Abstract
Telomerase is a ribonucleoprotein complex that adds telomeric DNA repeats to the ends of most eukaryotic chromosomes. The reverse transcriptase subunit of telomerase (TERT) differs from retroviral reverse transcriptases in having a long basic amino-terminal extension. We made a large library containing random mutations in the amino terminus of the EST2 gene, which encodes the Saccharomyces cerevisiae TERT, and selected functional alleles by their ability to rescue senescence of telomerase-negative cells. Through analysis of 265 mutations, the amino terminus of Est2p was found to contain at least four essential regions. This domain structure was verified by a combination of deletion and alanine-block mutations. Mutations within two essential domains of the protein reduced RNA binding, suggesting that the amino terminus of Est2p makes important contacts with the intrinsic RNA component of telomerase. A mutant close to the amino terminus retained RNA binding and in vitro enzymatic activity but was defective in vivo, suggesting a role in interaction with other macromolecular components of telomerase.
Keywords: Telomerase, reverse transcriptase, unigenic evolution, mutagenesis, telomerase RNA, Saccharomyces cerevisiae
The ends of most eukaryotic chromosomes are maintained by telomerase, a ribonucleoprotein complex that adds telomeric sequence to the 3′ end of the G-rich strand of the telomere (Greider and Blackburn 1985). The catalytic protein subunit of telomerase has been identified from a number of organisms including human, mouse, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and the ciliates Euplotes aediculatus, Tetrahymena thermophila, and Oxytricha trifallax (Lendvay et al. 1996; Lingner and Cech 1996; Harrington et al. 1997; Kilian et al. 1997; Lingner et al. 1997b; Meyerson et al. 1997; Nakamura et al. 1997; Bryan et al. 1998; Collins and Gandhi 1998; Greenberg et al. 1998). These related proteins are collectively known as TERTs (telomerase reverse transcriptases) because they show sequence homology to the reverse transcriptase (RT) family of DNA polymerases (Lingner et al. 1997b), including a triad of essential aspartates that participate in metal binding and are absolutely required for RT catalytic activity (Joyce and Steitz 1994). S. cerevisiae TERT (Est2p) was originally identified genetically as a mutation causing telomere shortening and senescence, a delayed lethal phenotype in which cells cease to grow after 50–75 generations (Lendvay et al. 1996). As predicted, mutations of the conserved aspartates of Est2p eliminate telomerase activity in vivo and in vitro (Lingner et al. 1997b). Unlike viral RTs, telomerase contains an intrinsic RNA molecule that serves as the template for addition of telomere sequences (Greider and Blackburn 1989). The S. cerevisiae telomerase RNA is encoded by the gene TLC1 (Singer and Gottschling 1994).
Although TERT and telomerase RNA comprise the catalytic center of the telomerase ribonucleoprotein particle, other proteins are constitutively or transiently associated with the enzyme (for review, see Nugent and Lundblad 1998; Bryan and Cech 1999). In S. cerevisiae, a genetic approach has been fruitful in identifying proteins that affect telomerase activity. The same genetic screen that revealed EST2 led to additional mutations with telomerase-negative phenotypes in the genes EST1, EST3, and EST4 (Lundblad and Szostak 1989; Lendvay et al. 1996). The est4 allele recovered in this screen was renamed cdc13-2est with the discovery that the mutant was an allele of the cell-division-cycle gene CDC13 (Garvik et al. 1995; Nugent et al. 1996).
Epistasis tests suggest that EST1, EST2, EST3, cdc13-2est, and TLC1 are all involved in a single pathway of telomere maintenance (Lendvay et al. 1996; Lingner et al. 1997a). Although elimination of the template RNA or catalytic subunit of telomerase results in a loss of in vitro telomerase activity, extracts from strains deleted for EST1 or EST3, or strains carrying the cdc13-2est mutation, have normal levels of telomerase activity in vitro (Lingner et al. 1997a). Est1p coimmunoprecipitates TLC1 RNA, suggesting that the protein is either constitutively or transiently associated with telomerase (Nugent and Lundblad 1998). Est1p binds in vitro to G-rich single-stranded telomere sequences in a 3′ end-dependent manner (Virta-Pearlman et al. 1996), suggesting that it may recruit or anchor telomerase to the 3′ end of the chromosome. Such an activity may be dispensable at the high DNA primer concentrations of the in vitro assay. The function of Est3p is unknown, though it shows EST2-dependent association with TLC1 RNA (Nugent and Lundblad 1998). Finally, recent experiments have indicated that telomerase may be processed and assembled in an analogous manner to the small nuclear ribonucleoprotein particles (snRNPs) involved in mRNA splicing. Two Sm proteins (involved in snRNA maturation) are associated with the majority of active telomerase, suggesting (by analogy with the snRNPs) that as many as seven Sm proteins may be integral components of the mature telomerase enzyme (Seto et al. 1999).
Previous work has focused on the similarities between TERT and other reverse transcriptases such as HIV-1 RT. However, the TERTs are larger than HIV-1 RT, primarily because of a substantial amino-terminal basic region that has no obvious homology to other proteins. The amino terminus does contain at least one sequence block that is conserved among TERT homologs—the T motif (Nakamura et al. 1997). Mutations within this motif in human TERT confirmed its important role in telomerase function, perhaps facilitating interaction with telomerase RNA (Weinrich et al. 1997). In this study, we made a combinatorial library of mutations and used an approach called unigenic evolution to explore the domain structure of the amino terminus of EST2. The domains identified by this approach contribute to the ability of Est2p to interact with TLC1 RNA and to maintain telomeres in vivo.
Results
System for unigenic evolution of EST2
To identify essential domains of Est2p, we applied a technique called unigenic evolution (Deminoff et al. 1995). Previous work indicated that statistical analysis of the distribution of mutations within complementing alleles of a gene could accurately distinguish essential and dispensable regions of the encoded protein. The plasmid-borne gene of interest is mutagenized, functional variants are selected by complementation in S. cerevisiae, and the resulting artificial phylogeny of mutant alleles is analyzed. Dispensable protein regions are assumed to tolerate mutations well, whereas essential regions are predicted to contain few or no amino acid substitutions. Hypomutability could arise either by selection against mutations within essential regions or through intrinsic differences in mutation rate. To distinguish these possibilities, the occurrence of missense mutations is normalized to that of silent mutations, because the latter should not be selected against during an assay for protein function.
