Abstract
The immunoglobulin intragenic μ enhancer region acts as a locus control region that mediates transcriptional activation over large distances in germ line transformation assays. In transgenic mice, but not in transfected tissue culture cells, the activation of a variable region (VH) promoter by the μ enhancer is dependent on flanking nuclear matrix attachment regions (MARs). Here, we examine the effects of DNA methylation, which occurs in early mouse development, on the function of the μ enhancer and the MARs. We find that methylation of rearranged μ genes in vitro, before transfection, represses the ability of the μ enhancer to activate the VH promoter over the distance of 1.2 kb. However, methylation does not affect enhancer-mediated promoter activation over a distance of 150 bp. In methylated DNA templates, the μ enhancer alone induces only local chromatin remodeling, whereas in combination with MARs, the μ enhancer generates an extended domain of histone acetylation. These observations provide evidence that DNA methylation impairs the distance independence of enhancer function and thereby imposes a requirement for additional regulatory elements, such as MARs, which facilitate long-range chromatin remodeling.
Keywords: MAR, enhancer, LCR, methylation, chromatin
Transcriptional activation of genes in mice has been shown to depend on enhancers or locus control regions (LCRs) (for review, see Dillon and Grosveld 1994; Martin et al. 1996). LCRs, described initially for the human β-globin locus, are required for the formation of an “open,” DNase I-sensitive chromatin domain before transcriptional activation (Forrester et al. 1987; Jimenez et al. 1992). In transgenic mice, LCRs are functionally defined as elements that mediate developmentally regulated expression of linked transgenes at physiological levels, independent of the site of chromosomal integration (Grosveld et al. 1987). In addition, these sequences overcome variegation of gene expression at the single cell level (Festenstein et al. 1996; Walters et al. 1996). LCRs have been identified in many genes and are composite sequence elements that typically contain an enhancer combined with auxiliary sequences. Although the role of enhancers in chromatin accessibility and transcriptional activation of linked promoters has been studied extensively (for review, see Blackwood and Kadonaga 1998), the functions of the auxiliary sequences remain obscure.
The immunoglobulin μ heavy chain locus contains an intragenic enhancer region that can function as an LCR to activate a distal variable region (VH) promoter or a heterologous promoter in germ-line transformation assays (Adams et al. 1985; Jenuwein and Grosschedl 1991). The 1-kb μ enhancer region includes a well-characterized transcriptional enhancer (for review, see Ernst and Smale 1995), the promoter for germ-line noncoding Iμ transcripts (Lennon and Perry 1985), and nuclear matrix attachment regions (MARs) that flank the enhancer on either side (Cockerill et al. 1987). In transgenic mice, the MARs augment the function of the μ enhancer in activating the VH promoter by a factor of 30–1000, whereas the enhancer-proximal Iμ promoter is significantly less dependent on the presence of MARs (Forrester et al. 1994). The dependence of μ gene expression on MARs in germ-line transformation assays also contrasts with the modest effects of MARs in transiently or stably transfected B cell lines (Forrester et al. 1994). In transfected mature B cell lines, no effects of the MARs are observed, whereas the MARs contribute to μ gene expression by a factor of five in immunoglobulin-secreting plasmacytomas (Herrscher et al. 1995). This effect is likely due to binding of the transcription factor Bright, which is expressed in activated or terminally differentiated B cells, to multiple sites in the MARs (Herrscher et al. 1995).
MARs were proposed initially to contain DNA sequences that mediate attachment to the proteinaceous scaffold in histone-depleted metaphase chromosomes (Paulson and Laemmli 1977). By virtue of these interactions, MARs have been hypothesized to represent the bases of large chromatin loops, which are anchored to the nuclear matrix (Mirkovitch et al. 1984). Consistent with this view, MARs have been found to colocalize with the boundaries of nuclease-sensitive chromatin domains (Loc and Stratling 1988). In addition, MARs can function as boundary elements to alleviate position effects in transgenic animals (McKnight et al. 1992; Kalos and Fournier 1995; Phi-Van and Stratling 1996). MARs also have been found to interfere with enhancer–promoter interactions when placed between these elements (Stief et al. 1989). However, in association with transcriptional enhancers, MARs may exert a different function. Together with flanking MARs, the μ enhancer can confer chromatin accessibility upon binding sites for bacteriophage RNA polymerases at positions 1 kb distal to the enhancer, whereas the enhancer alone mediates only localized accessibility (Jenuwein et al. 1993, 1997). Therefore, the function of MARs in extending or blocking enhancer function may be locus or context dependent.
One clue into the function of the μ MARs came from the observation that they appear to act predominantly in germ-line transformation, but not in transfection assays (Forrester et al. 1994). During early mammalian development, genome-wide CpG methylation, which provides a general repression of gene expression, occurs after the implantation stage (for review, see Brandeis et al. 1993; Tate and Bird 1993; Yoder et al. 1997). DNA methylation is reversible and genes that are expressed in differentiating somatic cells are regionally demethylated (Cedar 1988). A role for MARs in demethylation was suggested by studies in which immunoglobulin κ gene constructs, methylated before transfection, were found to be demethylated only in the presence of both MAR and intragenic κ enhancer region (Lichtenstein et al. 1994; Kirillov et al. 1996). However, these experiments did not examine whether MARs are required for enhancer function at a distance and they did not investigate the correlation between the methylation state and transcription. Recently, a direct link between DNA methylation and inaccessible chromatin structure was provided by the finding that the methyl-CpG-binding protein-2 (MeCP-2), which acts as a repressor when artificially tethered to a reporter gene, recruits the mSin3/histone deacetylase complex (Nan et al. 1997, 1998; Jones et al. 1998). Thus, the question arises as to whether MARs collaborate with the μ enhancer to overcome long-range repression of promoter activation by a mechanism involving DNA demethylation or histone acetylation.
Here, we describe experiments in which we methylate μ gene constructs at all CpG dinucleotides, before stable transfection into B cell lines, and examine the effects of MARs on the activity of the distal VH promoter, the methylation state of the transfected genes, and the acetylation of histones. We find that long-range, but not short-range, enhancer function is inhibited by DNA methylation. Moreover, we observe that extended histone acetylation in methylated μ genes requires both the MARs and the enhancer, providing a mechanistic basis for understanding the requirement for composite regulatory elements, such as LCRs, that act over large distances in nuclear chromatin.
Results
Methylation state of immunoglobulin transgenes
In transgenic mice, previously we have shown that the expression of a rearranged μ gene is dependent on the presence of both the μ enhancer and the flanking MARs (Fig. 1A; Forrester et al. 1994). To examine the methylation status of the transcriptionally active wild-type μ transgene and the transcriptionally inactive ΔMAR transgene, which lacks both MARs, we digested genomic DNA from transgenic pre-B lymphoid cells with BamHI and the methylation-sensitive restriction enzyme HpaII (H) or with the methylation-insensitive isoschizomer MspI (M; Fig. 1C). Demethylation of the transgene at a HpaII site 0.8 kb upstream of the enhancer, which is accompanied by the appearance of a 0.8-kb fragment, is observed in the μ wild-type but not the ΔMAR gene (Fig. 1C, left).
