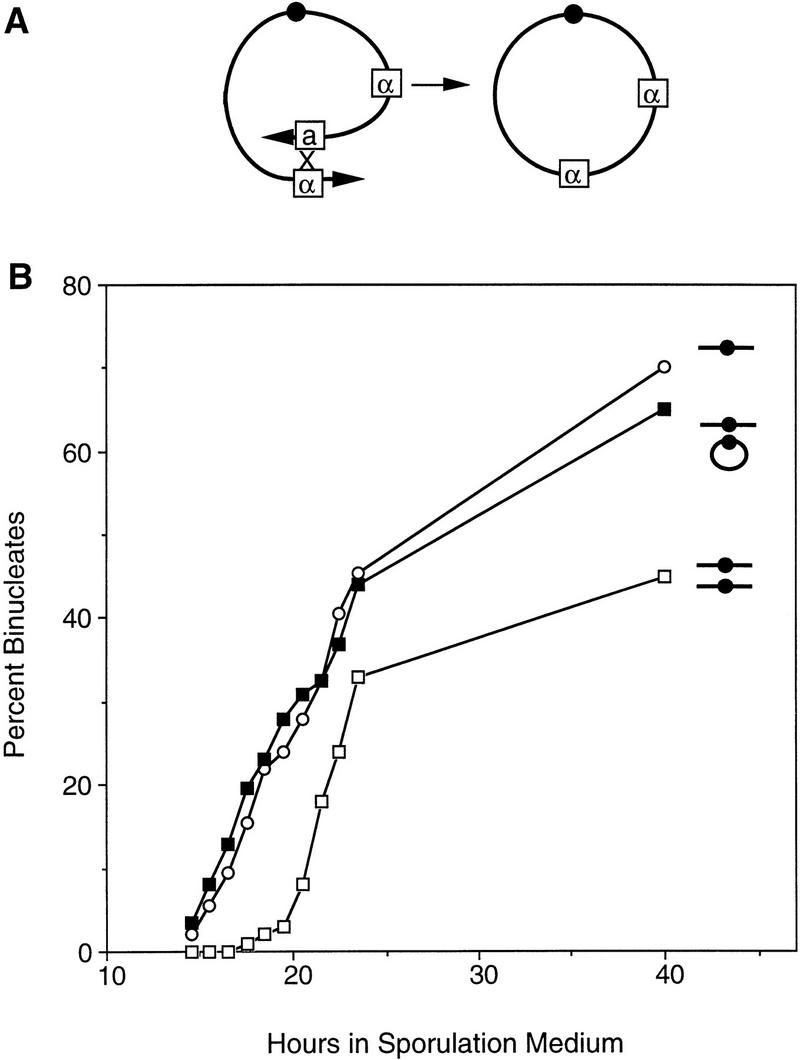
Construction and analysis of a disome containing a circular chromosome III. (A) Diagram of the formation of a circular derivative of chromosome III (see Materials and Methods). Boxes a and α indicate mating type information (see Fig. 3A for other symbols). (B) Binucleate formation was monitored in a haploid containing a normal chromosome III (BR3133, ○, T50 = 21.5 hr), a strain disomic for the normal chromosome III (BR3108, , T50 = 22.2 hr), and a disomic strain containing one linear and one circular version of chromosome III (BR3139, ▪, T50 = 21.5 hr). All strains are isogenic.
Telomeres promote homolog pairing in disomic strains
To determine whether the inability of a circular chromosome to delay sporulation reflects a defect in physical pairing with its homolog, fluorescent in situ hybridization (FISH) was used to measure chromosome pairing (Fig. 6). Homolog pairing was measured in disomic strains containing either two linear copies of chromosome III or one linear copy and one circular derivative. Disomic strains carrying a disruption of the MER2 gene were used for FISH analysis so that homolog pairing could be assessed in the absence of genetic recombination and SC formation. Under these conditions, pairing interactions appear to be unstable (Nag et al. 1995), so FISH provides a minimum estimate of the extent of pairing. The actual level of pairing in vivo may be much higher.
Figure 6.
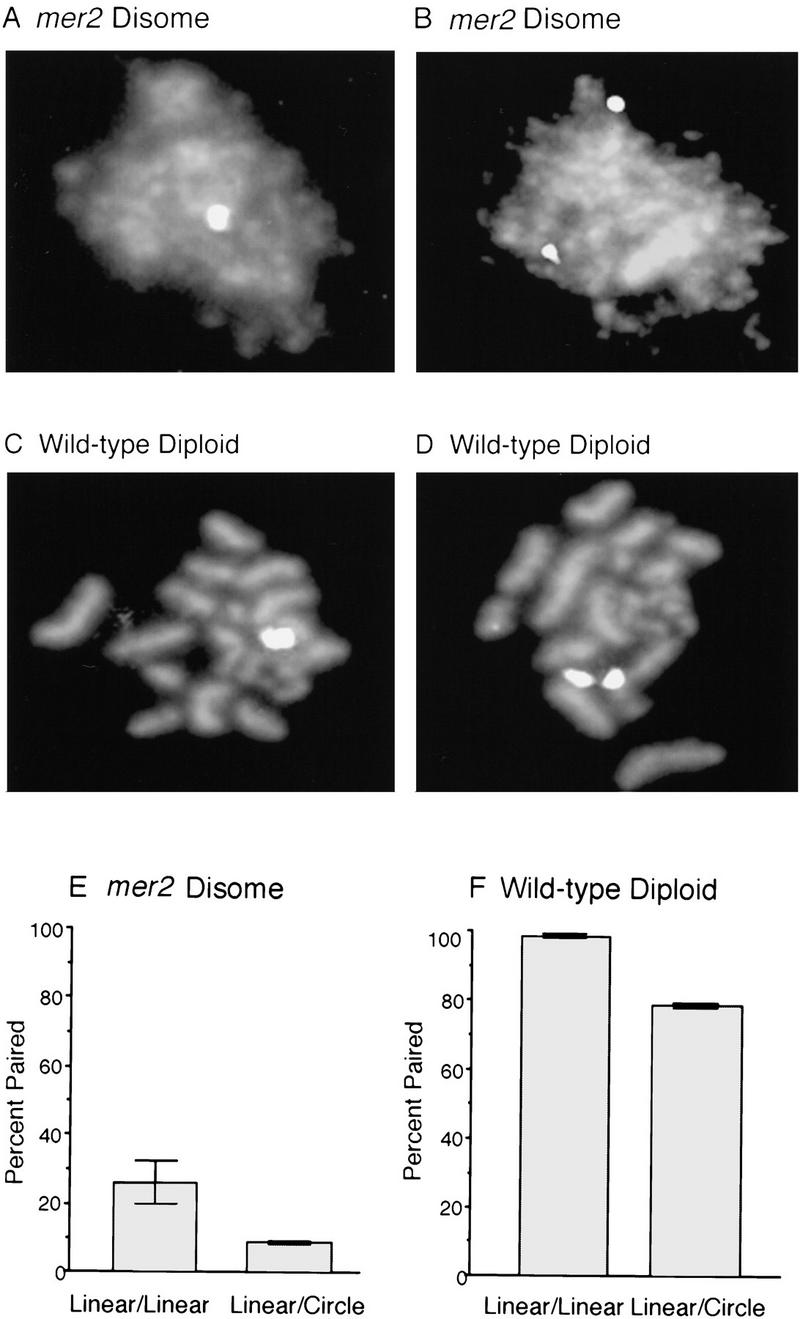
FISH Analysis of meiotic homolog pairing. (A–D) Examples of FISH data. Meiotic nuclear spreads were stained with DAPI to visualize chromosomes and hybridized with a probe for chromosome III (see Materials and Methods). (A,B) Nuclear spreads from a mer2 disome (BR3147) containing one linear and one circular version of chromosome III. Chromosomes III are paired in A and unpaired in B. (C,D) Spread nuclei from a wild-type diploid (BR3167) containing one linear and one circular version of chromosome III. Chromosomes III are paired in C and unpaired in D. (E,F) Quantitative analysis of chromosome pairing. Spread preparations of meiotic nuclei were hybridized with probes specific for chromosomes III and V, which were detected with fluorescent tags of different colors (see Materials and Methods). Homologous chromosomes were considered to be paired if a single hybridization signal (of a particular color) was observed, or if the two signals were less than 0.5 μm apart. Standard deviations are indicated. The frequency of fortuitous interactions between nonhomologous chromosomes was ∼1%. (E) FISH analysis of chromosome III pairing in nuclear spreads from a mer2 disome containing two linear copies of chromosome III (BR3136-9B) and an isogenic mer2 disome containing one linear and one circular copy of chromosome III (BR3147). FISH was carried out after 15 hr in sporulation medium, when most nuclei contain thread-like DAPI-stained chromosomes (representing condensed but unsynapsed chromosomes). (F) FISH analysis of chromosome III pairing in pachytene nuclei from a normal diploid (BR3168) and a diploid carrying one linear and one circular version of chromosome III (BR3167). FISH was carried out after 15 hr in sporulation medium, when most nuclei are in pachytene. Homolog pairing was measured only in nuclei at the pachytene stage as determined by DAPI staining (chromosome pairs that are condensed and synapsed appear as “sausages”). The linear chromosomes V were paired in nearly all (∼98%) nuclei from both diploid strains (data not shown).
The data presented in Figure 6E demonstrate that the chromosome III homologs are paired in ∼25% of nuclei from the disomic strain carrying two linear chromosomes. In contrast, the chromosome III homologs are paired in only 8% of nuclei from the disome in which the extra chromosome is circular. These results are consistent with the hypothesis that homolog pairing is the cause of the delay in sporulation.
Even nonhomologous chromosome ends facilitate homolog recognition
If telomeres promote homolog recognition exclusively by causing bouquet formation, then telomeres should facilitate alignment only if they are located at the same position on both homologs. We determined whether this is the case by constructing and analyzing a disomic strain in which the two copies of chromosome III are linear but permuted. This was achieved by relinearizing the circular version of chromosome III through the introduction of a telomere-seeding cassette adjacent immediately to the chromosome III centromere (Fig. 7A).
Figure 7.
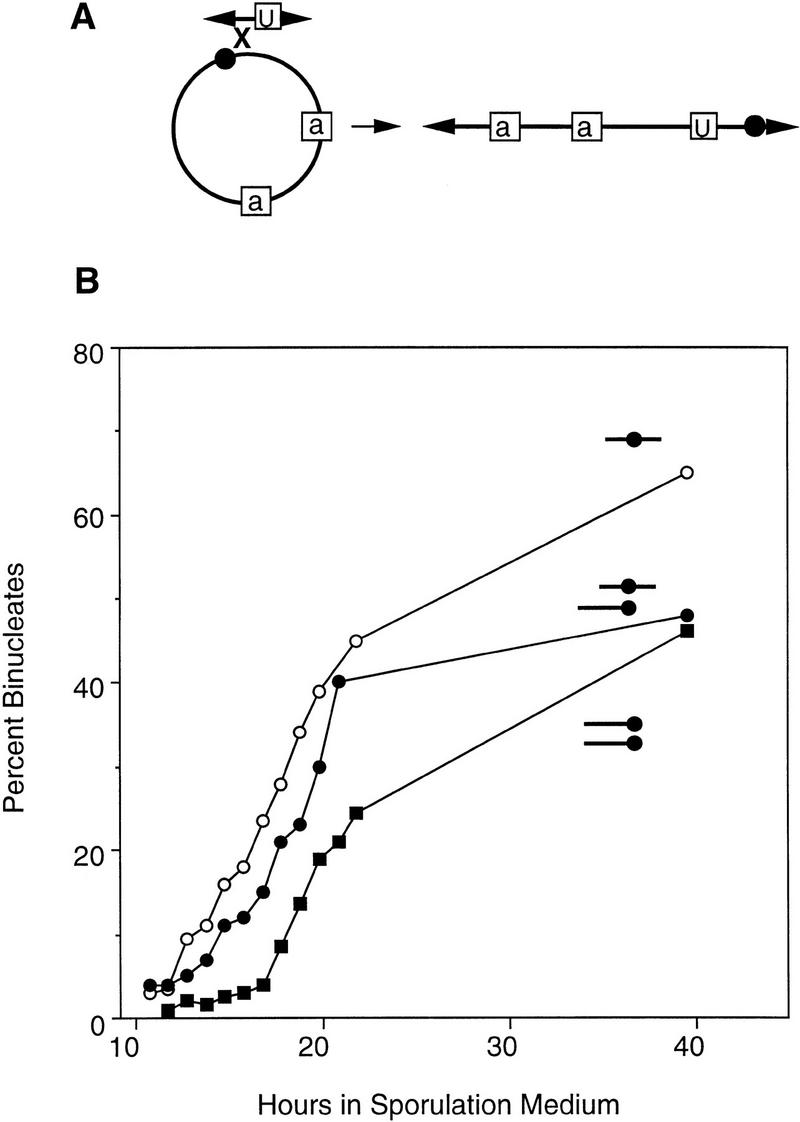
Construction and analysis of disomes containing permuted linear chromosomes. (A) Diagram of the formation of a permuted derivative of chromosome III (see Materials and Methods; see Figs. 3A and 5A for symbols). (B) Binucleate formation was monitored in a haploid containing a normal linear copy of chromosome III (BR3133, ○, T50 = 19.3 hr), in a disomic strain carrying two copies of the permuted version of chromosome III (BR3174, ▪, T50 = 21.9 hr), and in a disomic strain carrying one normal and one permuted copy of chromosome III (BR3125-1A, •, T50 = 20.4 hr).
