Abstract
Multivesicular body (MVB) formation is the result of invagination and budding of the endosomal limiting membrane into its intralumenal space. These intralumenal vesicles (ILVs) contain a subset of endosomal transmembrane cargoes destined for degradation within the lysosome, the result of active selection during MVB sorting. Membrane bending and scission during ILV formation is topologically similar to cytokinesis in that both events require the abscission of a membrane neck that is oriented away from the cytoplasm. The endosomal sorting machinery required for transport (ESCRTs) represents cellular machinery whose function makes essential contributions to both of these processes. In particular the AAA-ATPase Vps4 and its substrate ESCRT-III are key components that seem to execute the membrane abscission reaction. This review summarizes current knowledge about the Vps4-ESCRT-III system and discusses a model how the recruitment of Vps4 to the different sites of function might be regulated.
Keywords: ESCRT, MVB pathway, cytokinesis, endocytosis, protein trafficking
Introduction
The ESCRTs (Endosomal Sorting Complexes Required for Transport) are involved in a number of cellular processes, including degradation of transmembrane proteins via the multivesicular body (MVB) pathway and cytokinesis (Figure 1). In MVB sorting, the ESCRTs deliver ubiquitinated membrane proteins into vesicles that bud in to the lumen of the endosomal compartment, ultimately resulting in the degradation of these cargoes within the lysosome/vacuole. During cytokinesis, the ESCRTs are recruited to the midbody where they execute the final membrane abscission that disconnects the two dividing cells. The ESCRTs require the enzymatic activity of the AAA-ATPase Vps4 to accomplish these processes. While the precise role of Vps4 in maintaining ESCRT activity remains unclear, its essential role in ESCRT-mediated processes is further highlighted by the evolutionary conservation of Vps4 and a subset of the ESCRT-III subunits into archaea. The activity of Vps4 and ESCRT-III therefore appear to represent the core reaction of the ESCRT machinery and an important point of modulation. Here we discuss recent work that has provided insights into the function of Vps4 and its multiple regulators.
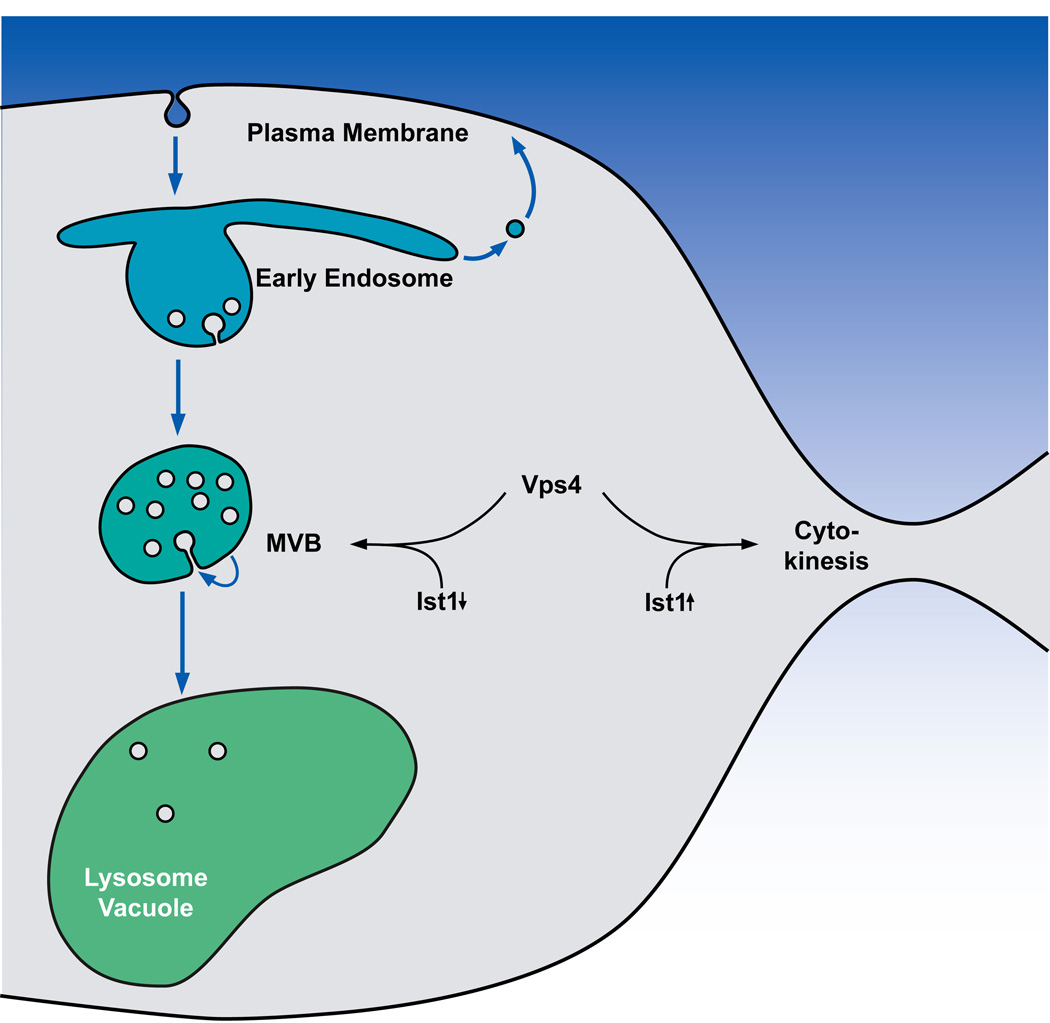
Figure 1.
