Abstract
Wnt target gene activation in C. elegans requires simultaneous elevation of β-catenin/SYS-1 and reduction of TCF/POP-1 nuclear levels within the same signal-responsive cell. SYS-1 binds to the conserved N-terminal β-catenin-binding domain (CBD) of POP-1 and functions as a transcriptional co-activator. Phosphorylation of POP-1 by LIT-1, the C. elegans Nemo-like kinase homolog, promotes POP-1 nuclear export and is the main mechanism by which POP-1 nuclear levels are lowered. We present a mechanism whereby SYS-1 and POP-1 nuclear levels are regulated in opposite directions, despite the fact that the two proteins physically interact. We show that the C terminus of POP-1 is essential for LIT-1 phosphorylation and is specifically bound by the diverged β-catenin WRM-1. WRM-1 does not bind to the CBD of POP-1, nor does SYS-1 bind to the C-terminal domain. Furthermore, binding of WRM-1 to the POP-1 C terminus is mutually inhibitory with SYS-1 binding at the CBD. Computer modeling provides a structural explanation for the specificity in WRM-1 and SYS-1 binding to POP-1. Finally, WRM-1 exhibits two independent and distinct molecular functions that are novel for β-catenins: WRM-1 serves both as the substrate-binding subunit and an obligate regulatory subunit for the LIT-1 kinase. Mutual inhibitory binding would result in two populations of POP-1: one bound by WRM-1 that is LIT-1 phosphorylated and exported from the nucleus, and another, bound by SYS-1, that remains in the nucleus and transcriptionally activates Wnt target genes. These studies could provide novel insights into cancers arising from aberrant Wnt activation.
Keywords: A-P polarity, C. elegans, NLK/LIT-1, TCF protein, Wnt signaling, β-Catenin
INTRODUCTION
Wnt signaling stabilizes β-catenin, which binds to TCF/LEF proteins, converting them from transcriptional repressors to activators of Wnt target genes (Bienz, 1998; Brantjes et al., 2002). In addition to β-catenin levels, nuclear levels of TCF proteins are also crucial determinants of Wnt signal strength, as they exhibit repressive function in the absence of β-catenin (Cavallo et al., 1998; Parker et al., 2007). It has been shown recently that selective nuclear export of a repressive TCF isoform but not of an activating isoform underlies aberrant activation of Wnt target genes in human colon cancer cells (Najdi et al., 2009). It is therefore important to understand how the balance between the nuclear levels of β-catenin and TCF is regulated, both as it relates to normal development as well as cancer (Clevers, 2006).
Regulation of Wnt signal strength through simultaneous control of both β-catenin and TCF levels is best exemplified in specification of the endoderm precursor in C. elegans embryos. Signal-induced elevation of co-activator β-catenin (SYS-1) levels and reduction of the single TCF protein (POP-1) within the same blastomere are both required for specification of endoderm fate (Huang et al., 2007; Meneghini et al., 1999; Phillips et al., 2007; Rocheleau et al., 1997; Shetty et al., 2005; Shin et al., 1999; Thorpe et al., 1997). In the four-cell C. elegans embryo, a signal from blastomere P2 to its neighbor, EMS, is required to specify E, the posterior daughter of EMS, as the sole founder for the entire endoderm (gut) (Fig. 1A,B) (Goldstein, 1992). In the absence of this P2-to-EMS signal, the E blastomere adopts the fate of its anterior sister, MS, and the affected embryo lacks all endoderm. Genetic and molecular analyses have identified the Wnt, MAP kinase and SRC tyrosine kinase signaling pathways as being crucial for the specification of E as the endoderm precursor (Bei et al., 2002; Meneghini et al., 1999; Rocheleau et al., 1997; Rocheleau et al., 1999; Shin et al., 1999; Thorpe et al., 1997). Individual mutations in most genes in these pathways result in partial penetrance for the lack of endoderm phenotype. Penetrance for the endoderm defect is enhanced when combining mutations in different pathways, suggesting that they function in parallel to specify endoderm (Bei et al., 2002; Rocheleau et al., 1997; Shin et al., 1999; Thorpe et al., 1997).
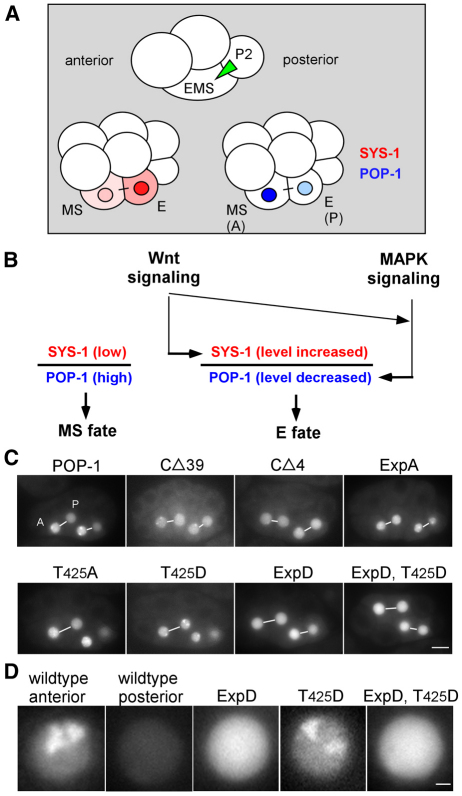
Fig. 1.
The POP-1 C-terminal domain is required for nuclear A-P symmetry. (A) Cartoon drawings of four-cell and eight-cell embryos, highlighting the P2-to-EMS signal (green triangle), and localization in MS and E blastomeres of SYS-1 (red) and POP-1 (blue). (B) Wnt and MAPK signal regulation of SYS-1 and POP-1 levels in MS and E. (C) Fluorescence in EMS lineage of GFP-tagged wild-type POP-1 and the indicated POP-1 mutants at a stage with two MS daughters (MSa, MSp; left-most pair) and two E daughters (Ea, Ep). A-P sisters in the same focal plane are joined by a white line. Embryos are oriented with anterior towards the left. The posterior sister of the posterior pair for embryos labeled T425A and T425D is not focused in the focal plane shown. (D) Higher magnification view of GFP fluorescence in typical wild-type anterior and posterior nuclei, compared with typical nuclei from the three indicated GFP-tagged POP-1 variants. Note the puncta observed in the wild-type anterior nucleus and the T425D nucleus. Scale bars: 10 γm in C; 1 γm in D.
Nuclear export is the major mechanism by which nuclear POP-1 levels are reduced in the E blastomere (Lo et al., 2004; Rocheleau et al., 1999). The MAP kinase LIT-1, the C. elegans NLK homolog, phosphorylates POP-1, its only known substrate, promoting its nuclear export (Lo et al., 2004; Rocheleau et al., 1999). We identified a cluster of five LIT-1 phosphorylation sites that are essential for POP-1 nuclear export (Lo et al., 2004).
The single vertebrate β-catenin is a multifunctional protein and a key regulator in many important biological processes (Harris and Peifer, 2005; Xu and Kimelman, 2007). C. elegans has four genes encoding diverged β-catenins: SYS-1, BAR-1, HMP-2 and WRM-1 (Costa et al., 1998; Eisenmann et al., 1998; Kidd et al., 2005; Rocheleau et al., 1997). SYS-1, BAR-1 and HMP-2 each perform a subset of the functions ascribed to the single β-catenin in vertebrates (Costa et al., 1998; Eisenmann et al., 1998; Kidd et al., 2005; Korswagen et al., 2000). Both BAR-1 and SYS-1 bind to the CBD of POP-1 and function solely as transcriptional co-activators of Wnt target genes at different times during development (Huang et al., 2007; Kidd et al., 2005; Korswagen et al., 2000; Natarajan et al., 2001; Phillips et al., 2007). HMP-2 does not bind to the CBD of POP-1, nor does it activate Wnt reporters in tissue culture cells (Korswagen et al., 2000; Natarajan et al., 2001). Instead, HMP-2 binds to α-catenin and cadherin, and functions in cell adhesion (Costa et al., 1998; Korswagen et al., 2000). However, recent results suggest that HMP-2 may also function in endoderm specification, although the molecular mechanism remains unclear (Putzke and Rothman, 2010; Sumiyoshi et al., 2011). The molecular function of the fourth β-catenin, WRM-1, has remained imprecise. Like HMP-2, WRM-1 does not bind to the CBD of POP-1 or activate Wnt reporters in tissue culture cells (Korswagen et al., 2000; Natarajan et al., 2001; Rocheleau et al., 1999). Nor does WRM-1 appear to function in cell adhesion (Korswagen et al., 2000).