Figure 1.
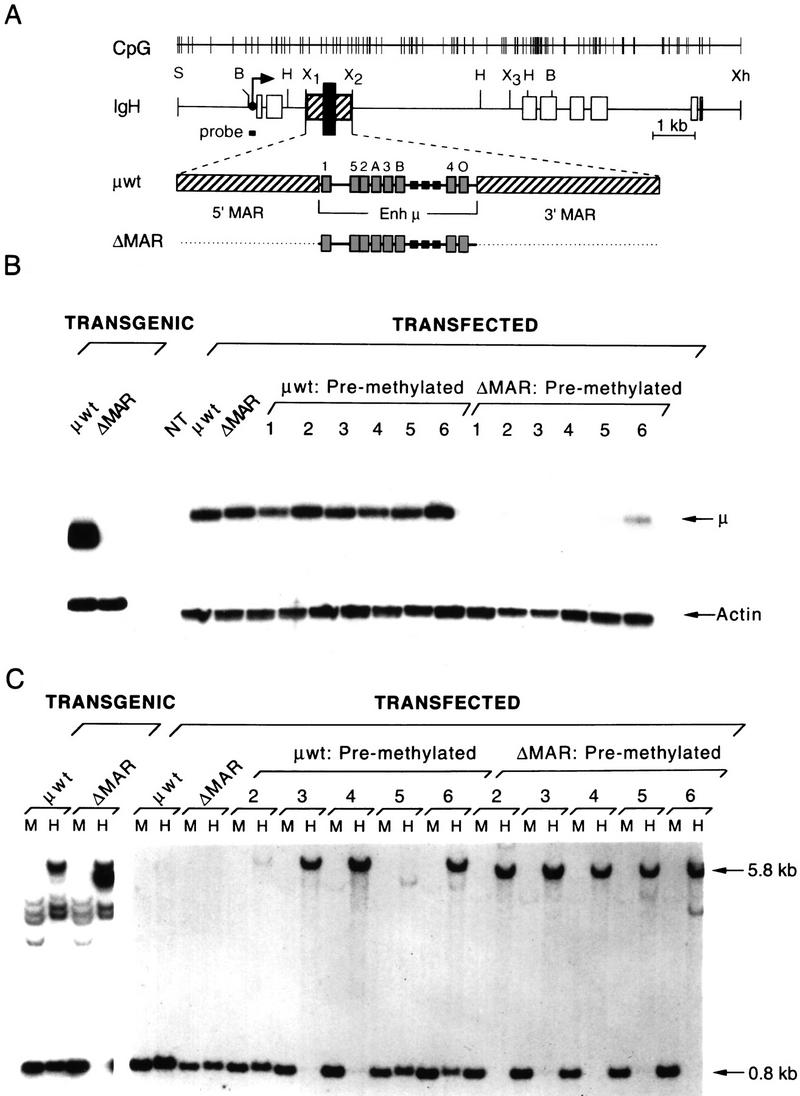
Analysis of the expression and methylation status of rearranged wild-type and ΔMAR μ genes in transgenic mice and stably transfected B cells. (A) Structure of the rearranged μ gene. Above the map of the μ gene, the positions of all CpG dinucleotides are indicated as vertical lines. The intragenic locus control region (LCR), enlarged below, contains the enhancer (Enh μ; black bar), flanked by matrix attachment regions (MARs; hatched bars). The exons are shown as open boxes, and the transcription start site of the VH promoter is indicated by an arrow. Transcription factor-binding sites are indicated as gray boxes with numbers 1–5 corresponding to binding sites for proteins of the E2A family, the A and B sites are recognized by Ets family proteins and PU.1, respectively, and the O site interacts with Oct proteins. Small black boxes represent SV40 enhancer core sequences (Ernst and Smale 1995). Relevant restriction sites: (S) Sal; (B) Bam; (H) HpaII/Msp; (X) Xba sites 1–3; and (Xh) Xho. (B) S1 nuclease protection assay detecting specific μ transcripts in transgenic and transfected M12 cells. μ wild-type and ΔMAR genes were stably transfected in an unmethylated or in vitro premethylated form. The positions of the specific μ transcripts and the endogenous β-actin transcripts are indicated. Numbers represent individual cell clones. (NT) nontransfected cell line. For the S1 nuclease protection assays, 10 and 20 μg of total cytoplasmic RNA were used to detect actin and μ-specific transcripts, respectively. (C) Analysis of the methylation pattern of the transgenic or transfected μ genes. Genomic DNA from the corresponding cells was digested to completion with BamHI and with either Msp (M) or HpaII (H), and blots were hybridized with a radiolabled probe that abuts the 5′ Bam site as shown in A.
In vitro methylation represses enhancer function in the absence of MARs
To establish a cause-and-effect relationship between the methylation state and the transcriptional activity of the genes, we adopted the approach of methylating DNA in vitro before transfection of tissue culture cells (Lichtenstein et al. 1994). The μ gene was removed from plasmid DNA backbone and incubated with the prokaryotic SssI methyltransferase, which will convert the cytosine within a CpG dinucleotide to the 5-methyl-C derivative, thereby reproducing the specificity of a mammalian de novo methyltransferase. We introduced methylated μ genes into M12 B cells and determined, by RNA analysis, the activity of the VH promoter in clones containing stably integrated μ genes (Fig. 1B). Transfectants containing the unmethylated wild-type and ΔMAR genes, generated similar numbers of specific transcripts initiating at the VH promoter, consistent with the previous finding that MARs are dispensable for enhancer function (Forrester et al. 1994). In contrast, the VH promoter activity of the premethylated ΔMAR gene in individual clones is reduced by a factor of 5–20 relative to the activity in clones containing the wild-type gene. Premethylation of the ΔMAR gene decreased both the frequency of μ-expressing clones as well as the levels of VH promoter activity in μ-expressing clones. Thus, methylation of the μ gene before transfection imparts a requirement for MARs similar to that observed in germ-line transformation assays (Forrester et al. 1994).
MARs contribute to demethylation of the transfected μ gene
We examined the methylation states of the transfected μ genes by analyzing genomic DNA as described above. In some clones containing the premethylated μ wild-type gene, quantitative demethylation was detected (clones 2 and 5), whereas partial demethylation was observed in clone 6, and no demethylation was detected in clones 3 and 4. In contrast, the ΔMAR μ gene remains fully methylated in all clones, including clone 6, which contains a low level of μ-specific transcripts. These results suggest that quantitative demethylation is not necessary for the active transcriptional state of the transfected μ gene. In transfectants containing the unmethylated μ and ΔMAR genes, we do not detect de novo methylation suggesting that in the time course of these experiments, the MARs are not acting to block de novo methylation.