Model for the MVB pathway and Ist1-regulated recruitment of Vps4 to MVBs and the midbody. Cell surface proteins are endocytosed and delivered to an early endosome where these proteins are either recycled or sorted into the MVB pathway for degradation. Ist1 levels are affected by the growth state of the cell. Fast growing cells contain high levels of Ist1 whereas in stationary cells Ist1 levels are low. These low levels of Ist1 inhibit cytokinesis but enhance Vps4 activity at the MVB pathway. As a result, protein degradation via the MVB pathway is increased, which might be part of the cells adaptation to the stationary conditions.
Vps4 functions as an oligomer
AAA-ATPases convert the chemical energy from ATP hydrolysis into mechanical energy for the purpose of remodeling a wide range of substrates. In the realm of protein trafficking, these substrates include proteins or protein complexes such as SNAREs (dissociated by NSF), microtubules (severed by Spastin and katanin), and proteins undergoing ER-associated degradation (unfolded by p97) (reviewed in (1–3)). For Vps4, the principal substrate appears to be ESCRT-III, a polymer comprised of a number of structurally similar, 25–35 kDa proteins (reviewed in (4))(Table 1). These ESCRT-III subunits are metastable molecules that adopt one conformation as soluble monomers in the cytoplasm and a distinct shape associated with polymerization (5–8) to facilitate intralumenal vesicle (ILV) budding in MVB sorting and membrane abscission in cytokinesis (Figure 2). Vps4 uses the energy of ATP to facilitate the conversion of ESCRT-III subunits from the assembled conformation back into their soluble, monomeric forms, although it remains unclear how this transition is accomplished (5, 9, 10).
Table 1.
 |
Proteins with ESCRT-III subunit fold | ||
|---|---|---|---|
| yeast | mammals | functions | |
| ESCRT-III subunits | Vps20 | CHMP6 | binds ESCRT-II |
| Snf7 | CHMP4A,B,C | binds Bro1-Doa4 | |
| Vps24 | CHMP3 | ||
| Vps2 | CHMP2A,B | activates Vps4 | |
| Vps4 regulators | Did2 | CHMP1A,B | activates Vps4-Vta1 |
| lst1 | hlst1 | inhibits Vps4 | |
| Vps60 | CHMP5 | activates Vps4 via Vta1 | |
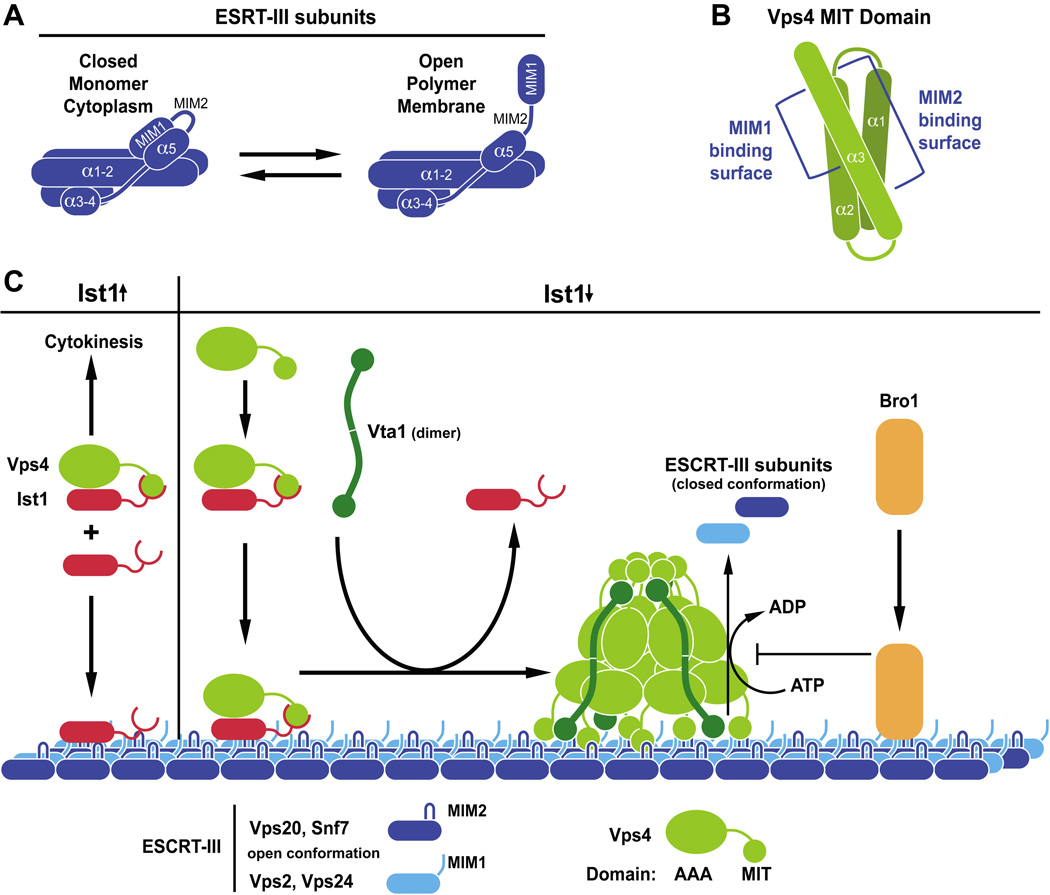
Figure 2.
