Abstract
The cytochrome P450 (CYP) gene products such as CYP3A and CYP2B are essential for the metabolism of steroid hormones and xenochemicals including prescription drugs. Nuclear receptor SXR/PXR (steroid and xenobiotic receptor/pregnenolone X receptor) has been shown both biochemically and genetically to activate CYP3A genes, while similar studies have established constitutive androstane receptor (CAR) as a CYP2B regulator. The response elements in these genes are also distinct, furthering the concept of independent regulation. Unexpectedly, we found that SXR can regulate CYP2B, both in cultured cells and in transgenic mice via adaptive recognition of the phenobarbital response element (PBRE). In a type of functional symmetry, orphan receptor CAR was also found to activate CYP3A through previously defined SXR/PXR response elements. These observations not only provide a rational explanation for the activation of multiple CYP gene classes by certain xenobiotics, but also reveal the existence of a metabolic safety net that confers a second layer of protection to the harmful effects of toxic compounds and at the same time increases the propensity for drug–drug interactions.
Keywords: CYP gene, nuclear receptor, SXR/PXR, CAR, metabolic safety
The liver cytochrome P450 (CYP) enzymes represent a supergene family of hemeproteins that catalyze the metabolic conversion to more polar derivatives of an amazing diversity of foreign chemicals (xenobiotics) including various environmental pollutants, carcinogens, and prescription drugs as well as endogenous substrates such as steroid hormones (for reviews, see Gonzalez 1992; Denison and Whitlock 1995). The levels of some CYP enzymes are typically induced by their xenobiotic substrates. For example, administration of glucocorticoids (both agonists such as dexamethasone [DEX] and antagonists such as RU486), rifampicin (RIF), or phenobarbital (PB) increases the levels of CYP3A, a family of medically significant isoenzymes involved in the metabolism of more than half of all prescription drugs as well as neutraceuticals and herbal medicines (Maurel 1996). Specificity studies reveal that some (but not all) CYP3A-inducing compounds also activate CYP2B genes (e.g., Strom et al. 1996; Honkakoski and Negishi 1997). The metabolic versatility of CYP3A and CYP2B coupled with their inducibility by xenobiotic substrates constitutes a molecular basis for many clinical drug–drug interactions. Such interactions pose one of the most vexing problems in drug development. These problems arise when P450 inducers such as glucocorticoids, PB, or RIF are administered concurrently with medications such as immunosuppressant cyclosporine A, oral contraceptives and antihypertensives that are normally metabolized by these CYP enzymes (Maurel 1996). Recently, St John's wort, a popular herbal remedy for depression, was found to trigger severe adverse drug interactions with oral contraceptives, the HIV protease inhibitor indinavir, and cyclosporine A as a consequence of activating the CYP3A system (Moore et al. 2000a; Fugh-Berman 2000; Piscitell et al. 2000; Ruschitzaka et al. 2000).
The human steroid and xenobiotic receptor (SXR) and its rodent homolog pregnenolone X receptor (PXR) were isolated as candidate xeno-sensors postulated to regulate CYP3A genes (Blumberg et al. 1998; Kliewer et al. 1998; Bertilson et al. 1998; and for reviews, see Blumberg and Evans 1998; Savas et al. 1999; Waxman 1999). Indeed, SXR/PXR bind to the IR-6 and DR-3 response elements localized to the 5' regulatory regions of the human CYP3A4 and rat CYP3A23 gene, respectively. Recently, we have established unequivocally that SXR and PXR function as xeno-sensors in vivo, by demonstrating that targeted disruption of the mouse PXR gene by homologous recombination abolishes the CYP3A response. In contrast, hepatic expression of an activated SXR transgene results in constitutive upregulation of CYP3A gene expression and enhanced protection against xenotoxicants (Xie et al. 2000). Having demonstrated that SXR and PXR mediate the hepatic CYP3A response, we noted that the presence of candidate DR-3 or IR-6 response elements in CYP2A, CYP2C, CYP2E, and glucouronosyl transferase genes (Blumberg et al. 1998) raised the potential for a broader physiologic function. All of these enzymes are involved in steroid and xenobiotic catabolism (for review, see Gonzalez 1992). However, whether these or other CYP genes can serve as in vivo targets for SXR and PXR is unclear. If so, this would have widespread implications in understanding the nature and properties of the adaptive hepatic response.
The orphan receptor CAR (constitutive androstane receptor) binds DNA as a heterodimer with RXR (retinoid × receptor) and activates gene transcription in a constitutive manner (Baes et al. 1994; Choi et al. 1997). As we have previously shown, the CAR-mediated transcriptional activation can be inhibited by androstane metabolites such as androstenol and androstanol (Forman et al. 1998). In addition to its activation of a DR-5 type of retinoid acid response element (βRARE), CAR also activates a distal 51-bp enhancer called the phenobarbital response element (PBRE) found in phenobarbital (PB)-inducible CYP2B genes (Trottier et al. 1995; Park and Kemper 1996; Honkakoski and Negishi 1997). Inspection of the PBRE reveals it to contain two nuclear receptor (NR) binding sites composed of imperfect direct repeats of AG(G/T)TCA-like half sites spaced by four nucleotides (DR-4 motif). The repeat sequences of these binding sites are conserved in PB-inducible rodent and human CYP2B genes, whereas they are divergent in the PB-nonresponsive mouse CYP2B9 gene (Honkakoski et al. 1998a). Recently, it has been suggested that CAR may regulate the CYP3A gene, on the basis of transient transfection experiments using isolated response elements (Honkakoski et al. 1998b; Tzameli et al. 2000; Moore et al. 2000b). Based on these observations, we wish to explore the potential of cross-regulation between these two xenobiotic sensors and a potential molecular means to integrate what is generally viewed as two distinct xenobiotic response systems.
