Abstract
Elimination of germ-line DNA segments is an essential step in the somatic development of many organisms and in the terminal differentiation of several specialized cell types. In binuclear ciliates, including Tetrahymena thermophila, DNA elimination occurs during the conversion of the germ-line micronuclear genome into the somatic genome of the new macronucleus. Little is known about molecular determinants and regulatory mechanisms involved in this process. Pdd2p is one of a small set of Tetrahymena polypeptides whose time of synthesis, nuclear localization, and physical association with sequences destined for elimination suggest an involvement in the DNA elimination process. In this study, we report that loss of parental expression of Pdd2p leads to the perturbation of several DNA rearrangement processes in developing zygotic macronuclei, including excision of internal eliminated sequences, excision of chromosome breakage sequences, and endoreplication of the new macronuclear genome and eventually results in lethality of the progeny. We demonstrate that excision and elimination of micronuclear-specific DNA occurs independently of endoreplication of the new macronuclear genome that takes place during the same period of time. Thus, our data indicate that parental expression of Pdd2p is required for successful DNA elimination and development of somatic nuclei.
Keywords: Programmed DNA elimination, chromosome breakage, DNA replication, Pdd2p, Tetrahymena macronuclear development
DNA rearrangements and partial elimination of the germ-line genome accompany differentiation of the soma in many organisms. For example, surface antigen variation in trypanosomes (Robertson and Meyer 1992), switching of mating type in yeast (Abraham et al. 1984), and V(D)J recombination in immunoglobulin genes in vertebrates (for review, see Gellert 1992) all occur by programmed DNA rearrangements. Selective loss of germ line-restricted chromatin has been demonstrated to take part in somatic development of several groups of flies (Fuge 1997; Glaser et al. 1997) and ascarid worms (for review, see Muller et al. 1996), and in differentiation of some human tissues (Bassnett and Mataic 1997). Thus, a wide range of organisms employs this strategy as a part of their normal differentiation of the soma. Programmed DNA rearrangements and elimination of the germ-line DNA segments are prominent steps in the somatic nuclear differentiation of ciliated protozoa (Prescott 1994; Coyne et al. 1996).
Like most ciliated protozoa, Tetrahymena thermophila contains both somatic (macro-) and germ-line (micro-) nuclei. The transcriptionally active macronucleus is polyploid (∼45C), whereas the micronucleus is diploid and transcriptionally inert (for review, see Gorovsky 1980). Despite large differences in structure and function of their genomes, both nuclei originate from the same zygotic micronucleus in the course of conjugation (Nanney 1986). Conjugation is a sexual process during which micronuclei from two mating cells undergo meiosis to produce gametic nuclei that are exchanged, fuse, and differentiate into two new micronuclei and two developing new macronuclei, often referred to as anlagen (Orias 1986). During this time, the original parental (old) macronucleus in each conjugating cell becomes pycnotic and finally is resorbed (Orias 1986).
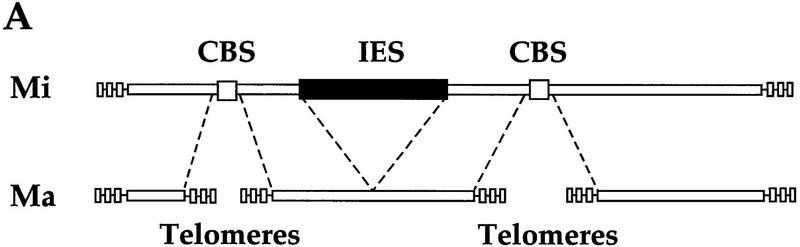
Conversion of the micronuclear genome into the genome of the new somatic macronucleus involves two major types of DNA rearrangements: elimination of micronuclear-specific DNA and processing of micronuclear chromosomes. In the course of DNA elimination, ∼6000 DNA segments, dispersed throughout the micronuclear genome, are precisely excised and subsequently degraded (for review, see Coyne et al. 1996). These DNA segments, termed internal eliminated sequences (IESs), constitute ∼10%–15% of micronuclear DNA (Yao and Gorovsky 1974). Sequences adjacent to IESs are rejoined and maintained during vegetative growth. During the same time period, each of the 10 micronuclear chromosomes in the early anlagen is fragmented into 40–60 DNA segments, each of which is flanked by telomeres added de novo (Yu and Blackburn 1991). Chromosome breakage sequences (CBSs) determine the sites of chromosome breakage and telomere addition (Yao et al. 1987). CBSs and ∼40 bp of DNA adjacent to them are eliminated (Godiska and Yao 1990). Finally, anlagen nuclei undergo a series of endoreplications (Allis and Dennison 1982). It is unclear to what extent any of these processes (DNA elimination, chromosome breakage and telomere addition, DNA replication, etc.) are dependent on another.
Until recently, little was known about the protein factors associated with DNA elimination in ciliates. However, using biochemical approaches, a small group of stage-specific polypeptides referred to as Pdds (for programmed DNA degradation) were discovered that are significantly enriched in developing macronuclei during the period corresponding to most, if not all, of the above processes (Madireddi et al. 1994). Pdd1p, the first member of this group, is an abundant 65-kD phosphoprotein possessing three chromodomains suggestive of a formational role in unique heterochromatin-like structures that are closely associated with DNA elimination (Madireddi et al. 1996). Pdd2p, a 43-kD phosphoprotein, can be cross-linked to Pdd1p during stages corresponding to DNA elimination, but is considerably less abundant than Pdd1p and does not reveal sequence similarities with any known proteins (Smothers et al. 1997b). In developing anlagen, both Pdd1p and Pdd2p are organized into specialized electron-dense structures that contain micronuclear-limited DNA as evidenced by in situ hybridization and immunofluorescence colocalization (Madireddi et al. 1996; Smothers et al. 1997b).
To elucidate the role of Pdd2p in Tetrahymena development, we eliminated its parental expression during conjugation by mating cells of two different mating types in which we had replaced all somatic copies of the PDD2 gene. Our data demonstrate that Pdd2p, like Pdd1p, localizes to the parental macronucleus well ahead of anlagen development in addition to uptake into developing anlagen later in the conjugation pathway (Madireddi et al. 1994, 1996). We demonstrate that excision of IESs and CBSs is severely impaired in exconjugants originating from cells lacking parental expression of Pdd2p, even though postzygotic expression of Pdd2p appears unaffected. Inability to eliminate DNA is also associated with the inhibition of DNA endoreplication from 4C to 8C and with lethality of the progeny. Our data also indicate that DNA elimination occurs independently of DNA replication. Thus, it seems likely that parental expression of Pdd2p has an important role in DNA elimination within the soma such that failure to carry out this process results in abortive macronuclear development and death of the progeny.
Results
Pdd2p localizes to the parental macronuclei during early stages of conjugation
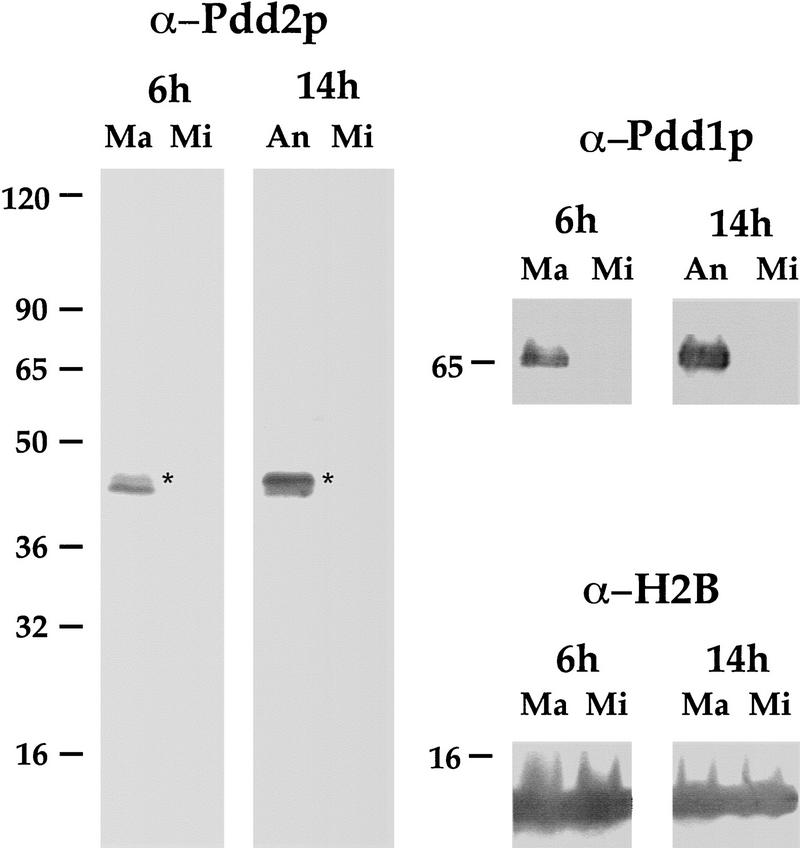
Expression of Pdd2p could be detected by Western blotting as early as 4 hr after cells of different mating types were mixed for conjugation, well ahead of the formation of new macronuclei (Smothers et al. 1997b). After formation of new macronuclei and concomitant programmed transcriptional inhibition of old macronucleus, Pdd2p expression is significantly increased (Madireddi et al. 1994; Smothers et al. 1997b), suggesting that during conjugation Pdd2p could be translated from messages sequentially originating from old macronucleus (preanlagen stages of conjugation) and from anlagen (late stages of conjugation). Intracellular localization of Pdd2p during these early stages of conjugation has not been described. To pursue this question, new antibodies were generated against a Pdd2p-specific peptide (see Materials and Methods for details) that displayed high selectivity for Pdd2p in Western blot analyses of total anlagen proteins purified from postzygotic (6 and 14 hr) conjugants (Fig. 1). Pdd2p was detected in parental macronuclei during early stages of conjugation (6 hr), but to a lesser extent than in 14 hr anlagen; no Pdd2p was detected in micronuclei isolated from cells at either time point.