Nevertheless, WRM-1 function is clearly required for endoderm specification through an intimate association with LIT-1 kinase activity. First, WRM-1 is required for autophosphorylation of LIT-1 when both proteins are expressed in mammalian tissue culture cells (Rocheleau et al., 1999). Second, WRM-1 is required for all LIT-1-mediated POP-1 phosphorylation, both in vitro and in embryos (Lo et al., 2004; Rocheleau et al., 1999). Finally, WRM-1 has been shown to form a stable complex with LIT-1 (Rocheleau et al., 1999).
The regulation of SYS-1 and POP-1 levels in opposite directions in the E nucleus, a requirement for driving transcriptional activation of endoderm genes (Huang et al., 2007), is problematic as these two proteins physically interact. That is, how do SYS-1 levels increase in the E nucleus when its binding partner, POP-1, is simultaneously being exported out of the nucleus? That this happens suggests either that SYS-1 can distinguish and selectively bind to a pool of POP-1 that is destined to stay in the nucleus, or that LIT-1 phosphorylation and nuclear export occurs only with POP-1 that is not bound by SYS-1.
Here, we show that WRM-1 binds to POP-1 via a newly identified C-terminal domain of POP-1, which is essential for all LIT-1/WRM-1-mediated phosphorylation of POP-1. We show that WRM-1 functions as both the substrate-binding subunit, and, independently, as a regulatory subunit for the LIT-1 kinase. We present evidence that the C-terminal domain of POP-1 bears sequence and structural similarity to the conserved N-terminal CBD. Structural modeling suggests that SYS-1 and WRM-1 bind to opposite termini of POP-1 via distinct structural modules. Most importantly, we show that the binding of these two β-catenins to POP-1 is mutually inhibitory, providing a molecular mechanism by which levels of SYS-1 and POP-1 can be regulated in opposite directions in Wnt signal-receiving cells. This results in two populations of POP-1: one, bound by WRM-1 but not bound by SYS-1, that is exported from the E nucleus; and another, bound by SYS-1 but not WRM-1, that remains in the nucleus and transcriptionally activates Wnt target genes.
MATERIALS AND METHODS
Strains
N2 was used as the wild-type strain. Genetic markers used are: LGI, pop-1(zu189), dpy-5(e61), hT1(I;V); LGIII, unc-119(ed3). All transgenic strains used were generated by injection and are non-integrated lines.
Plasmid construction
All expression clones used in this study were generated using Gateway cloning technology (Invitrogen). Mutations were generated with the QuikChange Site-Directed Mutagenesis Kit (Stratagene) or by PCR using Phusion polymerase (New England Biolabs). All clones expressed in HeLa cells were driven by the CMV promoter, whereas those expressed in embryos were driven by the med-1 promoter. Unless specified, all tags were added at the N terminus, although fusion to the C-terminus resulted in the same results for both WRM-1 and LIT-1. The FKBP (108 amino acids) and FRB (97 amino acids) domains were amplified from plasmids γ-AP-1-FKBP and Mito-EYFP-FRB (Robinson et al., 2010), respectively, and inserted into the appropriate GW expression clone. Expression clones used are listed in Table 1. We refer here to the POP-1 amino acid residue number according to the Wormbase entry, which is shortened by one methionine residue at the N terminus. D8, S117 and S126 were previously referred to as D9, S118 and S127.
Table 1.
Plasmids used in this study
Analysis of embryos and imaging
Imaging of live embryos was as described previously (Huang et al., 2007). Assay for rescue of the MS defect in pop-1 mutant embryos was as described previously (Gay et al., 2003; Lo et al., 2004). The formation of intestine was assayed with both DIC and polarizing optics, and that for pharyngeal tissues was scored by DIC.
Co-immunoprecipitation and western blots
HeLa cells were cultured in 100 mm petri dishes in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. Cells were transfected at ∼80-90% confluency with up to 12 γg of total DNA using Turbofect transfection reagent (Fermentas). Twenty-four hours post-transfection, cells were harvested in CelLytic M cell lysis reagent (Sigma) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Pierce) and centrifuged at ∼16,000 g for 10 minutes. The resulting supernatants were either used for immunoprecipitation or boiled in SDS sample buffer. For immunoprecipitation, anti-POP-1 (94I) plus protein A sepharose, anti-Myc agarose or anti-FLAG agarose, was added and incubated at 4°C for 2-3 hours. Beads were washed three times with IP wash buffer [50 mM HEPES (pH 7.4), 250 mM NaCl, 1 mM EDTA, 0.1% NP-40] and subjected to SDS-PAGE and western blot analyses. Rapamycin (Sigma, 300 nM) was added to the culture medium 24 hours post-transfection for the indicated time periods.
Embryo extract preparation, immunoprecipitation using the camelid anti-GFP antibody (GFP-Trap, Chromotek) and western blot analyses were performed as described previously (Lo et al., 2004). Antibodies used in western blots include: 94I (Lin et al., 1995) at 1:2000, anti-S117-P and anti-S126-P at 1:500 (Lo et al., 2004), anti-c-Myc (9E10, Santa Cruz) at 1:5000, anti-FLAG (M2, Sigma) at 1:2000 and anti-HA (3F10, Roche) at 1:1000. Secondary antibodies used were: donkey anti-rabbit IgG-HRP (GE Healthcare), goat anti-mouse IgG1-HRP (Santa Cruz), and goat anti-rat IgG-HRP (Santa Cruz), all at 1:10,000.
Molecular modeling of POP-1/WRM-1 interaction
The WRM-1 ARM repeat structure was predicted using the I-TASSER server at the University of Michigan (Roy et al., 2010; Zhang, 2008). I-TASSER generates a 3D structure from the primary amino acid sequence using multiple threading alignments and repeated assembly simulations. I-TASSER predicted one structure for WRM-1 that was highly preferred versus any of the other possible structure predictions. We then modeled the docking of the POP-1 C-terminal helix onto the ARM repeats 3-8 of WRM-1 using HADDOCK (de Vries et al., 2007; Dominguez et al., 2003), a leading program for modeling protein-protein interactions. HADDOCK generated a single model cluster for the docking between the POP-1 C-terminal helix and WRM-1 ARM repeats 3-8 (HADDOCK score of –105.7 and a cluster size of 198). The interaction in this model buries 1031.5±83.5 Å2 of protein surface, which is typical for a helix-protein docking interaction. This docking model places the POP-1 T425 side chain inside a hydrophobic pocket of the WRM-1 ARM repeat groove.