In this report, we demonstrate the presence of this hypothetical cross-regulatory response between SXR/PXR and CAR. In particular, cultured primary hepatocytes, transgenic mice, and natural and synthetic reporters were all used to show that CYP2B is an endogenous target of SXR and that CYP3A is an in vivo target for CAR. This establishes the existence of a simple and unique integrative mechanism to modulate the metabolism of endogenous steroids, bioactive dietary compounds, and xenobiotic substances.
Results
Binding of the PBRE by SXR
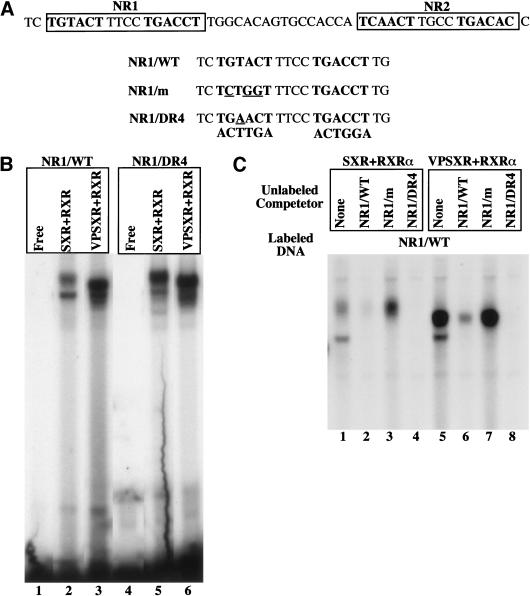
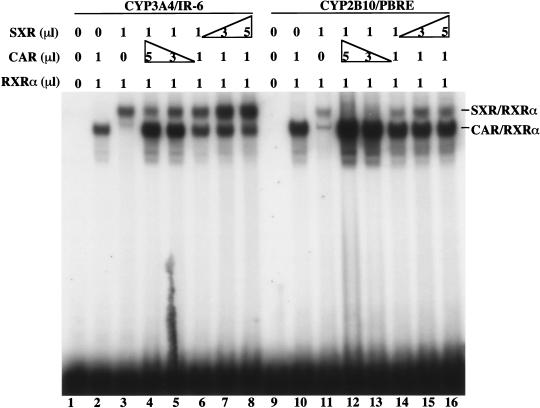
A 51-bp sequence termed the PBRE is conserved in the inducible rodent and human CYP2B genes (Trottier et al. 1995; Park and Kemper 1996; Honkakoski and Negishi 1997) and has been shown to be necessary and sufficient for phenobarbital induction of mouse CYP2B10 gene (Honkakoski et al. 1998b; Kawamoto et al. 1999; Sueyoshi et al. 1999; Tzameli et al. 2000; Moore et al. 2000b). Sequence analysis of the PBRE reveals two imperfect DR-4 motifs (NR1 and NR2) (Fig. 1A) that have previously been shown to bind CAR (Honkakoski et al. 1998b). To explore the potential cross-regulation of CYP2B, electrophoretic mobility shift assays (EMSA) were used to determine the ability of SXR to bind the PBRE. As shown in Figure 1B, both wild type SXR and its activated variant VPSXR (Xie et al. 2000) bound the NR1 efficiently. The binding was dependent on the presence of their heterodimerization partner RXR (Fig. 1B, lanes 2 and 3), while no DNA binding was seen in the absence of RXR as expected (data not shown). These results demonstrate that both SXR and VPSXR bind NR1 in a fashion similar to the binding of CAR/RXR to the PBRE (see below; Honkakoski et al. 1998b). This specific binding was abrogated when the first half site of NR1 was mutated to NR1/m TCTGGT (mutated nucleotides are underlined) (data not shown); while the binding was maintained and slightly improved when the NR1 was converted to a perfect DR-4 element (Fig. 1, NR1/DR4, lanes 5 and 6). The binding of NR1 by SXR and VPSXR was specific, inasmuch as efficient competition of binding was achieved by excess unlabeled wild type NR1 or NR1/DR4, but not by NR1/m (Fig. 1C)
Figure 1.
Binding of the PBRE by SXR. (A) The DNA sequence of the 51-bp PBRE derived from the mouse CYP2B10 gene. The two putative DR-4 type NR binding sites, NR1 and NR2, are boxed. The DNA sequences of the oligonucleotides corresponding to the wild type NR1 and its mutant variants used for EMSA are also shown, with the mutated nucleotides underlined. (B) SXR:RXR heterodimers bind to the PBRE. EMSA was performed using in vitro synthesized SXR, VPSXR, and RXRα proteins and radiolabeled oligonucleotides of NR1 (lanes 1–3) and its mutant NR1/DR4 (lanes 4–6) in which the imperfect DR-4 of NR1 was mutated to an AG(G/T)TCA type of DR-4. (C) The binding of NR1 by SXR/RXRα or VPSXR/RXRα can be efficiently competed away by excessive unlabeled NR1/WT (lanes 2 and 6), NR1/DR4 (lanes 4 and 8), but not by NR1/m (lanes 3 and 7). The free probes ran off the gel.