Figure 1.
Pdd1p and Pdd2p are detected in parental macronuclei as well as anlagen during conjugation. Total proteins from micro- and macronclei isolated from 6-hr conjugants and from micronuclei and developing new macronuclei from conjugating cells at 14 hr postmixing were resolved on 10% SDS–polyacrylamide gel, transferred to membrane, and probed with Pdd2p peptide-derived antibodies. Blots were stripped and reprobed with Pdd1p- and, as a loading control, histone H2B-specific antibodies. Asterisks denote slow-migrating isoforms of Pdd2p (see Madireddi et al. 1996; Smothers et al. 1997b).
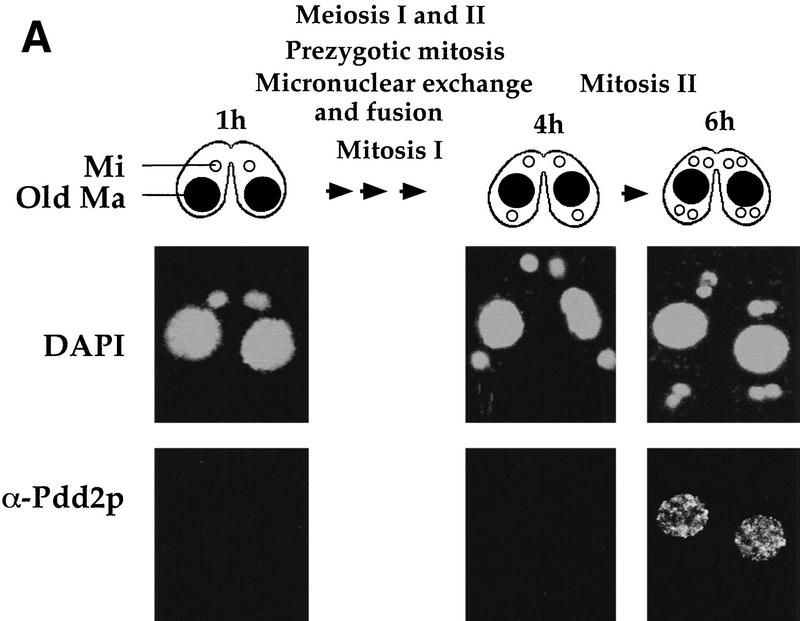
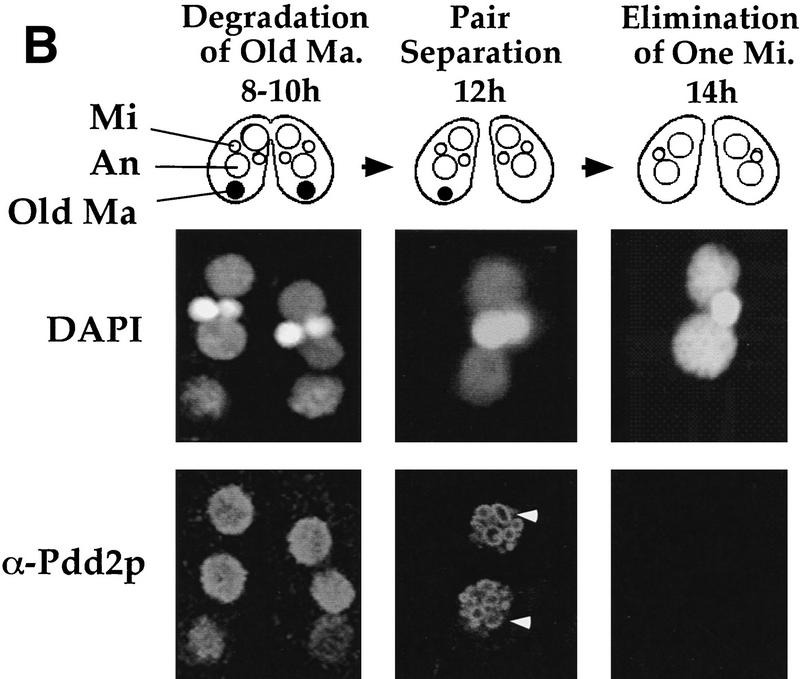
The Pdd2p peptide antibodies were then used to examine the subcellular localization of Pdd2p during both preanlagen (Fig. 2A) and postanlagen (Fig. 2B) stages of conjugation. Pdd2p localizes specifically to somatic, parental macronuclei of conjugating cells at 6 hr postmixing. During later stages, Pdd2p is detected in developing anlagen, and to a lesser extent, in old macronuclei. The protein is no longer detected in stages following DNA elimination (15 hr, characterized by two anlagen and one micronucleus). Similar, if not identical, distributions have been detected for Pdd1p (Coyne et al. 1999). Thus, these data document a complex pattern of localization suggesting involvement of the Pdd proteins in macronuclei processes during both pre- and postanlagen differentiation.
Figure 2.
Nuclear localization of Pdd2p during conjugation. Selected stages of conjugation are shown schematically and by micrographs of wild-type Tetrahymena fixed at indicated time points and stained by DAPI and Pdd2p antibodies. (A) Tetrahymena development before formation of anlagen; (B) late conjugation stages after anlagen formation. (Mi) Micronuclei; (An) anlagen; (Old Ma) old macronuclei (●). Several intranuclear structures, associated with DNA elimination (doughnut-like structures) are indicated by white arrowheads.
Creation of Tetrahymena strains with somatic knock out of PDD2 gene
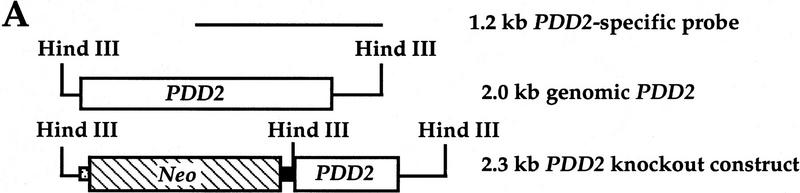
To elucidate the function of Pdd2p during early stages of Tetrahymena development we eliminated its prezygotic expression. A knockout construct was created in which the 5′ coding region of the PDD2 gene (380 bp) was replaced with the neo cassette (Gaertig et al. 1994) (see Fig. 3A). This construct was introduced into conjugating B2086 and CU428 cells at 9 hr postmixing using biolistic transformation, and transformants were selected using step-wise increments of paromomycin according to established procedures (see Materials and Methods for details).
Figure 3.