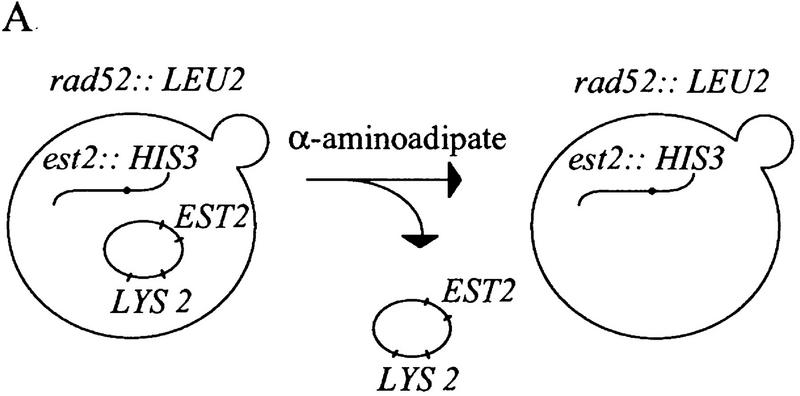
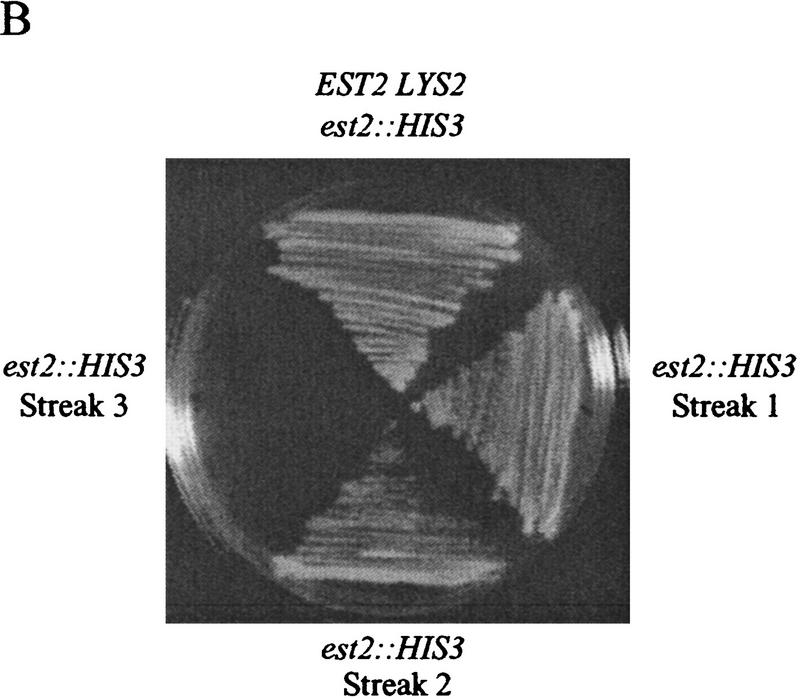
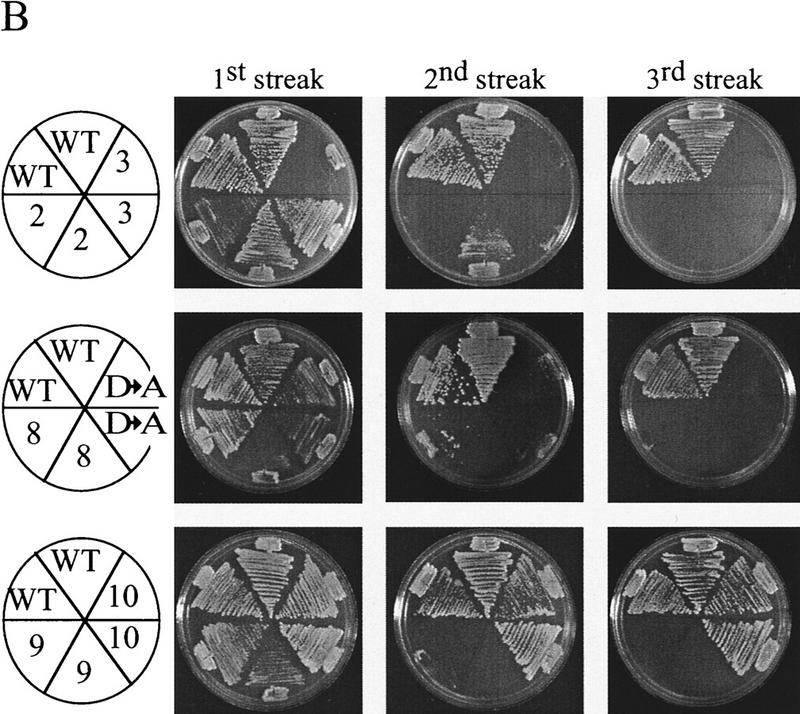
An est2::HIS3 disruption strain was constructed to allow rapid identification of complementing EST2 variants. This strain also carried a disruption of RAD52, because senescence of an est2 strain can be suppressed by a RAD52-dependent amplification of telomeric and subtelomeric sequences (Lundblad and Blackburn 1993; Lendvay et al. 1996). Cell growth was maintained by wild-type EST2 carried on a LYS2 plasmid. Loss of this complementing plasmid was selected by growth on α-aminoadipate, which is toxic in the presence of the LYS2 gene product (Fig. 1A; Winston and Bhattacharjee 1982). The resulting est2::HIS3 rad52::LEU2 strain underwent senescence after ∼50–75 generations, with no visible colonies after the third restreak (Fig. 1B). Senescing est2::HIS3 rad52::LEU2 cells were rescued from senescence by transformation with wild-type EST2, whereas transformation with a deleted version of EST2 lacking most of the open reading frame (est2 ΔHindIII–XbaI) gave few or no transformants (Fig. 1C). Small papillae that did arise could not be restreaked. This selection allowed the isolation of functional EST2 alleles from plasmid-borne libraries containing a large number of EST2 variants.
Figure 1.
(A) Selection scheme for functional EST2 alleles. An est2::HIS3 rad52::LEU2 strain is maintained by EST2 on a LYS2 plasmid. Cells are plated on α-aminoadipate to select for loss of the complementing plasmid. (B) A single lys2 colony is picked from α-aminoadipate and streaked to YPD (streak 1). During successive streaks (streaks 2 and 3), cells undergo senescence. (C) A colony is picked from streak 1 and grown in YPD for transformation. Transformation with wild-type EST2 results in a large number of transformants, whereas transformation with a deleted version of the gene is unable to rescue senescence.
Identification of essential domains in Est2p
Analysis concentrated on the amino-terminal half of EST2 because this region lacks homology with other RTs and may therefore contain functional domains specific to telomerase. Several libraries of multiply mutated est2 alleles, created by mutagenic PCR amplification of 1350 bp in the amino terminus, were transformed into senescing est2::HIS3 rad52::LEU2 cells. Plasmids were isolated from successful transformants and retransformed into senescing cells to confirm rescue. Telomere length was assayed in a sample (11) of the mutants. Telomere length was wild type in five clones, slightly reduced (∼50 bp) in five others, and more severely reduced (∼100 bp) in one clone (data not shown). Although short telomere length is indicative of some reduction in telomerase activity, the remaining activity was sufficient to allow cell growth, confirming that the mutant proteins were capable of performing essential Est2p functions. Alleles that support cell growth are referred to as functional in this study. Because the selection did not require wild-type telomere length, Est2p domains with sublethal effects on telomerase activity were not identified.
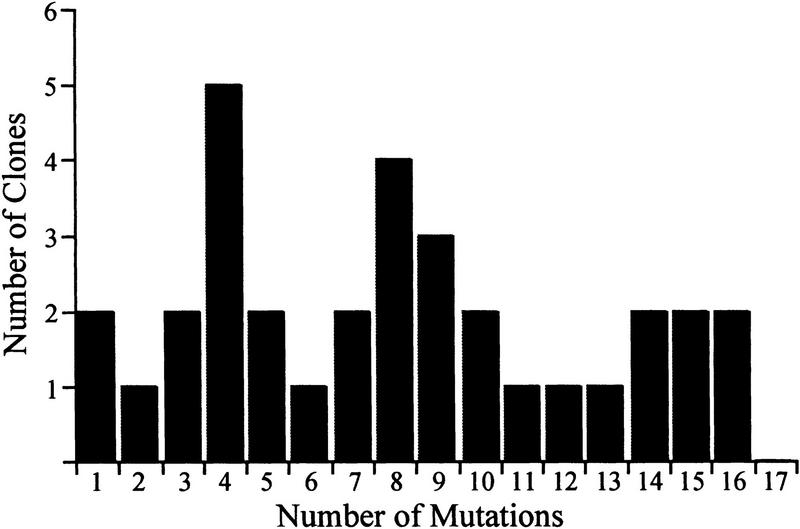
The distribution of mutation number in 33 functional clones is shown in Figure 2. Among these alleles were 265 bp changes, representing 99 silent and 166 missense mutations. The largest number of missense mutations recovered in a single allele was 11. Transversions (purine-to-pyrimidine changes) and transitions (purine-to-purine or pyrimidine-to-pyrimidine changes) were recovered nearly equally, with 47% transversions and 53% transitions. However, among transversions, T-A and A-T changes predominated.
Figure 2.
Mutation number among 33 functional est2 alleles. Mutations were created by random PCR mutagenesis across a 1350-nucleotide region comprising the amino terminus of EST2. The distribution of mutation number in 33 clones is shown.
Mutations from all 33 clones were pooled for statistical analysis. For each codon within the 450-amino-acid region, an expected ratio of silent to missense mutation was calculated based on the sequence of the codon and the transition/transversion ratio in the mutation data set. The actual ratio of silent to missense mutations observed was compared to this expected ratio in a 25-codon sliding window moved downstream one codon at a time (Fig. 3A). Regions with negative mutability values had decreased numbers of missense mutations and were candidates for essential protein domains.
Figure 3.