RESULTS
The C-terminal 39 amino acids of POP-1 are required for A-P asymmetry
During deletion and mutational analyses of POP-1, we identified the very C-terminal domain, amino acids 399-437, as being required for POP-1 A-P nuclear asymmetry. Wild-type GFP::POP-1 expressed from the med-1 promoter, which drives transgene expression specifically in the EMS lineage, recapitulates the POP-1 nuclear asymmetric pattern observed with POP-1 antibody staining (Fig. 1C) (Lin et al., 1995; Maduro et al., 2001). That is, GFP::POP-1 fluorescence levels are higher in the MS nucleus than in the E nucleus, and this asymmetry is reiterated between sisters at each subsequent A-P division. A GFP::POP-1 fusion protein with the last 39 amino acids of POP-1 deleted (POP-1 CΔ39) exhibits high POP-1 levels in both MS and E nuclei, and all subsequent A-P sisters (Fig. 1C). This POP-1 CΔ39 pattern is very similar to that reported earlier for POP-1 bearing non-phosphorylatable mutations at the five LIT-1 sites required for nuclear export (Lo et al., 2004). We will refer to these five export-promoting sites collectively as `Exp sites', and all alanine or all aspartate substitutions at these sites as ExpA and ExpD, respectively. To delineate further the region of the C terminus required for POP-1 A-P asymmetry in vivo, we generated a series of POP-1 deletions that deleted 2, 4, 6, 8 or 10 amino acids, respectively, from the C terminus. We observed that GFP::POP-1 with two amino acids deleted (POP-1 CΔ2) exhibited only a minor defect in A-P asymmetry in vivo (see Fig. S1A in the supplementary material). However, POP-1 CΔ4 exhibited a severe defect in A-P asymmetry, and deleting more than four amino acids completely abolished POP-1 A-P asymmetry (Fig. 1C; see Fig. S1A in the supplementary material).
GFP::POP-1 expressed in pop-1(zu189) mutant embryos rescues the MS defects (100%, n>100), demonstrating repression of Wnt target genes in MS (Gay et al., 2003; Huang et al., 2007; Maduro et al., 2001; Shetty et al., 2005). POP-1 CΔ39, which is similar to POP-1 ExpA (Lo et al., 2004), also fully rescued the MS defects (100%, n=28) in pop-1(zu189) mutant embryos, indicating that the C-terminal 39 amino acids are not required for POP-1 to function as a repressor. We were unable to assay the effect of CΔ39 on POP-1 activating activity in E, which requires lowering of POP-1 levels in the E nucleus.
All single residue or combinatorial mutations in the C-terminal domain tested in vivo produced either no, or only minor, defects in POP-1 A-P asymmetry, with one exception. When threonine 425 was changed to aspartate (T425D), POP-1 asymmetry was completely abolished, similar to POP-1 CΔ39 (Fig. 1C; see Fig. S1A in the supplementary material). This defect in POP-1 asymmetry was not observed when T425 was changed to alanine (T425A). However, changing T425 to another amino acid with a large side chain but no charge, asparagine (T425N), also abolished POP-1 asymmetry in vivo (see Fig. S1B in the supplementary material). This result suggests that an amino acid with a charge or a large side chain at position 425 interferes with regulation of POP-1 A-P asymmetry.
POP-1 T425D is defective in phosphorylation of Exp sites in C. elegans embryos
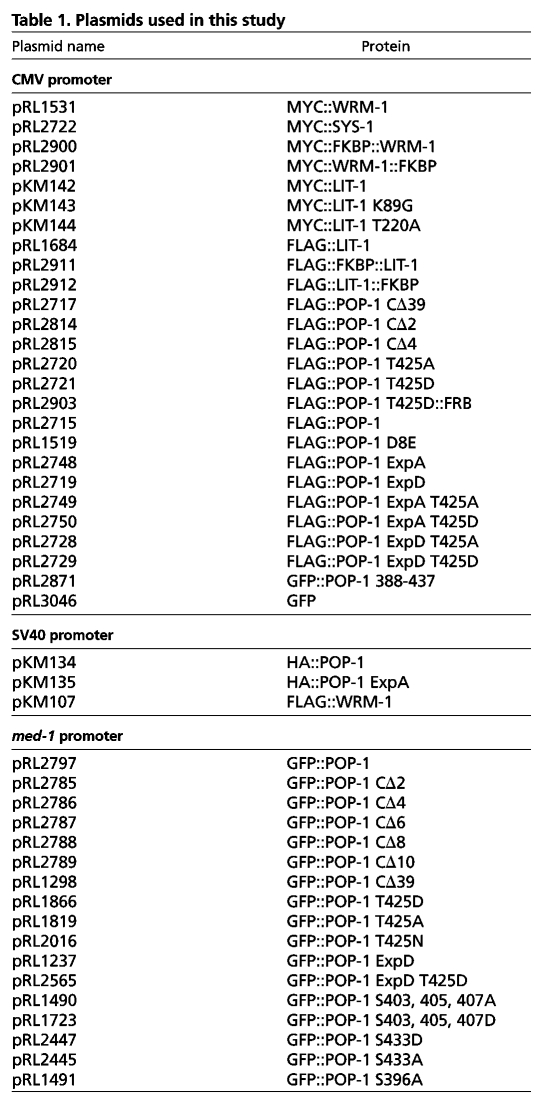
Antibodies against phosphorylated serine 117 and serine 126 showed previously that at least these two out of the five proposed POP-1 Exp residues were phosphorylated in embryos in a LIT-1- and WRM-1-dependent manner (Lo et al., 2004). Using these two phospho-specific antibodies, we investigated whether POP-1 T425D is defective in LIT-1/WRM-1-mediated phosphorylation in embryos. Wild-type GFP::POP-1 or GFP::POP-1 T425D was pulled down from embryonic extracts derived from the respective transgenic worm strains. Western blots were performed with these extract pulldowns using either anti-POP-1 (94I), anti-S117-P or anti-S126-P antibodies. GFP::POP-1 T425D migrates faster than GFP::POP-1 following SDS-PAGE when assayed with 94I, consistent with reduced phosphorylation for GFP::POP-1 T425D (Fig. 2A). We detected no phosphorylation at either S117 or S126 for GFP::POP-1 T425D, in stark contrast to the readily observed phosphorylation for wild-type GFP::POP-1 at these two residues (Fig. 2A). This result suggests that the C-terminal domain of POP-1 is important for LIT-1 phosphorylation.
Fig. 2.
The POP-1 C-terminal domain is required for LIT-1/WRM-1-mediated phosphorylation. (A) POP-1 T425D expressed in embryos is not phosphorylated at S117 or S126. Pulldowns with GFP-Trap were performed with extracts from worm embryos expressing either GFP-tagged wild-type POP-1 or GFP-tagged POP-1 T425D, and western blots probed with anti-POP-1 (94I), anti-POP-1 S117-P or anti-POP-1 S126-P. (B) Phosphorylation of POP-1 by the WRM-1/LIT-1 kinase complex in HeLa cells. POP-1 or the indicated POP-1 variants were expressed in HeLa cells either alone (top panel) or together with WRM-1 and LIT-1 (lower panel). Lines and curly brackets indicate baseline POP-1 phosphorylation and the varying degrees of POP-1 phosphorylation, respectively. Asterisks indicate POP-1 CΔ39.
We next examined the epistatic relationship between the T425D and ExpD mutations in embryos. If T425D prevents phosphorylation of POP-1 at the Exp sites in vivo, then any phenotype(s) caused by phosphomimicking mutations at the Exp sites (ExpD) should be epistatic to the phenotype(s) caused by T425D. That is, the POP-1 ExpD T425D phenotype should resemble the POP-1 ExpD phenotype. This analysis is possible because we are able to distinguish between the two mutant phenotypes. POP-1 T425D exhibits phenotypes that are normally associated with POP-1 in the anterior blastomere, a population presumably non- or hypo-phosphorylated by LIT-1, whereas POP-1 ExpD exhibits phenotypes characteristic of POP-1 in posterior cells, a population presumably phosphorylated by LIT-1. First, GFP::POP-1, and, to a lesser extent, endogenous POP-1, form nuclear puncta that are detected only in anterior nuclei, and lack of puncta in posterior nuclei depends on WRM-1 activity (Maduro et al., 2001) (S.H. and R.L., unpublished). POP-1 T425D exhibits prominent puncta in both anterior and posterior nuclei, whereas POP-1 ExpD forms no nuclear puncta (Fig. 1C,D). Second, POP-1 T425D rescues the MS defect in 78% (n=27) of pop-1(zu189) embryos examined, whereas POP-1 ExpD rescues only 41% (n=22) of pop-1(zu189) embryos. The double mutant, POP-1 ExpD T425D, does not form nuclear puncta and has a low rescuing activity (43%, n=44), similar to POP-1 ExpD alone (Fig. 1C,D). This result argues that the T425D phenotype results from defective LIT-1/WRM-1-mediated phosphorylation of POP-1 in vivo.
