Abstract
The rapid increase in the prevalence of chronic heart failure (CHF) worldwide underscores an urgent need to identify biomarkers for the early detection of CHF. Post-translational modifications (PTMs) are associated with many critical signaling events during disease progression and thus offer a plethora of candidate biomarkers. We have employed top-down quantitative proteomics methodology for comprehensive assessment of PTMs in whole proteins extracted from normal and diseased tissues. We have systematically analyzed thirty-six clinical human heart tissue samples and identified phosphorylation of cardiac troponin I (cTnI) as a candidate biomarker for CHF. The relative percentages of the total phosphorylated cTnI forms over the entire cTnI populations (%Ptotal) were 56.4±3.5%, 36.9±1.6%, 6.1±2.4%, and 1.0±0.6% for postmortem hearts with normal cardiac function (n=7), early-stage of mild hypertrophy (n=5), severe hypertrophy/dilation (n=4), and end-stage CHF (n=6), respectively. In fresh transplant samples, the %Ptotal of cTnI from non-failing donor (n=4), and end-stage failing hearts (n=10) were 49.5±5.9% and 18.8±2.9%, respectively. Top-down MS with electron capture dissociation unequivocally localized the altered phosphorylation sites to Ser22/23 and determined the order of phosphorylation/dephosphorylation. This study represents the first clinical application of top-down MS-based quantitative proteomics for biomarker discovery from tissues, highlighting the potential of PTM as disease biomarkers.
Keywords: Heart failure, Phosphorylation, Quantitative Proteomics, Top-Down Mass Spectrometry, Post-translational Modification, Cardiac troponin I
INTRODUCTION
Heart disease is the leading cause of mortality and morbidity for both men and women in developed countries.1 While treatments for acute coronary syndromes have dramatically improved in the past decades, chronic heart failure (CHF) has become an increasingly prevalent syndrome worldwide in aging populations.1–2 CHF is often diagnosed late during disease progression which substantially limits the options for therapeutic interventions.3 Early detection of CHF would allow implementation of early intervention strategies to delay or prevent disease progression, which will not only save costs for medical care but will also lead to improved prognosis and quality of life.3–4 Biomarker have become an increasingly useful tool in clinical practice for diagnosis, prognosis, and risk stratification of disease and for evaluation of therapy.5–8 However, very few clinically approved biomarkers are currently available for the diagnosis of CHF.9–10 Hence, there is an urgent need for the discovery of biomarkers with high specificity and accuracy for the early detection and treatment of CHF.3, 9–10
Protein post-translational modifications (PTMs) are associated with many critical signaling events during disease progression and thus offer a plethora of candidate biomarkers.11 PTMs are covalent modifications of a protein that can modulate its activity, stability, and function.12 Aberrant protein PTMs are found to be connected with a variety of human diseases, most notably cancer and heart diseases.11, 13–15 Recent studies have suggested that the profile of PTMs can be utilized as biological fingerprints for tracking and verifying the activity of key cellular signaling pathways, underscoring the potential of PTMs as a biomarker.8, 13, 15 Hence, a comprehensive assessment including the detection, identification, quantification and characterization of PTMs is essential for developing clinically useful biomarkers with high specificity.
"Top-down" mass spectrometry (MS) is emerging as a powerful technology for assessing protein modifications.16–27 Unlike the more traditional "bottom-up" method which requires in-gel or in-solution digestion of proteins prior to MS analysis, top-down MS analyzes whole proteins without proteolytic digestion therefore enabling detection of all existing modifications at the protein level including PTMs (i.e., phosphorylation, proteolysis, acetylation) and sequence variants (i.e., mutants, alternatively spliced isoforms) simultaneously in a single spectrum (a "bird's eye view”) without a priori knowledge.28–29 The specific modified form of interest can then be directly isolated in the mass spectrometer and subsequently fragmented by tandem MS (MS/MS) such as collision-induced dissociation (CID) and electron capture dissociation (ECD) for highly reliable mapping of the modification sites with full sequence coverage.16, 18–19, 21–25, 27 ECD30, a nonergodic MS/MS technique, is particularly suitable for the localization of labile PTMs since they are well-preserved during the ECD fragmentation process.20–24, 31–32 The top-down MS approach is especially valuable for quantification of the relative abundance of modified protein species, since the physico-chemical properties of whole proteins are much less affected by the presence of modifying groups in comparison with peptides.20, 33–34
Although serum and plasma have been the focus for clinical proteomic studies, the extremely low abundance of potential biomarkers present in blood against the huge complexity and dynamic range of serum/plasma proteome makes it very difficult to discover novel biomarkers directly from blood specimen.35 In contrast, damaged tissue closest to the disease source is known to contain the highest concentration of potential disease markers, and thus it is the preferred sample choice for biomarker discovery.5, 36–39 The identified biomarkers from tissues can be further validated in serum/plasma using targeted detection such as antibody-based immunoassays or MS-based multiple (or selected) reaction monitoring (MRM/SRM) methods.5, 35, 40 Nonetheless, extraction, separation/purification and MS analysis of whole proteins from tissues remain challenging.
Herein, we have employed a simple and robust top-down quantitative proteomics methodology featuring affinity chromatography and high-resolution MS for the comprehensive assessment of PTMs in whole proteins extracted from tissues for biomarker discovery. We have comprehensively evaluated the PTMs of cardiac troponin I (cTnI) purified from clinical human heart samples. cTnI is well recognized as the gold-standard biomarker for acute coronary syndrome since it is released into the general circulation following the necrosis of heart muscle tissues.41 However, whether cTnI can also be used as a biomarker for chronic heart diseases remains unclear. cTnI is the inhibitory subunit of the cardiac troponin complex (cTn) and its interactions with other cTn subunits, cTnC, cTnT and actin-tromopomyosin play pivotal roles in regulating Ca2+-dependent cardiac contraction and relaxation.42–43 cTnI is also known to exhibit PTMs under both physiological and pathological conditions.13, 43–45 PTMs, most notably phosphorylation, are known to modulate cardiac contractility, and altered PTMs/mutations of cTnI are believed to account for cardiac dysfunctions in various types of heart diseases.46–50 Hence, the status of PTMs in cTnI is likely to provide information related to disease etiology and prognosis, suggesting its potential as a disease biomarker. We have systematically analyzed a large set of postmortem (n=22) and transplant (n=14) human heart tissue samples with varying stages of CHF, together with healthy controls. We have unambiguously identified the phosphorylation status of cTnI as a reliable candidate biomarker in CHF with high potential for detection of CHF at the early stages. Moreover, we have localized the altered phosphorylation sites to Ser22/23, the bona fide substrates of protein kinase A (PKA), and determined the order of phosphorylation/dephosphorylation of these sites in normal and diseased myocardium, respectively. In contrast, no direct correlation could be established between the detected cTnI degradation products with the heart disease phenotypes.
EXPERIMENTAL PROCEDURES
Reagents
All reagents were purchased from Sigma Chemical Co (St Louis, MO, USA) unless noted otherwise. Complete protease and phosphatase inhibitor cocktail tablets were purchased from Roche Diagnostics Corporation (Indianapolis, IN, USA). All solutions were prepared in Milli-Q water (Millipore Corporation, Billerica, MA).
Human heart tissue samples
The postmortem (autopsy) heart tissue samples (n=22, clinical characteristics summarized in Supplemental Table S1) were collected within 34 hours from deceased patients in UW-hospital and clinics with the bodies being preserved at 4 °C prior to autopsy. All the fresh heart tissue samples (n=14, clinical characteristics summarized in Supplemental Table S2) were collected within 15 min after the heart stopped beating from explanted failing hearts of transplant recipients or patients receiving ventricular assist devices (VAD), as well as the healthy donor hearts with normal cardiac function. All tissue samples were excised from the free wall of left ventricle, snap-frozen in liquid nitrogen, and stored in −80 °C freezer. The use of postmortem and fresh human tissue samples from patients have been approved by the Institutional Review Board of the University of Wisconsin-Madison. Transplant tissue samples were obtained with informed consent from patients undergoing transplants or VAD surgery.
Immunoaffinity Purification of Human cTnI
Approximately 0.1–0.2 g of fresh transplant heart tissues or 1.0–1.5 g of postmortem heart tissues was excised and homogenized in tissue wash buffer (NaH2PO4 500 mM, Na2HPO4, 100 mM, MgCl2 100 mM, EGTA 100 mM, NaCl 0.1 M, Triton X-100 1%, DTT 5 mM, protease and phosphatase inhibitor cocktail tablet, PMSF 1 mM, leupeptin, 2 µg/mL, pH 7.4) using a Polytron electric homogenizer for 30 s on ice21, 24. The homogenate was centrifuged at 16,000 g for 10 min at 4 °C. The supernatant was discarded and the pellet was resuspended in 6 mL protein extraction buffer (0.7 M LiCl, 25 mM Tris, 5 mM EGTA, 0.1 mM CaCl2, 5 mM DTT, 1 mM PMSF, 2 µg/mL leupeptin, and 0.75 mg/mL protease and phosphatase inhibitor cocktail, pH 8.0). The protein extraction was performed with agitation on a nutating mixer (Fisher Scientific Inc., Pittsburgh, PA, USA) at 4 °C for 45 min. The sample was then centrifuged at 16,000 g (Centrifuge 5415R; Eppendorf, Hamburg, Germany) for 5 min to collect the supernatant. The collected supernatant was further centrifuged at 55,000 rpm (Beckman L-55 ultracentrifuge; Beckman Coulter, Fullerton, CA, USA) for 45 min to completely remove the tissue debris before affinity chromatography purification. The supernatant was incubated with 0.25 mL of CNBr-activated Sepharose CL-4B conjugated with 1.25 mg monoclonal cTnI antibody (anti-troponin I monoclonal antibodies 14G5 and MF4, Hytest, Finland) for 35 min at 4 °C to ensure the complete binding of the troponin complex to the antibody. After washing the column with 2 mL of extraction buffer, the bound troponin complex was eluted with 100 mM glycine at pH 2 into four 0.4 mL fractions and neutralized immediately with 40 µL of 1 M MOPS solution (pH 9). Fractions were analyzed for enriched protein content by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels stained with Coomassie blue. The analytical reproducibility was assessed with three technical replicates.