SXR activates CYP2B in cultured cells
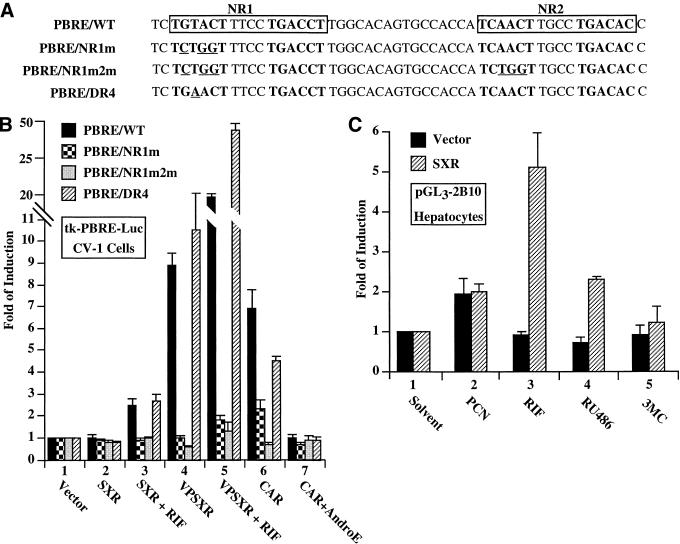
Transfection based assays were utilized to determine whether SXR can activate CYP2B. First, luciferase reporter plasmids containing the 51-bp PBRE or its mutant derivatives (Fig. 2A) upstream of a minimal thymidine kinase (tk) promoter were constructed and transfected into monkey kidney CV-1 cells together with expression vectors for SXR, VPSXR, or CAR. Modest but significant activation of the CYP2B reporter by SXR was seen when RIF, a known SXR specific activator, was added to the culture medium (Fig. 2B, lane 3). Expression of VPSXR resulted in a more potent induction of CYP2B without added ligand (lane 4), and RIF treatment promoted an additional two-fold of induction (lane 5). The CYP2B induction is SXR-specific, as another constitutively active orphan receptor, VPFXR (farnesoid X receptor) (Forman et al. 1995) has no effect on this reporter gene (data not shown). Consistent with DNA binding results, the PBRE-mediated activation was abrogated when either the NR1 (PBRE/NR1m) or both NRs (PBRE/NR1m2m) were mutated, and the activation was restored when the NR1 was converted to a perfect DR-4 site (Fig. 2B). Therefore, these NR binding sites are the putative mediators for both the binding and activation of the PBRE by SXR. As controls, CAR activates CYP2B in a ligand-independent manner as predicted, and androstenol inhibits this constitutive activation (lanes 6 and 7, respectively) (Forman et al. 1998).
Figure 2.
SXR activates CYP2B in cultured cells. (A) The DNA sequences of the wild type PBRE and its mutant variants in the synthetic tk-PBRE-Luc reporter genes. These reporters were used in the transfections shown in B. (B) Ligand-dependent activation of tk-PBRE-Luc by SXR and constitutive activation of tk-PBRE-Luc by VPSXR. The wild type (PBRE/WT) or mutant variants (PBRE/NR1m, PBRE/NR1m2m, and PBRE/DR4) of the synthetic tk reporter constructs were transfected into CV-1 cells in the absence (vector) or presence of expression vectors for SXR, VPSXR or CAR. The transfected cells were subsequently mock treated or treated with indicated compounds. Results are shown as fold induction over vector controls, and represent the averages and standard error from triplicate assays. RIF, rifampicin, 10 μM; AndroE, androstenol, 5 μM. (C) Ligand-dependent activation of the natural promoter of CYP2B10 gene. The CYP2B10 promoter driving luciferase construct was transfected into primary rat hepatocytes in the absence (vector) or presence of expression vector for SXR. Cells were subsequently mock treated or treated with indicated compounds. Results are shown as fold induction over solvent, and represent the averages and standard error from triplicate assays. PCN, pregnenolone-16α-carbonitrile; 3MC, 3-methylcholanthrene. The concentration of compound is 10 μM with the exception of 3MC (2 mM).
To explore this potential regulation of CYP2B in a more relevant system, we transfected the SXR or CAR vectors into primary cultures of rat hepatocytes and examined the effects of a panel of steroid and nonsteroid inducers on the expression of a co-transfected natural promoter of mouse CYP2B10 gene linked to firefly luciferase gene. Primary cultures were employed not only because CYP2B10 is a hepatic gene but also because this natural promoter is completely inactive in cultured cell lines (data not shown). In control rat hepatocytes without the human SXR, CYP2B10 was modestly induced by pregnenolone-16α-carbonitrile (PCN), presumably through the binding and activation of endogenous PXR (Fig. 2C, lane 2). In contrast, co-transfection of SXR resulted in a significant induction of CYP2B10 in respond to the known SXR activators RIF and RU486 (lanes 3 and 4), while the addition of PCN (lane 2), a specific activator for rodent PXR, or 3-methylcholanthrene (3MC) (lane 5), a known inducer of an unrelated cytochrome CYP1A1/1A2, had minimal effects. Taken together, these results provide compelling evidence that SXR is capable of inducing CYP2B gene expression in a ligand-dependent manner, and that this induction is mediated through the PBRE.
Binding of SXR/PXR response elements by CAR
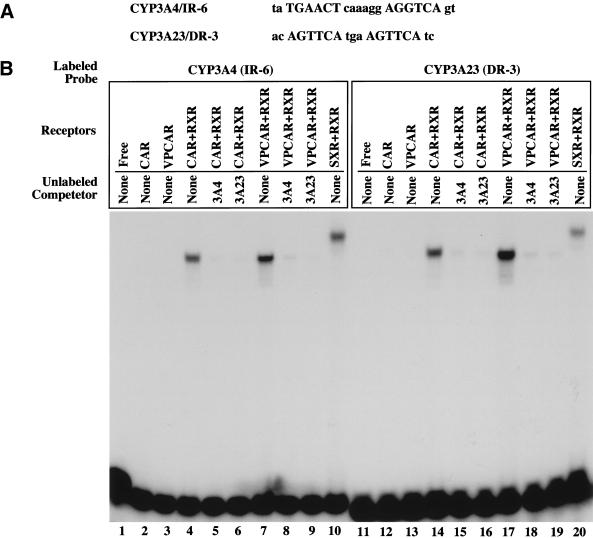
Based on the above observations, a set of reciprocal experiments were initiated to determine whether CAR might bind and activate SXR target genes. An IR-6 element from the human CYP3A4 gene and a DR-3 repeat from the rat CYP3A23 gene (Fig. 3A), have been identified as SXR/PXR response elements (Fig. 3, lanes 10 and 20; Blumberg et al. 1998; Kliewer et al. 1998). As shown in Figure 3B, EMSA analysis reveals that both CAR/RXR and VPCAR/RXR can bind the 3A4/IR-6 and the 3A23/DR-3. The VPCAR is an activated form of CAR generated by fusing the VP16 activation domain to the amino-terminal of CAR. In both cases, the binding was efficiently competed by excess unlabeled IR-6 or DR-3.