Methylation generates an inaccessible chromatin domain in a transfected μ gene
Previous analysis of the chromatin structure of the ΔMAR μ gene in transgenic B cells revealed that the μ enhancer alone was sufficient to establish DNase I hypersensitivity, although sequences distal to the enhancer were DNase I resistant, relative to the endogenous μ locus (Forrester et al. 1994). To address the role of DNA methylation in establishing a similar chromatin context, we incubated nuclei from transfected M12 cells with increasing amounts of DNase I and determined the sensitivity to digestion of the ΔMAR gene (Fig. 2). Similar to our observations with transgenic mice, we find that the enhancer of the transfected ΔMAR μ gene (EμT) is hypersensitive to DNase I digestion (Fig. 2A) regardless of the methylation state of the transfected DNA. The cross-reactivity of the DNA probe with a fragment containing the endogenous μ enhancer (EμE) serves as an internal control showing that both transfected and endogenous enhancers are similarly DNase I hypersensitive.
Figure 2.
Analysis of the chromatin structure of transfected ΔMAR genes by DNase I digestion. Nuclei from M12 cells stably transfected with an unmethylated or methylated ΔMAR gene were digested with increasing amounts of DNase I. Genomic DNA was digested with ScaI–BglII and hybridized with a 0.67-kb EcoRI–HindIII DNA probe. (A) DNase I hypersensitivity at the μ enhancer is indicated by arrow labeled EμT for the transfected and EμE for the endogenous μ locus. (B) General DNase I sensitivity of the transfected ΔMAR gene (IgHT) in comparison to transcriptionally active (IgHE and mb-1) and transcriptionally inactive (MyoD and the pseudogene ϑmb-1) endogenous gene loci. The sizes of the DNA fragments, in kilobases, are shown at right.
To examine the overall chromatin structures of the unmethylated and premethylated ΔMAR μ genes, we compared their rates of digestion by DNase I with that of the transcriptionally active endogenous μ and mb-1 genes, and the transcriptionally inactive MyoD gene (Fig. 2B). The unmethylated ΔMAR gene fragment is digested faster than that of the transcriptionally active mb-1 gene and endogenous μ locus fragments. In contrast, the digestion rate of the premethylated ΔMAR gene resembles more closely that of the inactive MyoD gene and the inactive mb-1 pseudogene (ϑmb-1; Kashiwamura et al. 1990), which is also detected with the mb-1 probe. As expected, the digestion rates of the endogenous gene loci are similar in both ΔMAR lines. Thus, the premethylated ΔMAR μ gene resides in an inaccessible chromatin structure, although the enhancer is locally hypersensitive to DNase I digestion.
Distal but not proximal enhancer function is repressed by DNA methylation
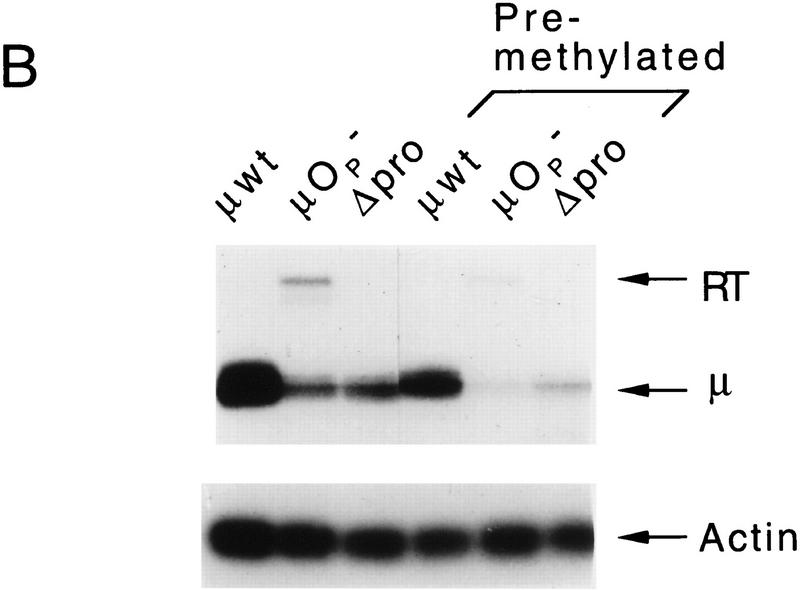
The DNase I hypersensitivity of the μ enhancer in transcriptionally inactive premethylated ΔMAR μ genes suggested that methylation may interfere with interactions between the enhancer and the distal VH promoter, but not with local factor binding at the μ enhancer. To examine short-range enhancer function in the absence of MARs, we placed the enhancer alone in a VH promoter-proximal position, 150 bp upstream of the transcription initiation site in a construct termed 5′Enh (Fig. 3A). This 5′Enh gene is expressed at levels comparable to those of the μ wild-type gene in both clones containing unmethylated and premethylated templates (Fig. 3B). These data suggest that methylation inhibits selectively long-range enhancer function and does not interfere with transcription factor binding and local chromatin remodeling, and with short-range enhancer function.
Figure 3.
Analysis of the expression and methylation status of the 5′Enh gene. (A) Structure of the 5′Enh gene in which the 220-bp enhancer (Enh) fragment lacking both MARs was inserted at a BamHI site 154 bp upstream of the VH transcription initiation site. (B) Analysis of the transcriptional state of unmethylated and premethylated 5′Enh genes in individual stably transfected M12 clones by S1 nuclease protection. The positions of VH-initiated transcripts (μ) and transcripts initiating upstream of the normal start sites (RT) are indicated. (C) Analysis of the methylation status is as described previously (Fig. 1C).
The methylation pattern of the 5′Enh genes before and after methylation was examined and indicated that enhancer-mediated VH transcription does not, by itself, produce demethylation. Partial demethylation of the distal HpaII site was detected in 4 out of 10 clones, whereas no significant demethylation was observed in the 6 other clones (Fig. 3C). In contrast, quantitative demethylation was observed at a HpaII site, introduced immediately adjacent to the enhancer (data not shown). These results resemble numerous examples showing that actively transcribed genes can retain methylated cytosines and argue against a passive role for transcription in the demethylation reaction.
Distal demethylation requires both MARs
MARs have been shown to augment transcription in late stage B cells by interaction with the protein Bright (Herrscher et al. 1995). To assess the repressive effects of DNA methylation in late stage B cells that contain Bright, we transfected unmethylated or methylated μ wild-type and ΔMAR genes into S194 plasmacytoma cells. For this experiment, in addition we used genes lacking either the 5′ or 3′ MAR (Fig. 4A). Analysis of pools of independent cell clones transfected with unmethylated μ genes indicated that deletion of both MARs reduced μ gene expression by a factor of 10, which is slightly more pronounced than the effect previously observed in transient transfection assays (Herrscher et al. 1995). Deletion of one MAR had no detectable effect (Δ5′MAR) or decreased gene expression by a factor of two (Δ3′MAR). However, premethylation of these genes revealed a marked dependence of μ gene expression on the presence of both MARs. Thus, the MARs may subserve two functions in plasmacytomas. One function, which requires both MARs, may antagonize methylation-mediated repression, whereas the other function, which requires only one MAR, appears to involve up-regulation of enhancer activity on unmethylated DNA templates and in cells containing the transcription factor Bright.
