Abstract
SR proteins are essential pre-mRNA splicing factors that act at the earliest stages of splice-site recognition and spliceosome assembly, as well as later in the splicing pathway. SR proteins consist of one or two RNA-recognition motifs and a characteristic arginine/serine-rich C-terminal RS domain. The RS domain, which is extensively phosphorylated, mediates the subcellular localization of individual SR proteins and also functions as a splicing activation module, apparently by engaging in protein–protein interactions. The RS domain of SF2/ASF is dispensable for the concentration-dependent effects of this SR protein on alternative splice-site selection. However, this RS domain is highly conserved phylogenetically, and was shown to be required for constitutive splicing in vitro and for cell viability. Here, we demonstrate that the RS domain of SF2/ASF is, in fact, dispensable for splicing of several substrates, including constitutive and enhancer-dependent pre-mRNAs. The requirement for this RS domain is substrate specific, and correlates with the strength of the splicing signals. When the 3′ splice site is weak, both the SF2/ASF RS domain and U2AF35 are required for splicing. These results show the existence of an RS domain-independent function of SR proteins in constitutive and enhancer-dependent splicing, and suggest mechanisms for their role in enhancer function besides U2AF recruitment.
Keywords: SR proteins, SF2/ASF, RS domain, U2AF, pre-mRNA splicing, splicing enhancer
Pre-mRNA splicing is a critical step in gene expression and requires both small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP protein factors. These factors assemble into a large complex on the pre-mRNA known as a spliceosome, which catalyzes splicing (for review, see Krämer 1996). Members of the SR protein family are well-studied non-snRNP protein factors required for pre-mRNA splicing (for review, see Valcárcel and Green 1996; Cáceres and Krainer 1997). SR proteins function early in spliceosome assembly to promote the formation of complexes containing U1 snRNP bound to the intron 5′ splice site and U2 snRNP bound to the branch site (Krainer et al. 1990b; Wu and Maniatis 1993; Kohtz et al. 1994; Staknis and Reed 1994; Jamison et al. 1995). SR proteins also function at subsequent stages of splicing by facilitating the recruitment of U4/U6·U5 tri-snRNP (Roscigno and Garcia-Blanco 1995; Tarn and Steitz 1995) and by promoting the second transesterification step (Chew et al. 1999). In the case of regulated splicing, SR proteins can modulate alternative splice-site selection (Ge and Manley 1990; Krainer et al. 1990a), and also can overcome weak splicing signals by recruiting components of the general splicing machinery to the intron (for review, see Blencowe 2000).
SR proteins share a distinctive domain structure, which consists of one or two copies of an RNA-recognition motif (RRM), followed by a characteristic C-terminal arginine/serine-rich (RS) domain (Birney et al. 1993). Although the RRMs mediate binding to degenerate sequence motifs, they determine the substrate specificity of individual SR proteins (Chandler et al. 1997; Tacke et al. 1997; Mayeda et al. 1999; Schaal and Maniatis 1999). The RS domains are thought to be required for protein–protein interactions of SR proteins with each other and with other components of the splicing machinery (Wu and Maniatis 1993; Kohtz et al. 1994), as well as to mediate subcellular localization (Cáceres et al. 1997). In vitro studies showed that the RS domain of the prototype SR protein SF2/ASF is essential for constitutive splicing, although the same domain is dispensable for concentration-dependent effects on alternative splice-site selection (Cáceres and Krainer 1993; Zuo and Manley 1993). Supporting and extending these initial observations, an in vivo study in chicken DT40 B-cells showed that the RS domain of SF2/ASF is essential for cell viability (Wang et al. 1998b), which is consistent with the high degree of conservation of this domain (Birney et al. 1993).
Reversible phosphorylation of SR proteins, mainly at the serine residues within the RS domains, can influence protein-RNA (Tacke et al. 1997) and protein–protein interactions (Xiao and Manley 1997, 1998), as well as localization of SR proteins and recruitment to transcriptionally active sites (for review, see Misteli 1999). The functional significance of this post-translational modification of SR proteins has been demonstrated in systems in which the activity of these proteins is under tight control, such as during early development in Ascaris lumbricoides, sex determination in Drosophila, and during adenovirus infection (Du et al. 1998; Kanopka et al. 1998; Sanford and Bruzik 1999).
Several distinct, but not mutually exclusive, functions have been ascribed to SR proteins. For example, SF2/ASF can cooperate with the U1 snRNP particle in recognition of the 5′ splice site. This effect is probably mediated by specific interactions between the RS domains of SF2/ASF and U1-70K protein (Kohtz et al. 1994; Jamison et al. 1995). SR proteins are also thought to promote splicing by bridging 5′ and 3′ splice sites through RS domain-mediated protein–protein interactions across an intron or an exon (intron definition or exon definition) (Robberson et al. 1990; Wu and Maniatis 1993). Components that are bound to the 5′ and 3′ splice sites, such as U1-70K and U2AF35, also have RS domains and can interact with the RS domains of SR proteins (Wu and Maniatis 1993; Xiao and Manley 1997). Finally, SR proteins can recognize exonic splicing enhancers (ESEs) and facilitate the removal of the adjacent intron(s) (for review, see Blencowe 2000).
The precise mechanism of ESE function is still unclear. Early experiments supported a U2AF65-recruitment model, according to which ESE-bound SR proteins facilitate U2AF65 binding to the 3′ splice site polypyrimidine tract via an RS domain-mediated interaction with U2AF35 (Wu and Maniatis 1993; Wang et al. 1995; Zuo and Maniatis 1996). In contrast, recent experiments showed that crosslinking of U2AF65 to the polypyrimidine tract of certain enhancer-dependent pre-mRNAs requires neither the enhancer sequences nor U2AF35 (Guth et al. 1999; Kan and Green 1999; Li and Blencowe 1999). Interestingly, depletion of U2AF35 from HeLa nuclear extract prevents the splicing of certain enhancer-dependent substrates, but not others. Thus, the roles of RS domain-mediated interactions and of U2AF in enhancer function and constitutive splicing are more complex than initially suggested.
We report the unexpected finding that the RS domain of SF2/ASF is, in fact, dispensable for in vitro splicing of several, but not all, pre-mRNAs. The requirement for this domain of SF2/ASF is related to the strength of the 3′ splice site and the requirement for U2AF35. These results extend our knowledge of the properties of SR proteins, and suggest the existence of RS domain-independent functions of these proteins in constitutive and enhancer-dependent splicing.
Results
SF2/ASF and mutant proteins
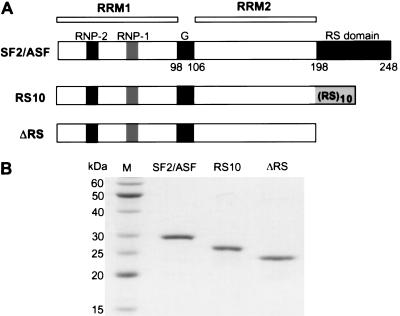
The SR protein SF2/ASF is an essential splicing factor in vitro when other SR proteins with overlapping functions are not present. The RS domain of SF2/ASF was thought to be essential for constitutive splicing, because deletion of all the RS or SR dipeptides within the RS domain, or deletion of the entire domain, greatly reduces splicing efficiency in an S100 extract complementation assay (Cáceres and Krainer 1993; Zuo and Manley 1993). For technical reasons, these and other SF2/ASF mutants analyzed in previous work were prepared as N-terminal his-tagged proteins. Such tags may, in some cases, interfere with a protein's normal functions, although in the case of SF2/ASF, the wild-type his-tagged protein has strong complementing activity. In this study, we made untagged SF2/ASF lacking the RS domain (ΔRS) to re-examine the RS domain requirement for in vitro splicing. Although RS domains from different SR proteins vary in length and sequence, they can be swapped without compromising splicing activity in vitro or cell viability (Chandler et al. 1997; Wang et al. 1998b; Mayeda et al. 1999). We therefore also designed a mutant form of SF2/ASF with only 10 consecutive RS dipeptides—RS10—to test whether a simplified RS domain can functionally substitute for the natural 51-amino-acid RS domain of SF2/ASF (Fig. 1A).
