Abstract
The SAGA complex of Saccharomyces cerevisiae is required for the transcription of many RNA polymerase II-dependent genes. Previous studies have demonstrated that SAGA possesses histone acetyltransferase activity, catalyzed by the SAGA component Gcn5. However, the transcription of many genes, although SAGA dependent, is Gcn5 independent, suggesting the existence of distinct SAGA activities. We have studied the in vivo role of two other SAGA components, Spt3 and Spt20, at the well-characterized GAL1 promoter. Our results demonstrate that both Spt3 and Spt20 are required for the binding of TATA-binding protein but not of the activator Gal4 and that this role is Gcn5 independent. These results suggest a coactivator role for Spt3 and Spt20 in the recruitment of TBP.
Keywords: SAGA complex, S. cerevisiae, transcription, Spt3, TBP
In eukaryotes, transcription initiation by RNA polymerase II (Pol II) requires not only the activities of the general transcription factors and gene-specific activators but also the activities of large coactivator complexes (Hampsey 1998; Hampsey and Reinberg 1999). The SAGA (Spt/Ada/Gcn5/acetyltransferase) complex of Saccharomyces cerevisiae is one such coactivator complex. It is required for the full transcriptional activity of a subset of Pol II-dependent genes (Hampsey 1997; Grant et al. 1998). SAGA contains >20 proteins, including Gcn5, a histone acetyltransferase (HAT) whose activity has been shown to be required for the transcription of a subset of genes in vivo (Kuo et al. 1998; Wang et al. 1998). Recent studies have implicated Gcn5 in the recruitment of activators to promoters by SAGA (Cosma et al. 1999).
In addition to the HAT activity of Gcn5, genetic and biochemical studies suggest that SAGA possesses other activities important for transcription (Horiuchi et al. 1997; Roberts and Winston 1997; Dudley et al. 1999; Sterner et al. 1999). In this paper we focus on two other components of SAGA, Spt20, and Spt3. Spt20 is required for the integrity of SAGA, as the complex cannot be detected in spt20Δ mutants (Grant et al. 1997). Spt3 is probably required for a subset of SAGA activities that are independent of Gcn5 and histone acetylation because SAGA purified from an spt3Δ mutant still possesses Gcn5-dependent HAT activity, yet spt3Δ mutants have multiple transcriptional defects (Roberts and Winston 1997; Dudley et al. 1999; Sterner et al. 1999). Previous genetic and biochemical studies have shown that Spt3 interacts with TATA-binding protein (TBP) (Eisenmann et al. 1992; Lee and Young 1998) and suggest that Spt3 is structurally similar to particular TBP-associated factors (TAFs) (Birck et al. 1998). These findings suggest a model in which Spt3 plays a role in the binding of TBP to particular promoters in vivo.
To better understand the mechanism by which the Spt components of SAGA facilitate the transcription of a subset of RNA Pol II-dependent genes, we chose to study the effects of SAGA mutations on activation by Gal4 at the well-characterized GAL1 promoter. Studies of activation of the GAL promoters by Gal4 have served as a model system for transcriptional regulation for many years (Johnston and Carlson 1992; Ptashne and Gann 1997; Zaman et al. 1998). Furthermore, the GAL1 promoter is useful for such studies because its regulation is well characterized and it has a relatively simple promoter structure. Under inducing conditions, two proteins, TBP and Gal4, are known to bind the GAL1 promoter (Johnston and Carlson 1992). Thus, of the genes known to be regulated by the Spt components of SAGA (Roberts and Winston 1997; Sterner et al. 1999), the GAL promoters are the only ones for which the Spt dependence of both TBP binding and activator binding can be measured.