The POP-1 C-terminal domain is required for all LIT-1/WRM-1-mediated phosphorylation in tissue culture cells
LIT-1 phosphorylates POP-1 at other sites in addition to the five Exp sites. Whereas POP-1 isolated from wrm-1(–) or lit-1(–) extracts has no reactivity against the two Exp phospho-specific antibodies, it remains partially phosphorylated based on its retarded migration following SDS-PAGE (Lo et al., 2004). In addition, POP-1 ExpA undergoes LIT-1/WRM-1-dependent phosphorylation when expressed in mammalian tissue culture cells (Fig. 2B). We examined whether the C-terminal domain is required for all LIT-1/WRM-1-mediated phosphorylation. Wild-type POP-1 was phosphorylated following transfection into mammalian tissue culture cells only if both LIT-1 and WRM-1 were co-transfected (Rocheleau et al., 1999). POP-1 CΔ39, POP-1 CΔ4 and POP-1 T425D were either very poorly phosphorylated or not phosphorylated at all in tissue culture cells when LIT-1 and WRM-1 were co-transfected (Fig. 2B). Phosphorylation of POP-1 CΔ2 by LIT-1 and WRM-1 was also reduced (not shown).
Although POP-1 T425D showed no phosphorylation when co-transfected with WRM-1 and LIT-1, both POP-1 ExpA and ExpD were phosphorylated (Fig. 2B). This suggests that LIT-1/WRM-1 phosphorylates POP-1 at other sites in addition to the Exp sites. Like POP-1 T425D, POP-1 T425D ExpA and POP-1 T425D ExpD were not phosphorylated when co-transfected with LIT-1 and WRM-1, indicating that T425D abolishes all LIT-1/WRM-1-dependent phosphorylation. Together, these results show that the C-terminal domain of POP-1 is required for most, if not all, LIT-1/WRM-1-dependent phosphorylation of POP-1.
WRM-1 binds to POP-1 C-terminal domain and T425D diminishes binding
We considered the possibility that the POP-1 C-terminal domain is required for LIT-1-mediated phosphorylation because it contains the binding site for the LIT-1/WRM-1 kinase complex. Although POP-1 has long been the only known substrate for the LIT-1/WRM-1 kinase, no stable interaction between POP-1 and WRM-1 or LIT-1 has been detected (Korswagen et al., 2000; Natarajan et al., 2001; Rocheleau et al., 1999). We now show that POP-1 and WRM-1 do in fact interact, and that this interaction requires the POP-1 C-terminal domain.
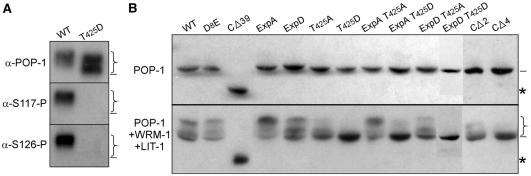
HeLa cells were transfected with plasmids expressing Myc-tagged full-length WRM-1, along with either FLAG-tagged POP-1 or POP-1 variants. Myc-WRM-1 was pulled down with anti-Myc antibody, and any POP-1 in the pulled down product was assayed following SDS-PAGE and western blot using the POP-1 antibody, 94I (Fig. 3A). We could detect a clear and specific interaction between full-length POP-1 and WRM-1. This interaction was abolished with POP-1 CΔ39, and dramatically decreased with POP-1 T425D. The POP-1 C-terminal domain is required specifically for WRM-1 binding, as SYS-1 binding to POP-1 CΔ39 or POP-1 T425D was unchanged. Furthermore, POP-1 carrying a mutation (D8E) that has previously been shown to abolish interaction with SYS-1 (Kidd et al., 2005; Liu et al., 2008; Siegfried and Kimble, 2002) had only a minor, if any, effect on POP-1 binding to WRM-1. Finally, a C-terminal 50 amino acid fragment of POP-1 is capable of binding to WRM-1, albeit less effectively than full-length POP-1 (see Fig. S2 in the supplementary material). These results show that the C-terminal and N-terminal domains of POP-1 are specific binding sites for WRM-1 and SYS-1, respectively. We will refer to the POP-1 C-terminal 39 amino acids as the WRM-1-binding domain (WBD). We detected no interaction between POP-1 and LIT-1 in similar co-immunoprecipitation assays (data not shown; Rocheleau et al., 1999).
Fig. 3.
Binding to WRM-1 requires the last 39 amino acids of POP-1. (A) POP-1 C terminus is required for WRM-1 binding. Co-expression of Flag-tagged POP-1 variants with either Myc-tagged WRM-1 or Myc-tagged SYS-1 in HeLa cells. Immunoprecipitation was performed using anti-Myc antibody and western blots were probed with 94I. Top panels, input; bottom panels, IP. Arrows indicate POP-1; asterisks indicate POP-1Δ39. (B) POP-1 binds to WRM-1 in embryos. Immunoprecipitation was performed using GFP-Trap and wild-type embryo extracts (lane 1), embryo extracts expressing GFP::WRM-1 (lane 2) or using a control antibody (RFP-Trap, which does not crossreact with GFP) and embryo extracts expressing GFP::WRM-1 (lane 3). Western blots were probed with anti-POP-1 (94I) or anti-GFP antibodies. POP-1 was detected only in lane 2 of immunoprecipitated products. (C) Phosphorylation of POP-1 by LIT-1 reduces the POP-1/WRM-1 interaction. FLAG-tagged WRM-1 co-transfected with HA-tagged POP-1 (wild-type or POP-1 ExpA) and Myc-tagged LIT-1 (wild-type or two kinase-dead variants). Input lysates and anti-FLAG pulldown products were probed with anti-HA or anti-FLAG antibodies as indicated. The SDS gel was run for a short period of time such that a migration difference due to phosphorylation was not observed.
Because assaying C. elegans proteins in human tissue culture cells is somewhat artificial, we sought to determine whether WRM-1 associates with POP-1 in the C. elegans embryo. To achieve this, we prepared an embryo extract from a transgenic GFP::WRM-1 strain, pulled down WRM-1, and analyzed for the presence of co-precipitated POP-1 by western blot (Fig. 3B). The POP-1 antibody detected a single band of the correct molecular weight. Control pulldowns, in which either a non-transgenic embryo extract, or a control antibody (RFP-Trap) was used, showed no POP-1. This result indicates that a specific WRM-1/POP-1 interaction occurs in C. elegans embryos.
We also observed that co-transfection of LIT-1 with POP-1 and WRM-1 resulted in reduced amounts of POP-1 being co-immunoprecipitated with WRM-1. This inhibition of the POP-1/WRM-1 interaction by LIT-1 is dependent on LIT-1 phosphorylation of POP-1 at the Exp sites, as it did not occur when kinase-dead versions of LIT-1, or POP-1 ExpA, were substituted for wild-type LIT-1 or POP-1, respectively (Fig. 3C). Therefore, LIT-1 phosphorylation of POP-1 appears to disengage the WRM-1/LIT-1 kinase complex from its phosphorylated substrate.
Together, these results demonstrate that WRM-1 binds to the POP-1 C-terminal WBD, thereby permitting LIT-1 phosphorylation of POP-1. The binding between WRM-1 and POP-1 is weakened once phosphorylation of the POP-1 Exp sites by LIT-1 has occurred.