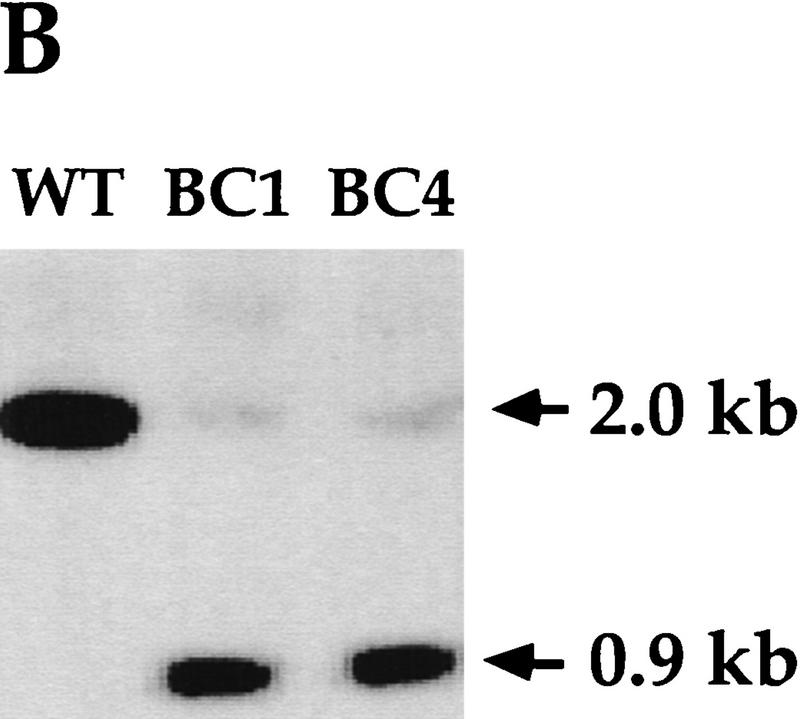
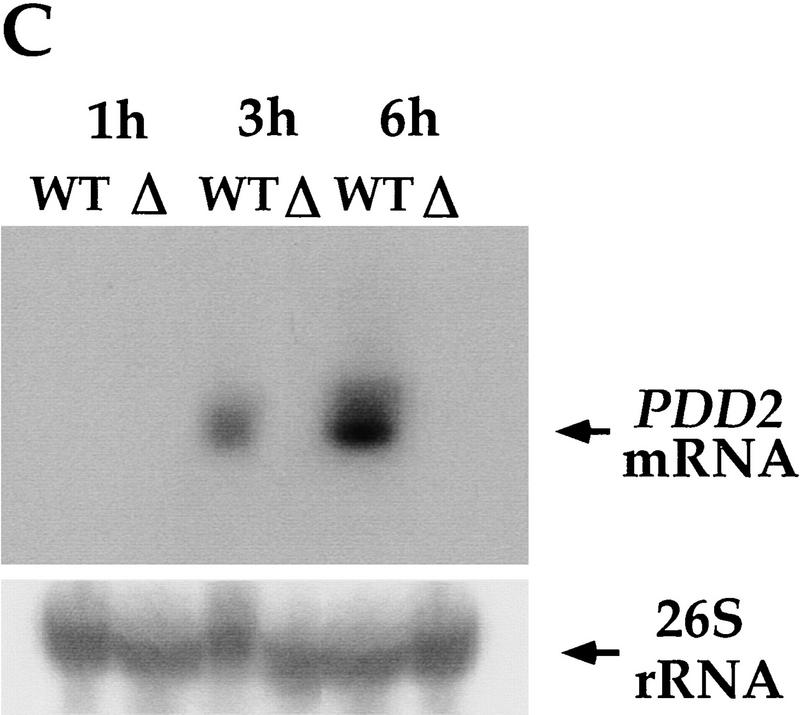
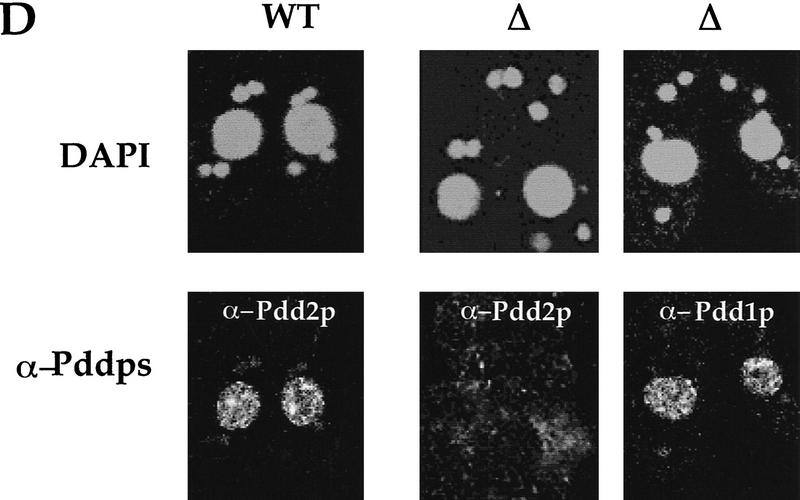
Disruption of the PDD2 gene eliminates parental expression during early stages of conjugation. (A) Schematic representation of the gene-disruption construct. The open box represents the coding region of the PDD2 gene. The hatched box is the coding region of the neo gene. The speckled box is a H4 histone promoter, and the solid box is a Btu2 terminator. A 1.2-kb PDD2-specific probe is shown at bottom. (B) Total genomic DNA (∼3μg) was digested with Hind III and probed by Southern blot hybridization with the probe shown above. The strains from which DNA was used are indicated at top. (C) Total cellular RNA isolated from wild-type (WT) and mutant (Δ) cells at the indicated time points was probed in Northern blot hybridization with the PDD2-specific PCR fragment (indicated in A, top), or as a loading control, with 28S ribosomal DNA (bottom). (D) Wild-type (WT) and mutant (Δ) cells fixed at 6 hr postmixing and stained with DAPI (top) or with antibodies against Pdd2p or Pdd1p (bottom). Note that the absence of Pdd2p-specific signal in PDD2 knockout cells cannot be attributed to a fixation artifact, as cells from the same source stain positively when probed with Pdd1p antibodies.
After selection, total genomic DNAs from two mutant clones of different mating type, BC1 and BC4, were subjected to Southern analysis to determine whether replacement of the PDD2 gene had occurred. DNA was probed with a PCR fragment corresponding to the 3′ part of the PDD2 coding region and 3′ flanking sequence (Fig. 3A). As shown in Figure 3B, most of the wild-type copies of the PDD2 gene were replaced in both clones. The small amount of wild-type band of 2 kb in mutant cells most likely came from micronuclear DNA, which presumably contains an intact unrearranged PDD2 gene. These results suggest that a replacement of wild-type copies of PDD2 gene occurred during selection for paramomycin resistance in two clones tested.
To determine whether the expression of the PDD2 gene was eliminated during the early stages of conjugation, we performed Northern blot analysis of total cellular RNA from conjugating mutant cells using the same PDD2-specific probe. As shown in Figure 3C, no expression of the PDD2 gene was detected during early stages of conjugation in the crosses of mutant cells (Δ). To further support these data, indirect immunofluorescent staining of wild-type and mutant conjugating cells was performed using the Pdd2p peptide-derived antibodies. As expected, no signal was found in the macronuclei of mutant cells (Fig. 3D). Thus, we conclude that two clones of different mating types were obtained in which somatic expression of PDD2 gene was eliminated. These clones were used in all further experiments.
Elimination of somatic expression of PDD2 gene is lethal for exconjugants
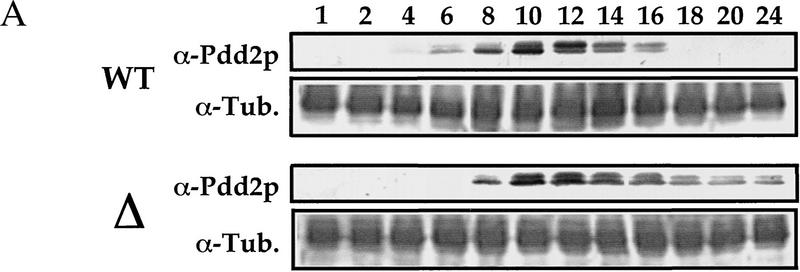
To determine to what extent postzygotic expression of Pdd2p is affected by somatic knockout of PDD2 gene, we performed developmental Western blot analyses using total cellular proteins from conjugating cells at different time points in the pathway. As shown in Figure 4A, although Pdd2p accumulation is delayed in mutant cells as expected because of the lack of parental expression of the PDD2 gene, no significant difference in the level of Pdd2p in wild-type (WT) and mutant (Δ) conjugating cells was detected between 8 and 14 hr. However, at later time points, a clear difference becomes apparent. Wild-type cells stop expressing Pdd2p between 16 and 18 hr, whereas Pdd2p expression in mutant cells persists until at least 24 hr. In addition, unlike wild-type cells, the ratio between phosphorylated (slower migrating) and unphosphorylated isoforms of Pdd2p is relatively constant with the progression of conjugation in mutant cells (Fig. 4A, 10–16 hr; see Smothers et al. 1997b for details). These data suggest that mutant cells undergo a developmental arrest or significant delay during late stages of conjugation.
Figure 4.
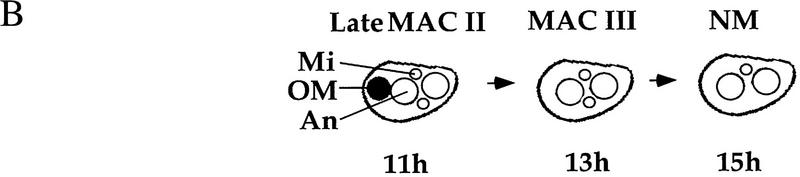
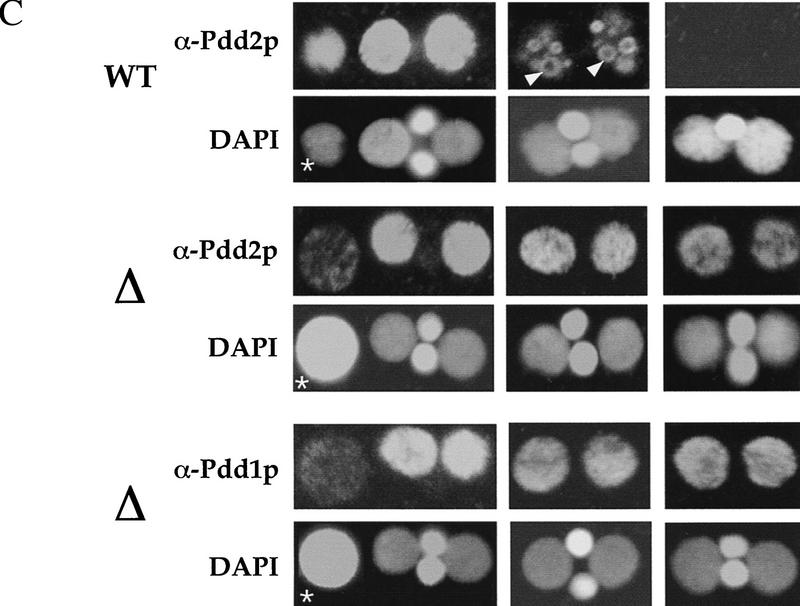
PDD2 knockout cells are arrested during late stages of conjugation. (A) Western blot analysis of total proteins from conjugating wild-type (WT) and mutant (Δ) cells at different times postmixing (indicated at top in hr) resolved on a 10% SDS–polyacrylamide gel and transferred to membrane. Distinct sections of the blots were then probed with previously obtained Pdd2p antibodies (Smothers et al. 1997b) and as a loading control tubulin (α-Tub.) antibodies. Note the differences in the relative ratio between the phosphorylated (slower-migrating) and unmodified isoforms of Pdd2p between wild-type and mutant cells starting at ∼12 hr. (B) Schematic representation of the late stages of conjugation. The following stages are shown: Macronuclear Development II MAC II; Macronuclear Development III MAC III; and New Macronuclei NM. (MI)Micronucleus; (AN) anlagen; (OM) old macronucleus (black). (C) Confocal sections of wild-type (WT) and mutant (Δ) cells fixed at the indicated time points and stained with DAPI and Pdd1p or Pdd2p antibodies as shown. Asterisks (*) indicate old macronuclei, stained with DAPI. Note the size difference between old macronuclei at 11 hr, suggestive of different extents of degradation in mutant and wild-type cells. White arrowheads indicate doughnut-like DNA degradation structures that are well organized in wild-type cells but not in mutants at 13 hr. Also, note the clear presence of Pdd1p and Pdd2p in mutants at 15 hr; these proteins are not detected in wild-type cells that have eliminated one micronucleus.
