Abstract
In higher plants, seed development requires maternal gene activity in the haploid (gametophytic) as well as diploid (sporophytic) tissues of the developing ovule. The Arabidopsis thaliana gene MEDEA (MEA) encodes a SET-domain protein of the Polycomb group that regulates cell proliferation by exerting a gametophytic maternal control during seed development. Seeds derived from female gametocytes (embryo sacs) carrying a mutant mea allele abort and exhibit cell proliferation defects in both the embryo and the endosperm. In this study we show that the mea mutation affects an imprinted gene expressed maternally in cells of the female gametophyte and after fertilization only from maternally inherited MEA alleles. Paternally inherited MEA alleles are transcriptionally silent in both the young embryo and endosperm. Mutations at the decrease in DNA methylation1 (ddm1) locus are able to rescue mea seeds by functionally reactivating paternally inherited MEA alleles during seed development. Rescued seeds are larger than the wild type and exhibit some of the abnormalities found in aborting mea seeds. Our results indicate that the maintenance of the genomic imprint at the mea locus requires zygotic DDM1 activity. Because DDM1 encodes a putative chromatin remodeling factor, chromatin structure is likely to be interrelated with genomic imprinting in Arabidopsis.
Keywords: Arabidopsis, genomic imprinting, MEDEA, DDM1, embryo, endosperm
The life cycle of higher plants alternates between a dominant diploid generation (sporophyte) and two sexually dimorphic haploid generations (gametophytes) that develop within the reproductive organs (Grossniklaus and Schneitz 1998). The delivery of two sperm cells into the multicellular female gametophyte ensures fertilization of both the egg cell and the binucleate central cell, the precursors of the embryo and the triploid endosperm, respectively. Viable seed formation depends on the coordinated development of the embryo, the endosperm, and the maternal seed coat. Although these interactions are poorly understood, seed development requires maternal gene activity in the gametophytic (Ohad et al. 1996; Chaudhury et al. 1997; Grossniklaus et al. 1998) as well as in the sporophytic tissues of the developing ovule (Ray et al. 1996; Colombo et al. 1997). Therefore, mutations displaying a maternal effect on seed development can be of either gametophytic or sporophytic nature (Grossniklaus and Schneitz 1998).
The Arabidopsis thaliana gene MEDEA (MEA) regulates cell proliferation by exerting a gametophytic maternal control during seed development (Grossniklaus et al. 1998). Seeds derived from embryo sacs carrying a mutant mea-1 allele (hereafter referred to as mea seeds) abort after delayed morphogenesis with excessive cell proliferation in the embryo and reduced free nuclear divisions in the endosperm. In addition, mea seeds are able to initiate endosperm development, seed coat differentiation, and fruit (silique) maturation in the absence of fertilization at a low frequency (Grossniklaus and Vielle-Calzada 1998; Kiyosue et al. 1999; Ming et al. 1999).
Five alleles of mea have been described (Castle et al. 1993; Chaudhury et al. 1997; Grossniklaus et al. 1998; Kiyosue et al. 1999; Ming et al. 1999), all of which are likely to be recessive loss-of-function mutations, although this has only been demonstrated for three alleles (Grossniklaus et al. 1998; Kiyosue et al. 1999). MEA encodes a SET-domain protein with homology to members of the Polycomb and trithorax group (Grossniklaus et al. 1998), which are believed to maintain active or repressed states of gene expression during development by modulating higher-order chromatin structure (Kennison 1995; Orlando and Paro 1995; Pirotta 1997). Because the endosperm inherits two maternal copies but only one paternal copy of the genome (Kermicle and Alleman 1990; Ray 1998) mea could affect a dosage-sensitive gene required for endosperm development. Alternatively, the mutation could disrupt a maternally produced gene product stored in the egg and/or central cell, which is subsequently required for seed development. As a third possibility, the mutation could affect an imprinted gene that is transcribed exclusively from the maternally inherited alleles after fertilization.
The expression of genes that are regulated by genomic imprinting is determined by the sex of the parent of origin (John and Surani 1996; Neumann and Barlow 1996). To date, imprinted genes have predominantly been found and studied in mammals (Tilghman 1999), where the disturbance of imprinting can result in dramatic developmental aberrations and cancer (Reik and Maher 1997). Parent-of-origin-specific differences in gene expression usually correlate with differential methylation, but there is no clear evidence for the involvement of methylation in establishing the initial imprint (Caspary et al. 1998; Jones et al. 1998). In higher plants the role of genomic imprinting in development is poorly understood. So far, no imprinted plant genes have been identified that are required for normal development. Only three genes which are expressed in the endosperm of maize are good candidates for regulation by genomic imprinting (Kermicle 1970; Chaudhuri and Messing 1994; Lund et al. 1995). In contrast to the situation in mammals, only specific alleles of these loci are subject to imprinting and none affects seed morphogenesis. Thus, a proposed role of imprinted genes for seed formation largely comes from interploidy crosses where entire parental genomes or individual chromosomes are manipulated (Kermicle and Alleman 1990). In maize, changes of the parental genome ratio lead to endosperm abortion but have little effect on embryo development (Lin 1984). In Arabidopsis, interploidy crosses have an effect on both endosperm and embryo proliferation (Scott et al. 1998a). The regulation of genomic imprinting in both animals and plants is poorly understood and regulatory factors involved in the establishment and maintenance of imprinting have yet to be identified.
Here we show that transcription of the mea locus is regulated by genomic imprinting and identify a modifier gene required to maintain this imprint during seed development. We show that MEA is expressed in the embryo sac before and after fertilization and demonstrate that the paternally inherited MEA allele is silenced in both products of fertilization. Similar to the situation in mammals and in contrast to the known imprinted genes in plants, the epigenetic regulation of the MEA locus by imprinting is not allele specific and the MEA gene product is required for normal seed morphogenesis. Finally, we show that mutations in ddm1 are able to rescue mea seeds by activating the paternally inherited MEA wild-type allele later during seed development. DDM1, is required for the maintenance but not the establishment of the imprint at the mea locus in Arabidopsis.
Results
MEA is expressed in the female gametophyte and in both products of fertilization
Although both the embryo and the endosperm are affected in seeds inheriting a mutant mea allele from the female gametophyte (Grossniklaus et al. 1998), the complex interactions between these tissues do not allow a distinction between primary and secondary effects based on a morphological characterization. To gain insight into the spatial and temporal pattern of MEA gene expression, we performed in situ hybridization (ISH) using digoxygenin-labeled MEA probes. In the ovule, MEA mRNA was detected in the eight-nucleated noncellularized female gametophyte (Fig. 1A). After cellularization, the MEA transcript was detected in several cells of the unfertilized embryo sac: the synergids, the egg cell, and the central cell (Fig. 1B,C). In contrast to the synergids where the signal is in the cytoplasm, the MEA transcript is in close association with the nuclei of the egg and central cell (Fig. 1C). MEA mRNA is not detected in any floral organs, including sepals, petals, stamens, or carpels at any stage of development; no signal could be detected in developing or mature pollen grains (data not shown). After fertilization, MEA mRNA was detected in all cells of the suspensor and the embryo proper (Fig. 1D,E). The transcript persists at a high level until the heart and torpedo stage (Fig. 1F–H) but gradually becomes weaker as embryos reach the cotyledonary stage (Fig. 1I). During the free nuclear phase of endosperm formation, MEA mRNA is abundant in dense regions of free nuclei, accumulating at the micropylar and chalazal poles of the embryo sac (Fig. 1D,J). The transcript becomes undetectable as the free nuclei start to cellularize at the periphery of the central cell (Fig. 1H). Thus, MEA mRNA is maternally transcribed in the female gametophyte, and MEA mRNA is detectable in both the embryo and the endosperm after fertilization. The high levels of MEA mRNA detected in late heart- and torpedo-stage embryos (Fig. 1H) and in free nuclear endosperm prior to cellularization (Fig. 1J) cannot be accounted for by maternal expression in the egg and central cell only, suggesting that MEA is zygotically transcribed.
Figure 1.
