Abstract
In bacteria with circular chromosomes, homologous recombination can generate chromosome dimers that cannot be segregated to daughter cells at cell division. Xer site-specific recombination at dif, a 28-bp site located in the replication terminus region of the chromosome, converts dimers to monomers through the sequential action of the XerC and XerD recombinases. Chromosome dimer resolution requires that dif is positioned correctly in the chromosome, and the activity of FtsK, a septum-located protein that coordinates cell division with chromosome segregation. Here, we show that cycles of XerC-mediated strand exchanges form and resolve Holliday junction intermediates back to substrate irrespective of whether conditions support a complete recombination reaction. The C-terminal domain of FtsK is sufficient to activate the exchange of the second pair of strands by XerD, allowing both intra- and intermolecular recombination reactions to go to completion. Proper positioning of dif in the chromosome and of FtsK at the septum is required to sense the multimeric state of newly replicated chromosomes and restrict complete Xer reactions to dimeric chromosomes.
Keywords: Xer recombination, Holliday junction, chromosome segregation, FtsK
The faithful replication and segregation of chromosomes is necessary to ensure the efficient transmission of genetic information. The replication of chromosomes depends on homologous recombination, which is used to rescue replication forks that have broken or stalled when encountering barriers such as DNA lesions or blocked transcription complexes (Seigneur et al. 1998; Cox et al. 2000; McGlynn and Lloyd 2000; Michel 2000). Crossing over by homologous recombination can lead to sister chromatid exchanges (SCEs), odd numbers of which result in dimerization of the two newly replicated chromatids when the chromosomes or replicons are circular, as is often the case in bacteria. Plasmid or chromosome dimers compromise stable plasmid inheritance and chromosome segregation (Austin et al. 1981; Summers and Sherratt 1984; Blakely et al. 1991; Clerget 1991; Kuempel et al. 1991; Hendricks et al. 2000). Hence, most bacteria with circular genomes possess the specialized Xer site-specific recombination system to convert chromosome and plasmid circular dimers to monomers (Recchia and Sherratt 1999). Xer site-specific recombination in Escherichia coli uses two recombinases of the tyrosine recombinase family, XerC (Colloms et al. 1990) and XerD (Blakely et al. 1993), which act on a 28-bp recombination site, dif, located in the replication terminus region of the chromosome (Blakely et al. 1991; Clerget 1991; Kuempel et al. 1991). When a chromosome dimer is present, a complete site-specific recombination reaction by XerCD introduces an additional SCE at dif, thus resolving the dimer into two monomers (Steiner and Kuempel 1998a).
Key questions about Xer site-specific recombination at dif concern how two dif sites separated by greater than 4 Mbp in a chromosome dimer can be efficiently synapsed together, and how the system is controlled to ensure resolution of chromosome dimers into monomers without risking the reverse reaction, which would create dimers from monomers. Xer-mediated recombination at dif, whether it be on the chromosome or on plasmids, depends on a functional homologous recombination system, apparently because complete Xer recombination reactions only occur in cells that contain chromosomal dimers (Steiner and Kuempel 1998b; Recchia et al. 1999). Furthermore, complete Xer recombination reactions require the C-terminal domain of FtsK (Recchia et al. 1999; Steiner et al. 1999), a septum-located protein that functions both in chromosome segregation and cell division (Begg et al. 1995; Diez et al. 1997; Draper et al. 1998; Liu et al. 1998; Wang and Lutkenhaus 1998; Yu et al. 1998a,b). These observations establish a link between Xer recombination, homologous recombination, and cell division. Finally, the 28-bp dif site is active only when located in a specific 30-kbp region of the chromosome (the dif activity zone or DAZ), where it acts independently of any other specific sequence in that region, thus indicating the importance of the structural and spatial organization of the chromosome inside the cell for Xer recombination (Leslie and Sherratt 1995; Tecklenburg et al. 1995; Cornet et al. 1996; Kuempel et al. 1996; Perals et al. 2000).
Here, we show that Xer recombination, independently of any other cellular element, can form Holliday junctions (HJs) between dif sites, thereby potentially anchoring the two sister dif sites together immediately after their replication. HJ intermediates result from the exchange of a first pair of strands by XerC. They are unstable and are converted back to substrate in cycles of XerC-mediated strand exchanges. We also show that the membrane-bound and septum-located FtsK protein serves as a molecular antenna, which activates, through its C terminus, the exchange of the second pair of strands by the XerD recombinase when a chromosome dimer is present. These results help us understand how bacterial cells can integrate information at the cellular organization level to control enzymatic processes at the molecular level.
Results
The C-terminal domain of FtsK supports Xer recombination at dif
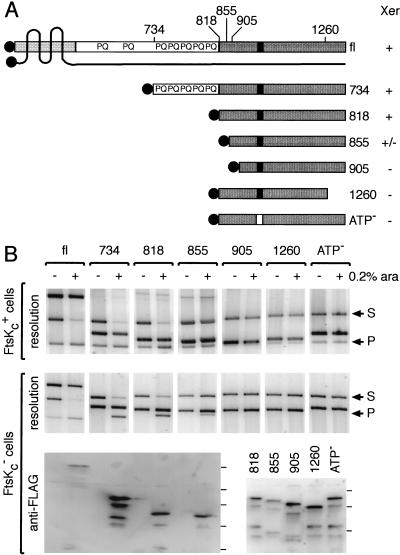
The 1329–amino acid FtsK protein contains three domains (Fig. 1A). The 200–amino acid N-terminal domain, domain 1, contains four transmembrane regions (Dorazi and Dewar 2000), localizes to the septum (Wang and Lutkenhaus 1998; Yu et al. 1998a) and is required for cell division (Diez et al. 1997; Draper et al. 1998). The 500–amino acid C-terminal domain, domain 3, is necessary for normal chromosome segregation (Liu et al. 1998; Yu et al. 1998b), at least in part because it is necessary for Xer recombination at dif (Recchia et al. 1999; Steiner et al. 1999). Cells lacking domain 3 form septate chains and filaments with aberrant and mispositioned nucleoids. Domain 3 is homologous with the C-terminal domain of SpoIIIE, a protein involved in DNA transfer from the mother cell to the prespore in Bacillus subtilis (Wu et al. 1995). Domains 1 and 3 are separated by a 600–amino acid linker region, domain 2, which is not generally conserved in FtsK homologs. Overall, it is abundant in proline and glutamine and contains seven PQ-rich repeats of 10 amino acids.
Figure 1.
Minimal region of FtsK required for Xer recombination. (A) FtsK derivatives used in this study. The Nterminal domain of FtsK, domain 1, is represented by a lightly shaded box, and the four putative transmembrane regions are shown as black lines crossing the protein. The extreme N terminus points toward the cytoplasm, as well as domain 2 and 3 of the protein. The C-terminal domain, domain 3, is shown as a dark box, and the putative nucleotide binding site as a black rectangle. The Flag peptide is shown as shaded circles. The N-terminal (734, 818, 855, 905) and C-terminal (1260) amino acid of each protein are indicated on top of the full-length protein (fl). The relative ability of each protein to support Xer recombination on plasmids is indicated by +/− signs on the right of the diagram. (B) Xer recombination between plasmid-borne dif sites in cells expressing FtsK deletions. FtsKc+ (DS941) and FtsKc− (DS9041) cells (as shown to the left of the gels) were transformed with the different ftsK expression vectors, as indicated along the top of the gels. Plasmid-containing cells were grown for 5 h in arabinose (+, 0.2% ara) or in glucose (−). Recombination between the two dif sites carried by the substrate (S) leads to the formation of two smaller product circles, of which one can replicate (P). The substrate and product bands are indicated by arrows to the right of the gels. The third band present in each lane is the ftsK expression vector. Expression of the FtsK derivatives was verified by Western blot analysis after resolution of the proteins on a 6% (fl, 734, 818, 855) or a 8% (818, 855, 905, 1260, ATP−) SDS-PAGE. Complete repression (−) was verified for fl, 734, 818, and 855. The position of the protein molecular weight markers are indicated on the right of each blot and correspond from bottom to top to 32, 47, 67, 83, and 175 kD. The relatively low signal with fl may reflect a poorer transfer to the membrane.