Other aspects of conjugation in mutant cells are consistent with this hypothesis. During late stages of conjugation wild-type cells typically undergo pair separation, degeneration of old macronuclei, and resorption of one micronucleus (see Fig. 4B). Examination of mutant cells indicates that, although they undergo pair separation at a normal time, some of them are delayed in degeneration (Fig. 4C, 11 hr) and subsequent resorption (data not shown) of their old macronuclei. Also, none of them resorb one of their two micronuclei as is always the case with wild-type cells (Fig. 4C, 15 hr). Strikingly, in anlagen of mutant cells, neither Pdd2p nor Pdd1p is organized into distinctive ring-like DNA elimination structures (compare anlagen nuclei in Fig. 4C, 13 hr). Finally, consistent with our immunoblotting analyses (Fig. 4A), the disappearance of Pdd2p and Pdd1p is clearly delayed in mutant cells (compare wild-type and mutant cells at 15 hr). Thus, we conclude that mutant cells indeed undergo arrest or a pronounced developmental delay during late stages of the conjugation process.
Analysis of several hundred individual pairs of mutant cells demonstrate that their exconjugant progeny are not able to divide upon return to nutrient conditions and eventually die (Table 1). Interestingly, mutant cells of either mating type do not undergo arrest at late macronuclear development II/III (MAC II/MAC III) stages and give rise to viable progeny when conjugated to wild-type partners (Table 1), suggesting that the PDD2 expression from parental macronuclei of the wild-type cell can rescue the mutant phenotype. Similar results were obtained in conjugation using four independent mutant clones (data not shown), ruling out the possibility that observed aberrant phenotypes were due to clonal variability.
Table 1.
PDD2 mutant cells do not give rise to viable progeny
| Type of mating cellsa
|
Number of individual pairs examined
|
Number of clones with more than 2 cells/drop
|
Number of clones completed conjugationb
|
|---|---|---|---|
| B2086 × CU427 | 100 | 80 ± 12 | 70 ± 10 |
| BC1 × BC4 | 100 | 17 ± 15 | 0 |
| BC1 × CU427 | 100 | 60 ± 13 | 54 ± 11 |
| BC4 × CU427 | 100 | 75 ± 15 | 63 ± 9 |
Wild-type and mutant cells were mated as indicated. Individual cell pairs were cloned in a drop of growth media at 10 hr postmixing. Only drops containing two cells able to move one hr after the transfer were counted. Cells were examined for the ability to divide and move inside drops at 24 hr after transfer.
Each lane represents results obtained in the course of four independent experiments.
To distinguish between cells that completed conjugation (progeny cells) and cells that have aborted from conjugation, all clones conjugated with the CU427 strain were tested for resistance to cycloheximide (only progeny cells should be resistant, see Material and Methods for the CU427 genotype). For the same purpose, cells from the BC1 × BC4 mating were checked for paromomycin resistance (only aborted cells should display resistance), and ability to mate with either CU427 or CU428 cells (only aborted cells should be able to participate in mating again, because progeny cells need to pass through multiple divisions before achieving sexual maturation).
Excision of internal eliminated sequences is affected in PDD2 knock out cells
To investigate potential cause(s) for arrest and subsequent death of PDD2-mutant cells, we attempted to analyze several molecular processes known to occur in anlagen during this time period. Physical association of Pdd2p with micronuclear-specific DNA sequences in wild-type cells (Smothers et al. 1997b), prompted us to check whether the process of DNA elimination is altered in mutant conjugants. To test this possibility, anlagen nuclei were isolated from conjugating wild-type and mutant cells at 24 hr postmixing, well after the time when DNA elimination is known to occur (i.e., 12–14 hr, Austerberry et al. 1984). To separate anlagen from contaminating micronuclei and old macronuclei, total nuclei were subjected to sedimentation at unit gravity according to Allis and Dennison (1982). To control for potential contamination of anlagen fractions with micronuclei, total protein from purified populations of analgen were probed in Western analyses with antibodies specific to micronuclear-specific linker histone (Chicoine et al. 1985). Only fractions that did not show any signal in the above assay (data not shown) were taken for further experiments.
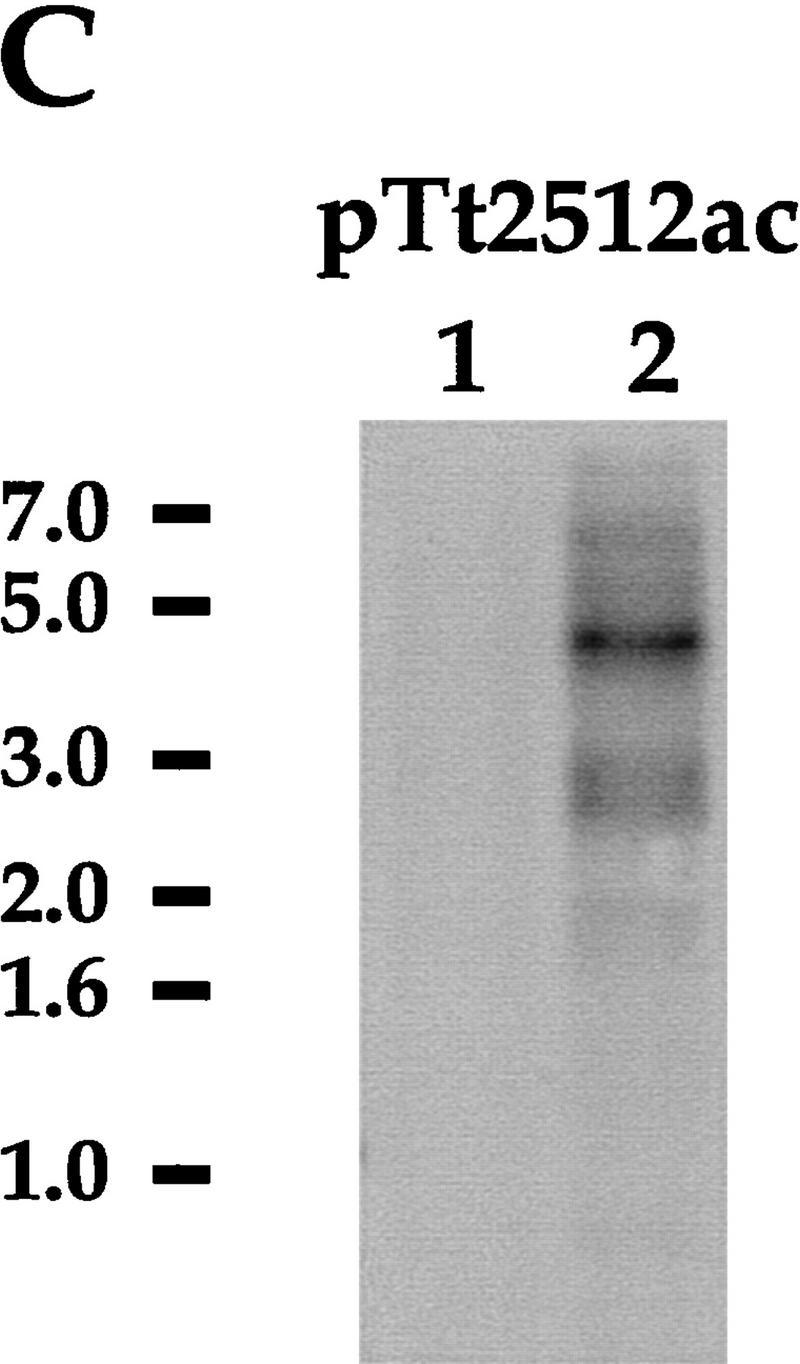
DNAs isolated from purified populations of nuclei from wild-type and mutant cells were then probed in Southern blot analysis using a fragment of micronuclear-specific DNA (pTt2512ac) that contains highly dispersed micronuclear-limited sequences repeated ∼200 times in the micronuclear genome (Yao 1982). Thus, this sequence represents a convenient probe to monitor global DNA elimination from a number of sites in the micronuclear germ line. As shown in Figure 5B, a very weak pTt2512ac hybridization is detected in wild-type macronuclear or anlagen DNA whereas a strong signal is detected in purified micronuclear populations. In contrast, 24-hr anlagen DNA from mutant cells displays a strong hybridization signal whose intensity and pattern is very similar to that of micronuclear DNA (Fig. 5B). In addition, as described above, Pdd1p and Pdd2p in mutant cells are not organized into the distinctive DNA elimination structures (Fig. 4C, 13 hr) that are closely associated with germ-line DNA segments in the process of DNA elimination (Madireddi et al. 1996; Smothers et al. 1997b). Thus, our data suggest that DNA elimination of at least some micronuclear-specific DNA sequences is inhibited in cells with somatic knockout of the PDD2 gene.
Figure 5.