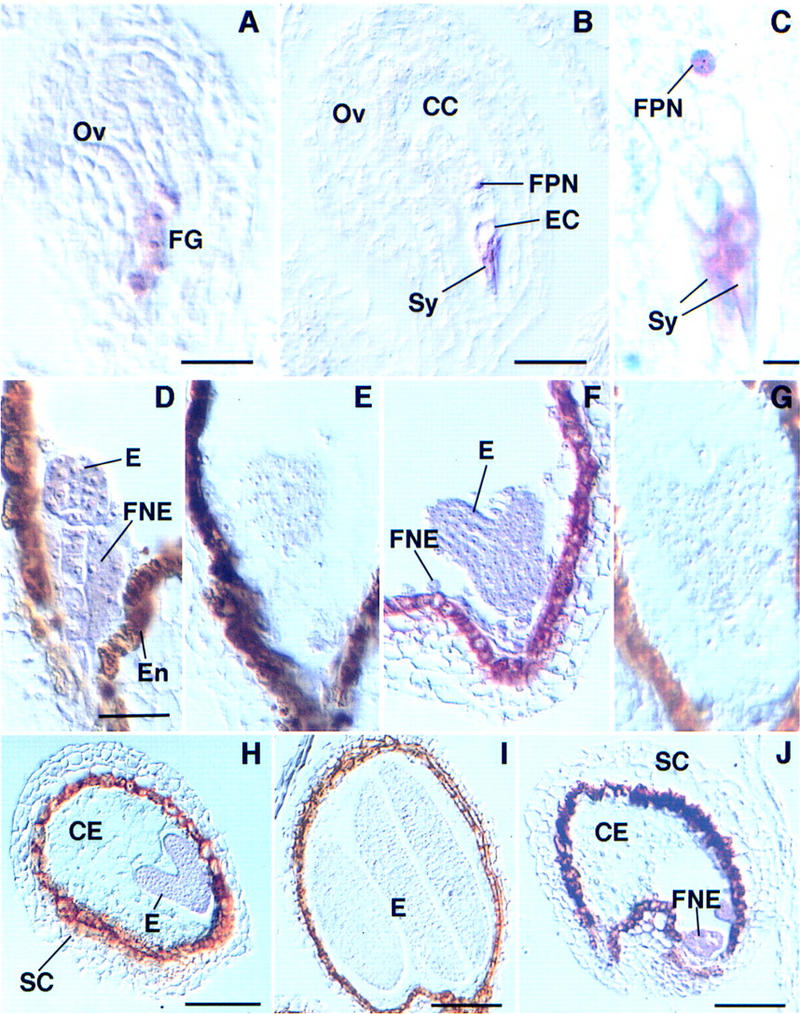
Localization of MEA mRNA in ovules and developing seeds of wild-type Arabidopsis. A-D, F, and H–J are hybridized with an anti-sense, E and G with a sense MEA probe. (A) Ovule (Ov) containing an eight-nucleated noncellularized female gametophyte (FG). The MEA transcript is present in the developing female gametophyte. (B) Mature ovule with unfertilized female gametophyte; MEA mRNA is present in the cytoplasm of the synergids (Sy), the egg cell (EC), and the central cell (CC) containing a homodiploid fused polar nucleus (FPN). (C) Detail of the synergids (Sy) and the fused polar nucleus (FPN) shown in B; the transcript is localized in the cytoplasm of the synergids, but appears closely associated with the fused polar nucleus. (D) MEA mRNA is localized in the globular embryo (E) and in the free nuclear endosperm (FNE) of a developing seed; artifactual staining in the endothelium (En) is seen in sense and anti-sense experiments. (E) Globular embryo hybridized with a sense MEA probe. (F) Heart-stage embryo hybridized with an anti-sense probe; the MEA transcript is localized in the embryo (E) and the free nuclear endosperm (FNE). (G) Heart-stage embryo hybridized with a sense MEA probe. (H) Seed showing an early torpedo embryo (E), the cellularized endosperm (CE), and the seed coat (SC); MEA mRNA is absent from the cellularized endosperm. (I) Seed containing a cotyledonary embryo hybridized with an anti-sense MEA probe. (J) Developing seed showing free nuclear endosperm (FNE), cellularized endosperm (CE), and the seed coat (SC); MEA mRNA is localized in the free nuclear endosperm. Bar, 17 μm in A; 22 μm in B; 4.3 μm in C; 35 μm in D–G; 51 μm in H–J.
The mea maternal effect is not caused by a dosage effect in the endosperm
We have previously shown that mea is not a haplo-insufficient locus by adding an extra wild-type MEA allele to both embryo and endosperm using a tetraploid pollen donor (Grossniklaus et al. 1998). However, endosperm derived from these crosses carried an equal number of mutant and wild-type alleles and it is possible that an excess of the wild-type product is required to ensure normal seed development. We tested this hypothesis by analyzing the seed phenotype in duplex tetraploid plants that carry two mutant mea-1 and two wild-type MEA alleles. The progeny of self-fertilized duplex tetraploids carries between zero and six mutant mea-1 copies in the hexaploid endosperm providing the material to differentiate between a dosage effect and maternal inheritance (Fig. 2). If an excess of wild-type MEA product was required for normal seed development, i.e., four or more wild-type MEA copies must be present in the hexaploid endosperm, we would expect 31.5% of the developing seeds to be normal. Alternatively, if the mea maternal effect depends on the maternal gametophytic genotype only, i.e., at least one wild-type MEA allele must be inherited from the mother, 78.8% of the developing ovules are expected to form normal seeds. Based on the pooled data of 15 duplex mea-1 plants where we characterized seed abortion, we found that 79.3% (1015/1280 = 0.793) of the developing seeds were normal, strongly suggesting that the ratio of mutant mea-1 to wild-type MEA alleles in the endosperm does not have an effect on seed development (X2 = 1356.5 > χ20.05[1] = 3.84), but that seed abortion is solely determined by the maternally inherited allele (X2 = 0.19 < χ20.05[1] = 3.84).
Figure 2.
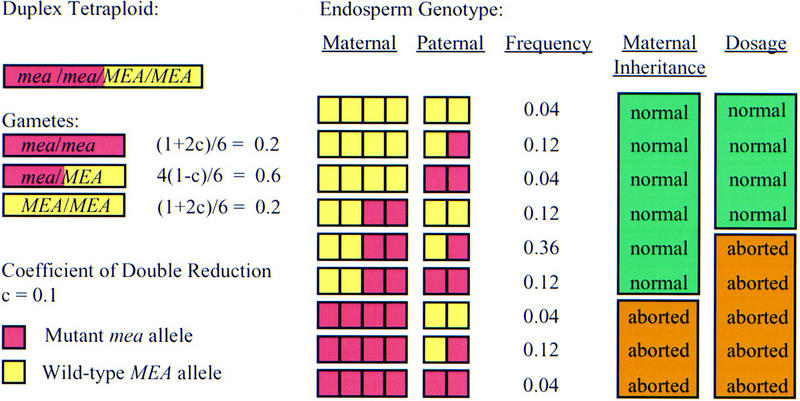
Segregation of mea alleles in a duplex tetraploid. In tetraploids, the frequencies of the different gamete classes depend on the coefficient of double reduction, c, which is the frequency at which the alleles of two sister chromatids are recovered in the same gamete (Burnham 1962). (Small boxes) Haploid genomes from mutant mea-1 (red) or wild-type MEA (yellow) alleles. For the chromosomal region containing the mea locus, c has been estimated as c = 0.1 (van der Veen et al. 1973). In duplex tetraploids, the frequencies of MEA/MEA, MEA/mea-1, and mea-1/mea-1 gametes are (1 + 2c)/6 = 0.2, 4(1 − c)/6 = 0.6, and (1 + 2c)/6 = 0.2, respectively. These values were used to calculate the frequencies of the different endosperm classes that receive two maternal and one paternal genome. If the maternal genotype determines the seed phenotype independent of gene dosage, we expect endosperm genotypes with at least one maternal wild-type MEA allele to develop normally amounting to 78.8% of the seeds [adjusting for a spontaneous seed abortion rate of 1.5% as determined for the parental tetraploid; (0.04 + 0.12 + 0.04 + 0.12 + 0.36 + 0.12) × (100 − 1.5)% = 78.8%]. If normal seed development depends on an excess of wild-type MEA product independent of parental origin in the endosperm we expect endosperm genotypes with MEA:mea-1 > 3:3 to develop normally amounting to 31.5% of the seeds [(0.12 + 0.04 +0.12 + 0.04) × (100 − 1.5)% = 31.52%].