Figure 3.
Binding of SXR/PXR response elements by CAR. (A) Sequences of the IR-6 and DR-3 elements found in human and rat CYP3A genes, respectively. (B) EMSA was performed using in vitro synthesized CAR, VPCAR, SXR, and RXRα proteins and radiolabeled oligonucleotides of IR-6 (lanes 1–10) from the human CYP3A4, or the DR-3 (lanes 11–20) from the rat CYP3A23 gene. One hundred-fold excess of the unlabeled 3A4/IR-6, or 3A23/DR-3 was used for binding competitions.
CAR activates CYP3A in cultured cells
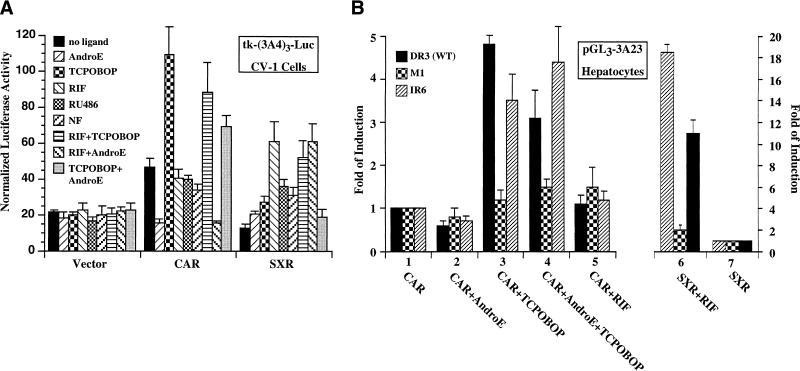
The activation of CYP3A gene by CAR was first examined in a transfection-based assay in which a synthetic CYP3A4/IR-6 containing reporter gene was introduced into CV-1 cells in the presence of CAR or SXR expression vectors. A panel of compounds were tested as shown in Figure 4A. As expected, this reporter was activated by CAR in the absence of ligand. The activation was inhibited by the antagonistic ligand androstenol but potentiated by the activating ligand 1,4-bis[2–(3,5 dichloropyridyloxyl)] benzene (TCPOBOP). In addition, TCPOBOP can reverse the inhibitory effect of androstenol when both ligands are added simultaneously (Honkakoski et al. 1998b; Tzameli et al. 2000). Three known SXR ligands, RIF, RU486, and nifedipine (NF) (Blumberg et al. 1998) have little effect on CAR activation. Furthermore, RIF has little effect on the effects of androstenol or TCPOBOP. Reciprocally, neither androstenol nor TCPOBOP significantly activates SXR, or interferes with the activation of SXR by RIF. These results demonstrated that CAR can clearly regulate CYP3A but only in response to its own (i.e., not SXR) ligands. In aggregate, these results expand the range of molecules that may function as activators of the CYP3A response, presumably via receptors other than SXR/PXR.
Figure 4.
CAR activates CYP3A in cultured cells. (A) The synthetic tk-(3A4)3-Luc reporter was transfected into CV-1 cells in the absence (vector) or presence of expression vectors for CAR or SXR. Cells were subsequently treated with individual compound or combination of compounds. Results are shown as normalized luciferase activity, and represent the averages and standard error from triplicate assays. AndroE, androstenol, 5 μM; TCPOBOP, 1,4-bis[2–(3,5-dichloropyridyloxy)] benzene, 250 nM; RIF, rifampicin; RU486; NF, nifedipine, 10 μM each. Similar results were obtained using the CYP3A23/DR-3 containing tk reporter (data not shown). (B) Ligand-dependent and DR-3/IR-6-mediated activation of the natural CYP3A gene promoter. The natural CYP3A23 promoter DR3 (WT) or its mutant variants (M1 and IR6) were transfected into primary rat hepatocytes in the presence of expression vectors for CAR or SXR. Cells were subsequently mock treated or treated with indicated compounds. Results are shown as fold induction over solvent, and represent the averages and standard error from triplicate assays. The concentrations of compound were the same as in A.
The activation of CYP3A by CAR (Fig. 4B, lanes 1–5) or VPCAR (data not shown) was also observed in the primary rat hepatocyte system with the natural rat CYP3A promoter reporter and its mutant variants (Xie et al. 2000). Primary hepatocytes were used because this natural CYP3A promoter is nonresponsive in CV-1 cells or liver carcinoma HepG2 cells (data not shown). Consistent with the observations in CV-1 cells, activation of the natural CYP3A promoter by CAR in rat hepatocytes was inhibited by androstenol (Fig. 4B, lane 2). This is the first time that a natural CYP promoter has been shown to be downregulated, establishing a direct link between the CAR, androstane metabolites and a target gene. TCPOBOP not only activates CAR by itself (lane 3) but also reverses the inhibitory effect of androstenol (lane 4). This is the first example in which the current natural CYP3A promoter has been active in a transfection-based system. In contrast, additions to the culture medium of the SXR activator RIF has little effect on CAR activity (lane 5). Having established a relevant physiologic context for the activation of CYP3A by CAR, we were in a position to examine the necessity of the DR-3 response element for this process. We showed previously that this element mediates the SXR response; mutation of this element (M1) abrogates while replacement of this element by the human IR-6 element rescues the induction (Fig. 4B, lanes 6 and 7; Xie et al. 2000). Remarkably, these mutations display the same effects on transactivation of CYP3A by CAR in that the mutant site (M1) fails to respond to TCPOBOP, and it is successfully rescued by the IR-6. These results provide compelling evidence that the SXR/PXR response elements are effective targets for transregulation by CAR.