In this study we use chromatin immunoprecipitation assays to determine which steps in transcriptional activation require Spt20 and Spt3 at the GAL1 promoter. Our results demonstrate a severe reduction in TBP binding at the GAL1 TATA in spt3Δ and spt20Δ mutants but only a modest decrease in a gcn5Δ mutant. These results correlate well with the effects on GAL1 transcription in these mutants. In contrast, the transcriptional activator Gal4 is bound to the GAL1 upstream activating sequence (UASG) in all three SAGA mutants. Furthermore, the acetylation of histone H3 at the GAL1 promoter is only slightly decreased in gcn5Δ, spt3Δ, and spt20Δ mutants. Taken together, our results suggest a Gcn5-independent activity for the Spt20 and Spt3 components of SAGA that is required for the recruitment of TBP, but not of activators, to a subset of Pol II-dependent promoters in vivo.
Results
GAL1 transcription is dependent on Spt3 and Spt20, but not Gcn5
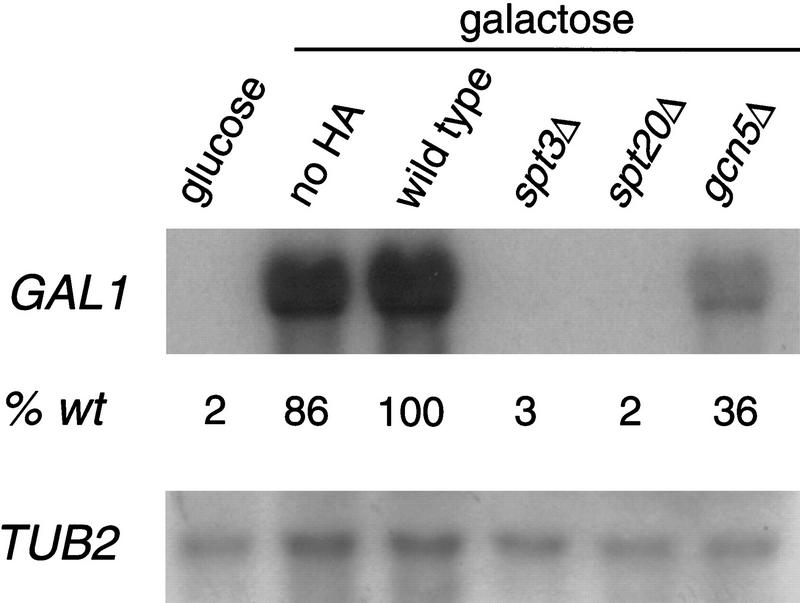
To understand the role of different SAGA functions, we examined the effects of mutations in three different classes of SAGA genes, SPT3, GCN5, and SPT20, on the well-studied GAL1 gene of S. cerevisiae. Spt3 and Gcn5 are probably required for distinct aspects of SAGA function, whereas Spt20 is believed to be required for all SAGA function (Grant et al. 1997; Sterner et al. 1999). The GAL1 promoter was particularly useful for analysis of Spt3 and Spt20 because gcn5Δ mutants are Gal+, whereas spt3Δ are moderately Gal−, and spt20Δ mutants are tight Gal− (Roberts and Winston 1997; Sterner et al. 1999). To study primary events of transcriptional activation at the GAL1 promoter, we analyzed cells at an early point in galactose induction, sufficient for the wild-type strain to have fully induced expression (Materials and Methods). At this point in galactose induction, GAL1 mRNA levels are decreased in the spt3Δ and spt20Δ mutants by ∼50-fold relative to wild-type levels (Fig. 1). In contrast, a gcn5Δ mutant shows only a modest 2.5-fold decrease in GAL1 mRNA levels. Thus, induction of the GAL1 promoter is strongly dependent on the Spt3 and Spt20 components of SAGA and only mildly dependent on the Gcn5 HAT.
Figure 1.
GAL1 mRNA levels in three classes of SAGA mutants. Northern hybridization analysis was performed on total yeast RNA prepared from glucose-repressed wild-type strains and galactose-induced wild-type, spt3Δ, spt20Δ, and gcn5Δ strains. Quantitation was performed by PhosphorImager analysis (Molecular Dynamics).