To characterize further the functions of the three FtsK domains, we constructed FtsK truncations tagged with a Flag peptide to allow for immunodetection (Fig. 1A). The ftsK derivatives were expressed from the arabinose promoter of plasmid pBAD24, the levels of expression being monitored by Western analysis. They were tested for Xer recombination between two dif sites carried by a plasmid (Fig. 1B). The full-length protein (fl), tagged at its N terminus, fully restored Xer recombination in cells that only expressed the ∼200 N-terminal amino acids of FtsK needed for viability (FtsKc−). Indeed, the level of recombination was higher (∼70% resolution) than that mediated by chromosomally located wild-type protein (∼30% resolution), thereby indicating that normal levels of wild-type protein are limiting for Xer recombination as assayed on a plasmid, either because of FtsK amount and/or location. Note that in the absence of arabinose, a low level of resolution is observed in FtsKc− cells carrying the fl plasmid derivative, presumably because of a low level of residual FtsK expression from the plasmid. Domains 1 and 2 were found not to be necessary for Xer recombination between dif sites: when overexpressed, the 734 and 818 FtsK derivatives, containing amino acid 734 and 818 to the normal C terminus, respectively, restored as much resolution as fl FtsK. The smaller 855 protein also showed activity although it was reduced compared with 818; this may be partly due to a lower level of protein. Further truncations of the N terminus (905) or of the C terminus (1260) of FtsK abolished Xer complementation, as did a lysine to alanine substitution at amino acid 997 in the putative nucleotide binding site (ATP−). C-terminally tagged proteins showed a decreased activity compared with the N-terminally tagged versions (data not shown). None of the FtsK derivatives that lacked the N terminus of the protein had any substantial dominant negative effect in cells expressing wild-type FtsK. This contrasts to a FtsK derivative containing all but the 65 C-terminal amino acids, which inhibited the action of the wild-type protein when expressed from a low-copy plasmid (Recchia et al. 1999). Thus, domain 1 or 2 of FtsK may be involved in the protein–protein interactions that allow a mutant protein to interfere with its wild-type counterpart, but the C-terminal domain of FtsK (FtsKc) is sufficient in itself to allow an Xer recombination reaction to go to completion.
FtsKc is cytoplasmic
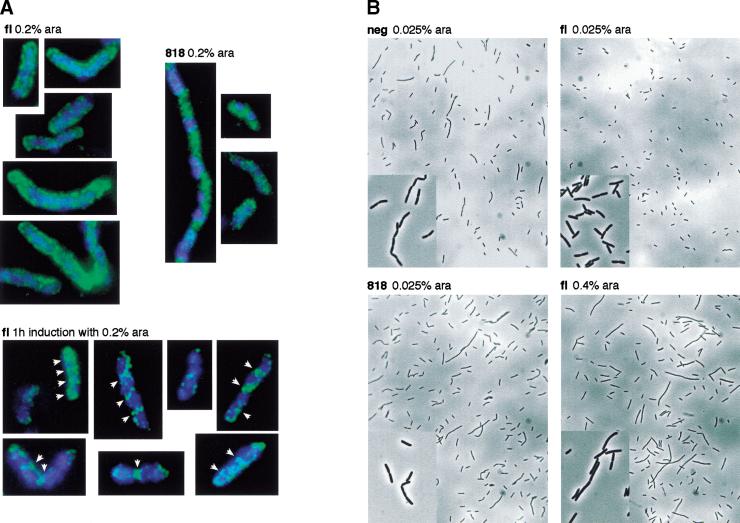
As septum localization of FtsK has been proposed to be important for its function in Xer recombination (Recchia et al. 1999; Steiner et al. 1999), we checked the localization of the fl and 818 FtsK derivatives under similar expression conditions to those used in the plasmid resolution assay. To ensure a steady state level of expression of the proteins in exponentially growing cells, we grew FtsKc− cells overnight in LB supplemented with 0.2% arabinose, diluted them in the morning with fresh medium, and grew them to an OD600 of ∼0.4 (∼25 generations). Vizualization of the 818 derivative revealed a cytoplasmic distribution without any specific localization (Fig. 2A, 818). Similarly, the fl FtsK covered the whole membrane of the cells and failed to localize specifically to septum rings (Fig. 2A, fl). Thus, a failure to position FtsKc specifically at the septum does not prevent its function in Xer recombination.
Figure 2.
(A) Cellular distribution of fl and 818 FtsK proteins. Induction with arabinose (0.2%) was for ∼25 generation in FtsKc− (DS9041) cells (top) or for 1 h in FtsKc+ (DS941) cells (bottom). The localization of the DNA and of the FtsK proteins is shown in blue and green pseudocolors, respectively. White arrows indicate septum ring-like structures. (B) Morphology of FtsKc− cells expressing various amounts of fl or 818 FtsK proteins or carrying a control expression vector (neg).
The observed membrane coating by fl FtsK is likely due to the excess number of fl FtsK molecules over the number of potential FtsK attachment sites at the septum. To confirm this hypothesis, we checked the localization of fl under conditions of expression that more nearly correspond to the level of FtsK in wild-type cells, in which septum localization has been shown (Wang and Lutkenhaus 1998; Yu et al. 1998a). Indeed, the fl FtsK derivative tended to localize to specific loci that are reminiscent of septum rings in exponentially growing FtsK+ cells when it was expressed for a shorter time (Fig. 2A, 1h induction). We could not detect the tagged proteins with lower, and even more physiological, levels of expression because the fluorescent signal decreased to background levels.
Septum localization of FtsKc is required for normal cell morphology
FtsKc− cells form filaments and characteristic septate chains (Fig. 2B). Most of the phenotype of FtsKc− cells can be accounted for by their inability to resolve chromosome dimers because FtsKc− RecA− cells have a similar morphology to RecA− cells (Recchia et al. 1999). To test if expression of the C-terminal domain of FtsK could complement the chromosome segregation/cell division mutant phenotype of FtsKc− cells, we examined the morphology of cells carrying the FtsK derivatives on plasmids in early exponential phase after growth in minimal medium containing different levels of arabinose. Expression of the FtsK derivatives was shown to increase with the amount of arabinose added as verified by Western analysis (data not shown). Cell morphology was restored to normal when fl FtsK was expressed at low concentration of arabinose (0.025%). High levels of expression (0.4% arabinose) resulted in the loss of the septate chains but increased filamentation. Low level expression (0.025% arabinose) of the 818 derivative did not complement the phenotype of FtsKc− cells. On the contrary, we had the impression that some longer septate chains were present. A higher concentration of arabinose did not result in complementation and did not induce filamentation as was the case with the full-length protein (data not shown). Thus, cells expressing domain 3 independently of domain 1 or cells overexpressing the full-length protein, both of which result in the loss of the specific localization of domain 3 at the septum, displayed an abnormal morphology. We conclude that septum localization of FtsKc is important for its cellular role in chromosome segregation.