Figure 4.
Analysis of the expression and methylation status of μ genes containing a single MAR. (A) Structure of genes lacking either the 3′MAR or the 5′MAR. The positions of BamHI and HpaII (H) sites are indicated. In these constructs, a HpaII site has been introduced at the 3′ end of the enhancer. (B) S1 nuclease protection assay of total cytoplasmic RNA isolated from stably transfected pools of S194 cells. (C) Analysis of the methylation status by digestion with BamHI and either MspI or HpaII. The size of the BamHI–HpaII fragment generated by cleavage at the enhancer-proximal HpaII site is 1.4 kb for the ΔMAR and Δ5′MAR, and it is 1.7 kb for the Δ3′MAR gene construct. The probe, shown in A, also hybridizes with an endogenous S194 DNA fragment indicated by an open arrow.
Analysis of the methylation state of the transfected genes indicated that the enhancer-distal HpaII site, 0.8 kb 3′ to the BamHI site in the VH promoter, is methylated in cells containing premethylated ΔMAR genes (Fig. 4C). In contrast, the enhancer-proximal HpaII sites, 1.4 or 1.7 kb 3′ of this BamHI site, are predominantly demethylated. The μ wild-type gene was demethylated quantitatively at both distal and proximal positions, consistent with previous observations (data not shown; Kirillov et al. 1996). Thus, MARs may facilitate extended demethylation by a process that is independent of transcription.
LCR-mediated demethylation is independent of VH promoter activity
To examine putative contributions of the VH promoter to the long-range interactions with the μ enhancer region, we tested the effects of a mutation in the octamer of the VH promoter (μOp−), and the deletion of all VH sequences upstream of the transcription initiation site (Δpro; Fig. 5A). The activity of the μOp− promoter in stably transfected S194 pools is reduced ∼10-fold relative to that of the μ wild-type gene (Fig. 5B). This mutant promoter yields a greater number of readthrough (RT) transcripts that initiate upstream of the major start site and resemble the germ-line transcripts described for unrearranged VH segments in immature B cells (Yancopoulos and Alt 1985). The Δpro μ gene is also transcribed, albeit at a 10-fold reduced level, suggesting that the initiator and downstream elements can direct transcription of this mutant gene (Ernst and Smale 1995). After methylation, the levels of transcription from the μ wild-type gene and both promoter mutants are similarly reduced by a factor of three relative to the unmethylated genes, suggesting that the VH promoter does not contribute to the effect of the LCR in overcoming methylation-dependent repression.
Figure 5.
The VH promoter is not necessary for μ LCR function. (A) Structure of genes containing point mutations in the VH promoter octamer site (μOp−) or a deletion of all sequences 5′ to the transcription initiation site (Δpro). (B) RNA anaysis by S1 nuclease protection. In μOp−, some transcripts, initiated at upstream start sites, read through the normal cap site (RT). In the Δpro gene, transcripts initiated at the VH start site or in the 5′ flanking mouse DNA will produce the same protected S1 fragment. (C) Analysis of the methylation status of transfected (Transf.) genes. The μOp− gene generates restriction fragments similar to those of the wild-type gene. In contrast, the digestion pattern of the Δpro gene is more complex because this analysis surveys genomic sequences at the junction of each chromosomal integration site. Endogenous cross-hybridizing restriction fragments (Endog.) are indicated.
Analysis of the methylation state of both premethylated VH promoter mutants indicated that the enhancer-distal HpaII site is predominantly demethylated, suggesting that demethylation is not dependent on full promoter activity (Fig. 6C). In the Δpro μ gene construct, the removal of the upstream BamHI site generates different junction fragments between the μ gene and flanking mouse DNA that reflect individual integration sites. Most of these fragments are demethylated, although at some integration sites this mutant μ gene is refractory to demethylation.
Figure 6.
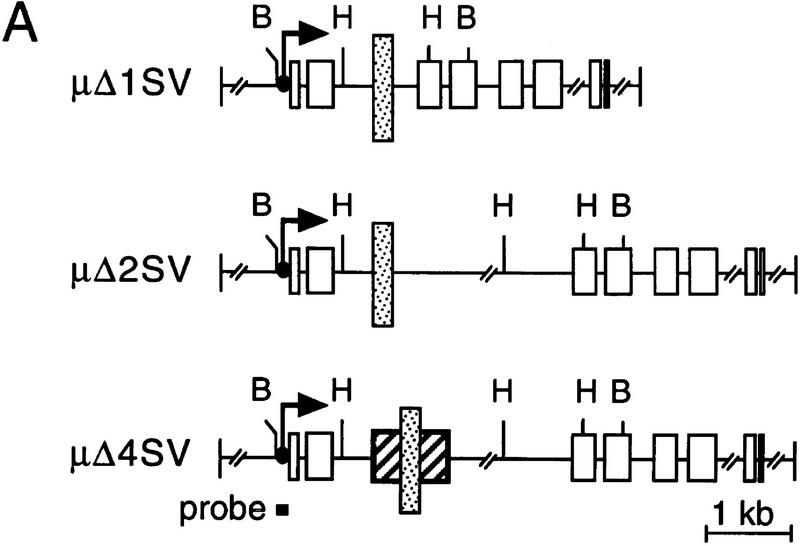
Specificity of enhancer–MAR interaction. (A) Structure of μ genes containing the SV40 enhancer (stippled box, see Materials and methods). In μΔ1SV, the SV40 enhancer is inserted between Xba sites 1 and 3, in μΔ2SV the SV40 enhancer was inserted between Xba sites 1 and 2. In μΔ4SV, the μ enhancer was replaced with SV40 enhancer without removing the flanking MARs. (B) RNA analysis by nuclease S1 nuclease protection assay. (C) Analysis of the methylation state of the transfected genes as described in Fig. 1C.
Specificity of enhancer–MAR combination
To examine the potential modular structure of the intragenic μ LCR, we replaced the μ enhancer with the simian virus 40 (SV40) enhancer (Fig. 6A). The μ and the SV40 enhancers share a similar composition of transcription factor-binding sites and are both highly active in transfected B cells (Ondek et al. 1987; Pettersson and Schaffner 1987). The SV40 enhancer was inserted alone (μΔ2SV), or together with the MARs (μΔ4SV) into the μ gene context to generate constructs analogous to the μ ΔMAR and wild-type gene, respectively. The μΔ1SV gene is a derivative in which the SV40 enhancer has replaced most sequences of the large intron.