Figure 1.
SF2/ASF and variant proteins. (A) Diagram of SF2/ASF, ΔRS, and RS10 proteins, showing relevant domains. The RNP-2 and RNP-1 submotifs of RRM1, the Gly-rich hinge, RRM2, and the RS domain are indicated, and the residue numbers at the boundaries of each module are shown. Changes in the RS domain relative to the wild-type protein are displayed. (B) Purity of recombinant SF2/ASF proteins. 0.5 μg of each protein was analyzed by SDS-PAGE and Coomassie blue staining.
Wild-type and mutant forms of SF2/ASF were expressed in Escherichia coli and purified to apparent homogeneity (Fig. 1B). Because of the denaturation and renaturation steps used during the protein purification process, a functional assay was used to ensure that at least the regions of the protein preceding the RS domain were correctly folded. The activities of wild-type or mutant forms of SF2/ASF were initially assayed with 5′D16X β-globin pre-mRNA. 5′D16X is a model substrate with a duplicated 5′ splice site, and it has been shown that SF2/ASF can promote selection of the proximal 5′ splice site of this pre-mRNA in an RS domain-independent manner (Krainer et al. 1990a; Cáceres and Krainer 1993). In HeLa nuclear extract, 5′D16X pre-mRNA spliced preferentially via the distal 5′ splice site. Use of the proximal 5′ splice site increased on addition of either wild-type SF2/ASF, ΔRS, or RS10 (data not shown). These results indicate that all three SF2/ASF proteins are functional, and therefore, the lack of activity of any of these proteins in other assays is unlikely to be due to misfolding or aggregation.
Phosphorylation of SF2/ASF mutant proteins
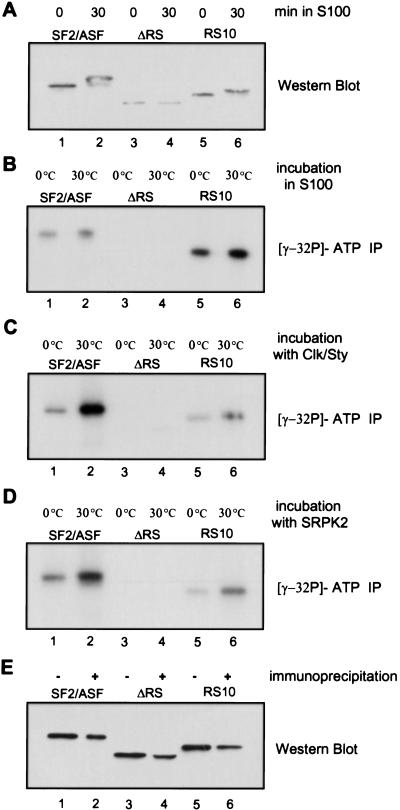
Proper protein phosphorylation and dephosphorylation within the RS domain of SR proteins is important for their activities in splicing (for review, see Misteli 1999). Sequence-specific RNA binding by SR proteins is also enhanced by RS domain phosphorylation (Tacke et al. 1997). Protein–protein interactions between SR proteins and other RS domain-containing proteins, as well as their activities in constitutive and enhancer-dependent splicing, are differentially affected by phosphorylation and dephosphorylation (Cao et al. 1997; Xiao and Manley 1997, 1998; Kanopka et al. 1998; Prasad et al. 1999). We therefore asked whether SF2/ASF RS10 and ΔRS can be phosphorylated under splicing conditions.
First, phosphorylation of the wild-type and mutant recombinant proteins by endogenous kinases present in the SR protein-deficient HeLa S100 extract was measured by Western blotting with an antibody specific for RRM1 of SF2/ASF (Hanamura et al. 1998). Phosphorylated SF2/ASF, which has decreased electrophoretic mobility compared with unphosphorylated SF2/ASF, was detected by 30 min of incubation under splicing conditions. RS10 was also phosphorylated, whereas no phosphorylated forms of ΔRS could be detected (Fig. 2A). It is possible that a substantial change in electrophoretic mobility requires phosphorylation of multiple residues. We therefore performed a more sensitive test, consisting of incubating the various SF2/ASF proteins in S100 extract under splicing conditions in the presence of [γ-32P]ATP, followed by immunoprecipitation with the anti-SF2/ASF antibody, SDS-PAGE, and autoradiography. Consistent with the Western blot result, no radiolabel was incorporated into the ΔRS protein. Interestingly, the RS10 protein was phosphorylated to a greater extent than wild-type SF2/ASF (Fig. 2B), which may be attributable to a variety of kinases present in the crude S100 extract.
Figure 2.
In vitro phosphorylation of SF2/ASF proteins. (A) 5 pmole of wild-type SF2/ASF (lanes 1,2), ΔRS (lanes 3,4), or RS10 (lanes 5,6) was incubated under splicing conditions for 0 (lanes 1,3,5) or 30 min (lanes 2,4,6). The reactions were carried out in S100 extract, fractionated by SDS-PAGE, and wild-type and mutant proteins were detected by Western blotting with a monoclonal antibody directed against the N terminus of SF2/ASF. The zero-time point reactions were immediately stopped with SDS sample buffer. (B) The reactions were carried out in S100 extract in the presence of [γ-32P]ATP, followed by immunoprecipitation with anti-SF2/ASF monoclonal antibody, SDS-PAGE, and autoradiography. The reactions were incubated on ice or at 30°C for 15 min, as indicated. (C) As in B, but the proteins were phosphorylated with recombinant Clk/Sty kinase instead of S100 extract. (D) As in C, but with recombinant SRPK2. (E) The efficiency of immunoprecipitation of the different SF2/ASF proteins was measured by IP/Western. The proteins before (−) and after (+) immunoprecipitation were detected by Western blotting with the same anti-SF2/ASF antibody used for immunoprecipitation.
Additional phosphorylation–immunoprecipitation experiments were done with two recombinant SR protein-specific kinases, SRPK2 and Clk/Sty (Colwill et al. 1996; Kuroyanagi et al. 1998; Wang et al. 1998a). In contrast to the kinases present in the extract, SRPK2 and Clk/Sty phosphorylated SF2/ASF to a greater extent than they did RS10 (Fig. 2C,D). This result confirms previous reports that both kinases specifically recognize the intact RS domain of an SR protein (Colwill et al. 1996; Wang et al. 1998a). In the case of ΔRS, a trace amount of phosphorylation was detected with Clk/Sty, but not SRPK2. These data are in agreement with the fact that Clk/Sty has a broader range of substrates than SRPK1 (which is closely related to SRPK2) and may phosphorylate regions of SF2/ASF other than the RS domain (Colwill et al. 1996). We conclude that the synthetic RS domain of RS10 is efficiently phosphorylated in the extract under splicing conditions, whereas ΔRS remains unphosphorylated.