Top-down mass spectrometry analysis
Immunoaffinity purified human cTn complexes were separated and desalted using an offline reverse phase protein microtrap (Michrom Bioresources, Inc., CA, USA), with a two-step reverse phase gradient elution method, first with 1% acetic acid in 50:50 methanol/water and then 1% acetic acid in 75:25 methanol/water. Desalted samples were analyzed using a 7T linear trap/Fourier transform ion cyclotron resonance (FTICR) (LTQ FT Ultra) hybrid mass spectrometer (Thermo Scientific Inc., Bremen, Germany) equipped with an automated chip-based nano electrospray (ESI) source (Triversa NanoMate; Advion BioSciences, Ithaca, NY, USA) as described previously.21, 24 The spray voltage was 1.2–1.6 kV versus the inlet of the mass spectrometer, resulting in a flow of 50–200 nL/min. Ion transmission into the linear trap and subsequently into the FTICR cell was automatically optimized for the maximal ion signal. The number of accumulated ions for the full scan linear trap (IT), FTICR cell (FT), MSn FTICR cell, and ECD were 3 × 104, 8 × 106, 8 × 106, and 8 × 106, respectively. The resolving power of the FTICR mass analyzer was typically set at 200,000 at m/z 400. For collisionally induced dissociation (CID) and electron capture dissociation (ECD) fragmentation, individual charge states of protein molecular ions were first isolated and then dissociated using 15% –25% of normalized collision energy for CID or 2% –3% electron energy for ECD with a 45–125 ms duration with no delay. Typically, 1000 to 3000 transients were averaged to ensure high quality ECD spectra.
All FTICR spectra were processed with Xtract Software (FT programs 2.0.1.0.6.1.4, Xcalibur 2.0.5, Thermo Scientific Inc., Bremen, Germany) using a S/N threshold of 1.5 and fit factor of 40% and validated manually. The resulting mass lists were further assigned using the in-house developed “Ion Assignment” software (version 1.0) based on the protein sequence of human cTnI (TNNI3_human) obtained from Swiss-Prot protein knowledgebase (UniProtKB/Swiss-Prot 2011-01). Allowances were made for possible PTMs such as the removal of the initial Met, acetylation of the N-terminus, and variable phosphorylation sites (residues Ser, Thr, and Tyr), using a 10 and 20 ppm tolerance for precursor and fragment ions. The assigned ions were manually validated to ensure the quality of assignments. For fragment ions containing possible phosphorylation sites, the masses of fragment ions were manually examined for 80 Da mass shifts to confirm or exclude the existence of phosphorylation. All reported masses were the most abundant masses.
Relative quantification of cTnI post-translational modifications
To quantitatively determine the detected levels of cTnI and its modified forms, the MS signal intensity values were used to calculate the relative ratios for all protein ions observed as described previously.21, 24 Briefly, the top five most abundant isotopomer peak heights were integrated to calculate the relative abundance of the intact proteins and their modified forms. The percentage of the total phosphorylated cTnI species (%Ptotal) is defined as the summed abundances of all phosphorylated cTnI species including both mono- and bis-phosphorylated over the entire cTnI population. The percentage of the mono- (%Pmono) or bis-phosphorylated species (%Pbis) is defined as the summed abundance of mono-(pcTnI) or bis-phosphorylated cTnI (ppcTnI) species, respectively, over the entire cTnI population. The percentage of the total degraded cTnI species (%Dtotal) is defined as the summed abundances of all degraded cTnI species over the summed abundances of entire cTnI population. The percentage of individual degraded cTnI species (%Dx) is defined as the summed abundances of each individual degraded cTnI species over the summed abundances of entire cTnI population.
Statistical analysis
Data were presented graphically using a Box-plot. Linear mixed effects models with random intercepts were used to compare the mean levels of phosphorylation and degradation between groups, accounting for the correlation between repeated measurements (technical replicates) for the same biological subject and determine the significance. P-values were given directly after fitting the linear mixed model. For all the analyses, a value of P≤ 0.05 was considered as statistically significant. Data are reported as mean ± S.E.M.
RESULTS
Top-down quantitative proteomics methodology
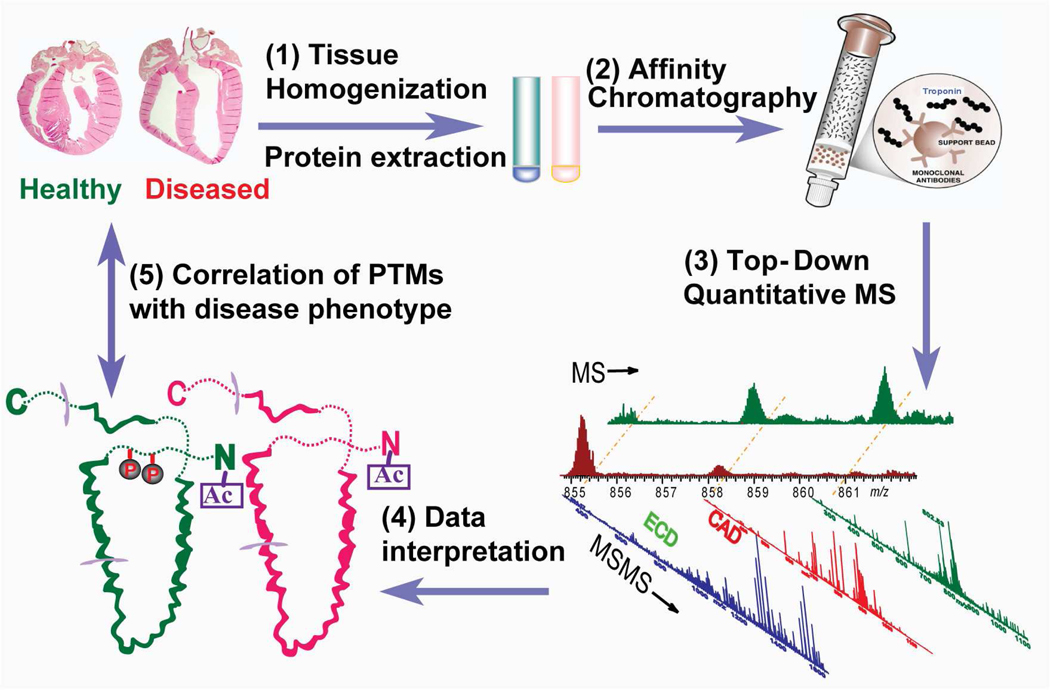
The top-down quantitative proteomics methodology combined the high specificity affinity chromatography and high-resolution MS for comprehensive analysis of PTMs in whole proteins extracted from normal and diseased tissues (Fig. 1). It was comprised of five steps: 1) Tissue homogenization and protein extraction. A cocktail of protease and phosphatase inhibitors were added to preserve the endogenous proteolysis and phosphorylation status. 2) Affinity chromatography purification of proteins. The purification procedures were performed at 4 °C where the enzymatic activities of phosphatase/kinase/protease were significantly reduced. 3) Top-down quantitative MS. The purified proteins were further separated and desalted via reverse-phase liquid chromatography (LC) and subject to in-depth high-resolution MS analysis. ECD was used for the localization of labile modifications. 4) Data interpretation. Protein sequences were characterized and their PTMs detected, identified, quantified, and mapped to single amino acid residues. 5) Correlation of PTMs with disease phenotypes. All protein PTMs in normal and diseased samples were quantified via a relative quantification method of the intact proteins. Altered PTM levels were correlated with disease phenotypes for the identification of potential biomarkers. Although this methodology was optimized for cTn analysis in this study, it can be adapted to other protein systems with proper modifications of the affinity purification procedures.
Fig. 1.
Schematic representation of top-down quantitative proteomics methodology featuring affinity-chromatography and high-resolution MS for comprehensive analysis of PTMs in whole proteins extracted from normal and diseased tissues.