(A) All ESCRT-III subunits seem to share a five-helix structure that is either in the closed conformation when localized to the cytoplasm or the open conformation when assembled into the ESCRT-III polymer. (B) The Vps4 MIT domain is composed of three α̃helixes that interact with MIM1 and MIM2 sites in ESCRT-III subunits via two distinct surfaces. (C) Model for recruitment and assembly of Vps4 on ESCRT-III. High Ist1 levels (Ist1↑) stabilize the cytoplasmic Ist1-Vps4 complex, thereby inhibiting the recruitment of Vps4 to MVBs and promoting Vps4 recruitment to the midbody, the site of cytokinesis. At low levels (Ist1↓) Ist1 promotes Vps4 recruitment to ESCRT-III. Vta1 supports the assembly of Vps4 into the active dodecameric complex that hydrolyses ATP in order to disassemble ESCRT-III. For simplicity, the green ball on each end of the Vta1 dimer represents two MIT domains per subunit. Bro1 binds to ESCRT-III and inhibits Vps4-dependent disassembly of ESCRT-III.
AAA-ATPases function as oligomers, commonly hexameric rings or stacked hexameric rings comprised of 6 or 12 subunits (11). This arrangement generates a central cavity, or pore, that has been implicated in facilitating substrate remodeling. Vps4 appears to adopt a similar stacked ring arrangement of 12 subunits (dodecamer) (12–14), although some studies have suggested alternative oligomers of 10 (9) or 14 subunits (15). While the precise oligomer composition may not be critical to our current understanding of Vps4 function, appreciating the limitations that have led to this ambiguity is helpful. Wildtype Vps4, both purified and in yeast lysates, has been characterized as a monomer or dimer (9, 16). Examination of higher order structures by size exclusion chromatography and electron microscopy has required the use of the ATP hydrolysis defective Vps4 mutant (Vps4E233Q) (9, 12, 14, 17). It is possible this mutant form of Vps4 adopts a conformation that has complicated appreciation of its oligomeric state. However, Vps4E233Q can form hetero-oligomers with wildtype Vps4 that retain ATP hydrolysis activity, suggesting Vps4E233Q is not grossly incapable of facilitating proper Vps4 oligomer formation (9, 10). While additional structural analyses will be required to resolve the precise higher order structure of Vps4, the Vps4 oligomer will be discussed as a dodecamer since variations in this parameter have minimal impact on present models pertaining to function.
While the precise number of subunits may not be important, the stability of the Vps4 oligomer appears to be a critical issue in models of Vps4 function. Some AAA-ATPases, such as NSF, are characterized by two AAA domains per subunit that form the stacked ring structure; within these AAA-ATPases, one AAA domain (and ring) exhibits less ATP hydrolysis activity, thereby facilitating stable oligomer formation, while the second domain exhibits higher ATP hydrolysis activity to remodel substrates (reviewed in (2, 3)). Although Vps4 appears to adopt a similar stacked ring structure, the presence of a single AAA domain per subunit seemed inconsistent with a similarly stabilized oligomer (12, 14). Moreover, the inability to characterize the higher order structure of wildtype Vps4 (in contrast to Vps4E233Q oligomers) led to a model in which synchronous ATP hydrolysis by all subunits necessitates dissociation of the Vps4 oligomer as an obligate step in the ATP hydrolysis cycle. This in turn suggested one model pertaining to Vps4 function: that dissociation of the Vps4 oligomer is linked to the disassembly of its substrate (ESCRT-III). However, more recent biochemical characterization has suggested that this model is incomplete. Vps4 exhibits concentration-dependent ATP hydrolysis activity, a characteristic attributed to the role of oligomer assembly in forming the active enzyme (9, 10, 17). Surprisingly, this concentration dependent ATP hydrolysis activity was recently demonstrated to be resistant to dilution (as long as ATP is maintained), indicating that the assembled oligomer is stable through multiple rounds of ATP hydrolysis (10). An in vitro ESCRT-III disassembly assay was also developed to further explore Vps4 function. Pre-incubation of Vps4E233Q with wildtype Vps4 disrupted in vitro ESCRT-III disassembly, consistent with the dominant negative effects of Vps4E233Q in vivo. However, wildtype Vps4 was resistant to inhibition by Vps4E233Q if the dominant negative mutant was added subsequent to initiation of the ESCRT-III disassembly reaction (10). This observation supported the model that Vps4 functions as a stable oligomer during ESCRT-III disassembly. This model is further supported by the previous observation that the Vps4 co-factor Vta1 stimulates Vps4 ATP hydrolysis activity in part through enhancing Vps4 oligomerization (17). Together these data suggest that a model of Vps4 function should incorporate stability of the Vps4 oligomer during ESCRT-III disassembly. By analogy to other AAA-ATPases, the stable Vps4 oligomer likely utilizes residues projecting into the central pore of the oligomer to remodel ESCRT-III subunits. Vps4 rearrangements that occur during ATP hydrolysis would be transduced via these pore residues to induce conformational changes in the substrate. Consistent with this idea, mutagenic studies have assigned functional significance to residues predicted to line the pore of the Vps4 oligomer (18, 19). It remains to be determined whether Vps4 is driving ESCRT-III disassembly by completely unfolding ESCRT-III subunits and translocating them through the central pore or by partially remodeling the ESCRT-III subunits to facilitate more subtle transitions from the assembled to monomeric conformations or if an alternative process is involved. While the Vps4 oligomer appears to be the relevant functional form catalyzing ESCRT-III disassembly, modulation of Vps4 assembly into this active state by other factors (including ESCRT-III itself) likely represents an important means of regulating Vps4 function in vivo, as will be discussed further within this review.
