Glucose monomycolate–loaded CD1b tetramers identify a subset of CD4+ T cells in patients with Mycobacterium tuberculosis infection.
Abstract
Microbial lipids activate T cells by binding directly to CD1 and T cell receptors (TCRs) or by indirect effects on antigen-presenting cells involving induction of lipid autoantigens, CD1 transcription, or cytokine release. To distinguish among direct and indirect mechanisms, we developed fluorescent human CD1b tetramers and measured T cell staining. CD1b tetramer staining of T cells requires glucose monomycolate (GMM) antigens, is specific for TCR structure, and is blocked by a recombinant clonotypic TCR comprised of TRAV17 and TRBV4-1, proving that CD1b–glycolipid complexes bind the TCR. GMM-loaded tetramers brightly stain a small subpopulation of blood-derived cells from humans infected with Mycobacterium tuberculosis, providing direct detection of a CD1b-reactive T cell repertoire. Polyclonal T cells from patients sorted with tetramers are activated by GMM antigens presented by CD1b. Whereas prior studies emphasized CD8+ and CD4−CD8− CD1b-restricted clones, CD1b tetramer-based studies show that nearly all cells express the CD4 co-receptor. These findings prove a cognate mechanism whereby CD1b–glycolipid complexes bind to TCRs. CD1b tetramers detect a natural CD1b-restricted T cell repertoire ex vivo with unexpected features, opening a new investigative path to study the human CD1 system.
Like most mammalian species, humans express several structurally distinct CD1 antigen-presenting molecules. The conservation of large CD1 gene families among most mammals suggests that each type of CD1 protein has distinct functions that confer selective advantage. Cellular studies of CD1 proteins increasingly explain how each CD1 protein differs from the others. CD1a, CD1b, CD1c, and CD1d have distinct antigen groove structures, patterns of expression in tissues, intracellular trafficking, and trigger T cells expressing diverse TCRs (Kasmar et al., 2009). CD1d (group 2) diverges most clearly from CD1a, CD1b, and CD1c (group 1) with regard to protein sequence. Also, group 1 and group 2 CD1 proteins show differing transcriptional responses to pathogens, suggesting that they function at different stages of the immune response (Roura-Mir et al., 2005b). Collectively, these cellular studies suggest that group 1 and group 2 CD1 proteins likely have differing roles in immune responses.
The majority of known foreign ligands for group 1 CD1 molecules are mycobacterial in origin, including dideoxymycobactin, mycolic acid, lipoarabinomannan, glucose monomycolate (GMM), glycerol monomycolate, diacylated sulfoglycolipid, phosphatidylinositol mannoside, and mannosyl phosphomycoketide (De Libero and Mori, 2005). Human T cells proliferate or produce interferon-γ in response to several types of mycobacterial lipid antigens presented by group 1 CD1 proteins during latent or active tuberculosis infection, suggesting a function in host response to mycobacteria (Moody et al., 2000b; Ulrichs et al., 2003; Gilleron et al., 2004; Layre et al., 2009; Montamat-Sicotte et al., 2011). However, existing experimental models for study of group 1 CD1 function rely on activation assays that destroy the responding cells or focus on a limited number of in vitro–derived human T cell clones, which may not accurately reflect the in vivo phenotype. Consequently, information about the precise frequencies, effector functions, and possible host-protective effects of group 1 CD1-restricted T cells remain unknown. In contrast, the biological functions of CD1d and NKT cells have been broadly studied through mice deficient in CD1d or invariant Vα14 or Jα18 T cell receptors, as well as CD1d tetramers (Benlagha et al., 2000; Matsuda et al., 2000; Karadimitris et al., 2001; Gumperz et al., 2002). Tetramers take advantage of multimerization to generate high avidity fluorescent staining reagents that bind to individual clonotypic TCRs and selectively track antigen-specific T cells within much larger T cell populations (Altman et al., 1996). Tetramers can identify even rare antigen-specific T cells (Moon et al., 2007) for functional analysis, and CD1d tetramers have allowed single-cell analysis of NKT cells during infection, autoimmunity, and cancer (Benlagha et al., 2000; Matsuda et al., 2000; Karadimitris et al., 2001; Gumperz et al., 2002; Lee et al., 2002; Jahng et al., 2004; Arrenberg et al., 2010). Germline deletion of group 1 proteins is not currently feasible, so development of CD1 tetramers represents a promising method to study fresh antigen-specific T cells at the population level.