Transient HJ formation by XerCD at the chromosomal dif site
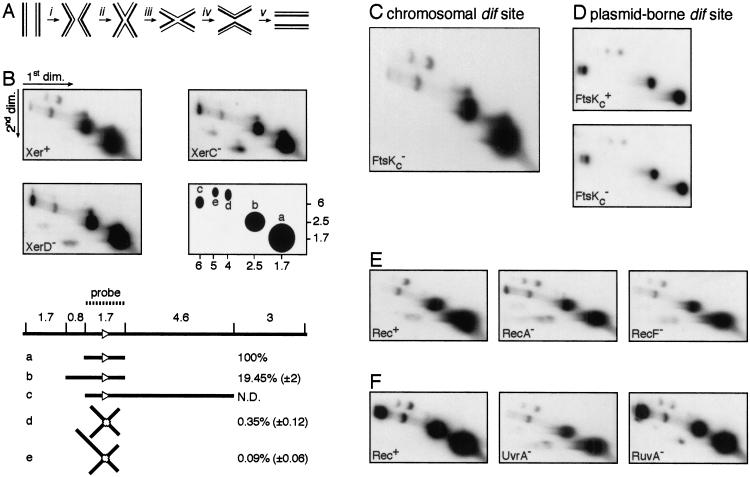
Xer site-specific recombination at dif can be viewed as a five-step process (Fig. 3A). After synapsis (Fig. 3A, i), a first pair of strand exchanges mediated by XerC (Fig. 3A, ii; Blakely et al. 2000; Hallet et al. 1999) leads to the formation of a HJ intermediate that needs to undergo a conformational change (Fig. 3A, iii) to be resolved by a pair of XerD-mediated strand exchanges (Fig. 3A, iv), before dissociation of the synaptic complex (Fig. 3A, v). FtsK has been proposed to be necessary for Xer recombination at dif either by facilitating synapsis (Fig. 3A, i) or by facilitating a conformational change (Fig. 3A, iii) of the HJ intermediate that would reciprocally activate XerD and inactivate XerC (Recchia et al. 1999). To differentiate these two models, we monitored in vivo Xer-dependent HJ formation at the chromosomal dif site. Earlier work that used a density label assay to measure Xer recombination at dif had failed to detect HJ intermediates (Steiner and Kuempel 1998a). As a consequence, we decided to use the highly sensitive technique of 2D gel electrophoresis, followed by DNA hybridization (Schwacha and Kleckner 1994, 1995). To prevent loss or branch migration of HJs, we cross-linked the DNA in cells by psoralen before isolation. In 2D gel electrophoresis, linear DNA migrates as an arc whereas X and Y branched structures migrate off the arc. Optimal separation between branched structures and their linear counterparts is obtained when their different arms are of equal length. As a consequence, EcoRV was used to digest the samples because it generates a 1.7-kb fragment centred on dif (Fig. 3B). This same fragment was used as a probe for specific detection of the dif region.
Figure 3.
In vivo detection of HJ intermediates at the chromosomal dif site. (A) The different steps of Xer site-specific recombination. (i) Synapsis; (ii) first pair of strand exchanges; (iii) HJ conformational change; (iv) second pair of strand exchanges; (v) dissociation. (B) Detection of HJs at dif in Xer+ (AB1157), XerC− (DS8029), and XerD− (GR18) cells. EcoRV was used to restrict the DNA samples. A simplified scheme of the hybridization signals is shown along with molecular weights in kilobase pairs. Five spots were detected that correspond to expected linear (a, b, and c) or joint (d and e) molecules. The EcoRV restriction digest map of the dif region and the molecules that can be attributed to the a, b, c, d, and e spots are represented below the blots. The dif site is shown as a white triangle. The relative intensity of each spot to the intensity of spot (a) was calculated in XerC+ XerD+ cells. Numbers correspond to the mean and standard deviation of values from eight independent experiments. (C) HJs at the chromosomal dif site in FtsKc− cells (MA3). (D) HJs in FtsKc+ (AB1157) and FtsKc− cells carrying a multicopy plasmid with a single dif site. (E) HJs at the chromosomal dif site in Rec+ (AB1157), RecA− (GR19), and RecF− (DS941) cells. (F) HJs at the chromosomal dif site in Rec+ (JJC40), UvrA− (N3137), and RuvA− (JJC662) cells. The smeary bands below the arc of linear DNA in some of the gels are blotting artefacts and contain small amounts of the major DNA species from the linear arc.
Three spots (a,b,c) could be detected that migrated as linear DNA in extracts from Xer+, XerC−, or XerD− strains. The more intense spot (a) migrated as the expected 1.7-kb fragment centred on dif. (b) and (c) migrated as a 2.5-kb and a 6.3-kb fragment, respectively, which correspond to partial digests. Those partial digests result from the specific inhibition of restriction enzymes by the presence of a cross-link at their cutting site (Barre et al. 2000), and therefore reflect the efficiency of cross-linking. A fourth and fainter fragment could be detected between (b) and (c) and is probably due to cross-hybridization of the dif probe with some other region of the genome. Two spots (d and e) are clearly visible above the arc of linear molecules extracted from Xer+ cells. Their sizes are in agreement with them being the expected 3.4-kb HJ at dif (d) and a 4.2-kb HJ (e) resulting from partial digest. No HJs were detected in XerC− or XerD− cells, as expected because the presence of both recombinases is required for catalysis by either (Blakely et al. 1993). Thus, the HJs observed at dif in the Xer+ strain result from the action of the Xer recombinases, and not from the action of other recombination systems.
A good approximation of the level of HJ-containing fragments in the psoralen-treated cell extracts can be obtained by quantitating the relative intensity of the HJ spot (d) to the intensity of the linear spot (a). We believe this value reflects the steady state level of HJs in exponentially growing cells, and therefore the equilibrium between the rates of HJ formation and resolution. A mean value of 0.3% of HJs was obtained in wild-type cells.
FtsKc is not required for HJ formation by the Xer recombinases at dif
We reasoned that if the C terminus of FtsK was implicated in synapsis, its absence would lead to a decrease in the level of HJs formed by XerCD at dif. In contrast, we observed as many HJs in DNA extracts from FtsKc− cells as in DNA extracts from FtsKc+ cells (Fig. 3C). This was also true when we looked at a dif site carried by a plasmid (Fig. 3D). We conclude that FtsKc is not needed for synapsis and for HJ formation by XerCD.
Homologous recombination is not required for HJ formation
In addition to FtsKc, homologous recombination is necessary for full Xer recombination activity at dif, presumably because the reaction requires chromosomal dimers (Steiner and Kuempel 1998b; Recchia et al. 1999). We found that HJ formation at dif was not impaired in a RecA− strain or in a RecF− strain (Fig. 3E), in which homologous recombination is abolished or impaired, respectively. Homologous recombination on plasmids is mostly dependent on the recF pathway (Summers and Sherratt 1984; Kolodner et al. 1985), but neither recA nor recF had any effect on HJ formation at a dif site carried by a plasmid (data not shown). We conclude that HJs are still formed by Xer recombination at dif in the absence of chromosome or plasmid dimers.