Global PTM profiling of cTnI in health and disease
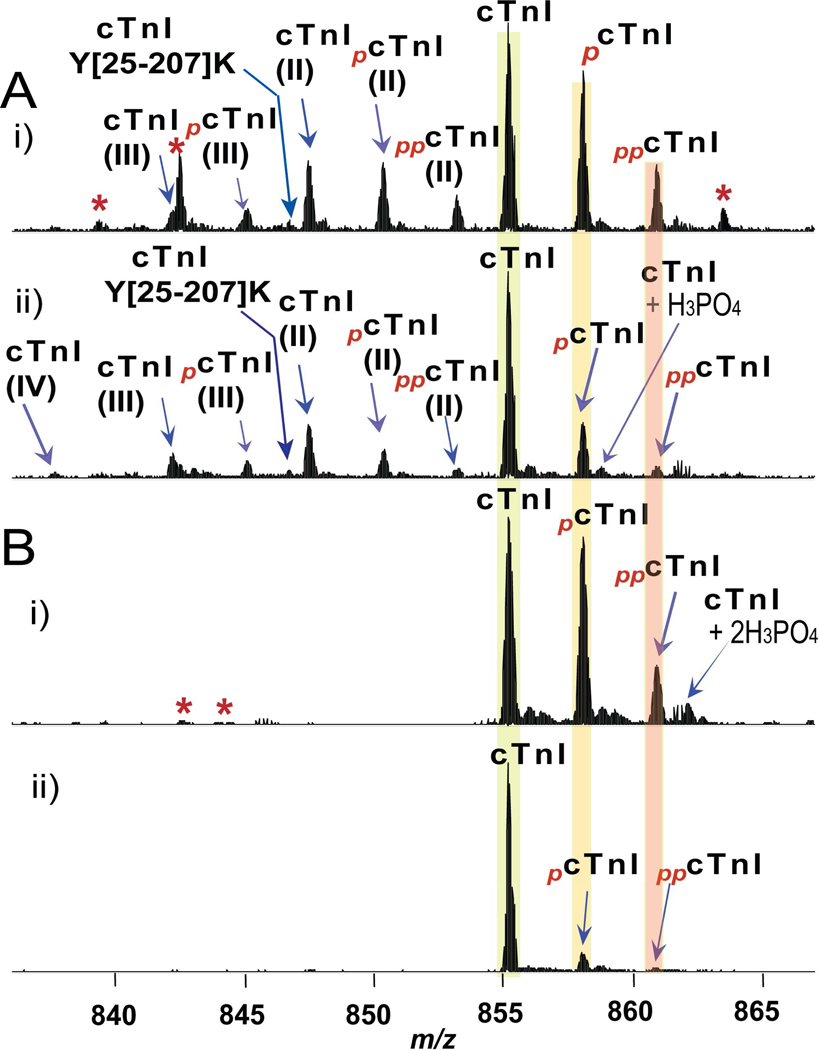
We have used human postmortem (autopsy) and fresh (transplant) myocardial tissues in this study. The postmortem samples were grouped into four categories according to the severity of the disease condition: control with normal cardiac functions (NOR, n=7), mild hypertrophy (HYP, n=5), severe hypertrophy/dilation (SHD, n=4) and CHF (n=6) (Table S1). The freshly collected transplant heart samples were divided into two groups: donor hearts with normal cardiac function (DOR, n=4), and end-stage failing hearts with ischemic/dilated cardiomyopathy (ICM/DCM, n=10) (Table S2). Human cTn was immunoaffinity purified from the heart tissue samples and MS analyzed using the established top-down quantitative proteomics methodology described above. The SDS-PAGE analysis of the elution from the affinity column revealed three relatively pure subunits of cTn: cTnI, cTnT, and cTnC (Supplemental Fig. S1). After the co-purified cTn proteins were separated by reverse-phase LC, the electrospray (ESI) FTICR/MS analysis of cTnI revealed a rich spectrum of PTMs corresponding to phosphorylation, proteolysis and acetylation in postmortem samples (Fig. 2A). Overall, we have identified 22 cTnI components from all postmortem cases (Table 1). Three major degradation isoforms of cTnI (II, 1–207; III, 1–206; IV, 1–205) resulting from C-terminal truncations from the full-length cTnI were detected, similar as observed previously.20 Both mono- and bis-phosphorylations were observed for the full length and the three isoforms (II–IV) of cTnI. In addition, five major degradation products (Y[28–206]K, Y[28–205]K, Y[25–209]S, Y[25–207]K, Y[25–206]K) and five minor ones (A[1–148]I, Y[28–209]S, Y[25–205]K, S[23–171]H, and P[32–209]S) were observed in some cases of postmortem samples resulting from both N- and C-terminal proteolysis. In contrast, minimal degradation products were observed in all 14 transplant samples. The only detected degradation products were Y[25–209] and P[32–209] at very low abundance (Table 1). Both mono- and bis-phosphorylations of cTnI were detected in fresh transplant samples (Fig. 2B), which was consistent with the postmortem samples (Fig. 2A). Representative high-resolution ESI/FTICR MS spectra of cTnI purified from healthy controls (Fig. 2A-i, B-i) and diseased (Fig. 2A-ii, B-ii) heart samples showed clear differences in PTM profiles regardless of the tissue source.
Fig. 2.
Representative high-resolution ESI/FTMS spectra of whole cTnI (M28+) purified from (A) postmortem and (B) fresh transplant human heart samples. A, i) NOR2; ii) HYP3; and B, i) DOR1; (ii) ICM3. Roman numerals (II–IV) indicate three different C-terminally truncated isoforms of N-terminally acetylated cTnI (II, 1–207; III, 1–206, IV, 1–205). Subscripts p and pp stand for mono and bisphosphorylation. +H3PO4, non-covalent adduct of phosphoric acid. Asterisks indicate co-purified minor cTnT related products. NOR, control heart with normal function; HYP, mild hypertrophy; DOR, donor heart; ICM, ischemic cardiomyopathy. Clinical details of the myocardial tissue samples were shown in Table S1–2.
Table 1.
Identification of cTnI components in ESI/FTMS spectra of postmortem (b) and fresh transplant (c) human heart tissue samples.
| Identification 1 | Observed Mr (Da) | Calculated Mr2(Da) | Error (ppm) |
|---|---|---|---|
| cTnIb,c | 23917.96 | 23917.83 | 5.4 |
| Major C-terminally truncated isoforms | |||
| cTnI(II)b | 23701.96 | 23701.75 | 8.9 |
| cTnI(III)b | 23554.86 | 23554.68 | 7.6 |
| cTnI(IV)b | 23426.72 | 23426.59 | 5.5 |
| Phosphorylation products | |||
| pcTnIb,c | 23997.95 | 23997.90 | 2.1 |
| ppcTnIb,c | 24077.84 | 24077.86 | −0.8 |
| pcTnI(II)b | 23781.60 | 23781.72 | −5.0 |
| ppcTnI(II)b | 23861.67 | 23861.68 | −0.4 |
| pcTnI(III)b | 23634.88 | 23634.65 | 9.7 |
| ppcTnI(III)b | 23714.68 | 23714.62 | 2.5 |
| pcTnI(IV)b | 23506.64 | 23506.55 | 3.8 |
| ppcTnI(IV)b | 23586.44 | 23586.52 | −3.4 |
| Major degradation products | |||
| cTnI Y[28–206]Kb | 20605.22 | 20605.19 | 1.5 |
| cTnI Y[28–205]Kb | 20477.12 | 20477.10 | 1.0 |
| cTnI Y[25–209]Sb,c | 21359.60 | 21359.59 | 0.5 |
| cTnI Y[25–207]Kb | 21143.46 | 21143.50 | −1.9 |
| cTnI Y[25–206]Kb | 20996.57 | 20996.56 | 0.5 |
| Minor degradation products | |||
| cTnI A[1–148]Ib | 16990.38 | 16990.46 | −4.7 |
| cTnI Y[28–209]Sb | 20969.32 | 20969.40 | −3.8 |
| cTnI Y[25–205]Kb | 20868.30 | 20868.32 | 1.0 |
| cTnI S[23–171]Hb | 17070.52 | 17070.27 | 14.6 |
| TnI P[32–209]Sc | 20504.48 | 20504.70 | −10.7 |
Roman numerals indicate four different C-terminal truncated isoforms of C-terminally acetylated cTnI (I, residues 1–209; II, residues 1–207; III, residues 1–206; IV, residues 1–205). Subscript p stands for monophosphorylation and pp stands for bisphosphorylation.
Calculated Mr is the most abundant isotopic mass based on the amino acid sequence of entry name TNNI3_human from the Swiss-Prot sequence database.
Quantification of cTnI phosphorylation
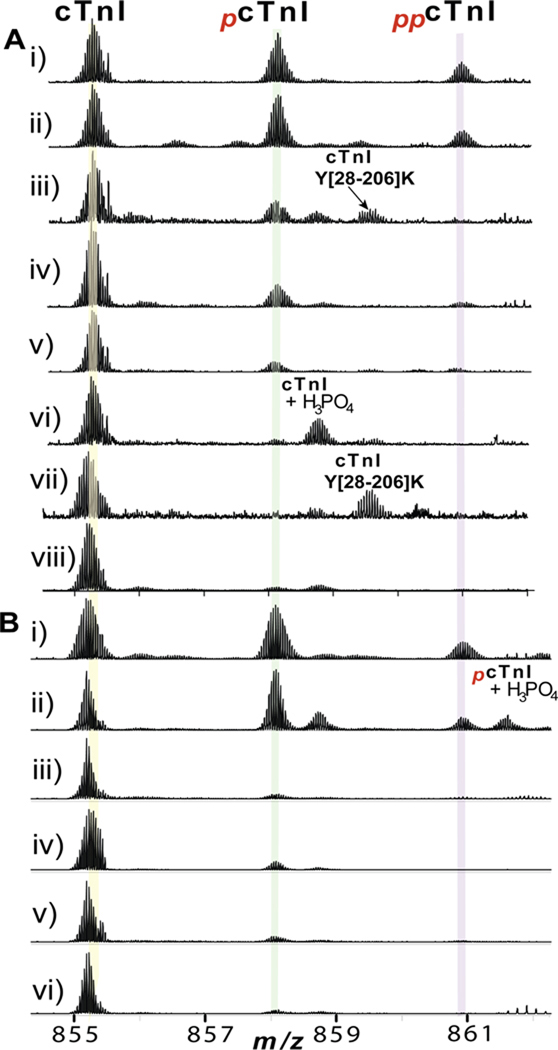
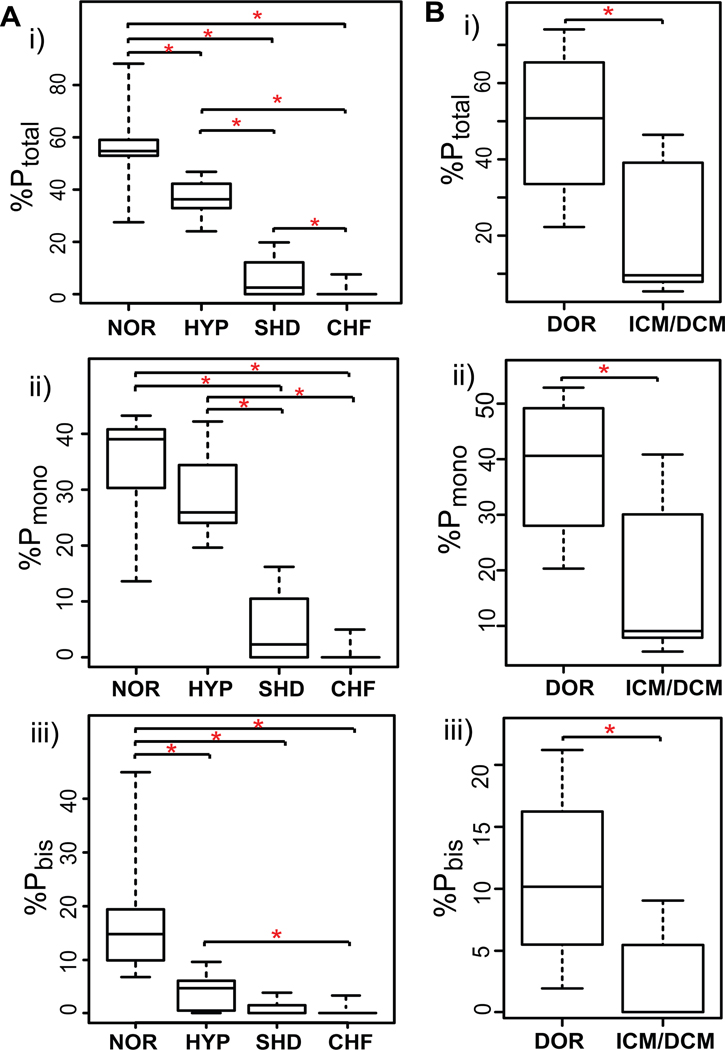
The phosphorylation levels of cTnI in a collection of 36 clinical human heart samples were quantified using the top-down MS approach. The technical reproducibility of this methodology was evaluated with highly consistent results among replicate analysis of the same biological sample (Supplemental Fig. S2). Two representative cases from each group were shown in Fig. 3 and the overall relative quantification of cTnI phosphorylation in all 36 heart samples were presented in box-whiskers plots (Fig. 4). cTnI mainly existed in un-, mono- (pcTnI), and bis-phosphorylated (ppcTnI) forms in both postmortem and fresh transplant hearts of normal cardiac function, NOR and DOR, respectively (Fig. 3A-i-ii, 3B-i-ii). ppcTnI was significantly reduced in the early-stage heart disease with HYP (Fig. 3A-iii-iv), whereas no significant difference was observed for pcTnI between HYP (Fig. 3A-iii-iv) and NOR (Fig. 3A-i-ii) groups. Both pcTnI and ppcTnI were significantly reduced in SHD (Fig. 3A-v-vi) and end-stage failing hearts with ICM/DCM (Fig. 3B-iii-vi), and nearly abolished in CHF (Fig. 3A-vii-viii). For the 22 postmortem heart samples studied, the relative percentages of the total phosphorylated cTnI species over the entire cTnI populations (%Ptotal) were 56.4±3.5%, 36.9±1.6%, 6.1±2.4%, and 1.0±0.6% for NOR, HYP, SHD, and CHF, respectively. The relative percentages of pcTnI (%Pmono) were 34.2±2.1%, 29.0±2.0%, 5.2±1.9, and 0.7±0.4; and those for ppcTnI (%Pbis) were 18.4±2.4%, 4.1±0.9%, 0.9±0.5, and 0.3±0.2, for NOR, HYP, SHD, and CHF, respectively (Fig. 4A-i-iii). For all of the 14 fresh transplant samples, the %Ptotal of 49.5±5.9% and 18.8±2.9%, %Pmono of 38.6±3.8% and 16.7±2.4%; %Pbis of 10.9±2.2% and 2.2±0.6%, were obtained for DOR and ICM/DCM, respectively (Fig. 4B-i-iii). Overall, the cTnI phosphorylation levels showed a clearly decreasing trend as the contractile dysfunction becomes increasingly severe in both the postmortem and the fresh transplant samples (Fig. 3, 4). Particularly, statistically significant P values were achieved between group comparisons in the case of %Ptotal, underscoring the potential of cTnI phosphorylation as a robust candidate biomarker for CHF with the potential for detection of CHF at the early stage of hypertrophy.