Competition of DNA binding by SXR and CAR
If, as we have found, CAR and SXR cross-regulate two classes of CYP genes, the relative DNA binding affinity of these two receptors toward these response elements may be important in establishing natural hierarchies. As shown in Figure 5, SXR and CAR exhibited surprisingly similar binding affinity toward IR-6 element found in human CYP3A4 (lanes 1–8), which is particularly obvious when expressed at equal molar ratios (lane 6). In contrast, the NR1 of CYP2B/PBRE can bind SXR, but it displays a clear preference for CAR/RXRα heterodimers.
Figure 5.
Competition of DNA binding by SXR and CAR. EMSA was performed using in vitro synthesized CAR, SXR, and RXRα proteins and radiolabeled oligonucleotides of 3A4/IR-6 (lanes 1–8), or the NR1 of CYP2B10/PBRE (lanes 9–16). The protein ratios of receptors were indicated.
Reciprocal target gene activation by SXR/PXR and CAR in vivo
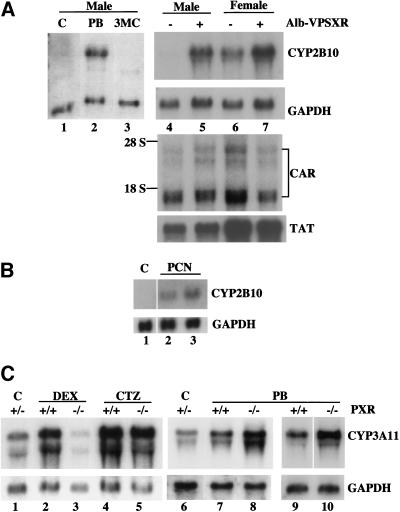
The binding of PBRE and activation of CYP2B by SXR in cultured cells prompted us to examine the hepatic expression of CYP2B in Alb-VPSXR mice in which an activated form of SXR has been expressed under the control of liver-specific albumin promoter/enhancer (Xie et al. 2000). For reasons that are not fully understood, expression of the mouse CYP2B10 gene is only detected in uninduced wild type female livers (Fig. 6, cf. lanes 4 and 6), but is inducible in both sexes in response to xenobiotics such as phenobarbital (e.g., lane 2). However, it is not induced by the CYP1A inducer 3MC (lane 3, and Noshiro et al. 1988). In clear results shown in Figure 6A lanes 4–7, we found that CYP2B10 is spontaneously induced in both male and female transgenic animals. This induction is a direct result of VPSXR transgene expression rather than the induction of other CYP2B10 regulator(s), because expression levels of CAR remain unchanged in transgenic animals. Furthermore, this effect is specific for SXR target genes since other hepatic genes such as the tyrosine aminotransferase (TAT) remain unchanged in the transgenic livers (Fig. 6A, cf. lanes 4 and 5, and lanes 6 and 7, respectively). The induction of CYP2B by PXR was also observed in wild type animals. Treatment with a PXR-specific ligand, PCN, resulted in elevated expression of CYP2B10 mRNA (Fig. 6B), consistent with our previous observations in cultured hepatocytes (Schuetz et al. 1988).
Figure 6.
Reciprocal target gene activation by SXR/PXR and CAR in vivo. (A) Constitutive upregulation of hepatic CYP2B10 mRNA in the liver of Alb-VPSXR mice. Mouse liver total RNAs were subjected to Northern blot analysis and probed for CYP3A11. The membranes were subsequently stripped and reprobed with cDNA probes of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH, as a loading control), CAR, and tyrosine aminotransferase (TAT). To show the CYP2B10 regulation in wild type animals, male mice were subjected to a single intraperitoneal injection of control solvent (lane 1), PB (lane 2), or 3MC (lane 3). (B) Induction of CYP2B10 expression by PXR-specific ligand PCN in wild type males. (C) CTZ and PB, but not DEX, efficaciously induce CYP3A11 in PXR null mice.
Next, the requirement for PXR versus CAR in regulation of CYP3A was examined in PXR null mice (Xie et al. 2000). Phenobarbitals have been shown to induce both CYP2B and CYP3A in cultured cells and in animals (Ramsden et al. 1993; Honkakoski et al. 1996). Clotrimazole (CTZ), a known SXR/PXR activator, has recently been shown to bind and activate CAR (Moore et al. 2000b). As shown in Figure 6C, even in the PXR null mice, CYP3A continues to be efficaciously induced both by CTZ (lane 5) and by PB (lanes 8 and 10), whereas its induction by DEX (lane 3) or PCN (data not shown; Xie et al. 2000) was completely abolished. Indeed, the PB-mediated induction of CYP2B10 was consistently higher in the PXR null mice than in wild type animals (Fig. 6C, cf. lanes 7 and 8, and 9 and 10, respectively). These results indicate that PB or CTZ induction of CYP3A genes can be achieved in the absence of PXR, suggesting that CAR may function as a fail-safe system, especially in the case where a single ligand can activate both receptors.