Vps4 and ESCRT-III function during membrane deformation
The dynamic assembly and disassembly of the ESCRT-III polymer plays a critical role in ESCRT-mediated membrane deformation events, and alterations in Vps4 activity disrupt the fidelity or execution of these events. The activation of Vps4 is inferred to be coordinated with ESCRT-III assembly to facilitate the proper timing of disassembly, so it is helpful to appreciate our current notion of ESCRT-III assembly. Studies in the yeast MVB sorting system have suggested that ESCRT-III subunits undergo ordered assembly into a polymer with the following sequence suggested: Vps20, multiple Snf7 subunits, Vps24, Vps2, Did2, and Ist1 (5, 20–22). In vitro studies using Giant Unilamellar Vesicles (GUV) and purified recombinant ESCRT-III subunits demonstrated that ESCRT-III itself can drive formation of vesicles within the GUV lumen and supported the order of assembly of the core ESCRT-III subunits (Vps20, Snf7, Vps24 and Vps2) suggested through studies in yeast (23). ESCRT-III itself is believed to drive ILV budding through forming a spiral fibril, as indicated by deep-etch cryoelectron microscopy visualization in mammalian cells (24). Fibrils have also been generated in vitro with a subset of truncated or chimeric ESCRT-III subunits (8, 25, 26) supporting the model that the aforementioned spiral fibrils represent ESCRT-III itself. These studies have suggested that ESCRT-III subunits undergo an ordered assembly into a spiral fibril to facilitate membrane deformation events.
The determinants permitting the specific ESCRT-III subunit assembly into this spiral fibril remain unclear, but one model has emerged that may explain how ESCRT-III assembly and regulation of Vps4 may be linked. Studies with Did2 and Ist1 suggest that the carboxyl-terminal helix of a preceding subunit may contribute to the recruitment of the next ESCRT-III subunit (27). The carboxyl-terminus is also implicated in maintaining the closed conformation of the soluble ESCRT-III subunit and inhibiting its aberrant inclusion into the polymer (6, 8). One model of assembly suggests that these functions of the carboxyl-terminus are linked: the carboxyl-terminus of Ist1 binds in cis to a pocket formed by the amino-terminal portion (the α1, α2 and α5 pocket) to prevent aberrant incorporation (8, 27) (Figure 2a); however, the presentation of the carboxyl-terminus of its preceding binding partner, Did2, permits replacement of this cis interaction with a trans interaction (carboxyl-terminus of Did2 into the α1/2/5 pocket of Ist1) (27), facilitating additional conformational changes and the formation of more extensive contacts to permit the incorporation of Ist1 into the ESCRT-III polymer; with incorporation, the carboxyl-terminus of Ist1 is now exposed. It is intriguing to speculate that this interaction between Did2 and Ist1 may represent a general intermediate for ESCRT-III subunits that contributes to the specificity of assembly. However, this model has not been confirmed with other ESCRT-III subunits, nor is it understood how the ESCRT-III subunits contact each other within the stably assembled polymer. It is interesting to note that ordered assembly of subunits into ESCRT-III may be impacted by this mode of interaction, as carboxyl-terminal truncations result in aberrant incorporation into ESCRT-III (6, 24). Furthermore, carboxyl -terminally truncated forms of ESCRT-III subunits can still generate fibrils in vitro (8, 26), suggesting the inherent ability of ESCRT-III subunits to form fibrils does not require this mode of interaction. Stable fibril assembly does require a region spanning the tip of the α1–α2 hairpin, suggesting that tip-to-tip contacts between ESCRT-III subunits contribute to fibril formation in vitro (8). One enticing aspect of a model wherein the carboxyl tail transiently participates in assembly is that the carboxyl-termini of ESCRT-III subunits have been implicated in coordinating Vps4 function (discussed later within this review). This model suggests that the exposure of these regulatory portions of ESCRT-III subunits could be linked with their incorporation into the ESCRT-III polymer to facilitate the proper recruitment and activation of Vps4.
The precise role of Vps4 in ESCRT-III-driven membrane budding remains unclear. Initial models suggested that Vps4 might hydrolyze ATP to directly provide the energy for membrane budding (9, 14). The reconstitution of vesicle budding in the GUV system indicated that ESCRT-III assembly is sufficient to drive this process, although multiple rounds of vesicle formation required Vps4 function (23). One interpretation of this data is that Vps4 plays a custodial role, cleaning up the ESCRT-III polymer after it has served its role in membrane deformation/scission (analogous to NSF function with regard to SNAREs). In this model, Vps4 transfers the energy of ATP hydrolysis into the closed/monomeric conformation of ESCRT-III subunits, and then this potential energy is released through polymerization to drive membrane budding. While this model of indirect participation is supported by the in vitro reconstitution system, multiple lines of evidence suggest that Vps4 may play a more active role in this membrane deformation process in vivo. Expression of dominant negative Vps4 alters the morphology of ESCRT-III-containing fibrils and, strikingly, induces membrane deformations coincident with these structures (24). This observation suggests that Vps4 binding to ESCRT-III may alter the fibril shape to facilitate membrane eversion independent of Vps4 ATP hydrolysis. Perturbation of Vps4 modulators such as Vta1 or Did2 has been demonstrated to impact the size of ILVs, suggesting that altered Vps4 function can disrupt the fidelity of vesicle budding without aborting the process entirely (28). This concept was further developed though studies with the ESCRT-III-associated factor Bro1, which is responsible for recruiting the deubiquitinating enzyme Doa4 (29). Excessive Bro1 induces the appearance of increased numbers of ILVs that had not yet undergone scission from the limiting membrane of MVBs, and this effect correlates with Bro1 inhibition of Vps4 disassembly of ESCRT-III (30). One possible interpretation of these results is that this particular perturbation of Vps4-dependent ESCRT-III disassembly results in a kinetic defect in ILV scission. While further study is required to understand this effect further, it is intriguing to speculate that Bro1 may be coordinating the assembly of ESCRT-III, sensed by Bro1 binding to incorporated Snf7 subunits, with the subsequent events of cargo deubiquitination, Vps4-mediated disassembly and membrane scission to insure that ubiquitin is removed from MVB cargoes before ILV budding is completed; disruption of this coordination with overexpression of Bro1 then results in the stabilization of the budding intermediates observed. These observations regarding effects of dominant negative Vps4 on ESCRT-III fibrils and the effects of altered Vps4 regulation on ILV budding or size may be related to the subtle disruptions in the dynamics and availability of ESCRT-III subunits; alternatively, these observations may be the initial indicators of an additional and more direct role for Vps4 in ESCRT-III-mediated membrane scission.