Fig. 3.
Correlation between the cTnI phosphorylation level and heart disease phenotypes. Representative cases from (A) postmortem heart samples: i–ii) NOR2,3; iii–iv) HYP2,3; v–vi) SHD1,2; vii–viii) CHF2,3; and (B) fresh transplant heart samples, i–ii) DOR1,4; iii–iv) ICM3, 6; v–vi) DCM 3,4. Clinical details of the myocardial tissue samples were shown in Supplemental Table S1–2.
Fig. 4.
Relative quantification of cTnI phosphorylation in normal and diseased human heart samples. (A) postmortem (NOR, control heart with normal function; HYP, mild hypertrophy, SHD, severe hypertrophy/dilation; CHF, congestive heart failure), and (B) fresh transplant (DOR, donor heart; ICM/DCM, ischemic/dilated cardiomyopathy) samples. i) Total phosphorylated cTnI percentage (%Ptotal); ii) mono-phosphorylated cTnI percentage (%Pmono); and iii) bis-phosphorylated cTnI percentage (%Pbis) over the entire cTnI populations. Box, median and interquartile range (25%, 75%); whiskers, minimum and maximum values. *p<0.05.
Phosphorylation sites localization and the order of phosphorylation/dephosphorylation
To locate the phosphorylation sites in pcTnI and ppcTnI and explore the potential order of phosphorylation/dephosphorylation, single charge states of pcTnI and ppcTnI from normal and hypertrophic hearts were isolated and dissociated by ECD (Fig. 5, and S3). In ECD spectra of pcTnI (Supplemental Fig. S3a–b), Ser22 was identified as the phosphorylation site for both NOR and HYP heart samples for the following reasons: (1) No phosphorylated product ions were detected for smaller c ions before Ser22 (c9 – c21). (2) The detection of monophosphorylated c22 (pc22) clearly identified the phosphorylation of Ser22, and the absence of unphosphorylated c22 indicated the nearly exclusive phosphorylation occupancy at Ser22 in pcTnI. (3) All the larger c ions were present only as phosphorylated forms, supporting the phosphorylation site assignment at Ser22 (Fig. 5A,B; S3a,b). ECD spectra of ppcTnI (Supplemental Fig. S3c) unambiguously determined Ser22/23 as the two basal phosphorylation sites in the normal human heart samples, based on the similar evidence stated above for pcTnI. Overall, four ECD spectra of pcTnI in normal heart samples provided consistent evidence for Ser22 being the mono-phosphorylation site, with 57 c ions, 55 z● ions and a full sequence coverage (Fig. 5A). Similarly, four ECD spectra of pcTnI in hypertrophic heart samples generated 53 c ions, 69 z● ions and a full sequence coverage which identified Ser22 as the same mono-phosphorylation site (Fig. 5B). Three ECD data of ppcTnI in normal heart samples generated 46 c ions, 83 z● ions with 100% sequence coverage supporting Ser22/23 being the only basal phosphorylation site observed in ppcTnI (Fig. 5C). The lack of phosphorylation in the major cTnI degradation products corresponds to the N-terminally truncated forms of cTnI with cleavage sites at Tyr25 or Tyr28 (Table 1) and also confirmed Ser22/23 as the only two basal phosphorylation sites in human cTnI.
Fig. 5.
Mapping cTnI phosphorylation sites by top-down MS with ECD. Fragmentation maps of pcTnI in (A) normal, (B) hypertrophic heart samples and (C) ppcTnI in normal heart samples. The identified phosphorylation site(s) of Ser22 in pcTnI and Ser22/23 in ppcTnI are highlighted in circles.
The fact that Ser22 was phosphorylated in both pcTnI and ppcTnI whereas Ser23 was phosphorylated only in ppcTnI indicates the phosphorylation occurs at Ser22 first and then Ser23 in NOR hearts. The facts that only pcTnI was observed in the ESI/FTMS spectra of HYP hearts (Fig. 3A-iv) and Ser22 was localized as the site of phosphorylation suggest that Ser23 was dephosphorylated first in HYP hearts (Fig. 5C, S3b).
Quantification of cTnI degradation
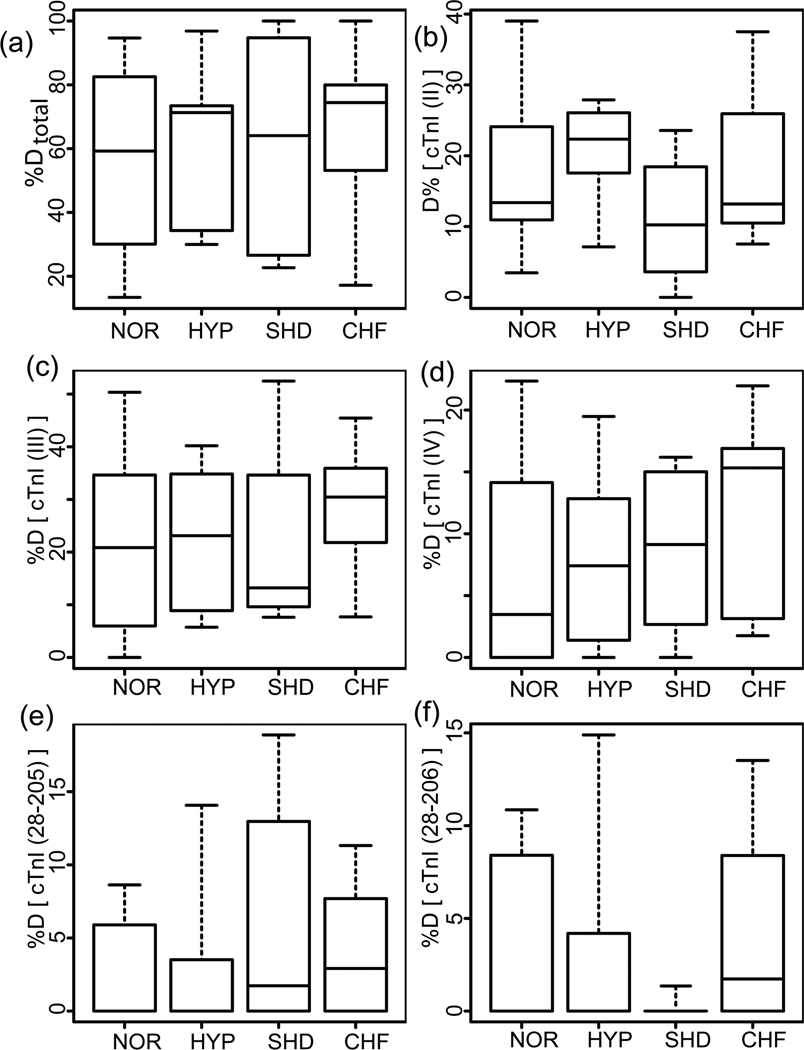
cTnI degradation was evaluated for all postmortem heart samples in a similar method used to evaluate the phosphorylation. Three representative cases from each group are shown in Fig. S4 and the relative quantification of cTnI degradation in four postmortem groups are presented in Fig. 6. The identities of major cTnI degradation products were confirmed by both high accuracy mass measurements and MS/MS (representative spectra shown in Fig. S5). In general, quantification results do not show a significant difference in the overall degradation product percentages in normal and diseased hearts (Fig. 6a). The relative percentages of all the degraded cTnI species over the entire cTnI populations were 55.1±6.3%, 61.6±6.6%, 61.3±10.1%, and 65.8±6.1% for NOR, HYP, SHD and CHF, respectively. The overall degradation is mainly contributed from three C-terminally truncated isoforms (II, III, IV) (Fig. 6). No significant differences in cTnI isoforms (Fig. 6b–d) and three major degradation products (Fig. 6e–f) were observed between normal and the diseased groups. For example, the relative percentages of cTnI (II) were 17.1±2.4%, 20.1±1.9%, 10.7±2.4%, and 17.8±2.3% for NOR, HYP, SHD and CHF, respectively. The relative percentages of cTnI [28–205] were 2.9±0.8%, 3.3±1.4%, 6.2±2.3%, and 3.7±0.9% in NOR, HYP, SHD and CHF, respectively. Additionally, there was not a substantial correlation between the degradation and the phosphorylation patterns or between the degradation level and the postmortem intervals (Fig. S6). Moreover, there were only minimal degradation products detected in freshly collected myocardium samples, including both healthy donors and explanted failing hearts.