The basic principle of tetramer staining requires that TCRs bind to the antigen-presenting molecule and that this physical interaction is mediated by a groove-bound cognate antigen that physically ligates CD1 to the TCR. For CD1d, lipids like synthetic α-galactosylceramides mediate the trimolecular complex of CD1d–antigen–TCR (Borg et al., 2007), so an analogous function of glycolipids in mediating TCR contact with group 1 CD1 proteins is a leading model. However, recent studies have emphasized three alternate mechanisms whereby TCRs bind to CD1 or activate T cells but do not physically ligate CD1 and TCR. Lipopolysaccharide stimulates iNKT cell activation, not as a CD1d-bound lipid antigen but by triggering release of cytokines such as IL-12, which augments CD1d–self-antigen–mediated reactivity (Brigl et al., 2003, 2011). Phosphatidylinositol mannoside might activate T cells indirectly by up-regulating CD1b surface expression (Roura-Mir et al., 2005b), and bacterial lipids induce more stimulatory self-ligands for CD1 proteins (De Libero et al., 2005; Paget et al., 2007). In each of the three scenarios, microbial glycolipids and CD1 proteins are both needed to activate T cells through indirect mechanisms in which the foreign glycolipid does not physically link CD1 to TCR. Therefore, tetramers represent both a test of cognate antigen recognition by CD1b and a potential tool to physically isolate and characterize a foreign glycolipid-reactive T cell repertoire for the first time. In this paper, we show that human CD1b tetramers loaded with a mycobacterial glycolipid antigen, GMM, selectively bind to GMM-specific TCRs and directly isolate a natural CD1b and glycolipid-reactive T cell repertoire in humans.
RESULTS AND DISCUSSION
Design of tetramerizable CD1b proteins
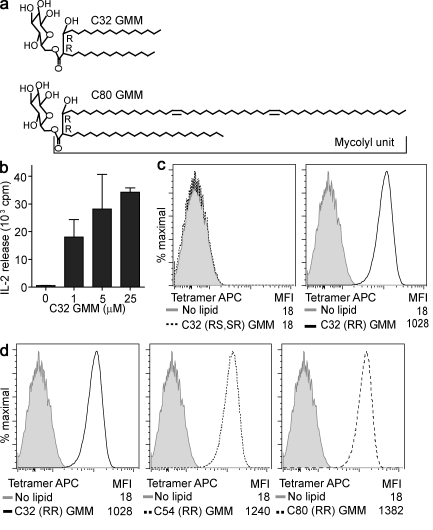
We produced a tetramerizable biotinylated CD1b monomer building on prior designs for MHC and CD1d tetramers (Altman et al., 1996; Gumperz et al., 2002). The extracellular domain of the human CD1b heavy chain was modified with a leucine zipper for binding to β-2 microglobulin and a Bir A sequence for biotinylation and complexed with streptavidin-allophycocyanin (CD1b tetramer–APC). CD1b tetramers were loaded with glucose-6-monomycolate, a natural mycobacterial glycolipid antigen comprised of glucose in 6-linkage with mycolyl groups that exist in an alkane series (Fig. 1 a). Three GMM preparations with an average mycolyl unit of C32, C54, or C80 (C32 GMM, C54 GMM, and C80 GMM) were separately loaded onto CD1b tetramers and used to stain the GMM-reactive CD1b-restricted T cell line LDN5. However, initial attempts to stain were unsuccessful, even after confirming monomer purity, biotinylation, and multimerization of CD1b proteins, as well as successful staining of NKT cells with control CD1d tetramers (Fig. S1 a and Fig. S2). Tetramer staining requires that key aspects of the cellular loading mechanism, which is particularly stringent for CD1b, be replicated in vitro. Therefore, we tested the sufficiency of in vitro conditions for antigen loading. In particular, the absence of cellular loading cofactors like saposin C (Winau et al., 2004) and the lack of essential cellular processing might alter structures in ways that are required for binding. After optimizing the time, pH, and chain length of the antigen, we were able to see high-level T cell activation with a plate bound CD1b monomer loaded with a C32 GMM antigen. This result confirmed that cellular processing and loading cofactors are not absolutely required and proved proper CD1b folding (Fig. 1 b).
Figure 1.
CD1b tetramers stain human αβ T cells. (a) Bacterial GMM is formed by glucose linked at the 6-position to a mycolyl unit that contains two chiral centers, which are in the R configuration at positions 2 and 3 (2R, 3R). (b) Tetramerizable CD1b monomers were used in plate-bound antigen presentation experiments to measure IL-2 release by the CD1b-restricted human T cell line LDN5 in response to C32 GMM loaded overnight at 37°C (mean + SEM). (c) CD1b was loaded with GMMs that are naturally formed with R configuration at C2 and C3 (R, R) or synthetic GMM prepared with an S configuration at C2 or C3 (2R,3S+2S,3R) and complexed to streptavidin-labeled APC (tetramer APC) and tested for staining LDN5 T cells. (d) CD1b tetramers were then loaded with GMMs of the indicated average chain length (C32, C54, or C80) and tested for staining LDN5. MFI is mean fluorescence intensity. Data are representative of three or more experiments.
CD1b tetramers bind to T cells
Using optimized conditions for loading CD1b with C32 GMM (Fig. S1 b), we observed CD1b tetramer staining of LDN5 (Fig. 1 c). Although CD1d tetramers bound to the synthetic superagonist α-galactosylceramide brightly stain CD1d-restricted T cells, self-antigens such as isogloboside 3 or sulfatide result in absent or moderate tetramer staining (Jahng et al., 2004; Zhou et al., 2004; Arrenberg et al., 2010). Therefore, it is notable that GMM, a natural foreign antigen, gives bright staining, such that the mean fluorescence intensity increases 10–100-fold after loading in optimized conditions (Fig. 1 c). To determine whether staining is specific for the structure of the antigen or is a result of nonspecific hydrophobic interactions resulting from the presence of lipids, we exposed CD1b to natural and synthetic antigens that recapitulate certain aspects of the C32 GMM structure. Whereas natural bacterial C32 GMM contains two chiral centers in the R conformation at the C2 and C3 positions of the meromycolate chain GMM (2R, 3R), synthetic C32 GMM diastereomers containing an S configuration at either position GMM (2R,3S + 2S,3R; Fig. 1 a) are nonantigenic (Moody et al., 2000a). Only C32 GMM (2R, 3R) mediated tetramer staining, indicating that chiral carbons, which determine the orientation of the glucose head group and β-hydroxyl unit relative to the TCR, are required for staining (Fig. 1 c).