The RS domain is not strictly required for constitutive splicing in vitro
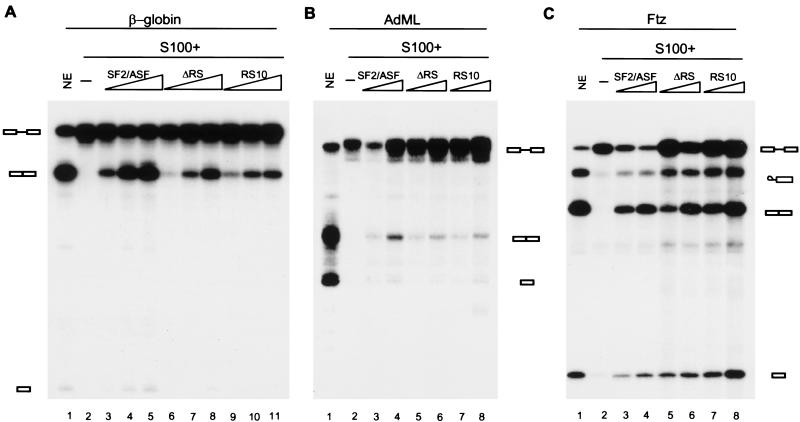
It has been reported that the RS domain of SR proteins is essential for both constitutive and enhancer-dependent splicing, as his-tagged SF2/ASF ΔRS cannot complement S100 in these assays (Cáceres and Krainer 1993; Zuo and Manley 1993; Mayeda et al. 1999). Surprisingly, when we added untagged ΔRS to S100 extract, it efficiently complemented β-globin pre-mRNA splicing (Fig. 3A). To extend this observation to additional substrates, adenovirus major late (AdML) and Drosophila fushi tarazu (ftz) pre-mRNAs were tested in S100 complementation assays. Both ΔRS and RS10 proteins promoted splicing of these substrates (Fig. 3B,C). Finally, spliceosome assembly was assayed on native gels, and no differences were found among the three SF2/ASF proteins (data not shown). These data show that SF2/ASF does not require its RS domain for constitutive splicing of several substrates, at least in vitro. Apparently, the presence of a his-tag at the N terminus of SF2/ASF used in previous studies somehow sensitizes the protein, such that the effect of the C-terminal RS domain deletion is exacerbated.
Figure 3.
ΔRS functions in constitutive splicing. (A) In vitro splicing of β-globin pre-mRNA in HeLa nuclear extract (lane 1), S100 extract alone (lane 2), S100 extract complemented by 2, 4, or 8 pmole of recombinant SF2/ASF (lanes 3–5), ΔRS (lanes 6–8), or RS10 (lanes 9–11). In vitro splicing of AdML pre-mRNA (B) or ftz pre-mRNA (C) in HeLa nuclear extract (lane 1), S100 extract alone (lane 2), S100 extract complemented by 4 or 8 pmole of recombinant SF2/ASF (lanes 3,4), ΔRS (lanes 5,6), or RS10 (lanes 7,8).
RS domain-dependent and RS domain-independent exonic splicing enhancers
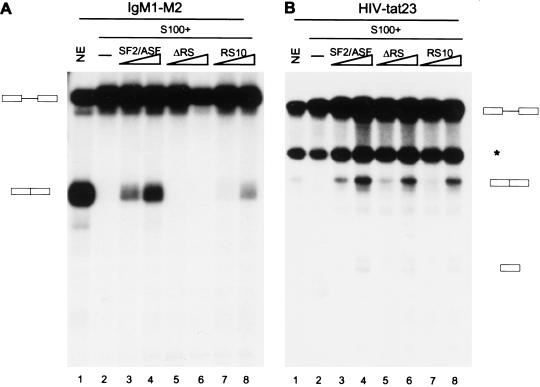
Two models have been proposed for ESE function. In the first model, binding of U2AF65 to the 3′ splice site polypyrimidine tract is stabilized through interactions between ESE-bound SR proteins and U2AF35 (Wu and Maniatis 1993; Zuo and Maniatis 1996). In the second model, ESEs promote splicing by antagonizing the effect of adjacent exonic splicing silencers (ESSs) (Guth et al. 1999; Kan and Green 1999). SR proteins, especially their RS domains, play a crucial role in the U2AF-recruitment model, whereas little is known about the mechanistic basis of the antagonism model. We therefore tested the SF2/ASF RS domain requirement with two well-characterized ESE-dependent substrates, IgM M1–M2 pre-mRNA and HIV tat23 pre-mRNA (Watakabe et al. 1993; Kan and Green 1999; Mayeda et al. 1999). ESE and ESS elements have been mapped in the 3′ exon of both pre-mRNAs. Interestingly, we observed that the RS domain of SF2/ASF is required for splicing of IgM M1–M2 pre-mRNA but not HIV tat23 pre-mRNA in S100 complementation assays (Fig. 4A,B, cf. lanes 3,4 and 5,6). We conclude that ESEs can function by different mechanisms, and can be categorized into RS domain-dependent and RS domain-independent. Moreover, although the RS domain of SF2/ASF is not required for constitutive splicing of all pre-mRNAs, it can have substrate-specific functions that make it essential for splicing of certain pre-mRNAs. Supporting this idea, the RS domain of SF2/ASF is required for several other substrates tested, as shown below in Figures 5 and 6.
Figure 4.
RS domain-dependent and RS domain-independent exonic splicing enhancers. In vitro splicing of IgM M1–M2 pre-mRNA (A) and HIV tat23 pre-mRNA (B) in HeLa nuclear extract (lane 1), S100 extract alone (lane 2), or S100 extract complemented by 4 or 8 pmole of recombinant SF2/ASF (lanes 3,4), ΔRS (lanes 5,6 ), and RS10 (lanes 7,8). The band marked with an asterisk is a cleavage product unrelated to splicing (Krainer et al. 1990b).
Figure 5.
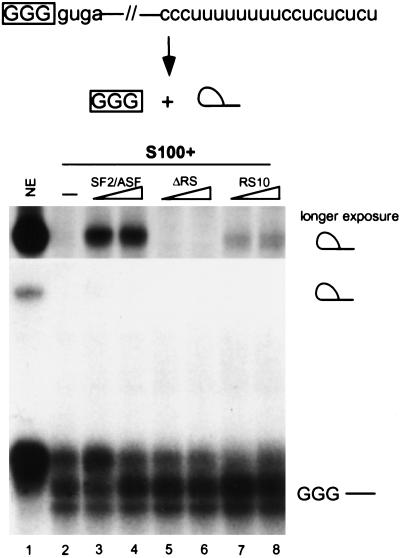
RS domain requirement for exon-independent splicing. Exon-independent lariat formation was tested in S100 extract complementation assays. A derivative of AdML pre-mRNA containing only 3 nucleotides of the 5′ exon and no 3′ exon was incubated in HeLa nuclear extract (lane 1), S100 extract alone (lane 2), or S100 extract complemented by 6 or 12 pmole of recombinant SF2/ASF (lanes 3,4), ΔRS (lanes 5,6), or RS10 (lanes 7,8). The intron lariat migrates above the precursor on the 12% denaturing polyacrylamide gel. The region corresponding to the lariat is shown above the main panel as a 10-fold longer exposure.
Figure 6.