Fig. 6.
Relative quantification of cTnI degradation levels in normal and diseased heart samples. NOR, normal controls, HYP, mild hypertrophy, SHD, severe hypertrophy/dilation, and CHF, congestive heart failure. (a) Total degraded cTnI percentage (%Dtotal); (b–f) percentages of degradation products cTnI(II), (III), (IV), [Y28–205K], [Y28–206K], over the entire cTnI populations, respectively. Calculated p values were not found to be statistically significant
DISCUSSION
cTnI phosphorylation as a potential candidate cardiac biomarker
Here we present a significant correlation between the heart disease phenotypes and the cTnI phosphorylation in both postmortem and fresh transplant myocardium samples. The use of relatively well-preserved postmortem samples allows us to explore the changes in cTnI at early-stages potentially during the progression to CHF, since the more readily available postmortem samples represent a comprehensive disease spectrum including not only the early-stage diseases (i.e., hypertrophy and dilation) but also the end-stage failure samples. As observed here, the total cTnI phosphorylation level was reduced in HYP, significantly reduced in SHD, and nearly abolished in CHF compared with NOR. To address the concern that postmortem proteolysis might occur before a tissue sample was collected, we have confirmed the results using fresh myocardial samples collected immediately after the hearts were explanted during transplantations. It revealed consistent results with greatly reduced cTnI phosphorylation in the end-stage failing hearts of ICM/DCM compared with DOR of brain-dead donor hearts. In addition, the highly comparable cTnI phosphorylation levels between postmortem and fresh control samples suggest that the stage-appropriate balance between phosphatase and kinase activities is still maintained in these postmortem heart tissues collected in this study. Nevertheless, we did observe more severe degradation in postmortem than in fresh tissues, possibly because proteolysis is an irreversible process whereas phosphorylation is reversible. To our knowledge, our study provides a direct comparison on the use of postmortem and freshly collected transplant heart samples for the first time and illustrates the potential value of widely available postmortem heart samples for the study of phosphorylation, but not proteolytic degradation.
The previous studies using the semi-quantitative Pro-Q Diamond phosphoprotein stain combined with immunoblotting also suggested reduced cTnI phosphorylation in end-stage explanted failing hearts in comparison to non-failing donor hearts.47–48, 51 The consistency between our top-down quantitative MS data of the affinity purified cTnI and SDS-PAGE with Pro-Q Diamond stain/immunoblotting of the entire cardiac muscle myofibrils demonstrates that quantitative changes after immunaffinity enrichment correlate to the actual abundance of total and phosphorylated cTnI in the intact muscle myofibril47–48, 51, thereby validate our top-down quantitative MS method. Nevertheless these studies used immunoblotting with phosphoprotein specific antibodies for phosphorylation site identification which were based on hypothesis of potential phosphorylation sites at Ser22/23 and therefore precluded comprehensive assessment of other existing phosphorylation events.47–48, 51 Here, we have comprehensively analyzed all modification state including all possible phosphorylation sites and unequivocally localized the altered phosphorylation sites in cTnI to Ser22/23 (Ser23/24 considering the N-terminal Met), which is consistent with our previous reports.20–21, 23–24 Subsequently we have determined the order of phosphorylation between these two sites (Ser22 phosphorylates prior to Ser23) in cTnI of normal hearts and the order of dephosphorylation (Ser23 dephosphorylates prior to Ser22) in diseased hearts. Phosphorylation of Ser22/23 is well-known to be mediated by PKA, the downstream kinase of β-adrenergic receptors.52–53 During exercise and stress, β-adrenergic stimulation is a major physiological mechanism to meet the increased demand of the body, leading to an increased level of intracellular cAMP which in turn activates cAMP-dependent PKA.51 The PKA phosphorylation of Ser22/23 reduces the Ca2+-sensitivity of myofilaments and enhances relaxation, both of which are considered to be beneficial in the diastolic function of the heart.51–52 Nevertheless, the chronic over-stimulation of the β-adrenergic receptor pathway to maintain the cardiac output in cardiac disease ultimately results in the desensitization of the β-adrenergic receptors which leads to reduced phosphorylation of the PKA target proteins.51–52 The chronic catecholamine stimulation is also known to promote cardiac hypertrophy and myocyte apoptosis and ultimately leads to a reduction in contractility.54 Indeed, we observed the maximal degree of cTnI phosphorylation in normal hearts at PKA sites and a continuous dephosphorylation from early-stage hypertrophy to end-stage failing hearts, underscoring the potential of cTnI phosphorylation as a candidate biomarker for early detection of CHF.
Recent research has shown evidence of reduced PKA phosphorylation in other myocardial proteins such as cardiac myosin binding protein C and myosin light chain 2, which suggests the role of Ser22/23 dephosphorylation in cardiac regulation may be a synchronized function with other contractile proteins.51, 55–56 CHF is a very complex disease in nature with varying etiologies, it is unlikely that a single biomarker can serve sensitively and specifically as the therapeutic or diagnostic biomarker for the disease. Biomarkers of the future are thus expected to be multi-marker panels characteristic of the complexity of the underlying pathophysiology of the disease process.9, 57 In our future work, we will extend the top-down methodology to systematically examine the correlation of the PTMs of other contractile proteins and the disease phenotypes toward developing multi-marker panel with improved specificity and accuracy for disease detection.
Lack of correlation between cTnI degradation and disease phenotype
cTnI is known to be highly susceptible to degradation.58–59 Consistently, we observed here a major category of proteolytically degraded cTnI forms in most of the postmortem clinical heart samples. Overall, we have identified twenty-two major cTnI components from all postmortem cases (Table 1). Three major degradation isoforms of cTnI (II–IV) were found resulting from truncations of the C-terminal Glu-Ser, Phe-Glu-Ser and Lys-Phe-Glu-Ser residues from the full-length cTnI (isoform I), which is consistent with our previous report.20 In addition, five major degradation products (Y[28–206]K, Y[28–205]K, Y[25–209]S, Y[25–207]K, Y[25–206]K) and five minor ones (A[1–148]I, Y[28–209]S, Y[25–205]K, S[23–171]H, and P[32–209]S) were observed in various samples resulting from both the N- and C-terminal proteolysis. No proteolytic fragments in the middle region between H[33–147]R were observed, possibly because of the protection by cTnC, in agreement with a previous study using human serum samples.60 The major degradation products have highly preferential cleavages at the N-terminal Tyr25 and Tyr28 since the enzyme most frequently considered to be involved in cTnI degradation is μ-calpain (also known as calpain 1) which is known to cleave Tyr preferably.61
No significant correlation could be established between the cTnI degradation level and the disease phenotype. Among all the major degradation products here, none of them can be used as a robust indicator of disease or disease progression in contrast to cTnI phosphorylation. Moreover, we only observed trace degradation products in freshly collected transplant heart samples. The only detected degradation products were Y[25–209]S and P[32–209]S with very low abundance (Table 1), which was consistent with those reported previously for mouse and swine cTnI: Y[26–210]G and P[33–210]G (mouse and swine cTnI sequences have one additional amino acid than that of human cTnI).21, 24 Nevertheless, every analytical method has its detection limit. It is possible that some degradation products of extremely low abundance (<1% of the total population) may not be detected here. Moreover, as each antibody has its specific epitope region, it is also possible that some degradation products lack those epitope regions may be missed during the immunoaffinity step. The antibodies we used in this study are 14G5 and MF4, with epitopes at N-terminal residues 1–23 and C-terminal residues 190–196 of cTnI, respectively.
Whether cTnI degradation can be considered a universal mechanism of contractile dysfunction is still of intense debate.62 Selective cTnI proteolysis has been proposed as a mechanism of contractile dysfunction mainly supported by a seminal report46 that the transgenic mouse line expressed cTnI[1–193], a cTnI degradation product first found in stunned rat myocardium44 exhibited ventricular dilation and decreased myofilament contractility. However, studies on large animals (in vivo canine and swine models of stunning) did not show such a degradation product associated with reversible stunning.62 The disparity among these studies suggests that the cTnI degradation might be species, disease-model, and experimental preparation dependent.62–64
cTnI degradation products have been reported in both human myocardium and serum but also with disparities. cTnI degraded products were detected in the myocardium of bypass patients with some patients experiencing an increase in cTnI modification and others experiencing a decrease.44 A recent study using human transplant samples reported no evidence of cTnI degradation between failing and non-failing myocardium.48 In contrast, Labugger et al. reported the detection of the extensive cTnI degradation (as many as 8 degradation products) in serum from patients with acute myocardial infarction (AMI), which seemed to correspond to the severity of the AMI.45 This also indicated that some cTnI modified products existing in the serum may also be present in the myocardium.65 It is believed that the degradation of cTnI can potentially occur as a part of a natural turnover of a protein or as a result of injury depending on the disease model and the sample origins.63 Overall, caution needs to be taken in consideration of cTnI degradation products as potential disease markers.