Vps4 and ESCRT-III disassembly
While Vps4 catalyzes ESCRT-III disassembly, ESCRT-III itself appears to regulate Vps4 ATPase activity during this process through multiple mechanisms. The amino-terminal Microtubule Interacting and Trafficking (MIT) domain of Vps4 plays critical roles in this regulation by ESCRT-III. One proposed mode of activation by ESCRT-III is the local concentration of Vps4 to facilitate oligomerization and activation. As previously discussed, Vps4 appears to exist as a dimer in the cytoplasm, and the MIT domain of Vps4 has been implicated in facilitating the recruitment of Vps4 to the ESCRT-III polymer on the membrane (9, 10, 31, 32). Two distinct modes of association appear to drive this recruitment: the MIT domain recognizes MIT Interacting Motifs 1 and 2 (MIM1, 2) present in the carboxyl-termini of various ESCRT-III subunits (Figure 2) (33–35). As mentioned previously, the availability of these MIM elements may be altered within different ESCRT-III subunit conformations, with the MIM more accessible upon subunit incorporation into ESCRT-III. Interestingly, different portions of the Vps4 MIT domain recognize MIM1 and MIM2 (Figure 2). The MIT domain is a 3-helix bundle (36), and the surface formed by helices 2 and 3 mediates MIM1 interaction whereas the helix 1–3 surface binds MIM2 (33–35). In addition to facilitating local concentration of Vps4, the MIM1 interaction appears able to enhance the ATPase activity beyond the maximal activity observed with Vps4 alone (stimulation was not observed with a MIM 2-containing ESCRT-III subunit under similar experimental conditions) (37). This observation suggests that ESCRT-III allosterically regulates Vps4 in addition to enhancing Vps4 oligomerization through concentration. The MIT domain has been implicated in auto-inhibiting Vps4 ATPase activity as mammalian Vps4 lacking the MIT domain (VPS4ΔMIT) displays increased activity relative to full-length Vps4, and a similar phenomenon has been observed with yeast Vps4 under certain conditions (9, 18). These observations suggest that ESCRT-III binding to the MIT domain via the MIM1 element may relieve this autoinhibition to enhance Vps4 activity. As MIM2 elements appear unable to similarly enhance Vps4 activity (37), the MIM2-Vps4 interaction has been speculated to be primarily involved in Vps4 recruitment to ESCRT-III. The structural basis of MIT interaction with MIM1 and MIM2 elements is known, but the means by which the MIT domain may autoinhibit the AAA domain and how MIM1 binding may modulate this intra-molecular regulation remain unresolved. A third mode of Vps4 activation by ESCRT-III appears to involve the region connecting the amino-terminal MIT domain with the AAA domain – the linker region. An acidic stretch present in all ESCRT-III subunits is capable of enhancing Vps4 ATP hydrolysis activity via the linker region, and a similar effect could be induced with poly-L-glutamic acid (18). The linker region is also implicated in autoinhibiting Vps4 activity, so once again, ESCRT-III association with this portion of Vps4 may stimulate activity through relieving intramolecular regulation. The similarity between the acidic stretch-linker activation and MIM1-MIT activation suggest that these regulatory mechanism may be one and the same, with both interactions inducing a similar rearrangement of the AAA domain to enhance ATP hydrolysis. However, direct assessment of this relation ship and resolution of this activation mechanism at the structural level are still forthcoming. In total, these observations suggest that ESCRT-III both facilitates Vps4 assembly and relieves an intramolecular inhibition to enhance Vps4 ATP hydrolysis activity during ESCRT-III disassembly, with the MIT and linker regions playing key roles in this activation.
Additional insights into the role of the MIT domain in Vps4 function have been gained through addressing its role in ESCRT-III disassembly more directly. Vps4ΔMIT is unable to support ESCRT-III disassembly in vitro or in vivo, consistent with the importance of the MIT domain in Vps4 function (9, 10, 31, 32). However, Vps4ΔMIT does not behave as a dominant negative in vivo or in vitro (10, 31). This observation has suggested that not all MIT domains within a Vps4 oligomer need to engage substrate for the oligomer to function. As mentioned previously, the catalytically inactive form of Vps4 (Vps4E233Q) is unable to support ESCRT-III disassembly and disrupts the activity of wildtype Vps4 both in vivo and in vitro (9, 10, 31). However, Vps4ΔMIT,E233Q lacks similar inhibitory activity, indicating that the MIT domain is required for the dominant negative activity of Vps4E233Q subunits (10, 31). These observations suggest that ESCRT-III engagement by the MIT domain and ATP hydrolysis by the AAA domain may be coordinated at the subunit level within the Vps4 oligomer. How it is that a substrate bound by the MIT is subsequently acted upon by the pore to drive remodeling is presently unclear, but it seems likely the MITs are playing a role in positioning substrate for remodeling as well as conveying regulatory cues from the substrate to the AAA domain.