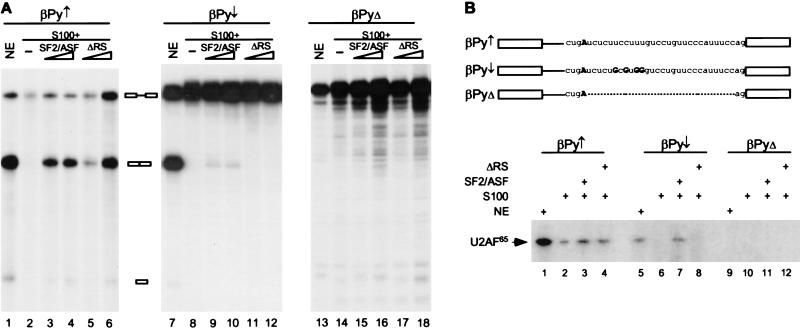
The RS domain of SF2/ASF is required for splicing of a substrate with a weak polypyrimidine tract. (A) In vitro splicing of β-globin pre-mRNA with an improved (βPy↑ ), a weakened (βPy↓ ), or no (βPyΔ) polypyrimidine tract in HeLa nuclear extract (lanes 1,7,13), S100 extract alone (lanes 2,8,14), or S100 extract complemented by 4 or 8 pmole of recombinant SF2/ASF (lanes 3,4,9,10,15,16) or ΔRS (lanes 5,6,11,12,17,18). (B) βPy↑ , βPy↓ or βPyΔ pre-mRNAs were incubated in HeLa nuclear extract (lanes 1,5,9), S100 extract alone (lanes 2,6,10), or S100 extract complemented by 8 pmole of either SF2/ASF (lanes 3,7,11) or ΔRS (lanes 4,8,12) under splicing conditions for 20 min. The reactions were then irradiated with UV light and digested with RNases A and T1. After immunoprecipitation with anti-U2AF65 monoclonal antibody, the cross-linked U2AF65 was detected by 12% SDS-PAGE and autoradiography. The sequence at the 3′ end of each intron is shown. The branchpoint A is shown in bold, and the four nucleotide changes in βPy↓ relative to βPy↑ are indicated.
Ten RS dipeptides can functionally substitute for the entire RS domain of SF2/ASF
As ΔRS is unable to complement an S100 extract for IgM M1–M2 pre-mRNA splicing, we further tested whether adding 10 consecutive RS dipeptides to the C terminus of ΔRS is sufficient to restore splicing activity. RS10 complemented splicing of IgM M1–M2 pre-mRNA, although not as efficiently as wild-type SF2/ASF (Fig. 4A). Without exception, RS10 promoted splicing of all substrates tested for which no activity was obtained with ΔRS (Fig. 5; data not shown). Therefore, 10 consecutive RS dipeptides can replace the entire natural RS domain of SF2/ASF in the context of splicing in vitro. These results indicate that a stretch of alternating arginine and serine residues, in which at least some of the serines are phosphorylated (Fig. 2), can serve as a minimal interface for the relevant protein–protein interactions.
An RS domain is required for exon-independent splicing
One of the proposed functions of SR proteins is to mediate protein–protein interactions across the intron during spliceosome assembly. Recent evidence showed that RNA substrates with only one nucleotide of exon sequence can undergo the first transesterification step of the splicing reaction in vitro; this reaction requires SR proteins, and natural RS domain alone has weak activity (Hertel and Maniatis 1999). Consistent with the previous report, wild-type SF2/ASF could activate the first step of exon-independent splicing in S100 extract (Fig. 5, lanes 3,4). The RS domain of SF2/ASF was required for splicing of the minimal exon substrate, as ΔRS protein failed to complement (Fig. 5, lanes 5,6). RS10 protein could also promote lariat formation, albeit much less efficiently than wild-type SF2/ASF (Fig. 5, lanes 7,8). We conclude that the RS domain of SF2/ASF is required to mediate protein–protein interactions across the intron in the absence of exon sequences, and conversely, exon sequences are required for SF2/ASF lacking an RS domain to promote splicing.
The RS domain of SF2/ASF is required for splicing of a pre-mRNA with a weak 3′ splice site
ESEs are thought to be capable of compensating for the presence of weak splice sites by promoting U2AF65 recruitment to the polypyrimidine tract (Wu and Maniatis 1993; Zuo and Maniatis 1996). If this model is correct, and RS domain-mediated protein–protein interaction between U2AF35 and SR proteins is necessary, weakening the polypyrimidine tract of an RS domain-independent substrate should result in the SR protein RS domain becoming indispensable. To test this prediction, we analyzed a series of β-globin pre-mRNAs with different polypyrimidine tract strengths (Reed 1989) in S100 complementation assays with SF2/ASF containing or lacking the RS domain. βPy↑, βPy↓, and βPyΔ are β-globin derivatives with an improved, a weakened, or without a polypyrimidine tract, respectively. As expected, no significant difference was observed between SF2/ASF and ΔRS for splicing of βPy↑ (Fig. 6A, lanes 3–6). However, the RS domain of SF2/ASF was required for splicing of βPy↓ pre-mRNA (Fig. 6A, lanes 9–12), although only low levels of splicing were observed in S100 extract. With both substrates, the levels of splicing obtained in the presence of RS10 were intermediate between those obtained with SF2/ASF and ΔRS (data not shown). No splicing of βPyΔ pre-mRNA was observed under any conditions, as expected (Fig. 6A, lanes 13–18). Because the RS domain requirement depends on the strength of the polypyrimidine tract, these results strongly suggest that the RS domain of SR proteins is required for stable binding of U2AF65 to a weak polypyrimidine tract.
To measure U2AF65 binding to the βPy substrates, UV cross-linking and immunoprecipitation experiments were carried out. Consistent with the observation that U2AF65 binds preferentially to consecutive pyrimidines, no U2AF65 cross-linking was detected in the absence of a polypyrimidine tract (Fig. 6B, lanes 9–12). In contrast, a 65-kD band was detected when cross-linking was performed in nuclear extract with either βPy↑ or βPy↓ pre-mRNAs (Fig. 6B, lanes 1,5). Striking differences in U2AF65 cross-linking between βPy↑ or βPy↓ were observed in S100 complementation reactions. In the absence of SR proteins, U2AF65 cross-linked to βPy↑ but not to βPy↓ (Fig. 6B, lanes 2,6). Adding either SF2/ASF or ΔRS to the S100 extract increased U2AF65 cross-linking to βPy↑, although not dramatically (Fig. 6B, lanes 3,4). In contrast, the RS domain of SF2/ASF was required to promote U2AF65 binding to a weak polypyrimidine tract, in that SF2/ASF but not ΔRS enhanced U2AF65 cross-linking to βPy↓ pre-mRNA (Fig. 6B, lanes 7,8). Consistent with the functional splicing data, the cross-linking results demonstrate that at least one RS domain-dependent function of SR proteins is to recruit U2AF65 to a weak polypyrimidine tract. Considering that ΔRS can function in the splicing of several substrates, we speculate that the splicing activity attributable to the RNA-binding domain of SF2/ASF is distinct from the interactions that result in U2AF65 binding. Because this portion of the protein comprises both RRMs, RNA binding is likely involved, although the precise mechanism of action is unclear.
U2AF35 is required for splicing of a substrate with a weak polypyrimidine tract
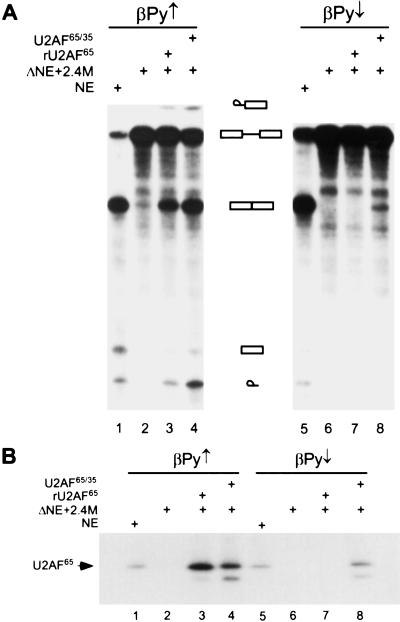
The requirement of U2AF35 for in vitro splicing has been controversial (Zuo and Maniatis 1996; Kan and Green 1999). Depletion of U2AF35 from HeLa nuclear extract prevents splicing of some enhancer-dependent substrates, but not others, and inconsistent results with the same substrate have been reported by different laboratories (Guth et al. 1999; Kan and Green 1999). Recently, site-specific cross-linking experiments showed that U2AF35 directly contacts the AG dinucleotide at the 3′ splice site during a very early step of spliceosome assembly (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). The requirement for U2AF35 in splicing appears to be substrate specific, but at least some introns with a weak polypyrimidine tract depend on the U2AF35-3′ splice site interaction to promote U2AF65 binding and splicing (Wu et al. 1999). The βPy↓ substrate, which is RS domain dependent, has a weak polypyrimidine tract, and we therefore tested whether splicing of βPy↓ is U2AF35 dependent by depletion and add-back experiments. Depleted nuclear extract (ΔNE) was made by passing the HeLa nuclear extract through a poly(U)-Sepharose column, which removes most of the U2AF65 and U2AF35, resulting in loss of splicing (Zamore and Green 1991; MacMillan et al. 1997). With the strong polypyrimidine tract substrate (βPy↑), either U2AF65 made in bacteria (rU2AF65) or baculovirus-expressed U2AF65/U2AF35 heterodimer (U2AF65/35) complemented ΔNE to restore splicing activity (Fig. 7A, lanes 3,4). In contrast, only U2AF65/35 heterodimer, but not rU2AF65 alone, restored splicing in ΔNE with the weak polypyrimidine tract substrate (βPy↓) (Fig. 7A, lanes 7,8; M. Hastings and A.R. Krainer, unpubl.). These data show that βPy↓ is an AG-dependent substrate and requires U2AF35 for efficient splicing.
