Abstract
Activation of p53-mediated transcription is a critical cellular response to DNA damage. p53 stability and site-specific DNA-binding activity and, therefore, transcriptional activity, are modulated by post-translational modifications including phosphorylation and acetylation. Here we show that p53 is acetylated in vitro at separate sites by two different histone acetyltransferases (HATs), the coactivators p300 and PCAF. p300 acetylates Lys-382 in the carboxy-terminal region of p53, whereas PCAF acetylates Lys-320 in the nuclear localization signal. Acetylations at either site enhance sequence-specific DNA binding. Using a polyclonal antisera specific for p53 that is phosphorylated or acetylated at specific residues, we show that Lys-382 of human p53 becomes acetylated and Ser-33 and Ser-37 become phosphorylated in vivo after exposing cells to UV light or ionizing radiation. In vitro, amino-terminal p53 peptides phosphorylated at Ser-33 and/or at Ser-37 differentially inhibited p53 acetylation by each HAT. These results suggest that DNA damage enhances p53 activity as a transcription factor in part through carboxy-terminal acetylation that, in turn, is directed by amino-terminal phosphorylation.
Keywords: CBP/p300, PCAF, histone acetyltransferase, DNA–PK, transcriptional activation, checkpoints
The p53 tumor suppressor is a critical component of cellular mechanisms that respond to certain stresses to preserve genomic integrity by arresting cell-cycle progression or by inducing apoptosis (Levine 1997; Agarwal et al. 1998). p53 normally is a short-lived protein that is maintained at low levels, but in response to DNA-damaging agents, nucleotide depletion, or hypoxia, the p53 protein is transiently stabilized and accumulates in the nucleus in which it functions in part to induce or repress the transcription of several genes including WAF1 (El-Deiry et al. 1993), SFN (Hermeking et al. 1997), and MDM2 (Juven et al. 1993; Perry et al. 1993) that regulate cell-cycle progression. The Mdm2 protein binds the amino terminus of p53 and functions in an autoregulatory loop that attenuates p53 activity by directly sequestering its transactivation domain (Oliner et al. 1993; Wu et al. 1993). Mdm2 also targets p53 for degradation and, therefore, may play a critical role in regulating its accumulation in response to DNA damage (Haupt et al. 1997; Kubbutat et al. 1997). The segment of p53 that interacts with Mdm2, which encompasses the region between Glu-17 and Pro-27, includes two potential phosphorylation sites, Thr-18 and Ser-20, and is surrounded by several other potential phosphorylation sites including Ser-9, Ser-15, Ser-33, and Ser-37 (Meek 1998). Phosphorylation of p53 by DNA–PK, a DNA-activated protein kinase that phosphorylates p53 at Ser-15 and Ser-37 in vitro (Lees-Miller et al. 1992), was reported to inhibit the interaction with Mdm2 (Shieh et al. 1997), as does phosphorylation of the amino terminus of Mdm2 by DNA–PK (Mayo et al. 1997). Phosphorylation of Ser-15 in vivo is stimulated by DNA-damaging agents including ionizing radiation (IR) and UV. These recent findings suggest that amino-terminal phosphorylation may be one of the signals regulating the p53 response to DNA damage (Shieh et al. 1997; Siliciano et al. 1997).
In addition to causing p53 to accumulate, DNA damage is widely believed to activate p53 as a transcription factor through post-translational mechanisms. In support of this hypothesis, microinjection of damaged DNA or a monoclonal antibody to the carboxyl terminus of p53 activated p53-specific transcription and induced p53-dependent cell-cycle arrest under conditions in which no increase in p53 protein was detected (Hupp and Lane 1995; Huang et al. 1996). Furthermore, in mouse fibroblasts, UV-C activated p53’s DNA-binding activity in G0 and G1 without causing p53 accumulation (Haapajärvi et al. 1997; Pitkänen et al. 1998). p53 has two DNA-binding domains, a sequence-specific DNA-binding domain encompassing amino acid residues ∼100–300 that interacts with the consensus sequence element 5′-RRRCWWGYY-(N)1-13-RRRCWWGYY-3′ (Funk et al. 1992), and a non-sequence-specific DNA-binding domain corresponding to the carboxy-terminal ∼100 amino acids (300–393) that include the tetramerization domain (Pavletich et al. 1993; Wang et al. 1993). Modifications to the carboxy-terminal domain, including deletion of the last 30 amino acids (Hupp et al. 1992), protein binding (Hupp et al. 1992), phosphorylation of Ser-315, Ser-378, and Ser-392 (Hupp and Lane 1995; Takenaka et al. 1995; Wang and Prives 1995), and most recently, acetylation by CREB-binding protein (CBP)/p300 (Gu and Roeder 1997) were shown to enhance sequence-specific DNA-binding of wild-type p53, possibly by inhibiting p53’s non-sequence-specific DNA-binding activity (Anderson et al. 1997). CBP and p300 are closely related histone acetyltransferases (HATs) that interact with p53 through its amino terminus and function as coactivators for p53-mediated transcription (Avantaggiati et al. 1997; Lill et al. 1997). CBP/p300 are complexed with another HAT, p300/CBP-associated factor PCAF (Yang et al. 1996). These recent findings suggest that activation of sequence-specific DNA-binding by p53 in response to DNA damage may be mediated through CBP/p300 and/or PCAF; however, a direct connection between DNA damage and activation of sequence-specific DNA-binding has not been made.
In this study, we present evidence that activation of sequence-specific DNA-binding by p53 after DNA damage depends on a signal transduction cascade involving phosphorylation-mediated p53 acetylation by the coactivator, p300. We confirm Gu and Roeder’s observation (1997) that p300 acetylates the carboxyl terminus of p53 in vitro, and show that PCAF acetylates p53 in vitro on Lys-320. Using antisera specific for each of the acetylated forms of p53, we show that Lys-382 is acetylated in response to both IR and UV light in vivo; p53 also became acetylated at Lys-320 after exposure of cells to UV. Acetylation by p300 and PCAF was strongly inhibited by phosphopeptides corresponding to the amino terminus of p53 phosphorylated at Ser-37 and/or Ser-33, and these residues were phosphorylated in response to UV and IR in vivo. These findings suggest that phosphorylations in response to DNA damage enhance the interaction of p300 and PCAF with p53, thereby driving p53 acetylation.
Results
PCAF and p300 acetylate p53
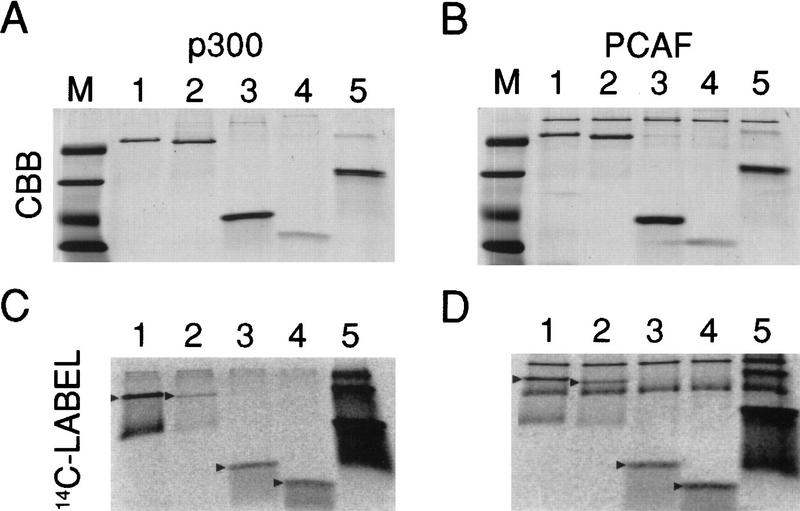
p53 is modified in vivo at amino- and carboxy-terminal sites by post-translational modifications including phosphorylation and acetylation. To determine whether the acetylation of lysine residues may contribute to regulating p53 activity, we incubated wild-type human p53 with recombinant PCAF or p300, two well-characterized HATs. In agreement with the findings of Gu and Roeder (1997), p300 acetylated full-length p53 (Fig. 1C). Although some acetylation was seen also with the fragment p53(1–355), the major site of acetylation was in the carboxy-terminal domain [i.e., in p53(283–393) and p53(318–393)], as observed by Gu and Roeder (1997). Interestingly, similar results were obtained with PCAF (Fig. 1D); full-length p53, p53(283–393) and p53(318–393) all were acetylated, whereas a construct lacking the carboxyl terminus, p53(1–355), was acetylated less efficiently.