A haploid containing the permuted chromosome III sporulates at the same time as a normal haploid, and a strain disomic for the permuted chromosome sporulates at the same time as a strain disomic for the normal chromosome (data not shown). Thus, the engineered telomeres do not interfere with homolog recognition. A disome that contains one normal copy of chromosome III and one linear permuted version of the chromosome undergoes meiotic nuclear division with timing intermediate between that of a simple haploid and that of a disome (Fig. 7B). Thus, chromosome ends appear to play a role in pairing even under conditions in which bouquet formation is not expected to bring homologous sequences into alignment. However, nonhomologous chromosome ends apparently promote pairing with reduced efficiency compared to telomeres located at homologous positions (Fig. 4D).
Proximity bypasses the requirement for telomeres
If telomeres promote pairing by bringing homologs into close proximity, then telomeres might not be necessary for pairing if homologs are physically attached to each other. To investigate this possibility, we constructed a strain carrying a circular chromosome that consists of two tandem copies of chromosome III (Fig. 8A). Meiotic nuclear division in a haploid strain carrying the dimeric copy of chromosome III is delayed by nearly 3 hr compared to a haploid carrying a circular chromosome III of normal size (Fig. 8B). The extent of the delay is similar to that observed in a disome containing two linear copies of chromosome III. These observations imply that the two copies of chromosome III in the dimeric circle efficiently recognize each other as homologs, despite the absence of telomeres, and this recognition is sufficient to effect a delay in meiotic prophase progression (Fig. 4E).
Figure 8.
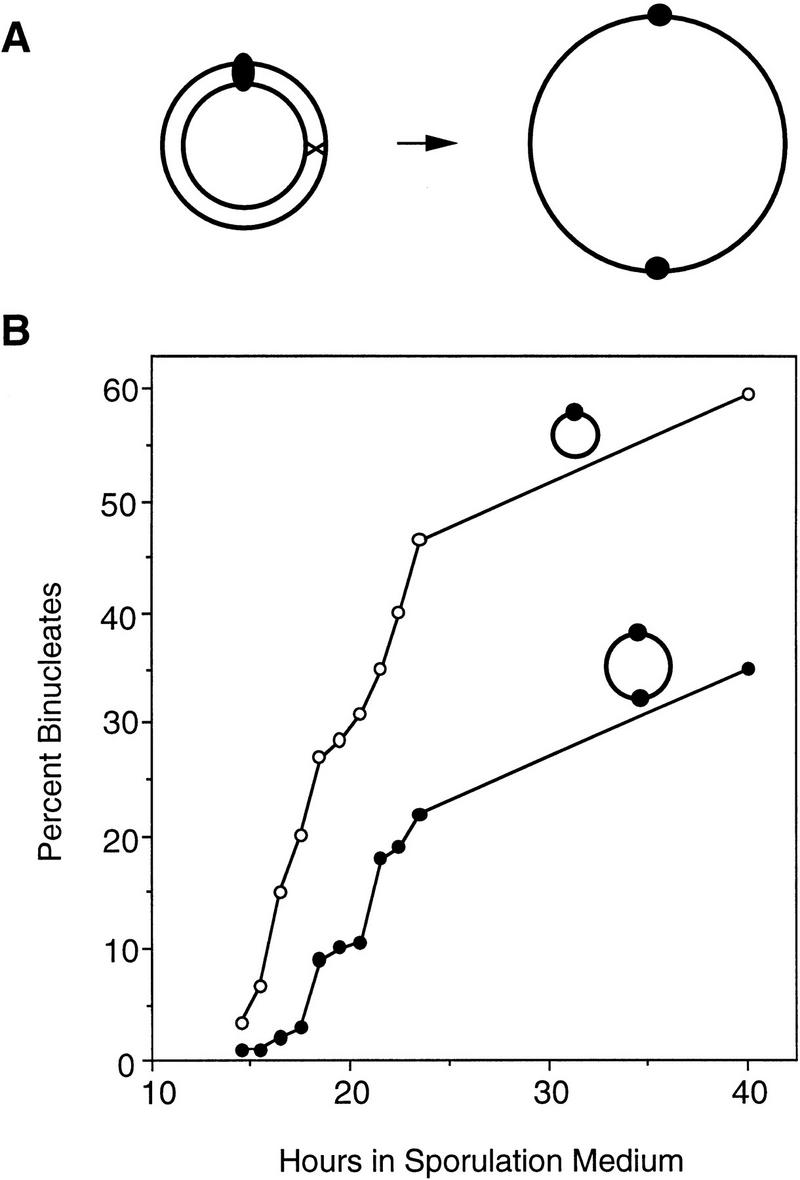
Construction and analysis of a haploid carrying a circular dimeric chromosome. (A) Diagram of the formation of a dimeric circular chromosome III (see Materials and Methods). (B) Binucleate formation was assayed in a haploid carrying either a circular chromosome III (BR3114, ○, T50 = 20.1 hr) or the double-sized circular version of chromosome III (BR3206, •, T50 = 22.8 hr).
Ndj1 is required to delay sporulation in disomes
The meiosis-specific Ndj1 protein (a.k.a. Tam1) localizes specifically to the ends of meiotic chromosomes (Chua and Roeder 1997; Conrad et al. 1997), making it a candidate for a protein involved in telomere-mediated homolog pairing. To determine whether the Ndj1 protein is required for the aspect of homolog recognition that leads to delayed sporulation in disomic strains, an ndj1 haploid and an isogenic mutant strain disomic for chromosome III were constructed and analyzed for the kinetics of meiotic nuclear division. The mutant haploid and the mutant disome undergo meiosis with the same timing (Fig. 1E), consistent with the hypothesis (Chua and Roeder 1997) that the Ndj1 protein promotes homolog recognition.
Telomeres promote chromosome pairing in diploids
If telomeres play an important role in chromosome pairing in a normal meiosis, then a circular chromosome should display a reduced capacity to pair and synapse with its homolog in wild-type diploids. To investigate this possibility, FISH was used to measure chromosome pairing in diploids carrying either two normal linear copies of chromosome III or one linear and one circular copy of the chromosome (Fig. 6). Homolog pairing was measured only in nuclear spreads at the pachytene stage of meiosis when SC formation is maximal.
In the diploid containing two linear copies of chromosome III, chromosome III failed to pair in only 2% of pachytene nuclei (Fig. 6F). In contrast, in a diploid containing one linear and one circular copy of the chromosome, the chromosome III homologs were unpaired in 22% of pachytene nuclei (Fig. 6F). Thus, telomeres play an important role in promoting interactions between homologous chromosomes even in wild-type diploids.