In pools of stably transfected S194 cells, the SV40 enhancer alone directed expression of the unmethylated μ gene construct at levels only fourfold lower than those observed with the unmethylated μ wild-type gene (Fig. 6B). The comparable μ enhancer-bearing construct ΔMAR is expressed at levels ∼2.5-fold lower (see Fig. 4B). In combination with the flanking MARs, the SV40 enhancer mediates μ gene expression at a level that exceeds that of the μ wild-type gene. Therefore, the SV40 enhancer is two to three times stronger than the μ enhancer. After methylation, however, all μ constructs containing the SV40 enhancer are transcriptionally inactive (Fig. 6B). Moreover, none of the premethylated templates containing the SV40 enhancer show demethylation at the distal HpaII site (Fig. 6C). These experiments suggest that the μ MARs act differently in unmethylated and methylated genes. Before methylation, the MARs act to modulate the activity of both μ and SV40 enhancers, whereas after methylation, the MARs facilitate long-range effects only in combination with the μ enhancer.
MARs induce long-range histone acetylation
Recently, the methyl-CpG-binding protein MeCP-2 has been shown to recruit a repressor complex containing mSin3 and histone deacetylase-1 (HDAC) to chromatin (Nan et al. 1997, 1998; Jones et al. 1998). This finding provides a potential mechanism for transcriptional repression by deacetylation of histones in the vicinity of methylated CpG dinucleotides. To examine whether the ability of the μ MARs to antagonize methylation-dependent repression of long-range enhancer function involves changes in the acetylation of histones, we used a cross-linking and chromatin immunoprecipitation assay (Belyaev et al. 1996; Kuo et al. 1998). M12 cells, stably transfected with premethylated μ wild-type or ΔMAR genes, were treated with formaldehyde, and sonicated nuclear chromatin fragments were immunoprecipitated with antibodies directed against the acetylated forms of histone H3 and H4 (Kuo et al. 1998). The precipitated (“bound”) DNA fragments were analyzed by PCR amplification with primers that detect either the VDJ exon of the transfected μ gene or the transcriptionally active endogenous mb-1 gene (Fig. 7). Serial dilutions of amplified DNA fragments indicated that the amount of the VDJ fragment of the μ wild-type gene that is precipitated by the anti-acetylated histone antibodies is ∼10-fold higher than that of the precipitated VDJ fragment of the ΔMAR gene. In contrast, similar amounts of mb-1 fragments were precipitated from wild-type and ΔMAR chromatin, although the mb-1 locus showed a preferential acetylation of histone H3 relative to histone H4. Together, these results suggest that the MARs facilitate the generation of an extended domain of histone acetyltation, which may allow for the long-range chromatin accessibility observed previously in the wild-type but not the ΔMAR μ gene (Forrester et al. 1994; Jenuwein et al. 1997).
Figure 7.
Analysis of histone acetylation in premethylated wild-type and ΔMAR μ genes. Formaldehyde-fixed chromatin extracts from M12 cells, transfected with the premethylated μ wild-type gene (clone 5) or the ΔMAR gene (clone 5), were immunoprecipitated using specific antiserum raised against acetylated histone H3, acetylated histone H4, and preimmune serum as a control. Bound chromatin was recovered and used as a template for PCR amplification. A series of fourfold dilutions of the immunoprecipitated DNA, starting with 10 ng, was used for the amplification and detection of VDJ exon sequences (300-bp product) and mb-1 promoter sequences (350-bp product) as internal control. Ten nanograms of total DNA “input” from each of the cell lines was used to assess the relative enrichment of specific sequences in the immunoprecipitations. Specific amplification products were analyzed by electrophoresis through a 3% agarose gel.
Discussion
Our studies with in vitro methylated μ genes provide several novel conclusions about the regulation of long-range gene control. First, methylation effectively inhibits enhancer function in a distance-dependent fashion. Second, the retention of local enhancer activity after methylation is manifested by the establishment of DNase I hypersensitivity, the ability to induce DNA demethylation, and by the activation of a proximal promoter. Third, the methylation-induced repression of long-range μ enhancer function is antagonized by MARs, which indicates that distance-dependent enhancer effects can be regulated. Fourth, MARs, in combination with the μ enhancer, are the first genetic elements shown to induce acetylation of nucleosomes at distal positions. Finally, methylation of genes before transfection may establish a cell culture model of LCR function and should provide additional insights into lineage-specific transcriptional control mechanisms.
Methylation-mediated repression and local enhancer competence
DNA methylation can inhibit gene expression either directly by interfering with DNA binding of specific proteins (Watt and Molloy 1988; Iguchi-Ariga and Schaffner 1989) or indirectly by recruiting repressor proteins such as the methyl-C binding proteins (MeCPs; Nan et al. 1997). The full transcriptional activity of the 5′Enh gene suggests that neither the VH promoter nor the μ enhancer is directly repressed as a consequence of CpG methylation. Rather, our data support an indirect mechanism that acts to interfere selectively with long-range enhancer function. Consistent with the recruitment of the Sin3/HDAC corepressor complex by the MeCP-2 protein (Jones et al. 1998; Nan et al. 1998), we find that the methylated ΔMAR μ gene is assembled into chromatin that is hypoacetylated and generally inaccessible to DNase I digestion, except at μ enhancer.
The recruitment of MeCP2 and transcriptional repression is a function of density of methylated CpG dinucleotides (Boyes and Bird 1992). In the region spanning the VH promoter and intragenic enhancer, the density of CpG dinucleotides is lower than that of one CpG per 126 nucleotides, which was found to be minimally required for repression by MeCP2 (Boyes and Bird 1992). However, MeCP2 can also bind specifically to MARs in the absence of methylated CpG dinucleotides suggesting that this protein may have two modes of DNA binding (Weitzel et al. 1997).
In premethylated DNA templates, the μ enhancer lacking both MARs is able to exert, at least, some functions. Specifically, the enhancer induces DNase I hypersentive sites and activates a proximal promoter, indicating that a functional nucleoprotein complex is formed. The LTR of murine mammary tumor virus has been shown to contain binding sites for the glucocorticoid receptor that serves as a “pioneer” protein to initiate localized chromatin remodeling by recruitment of the SWI/SNF complex (Cordingley et al. 1987; Yoshinaga et al. 1992). These changes are necessary for subsequent binding of nuclear factor-1 (NF-1) to sites located on the adjacent nucleosome (Fryer and Archer 1998) suggesting a hierarchical relationship similar to that described for the yeast HO promoter (Cosma et al. 1999). In the HO promoter, the Swi5 factor acts as a pioneer protein that sequentially recruits SWI/SNF and the SAGA acetyltransferase complex, which permits the binding of Swi4/6 to other sites in the promoter (Cosma et al. 1999). No pioneer proteins have yet been identified for the μ enhancer, and none of mutations in Oct, μB, or E2A-binding sites have been shown to abrogate enhancer function in transgenic mice (Jenuwein and Grosschedl 1991). However, the cooperative assembly of an enhancer complex during DNA replication may also induce a local perturbation in chromatin. Consistent with this view, the μ enhancer core forms an enhancer complex in assembled chromatin by cooperative binding of multiple proteins (Nikolajczyk et al. 1999).