Figure 1.
Acetylation of p53 and p53 fragments by p300 and PCAF. Wild-type human p53 or truncated p53 fragments were acetylated with either PCAF or p300 at 37°C for 20 min as described in Materials and Methods, and the reaction products were analyzed by SDS-PAGE. P300 acetylation (14C-Label) is depicted in the radioactive image in C; the corresponding Coomassie brilliant blue-stained image (CBB) is in A. PCAF acetylation is in D; the corresponding Coomassie brilliant blue-stained image is in B. Histone H1 served as a positive control for acetylation (Herrera et al. 1997). (Lanes M) Molecular weight markers; (lanes 1) full-length wild-type, baculovirus-produced human p53; (lanes 2) p53(1–355); (lanes 3) p53(283–393); (lanes 4) p53(318–393); (lanes 5) histone H1.
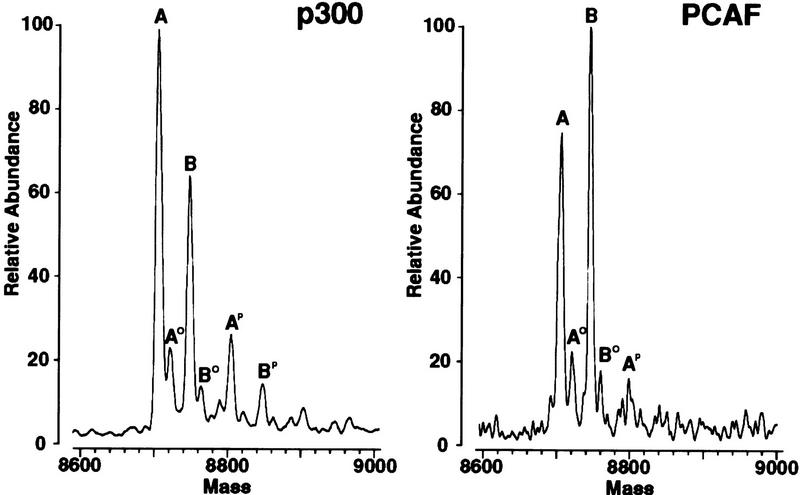
To identify the sites acetylated in the carboxyl terminus of p53, Escherichia coli-expressed p53(319–393) was acetylated with either p300 or PCAF, and the fragments were repurified. Ion spray mass spectrometry of the p53 fragment modified by either enzyme gave two major products that differed by 42 mass units and could be unambiguously interpreted (Fig. 2). The lower molecular weight product (A) in each case corresponded to the mass of the unmodified p53 product; the higher molecular weight component corresponded to the p53 product acetylated at a single site. Minor shoulders resulting from oxidation of methionine to the sulfoxide also were seen (A0 and B0). Notably, however, there was no product with a mass corresponding to a doubly acetylated product.
Figure 2.
p300 and PCAF each acetylate a single site in the carboxyl terminus of human p53. E. coli expressed p53(319–393) was acetylated with either p300 or PCAF, and the repurified fragments were analyzed by ion spray mass spectrometry. The mass profiles are shown. The major peak labeled A in each case corresponds to the mass of the unmodified p53 fragment (8707); the major peak labeled B is larger by 42 mass units, corresponding to the addition of one acetate residue. No mass was observed at the position expected for diacetylated or higher acetylated forms. Minor shoulders result from oxidation of methionine to the sulfone (A0 and B0) and formation of the phosphate salt (Ap and Bp).
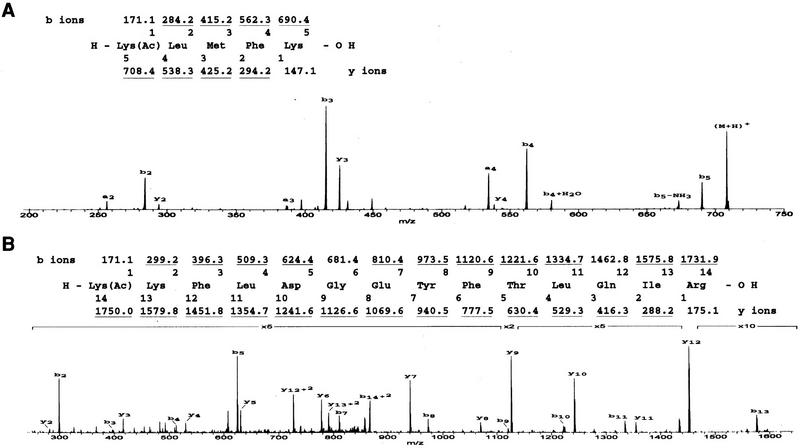
To identify the sites of acetylation, each product was digested with trypsin, and the tryptic fragments were analyzed by nanospray ion trap mass spectrometry (Fig. 3). For products acetylated by p300, we searched for fragments with masses corresponding to all predicted acetylated tryptic peptides. One such fragment was observed, corresponding to the tryptic fragment Lys-382–Lys-386 with a m/z [mass (in Daltons) divided by the charge] of 708 (Fig. 3A). All expected collision-induced ion products corresponding to the tryptic fragment acetylated at Lys-382 were observed, and these were inconsistent with the masses expected from acetylation at Lys-386. That the acetylation site is Lys-382 was confirmed by analyzing the synthetically prepared acetylated peptide, which gave a mass fragmentation pattern identical to that of the p300 acetylated product (data not shown). A similar analysis was carried out on the p53 fragment acetylated by PCAF. Only one fragment was observed with a mass corresponding to that expected for an acetylated tryptic fragment, and this mass (1750 m/z) corresponded to p53 tryptic fragment Lys-320–Arg-333 (Fig 3B). Although this peptide also contains two lysine residues, analysis of the collision-induced fragments was consistent only with acetylation of one position, corresponding to Lys-320 in full-length p53.
Figure 3.
Identification of the p53 sites acetylated by PCAF and p300. (A) p53(319–393) acetylated by p300 was digested with trypsin and the peptide products were analyzed by nanospray ion trap mass spectrometry. (Bottom) The fragmentation mass spectrum for the acetylated peptide of m/z 708. The mass of the acetylated tryptic product corresponds to that of monoacetylated p53(382–386). (Top) The sequence and expected m/z values of fragmentation products. Observed ions are underlined. The fragmentation pattern is consistent only with acetylation of Lys-382 and identical to synthetic p53(382–386) with Lys-382 acetylated. (B) p53(319–393) acetylated by PCAF was digested with trypsin and the peptide products were analyzed by nanospray ion trap mass spectrometry. (Bottom) The fragmentation mass spectrum for the acetylated peptide of doubly charged ion of m/z 876. Segments of the profile were expanded in thedirection of the y-axis by 2-, 5-, or 10-fold as indicated. The mass of the acetylated tryptic product corresponds to that of monoacetylated p53(320–333). (Top) The sequence and expected m/z values of fragmentation products. Observed ions are underlined. The fragmentation pattern is consistent only with acetylation of Lys-320 and identical to synthetic p53(320–333) with Lys-320 acetylated.