To confirm that chromosome dimer formation by homologous recombination is not necessary for synapsis and for the first pair of strand exchanges of the Xer recombination reaction, we monitored the level of HJs at dif in strains that should contain elevated numbers of chromosome dimers (Fig. 3F). UvrA− cells are deficient in nucleotide excision repair and therefore more heavily dependent on homologous recombination for the repair of DNA lesions; this should increase the probability of dimer formation and the need for Xer recombination. However, the level of HJ was not significantly increased in UvrA− cells. There is a bias toward noncrossover events during homologous recombination in E. coli, thereby minimizing dimer formation. The bias appears to occur at the level of HJ resolution and requires RuvABC (Michel et al. 2000). In the absence of Ruv, the bias is lost and Xer recombination becomes important for survival (Michel et al. 2000). However, inactivating the ruvA gene did not increase the level of HJ detected at dif. This was also true for a Rep− RuvA− strain (data not shown), which relies on Xer recombination for its viability (Michel et al. 2000). No HJs were observed in a XerC− RuvA− strain (data not shown), confirming that the HJs we detected are due to Xer recombination and not to homologous recombination.
DAZ does not define a zone of competence for HJ formation
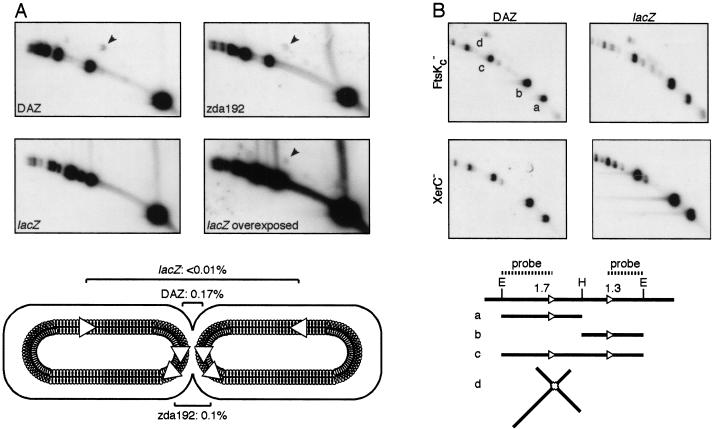
The location of dif in a ∼30-kbp zone of the E. coli chromosome, DAZ, is required for it to be an efficient substrate for a complete Xer recombination reaction (Tecklenburg et al. 1995; Cornet et al. 1996; Kuempel et al. 1996; Perals et al. 2000). To test if the location of dif in DAZ was important for synapsis and for the first pair of strand exchanges by XerCD, we compared the level of HJs in three strains carrying a single dif site at different locations in the chromosome. Displacing dif outside DAZ, ∼150 kbp away from its original position, results in a 10-fold decrease in its activity, as measured in a complete recombination reaction (Perals et al. 2000) but had only a ∼twofold effect on HJ formation (Fig. 4A; cf. DAZ with zda 192; HJ levels are shown in the bottom panel). Thus, XerCD can form HJs at dif outside DAZ. However, displacing dif far from the replication terminus region, to within lacZ, resulted in a dramatic decrease in the level of HJs detected, although a positive signal was observed with overexposure of the blot. A likely explanation to this dramatic decrease is that the opportunities for synapsis of the two sister dif sites are limited by the rapid segregation of the chromosome immediately after replication when dif is outside the replication terminus region.
Figure 4.
Effect of the location of dif on the level of HJs. (A) HJs at dif (indicated by arrowheads) were compared in three strains carrying a single site in DAZ (LN3080), at 150 kb to the left side of DAZ (zda192, LN3577) or at 1 Mb to the right side of DAZ (lacZ, LN3091). The positions of the two sister dif sites in dividing cells are represented below the blots. Numbers indicate the relative intensity of the HJ spot to the intensity of the smaller linear fragment (mean of two independent experiments). (B) Increased and location-independent levels of HJ in cells carrying two tandem dif sites. HJ levels were compared in FtsKc− and XerC− cells carrying a dif-Km-dif cassette in DAZ or in lacZ (FC313, FC354, FC235, FC284). EcoRV (E) and HindIII (H) were used to restrict DNA. The restriction digest map of the dif-Km-dif cassette and the molecules that can be attributed to the a, b, c, and d spots are represented below the blots. Note that the intensity of spot d must be compared with the sum of the intensities of spot a and b.
To confirm this hypothesis and rule out any effect of DAZ in HJ formation, we compared the level of HJs in strains carrying a tandem repeat of two dif sites inserted within DAZ or within lacZ (Fig. 4B). Because tandem repeats of dif within DAZ are unstable in FtsKc+ cells (Perals et al. 2000), levels of HJ formation were assayed in FtsKc− cells in which the Xer recombination reaction does not go to completion. The level of HJs reached 7.3% when the directly repeated dif sites were located within DAZ (Fig. 4B, DAZ). This corresponds to a greater than 10-fold increase when compared with strains with a single dif site. The observed HJs were due to Xer recombination because they were abolished in a XerC− background. The level of HJs remained high when the tandemly repeated dif sites were in lacZ (4.4%), HJ formation being also entirely due to Xer recombination. Despite this high level of HJs, tandem repeats at lacZ are stable in FtsK+ cells (data not shown). Thus, by providing a substrate in which two dif sites are readily accessible, we could overcome the need for dif to be located in the replication terminus region for HJ formation, but resolution to a complete recombinant product was still confined to DAZ.
In vitro HJ formation by XerCD at dif
We have shown that HJ formation at dif does not depend on chromosomal dimers, the location of dif in DAZ, or the activity of FtsK. Furthermore, ∼5% of HJs were detected between tandemly repeated dif sites. This high level of HJs was surprising because only very low levels of HJs had been observed in previous in vitro experiments with purified Xer recombinases (Blakely et al. 2000; Colloms et al. 1996). In those experiments, the catalytic activity of XerC but not XerD was shown to be necessary.
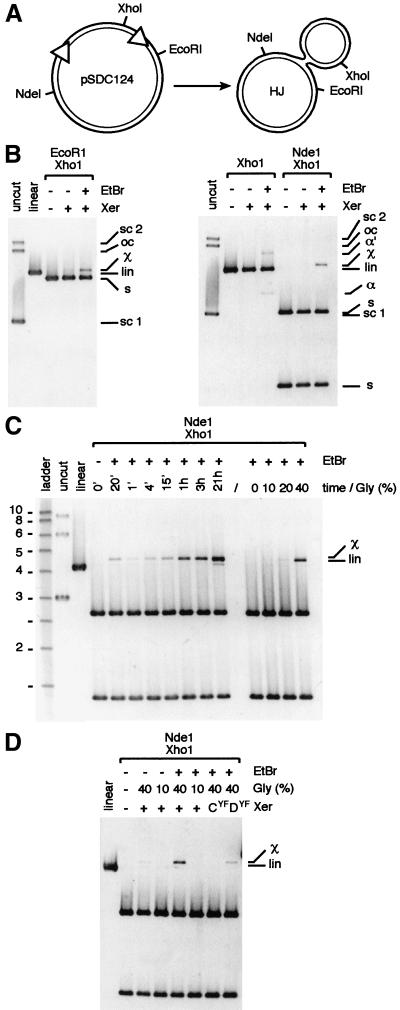
We reasoned that in vitro synapsed dif sites might also undergo cycles of Xer-mediated strand exchanges that lead to the formation of HJs and their conversion back to substrate, but that this results in steady state levels of HJs too low to be readily detected. We therefore tested whether ethidium bromide (EtBr), which inhibits the action of type I topoisomerases and many other DNA processing enzymes, blocks the conversion of HJs back to substrate in in vitro reactions performed on a supercoiled plasmid carrying two dif sites, for example, by preferentially binding to HJ DNA and thereby inhibiting the action of XerCD on HJ intermediates. This would increase the level of HJs by displacing the equilibrium between HJ formation and resolution.