Only two of the three MEA copies are transcribed in the endosperm after fertilization
To differentiate between a maternal effect of cytoplasmic or chromosomal (imprinting) nature, we characterized the expression from the mea locus during seed development. An improved ISH procedure allowed the detection of nuclear dots in the central cell (Fig. 3A). Nuclear dots of intense staining have not previously been reported in plants, but they have been observed in mammalian (Lawrence et al. 1989) and Drosophila nuclei where they were shown to be tightly associated with nascent transcripts of actively expressed genes (Shermoen and O'Farrell 1991). Because the nuclear dots are associated with a transcribed genomic locus, they allow an analysis of the transcriptional state of a particular locus at any given time in development. We confirmed that the nuclear dots represented nascent transcription sites associated with a genomic locus by hybridizing MEA riboprobes to ovules of a tetraploid plant. As expected, the number of nuclear dots was doubled in the nuclei of a tetraploid plant as compared to a diploid one (Fig. 3B), confirming that the nuclear dots are indeed correlated with the number of MEA loci present in these nuclei. The absence of nuclear dots in sections that were incubated with RNase prior to hybridization indicates that the signal is due to the presence of nascent RNA and not to hybridization to chromosomal DNA (data not shown). Thus, an analysis of nuclear dots allows a determination of the transcriptional state of all mea loci present in the central cell nucleus at a specific time in development.
Figure 3.

MEA transcription in the central cell before and after fertilization. (A) Two haploid polar nuclei in the unfertilized central cell; a single nuclear dot is localized in each nucleus. (B) A tetraploid nucleus (resulting from the fusion of two diploid polar nuclei in a tetraploid ovule) showing four nuclear dots prior to fertilization. (C) A diploid polar nucleus in the central cell of a diploid ovule prior to fertilization showing two nuclear dots. (D) The triploid nuclei resulting from the division of the primary endosperm nucleus, each showing two nuclear dots after fertilization. (E) Two nuclear dots in the triploid endosperm nuclei located at the chalazal pole of the embryo sac. Bar, ∼0.5 μm in A; 1.2 μm in B; 1.4 μm in C; 1.5 μm in D; 1.7 μm in E.
Nascent transcripts could only be visualized in the polar nuclei of the central cell. Nuclear dots were not observed in the smaller nuclei of the egg cell and synergids. To determine the transcriptional state of the mea loci in the central cell nuclei both before and after fertilization, we analyzed nuclear dots in wild-type diploid plants. Prior to fertilization, a single dot was detected in each of the two haploid polar nuclei present in the central cell (Fig. 3A). Following fusion of the two polar nuclei, two dots were detected in the resulting homodiploid nucleus (Fig. 3C). At fertilization this homodiploid nucleus fuses with a sperm nucleus to form the triploid primary endosperm nucleus. In this triploid nucleus only two dots persist after fertilization (results not shown) and following the first mitotic division (Fig. 3D,E). The nuclear dots are no longer detectable after a few free nuclear divisions as the nuclei decrease in size. Together, these results show that the mea locus is actively transcribed after fertilization during early endosperm development. However, only two nuclear dots are detectable both before and after fertilization, strongly suggesting that the paternally inherited MEA allele remains transcriptionally silent during early endosperm development.
Paternally inherited MEA alleles are not expressed in either embryo or endosperm
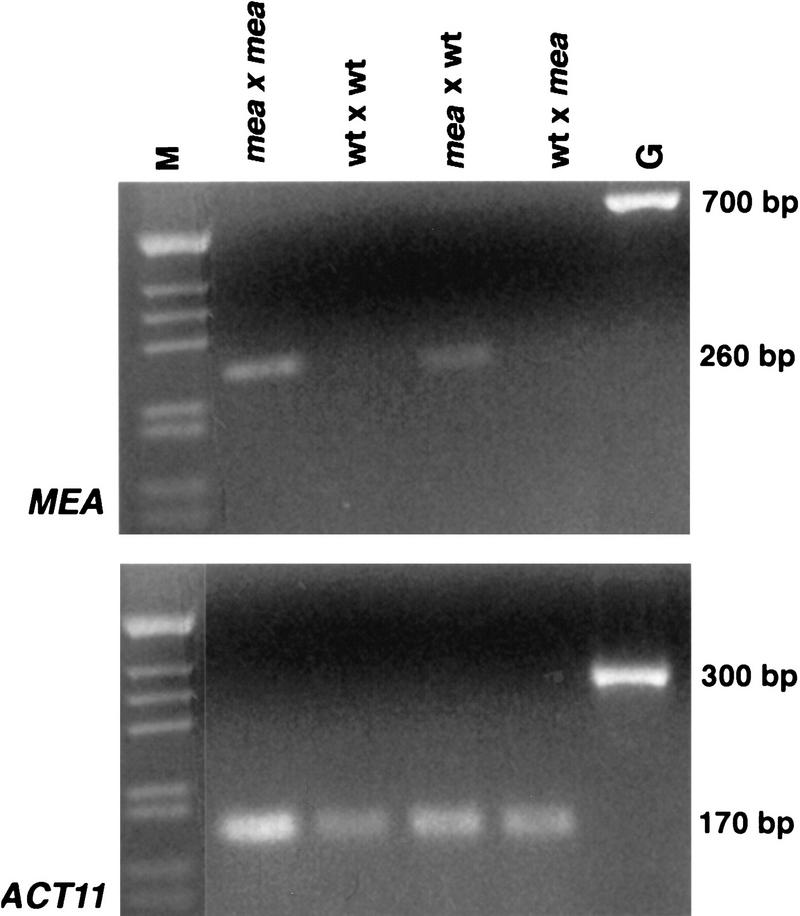
To independently confirm that postfertilization transcription of MEA is restricted to the maternally inherited alleles, we examined MEA expression by reverse transcription-polymerase chain reaction (RT–PCR) on RNA isolated from developing siliques derived from reciprocal crosses between wild-type and mea plants ∼54 hr after pollination (HAP) when the embryos have reached the midglobular stage. To obtain genetically homogeneous material resulting from reciprocal crosses, we generated plants that were homozygous for mea-1. Although mature desiccated mea seeds do not germinate, seedlings can be obtained by culturing embryos in vitro or by precocious germination of seeds prior to desiccation. Many of these seedlings show initial developmental aberrations but grow into adult plants that do not show any obvious mutant vegetative or floral phenotypes (J. Thomas and U. Grossniklaus, unpubl.). Plants homozygous for mea-1 are indistinguishable from heterozygotes except that they produce siliques containing 100% aborted seeds as compared with the 50% in a heterozygote. To distinguish between the mutant mea-1 and the wild-type MEA allele, we designed primers that specifically amplify the mea-1 allele under PCR conditions where the wild-type allele is undetectable (Fig. 4). In reciprocal crosses mea-1 mRNA can be detected if it is inherited maternally but not if it is inherited from the pollen donor. These results show that paternally inherited MEA alleles are not transcribed in either embryo or endosperm early during seed development, as both fertilization products were present in the tissue used for RNA isolation.
Figure 4.
Expression of the mea-1 allele during early seed development. Amplification of MEA-1 (top) and ACTIN-11 (ACT11) as a control for cDNA synthesis. RNA was isolated from siliques derived from self-pollinated mea-1 homozygous pistils (mea × mea), self-pollinated wild-type pistils (wt × wt), a cross between a homozygous mea-1 female and wild-type male (mea × wt), and a cross between wild-type female and homozygous mea-1 male (wt × mea). Primers that specifically amplify the mea-1 allele under these PCR conditions were used for RT–PCR. (M) Marker lane; (G) genomic DNA as a control.