Discussion
Our observations indicate that xenobiotics such as RIF and DEX, in addition to inducing human CYP3A, may function as primary inducers of both CYP3A and CYP2B isoenzymes (Strom et al. 1996; Zhou and Wilkinson 1990). The basis for this phenomenon appears to reside in the inherent capacity of both SXR and CAR to recognize each other's response elements. While these receptors were presumed to be distinct in both their ability to bind ligands and target DNA, some hints for adaptable DNA recognition were apparent in our previous studies. This idea was originally considered in Blumberg et al. (1998), where SXR and PXR were found to display measurable affinity for certain types of DR-4-like sequences. However, the ligand specificity of SXR and PXR, particularly for DEX, PCN and RIF clearly placed SXR/PXR as CYP3A regulators. The essential role of PXR/SXR in CYP3A regulation was confirmed by the PXR knockout and the humanized SXR transgenic mice (Xie et al. 2000). The initial rationale for examining the potential of SXR/PXR to activate CYP2B is the responsiveness of this CYP to structurally diverse xenochemicals that exceed the known recognition capacity of CAR (Lubet et al. 1992; Waxman and Azaroff 1992; Nims and Lubet 1995; Honkakoski et al. 1998b). Because different inducers bind to and activate these different nuclear receptors (e.g., SXR/PXR and CAR), our results indicate that the common target for induction must be the PBRE, rather than the receptors. The ability of SXR/PXR and CAR to each bind and activate the PBRE provides an explanation for the cross-regulation albeit little insight into its relevance. However, in the Alb-VPSXR mice, we cannot exclude the possibility that the activation of SXR could activate CYP2B10 by producing an endogenous ligand for CAR as a result of induced CYP enzyme systems. Nevertheless, this multiple receptor-mediated induction mechanism is distinct from that used by aryl hydrocarbon (AH) receptor. The AH receptor itself binds dioxin and other structurally similar polycyclic aromatic hydrocarbons to activate the cognate xenobiotic response element of the CYP1A and CYP1B1 genes (Hankinson 1995; Whitlock et al. 1996).
CAR/RXR heterodimers were originally shown to bind to the DR-5 type retinoid acid response elements (βRAREs) (Baes et al. 1994; Choi et al. 1997), leading to the transient view that it might be a variant retinoid receptor. More recently, CAR has been shown to preferentially bind to the imperfect DR-4 sites within the CYP2B PBRE (Honkakoski et al. 1998b; Tzameli et al. 2000). It was this observation, together with CAR-mediated PBRE activation by a PB-type inducer such as TCPOBOP, that established CAR as a potential mediator of the phenobarbital response. As we have shown here, CAR can also bind to the CYP3A4/IR-6 and the CYP3A23/DR-3 type of SXR/PXR response elements and regulate expression from the natural CYP3A promoter as well as reporter gene constructs containing these elements. Therefore, we conclude that CYP3A genes, in addition to CYP2B genes, are bona fide targets for CAR. CAR has also recently been reported to bind to the DR-4 type of liver × receptor (LXR) element from the MMTV promoter (Tzameli et al. 2000). However, whether CAR can activate LXR (Willy et al. 1995) target genes has yet to be seen.
The substantial overlap in DNA binding recognition by these receptors stands in contrast to their distinct specificities of ligand binding. Indeed, our results suggest an important functional parallel in addition to ligand binding. It is conceivable that for each specific element or target gene, the extent of this overlap would be dependent on a number of factors, such as relative affinity for various receptors, availability of endogenous or exogenous ligands, and levels of their expression as well as potential post-transcriptional regulation of CYP genes among different tissues. Indeed, in addition to their expression in the liver, SXR/PXR has been shown to express in the intestine, kidney (Blumberg et al. 1998; Kliewer et al. 1998) and mammary gland (Dotzlaw et al. 1999), whereas CAR also expresses in intestine, heart, muscle, kidney and lung (Baes et al. 1994). Nevertheless, the overlap in their response element recognition establishes a molecular basis for a regulatory network of CYP gene expression that expands the function of individual orphan receptors. As more is learned about these receptors, we expect that additional categories of target genes will be identified and that the concept of the metabolic safety net will be more clearly defined.
The reciprocal activation of CYP genes may also contribute to the apparently normal phenotype of PXR (Xie et al. 2000) or CAR (David Moore, personal communication) null mice. In the case of PXR null mice, although the inducibility of CYP3A by PCN and DEX is completely abolished, the basal expression level of CYP3A remains unchanged compared with wild type animals. This may result from the continued CAR expression and signaling in these animals. Indeed, retained regulation by the alternative receptor is supported by our observations that CYP3A inducibility by PB and CTZ, two shared ligands of SXR/PXR and CAR, was at least partially intact in the PXR null mice. This indicates that PB or CTZ induction of CYP3A genes can be achieved by a mediator other than SXR/PXR, presumably CAR. However, we cannot exclude the possibility that additional nuclear receptor(s) or other transcriptional regulators may be involved. The generation of mice deficient in both PXR and CAR genes will enable to further explore this problem in vivo. Moreover, we cannot exclude the possibility of “metabolic adaptation” in which an adaptive response may have been activated as a consequence of continuous stimulation (VPSXR) or absence (PXR null) of such a key regulatory factor.
Considering the diversity of drugs and xenobiotics that must be recognized by SXR and CAR (potentially thousands or more), it seems unlikely that the metabolic capacity of the enzymes should show the same recognition profile as the receptors themselves. After all, the protein folds for the ligand binding domain of nuclear receptors and the substrate recognition pockets for the cytochromes are completely different. Thus, the establishment of a metabolic safety net that enables dual enzyme activation seems advantageous by expanding the protective capacity of the xenobiotic response system. In summary, this work establishes a molecular basis for cross-regulation of the CYP regulatory network, increasing both the complexity and capacity of the response and perhaps most importantly providing a new way to think about both the regulation of the xenobiotic response and the realistic problem of drug–drug interactions.