In contrast, three preparations of natural bacterial GMMs containing a mean chain length of 32, 54, or 80 carbons and having 2R,3R configuration mediate bright staining (Fig. 1 d). Thus, C48 differences in overall lipid length can be tolerated, leading to high avidity binding. Whereas the length and conformation of the alkane chain hidden within the CD1d groove can significantly influence NKT cell activation (McCarthy et al., 2007), our results strongly suggest that CD1b-restricted TCR binding depends critically on head group positioning but can tolerate very large differences in lipid chain length. These results support and extend prior work suggesting that C80 lipids fill the entire groove, whereas shorter lipids partially fill the groove, allowing smaller spacer lipids to fill in the remaining volume (Gadola et al., 2002; Batuwangala et al., 2004; Garcia-Alles et al., 2006).
CD1b tetramers bind the TCR-αβ complex
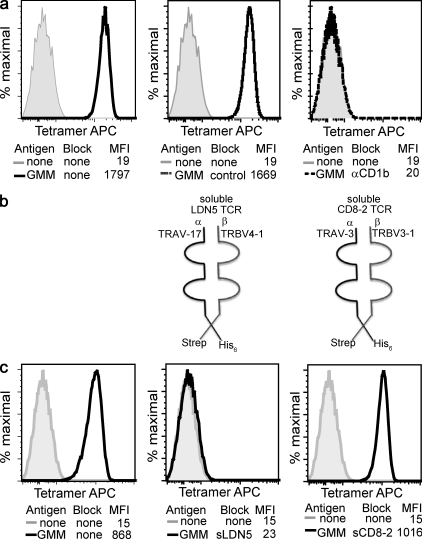
The cognate model predicts that the surface target of tetramer binding is the heterodimer of rearranged TCR-α and -β chains normally expressed on the LDN5 T cell clone, TRAV 17, and TRBV4-1. However, a physical interaction of TCRs with any group 1 CD1 protein has not been previously observed. In addition to any alternate surface ligands on T cells that are unknown and might bind to CD1b, NK receptors (Carbone et al., 2000) and immunoglobulin-like proteins (ILT; Li et al., 2009) have been implicated in binding CD1 proteins. Therefore, we designed experiments to test the presence and specificity of a proposed interaction between CD1b with the clonotypic αβ TCR. CD1d tetramers made from the same type of construct failed to stain LDN5 but did stain the CD1d-restricted T cell clone J3N.5, implicating CD1 isoform–specific sequences in tetramer staining (Fig. S2). Preincubation with anti-CD1b or anti–TRBV4-1 blocked tetramer staining to background (Fig. 2 a and Fig. S3 a). These studies were consistent with interaction between CD1b and TCR VB4-1, if binding of monoclonal antibodies directly interfered with contact between TCR-β and the distal surface of CD1b. However, antibodies might have blocked staining in indirect ways involving sequestration of TCRs or CD1b proteins. To definitively test the role of the distal domains of TRBV4-1 and TRAV17 in physical contact with CD1b-GMM complexes, we produced soluble leucine-zippered TCRs comprised of the distal domains of the TCR-α and -β chains from LDN5 (sLDN5) and from CD8-2 (sCD8-2), a TRAV3, TRBV3-1 heterodimer which recognizes CD1a (Fig. 2 b and Fig. S3 b). Preincubation with sCD8-2 TCR did not inhibit tetramer staining, but sLDN5 TCR reduced tetramer staining to background levels (Fig. 2 c). Thus, the clonotypic TCR is necessary for CD1b-GMM binding to cells, proving a cognate TCR interaction with the CD1b–antigen complex, which is TCR specific and necessary for cellular binding.
Figure 2.
Tetramer staining proves a specific trimolecular interaction among CD1b, GMM, and the clonotypic TCR. (a) The LDN5 T cell clone was stained with unloaded CD1b tetramers or CD1b tetramers loaded with C32 GMM. Tetramers were preincubated with 10 µg/ml isotype control antibody or anti-CD1b antibody. (b) Soluble TCR-α chains with hexahistidine tags and -β chains with streptavidin tags were formed into soluble TCR dimers. (c) Loaded and unloaded tetramers were preincubated with fivefold molar excess of soluble recombinant T cell receptors derived from CD1a-restricted (sCD8-2) or CD1b-restricted (sLDN5) T cell lines. Data are representative of three or more experiments.