In addition to the local perturbation of chromatin, the μ enhancer, but not the SV40 enhancer, can induce local DNA demethylation. Local demethylation at the μ enhancer region may be active, involving a “demethylase” (Weiss et al. 1996; Bhattacharya et al. 1999), or passive, reflecting the interference of maintenance methylation by an enhancer factor after DNA replication. Recent experiments have shown that demethylation of the Igκ locus occurs on one allele and precedes the rearrangement of the gene locus consistent with an active and targeted demethylation process (Mostoslavsky et al. 1998). Alternatively, it is also possible that an enhancer complex is assembled one allele at a time (Milot et al. 1996), leading to allele-specific demethylation.
MARs mediate long-range μ enhancer function and histone acetylation
Previously, we have shown that the μ enhancer, together with flanking MARs can confer accessibility on a distal T7 RNA polymerase promoter, independent of ongoing transcription by endogenous RNA polymerases (Jenuwein et al. 1993, 1997). These experiments, in which bacteriophage promoters were used instead of eukaryotic promoters, argue for a role of MARs in extending enhancer-induced accessibility and possibly demethylation in the absence of DNA looping. Thus, MAR-dependent effects may be propagated in cis along the DNA. We now find evidence that, in collaboration with the μ enhancer, the MARs are involved in extending local accessibility by inducing the acetylation of histones at distal positions. This extended acetylation of histones is reminiscent of the domain-wide histone acetylation that comaps with and may establish the general DNase I sensitivity across the globin locus (Hebbes et al. 1994). The domain of histone acetylation in the globin locus spans both transcriptionally active and inactive genes and encompasses both demethylated and methylated DNA. In our experiments, we also note that demethylation is neither necessary for nor a consequence of transcription, consistent with previous finding of partial demethylation of the endogenous μ locus in pre-B cells (Gerondakis et al. 1984). Thus, the extended histone acetylation in the premethylated μ gene may not be linked to DNA demethylation.
The regulation of long-range chromatin remodeling remains poorly understood. Histone acetylation is known to be targeted to specific sites by acetyltransferases that are associated with specific transcription factors and modify nucleosomes in a highly localized fashion (Kadosh and Struhl 1998; Kuo et al. 1998). In contrast to the β-interferon (IFNβ) enhancer, which induces histone acetylation only at proximal nucleosomes (Parekh and Maniatis 1999; this study), the μ enhancer/MAR region mediates extended histone modification.
Several mechanisms can be considered to underlie the propagation of histone acetylation and chromatin accessibility. Chromatin remodeling by MARs may reflect the mutually exclusive binding of histone H1 and high mobility group protein I/Y (HMG I/Y) to high affinity sites in the MARs, allowing for a switch between higher order and decondensed chromatin states (Zhao et al. 1993). HMG-I/Y also plays a role in mediating long-range transcriptional effects (Bagga and Emerson 1997) and facilitates the assembly of a multiprotein complex at the IFNβ enhancer (Thanos and Maniatis 1995). Another potential mechanism by which MARs influence the long-range function of enhancer complexes could involve changes in DNA topology. MARs contain DNA-unwinding elements as well as preferred sites for toposiomerases (Bode et al. 1992). Some chromatin-remodeling enzymes, such as CHRAC, have topoisomerase activity (Varga-Weisz et al. 1997), and recruitment of such protein complexes to MARs or the enhancer may allow for a propagation of an altered chromatin structure. MARs may also serve as preferred loading sites for chromatin remodeling or histone acetyltransferase complexes that are recruited to the μ enhancer after the binding of pioneer proteins or the assembly of a nucleoprotein complex. For example, recruitment of the histone acetyltransferase PCAF to DNA through a heterologous DNA-binding domain has been shown to mediate long-range activation of a linked promoter (Krumm et al. 1998). Finally, MARs may serve to maintain an open chromatin structure in transfection assays of premethylated DNA templates. The insulator of the chicken β-globin locus prevents transcriptional inactivation and maintains a transcriptionally permissive and hyperacetylated chromatin domain, but it does not protect against spreading of DNA methylation (Pikaart et al. 1998). Thus, multiple mechanisms may be used to overcome the repressive effects of DNA methylation and histone deacetylation.
Relationship of the μ enhancer/MAR region with LCRs
The combination of the μ enhancer and flanking MARs represents a simple LCR that controls both long-range chromatin remodeling leading to the acquisition of general DNase I sensitivity and transcriptional activation of the VH promoter. Several factors bind competitively to the same four sites in the μ MARs. Cux/CDP, previously named NFμNR, down-regulates the basal activity of the μ enhancer in early B and non-B cells (Scheuermann and Chen 1989; Wang et al. 1999), whereas the positive activator Bright increases μ enhancer function in terminally differentiated B cells (Herrscher et al. 1995). The role of these factors and associated proteins in mediating MAR-dependent changes in chromatin is unknown and it is possible that the transcription and chromatin effects are mediated either by distinct MAR-binding complexes or different MAR sequences. Multiple roles of MARs in transcriptional activation are also inferred from experiments showing that the μ MARs augment the function of both SV40 and μ enhancers in the context of unmethylated templates, whereas the MARs stimulate only μ enhancer function in methylated genes. Moreover, deletion of a single MAR has no effect in unmethylated μ genes but abrogates expression of premethylated DNA templates.
In the immunoglobulin and T-cell receptor loci, formation of an extended domain of accessible chromatin is a prerequisite for somatic gene rearrangements that precede high levels of V region promoter activity (for review, see Sleckman et al. 1996). A similar requirement for long-range remodeling of chromatin structure as a prerequisite for recombination has been observed in yeast. Recombination competence over the entire length of a chromosome arm has been found to be regulated by an LCR-like regulatory element that contains a cluster of factor-binding sites and flanking A-T rich domains (Wu and Haber 1996; Haber 1998). Given the strong dependence on the MARs in our transgenic and transfection experiments, it is surprising that no significant effects are observed in mice in which the MARs were deleted from one allele of the endogenous heavy chain locus (Sakai et al. 1999). One possible explanation is that one of the many MARs located elsewhere in the μ heavy chain locus compensates for the loss of the intronic μ MARs (Cockerill 1990). Redundancy of regulatory elements in the heavy chain locus was originally noted in variant B cell lines in which the entire intronic enhancer region has been deleted with little or no effect on immunoglobulin expression and rearrangement (Zaller and Eckhardt 1985). Redundancy was also observed in the native β-globin gene cluster, in which deletion of the LCR has only a modest effect on chromatin structure and transcription (Epner et al. 1998). Moreover, we cannot rule out the possibility that MARs can also act in trans to augment enhancer function.