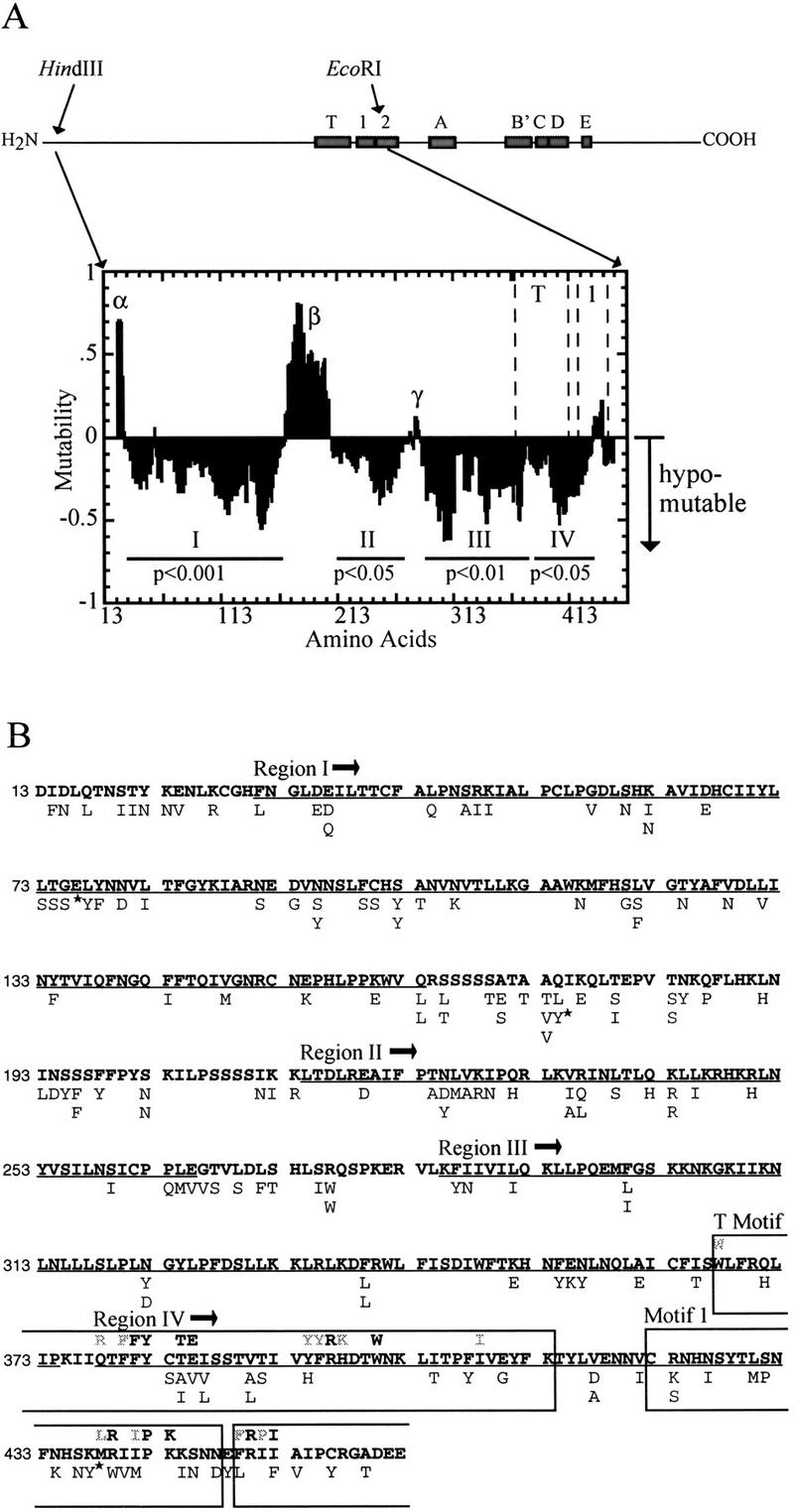
Domain structure of EST2 amino terminus as determined by unigenic evolution. Motif T is telomerase specific, whereas the sequence motifs 1, 2, and A–E are closely related to those of other RTs (Lingner et al. 1997b). Mutations between the HindIII and EcoRI sites were used to derive the mutability map shown below. Amino acids are numbered on the x-axis from the amino terminus. The average observed ratio of missense to silent mutation is compared with the expected ratio (based on the amino acid sequence) over successive 25 amino acid windows (Deminoff et al. 1995). Maximal hypomutability (no missense mutations) is normalized to a value of −1, and maximal hypermutability (all missense mutations) is assigned a value of +1. A value of 0 occurs when the observed and expected values are identical. The normalized mutability value is graphed at the center of the 25-amino-acid window. Regions showing hypomutability (negative values) may be essential for Est2p function. Four hypomutable regions are indicated (Region I, amino acids 31–163; Region II, 214–265; Region III, 285–374; Region IV, 378–432). P values for χ2 analysis of each region are indicated. The χ2 values are: Region I, 14.97; Region II, 4.00; Region III, 6.60; Region IV, 5.58. P values were determined for one degree of freedom (see Deminoff et al. 1995). Three nonessential regions (positive values) are labeled (α, β, γ). (B) Amino acid changes in the 33 functional clones. The 450 amino acids following the HindIII site near the amino terminus of Est2p are shown in standard single amino acid code. Amino acid changes are indicated below each position. Mutations indicated (star) produced a TAA stop codon; suppression of the nonsense mutation by SUP11 carried on a minichromosome in this strain background results in insertion of tyrosine (Y). Regions I–IV as described above are underlined. The boxes indicate (in order from amino to carboxyl terminus): the T motif, RT domain 1, and a portion of RT domain 2. The consensus sequence of each motif is shown above the Est2p amino acid sequence. Residues in bold-face type are invariant. Gray residues are conserved in at least 5 of 7 TERT sequences (Bryan et al. 1998).
As shown in Figure 3A, the amino terminus of EST2 contains several candidates for essential domains (labeled I–IV) that are separated by regions that appear to be dispensable for cell viability. All four hypomutable domains contained significantly fewer missense mutations than expected in the absence of selection as determined by χ2 analysis (Fig. 3A). Region IV overlaps two previously defined domains of the protein. Motif T is a telomerase-specific motif identified through sequence comparison among TERT proteins from diverse species (Nakamura et al. 1997). Several amino acids within this motif have been shown to be essential or important for human telomerase function (Weinrich et al. 1997). Region IV also overlaps with motif 1 of the reverse transcriptase domain. However, unigenic evolution did not cleanly define either motif. This result is not unexpected as motifs T and 1 contain only a small subset of residues that are conserved absolutely. Indeed, our results indicated that a large number of nonconserved residues within these motifs could be mutated in functional Est2p (Fig. 3B).
Three putative nonessential regions were evident from the unigenic evolution map (α, β, and γ; Fig. 3A). Although regions α and β actually contain more missense mutations than expected by chance, when subjected to a χ2 calculation, these regions do not show significant deviation from the expected ratio of silent to missense mutations. The boundary between regions III and IV was not well defined.
Verification of nonessential domains
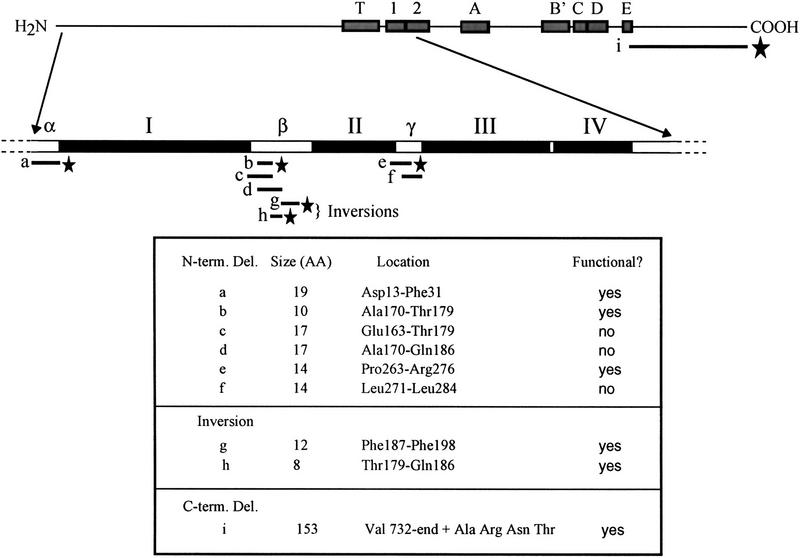
Because unigenic evolution is statistically based, the proposed domains were tested by a more direct approach. Deletions were created by site-directed mutagenesis within putative nonessential regions β and γ on a centromere plasmid bearing EST2 under the control of its endogenous promoter. As shown in Figure 4, plasmid-borne EST2 containing a 10-amino-acid deletion (deletion b) in region β rescued senescing est2::HIS3 rad52::LEU2 cells. However, additional deletions extending either upstream or downstream of deletion b were unable to transform, suggesting that these proteins provided insufficient telomerase function to rescue senescence. As a further test of the contribution of region β to Est2p function, the amino acid sequences in two blocks (g and h, Fig. 4) were inverted. The intention was to disrupt local sequence but maintain any spacing that may be critical for protein folding. Both inversion mutants complemented est2 in the senescence assay. Similarly, deletion e in region γ (Fig. 4) complemented est2, whereas the overlapping deletion f did not. The ability to create functional deletion or inversion mutations in regions β and γ is consistent with the existence of the nonessential domains defined by unigenic evolution.
Figure 4.