Lack of detection of PKC phosphorylation sites in human cTnI
Our results provide strong evidence that the phosphorylation at Ser22/23 (the bona fide PKA substrates) is a stable basal modification in healthy hearts, which is essential for normal cardiac function, whereas a decrease in the basal phosphorylation could be linked to the impaired contractile function in diseased hearts, underscoring the potential of cTnI phosphorylation as a candidate cardiac biomarker for chronic diseases with impaired cardiac contractile function. cTnI is also well-known to be regulated by protein kinase C (PKC) at three sites, Ser43/45 and Thr144, which were identified based on in vitro phosphorylation assays. However, we did not detect these PKC sites in this study, in agreement with other studies using transplant human heart tissues.48, 51 The lack of detection of PKC phosphorylation sites in cTnI purified from clinical human tissues is likely due to the following possibilities: 1) the potential fundamental differences between in vitro and in vivo experiments. 2) the well-known PKC sites (Ser43/45 and Thr144) have very low phosphorylation occupancy (<1% of total cTnI populations), which may be below the detection limit of our method;21 3) the potential PKC phosphorylation sites in cTnI (Ser43/45 and Thr144) might only be present in the hearts of other specific disease models which is not studied here; and 4) PKC phosphorylation might be offset by the increased activity of phosphatase in end-stage heart failure.48 Whereas PKA phosphorylation in cTnI is taken as a fact, the significance of PKC phosphorylation sites is still of intense debate due to the lack of in vivo evidence.66
An additional phosphorylation site, Ser76/Thr77, identified in commercially available human cTnI samples,20 was also not detected in the clinical samples here. This could be due to the sample source, as the commercially available cTnI sample was purified from a mixture of multiple hearts possibly without proper control or preservation. Nevertheless, the pathological implication of Ser76/Thr77 remains to be explored.
The effect of β-blockade on cTnI phosphorylation
Our study shows that there is no significant correlation between the reduced phosphorylation in cTnI and β-blocker treatment (Fig. S7). Moreover, there is no clear correlation between the β-blocker dosage and cTnI phosphorylation (Fig. S7B). Furthermore, the variability in cTnI phosphorylation within a group does not correspond to cardiac indices like left ventricular ejection fraction (LVEF) (Fig. S8). Although β-adrenergic antagonists are considered as an effective therapy in the treatment of heart failure, its actual effect on the β-adrenergic receptor systems is still not clear.67 Previous studies reported that β-blockers, such as metoprolol and atenolol, suppressed the hypophosphorylation of ryanodine receptor (RyR2) and restored channel function.68 In contrast, a recent paper demonstrated that the cTnI and cMyBP-C phosphorylation level was not altered in a pig myocardial infarction (MI) model with β-blocker therapy despite the relatively lower Ca2+ sensitivity in MI+β-blocker compared with untreated myocardium.69 Moreover, studies by Messer et al.48 and van der Velden et al.47 showed overall decreased phosphorylation of cTnI in human transplant heart tissues without β-blocker therapy, consistent with our results. Therefore, we reason that the decreased PKA phosphorylation of cTnI in diseased hearts may be independent of β-blocker treatment (in other words, reduced PKA phosphorylation may proceed with or without β-blocker therapy).
Tissue versus blood for biomarker discovery
Human blood (serum/plasma) has been the focus of clinical proteomics for biomarker discovery since it is readily available and is potentially a rich source for biomarkers of various diseases.70 However the blood sample also presents significant challenges for the current analytical technologies due to its extraordinary dynamic range (> 10 orders of magnitude) in protein concentration with several most abundant proteins dominating >99% of the total protein amount in blood.70 Thus the serious mismatch between the complexity and the huge dynamic range of proteins in blood against the extremely low abundant potential biomarker candidates makes the discovery of novel biomarkers in blood extremely difficult, and possibly with high false positive rate.35, 71–72 In contrast, damaged tissue closest to the disease source appears to be a preferred sample choice for biomarker discovery since it contains the highest concentration of potential disease markers.5, 36–39 Fresh frozen tissues are especially valuable for identification of biomarker with high accuracy and specificity. Here, we have used freshly collected heart tissue samples from transplant surgery within 15 min after the hearts stopped beating and our data revealed unambiguously the significant correlation between cTnI basal phosphorylation levels and the disease phenotypes, suggesting cTnI phosphorylation as a reliable candidate biomarker for CHF.
On the other hand, although tissue is ideal for biomarker discovery, it is not widely accessible as blood specimen, which limits its clinical utility in routine diagnostics. Conversely, the readily available blood specimen has the great advantage for developing a routine diagnostic assay since blood involves minimal risk and cost to obtain and thus is the preferred form of a clinical test.5, 35, 71 Hence, it is plausible to use tissue sample for biomarker discovery but use blood sample for biomarker validation.5 Once the candidate biomarkers have been discovered in tissue sample, it is practical and essential to validate them in serum/plasma using targeted experiments such as antibody-based immunoassays (by developing antibodies recognizing the protein and its specific modifications) or MRM/SRM-MS-based methods (by developing multiple reaction monitoring with stable isotope dilution methods).5, 35, 40 This serves as an essential biomarker validation step toward the ultimate goal of developing blood-based diagnostic assay for routine use in the clinic.
The cTnI modification products have been detected in serum.45 Thus, it was believed that some cTnI modification products necrotically released into the blood might be disease-related, which can reflect the disease progression and the functional status of the remaining viable myocardium with the potential to be the next-generation of cardiac biomarkers.13, 44 Therefore, the future efforts will be allocated to biomarker validation in human serum/plasma with a large sample size (n=500) by MRM/SRM for targeted detection of phosphorylated Ser22/23 and immunoassay with phosphoSer22/23 specific antibody to further demonstrate its utility in the clinical setting.
Top-down quantitative proteomics for assessing PTMs
The major advantages of our top-down methodology include: (a) highly effective and reproducible affinity purification of whole proteins from tissues. Affinity chromatography remains the most effective and specific protein purification method.73 Here we demonstrate the successful immunoaffinity purification of cTn complex from clinical human tissue samples with high specificity and reproducibility. (b) Global detection of all possible modifications (within the detection limit) without a priori knowledge. The traditional strategies for PTM characterization, such as Western blotting, require a priori knowledge of the modification types and seldom provide global information of a protein modification state. In contrast, high-resolution MS analysis provides a "bird’s eye view" of all detectable modifications (>1% of the total protein population) including various PTMs and sequence variants.29 Here, the top-down MS revealed a comprehensive spectrum of cTnI PTMs resulting from phosphorylation, proteolytic degradation and acetylation. (c) Identification and quantification of modifications that reflect tissue stress. Although other non-MS-based methods such as the recently developed Pro-Q diamond staining with 1D/2D PAGE can provide semi-quantification of phosphorylation levels in proteins,48, 51 they cannot provide the identification of proteins and their specific modification sites, which are essential for understanding the molecular mechanisms of the disease. In contrast, the top-down quantitative proteomics methodology not only detects and quantifies even subtle changes in the protein modification levels in response to stress, but also provides unambiguous identification of such altered modification site(s) regardless of the modification type. Our data unequivocally revealed reduced phosphorylation levels in cTnI in CHF and identified Ser22/23 as the phosphorylation sites by a unique nonergodic ECD method and determined the phosphorylation/dephosphorylation order in health and diseased states, respectively. (d) Characterization of the order of multiple PTMs. Since top-down MS directly analyzes the whole proteins, all the PTMs and their correlation are well preserved. Here we have unambiguously determined the phosphorylation/dephosphorylation order between the two sites in cTnI in normal and diseased myocardium, respectively. Such an in-depth PTM characterization can be hardly accounted for by a bottom-up proteomics strategy where the proteins are first digested into small pieces, thus disrupting the interconnection among multiple PTMs.28 (e) Highly reproducible quantification of all types of PTMs simultaneously. Compared with the bottom-up approach, the top-down method has a unique advantage in relative quantification of PTMs since the physico-chemical properties of whole proteins are much less affected by the presence of modifying groups in comparison with peptides.20, 33 In contrast to immunoblotting or Pro-Q diamond stain which can only provide quantification for a specific modification, top-down quantitative MS provides universal quantification of all types of modifications and thus can offer the correlation between different types of modifications. Here we investigated the correlation between phosphorylation and proteolytic degradation (Fig. S6). It also offers the possibility for global comparisons among multiple samples, providing insight into disease progression.