Discussion
The presence of homologs triggers a delay in meiotic progression
Our results demonstrate that meiotic nuclear division is delayed in disomic strains of S. cerevisiae, as compared to isogenic haploids. This effect is independent of the method used to achieve expression of both types of mating information (i.e., a sir2 mutation or integration of a MAT-containing plasmid). In addition, the delay is not because of an increase in the total number of chromosomes, because a strain in which chromosome III has been fragmented into two smaller chromosomes sporulates with the same timing as a haploid. Two observations indicate that the delay is not caused by an imbalance between the gene products encoded by the disomic chromosome and those specified by the rest of the genome. First, the same meiotic phenotype is observed in strains disomic for chromosome III or VIII, though neither of these strains displays any obvious defect in vegetative growth. Second, a disome containing one normal copy of chromosome III and one circular derivative of this chromosome sporulates with the same timing as a haploid. Thus, any problem with gene dosage (in a normal disome) would have to be caused by the small number of nonessential genes that lie distal to HML and HMR.
What aspect of homolog recognition is responsible for the delay in meiotic prophase progression? Our FISH analyses support the notion that recognition involves a direct physical association between homologs. Examination of disomic strains carrying meiotic mutations demonstrates that neither recombination intermediates nor SC are required. Homolog recognition does not involve any process or structure that is unique to telomeres, because it occurs in strains carrying the dimeric circular version of chromosome III. Weiner and Kleckner (1994) have suggested that early meiotic pairing involves the formation of unstable side-by-side joints between intact DNA duplexes; perhaps interactions of this type are able to promote the delay. Observations of pairing in premeiotic cells (Loidl et al. 1994; Weiner and Kleckner 1994) raise the possibility that the relevant aspect of homolog recognition occurs prior to the entry into meiosis. However, the requirement for the meiosis-specific Ndj1 protein for the disome-induced delay in sporulation argues against this possibility.
By what mechanism does homolog recognition effect a delay in meiotic progression? One possibility is that the delay reflects the operation of a cell cycle checkpoint (Hartwell and Weinert 1989) that monitors homolog pairing. Pairing of a single pair of homologs may impose a requirement that meiosis not proceed until most or all chromosomes are paired. Under these conditions, unpaired chromosomes might serve as negative regulators of meiotic progression. It is also possible that homolog pairing slows meiotic progression more directly. For example, there might exist a signaling pathway whereby pairing activates a protein that delays meiosis or inhibits a protein that promotes meiotic progression. The alternative to a delay that is regulatory in nature is that the early pairing process generates a product that serves as a necessary substrate for a downstream event and therefore for meiotic progression (Hartwell and Weinert 1989). This seems unlikely because no such product can be generated in simple haploids, and the requirement for this product would have to be bypassed (or a product would have to be generated faster) in ndj1 disomes and in disomes in which the extra chromosome is circular.
Chromosome ends play an important role in homolog recognition
A disomic strain in which the extra chromosome is circular sporulates with the same timing as a haploid, indicating that the aspect of homolog recognition responsible for delayed sporulation requires both chromosomes to have ends. The requirement for the telomere-associated Ndj1 protein provides additional evidence that telomeres participate in meiotic homolog pairing. A role for telomeres at an early stage of homolog pairing can be inferred from previous studies in which chromosome pairing in various meiotic mutants of yeast was assessed by FISH. Although spo11 strains display negligible amounts of pairing at internal sites on chromosomes, they display a significant level of pairing in subtelomeric regions (Weiner and Kleckner 1994). Thus, these previously published FISH data are consistent with an interaction between homologous telomeres that is independent of DSB formation and processing.
To determine whether homolog recognition requires chromosomes to be homologous at or near their termini, we constructed and analyzed a disomic strain in which the two copies of chromosome III are permuted with respect to each other. The telomeres of these chromosomes share the TG1–3 repeats required for telomeric function (for review, see Louis 1995); however, subtelomeric sequences that are located near the ends of the normal chromosome are positioned near the middle of the permuted chromosome and vice versa (thus, all four chromosome ends are different). Nuclear division in this strain is delayed compared to a haploid, but the delay is less pronounced than in a disome containing either two normal copies of chromosome III or two copies of the permuted chromosome. Thus, even nonhomologous chromosome ends have some ability to promote homolog pairing, though not to the same extent as homologous telomeres.
Physical attachment of homologs bypasses the requirement for telomeres
Meiotic nuclear division is delayed in a strain containing the dimeric circular version of chromosome III, suggesting that the two copies of chromosome III can find and recognize each other despite the absence of telomeres. An alternative explanation is that the dicentric circle causes a delay at metaphase (instead of prophase) because of the formation of chromosome bridges resulting from the attachment of a single chromosome to microtubules from opposite spindle poles. This possibility can be excluded based on two observations. First, a disome containing one linear and one circular copy of chromosome III undergoes meiosis with the same timing as a haploid, even though this strain generates a dicentric chromosome in most meioses (because of crossing over). Second, staining of spread nuclei with antitubulin antibodies demonstrates that a strain containing the double-sized chromosome III circle does not display an increase in the fraction of cells containing spindles (data not shown), which would be expected if cells were delayed at metaphase. Thus, the delay in sporulation caused by the dimeric circle is apparently due to homolog recognition. These observations imply that the role of telomeres is to bring homologous chromosomes into close physical proximity. They exclude models for telomere-mediated pairing in which the telomeres act as exclusive loading sites for proteins involved in homology searching.
Telomeres play a role in chromosome pairing and synapsis
To obtain physical evidence that telomeres play a role in pairing, we used FISH to measure chromosome pairing in disomic cells carrying either two linear copies of chromosome III or one linear chromosome III and one circular derivative. Disomic strains carrying a deletion of the MER2 gene were used in these experiments so that pairing could be assayed in the absence of recombination and synapsis (processes that are not required for homolog recognition). As expected, we observed a significant (about threefold) reduction in pairing between circular and linear homologs as compared to two linear homologs. These experiments demonstrate that telomeres play a role in establishing physical associations between homologs, and they are consistent with our hypothesis that the physical pairing of homologs promotes a delay in meiotic prophase progression.
Homolog pairing was also assessed in diploid cells, specifically at the pachytene stage of meiosis when chromosomes are fully condensed and SC formation is complete. In wild-type diploids containing two linear copies of chromosome III, the chromosomes were paired in nearly 100% of pachytene nuclei. In contrast, in diploids containing one linear and one circular copy, the chromosomes were paired in only 78% of nuclei. Thus, chromosome ends are important for pairing even in diploids, and the early telomere-dependent step in pairing appears to promote chromosome synapsis at a later stage.