CD1b tetramers detect GMM-specific T cells during TB infection
Development of tetramers for study of patient blood in the setting of an infectious disease requires low background among all types of cells present in PBMCs. To evaluate tetramer specificity, we mixed LDN5 T cells with CD1d-restricted NKT cells and found that GMM-loaded CD1b tetramers selectively stained the TRVB4-1+ clonotypic T cells, with no detectable staining over background of T cells with another TCR (Fig. S3 c). Also, titration of GMM-specific LDN5 T cells into fresh PBMC at known frequencies demonstrated that clonotypic T cells could be sensitively detected at the level of 0.01% of CD3+ cells. A discrete population of brightly staining cells was detected at frequencies near to their actual abundance when titrated into PBMC, so tetramers were not binding to T cells with diverse TCRs (Fig. S3 d). The potential problem of low but detectable background staining on CD3− cells was minimized by two-color flow cytometry to detect CD3+tetramer+ cells (Fig. S3 d), setting the stage for clinical studies of human CD1b-restricted T cells.
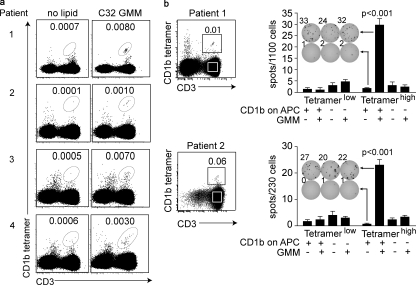
Several population studies have detected increased interferon-γ responses in tuberculosis patients, indicating that lipid-reactive T cells likely expand during infection (Moody et al., 2000a; Ulrichs et al., 2003; Gilleron et al., 2004; Layre et al., 2009; Montamat-Sicotte et al., 2011). However, group 1 CD1-restricted T cells have never been detected directly ex vivo without stimulation. Tetramer detection is desirable because it rules out false positive results from cytokine production by non-T cells, indirect stimulation of cells by lipid adjuvants, or activation of MHC-restricted cells by contaminating peptide antigens. Also, tetramer-based sorting allows live cell capture for diverse functional and phenotypic studies. Among four subjects with positive intradermal purified protein derivative tests, we observed a similar pattern: a small percentage of blood T cells (∼0.01%) stained brightly such that they were well separated from the pool of nonstaining cells (Fig. 3 a). The absolute frequency of cells was detected at similar rates among patients with latent (patients 1, 2, and 4) and active tuberculosis (patient 3), but staining was not observed in three healthy controls (Fig. S4 c). The detected frequency of T cells from individual patients was similar to one another and highly reproducible among experiments using blood from the same patient to assess different aspects of function and phenotype (Fig. 3, a and b; and Fig. 4, a and b).
Figure 3.
CD1b tetramers identify a mycobacterial glycolipid-reactive T cell repertoire in humans. (a) PBMCs of four subjects infected with Mycobacterium tuberculosis were stained with CD1b tetramers in addition to CD3 FITC, CD14 PercP-Cy5.5, CD19 PerCP-Cy5.5, and violet viability dye and gated on live lymphocytes. (b) PBMCs from patient 1 were cultured overnight in 0.2 ng/ml IL-15 before FACS sorting. PBMCs from patient 2 were expanded by stimulation with anti-CD3 in the presence of irradiated feeder cells and IL-2 before FACS sorting using CD3 FITC and CD1b tetramers. Equal numbers of cells were incubated with 20,000 CD1b or empty vector–transfected K562 APCs with or without 5 µg/ml GMM (mean + SEM).
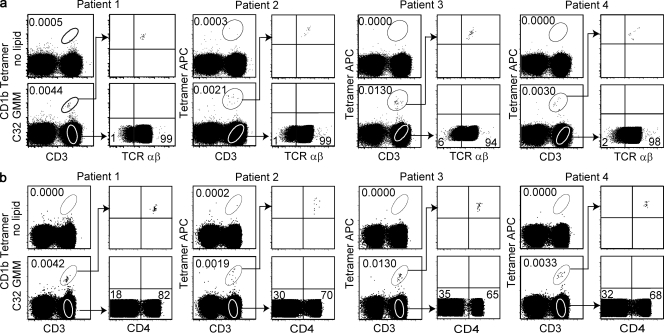
Figure 4.
CD1b-restricted T cell populations express the αβ TCR and CD4. PBMCs from four subjects infected with Mycobacterium tuberculosis were subjected to multicolor FACS analysis. Cells were stained with CD3, violet viability dye, CD14 and CD19-PercP-Cy5.5, loaded CD1b tetramers, and anti–TCR-αβ (a) and CD4 (b).
To determine whether cells staining with CD1b–GMM complexes functionally recognized CD1b and GMM, we sorted CD3+ cells into tetramerhigh and tetramerlow populations (Fig. 3 b and Fig. S4 b). After recovery, total cells were tested in γ-interferon ELISpot using K562 cells that do or do not express CD1b (de Jong et al., 2010). Only tetramerhigh cells produced interferon-γ in response to GMM, and this response required CD1b expression (Fig. 3 b). Thus, CD1b tetramers directly identify populations of foreign glycolipid–reactive T cells in the blood of human tuberculosis patients that constitute a natural sub-repertoire of human αβ T cells. A precursor frequency of 0.01% is similar to that of human NKT cells identified using CD1d tetramers (Gumperz et al., 2002). Despite limited numbers in the peripheral circulation, lipid-specific T cells have been proposed to act locally near the site as helper cells whose function is magnified by downstream responses of dendritic cells or other T cells (Vincent et al., 2002; Roura-Mir et al., 2005a,b). Sorting of blood-derived cells with CD1b tetramers can address core issues of CD1b-restricted T cell phenotype and function previously addressed in T cell clones which can now be studied ex vivo.