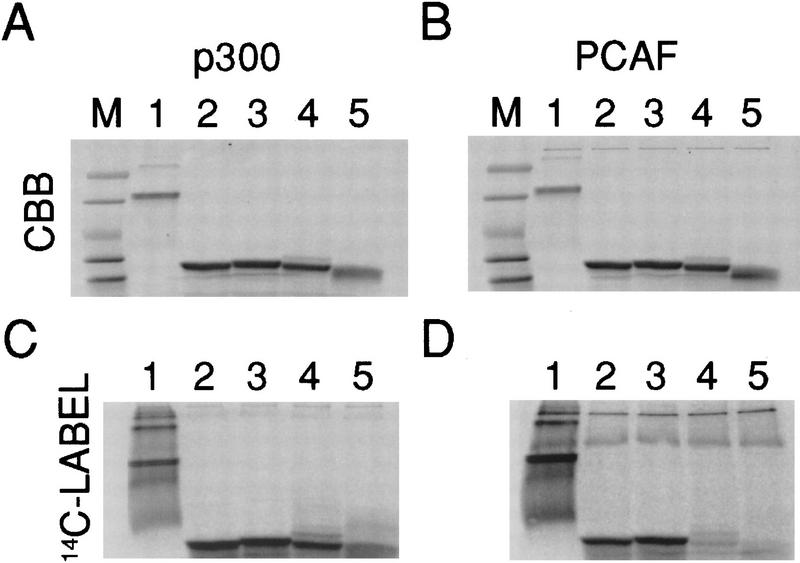
To determine whether acetylation was affected by the phosphorylation or oligomerization state of the p53 carboxyl terminus, chemically synthesized fragments were prepared with phosphate incorporated either at Ser-378, a predicted site of phosphorylation by protein kinase C (PKC) (Hupp and Lane 1995; Takenaka et al. 1995), or Ser-392, a site phosphorylated in vitro by casein kinase 2 (CK2) (Meek 1998). We also prepared peptides with alanines replacing residues Leu-323, Tyr-327, and Leu-330 [p53(319–393)AAA] in the tetramerization domain; these substitutions prevent the formation of p53 oligomers (Sakamoto et al. 1994). For p300, phosphorylation at either site had no effect on acetylation, and the monomer form of the carboxy-terminal fragment also was acetylated well by p300 (Fig. 4C, lane 4). In the case of PCAF, however, acetylation depended critically on the ability of the fragment to form tetramers (Fig 4D, lane 4), and phosphorylation at Ser-378 strongly inhibited acetylation (Fig. 4D, lane 5), whereas phosphorylation at Ser-392 had no significant effect (Fig. 4D, lane 3).
Figure 4.
Acetylation of mutant and phosphorylated p53 carboxy-terminal fragments by PCAF and P300. Chemically synthesized, unmodified p53(319–393), p53(319–393) with phosphoserine incorporated at either Ser-392 or Ser-378, or p53(319–393) with alanine replacing each of the three tetramerization domain residues Leu-323, Tyr-327, and Leu-330 [p53(319–393)AAA] were acetylated with p300. The radioactive image (14C-Label) is shown in C; the corresponding Coomassie brilliant blue (CBB)-stained image is in A. PCAF acetylation is in D; the corresponding Coomassie brilliant blue-stained image is in B. Histone H1 served as a positive control for acetylation. (Lanes M) Molecular weight markers; (lanes 1) histone H1; (lanes 2) unmodified p53(319–393); (lanes 3) p53(319–393) phosphorylated at Ser-392; (lanes 4) p53(319–393) phosphorylated at Ser-378; (lanes 5) tetramerization mutant p53(319–393)AAA.
Acetylation by p300 and PCAF activate sequence-specific DNA-binding
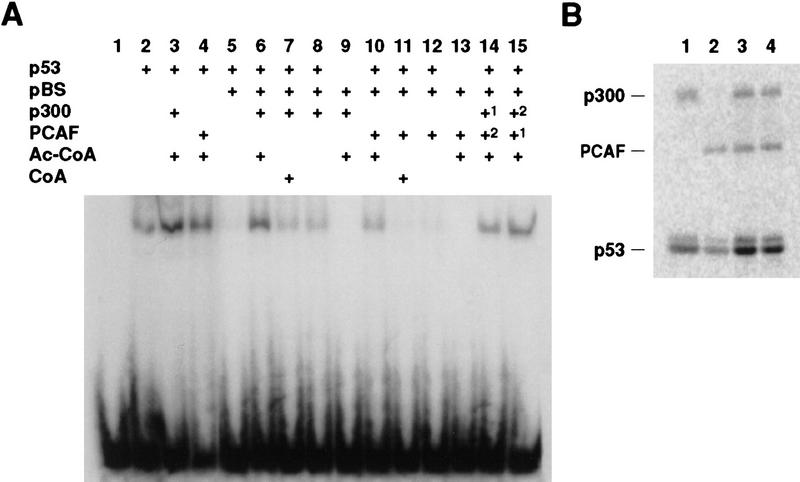
Several modifications to the carboxy-terminal region of p53, including phosphorylation by CK2, PKC, and Cdc2 kinases and truncation of the last 30 residues, modulate the ability of p53 to bind its recognition site through the central sequence-specific DNA-binding domain (Hupp et al. 1992; Hupp and Lane 1995; Takenaka et al. 1995). To determine whether acetylation by p300 and PCAF also affect sequence-specific DNA-binding, full-length p53 was incubated with either recombinant p300 or recombinant PCAF, and the reaction products were used in a DNA gel mobility shift assay described by Anderson et al. (1997) with a synthetic double-stranded probe containing the p53-binding site (Funk et al. 1992). In the absence of competitor DNA, some binding of the probe to wild-type p53 was observed (Fig. 5A, lane 2), but binding of untreated p53 was completely suppressed by the presence of nonspecific pBluescript competitor DNA (Fig. 5, lane 5). After incubation with either p300 or PCAF and acetyl–CoA, strong binding of p53 to the probe was observed both in the absence (Fig. 5, lanes 3,4) and the presence of competitor pBluescript, (Fig. 5, lanes 6,10). Strong binding also was observed with p53 that was incubated sequentially in either order with p300 and PCAF and acetyl–CoA (Fig. 5, lanes 14,15). Although a small amount of binding was observed either when acetyl–CoA was omitted or when it was replaced by unacetylated CoA (Fig. 5, lanes 7,8,11,12), this amount was substantially less than in the presence of acetyl–CoA, indicating that acetylation and not simply binding of either p300 or PCAF to p53 was necessary for the effect on DNA-binding. Analysis of the extent of acetylation in parallel reactions with [14C]acetyl–CoA showed that more acetate was incorporated in the presence of p300 than PCAF (Fig 5B). Quantitation of these results suggests that acetylation at either site enhances sequence-specific DNA binding to similar extents.
Figure 5.