Deletion and inversion mutations of Est2p. The essential regions I–IV as defined in Fig. 3 are shown as solid bars. Three nonessential domains defined by unigenic evolution are shown: an amino-terminal, 18-amino-acid region immediately flanking the HindIII site used as the boundary for mutagenesis (region α), a 50-amino-acid region between domains I and II (region β), and a 20-amino-acid region between domains II and III (region γ). Bars below the map delineate the locations of site-specific mutations created in plasmid-borne EST2. Mutations a-f are deletions, mutations g and h are inversions of the amino acid sequence over the indicated region, and mutation i is a carboxy-terminal deletion. Functional alleles are indicated (stars).
To assess the importance of sequences located at the amino and carboxyl termini of EST2, two libraries were constructed to generate deletions at or near the ends of the protein. As described in Materials and Methods, the amino-terminal deletions were slightly internal to the protein, extending inward from a HindIII site located at amino acid position 12. Transformation of these libraries into senescing est2::HIS3 rad52::LEU2 cells resulted in a small number of colonies that contained functional deletion derivatives of EST2. The largest deletions recovered are shown in Figure 4 (deletions a and i). Amino-terminal deletion a extended 19 amino acids from the HindIII site, a position that corresponds closely to the boundary of nonessential region α as defined by unigenic evolution. Restriction analysis of the original library indicated that larger deletions were present but not recovered in yeast, presumably because of their inability to complement the est2 mutation (data not shown).
The EST2 carboxyl terminus appears to be dispensable for survival. As shown in Figure 4, deletion i removes all 153 amino acids carboxy-terminal to the RT domain. Because this deletion removes the endogenous stop codon, four new amino acids are encoded by sequences in the EST2 3′ flanking region before termination occurs (Fig. 4). Surprisingly, approximately half of the region believed to correspond to RT motif E according to sequence alignment (Nakamura et al. 1997) is missing in this mutant. Although growth rate is slow, telomeres reach a stable, short length and senescence does not occur even in the absence of RAD52 (data not shown). Therefore, although the carboxyl terminus contributes to Est2p function, it is not required for cell survival.
Alanine-block mutations in putative essential domains
To further characterize the domains defined by unigenic evolution, 10 alanine-block mutations in which groups of adjacent amino acids were changed to alanine were made in the amino terminus of EST2 (Fig. 5A). An epitope-tagged version of EST2 was created to monitor mutant protein expression. The protein A (ProA) sequence was inserted at a HindIII site near the amino terminus of the protein and the tagged construct was integrated into the yeast genome at the endogenous EST2 locus. Cells containing ProA–Est2p as their sole source of Est2p appeared to grow normally (Fig. 5B) with a barely detectable decrease in telomere size in liquid culture (Fig. 5C, WT lanes), suggesting that the tagged protein retains nearly wild-type function. Although ProA–Est2p cannot be detected in extracts, a clear signal is visible when the protein is immunoprecipitated with IgG Sepharose beads (see Fig. 6A; ProA–Est2p).
Figure 5.
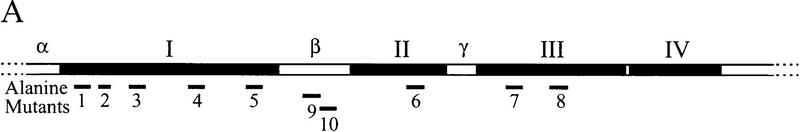
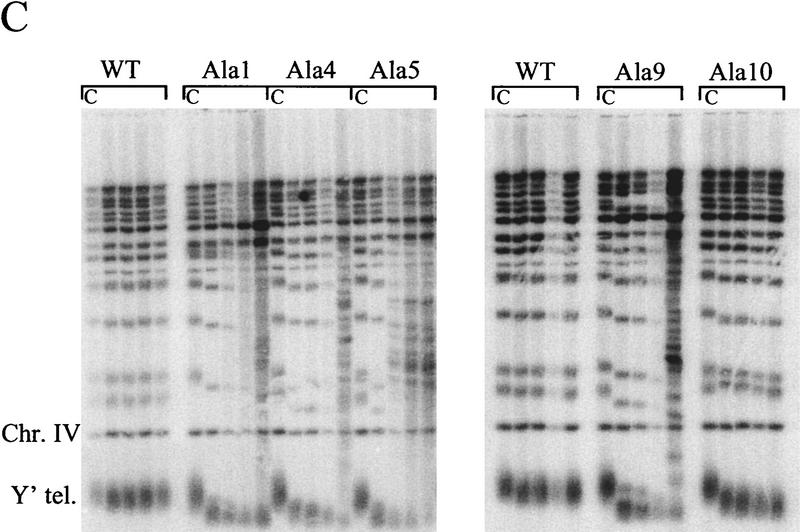
In vivo phenotypes of EST2 alanine-block mutations. (A) The region between the HindIII and EcoRI sites of EST2 is shown along with hypomutable regions I–IV and nonessential regions α, β, and γ. Locations of alanine-block mutations are shown below the map. The following amino acids were substituted with alanine in each mutant: Ala-1, Thr-40–Lys-49; Ala-2, Leu-55–His-61; Ala-3, Leu-73–Leu-82; Ala-4, Leu-110–His-119; Ala-5, Thr-145–Glu-154; Ala-6, Lys-243–Asn-252; Ala-7, Lys-303–Asn-312; Ala-8, Leu-330–Phe-339; Ala-9, Thr-179–Leu-188; Ala-10, His-189–Phe-198. Amino acid numbers refer to untagged Est2p. All mutants are ProA tagged and chromosomally integrated. (B) Senescence phenotype of representative alanine-block mutants. Cells containing the indicated allele in the chromosome and wild-type EST2 on a plasmid were plated on 5-FOA to select for loss of the complementing plasmid. Two single colonies were picked and successively streaked on complete medium to assay senescence. (WT) Wild-type ProA–Est2p. (D → A) Aspartate 670-to-alanine mutation in RT motif C. Alanine-block mutants Ala-1, Ala-4, Ala-5, Ala-6, and Ala-7 had identical senescence phenotypes in this assay (data not shown). (C) Telomere lengths of representative alanine-block mutants. Cells containing the indicated allele in the chromosome and wild-type EST2 on a plasmid were grown to saturation in complete media. DNA was prepared, cleaved with XhoI, and Southern blotted. (C) DNA of these complemented strain. Cells were subsequently plated on 5-FOA to select for loss of the complementing plasmid and a resulting single colony was picked into liquid culture. As each culture reached saturation, it was diluted 1000-fold for four successive serial dilutions. DNA from these cultures was run in the four lanes following lane C. Southern blots were probed for telomere sequences and for a 1.6-kb fragment of chromosome IV that served as an internal loading and size control. Chromosomes containing a Y′ element yield a heterogeneous telomere band as indicated (Y′ tel). Alanine-block mutants Ala-2, Ala-3, Ala-6, Ala-7, and Ala-8 gave telomere loss results similar to mutants Ala-1, Ala-4, Ala-5, and Ala-9 (data not shown).
Figure 6.
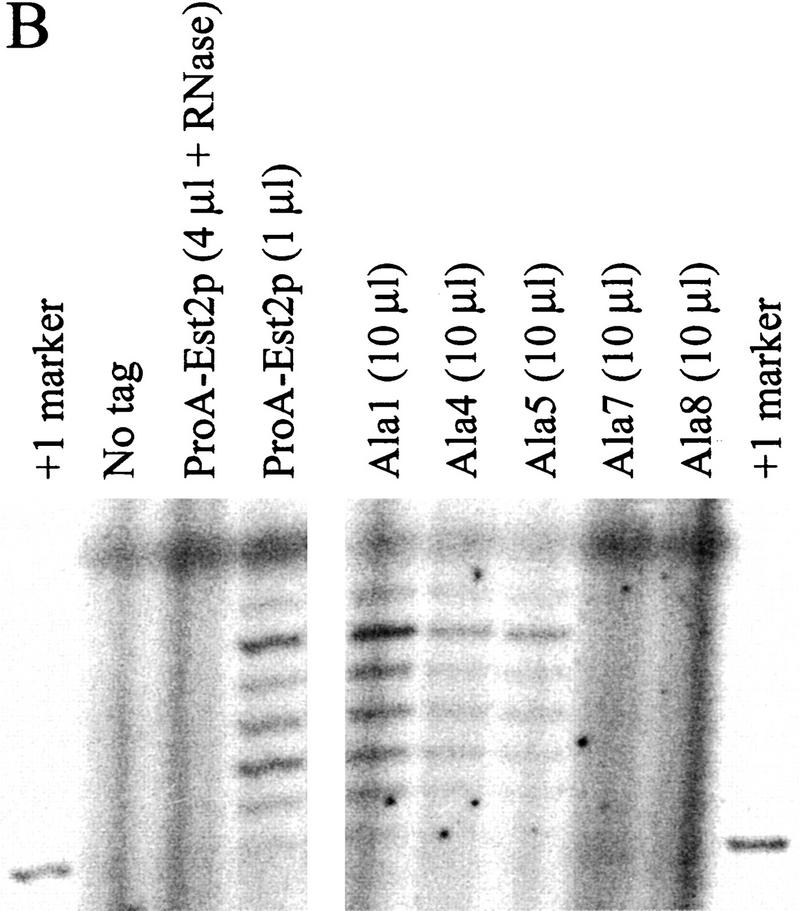
In vitro phenotypes of alanine-block mutants. (A, Top) Northern analysis showing coimmunoprecipitation of TLC1 RNA with Est2p. Whole-cell RNA (10 μg) from wild-type ProA–EST2 cells is run in the right lane. Locations of the polyadenylated (A+) and deadenylated (A−) forms of the TLC1 RNA are indicated. (Bottom) Western analysis of protein levels in strains carrying ProA–tagged wild-type Est2p or alanine-block mutants. Protein was immunoprecipitated from extracts using IgG Sepharose beads and protein from the indicated volume of beads was loaded on the gel. Coomassie staining of the starting extract indicated that protein concentration was approximately equivalent in all cases. (B) In vitro telomerase activity was measured by assaying the ability of immunoprecipitated ProA–Est2p to extend a telomeric primer. The same batch of immunoprecipitation beads was used in this assay and the experiments shown in A. The indicated amount of immunoprecipitation beads was pretreated with water or RNase A and telomerase was allowed to extend a 14-mer telomeric primer in the presence of [α-32P]dGTP. +1 marker is a 15-mer created by end-labeling the 14-mer telomeric oligonucleotide with terminal transferase in the presence of [33P]ddTTP.