Distinct features of the CD1b-GMM repertoire
The first and subsequent studies of group 1 CD1-restricted clones show expression of either γδ or αβ TCRs (Porcelli et al., 1992; Spada et al., 2000) in combination with CD4, CD8, or neither co-receptor. We found that CD1b tetramerhigh T cells uniformly stain with antibody against invariant components of αβ TCRs in all four patients tested (Fig. 4 a). CD4 and CD8 represent key subset markers for NKT cell and MHC-restricted T cells because they strongly influence thymic selection and, thereby, determine effector functions. CD1b-restricted T cell clones can express CD4 or CD8 or neither co-receptor (Porcelli et al., 1992; Moody et al., 1997; Stenger et al., 1998), but any general view of co-receptor expression is limited by the small number of clones studied and the possibility of selective outgrowth in vitro. Given the large number of CD4−CD8− and CD8+ clones isolated in early work on CD1b, it was unexpected to observe that CD4 single-positive cells dominate the population of tetramer+ T cell populations cells in all four patients studied (Fig. 4 b). The absence of CD4 positivity in early clone-based studies likely resulted from methods that depleted CD4 T cells in cultures to reduce MHC class II alloreactivity during cloning procedures. This was a key intervention that allowed the discovery of CD1-restricted T cells, but unbiased study of the CD1b and GMM repertoire now suggests that the CD4+ population dominates.
Identification of the CD1b and GMM reactive repertoire as TCR-αβ+CD4+ provides basic information about the CD1b-restricted T cell subset, which raises new questions about potential infection of these cells by HIV as well as a possible role of CD4 in development and effector function of this T cell subset. Furthermore, these results illustrate how any phenotypic question can be approached without confounders relating to in vitro growth or contaminating peptide antigens. Whereas NKT cells can be studied in CD1d- or Jα18-altered mice with human or mouse tetramers, there is no widely used small animal model for CD1b. Therefore, CD1b tetramers open a broad window for detailed study of the immunobiology of these cells. In contrast to highly polymorphic MHC proteins, which require haplotype matching for donors, the low rates of CD1 polymorphism in human populations allow one CD1b sequence in tetramer form to be readily applied to almost any human donor in ways that facilitate population studies. Prior clinical studies indicate that group 1 CD1 T cell responses are frequent in human tuberculosis patients (Moody et al., 2000a; Ulrichs et al., 2003; Gilleron et al., 2004; Layre et al., 2009; Montamat-Sicotte et al., 2011), so CD1 tetramers might be developed as a means of immunodiagnosis. Future studies will take advantage of this technology to determine whether the CD4+ T cell populations described in this paper may be expanded in the blood and tissues of tuberculosis patients and express effector functions that contribute to control of mycobacterial infection, like interferon-γ, TNF-α, and granulysin (Stenger et al., 1998), or instead have unexpected roles in immunosuppression or immunopathology.
MATERIALS AND METHODS
Generation of soluble CD1b proteins.
Soluble biotinylated CD1b monomers were produced in lentivirus-transduced HEK293 T cells by the National Institutes of Health Tetramer Core Facility (Emory University, Atlanta, GA) and tetramerized with fluorescently labeled streptavidin. In brief, human β-2-microglobulin and the extracellular domain of CD1b were cloned into the expression vector pCMJJ4 (gift from J. Jacob, Emory University, Atlanta, GA). Lentiviral particles were made in a second generation packaging system (Naldini et al., 1996). The light and heavy chains are expressed under control of the CMV promoter and are separated by the 2A-TaV peptide to generate two separate proteins from a single mRNA. The chains are followed by a C-terminal acidic or basic leucine zipper which stabilizes the complex and is used for affinity purification using the 2H11 monoclonal antibody (E. Reinherz, Harvard, Boston, MA). Purified monomers were enzymatically biotinylated at the BirA site at the C terminus of the heavy chain. Monomer purity and composition were confirmed by PAGE, and biotinylation was confirmed by streptavidin bead pulldown assay. Functional activity was assayed by affixing biotinylated monomers at final concentration of 5 µg/ml onto 96-well streptavidin plates (Thermo Fisher Scientific) in PBS, pH 7.4, for 24 h at 37°C. Lipid antigens were sonicated in PBS for 2 min, added to the wells, and incubated for 24 h at 37°C before washing three times with 200 µl/well sterile PBS. 105 LDN5 cells were added in a total volume of 200 µl T cell medium per well (RPMI). The plate was incubated for 24 h at 37°C after which culture supernatants were collected for HT2 bioassay.
Generation of soluble clonotypic TCR-αβ complexes.