Activation of sequence-specific binding by acetylation of p53 with p300 and PCAF. Baculovirus-produced wild-type p53 was acetylated with p300 or with PCAF as described in Materials and Methods, and the reaction products then were used in electrophoretic mobility shift assays as described by Anderson et al. (1997). (A) Radioactive images of the EMSA gels; the ingredients present during the p53 modification reactions are indicated at top. (Lanes 14,15) The order of p300 and PCAF additions are indicated by superscripts; (lane 7,11) unacetylated CoA was added in place of acetyl–CoA (Ac–CoA). The p53-shifted radioactive probe appears as a band near the top of the gel; free probe is at the bottom. (B) Parallel acetylation reactions were performed with 14C-labeled acetyl–CoA, and the reactions were fractionated by SDS-PAGE. Shown is the radioactive image of the gel. (Lane 1) Reaction with p53 and p300, (lane 2) reaction with p53 and PCAF; (lane 3) reaction with p53 incubated with PCAF and then also with p300; (lane 4) reaction with p53 incubated with p300 and then with PCAF.
The p300 and PCAF sites are acetylated in vivo in response to DNA damage
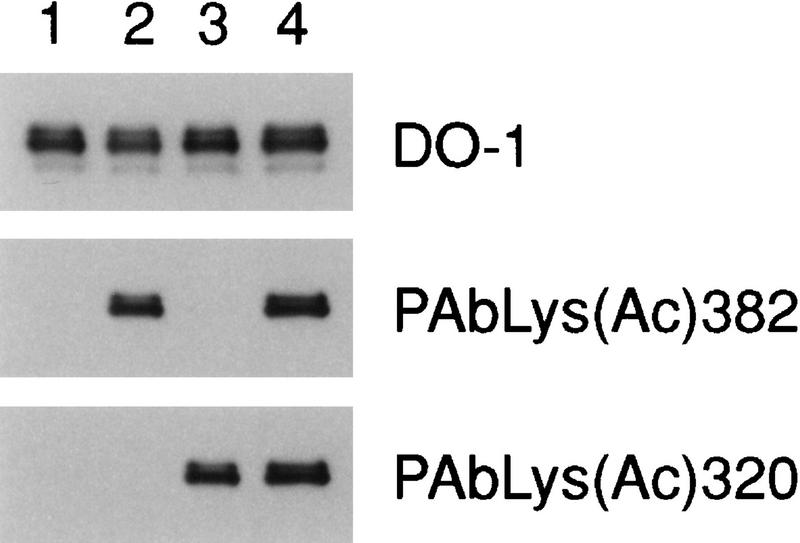
To determine whether p53 is acetylated in vivo, we prepared polyclonal antisera specific for each acetylation site (Materials and Methods). Figure 6 shows that the PAbLys(Ac)382 antibody, prepared with a peptide corresponding to p53(377–387)Cys acetylated at Lys-382, recognized p53 acetylated by p300 but not unacetylated p53 nor p53 acetylated by PCAF. Likewise, the PAbLys(Ac)320 antibody, prepared with a peptide corresponding to p53(315–325)Cys acetylated at Lys-320, recognized p53 acetylated by PCAF, but not unacetylated p53 nor p53 acetylated by p300. After incubation with both HATs and acetyl-CoA, p53 was recognized by both antibodies. These results indicated that the PAbLys(Ac)382 antibody is highly specific for the acetylated form of p53 produced by p300, and the PAbLys(Ac)320 antibody is highly specific for p53 acetylated by PCAF; neither antibody recognizes p53 acetylated at the nonhomologous site. The specificity of each antibody also was confirmed by ELISA assay with synthetically prepared p53 fragments (data not shown). Affinity-purified polyclonal antibodies specific for p53 phosphorylated at Ser-33 or Ser-37 were prepared similarly. By ELISA, the PAbSer(P)33 antibody reacted with p53[27–39(33P)Cys] and p53[1–39(33P)] but not with p53(25–65), p53(1–39), p53(1–39) phosphorylated at Ser-9, Ser-15, Ser-20, or Ser-37; the PAbSer(P)37 antibody reacted with peptides p53[1–39(37P)] and p53[25–65(37P)] but not with p53(1–39), p53[1–39(15P)], or p53(25–65) (data not shown).
Figure 6.
PAbLys(Ac)382 and PAbLys(Ac)320 recognize p53 acetylated by p300 and PCAF, respectively. Recombinant p53 was acetylated by incubation with p300, PCAF, or both, and a Western blot was prepared as described in Fig. 1. Affinity-purified rabbit polyclonal antibodies were prepared by use of acetylated peptides corresponding to sequences around Lys-382 and Lys-320 as described in Materials and Methods. The blot was probed sequentially with PAbLys(Ac)382, PAbLys(Ac)320, and DO-1, a monoclonal antibody specific for the amino terminus of p53. (Lane 1) Unacetylated p53; (lane 2) p53 incubated with p300 and acetyl–CoA; (lane 3) p53 incubated with PCAF and acetyl–CoA; (lane 4) p53 incubated with both p300 and PCAF and acetyl–CoA.
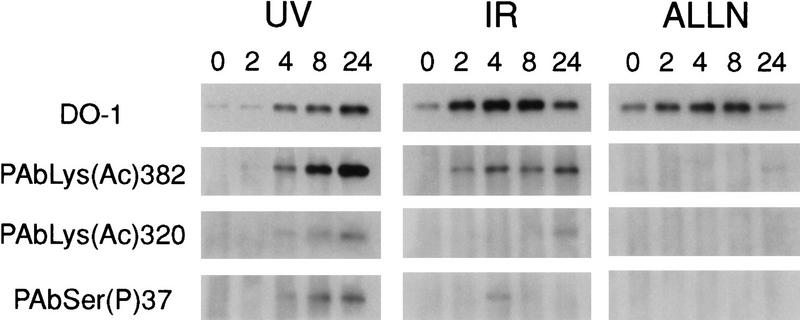
We used the p53 acetylation-specific antibodies to examine p53 from normal and DNA-damaged cells. The human cell lines A549 and HCT116, both of which have wild-type p53, were exposed to 25 J/m2 UV-C or 8 Gy γ-rays (IR), and extracts were prepared at different times after treatment. As a control, cells were treated with 20 μm ALLN, a proteasome inhibitor that partially stabilizes p53, thus causing its accumulation to levels similar to those obtained after treatment with DNA damage-inducing agents. To prevent rapid deacetylation of p53, trichostatin A (TSA), an inhibitor of histone deacetylases, was added at the time of exposure (Yoshida et al. 1990). After harvesting the cells, p53 was immunoprecipitated from the extracts and analyzed by Western blotting. Figure 7 shows the effect on A549 cells. As expected, exposure to UV light induced the accumulation of p53 in both cell lines, which was apparent by 4 hr and continued to increase until 24 hr after treatment. IR treatment induced a rapid accumulation of p53 that was readily evident at 2 hr and reached a maximum between 4 and 8 hr, and then declined. ALLN also induced an accumulation of p53, detected with the DO-1 monoclonal antibody, that was maximal in the 8 hr sample and then declined to approximately the level in untreated cells by 24 hr. No signal was visible from untreated cells with either the PAbLys(Ac)382 or the PAbLys(Ac)320 antibody, suggesting that neither site is significantly acetylated in undamaged cells. Furthermore, neither antibody gave a signal with p53 from ALLN-treated cells, indicating that the lack of a signal in untreated cells did not result from the lower amount of p53 in those samples and that stabilization and accumulation alone did not result in acetylation of p53. Significantly, a strong signal was obtained with the PAbLys(Ac)382 antibody in samples harvested 24 hr after exposure to either UV light or IR, and this signal was clearly visible as early as 2 hr after IR exposure and at 4 hr after exposure to UV. Similar results were obtained for HCT116 cells exposed to UV light (data not shown). For A549 cells, a similar but weaker signal was obtained with the PAbLys(Ac)320 antibody that was clearly visible 4 hr after exposure to UV light but was seen only at 24 hr after exposure to IR (Fig. 7). Phosphorylation at Ser-37, a site potentially phosphorylated in response to DNA damage (Lees-Miller et al. 1992; Shieh et al. 1997), was detected with the PAbSer(P)37 antibody at 4 hr after exposure to IR, but the signal rapidly decayed and was not visible by 8 hr. Phosphorylation at Ser-37 also was seen at 4 hr after exposure to UV light, coincident with the appearance of acetylation but, in this instance, the signal increased over the next 20 hr (Fig. 7, bottom). These results indicated that p53 is acetylated at Lys-382 in response to either IR or UV light and at Lys-320 after exposure to UV light. p53 also becomes phosphorylated at Ser-37 in response to both treatments.