Figure 7.
U2AF35 is necessary for splicing of a pre-mRNA with a weak 3′ splice site. (A) In vitro splicing of βPy↑ or βPy↓ pre-mRNAs in HeLa nuclear extract (NE; lanes 1,5), U2AF-depleted nuclear extract (ΔNE) plus 2.4 M NaCl wash (lanes 2,6), and ΔNE plus 2.4 M wash complemented with 0.3 μg rU2AF65 (lanes 3,7) or 0.5 μg U2AF65/35 (lanes 4,8). (B) UV cross-linking of U2AF65 to βPy↑ (lanes 1–4) or βPy↓ (lanes 5–8) under splicing conditions in HeLa nuclear extract (lanes 1,5), ΔNE plus 2.4 M wash (lanes 2,6), and ΔNE plus 2.4 M wash complemented with rU2AF65 (lanes 3,7) or U2AF65/35 (lanes 4,8). The faint band below U2AF65 is a degradation product of U2AF65 in the recombinant protein preparations.
To further address the relationship between a U2AF35 requirement for splicing and U2AF65 binding to the polypyrimidine tract, UV cross-linking and immunoprecipitation experiments were carried out in poly(U)-depleted nuclear extract, to which U2AF65 or U2AF65/35 were added back (Fig. 7B). No U2AF65 cross-linking was observed in ΔNE, as expected, because most of the U2AF65 and U2AF35 had been depleted (Fig. 7B, lanes 2,6). Adding back either rU2AF65 or U2AF65/35 to ΔNE restored U2AF65 cross-linking with the βPy↑ substrate (Fig. 7B, lanes 3,4). In contrast, with the βPy↓ substrate, cross-linking of U2AF65 was seen only when both U2AF65 and U2AF35 were present (Fig. 7B, lane 8). Although U2AF65 was made in bacteria and U2AF65/35 was made in baculovirus, the extent of U2AF65 cross-linking was comparable with both preparations in the case of the control βPy↑ substrate (Fig. 7B, lanes 3,4) but not the βPy↓ substrate (Fig. 7B, lanes 7,8). Likewise, both U2AF preparations had comparable splicing activity for the βPy↑ substrate (Fig. 7A, lanes 3,4) but not the βPy↓ substrate cross-linking (Fig. 7A, lanes 7,8).
These results confirm and extend the in vitro splicing data, establishing that although U2AF35 is dispensable in certain situations, it is crucial to stabilize U2AF65 binding to a weak polypyrimidine tract. Together with the observation that U2AF35 and SF2/ASF interact directly (Xiao and Manley 1998), the requirement for both the RS domain of SF2/ASF and U2AF35 for efficient U2AF65 binding suggests that these two factors act in a linear pathway. Although it would be of interest to test the requirement for U2AF35 in the presence of wild-type or RS domain-deleted SF2/ASF as the sole SR protein, our efforts to efficiently deplete U2AF from S100 extract and reconstitute splicing with recombinant proteins have not been successful.
Discussion
SR protein RS domain requirements for in vitro splicing
We report here that SF2/ASF lacking the RS domain (ΔRS) is active in S100 complementation assays with multiple splicing substrates, suggesting that the RS domain of this SR protein is not strictly required for in vitro splicing. This finding contradicts previous reports (Cáceres and Krainer 1993; Zuo and Manley 1993) and a likely explanation for the discrepancy stems from the previous use of N-terminal oligo-histidine tags. We speculate that an N-terminal tag has a subtle effect on the structure of RRM1 or the short segment immediately preceding it, interfering with an interaction that can also be mediated by the RS domain. For example, SF2/ASF may interact with another component through two separate contacts that are to some extent redundant. As a precedent, partially redundant signals that mediate the nuclear localization of SF2/ASF are also present within its RNA-binding and RS domains (Cáceres et al. 1998). Moreover, the crystal structure of an untagged UP1 fragment of hnRNP A1 shows that the N-terminal RRM1 is preceded by a short 310 helix that contacts bound nucleic acid (Ding et al. 1999), and N-terminal his-tagged hnRNP A1 is less active in splice-site switching in vitro than untagged protein (L. Manche and A.R. Krainer, unpubl.). Although protein tags can be very useful for purification and detection, their use in structure/function studies should be approached with caution, even when the wild-type tagged protein is active.
Although our results indicate that the biochemical requirement for the RS domain of SF2/ASF in splicing is not as stringent as previously thought, they do not imply that this domain plays no role in splicing. First, although it is difficult to compare the specific activities of SF2/ASF and ΔRS proteins, given the required renaturation steps, the overall trend showed that SF2/ASF is slightly more active than ΔRS in constitutive splicing. Second, ΔRS protein was inactive with certain enhancer-dependent or exon-independent substrates, as well as with a substrate with a weak 3′ splice site. These results argue that although the RS domain of SF2/ASF is dispensable under many circumstances, it is involved in the splicing mechanism, and the extent of its involvement is substrate specific. These data also support the notion that formation of functional spliceosomes can be achieved by multiple redundant mechanisms. It will be interesting to further investigate the similarities and differences between RS domain-dependent and RS domain-independent splicing complexes.
We cannot rule out the possibility that an SR protein RS domain is always required for splicing. If so, ΔRS protein may function in splicing by somehow recruiting another SR protein present in trace amounts in the S100 extract, with the latter protein providing its own RS domain. Although interactions between SR proteins appear to be mediated by the respective RS domains, the rest of the protein may contribute to weak protein–protein interactions. The presumptive weak interaction of SF2 ΔRS with trace endogenous SR proteins may be sufficient for splicing of some substrates, whereas other substrates clearly require SF2/ASF to have its own RS domain. In this sense, the mechanistic difference between RS domain-dependent and RS domain-independent substrates may be quantitative rather than qualitative.
Possible mechanisms for SR protein function in general splicing
The RS domain of SR proteins can participate in both protein–RNA and protein–protein interactions. Phosphorylation of serines within the RS domain influences RNA binding of SR proteins in vitro by preventing nonspecific interactions with RNA (Tacke et al. 1997). Protein–protein interactions between SR proteins and SR-related splicing factors, such as U1-70K, U2AF35, or SR protein-specific kinases, are mediated by the RS domains and regulated by reversible phosphorylation of these domains (Wu and Maniatis 1993; Cao et al. 1997; Xiao and Manley 1997, 1998; Koizumi et al. 1999; Prasad et al. 1999; Yeakley et al. 1999). However, the data presented here show that even without the RS domain, SF2/ASF ΔRS—which is not detectably phosphorylated—remains functional in S100 complementation assays with many, but not all, splicing substrates. Therefore, with these substrates, cycles of phosphorylation and dephosphorylation of the RS domain are less critical for splicing than originally proposed. It also follows that SR proteins can promote splicing by at least two distinct mechanisms, which are not mutually exclusive.