A disomic strain carrying one linear and one circular version of chromosome III sporulates with the same timing as a haploid, suggesting a complete failure of homolog recognition. Nevertheless, most pairs of circular and linear chromosomes are synapsed at pachytene in diploids. These observations imply that a defect at an early step in pairing can be partially compensated for by processes that occur later in meiotic prophase. For example, DSBs might be able to stimulate a homology search, as suggested by the observation that DSBs are repaired efficiently in vegetative cells even when the template for repair is located on a nonhomologous chromosome (Pâques et al. 1998). Our observations are consistent with the data of Haber et al. (1984), in which there was a significant increase in the fraction of chromosome III pairs that failed to recombine in a diploid containing one circular copy of chromosome III. Nevertheless, most chromosome III pairs underwent normal levels of recombination.
How do telomeres promote pairing?
How might telomeres play a role in homolog pairing? As indicated in the introductory section, meiotic chromosomes in many organisms form a bouquet in which telomeres are clustered together on the nuclear membrane (for review, see Dernburg et al. 1995). The base of the bouquet is juxtaposed to the microtubule-organizing center. Telomere clustering occurs in late leptotene or early zygotene, and telomeres then disperse during pachytene (Dernburg et al. 1995; Scherthan et al. 1996; Bass et al. 1997). Bouquet formation is coincident approximately with homolog pairing, and it precedes the initiation of chromosome synapsis (Dawe et al. 1994; Dernburg et al. 1995; Scherthan et al. 1996; Bass et al. 1997). Bouquet formation has been postulated to facilitate homolog alignment by bringing homologous subtelomeric sequences into close proximity and parallel alignment (Dernburg et al. 1995). Homolog alignment might initiate at telomeres and then proceed along each chromosome pair.
The extent and timing of bouquet formation in S. cerevisiae is unknown as most studies of yeast chromosomes have involved two-dimensional spread preparations in which three-dimensional aspects of nuclear architecture are not preserved. It is also difficult to assess the timing of homolog pairing in yeast relative to other events in meiotic prophase. FISH studies of homolog pairing indicate that at least some homolog recognition occurs in the absence of DSBs (Loidl et al. 1994; Weiner and Kleckner 1994; Nag et al. 1995; Rockmill et al. 1995). In studies of DSB induction, the level of breakage on one chromosome was found to be affected by sequences present on the homolog, suggesting that homologous chromosomes normally associate with each other prior to the formation of DSBs (Xu and Kleckner 1995; Bullard et al. 1996; Rocco and Nicolas 1996). Time-course analyses indicate that DSBs are formed early in meiotic prophase prior to the initiation of SC formation (Padmore et al. 1991). These observations suggest that homolog recognition occurs by the leptotene stage of meiotic prophase. Additional experiments (including the analysis of telomere clustering in intact cells) will be necessary to determine whether telomeres cluster early in meiotic prophase in yeast, as they do in many other organisms.
We have demonstrated that chromosome ends in which the subtelomeric sequences are nonhomologous can facilitate homolog recognition (to a limited extent). Thus, telomere clustering may not be the only factor contributing to telomere-mediated homolog recognition. If the normal and permuted copies of chromosome III engage in a bouquet configuration, homologous sequences would not be aligned with each other. It is possible that telomere-mediated chromosome movements during leptotene increase the frequency of random collisions between homologous sequences. In contrast to linear chromosomes, a circular chromosome might be immobile and possibly sequestered within a limited part of the nucleus. Thus, the circle would be inaccessible to its homolog, unless its homolog were physically attached. This model is suggested by studies of S. pombe in which telomere-mediated chromosome movements apparently promote interactions between homologs (Chikashige et al. 1994; Svoboda et al. 1995; Shimanuki et al. 1997).
Several studies have suggested a role for microtubules in homolog pairing (for review, see Loidl 1990; Dernburg et al. 1995). One study (Driscoll and Darvey 1970) employed a strain of wheat carrying an isochromosome (two homologous chromosome arms attached to a common centromere). Application of colchicine reduced the frequency of crossing over on the normal chromosome pairs, but not on the isochromosome. Thus, some aspect of the interaction between homologous chromosomes is sensitive to colchicine and therefore presumably involves microtubules. The requirement for this step in pairing is alleviated if homologs are physically attached to each other (analogous to our dimeric circular chromosome). We propose that the telomere-mediated step in homolog pairing demonstrated by our data corresponds to the colchicine-sensitive step demonstrated by the study of the wheat isochromosome.
Gene products involved in telomere-mediated homolog pairing
The Ndj1 protein is produced only in meiotic cells and localizes specifically to the ends of chromosomes (Chua and Roeder 1997; Conrad et al. 1997). An ndj1 mutation increases the frequency of chromosome pairs that fail to cross over and causes a delay in chromosome synapsis. The ndj1 mutation confers a defect in the distributive disjunction of linear, but not circular, artificial chromosomes (Conrad et al. 1997), consistent with the hypothesis that ndj1 effects on meiosis are mediated through chromosome ends. We have found that an ndj1 mutation causes a disome to undergo meiosis with the same timing as a haploid, indicating that the Ndj1 protein is required for the aspect of homolog recognition that delays sporulation in disomes. We propose that the Ndj1 protein promotes homolog pairing by mediating telomere-dependent chromosome movements and/or telomere clustering.
If chromosome movement is important for pairing, then proteins associated with microtubules and the spindle pole body might be involved. In this regard, it should be noted that loss of the microtubule-dependent motor protein, Kar3, causes defects in meiotic recombination and synapsis (Bascom-Slack and Dawson 1996). Mutations that affect chromatin structure at telomeres or telomere positioning within the nucleus might also have an effect on meiotic pairing (for review, see Gilson et al. 1993; Palladino and Gasser 1994). By screening for mutants that allow disomes to sporulate with the same timing as a haploid, we hope to identify novel genes whose products are required for homolog pairing.
Materials and methods
Yeast strains
Yeast strain genotypes are given in Table 1. All strains used are isogenic with BR1919-8Bl or BR3026-38D. To exchange genetic markers between transformants of BR1919-8Bl (or BR3026-38D), MATa derivatives were isolated. Mating type was switched by transformation with pJH84 (see below) containing the HO gene. Crosses were then carried out between isogenic MATa and MATα strains and haploid (or disomic) segregants were recovered.
Table 1.