Genomic imprinting at the mea locus is not allele specific
The genetic and molecular analysis of MEA expression strongly suggests that the mea locus is regulated by genomic imprinting. To date, only three plant genes have been identified that are likely to be imprinted, all of which are expressed in the endosperm of maize. These are the r gene involved in the regulation of anthocyanin biosynthesis (Kermicle 1970), the seed storage protein regulatory gene dzr1 (Chaudhuri and Messing 1994), and an α tubulin gene (Lund et al. 1995). In each case, only specific alleles are subject to epigenetic regulation by imprinting. In contrast, imprinting in mammals is generally locus specific and all alleles are subject to imprinting. Because our analysis of the transcriptional regulation of MEA was only based on a single allele, we tested whether other MEA alleles were also regulated by genomic imprinting. To this aim we crossed 21 Arabidopsis ecotypes (see Materials and Methods) to homozygous mea-1/mea-1 female plants to test whether any paternally derived MEA alleles could support normal seed development. Naturally occurring MEA alleles that are not paternally silenced would be expected to provide zygotic activity, which may be sufficient to rescue the mea phenotype. An average of 89 seeds were scored per ecotype cross (ranging from 42 to 150 seeds) and no cross produced a Mendelian ratio of wild-type to aborted seeds that would indicate a lack of paternal MEA silencing. Seventeen of the crosses produced exclusively aborted seeds whereas four produced a few seeds that reached maturity without any obvious phenotype. The latter occurred at a low frequency (between 1% and 10%) that we attribute to a genetic background effect but not to allele-specific differences in paternal silencing (see below). The production of a few wild-type seeds in these hybrids is likely to be due to instability of paternal silencing during seed development or may indicate that these four ecotypes harbor weakly penetrant modifiers of paternal MEA silencing. These findings suggest that none of the naturally occurring MEA alleles tested provides zygotic activity to support normal seed development, indicating that they are all paternally silenced. Thus, unlike the other imprinted loci in plants genomic imprinting of MEA is locus and not allele specific.
ddm1 is a zygotic modifier of genomic imprinting
Because MEA is expressed both maternally and zygotically, it is not clear whether parent-specific expression after fertilization is relevant to the phenotype observed in mea mutants. It is possible that MEA is only required during a short time before fertilization and that a lack of MEA activity in the female gametophyte causes seed abortion later in development. Alternatively, postfertilization expression of MEA, which is under the control of genomic imprinting, may be responsible for the mea phenotype. To distinguish between these possibilities we attempted to manipulate MEA expression in the developing seed. For instance, suppression of the mea seed abortion phenotype by postfertilization expression of MEA would suggest that zygotic MEA activity is sufficient for normal seed development. Ecotype hybrids gave a first indication that this might be the case since some hybrids produced a low percentage of wild-type seeds.
To further investigate the respective role of maternal and zygotic MEA expression we screened for potential modifiers of mea among mutants known to affect DNA methylation or gene silencing such as the ddm1 mutant (Vongs et al. 1993; Jeddeloh et al. 1998). Recently, DDM1, which reduces genomic DNA methylation to 30% (Vongs et al. 1993; Kakutani et al. 1999), has been shown to encode a chromatin remodeling factor of the SWI2/SNF2 family (Jeddeloh et al. 1999). Progeny from crosses involving a mea-1 plant and a ddm1-2/DDM1 heterozygote segregated two classes of mea-1 heterozygous plants that differed in their seed abortion frequency (Fig. 5A). One class showed an abortion frequency (47.5%) close to the one expected for mea-1/MEA (50%), whereas the other had a significantly lower abortion rate (39.7%). The two classes segregated in a 1:1 ratio (23:25, X2 = 0.08 < χ 20.05[1] = 3.84) suggesting that ddm1 acts as a suppressor of mea seed abortion. All plants produced slightly more normal seeds than expected, which we attribute to a weak suppression of mea seed abortion caused by a genetic background effect. The ddm1-2 allele is in the Columbia (Col) ecotype whereas mea-1 is in Landsberg erecta (Ler). Thus, the plants we analyzed were all Col/Ler hybrids. We had previously noted that Col acts as a weak suppressor of mea in outcrosses of mea-1/MEA with Col males. We observed that the mutant maternal mea-1 allele was transmitted to 3.4% (13/378 = 0.034) of the progeny from these crosses. This is in good agreement with the observed increase of wild-type seeds by 3.6% from the expected 50% to 53.6% (5025/9369 = 0.536) among all plants analyzed (Fig. 5A).
Figure 5.

ddm1 is a zygotic modifier of genomic imprinting. (A) Crossing scheme to test the effect of ddm1 on mea seed abortion. Progeny from the cross fell into two classes, characterized by a different frequency of aborted seeds (Ab) and the presence or absence of enlarged seeds (E). (# seeds) Numer of seeds scored. Data of six plants from each class were pooled because pairwise χ2 tests showed no significant differences in the frequency of aborted and viable [normal (N) plus enlarged seeds] produced by plants in either class. Among the plants carrying a mutant ddm1-2 allele one-eighth of the seeds that inherit a maternal mea-1m allele are expected to also carry a wild-type paternal MEAp allele and to be homozygous for ddm1-2 (0.5 × ¼ × ½ = 0.0625; X2 = 2.50 < χ20.05[1] = 3.84; without correction for the Col/Ler background effect). Given that 3.6% of the seeds that inherit a mutant maternal mea-1m allele survive in the Col/Ler hybrid background (see text), a frequency of 5.7% is in very good agreement with the expected 5.8% [(0.5 − 0.036) × 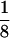 = 0.058; X2 = 0.86 < χ20.05[1] = 3.84; with correction for the Col/Ler background effect]. (B) Rescued mea-1m/MEAp; ddm1-2/ddm1-2 seeds (top) are larger than phenotypically wild-type siblings (bottom). (C) Embryos from dissected, rescued seeds (top) are larger than wild-type embryos (bottom). Bar, 400 μm.
= 0.058; X2 = 0.86 < χ20.05[1] = 3.84; with correction for the Col/Ler background effect]. (B) Rescued mea-1m/MEAp; ddm1-2/ddm1-2 seeds (top) are larger than phenotypically wild-type siblings (bottom). (C) Embryos from dissected, rescued seeds (top) are larger than wild-type embryos (bottom). Bar, 400 μm.
Remarkably, mea heterozygotes carrying a mutant ddm1-2 allele (mea-1/MEA; ddm1-2/DDM1) did not only show a decrease in the frequency of aborted seeds but also produced seeds that were considerably larger than their wild-type siblings (Fig. 5B). Embryos were still green when the wild-type seeds had lost their chlorophyll pigmentation suggesting a delay in seed development (Fig. 5C). Enlarged seeds were observed at a frequency of 5.7% (Fig. 5A), which is close to 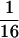 suggesting that they were homozygous for ddm1-2 (X2 = 2.50 < χ20.05[1] = 3.84; see legend to Fig. 5 for details). The observed segregation ratio strongly suggests that seeds inheriting a maternal mea-1 allele, which usually abort, are rescued provided they are homozygous for ddm1-2 and also carry a paternal wild-type MEA allele. That homozygosity of ddm1 in the developing seed is required for suppression of the mea phenotype was confirmed by outcrossing the mea-1/MEA; ddm1-2/DDM1 plants to mea-1/mea-1 homozygotes. If ddm1 acted in the male gametophyte and prevented the silencing of the paternal MEAp allele, 25% of the pollen from such a plant should carry a nonsilenced wild-type MEA allele (pollen genotype: MEAp; ddm1-2) expected to rescue 25% of the seeds. Among 259 seeds scored no normal or enlarged seeds were observed. Furthermore, we confirmed the predicted genotype of the enlarged seeds (mea-1m/MEAp; ddm1-2/ddm1-2) by scoring for kanamycin resistance associated with the mea-1 allele (Grossniklaus et al. 1998) and by confirming homozygosity for ddm1 by Southern analysis or scoring a cleaved amplified polymorphic sequences (CAPS) marker (Jeddeloh et al. 1999) linked to the ddm1-2 allele (data not shown). These analyses strongly suggest that ddm1 acts as a zygotic modifier of mea, i.e., seeds carrying a mutant maternal mea-1 allele and a paternal wild-type MEA allele survive to maturity if they are also homozygous for ddm1. In seeds of this genotype, MEA activity is likely to be provided by the paternally inherited MEA allele that gets reactivated during seed development because of a lack of DDM1 activity. This is supported by the fact that all 15 plants derived from enlarged seeds that we tested were heterozygous for mea-1 and, thus, had inherited a wild-type MEA allele from the father. If the effect of ddm1 was bypassing MEA rather than reactivating the paternal copy, we would expect half the plants to inherit a mutant mea-1 allele from the father and twice as many mea seeds should survive as observed. These findings strongly suggest that zygotic MEA activity provided from a reactivated paternal allele is sufficient to support seed development. Thus, postfertilization expression of MEA, which is subject to genomic imprinting, is responsible for the mea phenotype.