Figure 7.
Induction of p53 acetylation and phosphorylation by UV, γ-rays, and ALLN. A549 cells were exposed to 25 J/m2 of UV-C, 8 Gy γ-rays (IR), or treated with 20 μm calpain inhibitor I (ALLN). TSA was added to 5 μm immediately after DNA damage treatment. Samples were harvested at the indicated times after initiating treatment (top of lanes), and extracts were prepared for immunoprecipitation with Pab1801, a monoclonal antibody specific for human p53, followed by Western immunoblot analysis as described in Materials and Methods. The Western blots were probed sequentially with PAbLys(Ac)382, PAbLys(Ac)320, PAbSer(P)37, and DO-1.
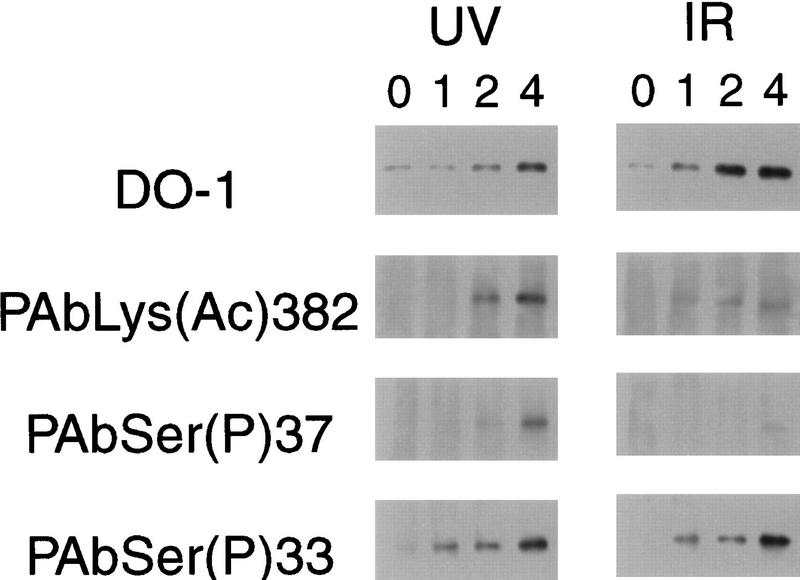
To examine how rapidly phosphorylation and acetylation occur after DNA damage, A549 cells were treated for 4 hr with ALLN to induce p53 accumulation; they then were treated with 25 J/m2 UV-C or 8 Gy γ-rays and TSA, and harvested for immunoprecipitation and Western blot analysis 0, 1, 2, or 4 hr later (Fig. 8). Acetylation of Lys-382 was clearly seen by 1 hr after IR treatment and by 2 hr after UV treatment. Phosphorylation of Ser-37 also was observed by 2 hr after UV treatment, but only weakly at 4 hr after IR treatment. However, phosphorylation of Ser-33 was strongly induced by both UV and IR as early as 1 hr after treatment. Little, if any, phosphorylation at Ser-33 was seen in untreated cells or in ALLNtreated cells.
Figure 8.
Phosphorylation and acetylation of p53 in response to UV and γ radiation after ALLN treatment. A549 cells were treated with 20 μm calpain inhibitor I (ALLN) and 4 hr later were exposed to 25 J/m2 of UV-C or 8 Gy γ-rays (IR). TSA was added to 5 μm immediately after DNA damage treatment. Samples were harvested at the indicated times after DNA damage treatment (Lanes 0,1,2,4 hr), and extracts were immunoprecipitated and analyzed by western immunobloting as described in Fig. 7 and Materials and Methods. Blots were probed sequentially with PAbLys(Ac)382, PAbSer(P)33, PAbSer(P)37, and DO-1.
Phosphorylation influences acetylation of p53 by p300 and PCAF
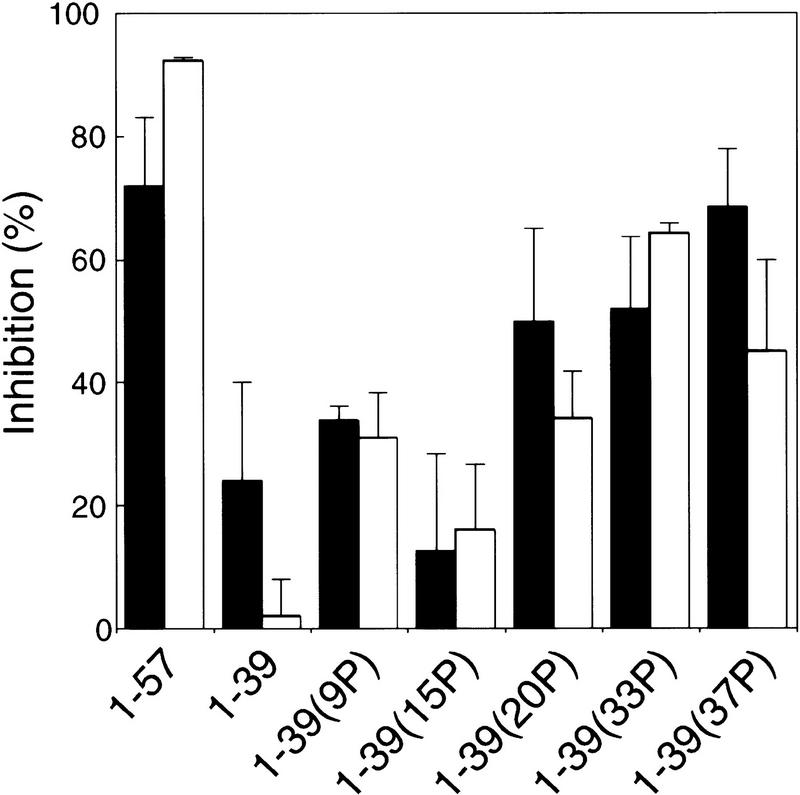
The above results, in conjunction with several recent studies, suggest that p53 acetylation might be triggered through amino-terminal DNA damage-induced phosphorylations. Our finding that full-length p53 is a better substrate for p300 than a tetrameric carboxy-terminal fragment (Fig. 1) is consistent with the importance of amino-terminal p53 residues for acetylation of the carboxyl terminus. To further explore the role of phosphorylation in modulating acetylation, we investigated the effect of phosphorylated amino-terminal p53 peptides on the ability of p300 and PCAF to acetylate full-length p53 in vitro (Fig. 9). We reasoned that if phosphorylation were important for the interaction between p53 and either HAT, the phosphorylated peptide would inhibit acetylation of p53 better than would the unphosphorylated peptide. As shown in Figure 9, p53(1–57) inhibited acetylation of full-length p53 by p300 or PCAF by 70%–90%, results consistent with the observation that Trp-53 and Phe-54 are important for the interaction between p53 and p300 (Scolnick et al. 1997). In contrast, p53(1–39) was a poor inhibitor (0%–15%) of acetylation by either HAT; however, p53(1–39) phosphorylated at Ser-33 or at Ser-37 inhibited p53 acetylation by either p300 or PCAF significantly better (40%–80%) than did the unphosphorylated peptide. Similar results were obtained with p53(25–65) phosphorylated at Ser-37 or at both Ser-33 and Ser-37 (data not shown). Phosphorylation of p53(1–39) at other amino-terminal sites including Ser-9 and Ser-20, but not Ser-15, inhibited acetylation by p300 or PCAF to lesser extents, but still more than the unphosphorylated peptide. In contrast, none of these p53 amino-terminal peptides inhibited acetylation of a carboxy-terminal fragment of p53 (data not shown). These results suggest that amino-terminal phosphorylation may enhance the interaction of p53 with p300 and PCAF, thereby potentiating acetylations that are critical for sequence-specific DNA-binding.