Addition of EtBr to in vitro reactions of XerCD with supercoiled DNA containing two directly repeated dif sites resulted in the detection of a high level of HJs, as confirmed by restriction analysis of the DNA with different enzymes (Fig. 5B). Forty percent glycerol was needed for maximum HJ detection (Fig. 5C,D). A time course experiment revealed that the level of HJs increased steadily in the plasmid population over time, and addition of EtBr at the beginning of the reaction had the same effect as addition of the EtBr at the end of the reaction (Fig. 5C, cf. the 20′ time point in which EtBr was added first, to the 15′ point in which it was added after 15′). We conclude that synapsis is the rate limiting factor in the reaction rather than XerC-mediated strand exchanges. The increasing level of HJs with time therefore reflects the increasing number of synaptic complexes formed in the plasmid population.
Figure 5.
In vitro HJ formation by XerCD on a supercoiled plasmid carrying two dif sites. (A) restriction map of plasmid pSDC124 and of the HJ created by action of XerCD. (B) One-hour reactions in 40% glycerol. (C) Time course experiment in 40% glycerol and effect of the glycerol concentration in 1-h reactions. Molecular weights are in kilobase pairs. (D) Determination of the Xer recombinase whose catalytic activity is necessary for HJ formation. One-hour reactions in 40% glycerol used catalytically inactive mutants of XerC (CYF) and XerD (DYF) mixed with their functional partner. pSCD124 was left untreated (uncut, linear) or treated with the Xer recombinases (Xer). EtBr was added at the end of reactions as indicated. For the 20′ time point, EtBr was added at the beginning of the reaction. sc 1, sc 2, lin, and oc, supercoiled monomer, supercoiled dimer, linear, open circle forms of pSDC124, respectively. s, restriction fragments from pSCD124; χ, χ-molecule containing a HJ; α and α′, supercoiled and nicked α-molecules containing a HJ.
To determine which recombinase was responsible for the pair of strand exchanges that led to HJ formation, we performed the reactions by using catalytically inactive XerC and XerD mutants (Fig. 5D). No HJs were detected with the XerCYF mutant whereas they were still formed with the XerDYF mutant. Thus, the HJs observed result from strand exchanges mediated by the XerC recombinase, as suggested by previous experiments (Hallet et al. 1999; Blakely et al. 2000).
Roles of DAZ and homologous recombination in Xer recombination
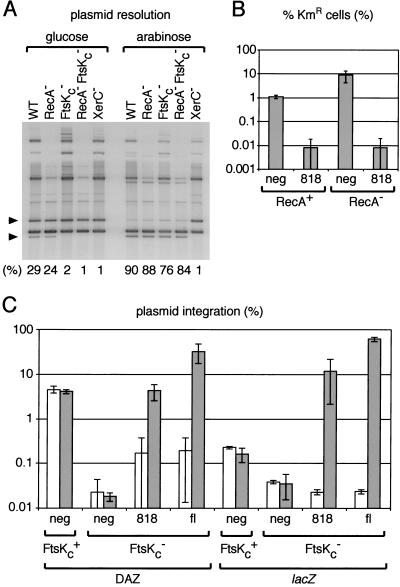
We noted that overexpression of the 818 FtsK derivative resulted in an increase in plasmid resolution from ∼30% to 90% independently of the status of the homologous recombination system (Fig. 6A), and independently of DAZ because this was observed on a plasmid.
Figure 6.
Overexpression of FtsKc abolishes the requirement for homologous recombination and for the location of dif in DAZ. (A) Plasmid resolution assay in WT (AB1157), RecA− (GR19), FtsKc− (MA3), RecA− FtsKc− (GR59), and XerC− (DS8029) cells under conditions of repression (glucose) or induction (0.4% arabinose) of 818 expression. The bands corresponding to the supercoiled substrate and supercoiled replicative product are indicated by the top and bottom black triangle, respectively. Numbers indicate the proportion of supercoiled product to total supercoiled substrate in each lane. (B) dif-Km-dif cassette resolution assay. RecA+ (FC235) and RecA− (FC217) cells were transformed with a control expression vector (neg) or with the vector expressing the 818 FtsK derivative. They then were transformed with pFC225, which carries a functional xerC gene. The proportion of kanamycin resistant cells in the cultures (KmR) was quantitated after overnight growth in the presence of 0.2% arabinose. Results from a typical experiment are shown. (C) Plasmid integration assay. FtsKc+ and FtsKc− cells carrying a single chromosomal dif site in DAZ or in lacZ (DS941, NL250, DS9041, MA2) were transformed with a control expression vector (neg), with the 818 expression vector or with the fl expression vector. They then were transformed with pFH127, which carries a single dif site. The proportion of cells having integrated this plasmid was quantitated after overnight growth at 30°C in the presence of glucose (white bars) or 0.2% arabinose (gray bars). Shown is the result of at least two independent experiments.
The dependence of Xer recombination at dif on the presence of chromosomal dimers is more obvious when the dif sites are in tandem repeat and positioned in chromosomal DAZ. Indeed, a KmR cassette placed between two tandem direct repeats of dif within DAZ is 10 times more stable in a RecA− strain than a RecA+ strain, as a consequence of the difference in the levels of complete Xer recombination reactions (Fig. 6B). However, overexpression of the 818 protein increased dramatically the level of recombination at dif in both strains, so that there were no more differences in Xer recombination levels, as measured by KmR stability, between the RecA− and the RecA+ strains.
Integration of a plasmid carrying a single dif site into chromosomal dif by Xer recombination depends on the proper location of dif inside DAZ and on the C terminus of FtsK (Recchia et al. 1999). Displacing dif from DAZ to lacZ decreases the integration frequency by ∼22.5-fold (Fig. 6C). In the absence of FtsKc, the remaining integration frequency decreases again to a 5.3-fold lower level. However, overexpression of the full-length FtsK protein (fl) or of the 818 protein restored the integration frequency to wild-type levels, irrespective of whether dif was in DAZ or in lacZ.
Thus, by overexpressing the C-terminal domain of FtsK so that it is distributed throughout the cell, we can overcome the dependence of Xer recombination at dif on conditions that generate chromosomal dimers (Fig. 6B) and on DAZ (Fig. 6C). We conclude that the influence of both homologous recombination and DAZ are linked to the spatial and temporal distribution of FtsKc.