suggesting that they were homozygous for ddm1-2 (X2 = 2.50 < χ20.05[1] = 3.84; see legend to Fig. 5 for details). The observed segregation ratio strongly suggests that seeds inheriting a maternal mea-1 allele, which usually abort, are rescued provided they are homozygous for ddm1-2 and also carry a paternal wild-type MEA allele. That homozygosity of ddm1 in the developing seed is required for suppression of the mea phenotype was confirmed by outcrossing the mea-1/MEA; ddm1-2/DDM1 plants to mea-1/mea-1 homozygotes. If ddm1 acted in the male gametophyte and prevented the silencing of the paternal MEAp allele, 25% of the pollen from such a plant should carry a nonsilenced wild-type MEA allele (pollen genotype: MEAp; ddm1-2) expected to rescue 25% of the seeds. Among 259 seeds scored no normal or enlarged seeds were observed. Furthermore, we confirmed the predicted genotype of the enlarged seeds (mea-1m/MEAp; ddm1-2/ddm1-2) by scoring for kanamycin resistance associated with the mea-1 allele (Grossniklaus et al. 1998) and by confirming homozygosity for ddm1 by Southern analysis or scoring a cleaved amplified polymorphic sequences (CAPS) marker (Jeddeloh et al. 1999) linked to the ddm1-2 allele (data not shown). These analyses strongly suggest that ddm1 acts as a zygotic modifier of mea, i.e., seeds carrying a mutant maternal mea-1 allele and a paternal wild-type MEA allele survive to maturity if they are also homozygous for ddm1. In seeds of this genotype, MEA activity is likely to be provided by the paternally inherited MEA allele that gets reactivated during seed development because of a lack of DDM1 activity. This is supported by the fact that all 15 plants derived from enlarged seeds that we tested were heterozygous for mea-1 and, thus, had inherited a wild-type MEA allele from the father. If the effect of ddm1 was bypassing MEA rather than reactivating the paternal copy, we would expect half the plants to inherit a mutant mea-1 allele from the father and twice as many mea seeds should survive as observed. These findings strongly suggest that zygotic MEA activity provided from a reactivated paternal allele is sufficient to support seed development. Thus, postfertilization expression of MEA, which is subject to genomic imprinting, is responsible for the mea phenotype.
mea seeds rescued by ddm1 show overgrowth of the embryo and persistent endosperm
At maturity, the viability of large seeds lacking DDM1 activity is comparable to the wild type, but their germination frequency is reduced by 18% after prolonged storage (124/157 = 0.79 for enlarged seeds vs. 126/130 = 0.97 for wild-type siblings). Except for their enlarged size, many embryos in these seeds are normal although some aberrations were observed. At maturity, three different classes of enlarged seeds can be distinguished (Fig. 6A–D). In the predominant class (50/111 = 0.45), most of the seed volume is occupied by a large, well-organized embryo that is normally curved, with both cotyledons parallel to each other (Fig. 6B). In these embryos aberrant cell proliferation in the hypocotyl and the cotyledons causes enlarged tissue sectors (Fig. 6E) with small irregular cells present in both epidermal and hypodermal layers (Fig. 6G). These cells appear to be poorly differentiated, but their presence does not compromise the overall morphology of the embryo. In contrast to wild-type seeds, a large portion of partially cellularized endosperm persists between the hypocotyl and the cotyledons (Fig. 6H). Because of their large size (diameter 1.6× larger than the wild type), these embryos sometimes break the seed coat in the cotyledonary region. The second class of rescued seeds (46/111 = 0.41) are characterized by the presence of massive amounts of partially cellularized endosperm (Fig. 6D) persisting in both the micropylar and chalazal chambers. The size of the tissue and its degree of cellularization appear to be variable. Embryos have a normal morphology with enlarged tissue sectors in the hypocotyl or the cotyledons; however, growth of the bent cotyledons appears to be limited and the embryos only reach the “walking stick” stage (Bowman 1994) of development. The region that is not occupied by the cotyledons is filled with persistent endosperm. Seeds of the third class (15/111 = 0.14) contain mature embryos that have aberrant morphology. In some seeds (6%) the cotyledons are perpendicular to the apical–basal axis (T-shaped embryos; Fig. 6C), whereas in other seeds (8%), the embryos are twisted in a spiral-like fashion that affects the whole length of the hypocotyl (data not shown). Because of these morphological aberrations, seeds are often swollen and round, with a large portion of partially cellularized endosperm surrounding the embryo. Taken together, these results show that rescued seeds have enlarged embryos that often complete morphogenesis. These defects suggest that they initially show the characteristic mea phenotype of delayed morphogenesis and abnormal cell proliferation as compared to wild-type siblings (Grossniklaus et al. 1998). However, and in contrast to mea seeds, enlarged embryos are able to resume embryogenesis and often complete their development. Differences in germination frequency between wild-type and enlarged seeds (18%) suggest that it is only the small proportion of enlarged seeds showing dramatic morphological defects (∼14%) that cannot withstand full desiccation to give rise to viable seedlings.
Figure 6.
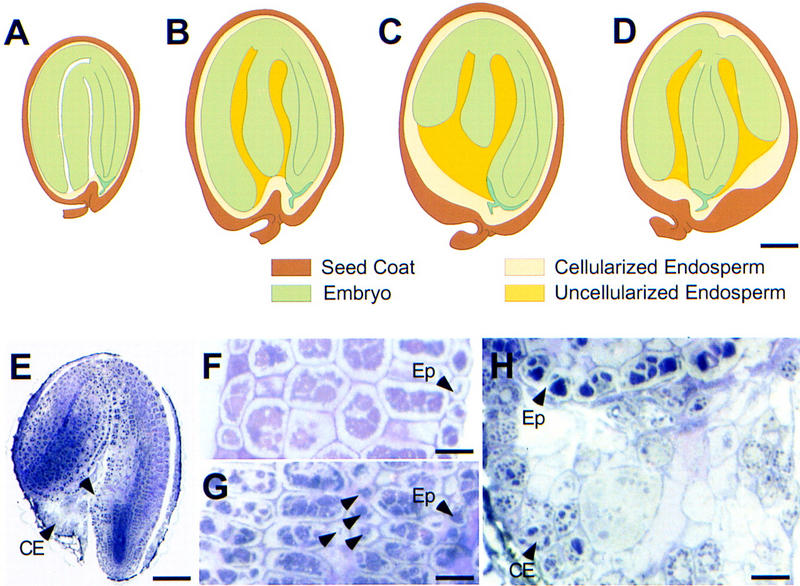
Morphology of mea-1m/MEAp; ddm1-2/ddm1-2 seeds. (A–D) Wild type and three different classes of rescued seeds; the diagrams are based on observations of freshly dissected, cleared, and sectioned specimens. (A) Wild-type seed; (B) rescued seed with giant embryo and partial endosperm; (C) rescued seed with partially bent embryo and massive endosperm; (D) rescued seed with T-shaped embryo and large volume of endosperm. (E) General morphology of a rescued seed belonging to class depicted in B; (arrowhead) an abnormal region of cell proliferation in the hypocotyl. (CE) Cellularized endosperm. (F–I) Morphological analysis of rescued embryos belonging to class depicted in B. (F) Sagital section of a wild-type cotyledon. (G) Sagital section of a rescued seed cotyledon with small undifferentiated cells (arrowheads). (H) Detail of E; partially cellularized endosperm persisting at the chalazal region of a rescued seed. (Ep) Epidermis. Bars, 100 μm in A–E; 20 μ in F; 28 μm in G,H.
Discussion
The MEA locus is regulated by genomic imprinting
Our genetic experiments and expression studies strongly suggest that the MEA locus is regulated by genomic imprinting with only maternally inherited alleles being transcribed following fertilization. To demonstrate transcriptional regulation by genomic imprinting requires evidence that (1) the locus is transcribed zygotically, and (2) maternal and paternal alleles are expressed differentially. Both of these criteria are met by MEA: First, the high levels of MEA mRNA in torpedo-stage embryos and free nuclear endosperm that are seen ∼4 days after pollination (DAP) (Fig. 1H,J) suggest zygotic transcription of MEA in addition to its maternal expression in the female gametophyte. Moreover, an analysis of nascent transcripts after fertilization has provided direct evidence for zygotic transcription in the endosperm (Fig. 3D,E). Second, RT–PCR analysis has shown that paternally inherited alleles are not transcribed in either the young embryo or endosperm (Fig. 4), which was also directly visualized in the endosperm by an analysis of nuclear dots during early endosperm development (Fig. 3D,E). Thus, the regulation of the mea locus by genomic imprinting is responsible for the mea maternal effect: Embryos inheriting a mutant mea allele from the mother abort because paternally inherited MEA alleles are silenced.