Figure 9.
Inhibition of in vitro acetylation by PCAF (open bars) and p300 (solid bars) of full-length, wild-type p53 by p53 peptides. Assays were performed as described in Materials and Methods and the radioactivity incorporated into wild-type p53 was quantitated with a PhosphorImager and Imagequant software. Amino-terminal p53 peptides and phosphopeptides were present at 100 μm; the p53 substrate concentration was 0.1 μm. The results shown are an average of three independent assays for p300 (error bars, s.d.) and two independent assays for PCAF (error bars, range). Acetylation of the carboxy-terminal fragment p53(319–393) by p300 was not inhibited by these same amino-terminal peptides (data not shown).
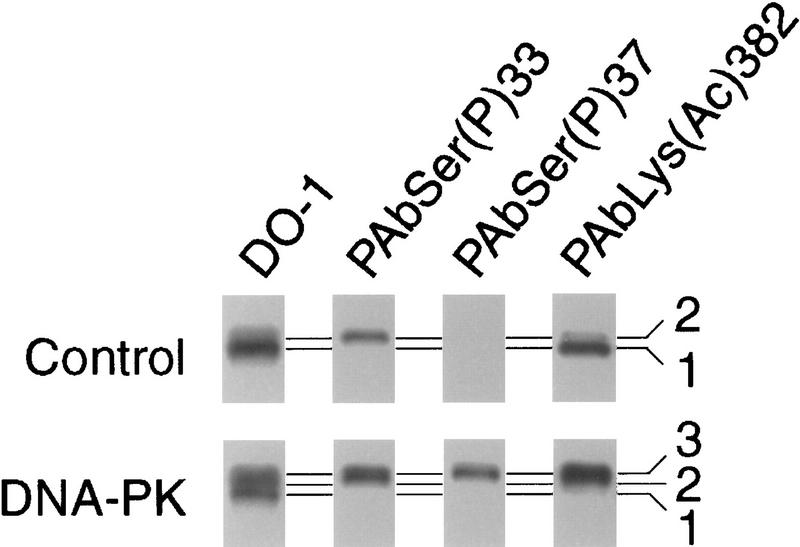
In support of the above hypothesis, phosphorylation of wild-type p53 by DNA–PK enhanced the acetylation of recombinant p53 (Fig. 10). For this experiment, full-length human p53 produced in insect cells, which was partially phosphorylated a several sites including Ser-33 (Patterson et al. 1996), but not to a significant extent at Ser-37 (Fig. 10), was incubated with DNA–PK or not, and then with p300 and acetyl–CoA. Incubation with DNA–PK produced a new, slower mobility p53 isoform (Fig 10, band 3) that is phosphorylated at Ser-33 and Ser-37. Although both the untreated (Fig 10, control) and DNA–PK-treated p53 were acetylated by catalytic amounts of p300, after DNA–PK treatment only the slower mobility isoform (Figure 10, band 3) was acetylated. This result indicates that the DNA–PK phosphorylated isoform was acetylated over p53 isoforms that were not phosphorylated by DNA–PK but were still present, as revealed by staining with DO-1.
Figure 10.
Phosphorylation by DNA–PK potentiates acetylation of human p53 by p300. Recombinant human p53 (250 ng in 20 μl) purified from insect cells was incubated with purified DNA–PK (40 units), DNA, and ATP at 30°C for 1 hr and then with recombinant p300 (∼0.5 ng) and acetyl–CoA at 37°C for 20 min. Western blots of the reaction products were probed sequentially with PAbLys(Ac)382, PAbSer(P)33, PAbSer(P)37, and DO-1. Note that incubation with DNA–PK produced a new p53 isoform (labeled 3) that is phosphorylated on Ser-37 as well as on Ser-33. This isoform was preferentially acetylated by p300.
Discussion
Here we report that p53 can be acetylated in vitro by two HATs, p300 and PCAF, that function as coactivators for p53-mediated transcription in vivo. Both HATs acetylated the carboxyl terminus of p53 at single sites, Lys-382 in the case of p300, and Lys-320 in the case of PCAF. Lys-382 was the major site of acetylation seen by Gu and Roeder (1997); the minor sites they identified may have resulted from their use of a relatively short peptide as the substrate for in vitro acetylation. The rate of acetylation by p300 was significantly reduced (∼80%) by removing the p53 amino terminus, but surprisingly, not by mutants that prevented tetramerization or by phosphorylation of Ser-378, which lies only six amino acids from the p300 acetylation site. In contrast, acetylation by PCAF was only modestly affected by removing the amino terminus, but was strongly influenced by conformation. Point mutants that prevent tetramerization dramatically decreased acetylation as did phosphorylation at Ser-378, a site potentially phosphorylated by PKC. Phosphorylation at Ser-392 had no significant effect. These findings suggest that recognition of p53 by p300 may depend in part on interactions with the p53 amino terminus, whereas at least in vitro, PCAF specifically recognized the tetrameric form of the carboxyl terminus, implying an interaction with this domain. In vivo p300 is tightly associated with the RNA polymerase II holoenzyme complex (Nakajima et al. 1997), and PCAF was reported to associate with p300 (Yang et al. 1996); thus, in vivo recognition of p53 by either HAT may be influenced by additional factors. Moreover, the general transcription factors for RNA polymerase II, TFIIEβ and TFIIF, are acetylated by PCAF, p300, and TAFII250, although considerable specificity in substrate preference was apparent (Imhof et al. 1997). Interestingly, the site of acetylation in TFIIEβ by PCAF was identified as the lysine at residue 52, which is in a sequence that is highly conserved among the human, Xenopus, and yeast homologs. Likewise, the sites acetylated by p300 and PCAF in p53 are in evolutionarily conserved segments, suggesting possible functional significance.
In vitro acetylation by either HAT activated sequence-specific DNA binding in the presence of a long competitor DNA, confirming the observations of Anderson et al. (1997) and suggesting that both acetylations may function, in part, by inactivating the nonsequence-specific binding activity of the p53 carboxyl terminus. Modifications to the carboxyl terminus that eliminate positively charged residues might be expected to have this effect. p300 and PCAF may further contribute differentially to transcriptional activation by weakening the interaction of chromosomal DNA with histones, thus facilitating the binding of transcription factors. Acetylation also could directly induce a conformational change in the carboxyl terminus that potentiates sequence-specific DNA binding by the central domain or influences interactions with other proteins that bind the carboxy-terminal domain, including TBP, XPB, XPD, CSB, S100b, CK2β, or Brca 1 (for review, see Ko and Prives 1996; Ouchi et al. 1998). These interactions might be affected directly by acetylation of the carboxyl terminus, or through an acetylation-induced conformational change. Both acetylation sites are located in nuclear localization signals; thus, acetylation might also affect nuclear import or export of p53.