Yeast strains
| Strain
|
Genotype
|
|---|---|
| BR1919-8B1 | MATα leu2-3,112 his4-260 trp1-289 ura3-1 thr1-4 ade2-1 lys2-100 |
| BR3001 | BR1919-8B1, but sir2::LEU2 spo13::URA3-1 |
| BR3002 | BR3001, but disomic for chromosome VIII |
| BR3090 | BR1919-8B1, but URA3::MATaspol3::ura3-1 |
| BR3092 | BR3090, but mer2::ADE2 |
| BR3231 | BR1919-8B1, but THR1::MATandj1::URA3 |
| BR3091 | BR1919-8B1, but disomic for chromosome III, MATa/MATα, spol3::ura3-1 |
| BR3093 | BR3091, but mer2::ADE2 |
| BR3200-12D | BR1919-8B1, but disomic for chromosome III, MATa/MATα, ndj1::URA3, spo13::ADE2 |
| BR3026-38D | MATα CEN3::TRP1 leu2::arg4-8, CUP1 his4-260 ura3-1 trp1-289 ade2-1 arg4-Δ thr1-4 cup1-Δ lys2 |
| BR3133 | BR3026-38D, but sir2::ura3-1 spo13::LEU2 |
| BR3110 | BR3133, but bisected chromosome III, CEN3::URA3 on chromosome IIIR |
| BR3111 | BR3110, but disomic for chromosome IIIL |
| BR3108 | BR3026-38D, but disomic for chromosome III, MATa/MATα spo13::LEU2 |
| BR3173 | BR3026-38D, but permuted chromosome III, CEN3::URA3 THR1::MATaspo13::LEU2 |
| BR3174 | BR3173, but disomic for permuted chromosome III |
| BR3125-1A | BR3173, plus unrearranged copy of chromosome III |
| BR3136-9B | BR3026-38D, but disomic for chromosome III, MATa/MATα, mer2::ADE2 |
| BR3139 | BR3026-38D, but disomic for chromosome III with one copy circular and marked with HIS4, MATa/MATα |
| BR3147 | BR3139, but mer2::ADE2 |
| BR3192 | BR3026-38D, but carrying the circular version of chromosome III marked with HIS4, THR1::MATaspo13::LEU2 |
| BR3206 | BR3026-38D, but carrying double-sized circular derivative of chromosome III, homozygous for HIS4, THR1::MATa, and spo13::LEU2 |
| BR3168 | diploid homozygous for all BR3026-38D markers, MATa/MATα |
| BR3167 | same as BR3168, but one copy of chromosome III is circular and marked with HIS4, SPO13/spo13::LEU2 |
Strains BR1919-8B1, BR3001, BR3002, BR3090, BR3092, BR3091, BR3093, BR3231, and BR3200-12D are isogenic with each other. Strains BR3026-38D, BR3133, BR3110, BR3111, BR3108, BR3173, BR3174, BR3125-1A, BR3136-9B, BR3139, BR3147, BR3192, BR3206, BR3168, and BR3167 are isogenic with each other. Chromosomes IIIL and IIIR result from the bisection of chromosome III; IIL represents the left arm of the chromosome and IIIR represents the right arm.
BR3002, which is disomic for chromosome VIII, was selected based on its resistance to copper (Rockmill and Fogel 1988). BR3091, which is disomic for chromosome III, was made by crosses to a disomic strain recovered during tetrad analysis of a diploid derivative of BR1919-8Bl that is homozygous for a zip1 mutation (Sym and Roeder 1994). The dmc1::LEU2 mutation was introduced into BR1919-8Bl by backcrossing as described by Rockmill et al. (1995). Derivation of the spo13::ura3-1 allele from the spo13::URA3 allele was described previously (Rockmill and Roeder 1990).
BR3026-38D carries an insertion of the ARG4 gene (carrying the temperature-sensitive arg4-8 allele) and the CUP1 gene at the LEU2 locus on chromosome III; ARG4 and CUP1 have been deleted from chromosome VIII. These markers were acquired in crosses to strain 6D (Spector and Fogel 1992). Both arg4-8 and CUP1 confer dosage-dependent phenotypes (Rockmill and Fogel 1988). Strains disomic for chromosome III were recovered by selecting for growth in the absence of arginine at 33°C. The sir2::ura3-1 allele was derived from a sir2::URA3 ura3-1 strain by selection on medium containing 5-fluoro-orotic acid.
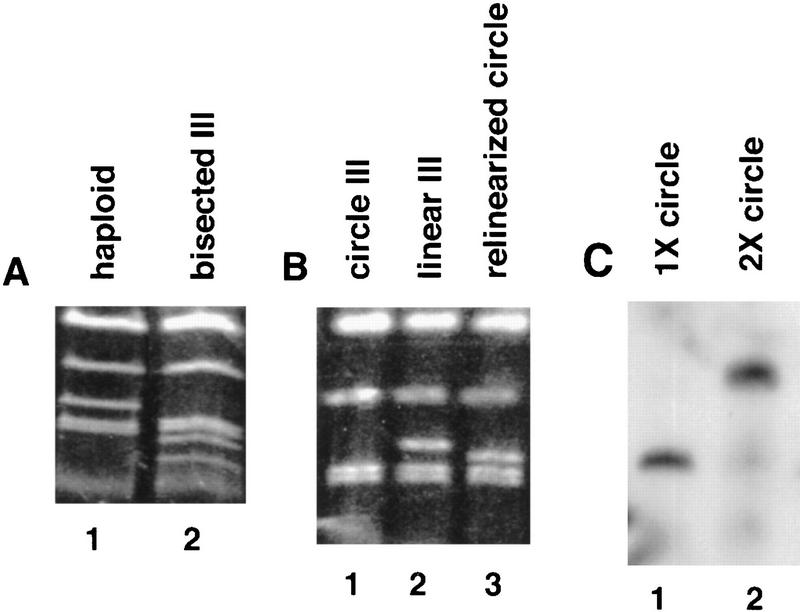
Chromosome III was bisected by transforming BR3133 with pB203 (see below). BR3110 was identified as a correct transformant by electrophoretic analysis of yeast chromosomes (Fig. 9A). BR3111 was derived from BR3110 by selecting for growth in the absence of arginine at 33°C. Strains disomic for the left arm of chromosome III are unstable and lose the extra chromosome during vegetative growth. To minimize chromosome loss, freshly selected disomes were immediately analyzed in meiotic time courses.
Figure 9.
Chromosome gel analysis of rearranged chromosomes III. (A) Crossed-field gel analysis (see Materials and Methods) of a normal haploid (BR3133, lane 1) and a haploid in which chromosome III has been bisected at the centromere (BR3110, lane 2). In lane 2, the full-length chromosome III is not detected, and two fragments of faster mobility are observed. (B) Crossed-field gel analysis of a haploid containing a circular version of chromosome III (BR3192, lane 1), a wild-type haploid (BR3133, lane 2), and a haploid in which the circular version of chromosome III has been relinearized to generate a permuted form of chromosome III (BR3173). The circular chromosome (lane 1) does not enter the gel. The permuted chromosome (lane 3) lacks the sequences distal to HML and HMR and thus migrates faster than the normal chromosome III (lane 2). (C) Southern blot analysis of strains containing either the circular version of chromosome III (lane 1) or the dimeric circle (lane 2). Strains BR3192 and BR3206 were made rad50S and then induced to undero double-strand breakage by introduction into sporulation medium. Electrophoretically separated chromosomes were probed with DNA from the THR4 locus on chromosome III. The linearized version of the dimeric chromosome (lane 2) migrates more slowly than the monomeric version (lane 1).