Mutual inhibition of WRM-1 and SYS-1 binding to the two termini of POP-1
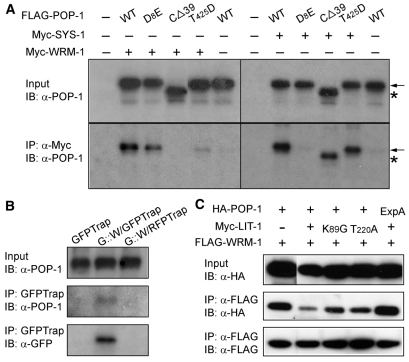
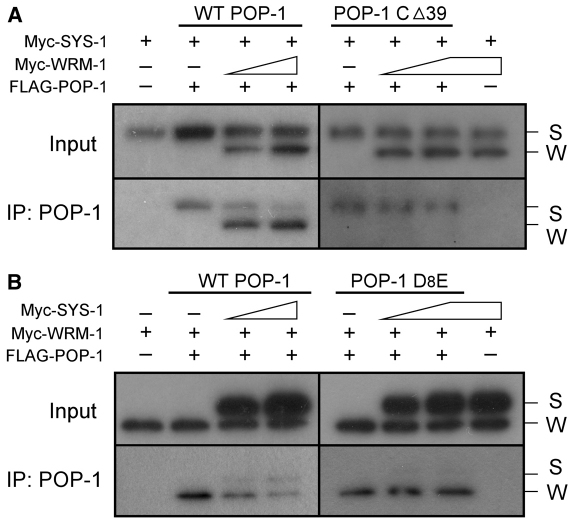
As noted earlier, the binding of both WRM-1 and SYS-1 to a single POP-1 protein would be counterproductive for the simultaneous lowering of POP-1 nuclear levels and elevation of SYS-1 nuclear levels that occurs in signal-responsive cells. We investigated therefore whether the same POP-1 molecule can be bound simultaneously by both SYS-1, via the CBD domain, and WRM-1, via the WBD domain. FLAG-tagged POP-1 was co-expressed in HeLa cells along with both Myc-tagged SYS-1 and Myc-tagged WRM-1. Following FLAG-POP-1 immunoprecipitation, the presence of coimmunoprecipitated Myc-SYS-1 and/or Myc-WRM-1 was analyzed by western blot using anti-Myc antibody (Myc-SYS-1 and Myc-WRM-1 resolve by SDS-PAGE due to their different molecular weights). Under transfection conditions that produced similar levels of SYS-1 and WRM-1 in the lysate, WRM-1 was preferentially co-immunoprecipitated along with POP-1. However, if FLAG-POP-1 CΔ39 was expressed in place of FLAG-POP-1, primarily SYS-1 was co-immunoprecipitated (Fig. 4A). This result suggests that POP-1, given equal amounts of WRM-1 and SYS-1, preferentially binds WRM-1, and that WRM-1 binding via the WBD interferes with SYS-1 binding to the CBD. However, if WRM-1 binding to POP-1 is blocked, SYS-1 then binds POP-1 via the N-terminal CBD. In addition, we also observed that SYS-1 was co-immunoprecipitated with POP-1, at the expense of WRM-1, when SYS-1 levels in the lysate were very high. These observations required SYS-1 binding to the POP-1 CBD, as increasing amounts of SYS-1 did not compete with WRM-1 for binding to POP-1 D8E (Fig. 4B). Taken together, these results demonstrate that, although WRM-1 and SYS-1 bind to different domains of POP-1 and with differing efficiencies, their bindings are nonetheless mutually inhibitory.
Fig. 4.
Binding of SYS-1 and WRM-1 to POP-1 are mutually inhibitory. HeLa cells were co-transfected with plasmids expressing Myc-SYS-1, Myc-WRM-1 and FLAG-POP-1. After pulldown with anti-POP-1 antibody (94I), western blots were probed with anti-Myc antibody. The positions of Myc-tagged SYS-1 (S) and WRM-1 (W) are indicated. (A) Co-transfection of constant amounts of plasmids expressing FLAG-tagged POP-1 (1.5 γg, wild type or POP-1 CΔ39) and Myc-tagged SYS-1 (2 γg), with varying amounts of plasmid expressing Myc-tagged WRM-1 (2 γg and 4 γg, respectively). (B) Co-transfection of constant amounts of plasmids expressing FLAG-tagged POP-1 (1.5 γg, wild type or POP-1 D8E) and Myc-tagged WRM-1 (2 γg), with varying amounts of plasmid expressing Myc-tagged SYS-1 (3 γg and 6 γg, respectively).
POP-1/WRM-1 binding resembles TCF-CBD/β-catenin binding
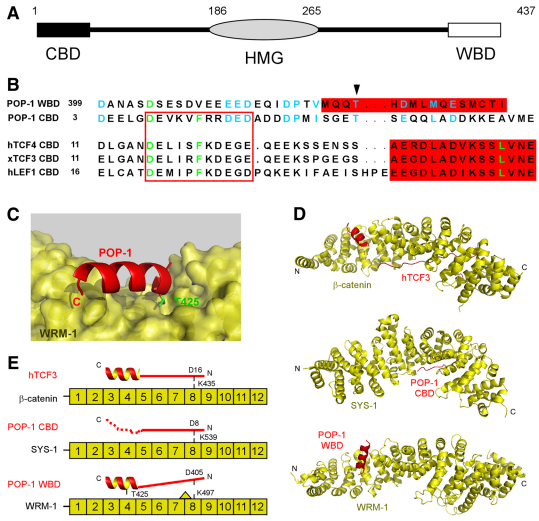
To better understand the specificity of POP-1 binding by the two divergent β-catenins, the physical basis for the POP-1 WBD/WRM-1 interaction, and the mechanism by which T425D abolishes this interaction, we undertook a structural modeling approach. The POP-1 CBD and WBD amino acid sequences exhibit limited, but significant, similarity (Fig. 5A,B), suggesting a possible structural similarity that could extend to the mode of interaction of each of these domains with their respective β-catenins.
Fig. 5.
POP-1/WRM-1 binding resembles TCF/β-catenin and POP-1/SYS-1 binding. (A) Schematic of POP-1 structure highlighting the N-terminal β-catenin-binding domain, CBD (black) and the C-terminal WRM-1-binding domain, WBD (white). Numbers above indicate amino acid residue number. (B) Amino acid sequence alignment of POP-1 CBD and WBD versus the CBD domains from human TCF4, Xenopus TCF3 and human LEF1. The highly conserved D, F and L residues shown previously to play crucial roles in TCF/β-catenin interaction are in green. Amino acids conserved between POP1 WBD and CBD are in blue. The extended region and alpha helix shown to be important in the interaction with β-catenin are boxed in red and shown in red highlight, respectively. The arrowhead indicates POP-1 T425. (C) The highly preferred HADDOCK prediction for the POP-1 WBD/WRM-1 interaction. POP-1 T425 is highlighted with its side chain occupying a pocket in WRM-1 at the proposed binding interface. (D,E) Comparison of TCF/β-catenin, POP-1/SYS-1 and POP-1/WRM-1 interactions (the first two determined by X-ray crystallography, the third predicted by computer modeling). TCF engages β-catenin using both the extended strand (solid line) and the alpha helix subdomains of the CBD, whereas POP-1 engages SYS-1 primarily through the extended strand of the CBD (from crystal structure) and WRM-1 primarily through the alpha helix of the WBD (predicted). (E) The location of amino acid residues and domains known to, or predicted to, play key roles in these interactions are marked. The second half of the POP-1 CBD was reported as unstructured (dotted line). The L491 near WRM-1 K497 (shown by a triangle) was shown to be incompatible with the extended strand of the POP-1 CBD and, presumably, the corresponding region of the POP-1 WBD.
The 12 armadillo repeats in β-catenin form a superhelix dominated by a long positively charged groove (Huber et al., 1997). Previous structural studies have shown that the TCF-CBD domain interacts with the β-catenin superhelix using two structural modules: an extended strand, which sits in the β-catenin groove and interacts with ARM repeats 5-9, primarily via charged amino acids; followed by an alpha helix, which interacts with ARM repeats 3-4, mostly through hydrophobic interactions (Fig. 5D,E; Graham et al., 2001; Graham et al., 2000; Kimelman and Xu, 2006; Xu and Kimelman, 2007). A conserved aspartate residue (D16 in human TCF4) forms a crucial salt bridge with a lysine residue in ARM repeat 8 (the `charged button', K435 in human β-catenin). The POP-1 CBD bound to SYS-1 adopts a conformation similar to that observed for TCF/β-catenin complexes, except that the extended strand is the primary mode of interaction (Liu et al., 2008). The crucial salt bridge between D16 and K435 (corresponding to D8 in POP-1 and K539 in SYS-1) is preserved. It has been suggested that the second half of the POP-1 CBD, which does not appear to form an alpha helix and was unstructured in the POP-1 CBD/SYS-1 crystals, plays little direct role in the interaction with SYS-1 (Liu et al., 2008).