TBP binding at GAL1 is defective in spt3Δ and spt20Δ mutants
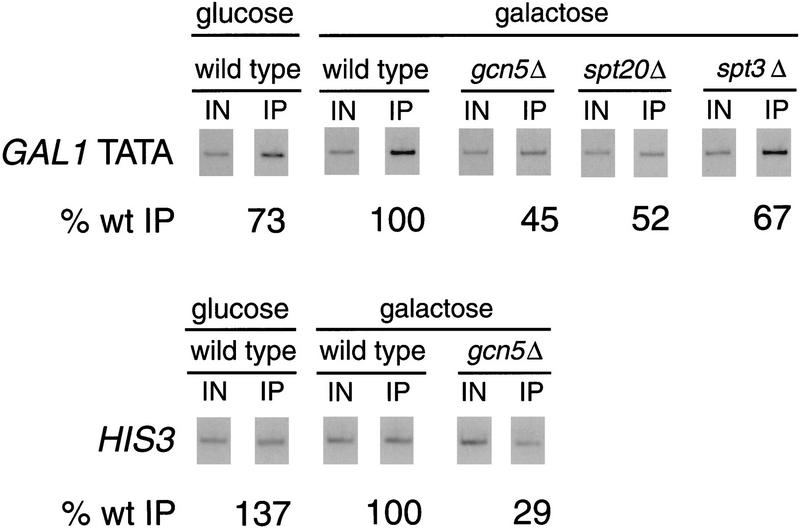
An important step in transcription initiation is the recruitment of TBP to the TATA region of a promoter. Previous studies of the GAL1 promoter in vivo demonstrated that TBP is absent from the GAL1 TATA region under conditions of both glucose repression or noninduction (raffinose or glycerol). However, upon induction with galactose, TBP associates strongly with the GAL1 TATA region (Selleck and Majors 1987a,b; Kuras and Struhl 1999; Li et al. 1999). To test whether SAGA mutations affect the binding of TBP to the GAL1 TATA in vivo, we used the chromatin immunoprecipitation assay (Dedon et al. 1991; Orlando and Paro 1993; Strahl-Bolsinger et al. 1997). We observed a strong (>10-fold) defect in the ability of TBP to bind the GAL1 TATA region in the spt3Δ and spt20Δ mutants, but only a modest (twofold) decrease in TBP binding in the gcn5Δ mutant (Fig. 2). These results strongly suggest that the spt3Δ and spt20Δ defects in GAL1 activation are caused by an inability to stably bind TBP to the promoter.
Figure 2.
TBP is not bound to the GAL1 TATA in spt3Δ and spt20Δ mutants. Chromatin immunoprecipitation was performed in parallel with RNA analysis (Fig. 1) from glucose-repressed wild-type strains and galactose-induced wild-type, spt3Δ, spt20Δ, and gcn5Δ strains. A galactose-induced wild-type strain that contained TBP without the triple HA tag (no HA) was included as a negative control for the immunoprecipitation. TBP was immunoprecipitated using the 12CA5 antibody against the HA epitope. PCR products correspond to the GAL1 TATA region (GAL1), the TUB2 TATA region (TUB2), or the POL1 open reading frame as a negative control. The percentage of DNA immunoprecipitated (% wt IP) in each of the mutants was normalized to the amount immunoprecipitated from the galactose-induced wild-type strain. One set of PCR reactions is shown, and the quantitation represents the average of several experiments. The average values with standard errors for the measurements of TBP binding to the GAL1 TATA on galactose-grown cells are as follows: spt3Δ, 9 ± 2; spt20Δ, 6 ± 2; gcn5Δ, 44 ± 3; No HA, 2 ± 1. The values for binding to the TUB2 TATA on galactose-grown cells are: spt3Δ, 87 ± 34; spt20Δ, 54 ± 10; gcn5Δ, 131 ± 22; No HA, 8 ± 2. The values for wild-type glucose-grown cells are as follows: GAL1, 6 ± 3; TUB2, 66 ± 8. The low level of DNA detected in the No-HA and the POL1 PCR reactions represents the low amount of TBP-independent DNA precipitated as background in this assay. The POL1 negative control was performed on all samples, and the results were essentially the same as the example shown.