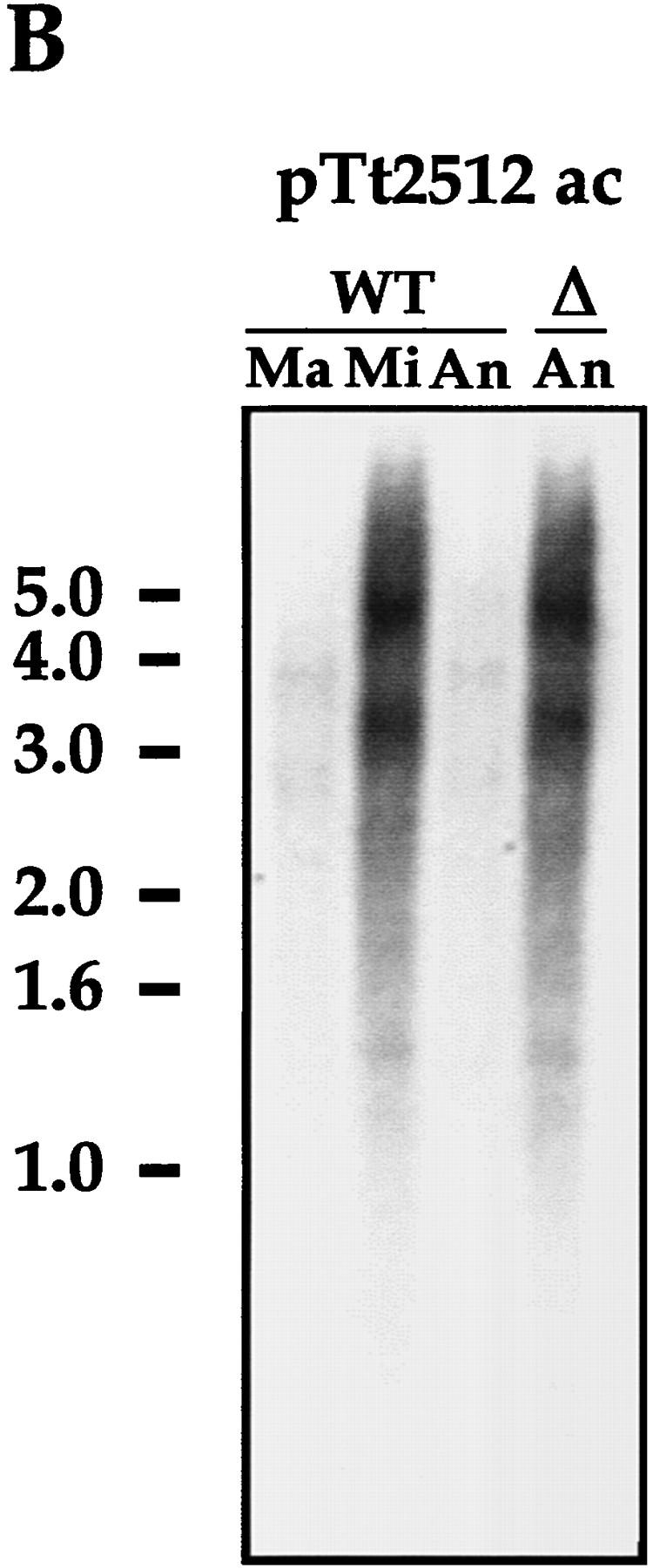
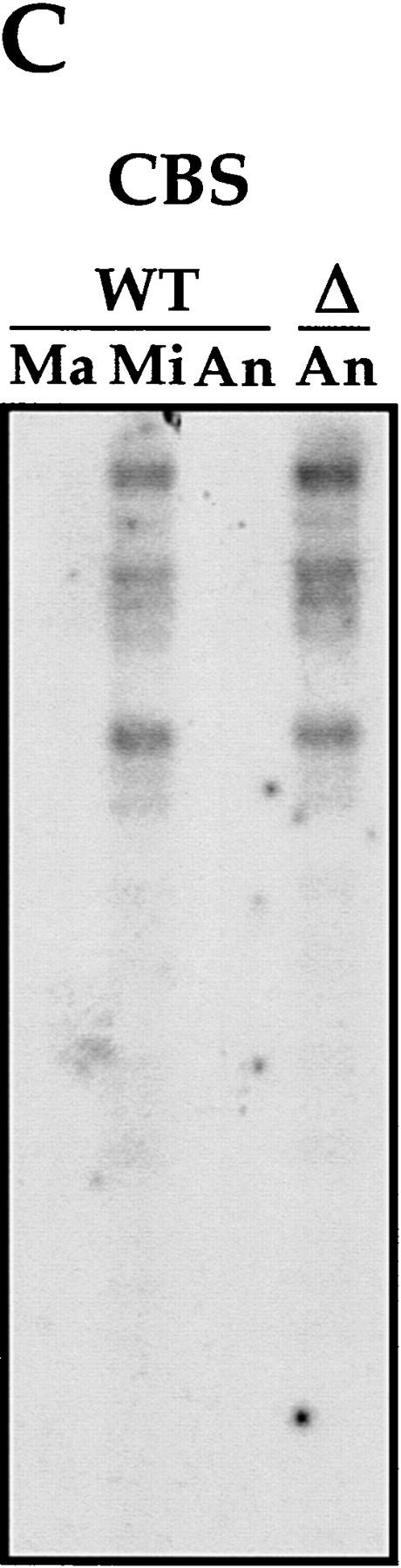
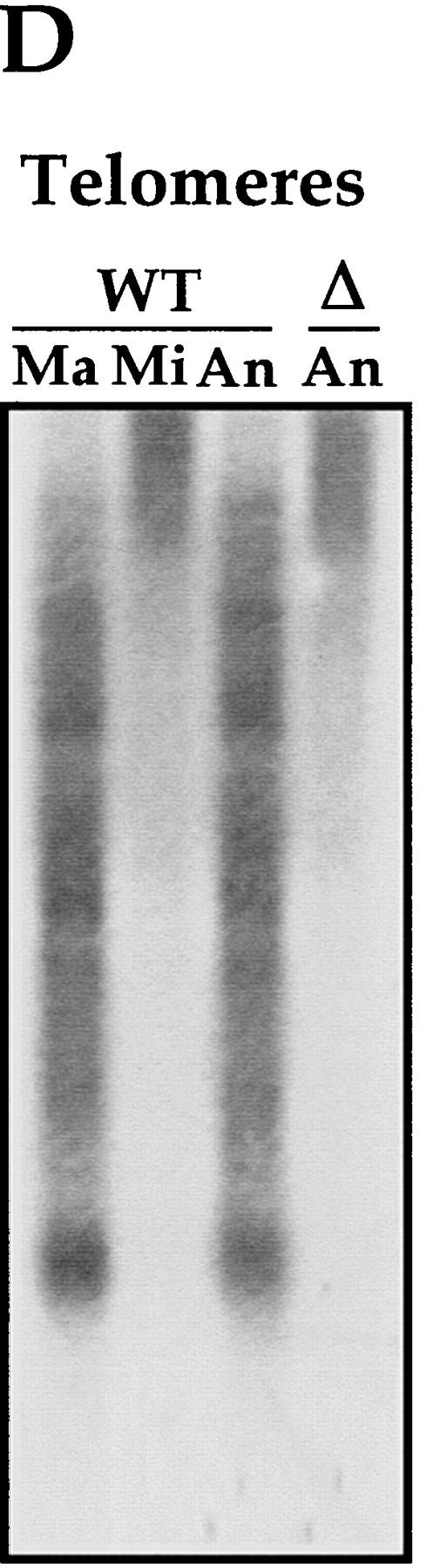
IES elimination, CBS elimination, and telomere addition are inhibited in PDD2 knockout cells. (A) Schematic representation of DNA rearrangements that occur during anlagen differentiation in wild-type cells. Open rectangles represent chromosomes, solid rectangles are IESs, open squares are CBSs; telomeres are shown as a group of rectangles. (Mi) Micronuclear chromosomes; (Ma) macronuclear chromosomes. (B–D) DNA (∼3 μg) isolated from macronuclei (Ma), micronuclei (Mi), or anlagen nuclei (An) from wild-type (WT) or mutant (Δ) cells was digested with HindIII enzyme and probed sequentially with the DNA sequences shown at the top of each blot. Anlagen from wild-type and mutant cells were isolated at 24 hr postmixing using sedimentation at unit gravity according to Allis and Dennison (1982). Purity of anlagen fractions was checked in Western blot analyses (not shown) using antibodies against micronuclear-specific linker histone (δ), a polypeptide known to be specific to micronuclei (Chicoine et al. 1985). DNA size standards are shown in kb.
Chromosome breakage and telomere addition are affected in PDD2 knockout cells
CBSs are also eliminated during anlagen development in the course of chromosome breakage and telomere addition (Yao et al. 1990). To determine whether CBSs are being excised in mutant cells, we performed Southern blot hybridization analysis as above, except that the DNA was probed with a 20-mer oligonucleotide CBS-1, which represents a consensus for CBSs (Yao et al. 1987). As expected, a strong hybridization signal is detected in DNA from wild-type micronuclei, but not from macronuclei or anlagen (Fig. 5). In contrast, strong hybridization is observed with DNA from 24-hr anlagen of PDD2 knockout cells, suggesting that chromosome breakage and CBS elimination did not take place in the absence of parental expression of Pdd2p.
Finally, to test whether the addition of telomeres was also inhibited in mutant cells, the blot in Figure 5C was stripped and reprobed with an oligonucleotide containing a triplet of the telomere sequence 5′-CCCCAA-3′ (King and Yao 1982). As shown in Figure 5D, the hybridization signal detected in DNA from mutant anlagen at 24 hr is very similar to that produced by wild-type micronuclar DNA, but differs markedly from that observed in DNA recovered from wild-type macronuclei and wild-type anlagen, suggesting that de novo telomeres have not been added to anlagen DNA in mutant cells. Collectively these data argue that excision of micronuclear-specific sequences and several associated DNA rearrangements do not occur in anlagen of mutant cells.