First, SR proteins function in an RS domain-dependent manner, and the requirement for this domain likely reflects the importance of RS domain-mediated protein–protein interactions in removal of certain introns. One example is exon-independent lariat formation (Hertel and Maniatis 1999) assayed by S100 extract complementation. With virtually no exon sequences present, we find that the RS domain of SF2/ASF is crucial to bridge the two splice sites together and to promote the first step of splicing. Similarly, the RS domain of SF2/ASF is required for recruiting U2AF65 to a β-globin substrate with a weak polypyrimidine tract (Fig. 6). Presumably, these interactions reflect an intron definition model (Robberson et al. 1990), and involve a network of RS domain-mediated interactions between SF2/ASF, U1-70K and U2AF65/35 (Wu and Maniatis 1993; Zuo and Maniatis 1996).
The second mechanism by which SR proteins promote splicing does not require the RS domain. In addition to our present results with SF2/ASF ΔRS, several observations support this notion. The RS domain of SF2/ASF was recently found not to be required for enhancing U1 snRNP binding to alternative 5′ splice sites (Eperon et al. 2000). Furthermore, using MS2–RS domain chimeric proteins to study the trans-activating properties of the RS domain, it was found that MS2–RS does not function in S100 extract complementation to enhance splicing via an MS2 RNA-binding site unless a recombinant SR protein is also added (Graveley and Maniatis 1998). The major difference between MS2–RS and genuine SR proteins resides in their RNA-binding domains. One explanation for the incomplete activity of MS2–RS is that the RNA-binding domain of SR proteins has specific functions in splicing besides binding to the pre-mRNA.
Little is known at present about the molecular basis for the RS domain-independent function of SR proteins. One possibility is that the RRMs of SF2/ASF, or the short flanking segments, comprise a surface(s) involved in specific protein–protein interactions, which may or may not overlap with the interactions mediated by the RS domain. For example, the RS domain of SF2/ASF is not sufficient for interaction with U1-70K, and a GST–SF2/ASF ΔRS fusion protein can interact with U1-70K, albeit more weakly than in the presence of the RS domain, in an apparently RNA-independent manner (Xiao and Manley 1997). The RRMs of SF2/ASF and SRp20 are capable of interacting with several proteins, although none of them are known to be involved in splicing (Ge et al. 1998; Elliott et al. 2000). A related question is why some SR proteins have only one RRM, whereas others have two. RRM2, which is somewhat atypical (Birney et al. 1993), may have evolved to mediate protein–protein interactions, as well as coordinate RNA binding with RRM1 (Chandler et al. 1997).
An alternative mechanism of RS domain-independent SF2/ASF function is that binding via the RRMs to the pre-mRNA is sufficient to promote splicing by competing with negative factors, such as hnRNPs (Eperon et al. 2000; for review, see Reed 2000). In the absence of other components, SF2/ASF lacking the RS domain can bind to the same sequences recognized preferentially by phosphorylated SF2/ASF (Tacke et al. 1997), and it can displace hnRNP A1 (Eperon et al. 2000). The sequence specificity of several individual SR proteins has been studied by binding or functional SELEX (Liu et al. 2000, and references therein). Recognition sites conform to degenerate, short consensus motifs unique to each SR protein. Some of these motifs are more prevalent in exons than in introns (for a given length of RNA) but they occur multiple times in most exons. Similarly, hnRNP A1, which antagonizes SF2/ASF in a concentration-dependent manner for alternative 5′ splice-site selection (Mayeda and Krainer 1992) and functions as a silencing factor for splicing of several pre-mRNAs (Caputi et al. 1999; Del Gatto-Konczak et al. 1999), can bind RNA promiscuously (Abdul-Manan and Williams 1996). These binding patterns imply that antagonism between positive and negative splicing factors may be derived from competition for overlapping pre-mRNA binding sites. Likewise, RSF1, a Drosophila splicing repressor, may compete with SR proteins for common binding sites (Labourier et al. 1999).
SR proteins and exonic splicing enhancers
The mechanism of ESE function has been controversial with respect to the role of U2AF binding (for review, see Blencowe 2000). Early experiments supported a U2AF-recruitment model, in which SR proteins bound to ESEs promote splicing by facilitating the binding of U2AF65 to the polypyrimidine tract through an interaction mediated by the RS domains of an SR protein and of U2AF35 (Wu and Maniatis 1993; Zuo and Maniatis 1996). However, more recent work argues that binding of U2AF65 to certain ESE-dependent pre-mRNAs does not require interactions mediated by an ESE or U2AF35. Instead, these ESEs may function in part by antagonizing juxtaposed silencers (Guth et al. 1999; Kan and Green 1999). Moreover, experiments with transgenic flies showed that a Drosophila U2AF small subunit lacking the RS domain is functional in vivo and can activate enhancer-dependent dsx pre-mRNA splicing, as long as the U2AF large subunit has its RS domain (Rudner et al. 1998). On the basis of our data ESEs can be divided into RS domain-dependent and RS domain-independent. It is possible that for substrates like βPy↑ and tat23 pre-mRNAs, U2AF65 binding to the polypyrimidine tract is not the rate-limiting step. Thus, the enhancers that function independently of the RS domain of SF2/ASF may promote splicing by counteracting certain silencers through competition with the cognate RNA-binding proteins for overlapping binding sites, and/or by protein–protein interactions between SR proteins and components of the general splicing machinery other than U2AF65/35. In contrast, when binding of U2AF65 to the polypyrimidine tract is rate limiting, the requirement for the RS domain of SF2/ASF correlates with the recruitment of U2AF65.
It was recently reported that U2AF35 recognizes the conserved 3′ splice site AG dinucleotide and might stabilize the binding of U2AF65 on specific substrates that are dependent on the 3′ AG for splicing (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). Supporting and extending this observation, our data show that both U2AF35 and the RS domain of SF2/ASF are required for splicing of a substrate with a weak polypyrimidine tract (βPy↓), and both factors function to stabilize U2AF65 binding to the weak splice site. Although the RS domain dependence and the U2AF35 dependence were assayed in different extracts (nuclear vs. S100) for technical reasons, these results are consistent with the idea that the RS domain-mediated protein network between an SR protein and U2AF35/65 is crucial for certain ESEs to overcome weak 3′ splice site signals (Zuo and Maniatis 1996; Guth et al. 1999).
The U2AF35 requirement for IgM M1–M2 pre-mRNA splicing was reported with conflicting results by different groups (Guth et al. 1999; Kan and Green 1999), although the positive result shows that U2AF35 is required at least under some conditions. Significantly, both studies showed that depletion of U2AF35 does not affect the extent of cross-linking of U2AF65 to the polypyrimidine tract. Because we now show that the RS domain of SF2/ASF is also required for splicing of this substrate, one possibility is that U2AF35 functions downstream to stabilize SR protein binding to the ESE, which in turn antagonizes the nearby splicing silencer.
Ten RS dipeptides can functionally substitute for the entire RS domain of SF2/ASF
The RS domains of SR proteins are well characterized splicing trans-activating domains. RS domains from individual SR proteins, when fused to the bacteriophage MS2 coat protein RNA-binding domain, can activate splicing of substrates with the MS2-binding site replacing an ESE (Graveley and Maniatis 1998). We report here that RS10, in which the 51-amino-acid RS domain of SF2/ASF was replaced by 10 consecutive RS dipeptides, is active in splicing of all substrates tested, including RS domain-dependent substrates. Moreover, RS10 can be phosphorylated either by endogenous kinases present in the S100 extract, or by the recombinant SR protein-specific kinases SRPK2 and Clk/Sty.