Gal4 binding is unaffected in spt3Δ and spt20Δ mutants
One condition that could account for both the defect in GAL1 transcription and in TBP binding is the inability of the transcriptional activator Gal4 to bind to UASG in the spt mutants. To test this hypothesis, we used chromatin immunoprecipitation assays to analyze the occupancy of the UASG by Gal4 (Fig. 3). Interestingly, in all the SAGA mutants tested we observed in vivo occupancy of the UASG by Gal4 at levels similar to that of wild type. Therefore, Spt3 and Spt20 are not required for Gal4 binding. Together with the defects in TBP binding in spt3Δ and spt20Δ mutants, these results strongly suggest that Gal4 cannot recruit TBP in the absence of Spt3 or Spt20.
Figure 3.
Gal4 occupies the UASG in SAGA mutants. Chromatin immunoprecipitation was performed in parallel with RNA analysis (Fig. 1) from glucose-repressed wild-type strains and galactose-induced wild-type, spt3Δ, spt20Δ, or gcn5Δ strains. A galactose-induced gal4Δ strain was included as a negative control for the immunoprecipitation. Gal4 was immunoprecipitated using the RK5C1 antibody against Gal4 (Santa Cruz Biotechnology). PCR products correspond to the UASG or the POL1 ORF as a negative control. The percentage of UASG DNA immunoprecipitated (% wt IP) from each of the mutants was normalized to the amount precipitated from the galactose-induced wild-type strain. One set of PCR reactions is shown, and the quantitation represents the average of several experiments. The average values with standard errors for the measurements of Gal4 binding to the UASGAL on galactose-grown cells are as follows: spt3Δ, 96 ± 6; spt20Δ, 69 ± 1; gcn5Δ, 146 ± 12; gal4Δ, 6 ± 1. The measurement for wild type grown on glucose was 32 ± 4. The low level of DNA detected in the gal4Δ and the POL1 PCR reactions represents the low amount of Gal4-independent DNA precipitated as background in this assay. The POL1 negative control was performed on all samples, and the results for all the other extracts were essentially the same as the example shown. In this experiment we detected a modest, but significant, threefold increase in Gal4 binding to the UASG in galactose-induced cells relative to glucose-repressed cells. These results differ slightly from previous in vivo footprint analyses that detected little or no Gal4 occupancy of the UASG in glucose-repression conditions (Giniger et al. 1985; Selleck and Majors 1987b), a result that highlights the sensitivity of the chromatin immunoprecipitation assay.