Mutations of 10 consecutive amino acids to alanine were created throughout hypomutable regions I, II, and III (Ala1–Ala8; Fig. 5A). Ala2 is slightly smaller and consists of a seven-amino-acid block. An additional mutant was created in which aspartic acid 670 was changed to alanine. This residue is located in RT motif C and is required for catalytic activity and cell viability (Lingner et al. 1997b). For comparison, two additional alanine-block mutations (Ala-9 and Ala-10) were created in nonessential region β (Fig. 5A). All mutations were integrated into the chromosome in a strain bearing ProA–Est2p, and cell viability was maintained by complementation with a URA3-marked centromere plasmid bearing wild-type EST2.
The ability of the alanine-block mutants to maintain cell growth was assayed by plating the mutants on 5-fluoroorotic acid (5-FOA) to select for loss of the EST2 plasmid. Single colonies containing either wild-type ProA–Est2p or tagged mutant versions of the protein were successively streaked on complete medium. Cells bearing wild-type ProA–Est2p or alanine-block mutant Ala-10 (located in nonessential region β) regrew after multiple restreaks (Fig. 5B). However, alanine-block mutants Ala-1-Ala-9 senesced in a manner indistinguishable from that of the catalytic-site mutant (Fig. 5B; summarized in Table 1). The Ala-9 mutant protein was not detectable by Western blot (data not shown), indicating that lack of complementation was due to destabilization of the protein.
Table 1.
Summary of in vivo and in vitro phenotypes of alanine–block mutants
|
|
Locationa
|
Viabilityb
|
Proteinc
|
RNA bindingd
|
In vitro activityd
|
|---|---|---|---|---|---|
| Ala-1 | I | n | + | + | + |
| Ala-2 | I | n | − | n/a | n/a |
| Ala-3 | I | n | − | n/a | n/a |
| Ala-4 | I | n | + | +/− | +/− |
| Ala-5 | I | n | + | +/− | +/− |
| Ala-6 | II | n | − | n/a | n/a |
| Ala-7 | III | n | + | − | − |
| Ala-8 | III | n | + | − | − |
| Ala-9 | β | n | − | n/a | n/a |
| Ala-10 | β | y | + | ND | ND |
Regions shown in Fig. 3.
Ability of alanine-block mutant to support cell growth when integrated at the endogenous locus. (n) Cells undergo senescence; (y) cells grow through multiple restreaks.
Expression levels determined by Western blot. (+) Protein level 10–50% of wild type. (−) Protein undetectable (<5% of wild type).
Coimmunoprecipitation of TLCI RNA with alanine-block mutant proteins and in vitro telomerase activity as assayed by ability of immunoprecipitated proteins to extend a telomeric oligo. (+) Comparable to wild type after adjustment for decreased mutant protein levels. (+/−) Decreased from wild type, but detectable. (−) Not detectable. (n/a) Not applicable. (ND) Not done.
Telomere length in the mutant strains provided a second measure of telomerase function. In contrast to wild-type ProA–Est2p (WT; Fig. 5C), alanine-block mutants Ala-1-Ala-9 underwent dramatic telomere loss during growth on 5-FOA and in the first liquid culture (Fig. 5C). Telomere length in these mutants continued to decrease with successive generations. By the fourth serial dilution, these mutants also showed a shift in the XhoI banding pattern that is indicative of recombination between Y′ elements, a survival mechanism that can occur in senescing cells containing functional RAD52 (Lundblad and Blackburn 1993; Lendvay et al. 1996). In contrast, the Ala-10 mutant, which did not undergo senescence, also did not undergo Y′ recombination. Telomere length was slightly decreased in this mutant after loss of the complementing plasmid but remained stable and was sufficient to maintain essentially normal cell growth.
Protein expression and RNA subunit binding by alanine-block mutants
To analyze expression levels of the mutant proteins, ProA-tagged proteins were precipitated using IgG Sepharose beads from extracts prepared from cells expressing alanine-block mutants 1–10 or wild-type ProA–Est2p. Immunoprecipitated material was Western blotted and probed with rabbit IgG antibody. Expression of mutant proteins Ala-2, Ala-3, Ala-6, and Ala-9 was undetectable (data not shown) and these mutants were not further characterized. The remaining mutants reproducibly showed protein levels reduced 2-to 10-fold from wild-type Est2p (Fig. 6A; data not shown for Ala-10). Immunoprecipitation of Ala-5 protein was inefficient in the experiment shown in Figure 6A; additional immunoprecipitation experiments showed that Ala-5 protein levels were equivalent to those in the Ala-4 mutant (summarized in Table 1). Note that the Ala-10 mutant retained telomerase function even though protein level was decreased 5- to 10-fold from wild type. Therefore, decreased protein levels per se are not responsible for the defects of these alanine-block mutants.
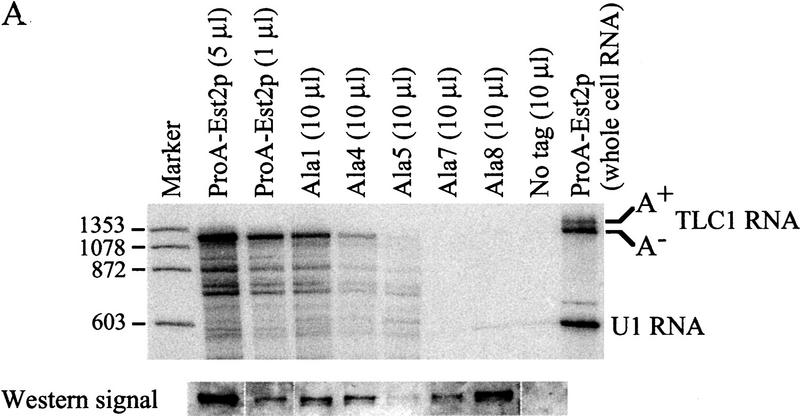
To begin to define the function of the amino-terminal domains of Est2p, the ability of the alanine-block mutant proteins to coimmunoprecipitate TLC1 RNA was assayed. Immunoprecipitated material was phenol extracted and the RNA was precipitated, Northern blotted, and probed for TLC1 RNA and U1 RNA. As a control, whole-cell RNA was prepared from wild-type cells grown to the same density as that used for extract preparation. As shown in the last lane of Figure 6A, TLC1 RNA runs as a doublet. The upper band consists of a polyadenylated form of the RNA, which is thought to be a precursor of the faster migrating, deadenylated form (Chapon et al. 1997; Seto et al. 1999).