Materials and methods
DNA-binding analysis
Electrophoretic mobility shift assays (EMSA) were performed using in vitro transcribed and translated protein (TNT, Promega). Proteins (2 μl each) were incubated for 10 min at room temperature with 200,000 cpm of 32P-labeled probes prepared by Klenow fill-in reactions. The binding buffer contained 10 mM Tris (pH8), 100 mM KCl, 6% glycerol, 1 mM PMSF, 1 mM DTT, and 100 ng/ml poly[d(I-C)] (Pharmacia) and was electrophoresed through a 6% polyacrylamide gel in 0.5x TBE (45 mM Tris-base, 45 mM boric acid, 1 mM EDTA) at 4°C. For competition binding, proteins plus unlabeled oligonucleotides at 100-fold molar excess were preincubated for 10 min on ice, labeled probes added, and further incubated for 20 min at room temperature. Oligonucleotides were the following: NR1/WT, TCTGTACTT TCCTGACCTTG; NR1/m, TCTCTGGTTTCCTGACCTTG; NR1/DR3, TCTGAACTTTCCTGACCTTG; CYP3A4/IR-6, TA TGAACTCAAAGGAGGTCAGT; CYP3A23/DR-3, ACAGTT CATGAAGTTCATC.
Plasmid constructs
The tk-PBRE-Luc and its mutant variants were generated by insertion of corresponding annealed oligonucleotides into the tk-Luc vector. The CYP2B10 cellular promoter reporter, PGL3-CYP2B10, was cloned by inserting the PCR-amplified 5' regulatory sequence of mouse CYP2B10 gene (nt -1406 to 24) (Honkakoski et al. 1996) into the PGL3 vector (Promega). The CYP3A reporters (tk-(3A4)3-Luc, and PGL3-CYP3A23 and its mutant variants) were described before (Xie et al. 2000). The expression vectors for the wild type SXR and an activated form of SXR (VPSXR) (Blumberg et al. 1998; Xie et al. 2000) and CAR (Forman et al. 1995) were as described. The expression vector for VPCAR was generated by replacing the cDNA of SXR in VPSXR with that of CAR.
Preparation of hepatocytes, DNA transfections and drug treatment
Primary cultures of rat hepatocytes were prepared as described previously (Barwick et al. 1996). The plates were coated with Vitrogen. Lipofectin (Gibco-BRL)-mediated DNA transfections were carried out as described (Barwick et al. 1996). For each transfection, we used 3.5 μg of PGL3-CYP2B10, or PGL3-CYP3A23 reporter with 50 ng of expression vectors for receptors. When necessary, cells were treated with RIF, DEX, PCN, NF, RU486 (10 μM each), PB, 3MC (2 mM each), TCPOBOP (250 nM), or the control solvent. Compounds except for TCPOBOP were purchased from Sigma. CV-1 cell transfections using 48-well-plates and DOTAP transfection reagent (Bohringer) were carried out as described by Blumberg et al. (1998).
Animals and drug treatment
The generation of Alb-VPSXR transgenic mice and PXR null mice has been described before (Xie et al. 2000). The animals were maintained ad libitum. When necessary, mice were subjected to a single intraperitoneal injection of DEX (50 mg/kg), PB (40 mg/kg), CTZ (50 mg/kg), 3MC (4 mg/kg), or PCN (40 mg/kg) 24h before sacrifice.
Northern blot analysis
Total RNA was prepared from liver tissues using TRIZOL Reagent (Gibco-BRL). Northern hybridization was carried out as described (Xie et al. 1999). The probe of CYP3A11 gene was described before (Xie et al. 2000). The cDNA probe of CYP2B10 (nt 652–1457) (Noshiro et al. 1988) was cloned by RT-PCR from wild type mouse liver mRNA.
Acknowledgments
We thank Dr. Chihcheng Tsai for his invaluable advice on EMSA and for his comments on the manuscript; Alex Shearer for his effort at the early stage of this study; Michael C. Nelson, Ardavan Arianpour and Henry Juguilon for technique assistance; Dr. Enrique Saez for expression vector pCMX-VPFXR; and Elaine Stevens and Lita Ong for administrative assistance. W.X. is supported by Susan G. Komen Breast Cancer Foundation. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by Mathers Foundation (R.M.E.), the Howard Hughes Medical Institute (R.M.E.), and grants for the National Institute of Health (ES05744 to P.S.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.846800.