Given that 10 consecutive RS dipeptides linked to the SF2/ASF RNA-binding domain are sufficient for trans-activating splicing in vitro, it is surprising to note that the common features of the C-terminal RS domains of different SR proteins are limited to the overall composition and the presence of numerous consecutive RS or SR dipeptides, even though the RS domains of individual SR proteins are as highly conserved between true orthologs as the rest of the protein (Birney et al. 1993). The very high degree of phylogenetic conservation of the RS domain of individual SR proteins is suggestive of a specific function in vivo for each family member. A similar situation was reported for U2AF65. Whereas a synthetic RS domain consisting of seven consecutive RS dipeptides is sufficient for human U2AF65 to recruit U2 snRNP in vitro (Valcárcel et al. 1996), the identical synthetic RS domain is insufficient for Drosophila U2AF large subunit activity in vivo in the absence of the small subunit RS domain (Rudner et al. 1998). As suggested previously, (Cáceres et al. 1997, 1998), the high conservation of RS domain sequences may reflect primarily the unique properties of individual SR proteins in subnuclear targeting and nucleo-cytoplasmic shuttling.
Materials and methods
Construction of SF2/ASF mutants
ΔRS was constructed by subcloning the NdeI–BamHI fragment of pET19b-RRMΘRRM of SF2/ASF (Cáceres and Krainer 1993) into the pET9c expression vector (Novagen). RS10 was constructed by partial gene replacement, as described (Cáceres and Krainer 1993). Two partially complementary oligonucleotides (RS10-1, 5′-catgggccccgctctggtagccgctccctgtctcgcagccgttcgcgc-3′; RS10-2, 5′-cttgaatccttagctacgggagcggctacgagagcgcgaacggctgcgag aag-3′) were annealed and filled in with Sequenase 2.0 (US Biochemical). After digestion with ApaI and BamHI, the resulting restriction fragment was gel purified and subcloned into the corresponding sites in pET9c-SF2(R/S) (Krainer et al. 1991). The procedure inadvertently introduced a 6-nucleotide insertion preceding the ApaI site, placing a His-Gly dipeptide between Pro197 and Arg198. In addition, the serine in the penultimate RS repeat was changed to a cysteine. Although the experiments in Figures 1–5 were done with the original RS10 preparation, we have recently reconstructed a correct version of the protein without insertions or substitutions. This bonafide RS10 preparation is at least as active as the original one.
Preparation of recombinant proteins
Expression and initial fractionation of untagged recombinant wild-type and mutant SF2/ASF were essentially as described (Krainer et al. 1991). Recombinant SF2/ASF was exclusively recovered in the pellet after separation by CsCl density gradient centrifugation and dialysis. The pellet was dissolved and denatured by sonication in buffer D (20 mM Hepes-NaOH at pH 8.0, 0.2 mM EDTA, 5% glycerol (v/v), 1 mM DTT, 0.5 mM PMSF) containing 0.1 M KCl and 6 M urea, followed by rocking for 1 hr at 4°C. The protein was purified by Perseptive HS chromatography in a Perseptive Biosystem under denaturing conditions in urea. The combined peak fractions were dialyzed against buffer D with 0.1 M KCl and 3 M urea and then against the same buffer without urea. The final protein concentration was determined by the dye-binding method (BioRad) with BSA as a standard. ΔRS protein was further purified by Perseptive HQ chromatography under urea-denaturing conditions. The final dialysis was against buffer D with 0.4 M KCl to maintain solubility.
Untagged recombinant U2AF65 was also expressed in the pET-9c vector and similarly fractionated by CsCl gradient centrifugation. The protein remained soluble after dialysis against buffer D with 0.1 M KCl. It was purified on a Perseptive HQ column eluted with a linear gradient from 0.1 M to 1 M NaCl in buffer D, and the combined peak fractions were dialyzed against buffer D with 0.1 M KCl. U2AF65/35 heterodimer, kindly provided by M. Hastings, was prepared as described (Graveley and Maniatis 1998) by use of a baculovirus stock generously provided by B. Graveley (University of Connecticut Health Center, Farmington).
Transcripts
7CH3GpppG-capped, 32P-labeled pre-mRNA substrates were made by runoff transcription from either linearized templates or purified PCR products with an SP6 or T7 promoter (Mayeda and Krainer 1999a). β-globin and derivative plasmid templates, except βPyΔ, have been described (Reed and Maniatis 1986; Reed 1989; Krainer et al. 1990a). HIV-tat, IgM M1–M2, Drosophila ftz, and AdML were transcribed from plasmids pSP64-HIV-1tat23, pμM1–M2, pSPftz, and pADML-PAR as described (Inoue et al. 1990; Krainer et al. 1990b; Watakabe et al. 1993; Chew et al. 1999). βPyΔ was constructed by overlap-extension PCR. Two sets of PCRs were performed with βPy↑ as template. The first PCR was carried out with primers βPy1 (5′-gaatacaagcttgctt ac-3′) and βPy4 (5′-caccaccagcgtccagtgcccaccag-3′). The second PCR used primer βPy2 (5′-ccggggatccacg-3′) and βPy3 (5′-ctg gtggggcactggacgctggtggtg-3′). The products from the two reactions were then combined and further amplified with primers βPy1 and βPy2. The resulting PCR product was digested with HindIII and BamHI and subcloned into the parent vector. The exon-independent substrate with only 3 nucleotide of upstream exon was made by PCR as described (Hertel and Maniatis 1999) using the primers T7E3, 5′-taatacgactcactataggggtgagtactcc ctctcaaaagc-3′ and AdMLPY, 5′-agagagagaggaaaaaaaagggaaag ggtcagc-3′, and was transcribed without a 5′ cap.
Phosphorylation and immunoprecipitation
Phosphorylation in extracts under splicing conditions was carried out in 10-μL reactions lacking polyvinyl alcohol and labeled pre-mRNA, and containing 10 pmole of SF2/ASF or mutant proteins and in some cases 0.5 μL [γ-32P]ATP (5 μCi, 6000 Ci/mmole). For Westerns and IP/Westerns, anti-SF2/ASF mAb96 was used as described (Hanamura et al. 1998). Phosphorylation with recombinant kinases was performed in 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM cold ATP, 0.5 μL [γ-32P]ATP, by use of a 1:10 enzyme to substrate ratio. The reactions were incubated at 30°C for 15 min. 12.5 μL of a 1:1 slurry of protein A-agarose coupled with anti-SF2/ASF mAb96 (Hanamura et al. 1998) was added and the total volume was brought up to 400 μL with IP100 buffer (50 mM Tris-HCl at pH 7.6, 2 mM MgCl2, 100 mM KCl, 0.5 mM DTT, 0.25 % NP40). After rocking at 4°C for 1 hr, the beads were pelleted in a microcentrifuge at 1000 rpm for 15 sec, followed by washing four times with IP100. Bound protein was released with SDS-sample buffer and analyzed by SDS-PAGE and autoradiography.
In vitro splicing assays
HeLa cell nuclear and S100 extracts were prepared as described (Mayeda and Krainer 1999b). Standard conditions were used for the splicing reactions (Mayeda and Krainer 1999a). Briefly, 10 fmole of 32P-labeled 7CH3GpppG-capped SP6 or T7 transcripts were incubated in 10-μL splicing reactions. Each reaction contained either 30% HeLa nuclear extract, or 40% S100 extract complemented with 5–10 pmole of SF2/ASF (wild-type or mutant) protein. The final MgCl2 concentration varied between 1.6–4.8 mM, depending on the substrate and previously described optima. After incubation at 30°C for 2–4 hr, the RNA was extracted and analyzed on 5.5% or 12% polyacrylamide denaturing gels, followed by autoradiography.