TLC1 RNA coimmunoprecipitated with ProA–Est2p but was not detected in immunoprecipitates from an untagged strain (Fig. 6A). The deadenylated form of TLC1 RNA was significantly enriched following immunoprecipitation, suggesting that only the deadenylated RNA is contained in the active telomerase complex. This result supports the proposal that the deadenylated form is the mature form of TLC1 RNA (Chapon et al. 1997). TLC1 RNA coimmunoprecipitated with the Ala-1 mutant at levels ∼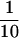 of wild type (cf. 1 μl of ProA–Est2p beads and 10 μl of Ala-1 beads). Because protein levels in this mutant were similarly decreased, the RNA binding ability of the Ala-1 mutant appears to be largely unaffected. The Ala-4 mutant, despite having a protein level similar to that of the Ala-1 mutant, showed decreased RNA binding. RNA binding in the Ala-5 mutant was decreased to the same extent as that of Ala-4 in experiments in which protein levels of the two mutants were similar (data not shown). Most strikingly, mutants Ala-7 and Ala-8 did not detectably bind TLC1 RNA, despite having protein levels comparable to or greater than those of the Ala-1 mutant. Whole-cell RNA was prepared from cells carrying wild-type ProA–Est2p, untagged Est2p, or alanine mutants 1, 4, 5, 7, and 8. The ratio of TLC1 RNA to U1 RNA in all strains was similar (data not shown), indicating that the lack of coimmunoprecipitation was not due to instability of TLC1 RNA in the mutant strains. Furthermore, TLC1 RNA was detected in the starting extract of all strains (data not shown). Interestingly, Northern analysis of RNA present in the supernatant following immunoprecipitation showed that TLC1 RNA is depleted from all of the mutant extracts but not from extract containing untagged Est2p (data not shown). This result suggests that TLC1 RNA is initially bound to the Ala-7 and Ala-8 mutant proteins but that binding is too weak to withstand the subsequent washes.
of wild type (cf. 1 μl of ProA–Est2p beads and 10 μl of Ala-1 beads). Because protein levels in this mutant were similarly decreased, the RNA binding ability of the Ala-1 mutant appears to be largely unaffected. The Ala-4 mutant, despite having a protein level similar to that of the Ala-1 mutant, showed decreased RNA binding. RNA binding in the Ala-5 mutant was decreased to the same extent as that of Ala-4 in experiments in which protein levels of the two mutants were similar (data not shown). Most strikingly, mutants Ala-7 and Ala-8 did not detectably bind TLC1 RNA, despite having protein levels comparable to or greater than those of the Ala-1 mutant. Whole-cell RNA was prepared from cells carrying wild-type ProA–Est2p, untagged Est2p, or alanine mutants 1, 4, 5, 7, and 8. The ratio of TLC1 RNA to U1 RNA in all strains was similar (data not shown), indicating that the lack of coimmunoprecipitation was not due to instability of TLC1 RNA in the mutant strains. Furthermore, TLC1 RNA was detected in the starting extract of all strains (data not shown). Interestingly, Northern analysis of RNA present in the supernatant following immunoprecipitation showed that TLC1 RNA is depleted from all of the mutant extracts but not from extract containing untagged Est2p (data not shown). This result suggests that TLC1 RNA is initially bound to the Ala-7 and Ala-8 mutant proteins but that binding is too weak to withstand the subsequent washes.
In vitro telomerase activity
Some mutants that undergo senescence and telomere shortening (est1, est3, and cdc13-2est) nevertheless display apparently normal levels of in vitro telomerase activity (Cohn and Blackburn 1995; Lingner et al. 1997a). We were interested in the in vitro telomerase activity of the alanine-block mutants, as they do not directly affect the RT domain and therefore may leave Est2p catalytic activity intact. Immunoprecipitated Est2p (the same material analyzed in Fig. 6A) was subjected to an in vitro telomerase assay that monitored the extension of a telomeric DNA primer. As shown in Figure 6B, the ladder of addition products characteristic of yeast telomerase (Cohn and Blackburn 1995; Lingner et al. 1997b; Prescott and Blackburn 1997) was visible when beads containing wild-type ProA–Est2p were used in the assay. As expected, activity was destroyed by pretreatment with RNase A. Furthermore, no activity was detectable on beads prepared from cells containing untagged Est2p. Ala-1 had in vitro telomerase activity that was reduced ∼10-fold from wild type (cf. 1 μl of wild-type ProA–Est2p beads with 10 μl of Ala-1 beads). This decrease in activity is roughly comparable to the reduction in protein levels seen by Western blot and suggests that the enzyme's specific activity is minimally affected by this mutation. Activity in the remaining mutants was reduced or absent as predicted by the ability of the mutants to bind TLC1 RNA (Fig. 6B and Table 1).
Because the Ala-1 mutant was not detectably defective in vitro, the lack of viability in vivo might have been due to low protein expression. Therefore, Ala-1 was expressed on a high-copy 2μ plasmid. Although the protein was expressed at levels approximately equal to wild-type ProA–Est2p (data not shown), the cells continued to senesce. This result suggests that the Ala-1 mutant uncovers a specific defect in Est2p function.
Discussion
A major feature that distinguishes the TERT proteins from many other RTs is the large, basic amino terminus. Sequence analysis of this region revealed no known sequence motifs. Although alignment of TERT family members in this region is problematic because of low primary sequence conservation, two regions of homology are evident. The first is the T motif, a region just amino-terminal to the RT domain that is contained in all TERT homologs (Nakamura et al. 1997). TERT proteins from the ciliated protozoa have an additional region of homology of unknown function (CP motif; Bryan et al. 1998). Although TERT proteins from other organisms seem to have a conserved cysteine and leucine present in the CP motif, lack of additional homology makes alignment of this region unconvincing in nonciliates. Our work suggests that the amino-terminal half of Est2p contains several additional domains that are essential for Est2p function. Certain mutations in regions I and III affect binding to TLC1 RNA. The Ala-1 mutation in region I shows discordance between in vivo and in vitro telomerase activity that resembles the est1 or est3 phenotype, suggesting that the amino-terminal portion of region I may contribute to binding of accessory proteins.
Unigenic evolution reveals domain structure of amino-terminal region
We explored the domain structure of Est2p through a powerful mutational approach termed unigenic evolution (Deminoff et al. 1995). Several libraries of est2 alleles were created that contained multiple point mutations within the amino-terminal half of the protein. Functional alleles were selected by their ability to rescue the senescence of cells lacking Est2p. The cells also lacked Rad52p, because it would have enabled survival through an alternative telomere recombination pathway (Lundblad and Blackburn 1993; Lendvay et al. 1996). This screen was specifically intended to find essential regions of the protein. Therefore, mutants identified in the screen were not required to have normal telomere length, and many clones showed reductions in telomere size. Statistical analysis of the distribution of 265 base pair changes in these functional alleles of Est2p indicated the existence of at least four regions (I–IV) that are essential for Est2p function (hypomutable regions), separated by several nonessential regions that are highly accepting of missense mutations (α, β, and γ). Hypomutable regions I and II were well defined by their borders with these nonessential domains, whereas the boundary between regions III and IV was not as clear. Regions α, β, and γ all tolerated large deletions or inversions, in agreement with the classification of these regions as nonessential.
The next step in verification of the unigenic evolution map was to test mutations within the putative essential regions of the amino terminus. Alanine-block mutations were created in the context of Est2p tagged with ProA, which allowed measurement of mutant protein expression and facilitated biochemical analysis of the mutant enzymes. Most importantly, this epitope tag allowed detection of Est2p at endogenous levels following immunoprecipitation. Because some mutations in Est2p have been shown to have dominant negative effects (Lingner et al. 1997b), it was advantageous to avoid overexpression of the mutant proteins. Therefore, all of the mutants were integrated in the chromosome at the endogenous locus to maintain normal transcription levels.
As summarized in Table 1, several mutant alleles had drastically reduced protein levels. Unfortunately, the only mutation in region II, Ala-6, had undetectable levels of Est2p. Unigenic evolution identifies domains that contribute to protein structure as well as those that contribute in essential ways to protein function. More subtle mutations in region II will be required to elucidate whether this domain is primarily structural or whether it contributes in a specific way to Est2p function.
Amino-terminal domains implicated in binding TLC1 RNA
Five alanine-block mutations were created within region I because its large size suggested that it might contain several separable functions. Although all of the mutants are incapable of maintaining telomere length and cell growth, they have differential abilities to bind TLC1 RNA (Table 1). The most amino-terminal mutant, Ala-1, appears to have virtually normal RNA binding levels, after adjustment for the reduction in Est2p expression. However, mutants Ala-4 and Ala-5, which are located at the carboxy-terminal end of region I, show reduced levels of RNA binding. Because Ala-2 and Ala-3 produced undetectable amounts of Est2p, it is impossible to draw conclusions about the effect of these mutations on TLC1 RNA binding.
Region III was targeted by alanine-block mutations Ala-7 and Ala-8. Despite expression of these mutant proteins at levels comparable to or greater than that of Ala-1, no detectable TLC1 RNA coimmunoprecipitates with these mutant enzymes (Table 1). However, binding is not completely abolished in vivo, as TLC1 RNA is depleted from the starting extract during immunoprecipitation. This instability of RNA binding may indicate that binding of Est2p to TLC1 RNA is directly weakened by the Ala-7 and Ala-8 mutations. Alternatively, loss of TLC1 RNA on the beads may result from a general loss of integrity of the higher-order structure of the telomerase complex.