The cDNAs of the α and β chains of TCR (LDN5 and CD8.2) were cloned into the baculovirus transfer vector pAcUW51 (BD). Honey Bee Melittin and envelope glycoprotein gp67 were used as signal peptides to optimize secretion of the α and β chains. The C terminus of the α chain has a thrombin cleavage site followed by an acidic zipper and hexahistidine tag. The β chain also has a thrombin cleavage site followed by a basic zipper and Strep-tag II (WSHPQFEK). The TCRs were expressed using the baculovirus cotransfection method and the protein was secreted by SF9 insect cells. The secreted TCR from the supernatant was purified using Nickel beads (QIAGEN), and a Strep-Tactin column (IBA), followed by gel-filtration chromatography. The pooled protein was concentrated to ∼1 mg/ml in 20 mM Tris-HCl and 100 mM NaCl, pH 8.0, confirmed for purity by gel electrophoresis, and stored at −80°C in small aliquots.
Loading CD1b monomers with GMM.
GMM with differing average chain lengths produced by Rhodococcus equi (C32), Nocardia farcinica (C54), or Mycobacterium phlei (C80) was isolated as previously described (Moody et al., 2002). Antigen identity and purity were confirmed by biochemical analysis including thin layer chromatography and electrospray ionization mass spectrometry in the positive mode (LXQ Linear Ion Trap Mass Spectrometer; Thermo Fisher Scientific). Loading conditions were guided by results from T cell activation by monomeric proteins and optimized by staining T cells after loading under conditions ranging in pH (5–7.4), temperature (20–37°C), concentration (10–100-fold excess antigen), and time (2–24 h). Optimal staining was seen with GMM sonicated into 50 mM sodium citrate at pH 5.0 for 2 min, added at 40-fold molar excess to CD1b monomers, and incubated in a 37°C water bath for 2 h with vortexing every 15 min, followed by incubation at room temperature for an additional 22 h before neutralization to pH 7.4 with 10 µl TRIS, pH 9. The duration of antigen loading and the purity of antigen preparations were critical for obtaining bright staining of T cells. After loading, CD1b monomers were multimerized using fluorescently labeled streptavidin (Invitrogen) at a 5:1 molar ratio.
CD1b tetramer staining of clones.
CD1b tetramers were validated by staining the clone LDN5 (Moody et al., 1997). In brief, 2 × 105 T cells were treated with human AB serum for 10 min, washed, and then suspended in FACS buffer (PBS with 2% fetal calf serum; Gemini) and stained with 1 µg of fluorescently labeled CD1b tetramer for 60 min at room temperature in the dark. Cells were acquired on a FACSCanto flow cytometer (BD) and analyzed using FlowJo (Tree Star) software with doublet exclusion based on forward and side scatter in the presence or absence of anti-CD1b or recombinant TCRs.
Tetramer staining of human PBMC.
After informed consent, 50 ml of blood were collected from healthy controls, asymptomatic tuberculin-positive subjects with no clinical or radiographical evidence of active tuberculosis, and active tuberculosis patients overseen by the institutional review boards of the Lemuel Shattuck Hospital (00000786) and Partners Healthcare (2002-P-000061) and the Harvard Committee on Microbiologic Safety (08–184). PBMCs were separated by Ficoll density gradient centrifugation. After thawing, one million PBMCs were treated with human AB serum and stained with 1 µg tetramer for 40 min at room temperature in the dark, after which they were stained with violet fluorescent reactive dye (Invitrogen) to exclude dead cells. Cells were stained with monoclonal antibodies including CD3 (BD), CD14 (BD), and CD19 (eBioscience) for an additional 20 min and then fixed in 2% formaldehyde before FACS analysis. Cells from patient 1 were stained in 12 experiments; cells from patients 2, 3, and 4 were each stained four times. For functional assays, unfixed tetramer-positive cells were sorted using a FACSAria flow cytometer and tested for antigen specificity using untransfected or CD1b-transfected K562 cells as antigen presenting cells in ELISpot assays (de Jong et al., 2010). Tetramer-positive cells were stained with TCR-αβ FITC (BD) or CD4-PE (BD).
Online supplemental material.
Fig. S1 shows optimization of tetramer staining of the T cell clone LDN5. Fig. S2 shows a comparison of CD1b and CD1d tetramer staining. Fig. S3 shows tetramers staining clonotypic T cell receptors. Fig. S4 shows FACS gating strategies and tetramer staining of healthy controls. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110665/DC1.