Histone H3 acetylation in gcn5Δ, spt3Δ, and spt20Δ mutants
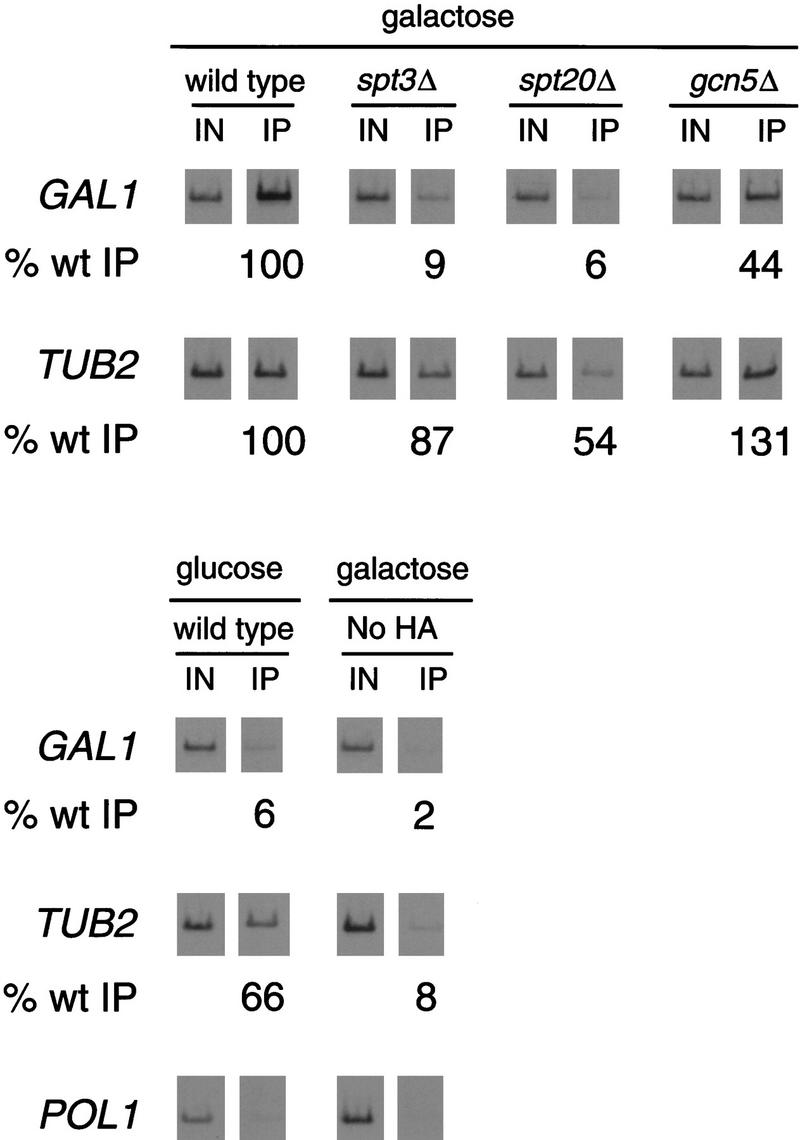
Previous studies have suggested that Spt3 functions in SAGA independently of the HAT activity of Gcn5 (Roberts and Winston 1997; Dudley et al. 1999; Sterner et al. 1999). To test directly whether spt3Δ, gcn5Δ, or spt20Δ mutations alter histone acetylation at the GAL1 promoter, we used chromatin immunoprecipitation assays to determine the histone H3 acetylation levels of GAL1 in these SAGA mutants (Fig. 4). As a control, we examined the H3 acetylation levels at HIS3 and observed a three-fold decrease in a gcn5Δ mutant, consistent with previously published results (Kuo et al. 1998). At GAL1, our results show that H3 acetylation is only mildly reduced in spt3Δ, gcn5Δ, and spt20Δ mutants. For the gcn5Δ mutant, this effect correlates with the weak effect on GAL1 transcription. However, for the spt3Δ and spt20Δ mutants, there is not a good correlation between H3 acetylation and the severe GAL1 transcriptional defects. This result strongly suggests that GAL1 transcription is not significantly dependent on histone acetylation by SAGA and that the role of Spt3 is unrelated to Gcn5-dependent histone acetylation.
Figure 4.
Histone H3 acetylation in spt3Δ, gcn5Δ, and spt20Δ mutants. Chromatin immunoprecipitation was performed in parallel with RNA analysis (Fig. 1) from glucose-repressed, wild-type strains and galactose-induced wild-type, spt3Δ, spt20Δ, or gcn5Δ strains. Histone H3 acetylated at Lys-9 and Lys-14 was immunoprecipitated using antisera previously described (Kuo et al. 1998). PCR products correspond to the GAL1 TATA or the HIS3 promoter as a control. The percentage of GAL1 TATA DNA immunoprecipitated (% wt IP) from each of the mutants was normalized to the amount precipitated from the galactose-induced wild-type strain. One set of PCR reactions is shown, and the quantitation represents the average of several experiments. The spt3Δ and spt20Δ mutants showed some variation, 40%–110% for spt3Δ and 36%–70% for spt20Δ. At HIS3, a threefold decrease in H3 acetylation level was observed in the gcn5Δ mutant compared with wild type, similar to previously reported results (Kuo et al. 1998).