The T motif has been hypothesized to be involved in interactions with telomerase RNA (Weinrich et al. 1997). In our study, there is a positive correlation between the proximity of the alanine-block mutations to the T motif and their effect on TLC1 RNA binding. Ala-1, the most distal mutation (327 amino acids from the T motif), has no effect on binding, whereas mutants Ala-7 and Ala-8, which are proximal to the T motif (64 and 37 residues, respectively), severely weaken binding. Ala-4 and Ala-5, which are located at an intermediate position, have an intermediate effect on TLC1 coimmunoprecipitation. It is tempting to speculate that the T motif and the neighboring region III may be primarily involved in interactions with TLC1 RNA, whereas a portion of region I contributes to that binding.
Ala-1 mutant phenocopies est1, est3, and cdc13-2est
In vitro telomerase activity of most of the mutants directly mirrored the integrity of the telomerase complex as assayed by coimmunoprecipitation of TLC1 RNA with ProA–Est2p (Fig. 6C). The Ala-1 mutation in region I is intriguing because the defect appears to be something other than Est2p–TLC1 RNA complex formation. Protein levels were reduced, but RNA binding and in vitro telomerase activity appeared to be largely intact. Expression of the mutant protein at levels comparable to wild-type Est2p did not support cell survival. Furthermore, the survival of the Ala-10 mutant suggested that protein levels reduced 5- to 10-fold below wild type still provide sufficient telomerase activity for cell growth. Therefore, the reduction in Ala-1 protein level does not appear to be primarily responsible for the in vivo, telomerase-negative phenotype of the mutant. The residues mutated in Ala-1 may affect a region of Est2p that is important for in vivo regulation of telomerase activity. The est1, est3, and cdc13-2est mutants have been shown to have a similar phenotype: The cells undergo telomere loss and senescence in vivo (Lendvay et al. 1996) but have apparently normal levels of in vitro telomerase activity (Lingner et al. 1997a). Est1p and Cdc13p both bind to the single-stranded region of the telomere (Lin and Zakian 1996; Nugent et al. 1996; Virta-Pearlman et al. 1996). Est1p has been shown to coimmunoprecipitate with TLC1 RNA and therefore is presumed to interact with the telomerase complex either transiently or as a component of the holoenzyme. Est1p may facilitate interactions between the DNA primer and telomerase, a function which would presumably be dispensable in vitro where primer concentrations are high (Nugent and Lundblad 1998). One intriguing possibility, therefore, is that the Ala-1 mutant is defective in an interaction of Est2p with Est1p or another component of telomerase. Additional biochemical and genetic approaches will help elucidate the defect in this mutant.
The unigenic evolution approach
The results presented here highlight the strength of the unigenic evolution approach in giving an overview of protein structure in a region for which sequence analysis is not helpful. A strong selection for functional alleles, such as the senescence phenotype of cells lacking Est2p and Rad52p, allows efficient isolation of functional variants containing multiple point mutations. Because the est2 alleles isolated in this study contained an average of more than eight mutations per gene, only 33 clones were required to obtain a large sample of tolerated amino acid changes. Furthermore, automated sequencing technology allowed rapid data collection, because only three primers were required to sequence the 1350-nucleotide region subjected to mutagenesis in its entirety.
A version of the unigenic evolution technique may also prove useful for analysis of smaller, defined regions such as the RT domain and T motif. Indeed, important residues in the RT domain of HIV-1 RT were identified by selecting alleles that complemented the temperature-sensitive phenotype of a DNA polymerase I mutant in Escherichia coli from a library containing mutations over a 36-amino-acid region (Kim et al. 1996). Although our study did not specifically target the RT domain or T motif, a number of interesting mutations were isolated that can be interpreted in the context of known conserved residues. For example, several functional alleles were isolated that contained mutations of conserved residues in the T motif (Fig. 3C). Threonine 384 was mutated either to alanine or isoleucine and glutamic acid 385 was changed to valine. Intriguingly, when the corresponding threonine was mutated to alanine in human TERT, in vitro activity was diminished but the enzyme maintained ∼10%–40% wild-type activity (Weinrich et al. 1997). Mutation of the homologous glutamic acid to alanine in human TERT had a negligible effect on in vitro telomerase activity (Weinrich et al. 1997). Our results suggest that these residues are not essential for S. cerevisiae TERT and that telomerase activity in the mutant strains is sufficient to maintain cell growth.
Through the unigenic evolution approach, we have identified several essential domains in the amino terminus of Est2p. At least two of these domains contribute to binding of the RNA component of telomerase, whereas a mutation near the amino terminus suggests that this region may interact with other components of the enzyme. These results indicate that regions outside of the catalytic RT domain in TERT proteins are necessary to provide functions not required for retroviral RTs, such as stable association with the RNA template and formation of a multisubunit holoenzyme.
Materials and methods
Senescence assay
A complete replacement of the EST2 open reading frame in strain TVL268 (MATa ura3-52 ade2-101 lys2-801 leu2-Δ1 trp1-Δ1 his3-Δ200/CF+ [TRP1 SUP11]; Vicki Lundblad, Baylor College of Medicine, Houston, TX) was accomplished as described (Baudin et al. 1993). The HIS3 gene from plasmid pRS313 (Sikorski and Hieter 1989) was amplified using primers oligopro est2 (5′-ATGACACAAGTGAAATAGAAAAGTGAAAAATTAAAAAAAAAAAAAAAAAAAAAAAAACTGATTTATACTCCTCTTGGCCTCCTCTAG) and oligoterm est2 (5′-CCTTATCAGCATCATAAGCTGTCAGTATTTCATGTATTATTAGTATCGTTCAGAATGACACG) and transformed into TVL268. After minimal growth, the est2 strain was transformed with plasmid pKF400 [LYS2 plasmid pRS317 containing the BamHI EST2 fragment (from plasmid pVL291; Vicki Lundblad)]. Strain YKF102 (est2 rad52) was created by transformation with a rad52::LEU2 fragment obtained from plasmid pSM20 (gift of C. Chapon). For selection of functional est2 alleles, YKF102 was plated on α-aminoadipate plates to select for loss of pKF400, and the resulting est2::HIS3 rad52::LEU2 cells were restreaked once. A 100-ml culture of these senescing cells was grown and transformed with a library of est2 alleles. Transformants were restreaked to verify rescue of senescence, DNA was isolated using the DNA-Pure Yeast Genomic Kit (CPG Inc.), and complementing plasmids were rescued by transformation into electrocompetent XL1 Blue E. coli.
Plasmid construction
Plasmid pKF404 served as the basis for mutant libraries used in this study. pKF404 contains the chromosomal BamHI–SacI fragment of EST2 cloned into YCplac33 (Gietz and Sugino 1988). Site-directed mutagenesis (Deng and Nickoloff 1987) was used to destroy the carboxy-terminal HindIII site and to create an XbaI site immediately following the stop codon. HindIII and XbaI sites in the polylinker were destroyed.
Unigenic evolution
Several libraries of multiply mutated est2 alleles were created by PCR mutagenesis. Primers Hprimer (5′-GAGTTCATTCAAGACAAGCTTG) and RIprimerB ( 5′-CTGCTTAAATTCTTTGATACGG) were used in mutagenic PCR using pKF404 template. PCR was as described (Fromant et al. 1995) with 0.056 mm dATP, 0.090 mm dCTP, 0.020 mm dGTP, and 0.140 mm dTTP and without Triton X. Annealing was at 52°C for 30 sec, extension at 72°C for 10 min over a total of 30 cycles. The PCR product was purified using the QIAquick gel extraction kit (Qiagen). A portion of the mutagenized DNA was cut with HindIII and EcoRI and cloned into pHRadaptor, a plasmid derived from pKF404 in which the HindIII-to-EcoRI fragment of EST2 was replaced by a small linker fragment. Each PCR product was used as the template for the next round of PCR mutagenesis, thereby generating three libraries with increasing extents of mutagenesis. Functional est2 clones were identified by transformation into senescing est2::HIS3 rad52::LEU2 cells as described above. After plasmid rescue, the mutagenized region between the HindIII and EcoRI sites was amplified from each individual plasmid and the PCR product was sequenced at the DNA Sequencing Facility, University of Colorado at Boulder. Statistical analysis of mutation distribution has been described (Deminoff et al. 1995).
Construction of deletion and inversion mutants
Deletions and inversions were created in EST2 on plasmid pKF404 by site-directed mutagenesis (Deng and Nickoloff 1987). Following mutagenesis, the HindIII–EcoRI fragment was sequenced and recloned into pHRadaptor (described above) to eliminate extraneous mutations. Function of the clones was assessed by transformation into senescing est2::HIS3 rad52::LEU2 cells as described above.