Admittedly, in comparison to the mature bottom-up proteomics technologies, the top-down MS is still in its early developmental stage and yet to become available for routine use. Nevertheless, the top-down MS has achieved substantial progress in the last few years and started to gain increasing space in modern proteomics.19, 22, 27, 74–76 Hence, the continuing development of new technologies with improvements in MS instrument sensitivity and detection limits, front end protein separation, and user-friendly data processing and automation software will ultimately promote wider use of top-down proteomics in the near future.75
CONCLUSION AND FUTURE DIRECTION
We have employed a top-down quantitative proteomics methodology for in-depth comparative analysis of PTMs in whole proteins extracted and purified from normal and diseased cardiac tissues for the discovery of candidate biomarkers with high reproducibility and specificity. Specifically, have analyzed 36 clinical human heart tissue samples and unambiguously identified the phosphorylation of cTnI as a robust candidate biomarker for CHF. We report, for the first time, the detection of reduced cTnI phosphorylation at the early-stage of CHF highlighting its potential as a diagnostic marker for the early detection. We have further mapped phosphorylation sites on cTnI to Ser22/23 and determined the order of phosphorylation/dephosphorylation between these sites in normal and diseased myocardium, respectively. In contrast, no correlation between detected proteolytic degradation products and disease phenotypes was established. This study represents the first clinical application of top-down quantitative proteomics for biomarker discovery from tissues. It demonstrates the advantages of the top-down MS methodology to detect, identify, characterize, and quantify protein PTMs for developing PTM-based disease biomarker. In our future work, we will extend this methodology to systematically examine the correlation of the PTMs of other contractile proteins and the disease phenotypes since the next generation biomarkers are expected to be multi-marker panels characteristic of the complexity of the underlying patho-physiology of the disease process. Moreover, the future efforts will be allocated to biomarker validation in serum/plasma with a large sample size via antibody-based and/or SRM/MRM MS-based targeted approach toward an ultimate goal of developing a sensitive blood-based assay for routine diagnostics.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Chad Dooley, Matthew Lawrence, and Dr. Jitandrakumar R. Patel for experimental assistance and Dr. Han Zhang for critical reading of this manuscript. We are grateful to Professors Tim Kamp, Marion Greaser, and Lingjun Li for helpful discussions. This work was supported by the Wisconsin Partnership Fund for a Healthy Future, NIH R37 HL82900 (to R.L.M.), and American Heart Association Scientist Development Grant 0735443Z (to Y.G.).
ABBREVIATIONS
- CHF
chronic heart failure
- PTMs
post-translational modifications
- ESI
electrospray
- LC
liquid chromatography
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- CID
collision-induced dissociation
- ECD
electron capture dissociation
- FTICR
Fourier transform ion cyclotron resonance
- MRM/SRM
multiple (or selected) reaction monitoring
- cTnI
cardiac troponin I
- cTn
cardiac troponin complex
- pcTnI
mono-phosphorylated cardiac troponin I
- ppcTnI
bis-phosphorylated cardiac troponin I
- PKA
protein kinase A
- PKC
protein kinase C
- LV
left ventricle
- RV
right ventricle
- S.E.M.
the standard error of the mean
- NOR
control postmortem hearts with normal cardiac function
- HYP
postmortem hearts of mild hypertrophy
- SHD
postmortem hearts of severe hypertrophy/dilation
- DOR
donor transplant hearts with normal cardiac function
- ICM
ischemic cardiomyopathy
- DCM
dilated cardiomyopathy
- CAD
coronary artery disease
- OCT
orthotopic cardiac transplantation
- VAD
ventricular assist devices
- LVEF
left ventricular ejection fraction
- ACE
angiotensin-converting enzyme
- ARB
angiotensin receptor blockers
- CCB
Ca2+ channel blocker
Footnotes
SUPPLEMENTAL MATERIALS
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451(7181):919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- 2.Yach D, Hawkes C, Gould CL, Hofman KJ. The global burden of chronic diseases - Overcoming impediments to prevention and control. J. Am. Med. Assoc. 2004;291(21):2616–2622. doi: 10.1001/jama.291.21.2616. [DOI] [PubMed] [Google Scholar]
- 3.de Couto G, Ouzounian M, Liu PP. Early detection of myocardial dysfunction and heart failure. Nat. Rev. Cardiol. 2010;7(6):334–344. doi: 10.1038/nrcardio.2010.51. [DOI] [PubMed] [Google Scholar]
- 4.Kullo IJ, Cooper LT. Early identification of cardiovascular risk using genomics and proteomics. Nat. Rev. Cardiol. 2010;7(6):309–317. doi: 10.1038/nrcardio.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surinova S, Schiess R, Hu¨ttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the Development of Plasma Protein Biomarkers. J. Proteome Res. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 6.Vasan RS. Biomarkers of cardiovascular disease - Molecular basis and practical considerations. Circulation. 2006;113(19):2335–2362. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- 7.Mayr M, Zhang J, Greene AS, Gutterman D, Perloff J, Ping PP. Proteomics-based development of biomarkers in cardiovascular disease - Mechanistic, clinical, and therapeutic insights. Mol. Cell. Proteomics. 2006;5(10):1853–1864. doi: 10.1074/mcp.R600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 9.Arab S, Gramolini AO, Ping PP, Kislinger T, Stanley B, van Eyk J, Ouzounian M, MacLennan DH, Emili A, Liu PP. Cardiovascular proteomics - Tools to develop novel biomarkers and potential applications. J. Am. Coll. Cardiol. 2006;48(9):1733–1741. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 10.Hochholzer W, Morrow DA, Giugliano RP. Novel biomarkers in cardiovascular disease: Update 2010. Am. Heart J. 2010;160(4):583–594. doi: 10.1016/j.ahj.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Krueger KE, Srivastava S. Posttranslational protein modifications - Current implications for cancer detection, prevention, and therapeutics. Mol. Cell. Proteomics. 2006;5(10):1799–1810. doi: 10.1074/mcp.R600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003;21(3):255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 13.McDonough JL, Van Eyk JE. Developing the next generation of cardiac markers: Disease-induced modifications of Troponin I. Prog. Cardiovasc. Dis. 2004;47(3):207–216. doi: 10.1016/j.pcad.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Mazur MT, Cardasis HL, Spellman DS, Liaw A, Yates NA, Hendrickson RC. Quantitative analysis of intact apolipoproteins in human HDL by top-down differential mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2010;107(17):7728–7733. doi: 10.1073/pnas.0910776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An HJ, Kronewitter SR, de Leoz MLA, Lebrilla CB. Glycomics and disease markers. Curr. Opin. Chem. Biol. 2009;13(5–6):601–607. doi: 10.1016/j.cbpa.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J. Am. Chem. Soc. 1999;121(4):806–812. [Google Scholar]
- 17.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, Park JH, Begley TP, McLafferty FW. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 2002;124(4):672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 18.Sze SK, Ge Y, Oh H, McLafferty FW. Top-down mass spectrometry of a 29-kDa protein for characterization of any posttranslational modification to within one residue. Proc. Natl. Acad. Sci. U. S. A. 2002;99(4):1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han XM, Jin M, Breuker K, McLafferty FW. Extending Top-Down Mass Spectrometry to Proteins with Masses Greater Than 200 Kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 20.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Unraveling Molecular Complexity of Phosphorylated Human Cardiac Troponin I by Top Down Electron Capture Dissociation/Electron Transfer Dissociation Mass Spectrometry. Mol. Cell. Proteomics. 2008;7(10):1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Ayaz-Guner S, Zhang J, Li L, Walker JW, Ge Y. In vivo phosphorylation site mapping in mouse cardiac troponin I by high resolution top-down electron capture dissociation mass spectrometry: Ser22/23 are the only sites basally phosphorylated. Biochemistry. 2009;48(34):8161–8170. doi: 10.1021/bi900739f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge Y, Rybakova IN, Xu QG, Moss RL. Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc. Natl. Acad. Sci. U. S. A. 2009;106(31):12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu F, Xu Q, Dong X, Guy M, Guner H, Hacker TA, Ge Y. Top-Down High-Resolution Electron Capture Dissociation Mass Spectrometry for Comprehensive Characterization of Post-translational Modifications in Rhesus Monkey Cardiac Troponin I. Int. J. Mass Spectrom. 2010 Epub ahead of print. [Google Scholar]
- 24.Zhang J, Dong X, Hacker TA, Ge Y. Deciphering Modifications in Swine Cardiac Troponin I by Top-Down High-Resolution Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2010;(21):940–948. doi: 10.1016/j.jasms.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn P, Xu QG, Cline E, Zhang D, Ge Y, Xu W. Delineating Anopheles gambiae coactivator associated arginine methyltransferase 1 automethylation using top-down high resolution tandem mass spectrometry. Protein Sci. 2009;18(6):1272–1280. doi: 10.1002/pro.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jebanathirajah JA, Pittman JL, Thomson BA, Budnik BA, Kaur P, Rape M, Kirschner M, Costello CE, O'Connor PB. Characterization of a new qQq-FTICR mass spectrometer for post-translational modification analysis and top-down tandem mass Spectrometry of whole proteins. J. Am. Soc. Mass Spectrom. 2005;16(12):1985–1999. doi: 10.1016/j.jasms.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping PP, Durazo A, Bowie JU, Hasan SS, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Post-translational Modifications of Integral Membrane Proteins Resolved by Top-down Fourier Transform Mass Spectrometry with Collisionally Activated Dissociation. Mol. Cell. Proteomics. 2010;9(5):791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chait BT. Mass Spectrometry: Bottom-up or Top-Down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 29.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 31.Shi SDH, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal. Chem. 2001;73(1):19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 32.Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom. Rev. 2005;24(2):201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 33.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: Human histone H4. Anal. Chem. 2006;78(13):4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 34.Zabrouskov V, Han XM, Welker E, Zhai HL, Lin C, van Wijk KJ, Scheraga HA, McLafferty FW. Stepwise deamidation of ribonuclease A at five sites determined by top down mass spectrometry. Biochemistry. 2006;45(3):987–992. doi: 10.1021/bi0517584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 36.Veenstra TD, Zhou M. Tissue proteomics and metabolomics: An excellent start and a promising future. J. Proteome Res. 2009;8(4):1617–1617. doi: 10.1021/pr900157d. [DOI] [PubMed] [Google Scholar]