Acknowledgments
This work was supported by grants from the Howard Hughes Medical Institute KwaZulu-Natal Research Institute for Tuberculosis and HIV, the Harvard University Initiative for Global Health, the Burroughs Wellcome Fund program in Translational Research, and the National Institutes of Health (T-32 AI 007306-22, T-32 AR 007530-23, R01 AI49313, R01AR 048632, K08 AI089858, and R01 CA58896).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- GMM
- glucose monomycolate
References
- Altman J.D., Moss P.A., Goulder P.J., Barouch D.H., McHeyzer-Williams M.G., Bell J.I., McMichael A.J., Davis M.M. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science. 274:94–96 10.1126/science.274.5284.94 [DOI] [PubMed] [Google Scholar]
- Arrenberg P., Halder R., Dai Y., Maricic I., Kumar V. 2010. Oligoclonality and innate-like features in the TCR repertoire of type II NKT cells reactive to a β-linked self-glycolipid. Proc. Natl. Acad. Sci. USA. 107:10984–10989 10.1073/pnas.1000576107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batuwangala T., Shepherd D., Gadola S.D., Gibson K.J., Zaccai N.R., Fersht A.R., Besra G.S., Cerundolo V., Jones E.Y. 2004. The crystal structure of human CD1b with a bound bacterial glycolipid. J. Immunol. 172:2382–2388 [DOI] [PubMed] [Google Scholar]
- Benlagha K., Weiss A., Beavis A., Teyton L., Bendelac A. 2000. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 191:1895–1903 10.1084/jem.191.11.1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg N.A., Wun K.S., Kjer-Nielsen L., Wilce M.C., Pellicci D.G., Koh R., Besra G.S., Bharadwaj M., Godfrey D.I., McCluskey J., Rossjohn J. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49 10.1038/nature05907 [DOI] [PubMed] [Google Scholar]
- Brigl M., Bry L., Kent S.C., Gumperz J.E., Brenner M.B. 2003. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat. Immunol. 4:1230–1237 10.1038/ni1002 [DOI] [PubMed] [Google Scholar]
- Brigl M., Tatituri R.V.V., Watts G.F.M., Bhowruth V., Leadbetter E.A., Barton N., Cohen N.R., Hsu F.-F., Besra G.S., Brenner M.B. 2011. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J. Exp. Med. 208:1163–1177 10.1084/jem.20102555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone E., Terrazzano G., Melián A., Zanzi D., Moretta L., Porcelli S., Kärre K., Zappacosta S. 2000. Inhibition of human NK cell-mediated killing by CD1 molecules. J. Immunol. 164:6130–6137 [DOI] [PubMed] [Google Scholar]
- de Jong A., Peña-Cruz V., Cheng T.-Y., Clark R.A., Van Rhijn I., Moody D.B. 2010. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat. Immunol. 11:1102–1109 10.1038/ni.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Libero G., Mori L. 2005. Recognition of lipid antigens by T cells. Nat. Rev. Immunol. 5:485–496 10.1038/nri1631 [DOI] [PubMed] [Google Scholar]
- De Libero G., Moran A.P., Gober H.J., Rossy E., Shamshiev A., Chelnokova O., Mazorra Z., Vendetti S., Sacchi A., Prendergast M.M., et al. 2005. Bacterial infections promote T cell recognition of self-glycolipids. Immunity. 22:763–772 10.1016/j.immuni.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Gadola S.D., Zaccai N.R., Harlos K., Shepherd D., Castro-Palomino J.C., Ritter G., Schmidt R.R., Jones E.Y., Cerundolo V. 2002. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat. Immunol. 3:721–726 10.1038/ni821 [DOI] [PubMed] [Google Scholar]
- Garcia-Alles L.F., Versluis K., Maveyraud L., Vallina A.T., Sansano S., Bello N.F., Gober H.J., Guillet V., de la Salle H., Puzo G., et al. 2006. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 25:3684–3692 10.1038/sj.emboj.7601244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron M., Stenger S., Mazorra Z., Wittke F., Mariotti S., Böhmer G., Prandi J., Mori L., Puzo G., De Libero G. 2004. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J. Exp. Med. 199:649–659 10.1084/jem.20031097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumperz J.E., Miyake S., Yamamura T., Brenner M.B. 2002. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J. Exp. Med. 195:625–636 10.1084/jem.20011786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahng A., Maricic I., Aguilera C., Cardell S., Halder R.C., Kumar V. 2004. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J. Exp. Med. 199:947–957 10.1084/jem.20031389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadimitris A., Gadola S., Altamirano M., Brown D., Woolfson A., Klenerman P., Chen J.L., Koezuka Y., Roberts I.A., Price D.A., et al. 2001. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc. Natl. Acad. Sci. USA. 98:3294–3298 10.1073/pnas.051604498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmar A., Van Rhijn I., Moody D.B. 2009. The evolved functions of CD1 during infection. Curr. Opin. Immunol. 21:397–403 10.1016/j.coi.2009.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layre E., Collmann A., Bastian M., Mariotti S., Czaplicki J., Prandi J., Mori L., Stenger S., De Libero G., Puzo G., Gilleron M. 2009. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem. Biol. 16:82–92 10.1016/j.chembiol.2008.11.008 [DOI] [PubMed] [Google Scholar]