Terminal deletion libraries were created by cutting pKF404 with HindIII or XbaI and treating with exonuclease III, S1, and Klenow polymerase as described (Sambrook et al. 1989). Resulting DNA was cleaved with EcoRI to create a collection of fragments with constant EcoRI ends and variable amino or carboxyl termini. These fragments were isolated by QIAquick gel extraction (Qiagen) and cloned into HRadaptor or RXadaptor, plasmids derived from pKF404 and containing a small DNA fragment replacing the HindIII-to-EcoRI or EcoRI-to-XbaI fragments, respectively. Plasmids containing functional deletion derivatives of EST2 were selected as described above.
Construction of ProA–EST2
The ProA sequence tag was amplified from plasmid pRIT2T (Nilsson et al. 1985) using primers containing terminal PvuII sites (5′-CAGGTAAGCCCCAGCTGGGGTGAAGCTCAAAAACTTAATG and 5′-ACGGTAAGCCCCAGCTGTATTTGTTATCTGCAGGTCGACG). The PCR product was cleaved with PvuII and cloned into the HindIII site (filled with Klenow polymerase) of pKF408. pKF408 contains the SacI–BamHI fragment of pKF404 in the URA3 integrating vector pRS306 (Sikorski and Hieter 1989). The resulting plasmid bearing ProA–Est2p (pKF409) was cleaved with ClaI and integrated in strain TVL268 by two-step gene replacement (Scherer and David 1979) to create strain YKF103.
Alanine-block mutations
Site-directed mutagenesis (Deng and Nickoloff 1987) was used to change blocks of 10 amino acids to alanine in pKF409 (the Ala-2 mutant changes seven amino acids). To create 10-amino-acid substitutions, the endogenous gene sequence (see legend to Fig. 4 for locations) was replaced by 5′-GCTGCAGCTGCTGCAGCGGCCGCAGCAGCT-3′. Nucleotides encoding the seven-amino-acid block of Ala-2 were replaced by 5′-GCTGCAGCTGCGGCCGCTGCT-3′. Following mutagenesis, the HindIII-to-EcoRI fragment was sequenced and recloned into the original pKF409 plasmid to verify that no other mutations were present. The resulting plasmids were cleaved with ClaI and placed in strain YKF103 by two-step gene replacement (Scherer and David 1979). A mutation (D670A) in RT motif C was created by an identical approach. Overexpression of Ala-1 was accomplished by cloning the insert from pKF409 into the URA3 2μ vector, pRS426 (Christianson et al.1992).
After minimal growth, strains were transformed with the complementing plasmid pKF404. Strains were plated on 5-FOA to select for loss of pKF404, and resulting single colonies were successively restreaked to YPD plates to assay senescence phenotype. Telomere length was assayed by picking a single colony from 5-FOA into liquid YPD. Saturated cultures were diluted 1000-fold and allowed to regrow. DNA was isolated using the DNA-Pure Yeast Genomic Kit (CPG Inc.), cleaved with XhoI, run in a 1% agarose gel, blotted, and probed with a random-primed yeast telomere sequence probe. A randomly selected probe to a 1621-bp fragment of chromosome IV was included as a loading control. The chromosome IV probe was amplified by PCR from genomic DNA using primers 5′-GTACCTCGGTTTAGTTAAGCG and 5′-CTCATTCGAATCCATACGACC.
Extract preparation and Western and Northern analyses
Strains were grown at 30°C in YPD to OD600 ∼1. Extracts were prepared by glass bead lysis in TMG (10 mm Tris-Cl at pH 8, 1 mm MgCl2, 10% glycerol, 0.1 mm DTT) with 300 mm NaCl. One Complete, Mini protease tablet (Boehringer) was added for each 50 ml of TMG. RNasin (Promega) (1 μl) was added to 500 μl of extract. Protein concentration was shown to vary less than twofold between samples by Coomassie stain. Extract was adjusted to 0.5% Tween-20 and incubated overnight with IgG Sepharose beads (Pharmacia) equilibrated with TMG (plus 200 mm NaCl, 0.5% Tween-20). Beads were washed three times with TMG (plus 200 mm NaCl, 0.5% Tween-20), once with TMG (plus 50 mm NaCl) and resuspended in 20 μl of TMG (plus 50 mm NaCl, 0.5 mm DTT).
Immunoprecipitation beads were mixed with 2× Laemmli loading buffer (125 mm Tris-HCl at pH 6.8, 4% SDS, 0.05% bromophenol blue, 20% glycerol, 5% β-mercaptoethanol), heated to 98°C for 5 min, and the supernatant was run on a 6% Tris-glycine gel (Novex). Protein was semi-dry transferred to Zetabind membrane (CUNO Laboratory Products). Primary antibody was rabbit IgG (Sigma). Secondary was peroxidase-conjugated goat anti-rabbit antibody (Boehringer Mannheim).
For Northern analysis of RNA coimmunoprecipitation, 10 μl of bound beads was mixed with 90 μl of 0.4 mg/ml proteinase K, 150 mm NaCl, 10 mm Tris-HCl at pH 7.5, 12.5 mm EDTA, 1% SDS and incubated at 37°C for 30 min. RNA was phenol/chloroform extracted, precipitated, and run in a 4% acrylamide/7 m urea gel. Following transfer to Hybond N+ membrane (Amersham), blots were probed with random-primed TLC1 fragment and end-labeled U1 oligonucleotide. Whole cell RNA was prepared as described (Chapon et al. 1997) from 50 ml cultures grown to OD600 ∼1. Gel conditions were as described above.
Telomerase assay
Packed beads (5 μl) from immunoprecipitation described above were mixed with 0.5 μl of water or 0.5 μl of 1 mg/ml RNase A and incubated 10 min at 30°C. Following addition of 5.5 μl of 2× reaction mixture consisting of 80 mm Tris-Cl at pH 8, 100 mm NaCl, 10% glycerol, 5 mm MgCl2, 1 mm spermidine, 1 mm DTT, 5 μm substrate oligonucleotide [5′-GTGTGGTGTGTGGG-3′], 200 μm TTP, 1.5 μl [α32P]dGTP (800 Ci/mm; 10 μCi/μl) per reaction, the reaction proceeded for 20 min at 30°C and was stopped by the addition of 1.5 μl of stop buffer (250 mm Tris at pH 8, 250 mm EDTA at pH 8, 2% SDS) and 1.75 μl of proteinase K (20 mg/ml) for 30 min at 60°C. Reaction products were precipitated and run in a 12% acrylamide/8 m urea sequencing gel. Oligonucleotide +1 marker was made by treating the telomeric 14-mer (see above) with terminal transferase (United States Biochemical) in the presence of [33P]ddTTP.
Acknowledgments
We thank V. Lundblad for generous gifts of strains and plasmids and for pointing out the utility of the unigenic evolution approach; S. Deminoff for help in statistical analysis of the mutation distribution; R. Weilbaecher and V. Lundblad for sharing their protocol for assaying telomerase activity on beads; J. Heit for technical assistance; Y. Han for automated DNA sequencing; D. Friedman, J. Sperger, T. Bryan, and T. Nakamura for critical reading of the manuscript; and M. Winey and colleagues for helpful suggestions and reagents. K.L.F. is an Associate and T.R.C. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Thomas.Cech@Colorado.Edu; FAX (303) 492-6194.
References
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C, Cech TR, Zaug AJ. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff SJ, Tornow J, Santangelo GM. Unigenic evolution: A novel genetics method localizes a putative leucine zipper that mediates dimerization of the Saccharomyces cerevisiae regulator Gcr1p. Genetics. 1995;141:1263–1274. doi: 10.1093/genetics/141.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WP, Nickoloff JA. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1987;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- ————— A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes & Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- Kim B, Hathaway TR, Loeb LA. Human immunodeficiency virus reverse transcriptase: Functional mutants obtained by random mutagenesis coupled with genetic selection in Escherichia coli. J Biol Chem. 1996;271:4872–4878. doi: 10.1074/jbc.271.9.4872. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-J, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci. 1997a;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997b;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nilsson B, Abrahmsen L, Uhlen M. Immobilization and purification of enzymes with staphylococcal protein A gene fusion vectors. EMBO J. 1985;4:1075–1080. doi: 10.1002/j.1460-2075.1985.tb03741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: Components and regulation. Genes & Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes & Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scherer S, David RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm snRNP. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Virta-Pearlman V, Morris DK, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes & Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Winston MK, Bhattacharjee JK. Growth inhibition by alpha-aminoadipate and reversal of the effect by specific amino acid supplements in Saccharomyces cerevisiae. J Bacteriol. 1982;152:874–879. doi: 10.1128/jb.152.2.874-879.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]