The Vps4 co-factor Vta1/SBP1/LIP5 also contributes to coordinating Vps4 activity for appropriate ESCRT-III disassembly. Vta1 binds to the Vps4 β-domain, an insert within the AAA domain (19, 38). Vta1 itself forms dimers via a carboxyl-terminal VSL domain, and the symmetric surfaces generated by the VSL dimer is thought to stabilize Vps4 inter-ring contacts by binding the β-domains of two non-neighboring Vps4 subunits (17, 39). Consistent with this model, Vta1 enhances Vps4 oligomerization and promotes Vps4 concentration-dependent activity (17). Vta1 enhances Vps4 ATPase activity beyond the apparent maximal activity of Vps4 alone, suggesting that Vta1 not only stabilizes the oligomeric state of Vps4 but activates Vps4 ATPase activity by an additional mechanism as well (17). Vta1 contains 2 MIT domains within its amino terminus, however these domains recognize a distinct subset of yeast ESCRT-III subunits (Vps60 and Did2) as compared to the yeast Vps4 MIT (37, 39). By contrast, mammalian SBP1/LIP5 exhibits more extensive overlap with the mammalian Vps4 MIT MIM1 associations (40). ESCRT-III binding to Vta1 in the context of Vps4-Vta1 hetero-oligomers stimulates Vps4 ATPase activity above the level observed with Vta1 alone, although in this case the MIT domain of Vps4 itself is not required (37). The mechanism by which Vta1-bound ESCRT-III stimulates Vps4 activity is presently unclear, but the fact that it does not require the Vps4 MIT domain suggests it is likely distinct from direct stimulation by ESCRT-III subunits. It is also interesting to observe that Vps60 recruitment to endosomes is dependent upon Vps4 and Vta1 (28, 37) and that the Vps60-Vta1 complex can be isolated from yeast lysates (17), suggesting that Vps60 may have a unique role among the ESCRT-III subunits as a Vta1-cofactor coordinating Vps4 function in ESCRT-III disassembly. Regardless, a theme that emerges from these studies is the ability of MIT domains to transduce ESCRT-III regulatory signals into the Vps4-Vta1 hetero-oligomer to promote ESCRT-III disassembly.
Ist1 as a coordinator of cellular physiology
Ist1 is unique among the ESCRT-III subunits in its ability to inhibit Vps4 ATPase activity. Structural determination revealed that Ist1 adopts an ESCRT-III fold with additional helices inserted (8, 27). However, Ist1 is unique in that it contains both MIM1 and MIM2 elements within its C-terminus that bind simultaneously Vps4 MIT with high affinity (20, 41, 42). Furthermore, the amino-terminal portion of Ist1 interacts with the AAA domain of Vps4 (20). T ogether, these combined interactions result in the formation of stable Vps4-Ist1 hetero-dimers in vitro that exhibit reduced ATPase activity as a result of defective assembly into higher-order Vps4 complexes (20). While these hetero-dimers have not been observed in vivo, support for their physiological relevance comes from the observation that over-expression of Ist1 interferes with Vps4 localization and activity in yeast cells (20). These studies have led to the conclusion that Ist1 is unique among ESCRT-III subunits in its ability to inhibit Vps4 ATPase activity. However, genetic studies revealed a synthetic interaction between loss of IST1 and VTA1, suggesting a positive role for Ist1 (20, 43). While deletion of IST1 alone has no apparent defect in MVB sorting this strain displays a defect in endosomal association of a reporter consisting of the MIT domain from Vps4 fused to GFP (31) and a subtle defect in Vps4 membrane association (20). Together, these data suggest a positive role for Ist1 in driving Vps4 association with ESCRT-III. Ist1 recruitment to ESCRT-III is dependent upon Did2, and the interaction of these factors has been documented, suggesting a model in which Ist1-Did2 interaction promotes Vps4 association with ESCRT-III (8, 20, 27, 31). The precise mechanism by which this occurs is unclear and the observed synthetic genetic interaction between vta1Δ and ist1Δ suggest that Ist1-Did2 and Vta1-Vps60 may be able to perform overlapping functions in the context of coordinating Vps4 activity. However, the notion that Ist1-Did2 interactions positively impact Vps4 activity represents a starting point for resolving these apparently contradictory Ist1 roles.
The stimulatory and inhibitory activities of Ist1 represent a potential point of modulation with regard to MVB sorting if Ist1 levels fluctuate: low Ist1 levels may facilitate Vps4 activity in combination with Did2 while high Ist1 levels may inhibit Vps4 function through the formation of inactive Vps4-Ist1 dimers. As yeast progress through the growth curve and transition into stationary phase Ist1 protein levels drop dramatically (M. Babst, personal communication). When nutrients are not limiting and rapid growth is occurring it may be advantageous for cells to suppress Vps4 activity and MVB sorting, thereby limiting the degradation rate of membrane proteins as an amino acid source. In contrast, as nutrients become limiting and cells transition into stationary phase decreased Ist1 levels may increase Vps4 activity and flux through the MVB pathway. Increased protein turnover via the MVB pathway could generate an increased pool of amino acids and represent a part of the cell’s ability to adapt to starvation conditions.