To investigate the relevance of acetylation to p53 activation by DNA damage in vivo, we used antibodies specific for acetylated Lys-382 and Lys-320 in Western blot analyses of lysates from undamaged, damaged, and ALLN-treated cells. p53 levels dramatically increased in response to UV-C, IR, and ALLN; however, only DNA damage-causing agents gave a signal with the acetylation-specific antibodies. The lack of immunoreactivity in lysates from ALLN treated cells cannot be caused by lower amounts of p53 because there was an equivalent amount of p53 in lysates from damaged and ALLN-treated cells. Kinetic analyses showed that acetylation is an early event in the DNA damage response. The signal obtained with the PAbLys(Ac)382 antibody was readily detected by 2 hr after IR, by 4 hr after UV, and by 1 hr after IR exposure in cells pretreated with ALLN to increase p53 levels. After UV irradiation, the signal continued to increase for 24 hr, consistent with the continued accumulation of p53 and the fact that UV-induced damage is repaired slowly (Dumaz et al. 1997). The signal obtained with the PAbLys(Ac)320 antibody after UV irradiation was weaker than that with the PAbLys(Ac)382 antibody, possibly indicating that less p53 becomes acetylated at Lys-320 compared with Lys-382. Nevertheless, the Lys-320 signal also increased with time after treatment; however, after IR, acetylation of Lys-320 was seen only at 24 hr, suggesting that it may not play a role in activating p53 in response to DNA strand breaks. These data clearly provide, for the first time, a potential mechanism for coupling the regulation of p53’s activity as a transcription factor with acetylation. Our data further show that p53 accumulation and acetylation are not directly coupled, and that agents which produce different types of DNA damage may differ in their ability to induce p53 acetylation at individual sites. Determining the exact role of acetylation in the cytostatic and apoptotic responses must await the creation of p53 mutants at the acetylated sites, stable cell lines that express them, and a comparison of p53-mediated functions with acetylation in response to stress-inducing agents.
p53 is stabilized and activated in response to DNA damage by two at least partially independent pathways, one of which responds to DNA double-strand breaks (DSBs), the major DNA damage caused by IR, the other of which is activated by bulky lesions in DNA such as those caused by exposure to UV light (for review, see Agarwal et al. 1998). Phosphorylation is widely believed to be a major mechanism for regulating these effects. The DSB activation pathway depends on the function of the ATM (ataxia telangiectsia) gene product, a member of the DNA–PK class of large serine/threonine protein kinases that have homology with phosphatidylinositol kinases (Khanna and Lavin 1993). p53 becomes phosphorylated on Ser-15 in response to both UV and IR (Siliciano et al. 1997), and ATM was shown recently to be activated in response to IR and to directly phospharylate Ser-15 of human p53, suggesting that it is likely to be involved in activating p53 in response to DNA strand breaks (Banin et al. 1998; Canman et al 1998). Detection of bulky lesions including pyrimidine dimers is independent of ATM and appears to be accomplished, at least in part, by components of the transcription apparatus (Dumaz et al. 1997). However, DNA damage-inducing agents such as UV light also activate other cellular response pathways, including the Jun amino terminal kinase (JNK) pathway. JNK kinases phosphorylate mouse p53 at Ser-34, and in vivo this serine is phosphorylated in response to exposing cells to UV-C (Milne et al. 1995), possibly implicating JNK kinases in a response to UV-C that is independent of direct DNA damage (Herrlich et al. 1997). The homologous human residue is Ser-33. Phosphorylation of Ser-15 and Ser-37 inhibits binding by Mdm2, implicating these modifications in stabilizing p53 in response to DNA damage (Haupt et al. 1997; Kubbutat et al. 1997; Shieh et al. 1997); however, p53 can be activated as a transcription factor without significantly accumulating (Huang et al. 1996; Haapajärvi et al. 1997; Chernov et al. 1998). Sequence-specific binding can be activated by phosphorylation of Ser-315, Ser-378, and Ser-392 (Hupp et al. 1992; Hupp and Lane 1995; Takenaka et al. 1995; Wang and Prives 1995; Kapoor and Lozano 1998); however, none of these modifications were shown to occur in response to DNA damage.
In vitro p53 interacts with p300 through its amino-terminal domain (Gu et al. 1997), and residues between Leu-22 and Phe-54 are critical for this interaction (Scolnick et al. 1997). p53 and p300 also colocalize within the nucleus and coexist as a stable DNA-binding complex (Lill et al. 1997). CBP/p300 are coactivators for p53-specific transcription, and activation of p53-specific transcription by p300 is potentiated by IR (Avantaggiati et al. 1997). Our finding that full-length p53 is a better substrate for p300 than is a tetrameric carboxy-terminal fragment (Fig. 1) is consistent with an importance of amino-terminal p53 residues for acetylation of the carboxyl terminus. To explore further a role for phosphorylation in modulating acetylation, we examined the effect of phosphorylated amino-terminal p53 peptides on the ability of p300 and PCAF to acetylate full-length p53 in vitro. Acetylation of full-length p53, but not a carboxy-terminal fragment, was significantly inhibited by phosphorylated p53 amino-terminal peptides compared with nonphosphorylated controls. Furthermore, p53 phosphorylated by DNA–PK was preferentially acetylated by p300. These data clearly suggest that the amino-terminal p53 phosphorylations affect the ability of these HATs to acetylate p53. We also show for the first time that p53 becomes phosphorylated at Ser-33 and Ser-37 in response to either IR or UV light, consistent with a role for these phosphorylations in promoting the acetylation of p53. Our finding that phosphorylation at Ser-37 of p53 in UV-irradiated cells is prolonged compared with cells treated with IR indicates that UV irradiation and γ irradiation may have different effects on the specific kinases that phosphorylate this site.
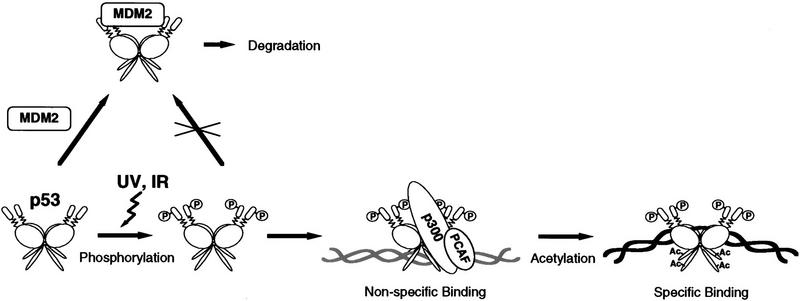
Our data, in conjunction with previous studies, suggest a model in which DNA damage activates p53 as a transcription factor through a cascade of post-translational modifications that include both phosphorylations and acetylations (Fig. 11). Phosphorylation of Ser-15 and Ser-37 at the amino terminus of p53 prevents Mdm2 binding, thus stabilizing p53. DNA damage also induces phosphorylation at additional amino-terminal residues, including Ser-33 (a JNK site) and Ser-37 (a DNA–PK site), which increase p53’s affinity for p300 and PCAF, thus promoting acetylation of carboxy-terminal sites including Lys-382 (by p300) and Lys-320 (by PCAF). Acetylation at either site inhibits nonspecific DNA-binding, thereby allowing sequence-specific DNA-binding. Alternatively or in addition, acetylation may cause a conformational change that promotes sequence-specific binding through the central domain. This simplified model shows one way that p53 may be activated as a transcription factor in response to a DNA damage-inducing agent. p53 is modified at several additional sites, and these may function independently or in conjunction with acetylation to activate p53 in response to UV or other agents; modifications to other components also may be important.
Figure 11.