DNA endoreplication is blocked in anlagen of PDD2 knockout cells
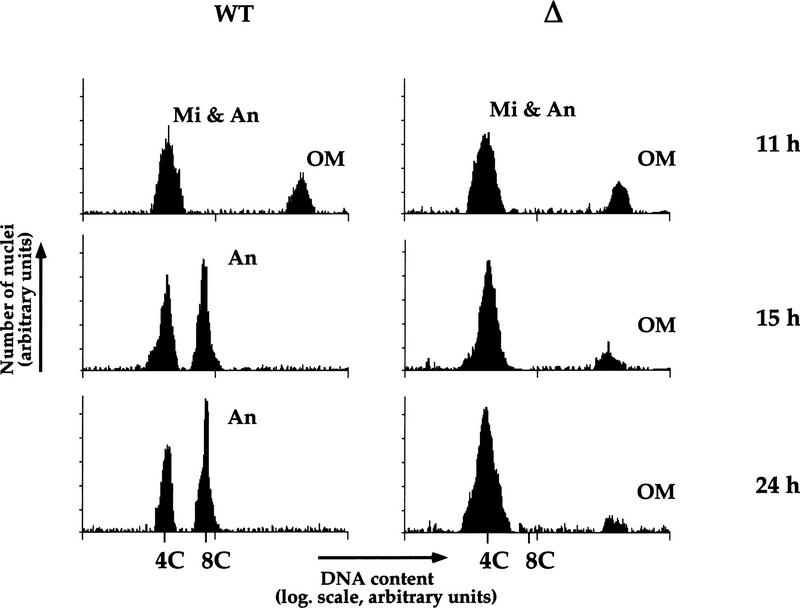
Another process that occurs in developing anlagen at about the same time as the processes discussed above (DNA elimination, chromosome fragmentation, and telomere addition) is DNA endoreplication (Austerberry et al. 1984). To determine whether this process is affected in anlagen of PDD2 knockout cells, we compared the relative DNA content of anlagen nuclei in mutant and wild-type cells using flow cytometry. Cells were collected at different time points of conjugation, lysed in buffer containing propidium iodide, and subjected to analyses (Fig. 6). In wild-type cells, two peaks corresponding to nuclei with relative ploidies of 4C and 8C are apparent at 15 hr and remain at least until 24 hr if cells are maintained under non-nutrient conditions (see Allis and Dennison 1982 for details). The peak of 8C DNA is not detected in anlagen purified from mutant cells at either 15 or 24 hr (Fig. 6, right) even when exconjugants are switched to nutrient conditions (not shown). These data demonstrate that anlagen in PDD2 knockout cells fail to endoreplicate from 4C to 8C.
Figure 6.
Endoreplication from 4C to 8C is blocked in anlagen in PDD2 knockout cells. Wild-type (WT) and mutant (Δ) cells were lysed at time points indicated at right, stained with propidium iodide, and the relative DNA contents of their nuclei were measured by flow cytometry. Relative ploidy values were determined from “standards” of micronuclei purified from starved cells that are known to be in G2 stage of the cell cycle with 4C DNA content (Allis and Dennison 1982). Peaks corresponding to micronuclei (Mi), anlagen (An), and old macronuclei (OM) are indicated.
The flow-cytometry data also support our earlier cytological observation that the elimination of old macronuclei does not occur efficiently in mutant cells as compared with wild-type cells.
DNA elimination occurs independently from anlagen endoreplication in wild-type cells
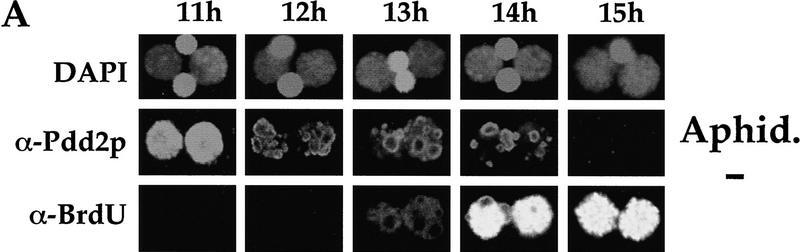
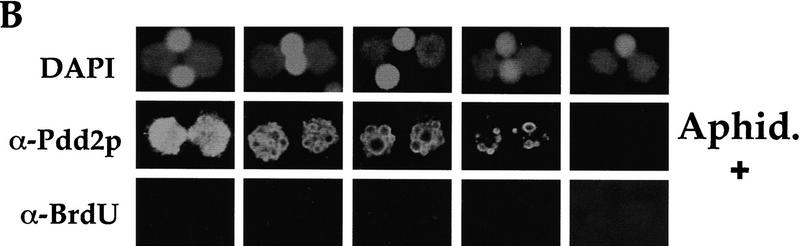
The apparent simultaneous block in DNA endoreplication and sequence elimination in anlagen of PDD2 mutant cells raises a question about cause–effect relationships between these two processes. To test whether inhibition of DNA replication abolishes DNA elimination in wild-type cells, aphidicolin, a specific DNA synthesis inhibitor (Ikegami et al. 1978), was added to wild-type conjugating cells at 10 hr postmixing to a final concentration of 5 μg/ml. At this time, the vast majority of cells (82%) were in the early MAC II stage (i.e., before DNA elimination and DNA replication have been reported to begin; see Austerberry et al. 1984 for details). To monitor DNA replication, 5-bromo-2′-deoxyuridine (BrdU) was added to cells together with the aphidicolin. Cells were fixed hourly between 11 and 15 hr and stained simultaneously with antibodies against BrdU to identify replicating nuclei and Pdd2p to monitor the formation of intranuclear DNA elimination structures.
As shown in Figure 7A, formation of the ring-like, DNA elimination structures occurs prior to the incorporation of BrdU in wild-type anlagen, which in this mating occurred maximally at 14–15 hr. Also, inhibition of BrdU incorporation (i.e., DNA replication) by treatment with aphidicolin (Fig. 7B) does not prevent formation and/or disappearance of these elimination structures, as was evidenced using morphological criteria. In addition, anlagen DNA isolated from wild-type cells treated with aphidicolin does not reveal any signal when probed with IESs or CBSs in Southern blot hybridization (Fig. 7C–E), demonstrating that DNA elimination and chromosome breakage can occur in wild-type Tetrahymena cells when anlagen DNA endoreplication from 4C to 8C is inhibited.
Figure 7.


DNA elimination in developing macronuclei occurs independently from 4C to 8C DNA endoreplication. Confocal sections of cells that had conjugated in media containing BrdU either in the absence (A) or presence (B) of 5 μg/ml of aphidicolin, fixed at indicated time points, and stained with DAPI and antibodies against Pdd2p and BrdU. (C–E) DNA (∼3 μg) from anlagen nuclei of cells treated with aphidicolin (lanes 1) and micronuclei of untreated cells (lanes 2) was digested with HindIII and probed sequentially with a pTt2512ac (C), CBS (D), or a PDD2-specific probe to control loading (E). DNA size standards are shown in kb.
Interestingly, inhibition of anlagen DNA endoreplication does not arrest cells in the MACIII stage; cells are clearly able to proceed to the new macronuclei (NM) stage, eliminating one micronucleus. Not surprisingly, treatment of conjugants with 5 μg/ml of aphidicolin for prolonged periods of time (>24 hr) is lethal (data not shown), in good agreement with previously reported results (Kapler and Blackburn 1994). Thus, these data suggest that the failure of anlagen to eliminate germ-line DNA segments and undertake other closely associated DNA rearrangements is not likely to be due to the failure of these cells to endoreplicate from 4C to 8C (Fig. 6).
Discussion
Phenotypes caused by the deficiency of parental expression of Pdd2p
Conjugating Tetrahymena lacking parental expression of Pdd2p display several abnormal developmental phenotypes culminating in arrest at late stages of anlagen differentiation (MAC II–MAC III stages) and eventually resulting in death of the progeny. Processes inhibited or delayed in mutant cells occur in all three types of nuclei: (1) mutant cells are unable to eliminate IES and CBS sequences, add telomeres de novo, or endoreplicate their genome in developing new macronuclei; (2) a delay in degradation of old macronuclei is detected in many mutant cells; and (3) none of the mutant cells eliminates one micronucleus and therefore they fail to advance to the NM stage of anlagen development. None of these aberrant phenotypes are observed in wild-type cells or in mutant cells conjugated with a wild-type partner. This is not surprising, as after pair formation conjugating cells share a common cytoplasm (Orias 1986), leading to the reasonable notion that Pdd2p synthesized in wild-type cell can migrate to parental nuclei in both wild-type and mutant cells.
On the other hand, several processes known to occur in wild-type cells during the same period of time are not affected in PDD2 mutants. For example, activation of transcription in anlagen, as evidenced by near-normal levels of Pdd2p and Pdd1p detected at late stages of conjugation (Fig. 4A,C; data not shown), endoreplication of anlagen to 4C (Fig. 6), and pair separation of mating cells all occur as in wild-type cells. Thus, our data underscore specificity of the phenotypic consequences in developing macronuclei associated with the disruption of somatic (parental) expression of Pdd1p. These consequences are likely to be caused by impaired DNA elimination and associated DNA rearrangements and by inhibition of DNA replication.