MEA activity is required during seed development
Because MEA is expressed both before and after fertilization, it is not clear whether maternal or zygotic expression is relevant to the mea phenotype. The fact that ddm1 acts as a zygotic suppressor of mea seed abortion strongly suggests that reactivation of a paternal wild-type MEA allele after fertilization allows mea embryos to resume morphogenesis and to form viable seeds. This is supported by the fact that mea seeds are only rescued if they are both homozygous for ddm1 and also carry a paternally inherited wild-type MEA allele. Because rescued, enlarged seeds show abnormal growth of both the embryo and the endosperm, it is likely that reactivation of the paternally inherited MEA allele occurs only after the manifestation of growth defects in mutant mea seeds. Cell proliferation defects and delayed morphogenesis of embryo and endosperm in mea seeds are not observed before the midglobular stage (Grossniklaus et al. 1998). The fact that not all rescued embryos have sufficient time to complete morphogenesis before maturity is in agreement with a late reactivation of the paternal MEA allele. These results suggest that zygotic MEA transcription, which is regulated by genomic imprinting, is required for normal seed morphogenesis. Therefore, the major regulatory role of MEA occurs after fertilization, during embryo and endosperm formation, and not during female gametophyte development, as one might infer from its early pattern of mRNA localization.
Genomic imprinting affects embryogenesis and endosperm development
The role of genomic imprinting in plant development is not well characterized as no imprinted genes affecting morphogenesis have been isolated (Messing and Grossniklaus 1999). Imprinting studies have been largely limited to whole-genome interploidy and interspecific crosses from which effects of imprinted genes on seed morphogenesis were inferred (Haig and Westoby 1991; Scott et al. 1998b). In higher plants, the natural occurrence of apomixis (Asker and Jerling 1992; Koltunow 1993; Vielle-Calzada et al. 1996) and the induction of androgenetic or gynogenetic plants in some species (Pelletier 1998) has led to the suggestion that imprinting is unlikely to act in the embryo (Walbot 1996; Martienssen 1998; Scott et al. 1998a). Instead, imprinted genes are conventionally thought to be required specifically for endosperm development. This conclusion has largely been drawn from experiments in maize where changes of the parental genome ratio lead to endosperm abortion but have little effect on embryo development (Lin 1984). In Arabidopsis, interploidy crosses have shown that an excess of maternal genomes leads to underproliferation of the endosperm and smaller embryos, whereas paternal excess promotes endosperm proliferation and yields larger embryos (Scott et al. 1998b). Although the parental genome ratio was changed concurrently in both tissues, the effect on embryo size was considered to be a result of changes in endosperm proliferation.
Such manipulations, which affect complete parental genomes, however, provide little information about the function of individual imprinted genes. Because of the interdependence of embryo and endosperm development and their identical genetic composition (except for the relative contribution of maternal and paternal genomes), it is difficult to distinguish between primary and secondary effects of a mutation affecting seed development. The different morphological classes observed in mea/MEA; ddm1/ddm1 seeds suggest that not all rescued embryos are able to complete growth and differentiation. In some cases, the large amounts of partially cellularized endosperm found in rescued seeds may indicate that the developing embryo plays an active role in reabsorption of the endosperm. Alternatively, overproliferation and partial cellularization of the endosperm may limit embryo growth in advanced stages of seed development. Such a reciprocal relationship between embryo and endosperm size was described for rice where mutants with large embryos have reduced endosperm and vice versa (Hong et al. 1996). Similarly, and in contrast to the effects observed in interploidy crosses, mea embryos grow to a larger size in the presence of a poorly developed endosperm indicating that at least some aspects of the embryo phenotype are direct. We have shown that MEA is expressed and regulated by genomic imprinting in both fertilization products early during seed development (Figs. 3 and 4) and that the formation of both the embryo and endosperm is affected in mutant mea seeds (Grossniklaus et al. 1998). Therefore, it is likely that MEA plays a direct role in both these tissues.
The regulation and function of MEA is similar to that of imprinted loci in mammals
We have shown that the mea locus is transcriptionally regulated by genomic imprinting. Unlike other imprinted genes in plants, mea exhibits many similarities to imprinted loci in mammals. In mice, correctly regulated expression of imprinted genes is essential for normal development (Jaenisch 1997; Tilghman 1999). Similarly, MEA is required for normal seed morphogenesis whereas the three genes from maize that are regulated by imprinting do not play an obvious role in development (Kermicle 1970; Chaudhuri and Messing 1994; Lund et al. 1995). The mea phenotype suggests that maternally controlled MEA expression restricts embryonic growth, which is in agreement with predictions of the parental conflict theory (Haig and Westoby 1989; Moore and Haig 1991; Grossniklaus et al. 1998). The theory predicts that imprinted genes regulate embryonic growth, with paternally controlled genes promoting growth and maternally controlled ones tending to reduce it, as has been found for most imprinted genes in mice (Haig and Graham 1991; Tilghman 1999). However, other authors have interpreted the mea phenotype differently and suggested that mea embryos show a reduction in growth (Scott et al. 1998a; Kiyosue et al. 1999). mea embryos do not complete morphogenesis and only reach the late heart or sometimes torpedo stage before the seeds abort due to desiccation (Grossniklaus et al. 1998). By this time sibling embryos have reached the cotyledonary stage and have fully grown cotyledons that account for the majority of the cells present in wild-type embryos. As compared with the delayed mea embryos that lack cotyledons, the wild-type siblings do indeed have a much higher cell number. If, however, the same developmental stage is compared, mea embryos at the late heart stage have about twice as many cell layers as wild-type embryos of the same stage and are ∼10 times larger in volume (Grossniklaus et al. 1998). The suppression of mea seed abortion by ddm1, which allows many embryos to complete morphogenesis to form giant embryos (Fig. 5C), confirms that mutations in the mea locus lead to increased embryo growth. When mea seeds can complete morphogenesis, they do form embryos much larger than the wild type. Because mea was shown to be a recessive loss-of-function mutation (Grossniklaus et al. 1998) the function of the maternally controlled wild-type MEA gene is to restrict growth of the embryo.
In addition, the epigenetic regulation of the mea locus by imprinting is not allele specific. As was found for the majority of imprinted genes in mammals, all MEA alleles we tested showed paternal silencing and, thus, were subject to genomic imprinting. In contrast, only individual alleles of each of the three imprinted genes of maize are regulated by genomic imprinting. This difference between the three imprinted loci in maize and MEA may reflect a difference in the mechanism underlying imprinting. It is possible that specific alleles can be subject to genomic imprinting because of their particular genomic organization, for instance the presence of a transposon that recruits the imprinting machinery to this specific allele (Martienssen 1996). Such loci are not necessarily expected to play a role in development and hence are not subject to the parental conflict theory. In contrast, imprinting at a locus involved in growth control during seed development is likely to have evolved as a consequence of a parental conflict and is expected to affect all allelic variants. In summary, MEA shows similarities to imprinted genes in mammals both at the functional and regulatory level and defines a novel class of imprinted genes in plants.
Maintenance of genomic imprinting at the mea locus requires DDM1 activity
Mutations in ddm1 rescue mea seeds by zygotically reactivating MEA function from the paternally inherited allele. With the exception of mammalian DNA methyltransferase, this is to our knowledge the first example of a trans-acting regulator of genomic imprinting. The absence of rescued seeds in outcrosses of mea homozygous plants with paternal genotypes lacking DDM1 activity suggests that silencing of the paternally inherited allele is maintained in the sperm cells prior to fertilization. These results suggest that DDM1 is not involved in establishing the imprint at the mea locus but that it is required for the maintenance of the imprint during seed development. Mutations in ddm1 result in the accumulation of a wide range of developmental abnormalities in Arabidopsis after prolonged inbreeding (Kakutani et al. 1996; Kakutani 1997). The induction of heritable changes in other loci by ddm1 is usually slow, with progressively more severe phenotypes in advancing generations (Richards 1997). Jeddeloh et al. (1999) reported that DDM1 encodes a putative chromatin remodeling factor of the SWI2/SNF2 family of DNA-dependent ATPases. It was recently shown the human SWI/SNF complex can remodel nucleosomes into a stable, altered chromatin state (Schnitzler et al. 1998).