Slight changes in MVB pathway activity can have strong effects on the overall turnover of plasma membrane proteins. There are at least two distinct recycling modes within the endocytic system that counterbalance the activity of the MVB pathway. Rapid recycling from the early endosome, although not well understood, seems to contribute to a large portion of endosomal recycling (44). The retromer machinery executes recycling from later endosomal structures. This recycling occurs from the same compartment as ESCRT-mediated sorting and thus is in direct competition to the MVB pathway (45). Ubiquitination and deubiquitination seem to maintain a dynamic endosomal sorting system that can direct cargo into either recycling or degradative pathways. Changes in ESCRT efficiency and MVB sorting, mediated by Ist1, could affect the balance between recycling and degradation of endocytosed cargo. A delay in sorting into MVB vesicles could provide the necessary time for cargoes to be de-ubiquitinated and recycled, thereby escaping degradation via the MVB pathway. In such a model, small changes in Vps4 activity could have a significant impact on the overall degradation rate of transmembrane MVB cargoes.
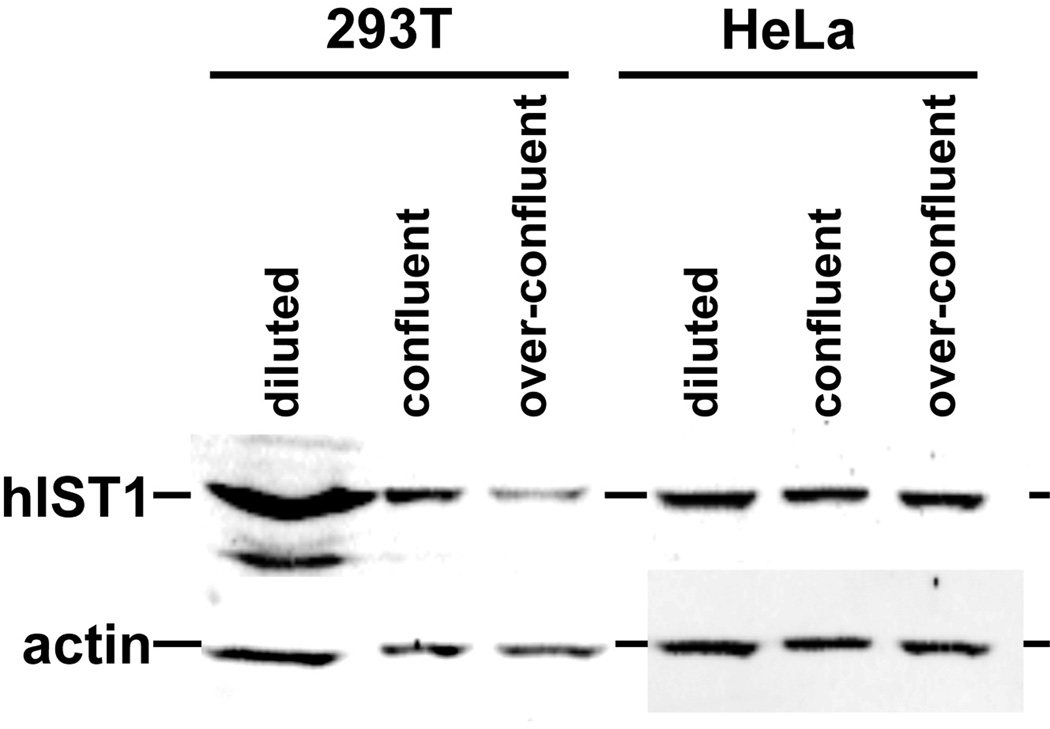
Vps4 and the ESCRT-III proteins, including Ist1, have been implicated in the final steps of mammalian cytokinesis (27, 41, 42, 46, 47). ESCRT factors seem to execute the membrane scission that separates the two cells, an activity that is evolutionary conserved and found in organisms as distant as archaea (see review by Anna Caballe and Juan Martin-Serrano in this issue). Similar to its proposed function during MVB sorting, Ist1 acts in cytokinesis together with Did2 as a recruiting factor for the proper localization of Vps4 to the midbody. This recruiting function at the midbody is essential for proper cell division, which is in contrast to the dispensable activity of Ist1 at the MVB. siRNA-based suppression of Ist1 levels results in a block of cytokinesis and the formation of multinucleated cells (8, 41). While alternate possibilities exist, one potential explanation would be an inverse relationship between these two Vps4 functions: high levels of Ist1 found in rapidly growing cells may direct Vps4 activity toward its function in cytokinesis, whereas, low Ist1 levels found in stationary cells may direct Vps4 toward its role in the MVB pathway. This growth-phase dependent regulation of Ist1 levels seems to be conserved in higher eukaryotes. Upon reaching confluency 293T cells show a dramatic drop in hIST1 protein levels. In contrast, HeLa cells, which exhibit no contact inhibition and thus continue to grow past confluency, showed constant hIST1 levels (Figure 3). Interestingly, a study found that more than 80% of primary lung cancer samples exhibited elevated levels of hIST1 compared to control samples and that hIST1 overexpression induced tumorigenesis in athymic mice (48). These observations might be explained by a model in which high Ist1 concentrations not only support cytokinesis but also suppress the downregulation of membrane proteins such as signaling cell surface receptors, thereby increasing the potency of pro-survival and proliferation signaling. Thus proper regulation of cellular Ist1 levels may play an important role in the transition from dividing to differentiated cells.
Figure 3.