In conclusion, our observation that the μ enhancer requires the collaboration with a flanking MAR to confer long-range action in methylated DNA templates provides insight into the complexity of regulation of gene expression by enhancers. Moreover, the pronounced similarity of the effects of μ enhancer mutations in transgenic mice and methylated DNA templates in transfected cells provides a strategy for studies of LCR function in cell culture transfection assays.
Materials and methods
Cell culture and transfections
All cells were propagated and electroporated as described previously (Forrester et al. 1994). S194 cells (Hyman et al. 1972) were grown in RPMI containing 5% heat-inactivated fetal bovine serum. Twenty-four hours after electroporation, G-418 (GIBCO-BRL) at 100 mg/ml (active fraction) in 100 mm HEPES (pH 7.4) was added to a final concentration of 1 mg/ml (active). Cells were either cloned by diluting to densities of 104–105 cells/ml and seeding of 1-ml aliquots into the wells of a 24-well plate or grown in culture as an uncloned pool. Ten days after plating, G-418-resistant clones were fed and grown thereafter in nonselective media (lacking G-418). S194 pools consisted of >100 independent transformants.
DNA constructs
To generate the 5′ Enh gene, the 220-bp Eμ enhancer was modified by the addition of Not linkers and inserted into the BamHI site, 154 bp 5′ to the transcription initiation, which had been converted to a Not site. The Δpro gene was prepared by digesting μ wild type with Nde, which cleaves uniquely at the transcription initiation site. Construction of the single MAR deletions, as well as the SV40 enhancer-containing genes involved Not linkering the appropriate fragments, which were then inserted into a common vector, μΔ2N1(Py), in which the region between Xba sites 1 and 2 (Fig. 1A) had been replaced with a NotI linker. The plasmids μΔ4SV, μΔ2SV, and μΔ1SV were prepared by inserting the SV40 enhancer into derivatives of the μ wild-type gene that lacked either Eμ (μΔ4), Eμ and the MARs (μΔ2), or most of the large intron (μΔ1), respectively. All plasmids were confirmed by sequencing.
Preparation of vector-free μ DNA and methylation in vitro
In all experiments, the immunoglobulin μ genes were released from the plasmid vector backbone by digestion with SalI and XhoI, or BstUI. The DNA was loaded onto a preformed, continuous 5–20% potassium acetate gradient in a SW 55.1 tube containing 1.5 μg/ml ethidium bromide and spun at 50 K for 3 hr at 4°C. DNA fragments were visualized under long-wave UV illumination and collected by bottom puncture. The ethidium bromide was removed by several extractions with butanol saturated with 10% potassium acetate and precipitated with 2.5 volumes of cold ethanol. Methylation of DNA fragments at all CpG dinucleotides was performed by incubating 20–40 μg of DNA with 10–20 units SssI methyltransferase (NEB) at 37°C for 3 hr. The extent of methylation is routinely monitored by the degree to which HpaII digestion is blocked.
Cross-linking and chromatin immunoprecipitations
Formaldehyde treatment of M12 cells resulting in covalent cross-links between DNA and proteins in close proximity, isolation of chromatin, and immunoprecipitations with anti-acetylated histone antibodies were performed essentially as described (Belyaev et al. 1996). Briefly, 2 × 107 M12 cells, stably transfected with premethylated μ or ΔMAR genes, were fixed in 1.1% formaldehyde. Cells were lysed in a 0.25% Triton solution and sonicated to yield DNA fragments of 0.5 kb average length. After centrifugation, the OD260 concentration of the supernatant was adjusted to six absorbance U/ml in IP buffer (NaCl 140 mm, Triton X-100 1% wt/vol, sodium deoxycholate 0.1% wt/vol, PMSF 1 mm, yeast tRNA 100 mg/ml, BSA 100 mg/ml) and preincubated for 1 hr at 4°C with 10 μl (per ml) of 50% (vol/vol) protein A–Sepharose (Pharmacia). After several washes, anti-AcH4, anti-AcH3 (Upstate Biotechnology) or a rabbit preimmune antiserum as a control was added to separate 600-μl aliquots at 1:100 dilution, and incubated overnight at 4°C. Immunocomplexes were isolated by retention on protein A beads, followed by centrifugation. Supernatants were kept as “unbound” fraction and protein A beads were washed repeatedly before being resuspended in 200 μl of elution buffer [Tris-Cl 50 mm (pH 8.0), EDTA 10 mm, SDS 1% wt/vol] and heated to 65°C for 15 min. After removing beads the unbound and bound samples were diluted by adding 300 μl of TE buffer, whereas input samples were adjusted to 0.5% SDS. All samples were incubated overnight at 65°C to reverse formaldehyde cross-links. Afterward 3 μl of RNase A (10 mg/ml) was added for 30 min at 37°C followed by 10 μl of proteinase K (12 mg/ml) for 2–3 hr at 37°C. Samples were extracted sequentially with phenol/chloroform and chloroform, and DNA was precipitated with two volumes of ethanol and 10 mg of glycogen (Sigma). Precipitated DNA was recovered by centrifugation, washed with 70% ethanol, and resuspended in 100 μl of TE. DNA concentration in bound samples ranged between 2 and 6 ng/ml, and in input and unbound fractions ranged from 0.1 to 0.5 μg/ml.
PCR amplifications
Template DNA from input and bound fractions was diluted by six serial, fourfold dilutions; DNA in the first dilution was 10 ng of DNA. PCR was performed in 50 μl of PCR buffer [Tris 10 mm (pH 8.3), 50 mm KCl, 250 mm each dNTPs, 0.001% gelatin (wt/vol), 0.5 mm each oligonucleotides 1 and 2, 1 unit Taq polymerase; MgCl2 was optimized for each primer set, being 3 mm for VDJ primers and 1.5 mm for mb-1 primers] using 25 cycles (94°C for 1 min; 55°C for 1 min; 72°C for 1 min). Fifteen microliters of this reaction was transferred to a new tube containing 50 μl of fresh PCR buffer, and cycled for an additional 25 times. Ten microliters was analyzed in a 3% agarose gel (Nusieve 3:1). The oligonucleotides used for amplification of the transfected VDJ DNA sequence were VDJ-1.2 (5′-GCCTCAGTCAAGTTGTCCT) and VDJ-2.2 (5′-GTAGTCCATAGCATAGTAA). For amplification of the endogenous mb-1 promoter we used the oligonucleotides mb-1-A (5′-AGGGATCCATGGTGATGAAC) and mb-1-B (5′-CAAACAGGCGTATGACAAGA).