All DNA remodeling processes affected in PDD2 knockout cells occur within a very short time period (Austerberry et al. 1984) making it difficult to determine their order or dissect cause–effect relationships. However, our data (Fig. 7) demonstrate that DNA endoreplication from 4C to 8C is not required for DNA elimination or for the scheduled formation and disappearance of DNA elimination structures. Thus, it seems likely that failure to eliminate DNA in PDD2 mutants is not a consequence of DNA endoreplication arrest.
Could the reciprocal be the case? In our study, inhibition of IES and CBS elimination correlate with a block in DNA endoreplication between 4C and 8C. Endoreplication to this ploidy level could be a first step in polyploidization of the new macronucleus. If correct, the presence of IESs, consisting largely of heterochromatic sequences (Madireddi et al. 1996), could be inhibitory for efficient polyploidization as has been suggested in studies addressing the under-representation of certain heterochromatic sequences during endoreplication of Drosophila polytene chromosomes (Glaser and Spradling 1994; Glaser et al. 1997). Alternatively, efficient amplification of the developing macronuclear genome could be impossible without first breaking it into smaller DNA fragments flanked with telomeres. Indeed, studies on amplification of a mutant nm11 rDNA locus in Tetrahymena suggested that this locus can only be partially amplified in the “micronuclear-type” form (i.e., retaining CBSs and lacking telomeres), and cells homozygous for this mutant locus eventually die (Kapler and Blackburn 1993; Ward et al. 1997).
Inhibition of DNA replication by treatment with aphidicolin results in cell death (Kapler and Blackburn 1993). In our study, mutant cells unable to replicate their DNA also die, although it remains unclear whether the arrest of DNA replication in PDD2 knockout cells is the primary (or the only) cause of cell lethality. Besides the presumed effect on DNA replication, retention of some or all IESs could be detrimental for progeny survival if they disrupt or misregulate expression of gene(s) essential for vegetative growth in the now-active somatic macronuclear genome.
We suspect that inhibition or delay of processes occurring in other nuclei of mutant cells (see above) is likely to be an indirect effect of general developmental arrest caused by the inhibition of DNA elimination and associated processing in anlagen.
Could parental Pdd2p be directly involved in nuclear differentiation of anlagen nuclei?
Lack of parental expression of Pdd2p does not have an apparent effect on Tetrahymena development at early stages of conjugation when somatic macronuclei are the only source for PDD2 mRNA. Indeed, mutant cells become arrested at later stages of conjugation, when there seems to be near-normal amounts of Pdd2p resulting from expression in developing anlagen. One model accounting for these data suggests that correct timing of Pdd2p deposition to old macronuclei is a crucial event for the development of new macronuclei. Another model would assume that parental Pdd2p contributes in a functionally significant way to the total amount of Pdd2p in anlagen. In this model, absence of parental Pdd2p results in a decrease in the total amount of Pdd2p at a critical stage of conjugation below a threshold level required to bring about DNA elimination in a manner that remains to be determined. If correct, this model presumes an extremely Pdd2p dose-dependent mechanism of regulation of nuclear differentiation. However, our immunoblotting data, while only semiquantitative, suggest that parental Pdd2p does not contribute substantially to the total pool of Pdd2p in anlagen, and matings to wild-type partners overcome any proposed deficiency in parental expression. Still, it remains a formal possibility, that a dose-sensitive mechanism regulates the efficient formation and assembly of DNA elimination structures by limiting key components involved in the assembly pathway such as Pdd2p.
Could parental Pdd2p be involved in transnuclear regulation of DNA elimination?
An alternative hypothesis that could explain the inhibition of DNA elimination in cells lacking parental expression of Pdd2p requires its involvement in parental transnuclear regulation of IES excision. Recent data demonstrate that introduction of an IES sequence into the parental macronuclei of Tetrahymena or Paramecium cells specifically inhibits excision of that element (but not others) in the genome of new macronuclei (Duharcourt et al. 1995, 1998; Chalker and Yao 1996). These observations suggest that parental macronuclei play a role in the excision of micronuclear-specific DNA by directing which sequences should be retained in the genome of the new macronucleus (for review, see Meyer and Duharcourt 1996). One of the models proposed to account for this phenomenon postulates the direct transport of DNA and/or RNA molecules between parental and new macronuclei (Duharcourt et al. 1995). In such a model, it is likely that proteins are involved in generation of the presumed nucleic acid “shuffling molecules” in parental macronuclei and/or in the proposed transport to anlagen. The properties of Pdd2p make it a candidate for such a role. Indeed, during the developmental program of conjugation, Pdd2p and Pdd1p are detected first in parental macronuclei and later in new macronuclei, where they are associated with DNA destined for elimination. Ultrastructural analyses of parental macronuclei immunogold stained with Pdd1p antibodies (Coyne et al. 1999) detected Pdd1p in discrete structures closely apposed to the cytoplasmic face of the nuclear envelope, suggestive of a docking mechanism that may be involved in an export pathway from parental macronuclei to anlagen. Neither of these models is mutually exclusive, and we look forward to biochemical and genetic tests of these and other possibilities. Although the detailed mechanisms remain to be determined, it is clear from our data and data recently collected from similar studies on the disruption of PDD1 gene (Coyne et al. 1999) that parental expression of Pdd family members is essential for DNA elimination and related nuclear events during differentiation of the somatic lineage in Tetrahymena.
Materials and methods
Strains, cell culture, and conjugation
T. thermophila strains CU428 [mpr1-1/mpr1-1 (MPR1, mp-s, VII)], CU427 [chx-1/chx-1 (CHX1, cy-s, VI)], and B2086 [mpr/mpr, chx/chx (mp-s, pm-s, cy-s, II)] were kindly provided by P.J. Bruns (Cornell University, Ithaca, NY). Cells were grown in SPP medium (1% proteose peptone, 0.2% dextrose, 0.1% yeast extract, 0.003% sequestrine) (Gorovsky et al. 1975). For conjugation, cells of different mating types were washed, starved (16–24 hr, 30°C), and mated in 10 mm Tris-HCl (pH 7.5), as described by Allis and Dennison (1982). Pairing efficiency was estimated by counting pairs at 2 hr postmixing. All experiments involving Tetrahymena conjugation were performed at pairing efficiencies of not <85%.
Plasmids
Plasmid p4T2-1-PDD2-KO was obtained by inserting a 327-bp PCR fragment corresponding to the 3′ untranslated region (UTR) of the PDD2 gene into the p4T2-1 plasmid (Gaertig et al. 1994) using SacII and BamHI enzymes, followed by the insertion of a 927-bp PCR fragment, corresponding to the 5′ portion of the PDD2 coding region and its 5′ UTR using XhoI and KpnI enzymes. The primers used in the PCR reaction for the 5′ UTR were 5UPFL: 5-AGACCGCGGCAAATTTTCTCTATTAAAACC-3′ and 3UPFL: 5′-AGAGGATCCTTTTCTATGTGTATTAATCAG-3′ and for the 3′ portion of the PDD2 gene and flank were 5DWNFL: 5′-AGACTCGAGAAATTCCAATTGATGATAGC-3′ and 3DWNFL: 5′-AGAGGTACCGATTACAGAGAAAAAGTAG-3′. For transformation of Tetrahymena cells the plasmid DNA was prepared according to Hai and Gorovsky (1997). Plasmid pTt2512 was a gift from Dr. Meng-Chao Yao (Fred Hutchinson Cancer Research Center, Seattle, Washington).
Gene replacement by biolistic transformation
CU428 and B2086 cells were mated and transformed at 9 hr postmixing with the p4T2-1-PDD2-KO construct using the DuPont Biolistic PDS-1000/He Particle delivery system (Bio-Rad), as described previously (Cassidy-Hanley et al. 1997). To select for transformants, cells were subjected to step-wise selection for paromomycin resistance starting from 120 μg/ml to a final concentration of 1.5 mg/ml, above which the cells failed to grow. Following drug selection, several single cells were cloned, propagated for ∼100 generations to achieve sexual maturity, and subjected to conjugation with each other and with CU427 cells. Clones able to conjugate with other clones and with CU427 cells were chosen for further experiments.
Southern and Northern blot hybridization
Total genomic DNA and total cellular RNA were isolated, resolved on standard or denaturing 1% agarose gels, respectively, and blotted as described previously (Richards 1988). A PCR fragment obtained in the reaction with 5PRB (5′-TAGGATGATCAAGGGTAATCAG-3′) and 3DWNFL primers (see above) was used as a PDD2-specific probe. Fragments a and c obtained by HaeIII restriction digestion of the insert from plasmid pTt2512 (Yao 1982) were used as a probe for Tetrahymena IES DNA. The insert from plasmid pBS26S (Engberg and Nielsen 1990), obtained by digestion with HindIII enzyme was used as a probe for Tetrahymena 26S ribosomal RNA. All probes were labeled with [α-32P]dATP by random priming (Richards 1988). Oligonucleotides CBS1: 5′-TAAAGAAAGAGGTTGGTTTA-3′ (Yao et al. 1987) and TEL1: 5′-CCCCAACCCCAACCCCAA-3′ (King and Yao 1982) were used as probes for CBSs and telomere sequences, respectively. Oligonucleotides were labeled with [γ-32P]ATP and hybridized as described previously (Yao et al. 1987).
Generation of antibodies
A Pdd2p-specific peptide D234GNKQNNQEQPYLPN248C, was synthesized containing an artificial cysteine on the carboxyl terminus. The peptide was conjugated to keyhole limpet hemocyanin via cysteine using standard protocols. Rabbits were then immunized with injections of 1 mg of conjugated peptide (for details, see Lin et al. 1989).