Ist1 levels in human cell lines at different growth states. HEK 293T and HeLa cells were grown to different densities and the cell extracts were analyzed by Western blot for the presence of hIST1 and actin (control) (courtesy of Virginie Sandrin and Wesley I. Sundquist, University of Utah).
Taken together, it would appear that Ist1 regulates Vps4 activity at both MVBs and the midbody. These functions appear to require distinct levels of Ist1 protein. Low levels of Ist1 may enhance MVB sorting under nutrient-limiting conditions whereas high levels of Ist1 may enhance Vps4 function during cytokinesis under rapid growth conditions. Whether Ist1 exhibits distinct modes of Vps4 recruitment to these two sites of action or plays a role in recruiting other factors critical for abscission of the midbody remains to be determined.
Concluding remarks
In spite of the fact that Vps4 was the first ESCRT factor described more than 14 years ago, the study of its relevant function in vivo is ongoing. While recent advances in our understanding of its activity and regulation have occurred, key questions with regard to the structure of the Vps4 oligomer and its ESCRT-III disassembly function remain open. In addition new Vps4-dependent activities, such as the regulation of spindle poles (49), have been discovered, adding to the complexity of understanding the role of Vps4 in cellular homeostasis. These observations support the view that Vps4 is an evolutionary ancient mechanoenzyme that has been integrated into several cellular functions, some of which might still wait discovery.
Acknowledgements
The research conducted in the Katzmann and Babst laboratories on the ESCRT proteins is supported by NIH Grants R01 GM073024 and R01 GM074171, respectively. B.A. Davies is supported by a grant from the Fraternal Order of the Eagles. We thank Andrew Norgan, Wes Sundquist, Jack Skalicky and Zhaohui Xu for helpful discussions.
Bibliography
- 1.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 2.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 3.White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8(12):1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 4.Davies BA, Lee JR, Oestreich AJ, Katzmann DJ. Membrane protein targeting to the MVB/lysosome. Chem Rev. 2009;109(4):1575–1586. doi: 10.1021/cr800473s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3(2):271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 6.Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8(8):1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 7.Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378(4):818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16(7):754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. Embo J. 1998;17(11):2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies BA, Azmi IF, Payne J, Shestakova A, Horazdovsky BF, Babst M, Katzmann DJ. Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol Biol Cell. 2010;21(19):3396–3408. doi: 10.1091/mbc.E10-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer RT, Baker TA. AAA+ Proteases: ATP-Fueled Machines of Protein Destruction. Annu Rev Biochem. 2010 doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 12.Yu Z, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J Mol Biol. 2008;377(2):364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue M, Kamikubo H, Kataoka M, Kato R, Yoshimori T, Wakatsuki S, Kawasaki M. Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B. Traffic. 2008;9(12):2180–2189. doi: 10.1111/j.1600-0854.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- 14.Landsberg MJ, Vajjhala PR, Rothnagel R, Munn AL, Hankamer B. Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure. 2009;17(3):427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann C, Chami M, Zachariae U, de Groot BL, Engel A, Grutter MG. Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J Mol Biol. 2008;377(2):352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. Biochemical and structural studies of yeast Vps4 oligomerization. J Mol Biol. 2008;384(4):878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172(5):705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrill SA, Hanson PI. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J Biol Chem. 2010;285(46):35428–35438. doi: 10.1074/jbc.M110.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Structural and mechanistic studies of VPS4 proteins. Embo J. 2005;24(20):3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates Vps4 localization and assembly. Mol Biol Cell. 2008;19(2):465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175(5):715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. Embo J. 2010;29(5):871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458(7235):172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180(2):389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16(9):1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321(5894):1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell. 2009;20(15):3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nickerson DP, West M, Henry R, Odorizzi G. Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21(6):1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166(5):717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wemmer M, Azmi I, West M, Davies B, Katzmann D, Odorizzi G. Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J Cell Biol. 2011;192(2):295–306. doi: 10.1083/jcb.201007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell. 2010;21(6):1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vajjhala PR, Catchpoole E, Nguyen CH, Kistler C, Munn AL. Vps4 regulates a subset of protein interactions at the multivesicular endosome. Febs J. 2007;274(8):1894–1907. doi: 10.1111/j.1742-4658.2007.05736.x. [DOI] [PubMed] [Google Scholar]
- 33.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15(1):62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 34.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449(7163):735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 35.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449(7163):740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 36.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci U S A. 2005;102(39):13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14(1):50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 38.Yang D, Hurley JH. Structural role of the Vps4-Vta1 interface in ESCRT-III recycling. Structure. 2010;18(8):976–984. doi: 10.1016/j.str.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao J, Xia H, Zhou J, Azmi IF, Davies BA, Katzmann DJ, Xu Z. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev Cell. 2008;14(1):37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shim S, Merrill SA, Hanson PI. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRTIII disassembly. Mol Biol Cell. 2008;19(6):2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20(5):1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20(5):1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19(2):475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20(4):427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strochlic TI, Schmiedekamp BC, Lee J, Katzmann DJ, Burd CG. Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol Biol Cell. 2008;19(11):4694–4706. doi: 10.1091/mbc.E08-03-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316(5833):1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 47.Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Muller S, Hanisch FG, Hulskamp M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133(23):4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- 48.Yuan J, Ma J, Zheng H, Shi T, Sun W, Zhang Q, Lin D, Zhang K, He J, Mao Y, Gao X, Gao P, Han N, Fu G, Xiao T, et al. Overexpression of OLC1, cigarette smoke, and human lung tumorigenesis. J Natl Cancer Inst. 2008;100(22):1592–1605. doi: 10.1093/jnci/djn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A. 2010;107(29):12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]