Discussion
XerC mediates cycles of strand exchange between substrate and HJ intermediate
Taken together, our in vivo and in vitro experiments provide strong support for the view that XerC continuously mediates the formation of HJs and their resolution back to initial substrate whenever the opportunity for synapsis between dif sites arises. We propose that this cycling continues from the time of synapsis immediately after dif replication until the time of chromosome segregation away from midcell and subsequent cell division some 10–20 min later. This process only requires the catalytic activity of XerC (Fig. 5) and is independent of the requirements for a complete Xer recombination reaction, in particular the activity of FtsKc, conditions that influence the presence and level of chromosome dimers at the initiation of cell division, and the correct location of dif within DAZ (Figs. 3, 4). The stability of synapsed dif sites in vivo may be facilitated by the cycles of XerC-mediated HJ formation and resolution. Once formed, a synaptic complex is stable in vitro, and under appropriate EtBr treatment conditions, a substantial level of HJ intermediate can be detected (Fig. 5). Indeed, we now have an explanation of a previous puzzling observation: dif-containing HJ molecules are excellent recombination substrates (Arciszewska and Sherratt 1995) and XerCD bind more tightly to duplex dif than to the plasmid substrates cer and psi (Blake et al. 1997), which themselves undergo efficient recombination on supercoiled plasmids in vitro (Colloms et al. 1997). However, in the absence of EtBr, XerCD appeared to generate no recombinant product and little or no HJs when they were reacted with a supercoiled molecule containing two dif sites (Colloms et al. 1996). Therefore, the presence of EtBr reveals the ability of XerCD to synapse dif sites and form HJs.
The presence of a stable synaptic complex between the two newly replicated sister dif sites may provide an answer to the potential problem of how two dif sites separated by more than 4 Mbp in a dimeric chromosome find each other before Xer recombination. Nevertheless, a stable synaptic complex may impose some constraints on the local DNA topology. This could explain the apparent interaction between the Xer recombination system and topoisomerase IV, the dif site being a hot spot for Topo IV activity in vivo (Hojgaard et al. 1999). The presence of XerCD is necessary for this, but not FtsK or cell division, consistent with the effect being mediated at the HJ stage. Topo IV is required to remove any catenation of the newly replicated sister monomer chromosomes, before their segregation. Note that such catenation can be converted into knotting during formation of a dimeric chromosome. We propose that after replication Topo IV will remove all catenation/knotting other than the crossings entrapped in the Xer synaptic complex. These final crossings can only be removed when the XerCD synaptic complex has finally dissociated. It is this interaction that may have been assayed in the experiments of Hojgaard et al. (1999).
The C-terminal domain of FtsK is sufficient to support a complete Xer recombination reaction at dif
Expression of the C-terminal domain of FtsK (FtsKc) restores Xer recombination between dif sites in FtsKc− cells (Fig. 1). The minimal domain necessary for complementation of Xer recombination at dif starts between amino acid residues 855 and 905 and ends between residues 1260 and 1329. Interestingly, the homology between domain 3 of FtsK and its homologs in other species starts at residue 839 at a predicted β-sheet and ends at residue 1323, after a predicted α-helix. The importance of the extreme C terminus of FtsK is underlined by our observation that proteins with Flag at their C terminus have decreased biological activity. Finally, the putative nucleotide binding site of FtsK was critical for its role in Xer recombination. It had been previously reported that two FtsK N-terminal truncations (starting at residue 825 and 863) fused to a polyhistidine tag (Boyle et al. 2000), and a protein containing domain 2 and 3 of FtsK (Recchia et al. 1999) did not complement Xer recombination at dif. However, we believe that these negative results may reflect a problem of expression or a lack of activity of the tagged proteins.
Overexpressing FtsKc increased the level of Xer recombination between dif sites carried by plasmids from ∼30% to 90% (Fig. 6A). It allowed Xer-mediated integration of a plasmid carrying a single dif site into chromosomal dif and rendered the reaction independent of the location of dif on the chromosome (Fig. 6C). Finally, it supported Xer recombination between two dif sites carried in tandem on the chromosome and rendered the reaction independent of conditions that lead to chromosomal dimers (Fig. 6B). We conclude that FtsKc can activate Xer recombination independently of its location or of the location of dif in the cell, and that it can only activate Xer recombination on sites that are in its vicinity. Low, physiologically relevant, amounts of the full-length FtsK protein are localized at the septum (Fig. 2A; Wang and Lutkenhaus 1998; Yu et al. 1998a). Therefore, multicopy plasmids, which are distributed throughout the cytoplasm (data not shown), may only access the C terminus of FtsK when they are located near the septum. Overexpression of the full-length FtsK protein, or of its C-terminal part, leads to an even distribution of the protein throughout the cell (Fig. 2A). Under these conditions, Xer recombination can then be activated by FtsKc whatever the location of dif.
Thus, our data support the view that FtsKc has a direct role in Xer recombination at dif. If its role were indirect, for example, by triggering a cascade of events leading to Xer activation, the unknown components of this cascade would have to be widely distributed and in excess amounts, because FtsK is the limiting factor in the Xer reaction.
Xer recombination at dif is controlled by FtsK at the XerD-mediated strand exchange step
Because the level of HJ formed in vivo at dif by XerCD is not influenced by the FtsK status of the cell (Fig. 3), FtsK must act after HJ formation, presumably by stimulating the second pair of strand exchanges mediated by XerD. Our previous work has led to a model in which a HJ conformational change is responsible for reciprocal activation of XerD and inactivation of XerC (Hallet et al. 1999; Recchia et al. 1999; Arciszewska et al. 2000). This is consistent with the observation that a recombinase variant combination that favors strand exchange by XerD can bypass the requirement for FtsK during recombination at dif (Recchia et al. 1999). We propose that FtsKc is directly implicated in the change of conformation of the HJ intermediate by remodeling the local structure of the DNA in a DNA sequence-independent and ATP-dependent reaction. Precedents already exist for such processes. For example, HJ resolvases RuvC and T7 Endo1 switch their direction of resolution when catenation entrapped by a HJ is removed (Zerbib et al. 1997). Furthermore, PepA, which directs the geometry of the synaptic complexes during recombination at cer and psi (Colloms et al. 1997), is also required to facilitate the conformational change in the HJ formed by XerC to allow strand exchange by XerD (M. Robertson and D.J. Sherratt, unpubl.).
Spatial distribution of FtsKc and chromosome segregation
We observed that the expression of the full-length FtsK protein could restore a wild-type phenotype to FtsKc− cells, whereas its overexpression led to filamentation (Fig. 2B). In addition, expression of the C-terminal domain had no effect on the morphology of FtsKc− cells. These observations correlate with the spatial distribution of the FtsK proteins inside the cell (Fig. 2A). We conclude that the preferential spatial localization of FtsK to the septum is critical for its role in chromosome segregation.
We propose that FtsK and DAZ are the two major components of a system designed to sense the presence of chromosome dimers at cell division, that is, to integrate information at the cellular level, and regulate on the basis of this information the activity of the Xer recombination machinery at the molecular level. DAZ could define a region of the chromosome that is actively maintained at midcell and thus is located close to the septum at cell division. Synaptic complex between dif sites in DAZ therefore would be the only ones that can be in contact with the C-terminal domain of wild-type FtsK. Supporting this idea, DAZ has been shown to stay preferentially localized at midcell in a strain containing a large chromosome inversion (Niki et al. 2000). As segregation of the chromosomes starts rapidly after replication, dif sites carried by monomeric chromosomes may be segregated away from each other before septum closure, therefore breaking the synaptic complex before access to septum-located FtsK is possible. In this case, only dif sites carried by a dimer will be accessible to FtsK. Thus, FtsK can be viewed as a molecular antenna that sits at the septum and activates the Xer recombination system when in the presence of a chromosome dimer.