UV cross-linking and immunoprecipitation
UV crosslinking and immunoprecipitation were carried out essentially as described (Guth et al. 1999; Kan and Green 1999). Briefly, 37.5-μL standard splicing reactions lacking polyvinyl alcohol were incubated at 30°C for 20 min, placed on ice, and exposed to 254-nm UV light at 0.56 J/cm2 in a Spectronics XL-1000 instrument. RNases A (10 μg) and T1 (100 units) were added and the reactions were incubated for 15 min at 37°C. The reactions were then incubated with 48 μL of MC3 monoclonal antibody culture supernatant (a generous gift from J. Valcárcel, EMBL, Heidelberg, Germany) for 1 hr on ice. Twelve microliters of anti-mouse IgG agarose beads (1:1 suspension) was added and the total volume brought up to 160 μL with IP100 buffer. After rocking at 4°C for 1hr, the samples were spun at 1000 rpm for 30 sec to pellet the beads, and the supernatant was removed. The beads were washed two times with IP500 (same as IP100 but with 500 mM KCl) and two times with IP100 buffer. Immunoprecipitated proteins were released by boiling in SDS-sample buffer, and analyzed by electrophoresis on a 12% SDS-polyacrylamide gel followed by autoradiography.
Depletion of HeLa nuclear extract
U2AF65/35 was depleted from HeLa nuclear extract by poly(U)-Sepharose chromatography (Zamore and Green 1991; MacMillan et al. 1997). Briefly, HeLa nuclear extract was first adjusted to 1 M KCl by dialyzing against buffer F (20 mM HEPES-KOH at pH 7.9, 1 M KCl, 3 mM MgCl2, 0.05 % NP-40, 1 mM DTT) with 20% glycerol. Poly(U)-Sepharose 4B (Pharmacia) was washed with buffer D and equilibrated with buffer F with 10% glycerol. Dialyzed nuclear extract was loaded on the column and washed with 10 column volumes of buffer F with 10% glycerol. The protein-peak fractions in the flowthrough were pooled to give U2AF65/35-depleted extract (ΔNE). The column was eluted sequentially with buffer F with 10% glycerol and 2.4 M KCl, and buffer F with 0.1 M KCl and 2 M guanidine hydrochloride, and the protein-peak fractions, designated 2.4 M and hU2AF, respectively, were pooled. All three fractions were dialyzed against buffer D with 0.1 M KCl and stored frozen.
Acknowledgments
We thank Lisa Manche for constructing the RS10 plasmid and Ikuko Watakabe for the ΔRS plasmid. We are grateful to Brent Graveley for the U2AF65/35 baculovirus stock and for useful discussions, Michelle Hastings for U2AF65/35 recombinant protein and helpful advice, Masatoshi Hagiwara for SRPK2 and Clk/Sty kinases, Robin Reed for β-globin derivative plasmids, Klemens Hertel for advice on exon-independent splicing, and Juan Valcárcel for the MC3 antibody and for helpful suggestions. We thank Shern Chew, Michelle Hastings, and Akila Mayeda for helpful comments on the manuscript. This work was supported by grant no. GM42699 from the NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL krainer@cshl.org; FAX (516) 367-8453.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.189500.
References
- Abdul-Manan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: Utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . Mammalian pre-mRNA splicing factors. In: Krainer AR, editor. Eukaryotic mRNA processing. New York, NY: Oxford University Press; 1997. pp. 174–212. [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler SD, Mayeda A, Yeakley JM, Krainer AR, Fu X-D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SL, Liu H-X, Mayeda A, Krainer AR. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc Natl Acad Sci. 1999;96:10655–10660. doi: 10.1073/pnas.96.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakley JM, Gish GD, Cáceres JF, Pawson T, Fu X-D. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes & Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, McGuffin ME, Dauwalder B, Rabinow L, Mattox W. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol Cell. 1998;2:741–750. doi: 10.1016/s1097-2765(00)80289-0. [DOI] [PubMed] [Google Scholar]
- Elliott DJ, Bourgeois CF, Klink A, Stévenin J, Cooke HJ. A mammalian germ cell-specific RNA-binding protein interacts with ubiquitously expressed proteins involved in splice site selection. Proc Natl Acad Sci. 2000;97:5717–5722. doi: 10.1073/pnas.97.11.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Cáceres JF, Hayward DG, Krainer AR. Selection of alternative 5′ splice sites: Role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H, Si Y, Wolffe AP. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell. 1998;2:751–759. doi: 10.1016/s1097-2765(00)80290-7. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- Guth S, Martinez C, Gaur RK, Valcárcel J. Evidence for substrate-specific requirement of the splicing factor U2AF(35) and for its function after polypyrimidine tract recognition by U2AF(65) Mol Cell Biol. 1999;19:8263–8271. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Cáceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Maniatis T. Serine-arginine (SR)-rich splicing factors have an exon-independent function in pre-mRNA splicing. Proc Natl Acad Sci. 1999;96:2651–2655. doi: 10.1073/pnas.96.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Sakamoto H, Shimura Y. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990;344:461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- Jamison SF, Pasman Z, Wang J, Will C, Lührmann R, Manley JL, Garcia-Blanco MA. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: Characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan JL, Green MR. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes & Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka A, Mühlemann O, Petersen-Mahrt S, Estmer C, Öhrmalm C, Akusjärvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) J Biol Chem. 1999;274:11125–11131. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990a;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- ————— Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes & Dev. 1990b;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi N, Onogi H, Wakabayashi T, Hagiwara M. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem Biophys Res Commun. 1998;242:357–364. doi: 10.1006/bbrc.1997.7913. [DOI] [PubMed] [Google Scholar]
- Labourier E, Bourbon HM, Gallouzi IE, Fostier M, Allemand E, Tazi J. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes & Dev. 1999;13:740–753. doi: 10.1101/gad.13.6.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Blencowe BJ. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J Biol Chem. 1999;274:35074–35079. doi: 10.1074/jbc.274.49.35074. [DOI] [PubMed] [Google Scholar]
- Liu H-X, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan AM, McCaw PS, Crispino JD, Sharp PA. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc Natl Acad Sci. 1997;94:133–136. doi: 10.1073/pnas.94.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- ————— Mammalian in vitro splicing assays. Methods Mol Biol. 1999a;118:315–321. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- ————— Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol Biol. 1999b;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Screaton GR, Chandler SD, Fu X-D, Krainer AR. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcárcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- Misteli T. RNA splicing: What has phosphorylation got to do with it? Curr Biol. 1999;9:R198–R200. doi: 10.1016/s0960-9822(99)80128-6. [DOI] [PubMed] [Google Scholar]
- Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: Both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes & Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- ————— Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Robberson BL, Cote GJ, Berget SM. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Breger KS, Rio DC. Molecular genetic analysis of the heterodimeric splicing factor U2AF: The RS domain on either the large or small Drosophila subunit is dispensable in vivo. Genes & Dev. 1998;12:1010–1021. doi: 10.1101/gad.12.7.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Bruzik JP. Developmental regulation of SR protein phosphorylation and activity. Genes & Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: Creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc Natl Acad Sci. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu X-D. SRPK2: A differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998a;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xiao SH, Manley JL. Genetic analysis of the SR protein ASF/SF2: Interchangeability of RS domains and negative control of splicing. Genes & Dev. 1998b;12:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hoffmann HM, Grabowski PJ. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes & Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes & Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- ————— Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Tronchère H, Olesen J, Dyck JA, Wang HY, Fu X-D. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: An essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- Zuo P, Manley JL. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]