Discussion
Coactivators are believed to act as intermediaries between gene-specific activators and TBP or other general transcription factors. However, the molecular mechanisms by which coactivators function are not well understood (Hampsey 1998; Hampsey and Reinberg 1999). Our experiments have defined an in vivo coactivator function for Spt3 and Spt20 of the SAGA complex. In both spt3Δ and spt20Δ mutants, Gal4 binding is normal, but TBP fails to bind to the GAL1 promoter, and transcription is reduced >50-fold. Thus, the Spt3 and Spt20 proteins are required for Gal4 to recruit TBP to the GAL1 promoter. These results constitute the first in vivo demonstration of such a coactivator role. Our results fit well with other studies of Gal4 and TBP binding in vivo that demonstrated that TBP binding requires the binding and activation of Gal4 (Selleck and Majors 1987a,b; Kuras and Struhl 1999; Li et al. 1999).
The model most consistent with these results is one in which TBP is physically recruited to the GAL1 promoter by the Spt components of SAGA. Previous analysis suggests a physical interaction between Spt3 and TBP (Eisenmann et al. 1992; Lee and Young 1998). In addition, the positions of amino acid changes of both TBP and Spt3 mutants that alter their functional interaction suggest specific regions of each protein that may be involved in TBP–Spt3 interactions (Eisenmann et al. 1992). These results are supported by recent studies of TBP–TAFII28 interactions (Lavigne et al. 1999), as Spt3 is predicted to have structural similarity to both TAFII18 and TAFII28 (Birck et al. 1998). Recent results have shown that SAGA can be recruited by several transcriptional activators including VP16 (Ikeda et al. 1999), Gcn4 (Natarajan et al. 1998), and Rtg3 (Massari et al. 1999). Taken together with these results, our data suggest a model for coactivation in which SAGA is targeted to the GAL1 promoter by interactions with Gal4, followed by recruitment of TBP via interactions with Spt3. The recruitment of TBP by the Spt proteins may be followed by the establishment of previously demonstrated interactions between Gal4 with certain general transcription factors, including TBP, TFIIB, and Srb4 (Melcher and Johnston 1995; Wu et al. 1996; Koh et al. 1998).
Our results, together with previous studies, suggest that other SAGA components may be required for this coactivator function. Spt20, Spt7, and Ada1 are all probably required for the integrity of SAGA (Grant et al. 1997; Sterner et al. 1999). Therefore, the defect observed in spt20Δ mutants is probably caused by the loss of all SAGA functions, including Spt3, although we cannot rule out a more direct role. In addition to Spt3, the Spt8 protein has been implicated in SAGA–TBP interactions, and in vitro data suggest a direct role for Spt8 (Sterner et al. 1999). These results suggest that Spt8, along with Spt3, might be required for TBP binding at the GAL1 promoter. However, an spt8Δ mutant is Gal+, suggesting that Spt8 plays, at most, a minor role at this promoter. Furthermore, previous studies have demonstrated that a particular mutation in SPT3 can partially bypass the requirement for Spt8, suggesting that Spt8 plays a more auxiliary role (Eisenmann et al. 1994). Possibly, both Spt3 and Spt8 assist TBP recruitment, but they contribute to different degrees in a promoter-specific fashion. In addition to these Spt proteins, other SAGA components, including the Ada and Taf proteins, may play related roles in the assembly of the preinitiation complex. Finally, recent evidence suggests that the Snf/Swi complex also helps to activate transcription at a step subsequent to activator binding (Ryan et al. 1998).