In conclusion, our studies distinguish three important features of the Xer recombination reaction at dif. XerCD can form and resolve HJs back to substrate independently of any other cellular element. The C terminus of FtsK is sufficient to activate XerD, allowing intra- and intermolecular recombination reactions to go to completion. Finally, dif needs to be within a specific region of the chromosome, DAZ, and the C-terminal domain of FtsK needs to be correctly anchored (through its N terminus) at the septum for the Xer reaction to show directionality, that is, to resolve dimers into monomers without risking the reverse reaction.
Materials and methods
Bacterial strains and plasmids
E. coli strains and plasmids are listed in Table 1. Deletions of ftsK were made by PCR and inserted into pBAD24. The lysine to alanine substitution of the ATP− mutant was generated by site-directed mutagenesis. All constructions were verified by direct sequencing.
Table 1.
Strains and plasmids
| Strain
|
Relevant genotype
|
Source/reference
|
|---|---|---|
| AB1157 | Rec+ laboratory strain | Summers and Sherratt 1988 |
| DS941 | AB1157 recF | Summers and Sherratt 1988 |
| DS9041 | DS941 ftsK∷CmR | Recchia et al. 1999 |
| DS8029 | AB1157 xerC∷KmR | G. Blakely |
| GR18 | AB1157 xer∷KmR | G. Recchia |
| MA3 | AB1157 ftsK∷CmR | Recchia et al. 1999 |
| GR19 | AB1157 recA∷CmR | Recchia et al. 1999 |
| N3137 | AB1157 uvrA∷Tn10 | McGlynn and Lloyd 2000 |
| JJC40 | AB1157 hsdR | Seigneur et al. 1998 |
| JJC662 | JJC40 ruvA60∷Tn10 | Michel et al. 2000 |
| JJC494 | JJC662 Δrep∷KmR | Seigneur et al. 1998 |
| JJC733 | JJC662 xerC∷KmR | Michel et al. 2000 |
| LN3080 | LN2666 (thy−) dif in DAZ | Cornet et al. 1996 |
| LN3091 | LN2666 dif in LacZ | Perals et al. 2000 |
| LN3577 | LN2666 dif in zda192 | Perals et al. 2000 |
| FC313 | AB1157 ftsK dif-Km-dif in DAZ | F. Cornet |
| FC354 | AB1157 ftsK dif-Km-dif in lacZ | F. Cornet |
| FC235 | LN2666 xerC dif-Km-dif in DAZ | F. Cornet |
| FC284 | LN2666 xerC dif-Km-dif in lacZ | F. Cornet |
| GR59 | MA3 recA56 srl∷Tn10 | Recchia et al. 1999 |
| FC217 | FC235 recA | F. Cornet |
| NL250 | DS941 difΔ6 dif in lacZ | Leslie and Sherratt 1995 |
| MA2 | NL250 ftsK | Recchia et al. 1999 |
| Plasmid | Description | Source/reference |
| pBAD24 | Expression vector | Guzman et al. 1995 |
| pFX83 | Flag fusion vector | This study |
| pFX88 | fl FtsK N-terminally tagged | This study |
| pFX89 | 734 N-terminally tagged | This study |
| pFX90 | 818 N-terminally tagged | This study |
| pFX91 | 855 N-terminally tagged | This study |
| pFX99 | 906 N-terminally tagged | This study |
| pFX101 | 1260 N-terminally tagged | This study |
| pFX102 | ATP− N-terminally tagged | This study |
| pSDC124 | pUC18-based dif reporter | Blakely et al. 1991 |
| pFC238 | pGB2-based dif reporter | Recchia et al. 1999 |
| pFH127 | pGB2ts carrying one dif site | Hayes and Sherratt 1997 |
| pFC225 | p15a-based XerC expression vector | F. Cornet |
DAZ, dif activity zone; fl, full length.
Growth conditions
Cells were grown in LB supplemented with thymine when necessary. M9 minimal medium containing 1% glucose and 0.5% casamino acids was used for cytological examination. Glucose (1%) was used for repression of the arabinose promoter controlling the expression of the FtsK proteins. L-arabinose (as indicated) was added to media for induction.
In vivo plasmid resolution assays
Plasmid pFC238 carrying two dif sites was used as a substrate for in vivo resolution assays (Recchia et al. 1999).
Immunohistochemistry
Western analysis was performed using the M2 anti-Flag antibody (Sigma) and an ECF kit (Amersham). Blots were scanned on a Molecular Dynamics Fluorimager 575.
Cells were prepared for immunofluorescence as described previously (Pedersen et al. 1999). DAPI was used as a DNA staining agent. The M2 anti-Flag antibody was used as a first layer and an antimouse antibody coupled to fluorescein as a second layer. Phase-contrast and fluorescent images were acquired using a cooled CCD camera (Princeton Instruments) and Metamorph image acquisition software (Universal Imaging Corporation) from an Olympus BX60 fluorescence microscope.
2D gel electrophoresis and hybridization
Ten milliliters of exponentially growing cells was collected by centrifugation, washed in PBS, and resuspended in a solution of 100 μg/mL 4,5′,8-trimethylpsoralen (Sigma) and 20% ethanol in PBS. Cells were exposed to UV light (365 nm, 40 kJ/m2) by using a Stratalinker (Stratagene). Cross-linking extent was measured by the ability of cross-linked DNA to renature under conditions of RAGE analysis (Barre et al. 2000), as well as by the inhibition of cleavage of the EcoRV restriction enzyme. The first dimension of electrophoresis was performed at 0.5 V/cm in a 0.7% agarose TBE gel. The second dimension was performed at 5 V/cm at 4°C in a 1.2% agarose TBE gel containing 0.3 μg/mL of ethidium bromide. DNA was alkali transferred onto a Hybond-N + membrane (Amersham). Hybridization was performed using 32P-probes labeled with a random priming kit (Roche) in Church buffer. DNA species were visualized after exposure to blue sensitive X-ray films (Kodak) with one intensifying screen for 0.2–3 wk at −70°C. They were quantitated using a Molecular Dynamic PhosphorImager and the ImageQuant software (Molecular Dynamics).
In vitro HJ formation on plasmids
In vitro reactions were performed in 50 mM Tris (pH 7.5), 50 mM NaCl, 5mM spermidine, 2 mM EDTA, 0.1 mM DTT, 10 μg/mL BSA, and glycerol (as indicated). Ethidium bromide (0.25 mg/mL final concentration) was used to trap HJ intermediates. Reactions were stopped by extraction with phenol. The supercoiled substrate and the XerC and XerD recombinases were purified as described elsewhere (Colloms et al. 1996).
Plasmid integration assays
The level of intermolecular recombination between a plasmid-borne dif site and a chromosomal dif site was assayed as described previously (Leslie and Sherratt 1995; Hayes and Sherratt 1997).
dif-Km-dif cassette resolution assays
XerC− strains carrying chromosomal insertions of the dif-Km-dif cassette were induced for dif recombination by transformation with the XerC expression vector pFC225, which carries a spectinomycin resistance cassette. After overnight growth, the percentage of kanamycin-resistant bacteria was determined by plating serial dilutions on spectinomycin plates with or without kanamycin.