The genetic interaction between ddm1 and mea suggests that changes in chromatin structure have an effect on genomic imprinting. The remodeling of nucleosomes into a new chromatin state may affect the imprint at the mea locus and allow transcription of the paternally inherited allele. It has been shown that chromatin remodeling by SWI/SNF facilitates transcription factor access (e.g., Imbalzano et al. 1994; Utley et al. 1997). Whether any changes in chromatin state affecting the genomic imprint at the mea locus are directly linked to changes in methylation remains to be seen (Jeddeloh et al. 1999). It is possible that DDM1 affects primarily chromatin organization and that the changes in methylation observed in ddm1 mutants are a secondary effect because the altered chromatin state is inaccessible to the methylation machinery. Alternatively, chromatin remodeling and DNA methylation could be intimately related to each other with DDM1 acting as a cofactor of the methylation machinery causing immediate hypomethylation and reactivation of the paternally inherited MEA allele. Strikingly, and unlike the effects of ddm1 on other single-copy genes, the reactivation of the paternally inherited MEA allele by loss of DDM1 activity occurs in the first generation. This may suggest that imprinted genes and repeated DNA sequences, in which the effects of ddm1 have also been shown to occur in the first generation (Katukani et al. 1999), share similarities in terms of their chromatin organization. An elucidation of the mechanism of DDM1 action with respect to chromatin remodeling and methylation promises to improve our knowledge of the mechanisms that mediate genomic imprinting in both plants and mammals.
Materials and methods
Plant material and growth conditions
The mea-1 mutant used in this study was described by Grossniklaus et al. (1998). The duplex tetraploid plants carrying two copies of mea-1 were obtained in the progeny of a self-pollinated simplex mea-1 plant described previously (Grossniklaus et al. 1998). The ddm1-2 mutant was kindly provided by Eric Richards (Vongs et al. 1993). The ecotypes used in this study were obtained from the Arabidopsis Biological Resource Center at Ohio State University (stock numbers in parentheses): Aa-0, Aua/Rhîn-Germany (CS0900); C24 (CS0906); Da(1)-12, Darmstadt-CSFR (CS0917); En-D, Donetsk-Ukraine (CS0920); Litva, Lituania (CS0925); Ca-0, Camberg/Taunus-Germany (CS1060); Ct-1, Catania-Italy (CS1094); Db-0, Dombachtal/Ts-Germany (CS1100); Di-0, Dijon-France (CS1106); Ga-0, Gabelstein-Germany (CS1180); Jl-1, Vranov u Brno-CSFR (CS1248); Kn-0, Kaunas-Russia (CS1286); Lc-1, Loch Ness-Scotland (CS1306); Mt-0, Martuba/Cyrenaika-Libya (CS1380); Nd-0, Niederzenz-Germany (CS1390); Tsu-0, Tsu-Japan (CS1564); Wil-1, Wilna-Lituania (CS1594); Oy-1, Oyetese-Norway (CS1643); Wei-0, Weiningen-Switzerland (CS3110); Shahdara, Pamiro/Alay-Tadjikistan (CS6180); Berkeley, Berkley-USA (CS8068). Growth conditions were as described in Moore et al. (1997).
ISH
ISH was performed as described (Coen et al. 1990; Jackson 1991) with modifications needed for improved resolution in developing female gametophytes of Arabidopsis. For synthesis of sense and anti-sense 11-digoxigenin-UTP labeled probes, a plasmid pGEM7Z (Promega) containing a 290-bp fragment (5′ end of the MEA cDNA) was linearized with restriction enzymes cutting in the polylinker (XhoI and BamHI, respectively), and 1 μg was used as a template for probe synthesis. Whole inflorescences, dissected carpels, and developing siliques were fixed in 4% paraformaldehyde and embedded in Paraplast. Sections at 12–15 μg thickness were cut using a Reichert microtome and attached to ProbeOnPlus slides (Fisher Biotech). After dewaxing and hydration, slides were digested with 0.125 mg/ml of Pronase E (SIGMA) for 28 min. at 37°C. Following a 2-min postfixation in 4% paraformaldehyde, slides were dehydrated and immediately processed for hybridization. RNA probes were hydrolyzed as described (Jackson 1991), and 3%–6% of each labeling reaction (400–800 ng of RNA) were mixed with 40 μl of 50% formamide, added to 200 μg of hybridization buffer, and used as a probe for a pair of slides. Following overnight hybridization at 55°C, the slides were washed twice with gentle agitation in 0.2× SSC for 1 hr at 55°C, followed by two rinses at room temperature (25°C) in NTE (0.5 m NaCl, 10 mm Tris at pH 7.5, 1 mm EDTA), and treated with 20 μg/ml of RNase in the same buffer at 37°C for 30 min. They were subsequently rinsed in fresh NTE and washed again in 0.2× SSC for 1 hr at 55°C. For immunological detection, slides were incubated twice (45 and 30 min) with gentle agitation in 0.5% blocking agent (Boehringer-Mannheim) in TBS (100 mm Tris-HCl at pH 7.5, 150 mm NaCl), followed by 45 min in 1% BSA, 0.3% Triton X-100 in TBS. This was followed by a 2-hr incubation in anti-digoxygenin conjugated antibody diluted 1:1250 in 1% BSA, 0.3% Triton X-100 in TBS, and four washes of 20 min in the same buffer. The slides were washed twice for 15 min in buffer C (100 mm Tris at pH 9.5, 50 mm MgCl2, 100 mm NaCl), and incubated for 2–5 days in 0.34 mg/ml of nitroblue tetrazolium salt (NBT) and 0.175 mg/ml of 5-bromo-4-chloro-3-indyl phosphate toluidinium salt (BCIP) in buffer C containing 7.6 mm levamisole (SIGMA). After stopping the reaction with 10 mm Tris-HCl, 1 mm EDTA, slides were dehydrated through an ethanol series, mounted in Cytoseal (Stephens Scientific), and analyzed with a Leica DMRB microscope under brightfield or Nomarski optics.
RT–PCR analysis
For RNA preparation young siliques were harvested between 52 and 56 HAP in liquid nitrogen. For accurate comparisons control siliques of self-fertilized Ler wild-type plants and mea-1 homozygous plants (mea-1/mea-1) were emasculated and hand pollinated in the same manner as the reciprocal crosses between Ler and mea-1/mea-1. RNA was prepared using the Trizol LS reagent (GIBCO-BRL). For RT–PCR, ∼5 μg of total RNA were treated with 5 units of RNase-free DNase (Boehringer-Mannheim) in 1× PCR buffer (GIBCO-BRL) containing 2.5 mm MgCl2 at 37°C for 30 min. After heat inactivation at 80°C for 5 min, samples were extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and precipitated with ethanol. The RNA was reverse transcribed using 5 pmoles of random hexamers (Pharmacia Biotech) in a 12-μl reaction containing 1× PCR buffer (GIBCO-BRL), 2.1 mm MgCl2, 0.5 mm of each deoxynucleotide triphosphate (dNTP), 10 mm dithiothreitol, and 120 units of Superscript II reverse transcriptase (GIBCO-BRL) by incubating at 25°C for 10 min followed by 42°C for 55 min and heat inactivation at 70°C for 20 min. One-fourth of the cDNA samples were used for PCR amplification of MEA, 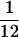 of the samples was used to amplify ACT11, and 0.5 ng of genomic Ler DNA for the controls. PCR was performed in 1× PCR buffer (Perkin Elmer) containing 2 mm MgCl2, 0.2 mm of each dNTP, 1 unit of Taq DNA polymerase (Perkin-Elmer/Cetus), TaqStart antibody (Clontech) in a molar ratio of 28:1 relative to Taq DNA polymerase, and 20 pmoles of each gene specific primer for 30 cycles at an annealing temperature of 58°C. The primers used for amplification of ACT11 were as described (Grossniklaus et al. 1998). For MEA , the primers which specifically amplify the mea-1 allele under these PCR conditions, were meaS12 (5′-CTCATGATGAAGCTAATGAGC-3′) and meaAS11 (5′-GCATGTTCTGGTCCATAGC-3′).
of the samples was used to amplify ACT11, and 0.5 ng of genomic Ler DNA for the controls. PCR was performed in 1× PCR buffer (Perkin Elmer) containing 2 mm MgCl2, 0.2 mm of each dNTP, 1 unit of Taq DNA polymerase (Perkin-Elmer/Cetus), TaqStart antibody (Clontech) in a molar ratio of 28:1 relative to Taq DNA polymerase, and 20 pmoles of each gene specific primer for 30 cycles at an annealing temperature of 58°C. The primers used for amplification of ACT11 were as described (Grossniklaus et al. 1998). For MEA , the primers which specifically amplify the mea-1 allele under these PCR conditions, were meaS12 (5′-CTCATGATGAAGCTAATGAGC-3′) and meaAS11 (5′-GCATGTTCTGGTCCATAGC-3′).