Selection of the circular derivative of chromosome III was based on the observation that recombination between HML and HMR is elevated in a Sir− strain (Klar et al. 1983). A MATα strain was made sir2::LEU2 and HIS4 and force mated to an isogenic MATa spo13::URA3 strain. One of 15 diploids isolated and analyzed by tetrad analysis displayed the patterns of spore inviability and segregation expected for a diploid carrying one circular and one linear copy of chromosome III. (Recombination between a linear and a circular chromosome produces a dicentric chromosome. When this chromosome is recovered, the result is a three-spore viable tetrad in which the spore carrying the dicentric chromosome is Arg+ at 33°C. When the dicentric chromosome is lost, a two-spore viable tetrad results, and both spores are Arg− at 33°C.) Several segregants were analyzed by crossed-field gel electrophoresis, and the presumptive circle failed to enter the gel, as expected (Fig. 9B). Southern blot analysis confirmed the presence of the expected HML/HMR fusion band (Klar et al. 1983). In addition, the presence of the circle was confirmed by crossed-field gel analysis of meiotic chromosomes. A rad50S mutation was introduced into the circle-bearing strain, BR3192; rad50S prevents the processing of meiosis-specific double-strand breaks (Alani et al. 1990). The rad50S derivative of BR3192 was induced to enter meiosis, and chromosomal DNA was analyzed by crossed-field gel electrophoresis followed by Southern blot analysis as described previously (Rockmill et al. 1995). The linearized circle resulting from double-strand breakage entered the gel and migrated slightly faster than the normal chromosome III (Fig. 9C).
Disomic strains carrying one linear and one circular copy of chromosome III were constructed in two steps. First, a MATα strain carrying the circular derivative of chromosome III was crossed to a BR3036-38D derivative that is a MATa/MATa disome. Second, the resulting trisomic strain was sporulated and tetrads were dissected. The segregants of interest are disomic (Arg+ at 33°C); they carry the circular chromosome (His+) and are heterozygous at MAT (proficient in sporulation). Such disomic strains are very unstable. To minimize the opportunity for chromosome loss during vegetative growth, disomes were freshly generated immediately prior to meiotic time-course analyses.
The permuted chromosome III was created by transforming a circle-bearing strain with pB202 (see below). Transformants were analyzed by crossed-field gel electrophoresis; the permuted chromosome enters the gel, but migrates slightly slower than the unrearranged version of chromosome III (Fig. 9B). A disome carrying one permuted and one normal copy of chromosome III (BR3125-1A) was generated by crossing a MATα strain carrying the permuted chromosome to an isogenic MATa/MATa disome and identifying a nonmating Ura+ meiotic segregant. The chromosome complement was confirmed by crossed-field gel electrophoresis.
Strain BR3206, carrying a double-sized circular chromosome III, was recovered from a haploid containing the circular version of this chromosome by selection on medium lacking arginine at 33°C. (The double-sized circle results from sister chromatid exchange.) To confirm the presence and size of the circular chromosome, BR3206 was made rad50S and DNA from meiotic cells was analyzed as described above for BR3192. After meiosis-specific double-strand breakage, the double-sized circular chromosome enters the gel, but migrates considerably slower than the normal chromosome III (Fig. 9C).
Plasmids
pB211 and pJH2 were used to integrate MATa at THR1 and URA3, respectively. pB211 was constructed by inserting a 1.8-kb HindIII fragment of THR1 from pCB156 (Chua and Roeder 1995) at the HindIII site in pJH3, which is pBR322 containing a 1.2-kb EcoRI–HindIII fragment of MATa between the EcoRI and HindIII sites. pJH2 is pBR322 with a 3.6-kb EcoRI–HindIII fragment of MATa inserted between the EcoRI and HindIII sites and a 1.2-kb HindIII fragment containing the URA3 gene inserted at the HindIII site. pJH2 and pJH3 were provided by James Haber (Brandeis University, Waltham, MA). pB211 and pJH2 were cut with MscI prior to transformation into yeast.
pB203 was used to bisect chromosome III at its centromere. To construct this plasmid, the 3.5-kb EcoRI–HindIII fragment containing CEN3 was inserted between the EcoRI and HindIII sites of YIp5 (Parent et al. 1985). The BamHI fragment of the resulting plasmid was deleted, and the BamHI site was destroyed by filling in the sticky ends. Then, a 1.5-kb XhoI fragment containing a telomere-seeding cassette was excised from pKR53 (provided by Kurt Runge and Virginia Zakian, Princeton University, NJ) and inserted at the SalI site. Prior to transformation with pB203, the telomeres were released by cutting with BamHI; Ura+ transformants were selected.
pB202, which was used to linearize the circular derivative of chromosome III, was constructed in three steps. First, the 1.5-kb EcoRI–HindIII fragment immediately to the right of CEN3 was inserted between the EcoRI and HindIII sites of YIp5 (Parent et al. 1985). Second, the BamHI site in YIp5 DNA was destroyed by filling in the ends. Third, an SphI fragment containing the telomere-seeding cassette from pKR53 was inserted at the SphI site. Prior to transformation with pB202, the telomeres were released by cutting with BamHI.
pR978 and pCB374 were used to introduce the spo13::ADE2 and spo13::LEU2 disruptions, respectively. To construct pR978, an EcoRI–PstI fragment carrying the SPO13 gene was inserted between EcoRI and PstI sites of pGEM (Sambrook et al. 1989); a 3.6-kb BamHI fragment carrying the ADE2 gene from Yp3.6 ADE (Engebrecht and Roeder 1990) was then inserted at the HpaI site in SPO13. This plasmid was digested with PstI prior to transformation into yeast. To construct pCB374, the PstI–HpaI fragment of LEU2 from YEp351 (Hill et al. 1986) was inserted between the PstI and EcoRV sites of SK− (Stratagene). The HindIII fragment containing LEU2 was then removed from this plasmid and inserted at the HindIII site in p(SPO13)16 (Wang et al. 1987). pCB374 was digested with AatII and PstI prior to transformation into yeast.
A derivative of BR3026-38D was rendered HIS4 by transformation with pR78 cut with PstI. pR78 contains the PstI fragment containing the wild-type HIS4 gene inserted at the PstI site in pBR322. The mating type of BR3026-38D and BR1919-8B was switched by transformation with pJH84 (provided by James Haber), which carries a 2.5-kb HindIII fragment containing the HO gene at the HindIII site in YEp13.
Other mutations were introduced by transformation with the following plasmids: pR1400 for mer2::ADE2 (Engebrecht et al. 1990), p(SPO13)16 for spo13::URA3 (Wang et al. 1987), pNKY349 for rad50S (Alani et al. 1990), pJ303-4 for CEN3::URA3 (Clarke and Carbon 1983), pJH103.1 for sir2::LEU2 (Ivy et al. 1986), pJH1088 for sir2::URA3 (provided by Jim Haber, Brandeis University, Waltham, MA) and pCB249 for ndj1/tam1::URA3 (Chua and Roeder 1997).