- Lee P.T., Benlagha K., Teyton L., Bendelac A. 2002. Distinct functional lineages of human V(alpha)24 natural killer T cells. J. Exp. Med. 195:637–641 10.1084/jem.20011908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Wang L., Yu L., Freundt E.C., Jin B., Screaton G.R., Xu X.N. 2009. Ig-like transcript 4 inhibits lipid antigen presentation through direct CD1d interaction. J. Immunol. 182:1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda J.L., Naidenko O.V., Gapin L., Nakayama T., Taniguchi M., Wang C.R., Koezuka Y., Kronenberg M. 2000. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 192:741–754 10.1084/jem.192.5.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy C., Shepherd D., Fleire S., Stronge V.S., Koch M., Illarionov P.A., Bossi G., Salio M., Denkberg G., Reddington F., et al. 2007. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J. Exp. Med. 204:1131–1144 10.1084/jem.20062342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montamat-Sicotte D.J., Millington K.A., Willcox C.R., Hingley-Wilson S., Hackforth S., Innes J., Kon O.M., Lammas D.A., Minnikin D.E., Besra G.S., et al. 2011. A mycolic acid-specific CD1-restricted T cell population contributes to acute and memory immune responses in human tuberculosis infection. J. Clin. Invest. 121:2493–2503 10.1172/JCI46216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody D.B., Reinhold B.B., Guy M.R., Beckman E.M., Frederique D.E., Furlong S.T., Ye S., Reinhold V.N., Sieling P.A., Modlin R.L., et al. 1997. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 278:283–286 10.1126/science.278.5336.283 [DOI] [PubMed] [Google Scholar]
- Moody D.B., Guy M.R., Grant E., Cheng T.-Y., Brenner M.B., Besra G.S., Porcelli S.A. 2000a. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J. Exp. Med. 192:965–976 10.1084/jem.192.7.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody D.B., Ulrichs T., Mühlecker W., Young D.C., Gurcha S.S., Grant E., Rosat J.-P., Brenner M.B., Costello C.E., Besra G.S., Porcelli S.A. 2000b. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 404:884–888 10.1038/35009119 [DOI] [PubMed] [Google Scholar]
- Moody D.B., Briken V., Cheng T.Y., Roura-Mir C., Guy M.R., Geho D.H., Tykocinski M.L., Besra G.S., Porcelli S.A. 2002. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat. Immunol. 3:435–442 [DOI] [PubMed] [Google Scholar]
- Moon J.J., Chu H.H., Pepper M., McSorley S.J., Jameson S.C., Kedl R.M., Jenkins M.K. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 27:203–213 10.1016/j.immuni.2007.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 272:263–267 10.1126/science.272.5259.263 [DOI] [PubMed] [Google Scholar]
- Paget C., Mallevaey T., Speak A.O., Torres D., Fontaine J., Sheehan K.C.F., Capron M., Ryffel B., Faveeuw C., Leite de Moraes M., et al. 2007. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity. 27:597–609 10.1016/j.immuni.2007.08.017 [DOI] [PubMed] [Google Scholar]
- Porcelli S., Morita C.T., Brenner M.B. 1992. CD1b restricts the response of human CD4-8- T lymphocytes to a microbial antigen. Nature. 360:593–597 10.1038/360593a0 [DOI] [PubMed] [Google Scholar]
- Roura-Mir C., Catálfamo M., Cheng T.-Y., Marqusee E., Besra G.S., Jaraquemada D., Moody D.B. 2005a. CD1a and CD1c activate intrathyroidal T cells during Graves’ disease and Hashimoto’s thyroiditis. J. Immunol. 174:3773–3780 [DOI] [PubMed] [Google Scholar]
- Roura-Mir C., Wang L., Cheng T.-Y., Matsunaga I., Dascher C.C., Peng S.L., Fenton M.J., Kirschning C., Moody D.B. 2005b. Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR-2. J. Immunol. 175:1758–1766 [DOI] [PubMed] [Google Scholar]
- Spada F.M., Grant E.P., Peters P.J., Sugita M., Melián A., Leslie D.S., Lee H.K., van Donselaar E., Hanson D.A., Krensky A.M., et al. 2000. Self-recognition of CD1 by γ/δ T cells: implications for innate immunity. J. Exp. Med. 191:937–948 10.1084/jem.191.6.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger S., Hanson D.A., Teitelbaum R., Dewan P., Niazi K.R., Froelich C.J., Ganz T., Thoma-Uszynski S., Melián A., Bogdan C., et al. 1998. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 282:121–125 10.1126/science.282.5386.121 [DOI] [PubMed] [Google Scholar]
- Ulrichs T., Moody D.B., Grant E., Kaufmann S.H., Porcelli S.A. 2003. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect. Immun. 71:3076–3087 10.1128/IAI.71.6.3076-3087.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.S., Leslie D.S., Gumperz J.E., Xiong X., Grant E.P., Brenner M.B. 2002. CD1-dependent dendritic cell instruction. Nat. Immunol. 3:1163–1168 10.1038/ni851 [DOI] [PubMed] [Google Scholar]
- Winau F., Schwierzeck V., Hurwitz R., Remmel N., Sieling P.A., Modlin R.L., Porcelli S.A., Brinkmann V., Sugita M., Sandhoff K., et al. 2004. Saposin C is required for lipid presentation by human CD1b. Nat. Immunol. 5:169–174 10.1038/ni1035 [DOI] [PubMed] [Google Scholar]
- Zhou D., Mattner J., Cantu C., III, Schrantz N., Yin N., Gao Y., Sagiv Y., Hudspeth K., Wu Y.P., Yamashita T., et al. 2004. Lysosomal glycosphingolipid recognition by NKT cells. Science. 306:1786–1789 10.1126/science.1103440 [DOI] [PubMed] [Google Scholar]