Structure modeling for WRM-1 predicted 12 central ARM repeats and a positively charged groove (Liu et al., 2008). The `charged button' (K497) and several key nearby residues were also predicted to be conserved in WRM-1. However, the presence near K497 of a large side chain from L491 of WRM-1 was predicted to be incompatible with the glutamate at position 9 (E9) of POP-1, which provided a plausible structural explanation for the lack of binding between WRM-1 and the POP-1-CBD. The residue in SYS-1 corresponding to WRM-1 L491 has a small side chain (A533), which, if mutated to leucine, abolished the SYS-1/POP-1 interaction (Liu et al., 2008).
The C-terminal region of the POP-1 WBD (amino acids 422-437) is predicted to form a stable helix. We performed computer modeling to determine whether POP-1 could interact with WRM-1 using this helix (will be termed the C-helix) in a manner similar to the interaction between the second structural module of the TCF-CBD and β-catenin. We first used I-TASSER, which uses multiple threading alignments and repeated assembly simulations, to predict the structure of the WRM-1 ARM repeat domain. We then simulated the docking of the POP-1 C-helix onto WRM-1 ARM repeats 3-9 using HADDOCK, a leading protein-docking program (de Vries et al., 2007; Dominguez et al., 2003). The single docking mode that was clearly most favored by the HADDOCK server places the POP-1 C-helix at ARM repeats 3-4, with the POP-1 T425 side-chain positioned in a pocket formed in WRM-1 at the proposed binding interface (Fig. 5C). A change of T425 to D or N was predicted to significantly compromise the positioning of the amino acid residue 425 side chain in this pocket, and was therefore considered incompatible with this highly preferred docking mode. In addition, T425D and T425N, as well as deletion of four or more C-terminal residues, are predicted to decrease the helical propensity of the POP-1 WBD. Any destabilization of the WBD C-helix is also likely to contribute to reduced WRM-1 binding.
WRM-1 has dual functions in the LIT-1/WRM-1 complex: POP-1 binding and LIT-1 kinase activation
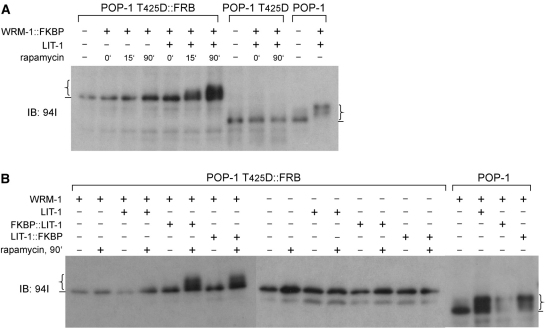
The simplest explanation for the essential function of WRM-1 in Wnt signaling/endoderm specification is that it serves as the substrate-binding subunit of the LIT-1/WRM-1 kinase complex. We used small protein domains as tags to drive interaction between WRM-1 and POP-1 T425D, which would otherwise not normally occur. This allowed us to test whether the WBD, which is absolutely required for LIT-1/WRM-1-mediated phosphorylation of POP-1, can be by-passed if POP-1 and WRM-1 are forced to interact through artificial dimerization domains. FKBP and FRB domain-containing proteins form a heterodimer, but only when the drug rapamycin is present (Ho et al., 1996). We co-expressed FKBP::WRM-1, POP-1 T425D::FRB and LIT-1 in HeLa cells. In the absence of rapamycin, POP-1 T425D::FRB remained unphosphorylated (Fig. 6A). Addition of rapamycin resulted in detectable phosphorylation of POP-1 T425D::FRB in as little as 15 minutes. After 90 minutes of rapamycin treatment, we detected POP-1 T425D::FRB phosphorylation levels similar to those observed with wild-type POP-1 phosphorylated by LIT-1/WRM-1. POP-1 CΔ39::FRB is also phosphorylated in a similar assay but to a lesser extent compared with POP-1 T425D::FRB (not shown). Phosphorylation of POP-1 T425D::FRB is dependent upon FKBP::WRM-1, addition of rapamycin and LIT-1. This result demonstrates that WRM-1 serves as the substrate-binding subunit in the LIT-1 kinase complex.
Fig. 6.
WRM-1 functions in the WRM-1/LIT-1 complex as both substrate (POP-1) binding subunit as well as LIT-1 kinase regulatory subunit. (A,B) Plasmids expressing the indicated proteins were co-transfected into HeLa cells in the combinations shown. Rapamycin was added to the culture medium for the times indicated immediately prior to lysate preparation. Western blots of HeLa cell extracts using anti-POP-1 antibody (94I), showing (A) that WRM-1 binding to POP-1 is required for POP-1 phosphorylation by LIT-1, and (B) that WRM-1 is required to activate LIT-1 kinase activity independently of its binding to POP-1. Lines and curly brackets indicate baseline POP-1 phosphorylation and the varying degrees of POP-1 phosphorylation, respectively.
The inducible FKBP/FRB heterodimerization system also allowed us to ask whether WRM-1 functions solely as the substrate-binding subunit in the LIT-1/WRM-1 kinase complex. We reasoned that if WRM-1 functions solely to bring POP-1 and LIT-1 together, then forced heterodimerization between POP-1 and LIT-1 might make WRM-1 dispensable for POP-1 phosphorylation. When expressed in HeLa cells, POP-1 T425D::FRB was not phosphorylated following rapamycin induction and binding to FKBP::LIT-1 (Fig. 6B). However, addition of non-tagged WRM-1, which is itself unable to bind to POP-1 T425D::FRB, restored the ability of FKBP-LIT-1 to phosphorylate POP-1 T425D::FRB. Together, our results demonstrate that WRM-1 has two distinct and separable functions in the WRM-1/LIT-1 kinase complex that phosphorylates POP-1. First, WRM-1 is the substrate-binding subunit of the complex. Second, WRM-1 also functions as a kinase regulatory subunit, absolutely required for kinase activity, a function completely independent of its ability to bind POP-1.
DISCUSSION
We show here that WRM-1 is an obligate subunit of the LIT-1 kinase, functioning both as a substrate-binding subunit and as a regulatory subunit. WRM-1 binds to POP-1 via a domain at the C terminus of POP-1, the WBD, that is essential for LIT-1 phosphorylation of POP-1 and for POP-1 localization in vivo. We show that the binding of WRM-1 to WBD and the binding of SYS-1 to CBD of POP-1 are mutually inhibitory. Although the WRM-1 interaction is favored, we show that increasing levels of SYS-1 relative to WRM-1 promotes SYS-1 interaction. These results provide a molecular mechanism by which signaling from the P2 blastomere can simultaneously regulate, in opposite directions, the levels of two interacting proteins, β-catenin/SYS-1 and TCF/POP-1, in the same signal-receiving cell.