Nuclei preparation
Macronuclei, micronuclei, and anlagen nuclei were isolated from Tetrahymena as described by Gorovsky et al. (1975) except that the nucleus isolation buffer contained 1 mm iodoacetamide, 1 mm phenylmethylsulfonyl fluoride (PMSF), 10 mm sodium butyrate, but not spermidine. Nuclei purification using sedimentation at a unit gravity was performed according to Allis and Dennison (1982).
Electrophoresis and immunoblotting
SDS-PAGE and immunoblotting analyses were performed as described previously (Lin et al. 1989). When whole-cell SDS lysates were used, aliquots of 1×106 cells were removed from a mating-cell culture and processed as described previously (Guttman et al. 1980). Antibodies against Pddp proteins and Tetrahymena α-tubulin antibodies were used as described previously (Van de Water et al. 1982; Smothers et al. 1997b)
Indirect immunofluorescence analysis
Conjugating cells were fixed and processed for indirect immunofluorescence as described previously (Smothers et al. 1997b). Cells were stained with the DNA-specific dye diamidinophenolindole (DAPI) at 0.3 μg/ml in Tris-buffered saline (TBS). Pdd1p antibodies were used as described previously (Smothers et al. 1997b). Pdd2p peptide-derived polyclonal rabbit antibodies were used at 1:100 dilution. Antibodies against BrdU were purchased from Amersham and used according to the manufacturer's guidelines. For simultaneous staining with Pdd2p and BrdU antibodies, cells were fixed according to the protocol for Pdd2p antibodies, dropped on microscope slides, treated with 2N hydrochloric acid for 30 min at room temperature, followed by several washes in TBS, and stained according to the protocol for Pdd2p antibodies.
Fluorescent and confocal microscopy
Fluorescent microscopy was performed using an Olympus BH-2 microscope and photographed on Kodak TMAX 400 film with a mounted Olympus C-35AD-2 camera. Confocal images were obtained with a Leica TCS NT microscope, and digital images were processed using Adobe Photoshop (Adobe Systems, Inc., San Jose, CA).
Flow cytometry analysis
Tetrahymena cells were lysed in a buffer containing 0.25 m sucrose, 10 mm MgCl2, and 0.5% NP-40 at a concentration of 1.0 × 106 cells/ml. The DNA-specific dye propidium iodide was then added to 50 μg/ml and nuclei were stained for 1 hr before flow cytometry analysis using either a Coulter Profiler or a Becton Dickinson Calibur flow cytometer.
Acknowledgments
We are grateful to Yi Wei and Yali Dou for their technical help, Nataliya Shulga for her help with confocal microscopy and critical reading of the manuscript, Jody Bowen for critical reading of the manuscript, and Jianxin Zhou for constant encouragement and suggestions throughout the work. M.A.N. was a recipient of Oncology Research Faculty Development Program Fellowship from the National Cancer Institute, National Institutes of Health (NIH). This research was supported by grants from the NIH to C.D.A. and M.A.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL allis@virginia.edu; FAX (804) 924-5069.
References
- Abraham J, Nasmyth KA, Strathern JN, Klar AJS, Hicks JB. Regulation of mating type information in yeasts. Negative control requiring sequences both 5′ and 3′ to the regulated regions. J Mol Biol. 1984;176:307–311. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- Allis CD, Dennison DK. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Devel Biol. 1982;93:519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- Austerberry CF, Allis CD, Yao MC. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci. 1984;81:7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Mataic D. Chromatin degradation in differentiating fiber cells of the eye lens. J Cell Biol. 1997;137:37–49. doi: 10.1083/jcb.137.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee J, Cole ES, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol Cell Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine LG, Wenkert D, Richman R, Wiggins JC, Allis CD. Modulation of linker histones during development in Tetrahymena: Selective elimination of linker histone during the differentiation of new macronuclei. Devel Biol. 1985;109:1–8. doi: 10.1016/0012-1606(85)90339-2. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao MC. Genome downsizing during ciliate development: Nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- Coyne, R.S., M.A. Nikiforov, J.F. Smothers, C.D. Allis, and M-C. Yao. 1999. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell (in press). [DOI] [PubMed]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes & Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg J, Nielsen H. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 1990;18:6915–6919. doi: 10.1093/nar/18.23.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuge H. Nonrandom chromosome segregation in male meiosis of a sciarid fly: Elimination of paternal chromosomes in first division is mediated by non-kinetochore microtubules. Cell Motil Cytoskeleton. 1997;36:84–94. doi: 10.1002/(SICI)1097-0169(1997)36:1<84::AID-CM8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Gu L, Hai B, Gorovsky MA. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination gets a break. Trends Genet. 1992;8:408–412. doi: 10.1016/0168-9525(92)90322-u. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Spradling AC. Unusual properties of genomic DNA molecules spanning the euchromatic-heterochromatic junction of a Drosophila minichromosome. Nucleic Acids Res. 1994;22:5068–5075. doi: 10.1093/nar/22.23.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser RL, Leach TJ, Ostrowski SE. The structure of heterochromatic DNA is altered in polyploid cells of Drosophila melanogaster. Mol Cell Biol. 1997;17:1254–1263. doi: 10.1128/mcb.17.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiska R, Yao MC. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell. 1990;61:1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- Gorovsky MA. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- Gorovsky MA, Yao MC, Keevert JB, Pleger GL. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Guttman SD, Glover CV, Allis CD, Gorovsky MA. Heat shock, deciliation and release from anoxia induce the synthesis of the same set of polypeptides in starved T. pyriformis. Cell. 1980;22:299–307. doi: 10.1016/0092-8674(80)90177-4. [DOI] [PubMed] [Google Scholar]
- Hai B, Gorovsky MA. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc Natl Acad Sci. 1997;94:1310–1315. doi: 10.1073/pnas.94.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- Kapler GM, Blackburn EH. A weak germ-line excision mutation blocks developmentally controlled amplification of the rDNA minichromosome of Tetrahymena thermophila. Genes & Dev. 1994;8:84–95. doi: 10.1101/gad.8.1.84. [DOI] [PubMed] [Google Scholar]
- King BO, Yao MC. Tandemly repeated hexanucleotide at Tetrahymena rDNA free end is generated from a single copy during development. Cell. 1982;31:177–182. doi: 10.1016/0092-8674(82)90417-2. [DOI] [PubMed] [Google Scholar]
- Lin R, Leone JW, Cook RG, Allis CD. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madireddi MT, Davis MC, Allis CD. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev Biol. 1994;165:418–431. doi: 10.1006/dbio.1994.1264. [DOI] [PubMed] [Google Scholar]
- Madireddi MT, Coyne RS, Smothers JF, Mickey KM, Yao MC, Allis CD. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- Meyer E, Duharcourt S. Epigenetic programming of developmental genome rearrangements in ciliates. Cell. 1996;87:9–12. doi: 10.1016/s0092-8674(00)81317-3. [DOI] [PubMed] [Google Scholar]
- Muller F, Bernard V, Tobler H. Chromatin diminution in nematodes. BioEssays. 1996;18:133–138. doi: 10.1002/bies.950180209. [DOI] [PubMed] [Google Scholar]
- Nanney DL. Introduction. In: Gall JG, editor. Molecular biology of ciliated protozoa. Orlando, FL: Academic Press; 1986. pp. 1–21. [Google Scholar]
- Orias E. Ciliate conjunction. In: Gall JG, editor. Molecular biology of ciliated protozoa. Orlando, FL: Academic Press; 1986. pp. 45–84. [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. Preparation and analysis of DNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York, NY: Wiley Interscience; 1988. pp. 2.0.1–2.12.5. [Google Scholar]
- Robertson BD, Meyer TF. Genetic variation in pathogenic bacteria. Trends Genet. 1992;8:422–427. doi: 10.1016/0168-9525(92)90325-x. [DOI] [PubMed] [Google Scholar]
- Smothers JF, Madireddi MT, Warner FD, Allis CD. Programmed DNA degradation and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. J Eukaryot Microbiol. 1997a;44:79–88. doi: 10.1111/j.1550-7408.1997.tb05942.x. [DOI] [PubMed] [Google Scholar]
- Smothers JF, Mizzen CA, Tubbert MM, Cook RG, Allis CD. Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development. 1997b;124:4537–4545. doi: 10.1242/dev.124.22.4537. [DOI] [PubMed] [Google Scholar]
- Van De Water L, Guttman SD, Gorovsky MA, Olmsted JB. Production of antisera and radioimmunoassays for tubulin. Methods Cell Biol. 1982;24:79–96. doi: 10.1016/s0091-679x(08)60649-4. [DOI] [PubMed] [Google Scholar]
- Ward JG, Blomberg P, Hoffman N, Yao MC. The intranuclear organization of normal, hemizygous and excision-deficient rRNA genes during developmental amplification in Tetrahymena thermophila. Chromosoma. 1997;106:233–242. doi: 10.1007/s004120050244. [DOI] [PubMed] [Google Scholar]
- Yao MC. Elimination of specific DNA sequences from the somatic nucleus of the ciliate Tetrahymena. J Cell Biol. 1982;92:783–789. doi: 10.1083/jcb.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Gorovsky MA. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48:1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- Yao MC, Zheng K, Yao CH. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell. 1987;48:779–788. doi: 10.1016/0092-8674(87)90075-4. [DOI] [PubMed] [Google Scholar]
- Yao MC, Yao CH, Monks B. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- Yu GL, Blackburn EH. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]