Our results cannot rule out a model in which the Spt components of SAGA act at another step in transcriptional activation that results in the recruitment of TBP. For example, the Spt proteins could facilitate some aspect of Gal4 activation, subsequent to its DNA binding, that allows Gal4 to recruit TBP to the GAL1 promoter. Such a role could include helping to determine the correct interactions of Gal4 with either Gal80 or Gal3, both known to interact with Gal4 under inducing conditions (Blank et al. 1997; Yano and Fukasawa 1997). Given that only particular promoters are dependent on the Spt proteins, it is likely that multiple factors determine both their requirement and allowing their function at any Spt-dependent promoter.
Finally, our results demonstrate that the function of SAGA at GAL1 is largely independent of the Gcn5 HAT. These results provide an interesting contrast to those from a recent study that demonstrated a strong Gcn5 requirement for the binding of the Swi4 activator to the HO promoter in vivo (Cosma et al. 1999). The differences between these two sets of results demonstrate that different promoters can require distinct SAGA components for mechanistically distinct functions. The determinants of these requirements at any promoter are an issue for future investigation.
Materials and methods
S. cerevisiae strains
All S. cerevisiae strains are isogenic to a GAL2+ derivative of S288C (Winston et al. 1995). Strains were constructed with the following relevant genotypes: wild type (FY1887 and FY1888), spt3-202 (FY1889 and FY1890), spt20Δ100::URA3 (FY1891 and FY1892), and gcn5Δ::HIS3 (FY1893 and FY1894). Each of these strains also contained an spt15Δ::LEU2 mutation in the genome and an HA3–SPT15 TRP1 CEN plasmid (Kuras and Struhl 1999) as the only source of wild-type TBP. The HA3–SPT15 construct fully complemented the spt15Δ102::LEU2 mutation for all phenotypes tested, including growth on galactose (data not shown). The control strain for the Gal4 chromatin immunoprecipitation experiments, FY760 (gal4Δ::LEU2), was SPT15+ in the genome and contained no plasmid. The control strain for the TBP chromatin immunoprecipitation experiments, FY1886 (the “no-HA” control), contained spt15Δ102::LEU2 in the genome and contained SPT15+ on a plasmid (Kuras and Struhl 1999) as the only source of wild-type TBP. Glucose-repressed strains were grown in YPD (2% glucose). Galactose-induced strains were grown in YPRaf (2% raffinose) and induced for 20 min by the addition of galactose to 2%. RNA for Northern analysis and chromatin extracts were prepared from the same cultures grown to cell densities of 1 × 107–2 × 107 cells/ml. Northern analysis and chromatin immunoprecipitation assays were performed on both sets of isogenic strains.
Northern hybridization analysis
Total yeast RNA was prepared as described previously (Swanson et al. 1991). Northern blot analysis was performed on both sets of strains used for chromatin immunoprecipitations. One of the experiments is shown (Fig. 1), and the quantitation represents the average of both sets of strains. The GAL1 (St. John and Davis 1981) and TUB2 (Som et al. 1988) probes have been described previously.