It has been suggested that the multiple β-catenins in C. elegans derive from a common β-catenin ancestor (Korswagen et al., 2000; Liu et al., 2008; Natarajan et al., 2001). Mutations have then been acquired that result in these duplicated β-catenins retaining only a subset of β-catenin functions. It would appear that BAR-1 and SYS-1 lost the ability to bind to α-catenin and cadherin, while maintaining the ability to bind with the POP-1 N-terminal domain (Korswagen et al., 2000; Liu et al., 2008; Natarajan et al., 2001). On the other hand, HMP-2 appears to have retained the ability to bind to α-catenin and cadherin, but acquired mutations that prevent or weaken its binding to POP-1 (Costa et al., 1998). Recent results suggest a function in endoderm specification for HMP-2 when it is overexpressed or expressed in a sensitized background. HMP-2, therefore, might retain, albeit weakly, an ability to bind to either, or both, ends of POP-1 in vivo (Putzke and Rothman, 2010; Sumiyoshi et al., 2011). During evolution, as WRM-1 diverged and progressively lost the ability to function as a POP-1 co-activator, continued binding to the POP-1 CBD would have competed with SYS-1 and BAR-1 for binding and would have converted POP-1 into a dominant-negative repressor. This is clearly counterproductive for the activation of Wnt target genes in E and specification of the endoderm fate. One possible scenario is that, in C. elegans, co-evolution of POP-1, SYS-1 and WRM-1 resulted in two distinct β-catenin interactions, each favoring a different module of the canonical TCF/β-catenin interaction. By having SYS-1 and WRM-1 bind to two distinct regions of POP-1, this permits separate optimization and regulation of WBD/WRM-1 versus CBD/SYS-1 binding. The similarity, albeit weak, between the primary sequence and secondary structure of the WBD and CBD of POP-1, and the predicted interaction complex of WRM-1/WBD and SYS-1/CBD is very intriguing. It raises the possibility that the WBD arose as a result of a reorganization at the C. elegans pop-1 genomic locus, in which the CBD, contained within a small first exon, was duplicated to the 3′ end of the gene.
Computer modeling suggests that POP-1/SYS-1 and POP-1/WRM-1 binding each employs primarily one of the two structural modules identified in the TCF/β-catenin structure. Optimization of these two distinct interactions would probably involve changes in all four interaction domains (extended strand and alpha helix of POP-1, ARM repeats 3-4 and 5-9 of WRM-1 and SYS-1). The POP-1 CBD maintains binding to SYS-1, primarily through its extended strand, but has apparently lost a propensity towards alpha helix formation (Liu et al., 2008). Specific amino acids in the CBD that are crucial for the interaction of the CBD extended strand with SYS-1 are lacking in the WBD, which could account for the inability of the WBD to bind to SYS-1. WRM-1, unlike SYS-1, has a residue with a large side chain near the conserved K497, which renders it incapable of binding either extended strand (Liu et al., 2008). However, WRM-1 maintains its ability to bind to the WBD, primarily through the WBD alpha helix. SYS-1 appears to have lost the ability to interact with the WBD alpha helix, presumably because it has been optimized for binding to the POP-1 CBD extended strand. This provides a structural basis for the binding of these two very divergent β-catenin-like proteins to distinct domains of POP-1.
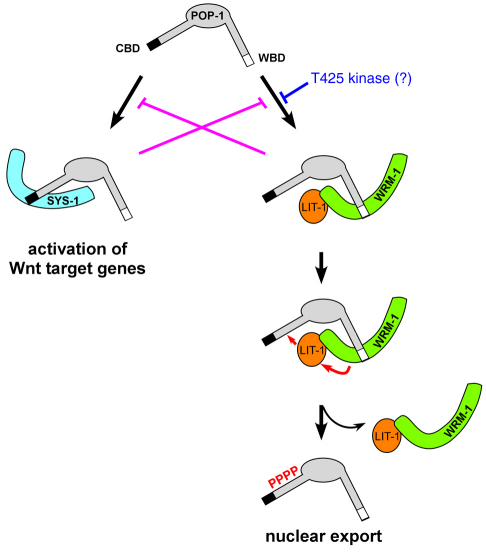
A mechanism must be in place to prevent POP-1 that is bound to SYS-1 from being exported out of the nucleus, or to prevent SYS-1 from binding to POP-1 that is destined to be exported. We propose two mechanisms by which this can be achieved (Fig. 7). First, inhibition of SYS-1 binding to the CBD when the POP-1 WBD is bound by WRM-1, and vice versa This will create two populations of POP-1 in the nucleus of Wnt-responsive cells: one that is bound to WRM-1 and cannot be bound by SYS-1, which is phosphorylated by LIT-1 and exported; and another bound by SYS-1, which is not phosphorylated by LIT-1, and remains in the nucleus functioning as a transcriptional activator. Second, our result that T425D dramatically decreases the WRM-1/WBD interaction suggests that a yet to be identified kinase could regulate the interaction between POP-1 and WRM-1. Phosphorylation at T425 of POP-1 bound to WRM-1 is unlikely, given the predicted location of the T425 side chain in the complex. Phosphorylation of T425 prior to WRM-1 binding would promote SYS-1 binding by generating a pool of POP-1 that would not be destined for export from the nucleus. We currently have no evidence that T425 is phosphorylated in embryos. We believe that, as a result of both of these processes, reception of the signal from P2 results in POP-1 levels being lowered in the E nucleus without jeopardizing the simultaneous increase of SYS-1 in the same nucleus.
Fig. 7.
How reciprocal binding of two β-catenins to POP-1 drives target gene activation versus nuclear export. Proposed mechanisms by which two structurally and functionally distinct populations of POP-1 are generated in the nucleus of the signal-responsive cell. One population of POP-1 binds to SYS-1 leading to activation of Wnt target genes, and the other binds to WRM-1, leading to LIT-1 phosphorylation and POP-1 nuclear export. The first mechanism involves mutual inhibition of POP-1/WRM-1 versus POP-1/SYS-1 interactions. The second mechanism invokes phosphorylation of POP-1 T425 by an as yet unknown kinase, favoring a POP-1/SYS-1 interaction. These two mechanisms are not mutually exclusive.
Our findings suggest, furthermore, that an ancestral single form of β-catenin may have also functioned during phosphorylation of TCF, a function retained only by WRM-1 in C. elegans. The recent reports that β-catenin binding to TCF proteins is required for phosphorylation of TCFs by the HIPK2 kinase, with β-catenin probably serving as a scaffold protein (Hikasa et al., 2010; Hikasa and Sokol, 2011), support this idea. HIPK2 phosphorylation, like NLK phosphorylation, was shown to inhibit TCF binding to DNA. Our findings regarding the function of C. elegans WRM-1 may reveal yet another function associated with the multifunctional single β-catenin in vertebrates and flies.
It is well established that TCF protein levels are crucial for the outcome of Wnt signaling. In Drosophila, the wingless mutant phenotype is partially suppressed by reduction of dTcf levels, or enhanced by overexpression of dTcf (Cavallo et al., 1998). Regulation of nuclear TCF protein levels as a means to modulate TCF transcriptional activity and Wnt signal strength may be more widespread than is currently appreciated. Although an increase in β-catenin levels and simultaneous decrease in nuclear TCF protein levels is likely to increase Wnt signal strength in many situations, it might be difficult to achieve because these two proteins physically interact. Our findings suggest that if Wnt signal promotes nuclear export of TCF proteins in mammalian cells, regardless of the mechanism, it is likely to be mutually exclusive with TCF binding by β-catenin. This can be achieved by selectively exporting a non-β-catenin-binding isoform or requiring mutual exclusivity between export and β-catenin binding. It is interesting to note that HIPK2 phosphorylates all TCF proteins except TCF1, the only TCF family member that was shown to undergo nuclear export in human colon cancer cells, although the reason remains unclear (Hikasa and Sokol, 2011; Najdi et al., 2009). Two TCF proteins are expressed in normal human colon cells, TCF4 and TCF1 (Najdi et al., 2009). The TCF1 isoform expressed in human colon cells lacks the β-catenin-binding domain, and therefore functions as a strong repressor. In human colon cancer cells, in addition to the continuing expression of TCF4 and induced expression of LEF1, TCF1 expression switches from the truncated repressive isoform to the full-length activating isoform. In colon cancer cells, the full-length TCF1, but not the other TCF proteins, is still selectively exported out of the nucleus, which dampens somewhat the magnitude of Wnt signal strength that would result from expression of three activating TCF/LEF isoforms. It would be interesting to know whether ineffective β-catenin binding underlies the lack of HIPK2 phosphorylation and preferential export of only TCF1, and not the other TCF proteins, in colon cancer. Because a number of different cancers can arise from an aberrantly activated Wnt pathway, our findings provide new mechanistic insights into how Wnt activation via independent regulation of nuclear levels of β-catenin and TCF proteins could occur in cancer cells.