Chromosome gels
Agarose plugs containing yeast chromosomes were prepared according to Sherman and Wakem (1991) in InCert agarose (FMC Bioproducts, Rockland, Maine). Gels were run on a crossed field gel apparatus at 150 volts with a 50-sec. pulse time for 17 hr.
Cytology and meiotic time-course analyses
For time-course analyses of meiotic nuclear division, cells were grown in YEPD medium (Sherman et al. 1986) supplemented with 0.3 mm adenine and 0.2 mm uracil. At early stationary phase, 1.5 ml of cells were spun down and resuspended in 10 ml of 2% potassium acetate and shaken vigorously at 30°C. At appropriate times, 200-μl aliquots were removed, added to 20 μl of 37% formaldehyde, and stored at 4°C for 3 days (to decrease the fluorescence of dityrosine); 5 μl of cells were then applied per well of 18-well glass slides. After incubation at 25°C for 20 min, the slides were washed once with PBS (150 mm NaCl, 50 mm KPO4 at pH 7.2) and a mounting solution (1 μg/ml p-phenylenediamine, 80% glycerol, 0.1 × PBS, 1 μg/ml DAPI) was applied. In Figures 1, 3, 5, 7, and 8, each graph represents strains analyzed in the same experiment. All strains were examined in at least two meiotic time courses, and at least 200 cells were scored for nuclear division at each time point. Although the absolute T50 value for a given strain varied somewhat among experiments, the relative order of the T50 values for different strains was constant among experiments.
Chromosome spreads were prepared, and immunofluorescence and FISH were carried out as described by Chua and Roeder (1998). The chromosome III probe consists of a 25-kb segment of chromosome III DNA including the CRY1 gene. The chromosome V probe contains ∼15 kb of chromosome V DNA including the CYC7 and RAD23 genes.
Acknowledgments
We are grateful to Penelope Chua, Jennifer Fung, Janet Novak, Pedro San Segundo, and Albert Smith for helpful comments on the manuscript. This work was supported by American Cancer Society grant VM-7G to G.S.R. and by the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shirleen.roeder@yale.edu; FAX (203) 432-3263.
References
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Bascom-Slack CA, Dawson DS. The yeast motor protein, Kar3p, is essential for meiosis I. J Cell Biol. 1996;139:459–467. doi: 10.1083/jcb.139.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: A 3-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard SA, Kim S, Galbraith AM, Malone RE. Double-strand breaks at the HIS2 recombination hot spot in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1996;93:13054–13059. doi: 10.1073/pnas.93.23.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Yanagida S, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y. Meiotic nuclear reorganization: Switching the position of centromeres and telomeres in fission yeast Schizosaccharomyces pombe. EMBO J. 1997;16:193–200. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P, Roeder GS. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol. 1995;15:3685–3696. doi: 10.1128/mcb.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes & Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature. 1983;305:23–28. doi: 10.1038/305023a0. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Dominguez AM, Dresser ME. Ndj1, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Dawe KR, Sedat JW, Agard DA, Cande WZ. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell. 1994;76:901–912. doi: 10.1016/0092-8674(94)90364-6. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Cande WZ, Bass HW. Cytology of telomeres. In: Blackburn EH, Grieder CW, editors. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 295–337. [Google Scholar]
- Driscoll CJ, Darvey NL. Chromosome pairing: Effect of colchicine on an isochromosome. Science. 1970;169:290–291. doi: 10.1126/science.169.3942.290. [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Roeder GS. MER1,a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol Cell Biol. 1990;10:2379–2389. doi: 10.1128/mcb.10.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J, Hirsch J, Roeder GS. Meiotic gene conversion and crossing over: Their relationship to each other and to chromosome synapsis and segregation. Cell. 1990;62:927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- Gilson E, Laroche T, Gasser SM. Telomeres and the functional architecture of the nucleus. Trends Cell Biol. 1993;3:128–134. doi: 10.1016/0962-8924(93)90175-z. [DOI] [PubMed] [Google Scholar]
- Giroux CN, Dresser ME, Tiano HF. Genetic control of chromosome synapsis in yeast meiosis. Genome. 1989;31:88–94. doi: 10.1139/g89-017. [DOI] [PubMed] [Google Scholar]
- Haber JE, Thorburn PC, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984;106:185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. colishuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Ivy JM, Klar AJS, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S, Esposito RE. Recombination and chromosome segregation during the single division meiosis in spo12-1 and spo13-1diploids. Genetics. 1980;96:589–611. doi: 10.1093/genetics/96.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJS, Strathern JN, Hicks JB, Prudente D. Efficient production of a ring derivative of chromosome III by the mating-type switching mechanism in Saccharomyces cerevisiae. Mol Cell Biol. 1983;3:803–810. doi: 10.1128/mcb.3.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. The initiation of meiotic chromosome pairing: The cytological view. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Palladino F, Gasser SM. Telomere maintenance and gene repression: A common end? Curr Opin Cell Biol. 1994;6:373–379. doi: 10.1016/0955-0674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Pâques F, Leung W-Y, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent SA, Fenimore CM, Bostian KA. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985;1:83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Rocco V, Nicolas A. Sensing of DNA non-homology lowers the initiation of meiotic recombination in yeast. Genes Cells. 1996;1:645–661. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Fogel S. DIS1:A yeast gene required for proper meiotic chromosome disjunction. Genetics. 1988;119:261–272. doi: 10.1093/genetics/119.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Engebrecht J, Scherthan H, Loidl J, Roeder GS. The yeast MER2gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics. 1995;141:49–59. doi: 10.1093/genetics/141.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scherthan H, Bahler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Wakem P. Mapping yeast genes. Methods Enzymol. 1991;194:38–57. doi: 10.1016/0076-6879(91)94006-x. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics: A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shimanuki M, Miki F, Ding D-Q, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol & Gen Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector LM, Fogel S. Mitotic hyperploidy for chromosome VIII and III in Saccharomyces cerevisiae. Curr Genet. 1992;21:309–318. doi: 10.1007/BF00351688. [DOI] [PubMed] [Google Scholar]
- Svoboda A, Bahler J, Kohli J. Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma. 1995;104:203–214. doi: 10.1007/BF00352185. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Wagstaff JE, Klapholz S, Esposito RE. Meiosis in haploid yeast. Proc Natl Acad Sci. 1982;79:2986–2990. doi: 10.1073/pnas.79.9.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Frackman S, Kowalisyn J, Esposito RE, Elder R. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol Cell Biol. 1987;7:1425–1435. doi: 10.1128/mcb.7.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]