Histological Analysis
For whole-mount specimens, siliques at various stages of development were dissected with hypodermic needles (Becton-Dickinson, 1 cc insulin syringes), mounted for direct observation or cleared in Hoyer's medium (Grossniklaus et al. 1998). Individual seeds and embryos were isolated and mounted for photography under a Leica dissecting stereoscope. For sectioned material, individual seeds were fixed in 3% glutaraldehyde for 2 hr, rinsed in 50 mm cacodylate buffer (EMS), and postfixed in 2% osmium tetroxyde (in same buffer). After dehydration in an acetone series, specimens were embedded in Spurr's resin. Specimens were sectioned on an UltracutE ultramicrotome (Reichert-Jung) and observed on a Leica DMR microscope under bright-field or Nomarski optics.
Acknowledgments
We are grateful to Eric Richards for providing ddm1 seeds and for his help and advice with respect to genotyping ddm1 mutants. We thank Winship Herr and Imran Siddiqi for comments on the manuscript, Tim Mulligan for plant care, and Jim Duffy for his help with the artwork in Figure 6. This work was supported by the CSHL President's Council, a gift from David Luke III, and a Competitive Grant Award from Pioneer Hi-Bred International. J.-P.V.-C. is supported by the Fonds National Suisse de la Recherche Scientifique, and U.G. was supported by the Human Frontier Science Program, the Demerec-Kaufmann-Hollaender Fellowship in Developmental Genetics, and is a Searle Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL grossnik@fmi.ch; FAX +41 61 697 39 76.
References
- Asker S, Jerling L. Apomixis in Plants. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- Bowman J. Arabidopsis, an atlas of morphology and development. New York, NY: Springer Verlag; 1994. [Google Scholar]
- Burnham CR. Discussions in cytogenetics. Minneapolis, MN: Burgess Publishing Company; 1962. [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan X-J, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;5:363–369. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Messing J. Allele-specific parental imprinting of dzr-1, a post-transcriptional regulator of zein accumulation. Proc Natl Acad Sci. 1994;91:4867–4871. doi: 10.1073/pnas.91.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ. Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Colombo L, Franken J, van der Krol AR, Wittich PE, Dons HJ, Angenent GC. Downregulation of ovule specific MADS box genes from petunia results in maternally controlled defects in seed development. Plant Cell. 1997;9:703–715. doi: 10.1105/tpc.9.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Schneitz K. The molecular and genetic basis of ovule and megagametophyte development. Sem Cell Dev Biol. 1998;9:227–238. doi: 10.1006/scdb.1997.0214. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada J-P. ··· response: Parental conflict and infanticide during embryogenesis. Trends Plant Sci. 1998;3:328. [Google Scholar]
- Grossniklaus U, Vielle-Calzada J-P, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Haig D, Graham C. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell. 1991;54:1045–1046. doi: 10.1016/0092-8674(91)90256-x. [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M. Parent specific gene expression and the triploid endosperm. Am Nat. 1989;134:147–155. [Google Scholar]
- ————— Genomic imprinting in endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Phil Trans R Soc Lond. 1991;333:1–13. [Google Scholar]
- Hong SK, Kitano H, Satoh H, Nagato Y. How is embryo size genetically regulated in rice? Development. 1996;7:2051–2058. doi: 10.1242/dev.122.7.2051. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Schnitzler GR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Jackson DP. In situ hybridization in plants. In: Bowles DJ, Gurr SJ, McPhereson M, editors. Molecular plant pathology: A practical approach. Oxford, UK: Oxford University Press; 1991. pp. 163–181. [Google Scholar]
- Jaenisch R. DNA methylation and imprinting: Why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes & Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- John RM, Surani MA. Imprinted genes and regulation of gene expression by epigenetic inheritance. Curr Opin Cell Biol. 1996;3:348–353. doi: 10.1016/s0955-0674(96)80008-1. [DOI] [PubMed] [Google Scholar]
- Jones BK, Levorse J, Tilghman SM. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes & Dev. 1998;12:2200–2207. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T. Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. Plant J. 1997;12:1447–1451. doi: 10.1046/j.1365-313x.1997.12061447.x. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci. 1996;29:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Munakata E, Richards EJ, Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151:831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- Kermicle J. Dependence of the R-mottled aleurone phenotype in maize on the mode of sexual transmission. Genetics. 1970;66:69–85. doi: 10.1093/genetics/66.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J, Alleman M. Gametic imprinting in maize in relation to the angiosperm life cycle. Development (Suppl) 1990;1:9–14. [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB. Control of fertilization independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM. Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell. 1993;5:1425–1437. doi: 10.1105/tpc.5.10.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lin B-Y. Ploidy barrier to endosperm development in maize. Genetics. 1984;107:103–115. doi: 10.1093/genetics/107.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund G, Messing J, Viotti A. Endosperm-specific demethylation and activation of specific alleles of α-tubulin genes of Zea mays L. Mol Gen Genet. 1995;246:716–722. doi: 10.1007/BF00290717. [DOI] [PubMed] [Google Scholar]
- Martienssen RA. Epigenetic phenomena: Paramutation and gene silencing in plants. Curr Biol. 1996;6:810–813. doi: 10.1016/s0960-9822(02)00601-2. [DOI] [PubMed] [Google Scholar]
- Martienssen RA. Chromosomal imprinting in plants. Curr Opin Gen Dev. 1998;8:240–244. doi: 10.1016/s0959-437x(98)80147-0. [DOI] [PubMed] [Google Scholar]
- Messing J, Grossniklaus U. Genomic imprinting in plants. In: Ohlsson R, editor. Results and problems in cell differentiation. Berlin-Heidelberg, Germany: Springer Verlag; 1999. pp. 23–40. [DOI] [PubMed] [Google Scholar]
- Ming L, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Neumann B, Barlow DP. Multiple roles for DNA methylation in gametic imprinting. Curr Opin Genet Dev. 1996;6:159–163. doi: 10.1016/s0959-437x(96)80045-1. [DOI] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu Y-C, Williams C, Repetti P, Fischer RL. A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995;5:174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- Pelletier G. Use of haplo-diploidization for plant breeding. In: Chupeau Y, Caboche M, Henry Y, editors. Androgenesis and haploid plants. Heidelberg, Germany: Springer Verlag; 1998. p. 104. [Google Scholar]
- Pirrotta V. Pc-G complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- Ray A. New paradigms in plant embryogenesis: Maternal control comes in different flavors. Trends Plant Sci. 1998;3:325–327. [Google Scholar]
- Ray S, Golden T, Ray A. Maternal effects of the short integument mutation on embryo development in Arabidopsis. Dev Biol. 1996;180:365–369. doi: 10.1006/dbio.1996.0309. [DOI] [PubMed] [Google Scholar]
- Reik W, Maher ER. Imprinting in clusters: Lessons from Beckwith-Wiedemann syndrome. Trends Genet. 1997;13:330–335. doi: 10.1016/s0168-9525(97)01200-6. [DOI] [PubMed] [Google Scholar]
- Richards EJ. DNA methylation and plant development. Trends Genet. 1997;13:319–323. doi: 10.1016/s0168-9525(97)01199-2. [DOI] [PubMed] [Google Scholar]
- Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- Scott R, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998a;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- Scott R, Vinkenoog R, Spielman M, Dickinson HG. Medea: Murder or mistrial? Trends Plant Sci. 1998b;3:460–461. [Google Scholar]
- Shermoen AW, O'Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman SM. The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- Utley RT, Cote J, Owen-Hughes T, Workman JL. SWI/SNF stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem. 1997;272:12642–12649. doi: 10.1074/jbc.272.19.12642. [DOI] [PubMed] [Google Scholar]
- van der Veen JH, Blankenstijn-de Vries H. Double reduction in tetraploid Arabidopsis thaliana, studied by means of a chlorophyll mutant with a distinct simplex phenotype. Arabidopsis Inf Serv. 1973;10:11–12. [Google Scholar]
- Vielle-Calzada J-P, Crane CF, Stelly DM. Apomixis: The asexual revolution. Science. 1996;274:1322–1323. [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards, E.J EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Walbot V. Sources and consequences of phenotypic and genotypic plasticity in flowering plants. Trends Plant Sci. 1996;1:27–32. [Google Scholar]