Supplementary Material
Acknowledgments
The authors thank the Lin laboratory members for discussions, CGC for strains, Margaret Robinson for plasmids γ-AP-1-FKBP and Mito-EYFP-FRB, and Andrea McReynolds and Jessica Medina for technical support.
Footnotes
Funding
This work was supported by NIH grants (HD37933 and GM84198) to R.L. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.069054/-/DC1
References
- Bei Y., Hogan J., Berkowitz L. A., Soto M., Rocheleau C. E., Pang K. M., Collins J., Mello C. C. (2002). SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev. Cell 3, 113-125 [DOI] [PubMed] [Google Scholar]
- Bienz M. (1998). TCF: transcriptional activator or repressor? Curr. Opin. Cell Biol. 10, 366-372 [DOI] [PubMed] [Google Scholar]
- Brantjes H., Barker N., van Es J., Clevers H. (2002). TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 383, 255-261 [DOI] [PubMed] [Google Scholar]
- Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., Peifer M., Bejsovec A. (1998). Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395, 604-608 [DOI] [PubMed] [Google Scholar]
- Clevers H. (2006). Wnt/beta-catenin signaling in development and disease. Cell 127, 469-480 [DOI] [PubMed] [Google Scholar]
- Costa M., Raich W., Agbunag C., Leung B., Hardin J., Priess J. R. (1998). A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141, 297-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007). HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins: Struc. Funct. Bioinformatic. 69, 726-733 [DOI] [PubMed] [Google Scholar]
- Dominguez C., Boelens R., Bonvin A. M. (2003). HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731-1737 [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Maloof J. N., Simske J. S., Kenyon C., Kim S. K. (1998). The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125, 3667-3680 [DOI] [PubMed] [Google Scholar]
- Gay F., Calvo D., Lo M. C., Ceron J., Maduro M., Lin R., Shi Y. (2003). Acetylation regulates subcellular localization of the Wnt signaling nuclear effector POP-1. Genes Dev. 17, 717-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. (1992). Induction of gut in Caenorhabditis elegans embryos. Nature 357, 255-257 [DOI] [PubMed] [Google Scholar]
- Graham T. A., Weaver C., Mao F., Kimelman D., Xu W. (2000). Crystal structure of a beta-catenin/Tcf complex. Cell 103, 885-896 [DOI] [PubMed] [Google Scholar]
- Graham T. A., Ferkey D. M., Mao F., Kimelman D., Xu W. (2001). Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat. Struct. Biol. 8, 1048-1052 [DOI] [PubMed] [Google Scholar]
- Harris T. J., Peifer M. (2005). Decisions, decisions: beta-catenin chooses between adhesion and transcription. Trends Cell Biol. 15, 234-237 [DOI] [PubMed] [Google Scholar]
- Hikasa H., Sokol S. Y. (2011). Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J. Biol. Chem. 286, 12093-12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikasa H., Ezan J., Itoh K., Li X., Klymkowsky M. W., Sokol S. Y. (2010). Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev. Cell 19, 521-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Biggar S. R., Spencer D. M., Schreiber S. L., Crabtree G. R. (1996). Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature 382, 822-826 [DOI] [PubMed] [Google Scholar]
- Huang S., Shetty P., Robertson S. M., Lin R. (2007). Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development 134, 2685-2695 [DOI] [PubMed] [Google Scholar]
- Huber A. H., Nelson W. J., Weis W. I. (1997). Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90, 871-882 [DOI] [PubMed] [Google Scholar]
- Kidd A. R., 3rd, Miskowski J. A., Siegfried K. R., Sawa H., Kimble J. (2005). A beta-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell 121, 761-772 [DOI] [PubMed] [Google Scholar]
- Kimelman D., Xu W. (2006). beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene 25, 7482-7491 [DOI] [PubMed] [Google Scholar]
- Korswagen H. C., Herman M. A., Clevers H. C. (2000). Distinct beta-catenins mediate adhesion and signalling functions in C. elegans. Nature 406, 527-532 [DOI] [PubMed] [Google Scholar]
- Lin R., Thompson S., Priess J. R. (1995). pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83, 599-609 [DOI] [PubMed] [Google Scholar]
- Liu J., Phillips B. T., Amaya M. F., Kimble J., Xu W. (2008). The C. elegans SYS-1 protein is a bona fide beta-catenin. Dev. Cell 14, 751-761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. C., Gay F., Odom R., Shi Y., Lin R. (2004). Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell 117, 95-106 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Meneghini M. D., Bowerman B., Broitman-Maduro G., Rothman J. H. (2001). Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell 7, 475-485 [DOI] [PubMed] [Google Scholar]
- Meneghini M. D., Ishitani T., Carter J. C., Hisamoto N., Ninomiya-Tsuji J., Thorpe C. J., Hamill D. R., Matsumoto K., Bowerman B. (1999). MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature 399, 793-797 [DOI] [PubMed] [Google Scholar]
- Najdi R., Syed A., Arce L., Theisen H., Ting J. H., Atcha F., Nguyen A. V., Martinez M., Holcombe R. F., Edwards R. A., et al. (2009). A Wnt kinase network alters nuclear localization of TCF-1 in colon cancer. Oncogene 28, 4133-4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan L., Witwer N. E., Eisenmann D. M. (2001). The divergent Caenorhabditis elegans beta-catenin proteins BAR-1, WRM-1 and HMP-2 make distinct protein interactions but retain functional redundancy in vivo. Genetics 159, 159-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. S., Blauwkamp T., Cadigan K. M. (2007). Wnt/beta-catenin-mediated transcriptional regulation. In Wnt Signaling in Embryonic Development (ed. Sokol S.), vol. 17, pp. 1-61 San Diego, CA: Elsevier; [Google Scholar]
- Phillips B. T., Kidd A. R., 3rd, King R., Hardin J., Kimble J. (2007). Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104, 3231-3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzke A. P., Rothman J. H. (2010). Repression of Wnt signaling by a Fer-type nonreceptor tyrosine kinase. Proc. Natl. Acad. Sci. USA 107, 16154-16159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. S., Sahlender D. A., Foster S. D. (2010). Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev. Cell 18, 324-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau C. E., Downs W. D., Lin R., Wittmann C., Bei Y., Cha Y. H., Ali M., Priess J. R., Mello C. C. (1997). Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90, 707-716 [DOI] [PubMed] [Google Scholar]
- Rocheleau C. E., Yasuda J., Shin T. H., Lin R., Sawa H., Okano H., Priess J. R., Davis R. J., Mello C. C. (1999). WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell 97, 717-726 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty P., Lo M. C., Robertson S. M., Lin R. (2005). C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev. Biol. 285, 584-592 [DOI] [PubMed] [Google Scholar]
- Shin T. H., Yasuda J., Rocheleau C. E., Lin R., Soto M., Bei Y., Davis R. J., Mello C. C. (1999). MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol. Cell 4, 275-280 [DOI] [PubMed] [Google Scholar]
- Siegfried K. R., Kimble J. (2002). POP-1 controls axis formation during early gonadogenesis in C. elegans. Development 129, 443-453 [DOI] [PubMed] [Google Scholar]
- Sumiyoshi E., Takahashi S., Obata H., Sugimoto A., Kohara Y. (2011). The beta-catenin HMP-2 functions downstream of Src in parallel with the Wnt pathway in early embryogenesis of C. elegans. Dev. Biol. 355, 302-312 [DOI] [PubMed] [Google Scholar]
- Thorpe C. J., Schlesinger A., Carter J. C., Bowerman B. (1997). Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90, 695-705 [DOI] [PubMed] [Google Scholar]
- Xu W., Kimelman D. (2007). Mechanistic insights from structural studies of beta-catenin and its binding partners. J. Cell Sci. 120, 3337-3344 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.