Chromatin immunoprecipitation
Formaldehyde cross-linking extracts were prepared essentially as described previously (Kuras and Struhl 1999) with the following exceptions: First, all centrifugations to pellet the chromatin extract were performed for 1 min at 14,000 rpm in an Eppendorf centrifuge. Second, the separation of soluble chromatin following sonication was accomplished by a 1-hr centrifugation at 14,000 rpm in an Eppendorf centrifuge. Immunoprecipitations of HA3-tagged TBP were performed as described previously (Kuras and Struhl 1999). Gal4 immunoprecipitations were performed by the same method used for the HA3-tagged TBP with the exception that binding was done in FA lysis buffer containing 150 mm NaCl (Kuras and Struhl 1999) and washes were done three times in the same buffer and once in TE (10 mm Tris-HCl, 1 mm EDTA at pH 8.0). Immunoprecipitation of the hyperacetylated form of histone H3 was performed as described previously (Kuo et al. 1998). PCR reactions were performed essentially as described previously (Kuras and Struhl 1999), with the exception that PCR products were detected by the incorporation of [33P]dATP in the reaction. The PCR primers amplify the following regions whose coordinates are given relative to the ATG (+1): GAL1 TATA primers amplify a 244-bp region from −190 to +54; GAL1 UAS primers amplify a 260-bp region from −536 to −276; TUB2 TATA primers amplify a 273-bp region from −186 to +87; POL1 ORF primers amplify a 219-bp region from +2499 to +2717; and HIS3 primers amplify a 105-bp region from −28 to +77.
Acknowledgments
We are extremely grateful to Laurent Kuras and Kevin Struhl for providing the triple HA1-tagged SPT15 plasmid, for sharing results prior to publication, and for invaluable assistance with the chromatin immunoprecipitation technique. We are also extremely grateful to Min-Hao Kuo and David Allis for providing the antiacetylated H3 antibodies and for helpful discussions. We thank Mary Bryk and Ting Wu for critical reading of the manuscript. This work was supported by a grant to F.W. from the National Institutes of Health (GM45720).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Winston@rascal.med.Harvard.edu; FAX (617) 432-3993.
References
- Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne AC, Davidson I, Moras D. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- Blank TE, Woods MP, Lebo CM, Xin P, Hopper JE. Novel Gal3 proteins showing altered Gal80p binding cause constitutive transcription of Gal4p-activated genes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2566–2575. doi: 10.1128/mcb.17.5.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- Dudley AM, Gansheroff LJ, Winston F. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912δ promoter in Saccharomyces cerevisiae. Genetics. 1999;151:1365–1378. doi: 10.1093/genetics/151.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: Convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- Hampsey M. A SAGA of histone acetylation and gene expression. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- ————— Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Pina B, Marcus GA, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Steger DJ, Eberharter A, Workman JL. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Carlson M. The molecular and cellular biology of the yeast Saccharomyces: Gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. Regulation of carbon and phosphate utilization; pp. 193–281. [Google Scholar]
- Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes & Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Lavigne AC, Gangloff YG, Carre L, Mengus G, Birck C, Poch O, Romier C, Moras D, Davidson I. Synergistic transcriptional activation by TATA-binding protein and hTAFII28 requires specific amino acids of the hTAFII28 histone fold. Mol Cell Biol. 1999;19:5050–5060. doi: 10.1128/mcb.19.7.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Regulation of gene expression by TBP-associated proteins. Genes & Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- Li X-Y, Virbasius A, Zhu X, Green M. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- Melcher K, Johnston SA. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Rhee E, Hinnebusch AG. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MP, Jones R, Morse RH. Swi-Snf complex participation in transcriptional activation at a step subsequesnt to activator binding. Mol Cell Biol. 1998;18:1774–1782. doi: 10.1128/mcb.18.4.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck SB, Majors J. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature. 1987a;325:173–177. doi: 10.1038/325173a0. [DOI] [PubMed] [Google Scholar]
- ————— In vivo DNA-binding properties of a yeast transcription activator protein. Mol Cell Biol. 1987b;7:3260–3267. doi: 10.1128/mcb.7.9.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som T, Armstrong KA, Volkert FC, Broach JR. Autoregulation of 2 micron circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- St. John TP, Davis RW. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981;152:285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes & Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wu Y, Reece RJ, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- Yano K, Fukasawa T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman Z, Ansari AZ, Gaudreau L, Nevado J, Ptashne M. Gene transcription by recruitment. Cold Spring Harbor Symp Quant Biol. 1998;63:167–171. doi: 10.1101/sqb.1998.63.167. [DOI] [PubMed] [Google Scholar]