Acknowledgments
We thank H. Capiaux, B. Lloyd, and B. Michel for the kind gift of strains; N. Hunter and V. Regnier for technical advice; M. Chandler and J. Roth for helpful discussions. M.A. was supported by an MRC studentship, A.H. by a Socrates award, and F.-X.B. by ARC and EMBO fellowships. S.D.C is supported by a Wellcome Trust Senior Fellowship. D.J.S. is supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sherratt@bioch.ox.ac.uk; FAX 44-1865-275297.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.188700.
References
- Arciszewska LK, Sherratt DJ. Xer site-specific recombination in vitro. EMBO J. 1995;14:2112–2120. doi: 10.1002/j.1460-2075.1995.tb07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska LK, Baker RA, Hallet B, Sherratt DJ. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J Mol Biol. 2000;299:391–403. doi: 10.1006/jmbi.2000.3762. [DOI] [PubMed] [Google Scholar]
- Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981;25:729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Barre FX, Ait-Si-Ali S, Giovannangeli C, Luis R, Robin P, Pritchard LL, Helene C, Harel-Bellan A. Unambiguous demonstration of triple-helix-directed gene modification. Proc Natl Acad Sci. 2000;97:3084–3088. doi: 10.1073/pnas.050368997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Ganguly N, Sherratt DJ. DNA sequence of recombinase-binding sites can determine Xer site-specific recombination outcome. Mol Microbiol. 1997;23:387–398. doi: 10.1046/j.1365-2958.1997.2261600.x. [DOI] [PubMed] [Google Scholar]
- Blakely G, Colloms S, May G, Burke M, Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- Blakely G, May G, McCulloch R, Arciszewska LK, Burke M, Lovett ST, Sherratt DJ. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Blakely GW, Davidson AO, Sherratt DJ. Sequential strand exchange by XerC and XerD during site-specific recombination at dif. J Biol Chem. 2000;275:9930–9936. doi: 10.1074/jbc.275.14.9930. [DOI] [PubMed] [Google Scholar]
- Boyle DS, Grant D, Draper GC, Donachie WD. All major regions of FtsK are required for resolution of chromosome dimers. J Bacteriol. 2000;182:4124–4127. doi: 10.1128/jb.182.14.4124-4127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerget M. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 1991;3:780–788. [PubMed] [Google Scholar]
- Colloms SD, Sykora P, Szatmari G, Sherratt DJ. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J Bacteriol. 1990;172:6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloms SD, McCulloch R, Grant K, Neilson L, Sherratt DJ. Xer-mediated site-specific recombination in vitro. EMBO J. 1996;15:1172–1181. [PMC free article] [PubMed] [Google Scholar]
- Colloms SD, Bath J, Sherratt DJ. Topological selectivity in Xer site-specific recombination. Cell. 1997;88:855–864. doi: 10.1016/s0092-8674(00)81931-5. [DOI] [PubMed] [Google Scholar]
- Cornet F, Louarn J, Patte J, Louarn JM. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes & Dev. 1996;10:1152–1161. doi: 10.1101/gad.10.9.1152. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Diez AA, Farewell A, Nannmark U, Nystrom T. A mutation in the ftsK gene of Escherichia coli affects cell–cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorazi R, Dewar SJ. Membrane topology of the N-terminus of the Escherichia coli FtsK division protein. FEBS Lett. 2000;478:13–18. doi: 10.1016/s0014-5793(00)01820-2. [DOI] [PubMed] [Google Scholar]
- Draper GC, McLennan N, Begg K, Masters M, Donachie WD. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol. 1998;180:4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: An enzymatic switch in site-specific recombination. Mol Cell. 1999;4:949–959. doi: 10.1016/s1097-2765(00)80224-5. [DOI] [PubMed] [Google Scholar]
- Hayes F, Sherratt DJ. Recombinase binding specificity at the chromosome dimer resolution site dif of Escherichia coli. J Mol Biol. 1997;266:525–537. doi: 10.1006/jmbi.1996.0828. [DOI] [PubMed] [Google Scholar]
- Hendricks EC, Szerlong H, Hill T, Kuempel P. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 2000;36:973–981. doi: 10.1046/j.1365-2958.2000.01920.x. [DOI] [PubMed] [Google Scholar]
- Hojgaard A, Szerlong H, Tabor C, Kuempel P. Norfloxacin-induced DNA cleavage occurs at the dif resolvase locus in Escherichia coli and is the result of interaction with topoisomerase IV. Mol Microbiol. 1999;33:1027–1036. doi: 10.1046/j.1365-2958.1999.01545.x. [DOI] [PubMed] [Google Scholar]
- Kolodner R, Fishel RA, Howard M. Genetic recombination of bacterial plasmid DNA: Effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985;163:1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuempel PL, Henson JM, Dircks L, Tecklenburg M, Lim DF. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- Kuempel P, Hogaard A, Nielsen M, Nagappan O, Tecklenburg M. Use of a transposon (Tndif) to obtain suppressing and nonsuppressing insertions of the dif resolvase site of Escherichia coli. Genes & Dev. 1996;10:1162–1171. doi: 10.1101/gad.10.9.1162. [DOI] [PubMed] [Google Scholar]
- Leslie NR, Sherratt DJ. Site-specific recombination in the replication terminus region of Escherichia coli: Functional replacement of dif. EMBO J. 1995;14:1561–1570. doi: 10.1002/j.1460-2075.1995.tb07142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Draper GC, Donachie WD. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- Michel B, Recchia GD, Penel-Colin M, Ehrlich SD, Sherratt DJ. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV irradiated cells. Mol Microbiol. 2000;37:181–191. doi: 10.1046/j.1365-2958.2000.01989.x. [DOI] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes & Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Angert ER, Setlow P. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J Bacteriol. 1999;181:3201–3211. doi: 10.1128/jb.181.10.3201-3211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perals K, Cornet F, Merlet Y, Delon I, Louarn JM. Functional polarization of the Escherichia coli chromosome terminus: The dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol Microbiol. 2000;36:33–43. doi: 10.1046/j.1365-2958.2000.01847.x. [DOI] [PubMed] [Google Scholar]
- Recchia GD, Sherratt DJ. Conservation of Xer site-specific recombination genes in bacteria. Mol Microbiol. 1999;34:1146–1148. doi: 10.1046/j.1365-2958.1999.01668.x. [DOI] [PubMed] [Google Scholar]
- Recchia GD, Aroyo M, Wolf D, Blakely G, Sherratt DJ. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Kuempel PL. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol Microbiol. 1998a;27:257–268. doi: 10.1046/j.1365-2958.1998.00651.x. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998b;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W, Liu G, Donachie WD, Kuempel P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- Summers DK, Sherratt DJ. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984;36:1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- Summers DK, Sherratt DJ. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 1988;7:851–858. doi: 10.1002/j.1460-2075.1988.tb02884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecklenburg M, Naumer A, Nagappan O, Kuempel P. The dif resolvase locus of the Escherichia coli chromosome can be replaced by a 33-bp sequence, but function depends on location. Proc Natl Acad Sci. 1995;92:1352–1356. doi: 10.1073/pnas.92.5.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes & Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- Yu XC, Tran AH, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998a;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Weihe EK, Margolin W. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol. 1998b;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D, Colloms SD, Sherratt DJ, West SC. Effect of DNA topology on Holliday junction resolution by Escherichia coli RuvC and bacteriophage T7 endonuclease I. J Mol Biol. 1997;270:663–673. doi: 10.1006/jmbi.1997.1157. [DOI] [PubMed] [Google Scholar]