Acknowledgments
W. Forrester would like to dedicate this paper to the memory of Hal Weintraub, whose work helped establish many paradigms in chromatin structure and gene regulation. We thank H. Cedar and Mark Groudine for discussions. We are also grateful to Nancy Biles for preparation of the manuscript. W. Forrester was supported by a special fellowship from the Leukemia Society of America. L. Fernández was supported by a postdoctoral fellowship from the Ministerio de Educacion y Ciencia of Spain. This work was supported by a National Institutes of Health grant to R.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rgross@lmb.uni-muenchen.de; FAX 49-89 21806949.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Bagga R, Emerson BM. An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes & Dev. 1997;11:629–639. doi: 10.1101/gad.11.5.629. [DOI] [PubMed] [Google Scholar]
- Belyaev ND, Keohane AM, Turner BM. Histone H4 acetylation and replication timing in Chinese hamster chromosomes. Exp Cell Res. 1996;225:277–285. doi: 10.1006/excr.1996.0177. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: A current view of enhancer action. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Ariel M, Cedar H. Dynamics of DNA methylation during development. Bioessays. 1993;15:709–713. doi: 10.1002/bies.950151103. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Cockerill PN. Nuclear matrix attachment occurs in several regions of the IgH locus. Nucleic Acids Res. 1990;18:2643–2648. doi: 10.1093/nar/18.9.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill PN, Yuen M-H, Garrard WT. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- Cordingley MG, Riegel AT, Hager GL. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987;48:261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Dillon N, Grosveld F. Chromatin domains as potential units of eukaryotic gene function. Curr Opin Genet Dev. 1994;4:260–264. doi: 10.1016/s0959-437x(05)80053-x. [DOI] [PubMed] [Google Scholar]
- Epner E, Reik A, Cimbora D, Telling A, Bender M, Fiering S, Enver T, Martin D, Kennedy M, Keller G, Groudine M. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- Ernst P, Smale ST. Combinatorial regulation of transcription II: The immunoglobulin mu heavy chain gene. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- Forrester W, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Evidence for a locus activation region: The formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin μ gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Boyd A, Bernard O, Webb E, Adams J. Activation of immunoglobulin μ gene expression involves stepwise demethylation. EMBO J. 1984;3:3013–3021. doi: 10.1002/j.1460-2075.1984.tb02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F, v. Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Haber JE. A locus control region regulates yeast recombination. Trends Genet. 1998;14:317–321. doi: 10.1016/s0168-9525(98)01501-7. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: A B cell-specific trans-activator that describes a new DNA-binding protein family. Genes & Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- Hyman R, Ralph P, Sarkar S. Cell-specific antigens and immunoglobulin synthesis of murine myeloma cells and their variants. J Nat Cancer Inst. 1972;48:173–184. [PubMed] [Google Scholar]
- Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes & Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Grosschedl R. Complex pattern of immunoglobulin μ gene expression in normal and transgenic mice: Nonoverlapping regulatory sequences govern distinct tissue specificities. Genes & Dev. 1991;5:932–943. doi: 10.1101/gad.5.6.932. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Forrester W, Qiu R, Grosschedl R. The immunoglobulin m enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes & Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Fernández-Herrero LA, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Griffiths SD, Ford AM, Greaves MF, Enver T. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc Natl Acad Sci. 1992;89:10618–10622. doi: 10.1073/pnas.89.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Targeted recruitment of the Sin3–Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalos M, Fournier RE. Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol Cell Biol. 1995;15:198–207. doi: 10.1128/mcb.15.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwamura S-I, Koyama T, Matsuo T, Steinmetz M, Kimoto M, Sakaguchi N. Structure of the murine mb-1 gene encoding a putative sIgM-associated molecule. J Immunol. 1990;145:337–343. [PubMed] [Google Scholar]
- Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Ig kappa locus. Nature Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- Krumm A, Madisen L, Yang XJ, Goodman R, Nakatani Y, Groudine M. Long-range transcriptional enhancement by the histone acetyltransferase PCAF. Proc Natl Acad Sci. 1998;95:13501–13506. doi: 10.1073/pnas.95.23.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes & Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G, Perry R. Cμ-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature. 1985;318:475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: A novel role for the intronic kappa chain enhancer sequence. Cell. 1994;76:913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- Loc P, Stratling W. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988;7:655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Fiering S, Groudine M. Regulation of beta-globin gene expression: straightening out the locus. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- McKnight RA, Shamay A, Sankaran L, Wall RJ, Hennighausen L. Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc Natl Acad Sci. 1992;89:6943–6947. doi: 10.1073/pnas.89.15.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Mirault M-E, Laemmli UK. Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes & Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nikolajczyk B, Aquiles Sanchez J, Sen R. Ets protein-dependent accessibility changes at the immunoglobulin μ heavy chain enhancer. Immunity. 1999;11:11–20. doi: 10.1016/s1074-7613(00)80077-1. [DOI] [PubMed] [Google Scholar]
- Ondek B, Shepard A, Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987;6:1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Pettersson M, Schaffner W. A purine-rich DNA sequence motif present in SV40 and lymphotropic papovavirus binds a lymphoid-specific factor and contributes to enhancer activity in lymphoid cells. Genes & Dev. 1987;1:962–972. doi: 10.1101/gad.1.9.962. [DOI] [PubMed] [Google Scholar]
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes & Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi-Van L, Stratling WH. Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry. 1996;35:10735–10742. doi: 10.1021/bi9603783. [DOI] [PubMed] [Google Scholar]
- Sakai E, Bottaro A, Davidson L, Sleckman BP, Alt FW. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc Natl Acad Sci. 1999;96:1526–1531. doi: 10.1073/pnas.96.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R, Chen U. A developmental-specific factor binds to suppressor sites flanking the immunoglobulin heavy chain enhancer. Genes & Dev. 1989;3:1255–1266. doi: 10.1101/gad.3.8.1255. [DOI] [PubMed] [Google Scholar]
- Sleckman BP, Gorman JR, Alt FW. Accessibility control of antigen-receptor variable-region gene assembly: Role of cis-acting elements. Annu Rev Immunol. 1996;14:459–489. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- Stief A, Winter D, Stratling WH, Sippel AE. A nuclear DNA attachment element mediates elevated and position- independent gene activity. Nature. 1989;341:343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Walters MC, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin DI. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes & Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- Wang Z, Goldstein A, Zong RT, Lin D, Neufeld EJ, Scheuermann RH, Tucker PW. Cux/CDP homeoprotein is a component of NF-muNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription activator. Mol Cell Biol. 1999;19:284–295. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes & Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: Involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- Weitzel JM, Buhrmeister H, Stratling WH. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol Cell Biol. 1997;17:5656–5666. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Haber J. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Alt FW. Developmentally controlled and tissue- specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Zaller DM, Eckhardt LA. Deletion of a B-cell-specific enhancer affects transfected, but not endogenous, immunoglobulin heavy-chain gene expression. Proc Natl Acad Sci. 1985;82:5088–5092. doi: 10.1073/pnas.82.15.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Kas E, Gonzalez E, Laemmli UK. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]