References
- Barwick JL, Quattrochi LC, Mills AS, Potenza C, Tukey RH, Guzelian PS. Trans-species gene transfer for analysis of glucocorticoid-inducible transcriptional activation of transiently expressed human CYP3A4 and rabbit CYP3A6 in primary cultures of adult rat and rabbit hepatocytes. Mol Pharmacol. 1996;50:10–16. [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi H-S, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1552. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter CM, Ong ES, Evans RM. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes & Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Evans RM. Orphan nuclear receptors–new ligands and new possibilities. Genes & Dev. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, Moore DD. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J Biol Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- Denison MS, Whitlock JP., Jr Xenobiotic-inducible transcription of cyctochrome P450 genes. J Biol Chem. 1995;270:18175–18178. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]
- Dotzlaw H, Leygue E, Watson P, Murphy LC. The human orphan receptor PXR messenger RNA is expressed in both normal and neoplastic breast tissue. Clin Cancer Res. 1999;5:2103–2107. [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is identified by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-β. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Fugh-Berman A. Herb–drug interactions. Lancet. 2000;355:134–138. doi: 10.1016/S0140-6736(99)06457-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ. Human cytochromes P450: problems and prospects. Trends Pharmacol Sci. 1992;13:346–352. doi: 10.1016/0165-6147(92)90107-h. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. Characterization of a phenobarbital-responsive enhancer module in mouse P450 Cyp2b10 gene. J Biol Chem. 1997;272:14943–14949. doi: 10.1074/jbc.272.23.14943. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Moore R, Gynther J, Negishi M. Characterization of phenobarbital-inducible mouse Cyp2b10 gene transcription in primary hepatocytes. J Biol Chem. 1996;271:9746–9753. doi: 10.1074/jbc.271.16.9746. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Moore R, Washburn KA, Negishi M. Activation by diverse xenochemicals of the 51-base pair phenobarbital-responsive enhancer module in the CYP2B10 gene. Mol Pharmocol. 1998a;53:597–601. doi: 10.1124/mol.53.4.597. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998b;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Lubet RA, Dragnev KH, Chauhan DP, Nims RW, Diwan BA, Ward JM, Jones CR, Rice JM, Miller MS. A pleiotropic response to phenobarbital-type enzyme inducers in the F344/NCr rat: Effects of chemicals of varied structure. Biochem Pharmacol. 1992;43:1067–1078. doi: 10.1016/0006-2952(92)90614-o. [DOI] [PubMed] [Google Scholar]
- Maurel P. In: Cytochrome P450: Metabolic and Toxicological Aspects. Ioannides C, editor. Boca Raton, FL: CRC Press; 1996. pp. 241–270. [Google Scholar]
- Moore LB, Goodwin B, Jones SA, Wisely GB, Serabjit-Singh CJ, Willson TM, Collins JL, Kliewer SA. St. John's wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc Natl Acad Sci USA. 2000a;97:7500–7502. doi: 10.1073/pnas.130155097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000b;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Nims RW, Lubet RA. Induction of cytochrome P-450 in the Norway rat, Rattus norvegicus, following exposure to potential environmental contaminants. J Toxicol Environ Health. 1995;46:271–292. doi: 10.1080/15287399509532035. [DOI] [PubMed] [Google Scholar]
- Noshiro M, Lakso M, Kawajiri K, Negishi M. Rip locus: Regulation of female-specific isozyme (I-P-450(16α) of testosterone 16α-hydroxylase in mouse liver, chromosome localization, and cloning of P-450 cDNA. Biochemistry. 1988;27:6434–6443. doi: 10.1021/bi00417a035. [DOI] [PubMed] [Google Scholar]
- Park Y, Li H, Kemper B. Phenobarbital induction mediated by a distal CYP2B2 sequence in rat liver transiently transfected in situ. J Biol Chem. 1996;271:23725–23728. doi: 10.1074/jbc.271.39.23725. [DOI] [PubMed] [Google Scholar]
- Piscitelli SC, Burstein AH, Chaitt D, Alfaro RM, Falloon J. Indinavir concentrations and St John's wort. Lancet. 2000;355:547–548. doi: 10.1016/S0140-6736(99)05712-8. [DOI] [PubMed] [Google Scholar]
- Ramsden R, Sommer KM, Omiecinski CJ. Phenobarbital induction and tissue-specific expression of the rat CYP2B2 gene in transgenic mice. J Biol Chem. 1993;29:21722–21726. [PubMed] [Google Scholar]
- Ruschitzka F, Meier PJ, Turina M, Luscher TF, Noll G. Acute heart transplant rejection due to Saint John's wort. Lancet. 2000;355:548–549. doi: 10.1016/S0140-6736(99)05467-7. [DOI] [PubMed] [Google Scholar]
- Savas U, Griffin KJ, Johnson EF. Molecular mechanisms of cytochrome P-450 induction by xenobiotics: an expanded role for nuclear hormone receptors. Mol Pharmacol. 1999;56:851–857. doi: 10.1124/mol.56.5.851. [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Li D, Omiecinski CJ, Muller-Eberhard U, Kleinman H, Elswick B, Guzelian PS. Regulation of gene expression in adult rathepatocytes cultured on extracellular matrix. J Cell Physiol. 1988;134:309–323. doi: 10.1002/jcp.1041340302. [DOI] [PubMed] [Google Scholar]
- Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Methods Enzymol. 1996;272:388–400. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Trottier E, Belzil A, Stoltz C, Anderson A. Localization of a phenobarbital-responsive element (PBRE) in the 5'-flanking region of the rat CYP2B2 gene. Gene. 1995;158:263–268. doi: 10.1016/0378-1119(94)00916-g. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound1,4-Bis[2–(3,5-Dichloropyridyloxy)]Benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ, Azaroff L. Phenobarbital induction of cytochrome P-450 gene expression. Biochem J. 1992;281:577–582. doi: 10.1042/bj2810577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- Whitlock JP, Okino ST, Dong L, Ko HP, Clarke-Katzenberg R, Ma Q, Li H. Induction of cytochrome P4501A1: A model for analyzing mammalian gene transcription. FASEB J. 1996;10:809–818. doi: 10.1096/fasebj.10.8.8666157. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes & Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Xie W, Chow LT, Paterson AJ, Chin E, Kudlow JE. Conditional expression of erbB2 oncogene in transgenic mice elicits striking hyperplasia in stratified epithelia, and up-regulation of TGFα expression. Oncogene. 1999;18:3593–3607. doi: 10.1038/sj.onc.1202673. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- Zhou HH, Wilkinson GR. Induction of polymorphic 4'-hydroxylation of S-mephenytoin by rifampicin. Br J Clin Pharmacol. 1990;30:471–475. doi: 10.1111/j.1365-2125.1990.tb03799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]