- 37.Zhou JY, Afjehi-Sadat L, Asress S, Duong DM, Cudkowicz M, Glass JD, Peng J. Galectin-3 Is a candidate biomarker for amyotrophic lateral sclerosis: Discovery by a proteomics approach. J. Proteome Res. 2010;9(10):5133–5141. doi: 10.1021/pr100409r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol. Cell. Proteomics. 2005;4(4):384–393. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Guo T, Lee CS, Wang WJ, DeVoe DL, Balgley BM. Capillary separations enabling tissue proteomics-based biomarker discovery. Electrophoresis. 2006;27(18):3523–3532. doi: 10.1002/elps.200600094. [DOI] [PubMed] [Google Scholar]
- 40.Fu Q, Schoenhoff FS, Savage WJ, Zhang PB, Van Eyk JE. Multiplex assays for biomarker research and clinical application: Translational science coming of age. Proteomics Clin. Appl. 2010;4(3):271–284. doi: 10.1002/prca.200900217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babuin L, Jaffe AS. Troponin: the biomarker of choice for the detection of cardiac injury. Can. Med. Assoc. J. 2005;173(10):1191–1202. doi: 10.1503/cmaj.050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature. 2003;424(6944):35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 43.Solaro RJ, Rosevear P, Kobayashi T. The unique functions of cardiac troponin I in the control of cardiac muscle contraction and relaxation. Biochem. Biophys. Res. Commun. 2008;369(1):82–87. doi: 10.1016/j.bbrc.2007.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonough JL, Labugger R, Pickett W, Tse MY, MacKenzie S, Pang SC, Atar D, Ropchan G, Van Eyk JE. Cardiac troponin I is modified in the myocardium of bypass patients. Circulation. 2001;103(1):58–64. doi: 10.1161/01.cir.103.1.58. [DOI] [PubMed] [Google Scholar]
- 45.Labugger R, Organ L, Collier C, Atar D, Van Eyk JE. Extensive troponin I and T modification detected in serum from patients with acute myocardial infarction. Circulation. 2000;102(11):1221–1226. doi: 10.1161/01.cir.102.11.1221. [DOI] [PubMed] [Google Scholar]
- 46.Murphy AM, Kogler H, Georgakopoulos D, McDonough JL, Kass DA, Van Eyk JE, Marban E. Transgenic mouse model of stunned myocardium. Science. 2000;287(5452):488–491. doi: 10.1126/science.287.5452.488. [DOI] [PubMed] [Google Scholar]
- 47.van der Velden J, Papp Z, Zaremba R, Boontje NM, de Jong JW, Owen VJ, Burton PBJ, Goldmann P, Jaquet K, Stienen GJM. Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins. Cardiovasc. Res. 2003;57(1):37–47. doi: 10.1016/s0008-6363(02)00606-5. [DOI] [PubMed] [Google Scholar]
- 48.Messer AE, Jacques AM, Marston SB. Troponin phosphorylation and regulatory function in human heart muscle: Dephosphorylation of Ser23/24 on troponin I could account for the contractile defect in end-stage heart failure. J. Mol. Cell. Cardiol. 2007;42(1):247–259. doi: 10.1016/j.yjmcc.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 49.Gomes AV, Liang JS, Potter JD. Mutations in human cardiac Troponin I that are associated with restrictive cardiomyopathy affect basal ATPase activity and the calcium sensitivity of force development. J. Biol. Chem. 2005;280(35):30909–30915. doi: 10.1074/jbc.M500287200. [DOI] [PubMed] [Google Scholar]
- 50.Bodor GS, Oakeley AE, Allen PD, Crimmins DL, Ladenson JH, Anderson PAW. Troponin I phosphorylation in the normal and failing adult human heart. Circulation. 1997;96(5):1495–1500. doi: 10.1161/01.cir.96.5.1495. [DOI] [PubMed] [Google Scholar]
- 51.Kooij V, Saes M, Jaquet K, Zaremba R, Foster DB, Murphy AM, dos Remedios C, van der Velden J, Stienen GJM. Effect of troponin I Ser23/24 phosphorylation on Ca2+-sensitivity in human myocardium depends on the phosphorylation background. J. Mol. Cell.Cardiol. 2010;48(5):954–963. doi: 10.1016/j.yjmcc.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Layland J, Solaro RJ, Shah AM. Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc. Res. 2005;66(1):12–21. doi: 10.1016/j.cardiores.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 53.Solaro RJ. Multiplex kinase signaling modifies cardiac function at the level of sarcomeric proteins. J. Biol. Chem. 2008;283(40):26829–26833. doi: 10.1074/jbc.R800037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang Y, Kobilka BK. Myocyte adrenoceptor signaling pathways. Science. 2003;300(5625):1530–1532. doi: 10.1126/science.1079206. [DOI] [PubMed] [Google Scholar]
- 55.El-Armouche A, Pohlmann L, Schlossarek S, Starbatty J, Yeh YH, Nattel S, Dobrev D, Eschenhagen T, Carrier L. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. J. Mol. Cell.Cardiol. 2007;43(2):223–229. doi: 10.1016/j.yjmcc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Hamdani N, Kooij V, van Dijk S, Merkus D, Paulus WJ, dos Remedios C, Duncker DJ, Stienen GJM, van der Velden J. Sarcomeric dysfunction in heart failure. Cardiovasc. Res. 2008;77(4):649–658. doi: 10.1093/cvr/cvm079. [DOI] [PubMed] [Google Scholar]
- 57.Zethelius B, Berglund L, Sundstrom J, Ingelsson E, Basu S, Larsson A, Venge P, Arnlov J. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N. Engl. J. Med. 2008;358(20):2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 58.Van Eyk JE, Powers F, Law W, Larue C, Hedges RS, Solaro RJ. Breakdown and release of myofilament proteins during ischemia and ischemia/reperfusion in rat hearts - Identification of degradation products and effects on the pCa-force relation. Circ. Res. 1998;82(2):261–271. doi: 10.1161/01.res.82.2.261. [DOI] [PubMed] [Google Scholar]
- 59.McDonough JL, Arrell DK, Van Eyk JE. Troponin I degradation and covalent complex formation accompanies myocardial ischemia/reperfusion injury. Circ. Res. 1999;84(1):9–20. doi: 10.1161/01.res.84.1.9. [DOI] [PubMed] [Google Scholar]
- 60.Katrukha AG, Bereznikova AV, Filatov VL, Esakova TV, Kolosova OV, Pettersson K, Lovgren T, Bulargina TV, Trifonov IR, Gratsiansky NA, Pulkki K, Voipio-Pulkki LM, Gusev NB. Degradation of cardiac troponin I: implication for reliable immunodetection. Clin. Chem. 1998;44(12):2433–2440. [PubMed] [Google Scholar]
- 61.Croall DE, Demartino GN. Calcium-activated neutral protease (calpain) system: structure, function, and regulation. Physiol. Rev. 1991;71(3):813–847. doi: 10.1152/physrev.1991.71.3.813. [DOI] [PubMed] [Google Scholar]
- 62.Canty JM, Lee TC. Troponin I proteolysis and myocardial stunning: Now you see it - Now you don't. J. Mol. Cell. Cardiol. 2002;34(4):375–377. doi: 10.1006/jmcc.2002.1531. [DOI] [PubMed] [Google Scholar]
- 63.Marston SB, Redwood CS. Modulation of thin filament activation by breakdown or isoform switching of thin filament proteins - Physiological and pathological implications. Circ. Res. 2003;93(12):1170–1178. doi: 10.1161/01.RES.0000105088.06696.17. [DOI] [PubMed] [Google Scholar]
- 64.Colantonio DA, Van Eyk JE, Przyklenk K. Stunned peri-infarct canine myocardium is characterized by degradation of troponin T, not troponin I. Cardiovasc. Res. 2004;63(2):217–225. doi: 10.1016/j.cardiores.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 65.Labugger R, Simpson JA, Quick M, Brown HA, Collier CE, Neverova I, Van Eyk JE. Strategy for analysis of cardiac troponins in biological samples with a combination of affinity chromatography and mass spectrometry. Clin. Chem. 2003;49(6):873–879. doi: 10.1373/49.6.873. [DOI] [PubMed] [Google Scholar]
- 66.Solaro RJ, van der Velden J. Why does troponin I have so many phosphorylation sites? Fact and fancy. J. Mol. Cell. Cardiol. 2010;48(5):810–816. doi: 10.1016/j.yjmcc.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lohse MJ, Engelhardt S, Eschenhagen T. What is the role of beta-adrenergic signaling in heart failure? Circ. Res. 2003;93(10):896–906. doi: 10.1161/01.RES.0000102042.83024.CA. [DOI] [PubMed] [Google Scholar]
- 68.Salazar NC, Chen J, Rockman HA. Cardiac GPCRs: GPCR signaling in healthy and failing hearts. Biochim. Biophys. Acta. 2007;1768(4):1006–1018. doi: 10.1016/j.bbamem.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duncker DJ, Boontje NM, Merkus D, Versteilen A, Krysiak J, Mearini G, El-Armouche A, de Beer VJ, Lamers JMJ, Carrier L, Walker LA, Linke WA, Stienen GJM, van der Velden J. Prevention of myofilament dysfunction by beta-Blocker therapy in postinfarct remodeling. Circ.-Heart Failure. 2009;2(3):233–242. doi: 10.1161/CIRCHEARTFAILURE.108.806125. [DOI] [PubMed] [Google Scholar]
- 70.Qian WJ, Jacobs JM, Liu T, Camp DG, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics. 2006;5(10):1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson NL, Anderson NG. The human plasma proteome - History, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 72.Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: A program to improve protein biomarker discovery for cancer. J. Proteome Res. 2005;4(4):1104–1109. doi: 10.1021/pr050027n. [DOI] [PubMed] [Google Scholar]
- 73.Hage DS. Affinity chromatography: A review of clinical applications. Clin. Chem. 1999;45(5):593–615. [PubMed] [Google Scholar]
- 74.Collier TS, Sarkar P, Rao B, Muddiman DC. Quantitative top-down proteomics of SILAC labeled human embryonic stem cells. J. Am. Soc. Mass Spectrom. 2010;21(6):879–889. doi: 10.1016/j.jasms.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 75.Kellie JF, Tran JC, Lee JE, Ahlf DR, Thomas HM, Ntai I, Catherman AD, Durbin KR, Zamdborg L, Vellaichamy A, Thomas PM, Kelleher NL. The emerging process of Top Down mass spectrometry for protein analysis: biomarkers, protein-therapeutics, and achieving high throughput. Mol. Biosys. 2010;6(9):1532–1539. doi: 10.1039/c000896f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Armirotti A, Damonte G. Achievements and perspectives of top-down proteomics. Proteomics. 2010;10(20):3566–3576. doi: 10.1002/pmic.201000245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.