Model for the activation of p53 in response to DNA damage. Unmodified p53 interacts with the Mdm2 protein, which targets it for rapid degradation. p53 that enters the nucleus interacts with chromatin through the carboxy-terminal domain that prevents sequence-specific DNA binding. DNA damage induces phosphorylation of amino-terminal p53 residues that increase p53’s affinity for p300 and PCAF, thus promoting acetylation of carboxy-terminal sites thereby allowing sequence-specific DNA binding. The model incorporates recent findings from other studies including the role of phosphorylation in regulating p53’s interaction with mdm2 (Shieh et al. 1997), induction of amino-terminal phosphorylation by UV, IR, and other agents (Siliciano et al. 1997), and the effect of p53’s nonspecific interaction with DNA on sequence-specific DNA binding (Anderson et al. 1997).
Material and methods
Cell lines
A549 (ATCC CCL-185), a human lung carcinoma cell line with wild-type p53, and HCT116 (ATCC CCL-247), a human colon carcinoma cell line with wild-type p53, were obtained from the American type culture collection (Rockville, MD). The cells were grown in Dulbecco’s modified Eagle medium (GIBCO-BRL, Bethesda, MD) supplemented with 10% fetal bovine serum (Biowhittaker, Wakerville, MD) and penicillin/streptomycin (GIBCO-BRL) in a humidified atmosphere with 5% CO2.
Enzymes, substrates, and peptides
Recombinant Flag-tagged human PCAF and p300 were prepared as described (Yang et al. 1996); acetyl–CoA and CoA were from Sigma Chemical Co.; [1-14C]acetyl–CoA (55 mCi/mMole) was from Amersham, Inc. Full-length, His-tagged wild-type human p53 protein and p53 fragments p53(283–393) and p53(318–393) were produced by use of a baculovirus expression system (Anderson et al. 1997) and were provided by M. Anderson and P. Tegtmeyer (SUNY, Stony Brook). The carboxy-terminal p53 fragment p53(319–393) and modified versions thereof were synthesized by the fragment condensation method by use of thioesters (Sakamoto et al. 1996). All other peptides were synthesized by the solid-phase method with Fmoc chemistry by an Applied Biosystems 430A peptide synthesizer (Foster City, CA). Phos phoserine residues were incorporated as Fmoc–Ser[PO(OBzl)OH]– OH (Novabiochem, San Diego, CA). The peptide was cleaved from the resin and side-chain-protecting groups were removed with reagent K (TFA/phenol/thioanisole/H2O/EDT = 82.5:5:5:5:2.5) for 3 hr at room temperature. The p53(1–39) peptides were purified by HPLC on a pH-stable Vydac C-8 column (Hesperia, CA) with 0.2% hexafluoroacetone–NH4OH at pH 7.0/acetonitrile. The mass of peptides were confirmed by electrospray ionization mass spectrometry on a Finnigan MAT SSQ 7000 (Finnigan MAT, San Jose, CA).
Acetylation- and phosphorylation-specific p53 antibodies
Rabbit polyclonal antibodies specific for p53 acetylated at Lys-320 [PAbLys(Ac)320] or Lys-320 [PAbLys(Ac)382] were raised against the human p53 sequence Ac-377–387(382Ac)Cys [i.e., Ac-SPQPKK(Ac)-KPLDGC] or Ac-315–325(320Ac)Cys [i.e., Ac-TSRHKK(Ac)-LMFKTC] conjugated through the added carboxy-terminal cysteine to KLH (keyhole limpet hemocyanin). Sera from immunized rabbits were affinity purified by use of each acetylated peptide coupled with SulfoLink (Pierce). The purified antibodies then were passed through a column coupled with the respective unacetylated peptide to deplete antibodies that react with unacetylated p53. Specificity was shown by use of unacetylated, baculovirus-produced, wild-type p53 or p53 acetylated in vitro (Fig. 6). Antibodies specific for p53 phosphorylated at Ser-33 or Ser-37 were prepared similarly. Rabbits were immunized with the p53 phosphopeptide Ac-27–39(33P)Cys [i.e., Ac-PENNVLS(P)-PLPSQAC] or Ac-32–43(37P)Cys [i.e., Ac-LSPLPS(P)-QAMDDLC], and phosphorylation site-specific antibodies were affinity purified by use of the corresponding SulfoLinked phosphorylated peptides. In ELISA and immunoblot assays, the purified PAbSer(P)33 antibody recognized the immunizing peptide and p53(1–39) phosphorylated at Ser-33 but not p53(1–39) phosphorylated at Ser-9, Ser-15, Ser-20, or Ser-37, whereas the PAbSer(P)37 antibody recognized p53(1–39) and p53(25–65) when phosphorylated at Ser-37 but not either unphosphorylated peptide nor p53(1–39) phosphorylated at ser-15 (data not shown).
Enzymatic assay of HAT
All assays were performed in buffer A [50 mm Tris-HCl (pH 8.0), 10% glycerol (vol/vol), 1 mm DTT, 0.1 mm EDTA, 10 mm Na-butyrate] as described (Herrera et al. 1997). Substrate concentrations were 0.1–0.25 mg/ml and [1-14C]acetyl–CoA concentrations were at 9–15 μm, unless otherwise indicated. Acetyltransferase reactions for mass spectral sequence analyses and for DNA-binding studies were performed in the same manner except that the acetyl–CoA was not radioactive. Assays were incubated at 37°C and initiated by adding either the protein substrate to a mixture containing the acetyltransferase and acetyl–CoA in buffer A or by adding the acetyltransferase to a mix containing the protein substrate and acetyl–CoA in buffer A. Incorporation of [1-14C]acetyl–CoA was quantitated after SDS-PAGE with a PhosphorImager (Molecular Dynamics).
Mass spectrometric analysis
Bacterium-expressed p53(319–393) was acetylated by either PCAF or p300 in buffer A as described above, except that the reaction time was extended to 4 hr, adding fresh enzyme every hour and also 10 μm of acetyl–CoA with the final addition of enzyme. After acetylation, the peptide was purified by HPLC and the mass of the peptide was analyzed by use of a single quadrupole mass spectrometer (Finnigan model SSQ-7000) equipped with an electrospray ion source.
To identify the acetylated site on p53(319–393), the purified peptide was digested with trypsin (Promega), and the resulting fragments were analyzed by ion-trap mass spectrometry (Finnigan model LCQ) equipped with a nanospray ion source. Purified synthetic peptides were used to confirm the molecular masses.
DNA-binding assay
DNA-binding assays were performed as described (Anderson et al. 1997) with 2 ng per 20-μl assay of labeled, 30-bp double-stranded, p53-specific probe sequence (5′-ctagAGCGGACATGCCCGGGCATGTCCGCG-3′/3′-TCGCCTGTADGGGCCCGTACAGGCGCagct-5′) (Funk et al. 1992), and 10 ng of pBluescript plasmid DNA as a nonspecific competitor.
Peptide inhibition studies
Assays were performed as described above except that the enzyme(s) were added to a mixture containing wild-type p53 at 0.1 μm and competing peptides were at 100 μm, and the concentration of enzyme was reduced to 0.01 μg/μl for PCAF and 5 ng/μl for p300. The incorporation of [14C]acetate into p53 was measured by use of PhosphorImager analysis of the Enhance-impregnated (Dupont) SDS–polyacrylamide gels by use of Imagequant software (Molecular Dynamics, Inc.).
Induction of DNA damage and detection of p53 post-translational modifications
Cells were seeded 24 hr prior to treatment and were 60%–70% confluent at the time of treatment. After washing twice with PBS, cells were exposed to UV irradiation with a Stratagene UV Stratalinker 2400. IR experiments were performed with a Shepherd Mark I 137Cs irradiator at a dose rate of 3.2 Gy per minute. The proteosome inhibitor ALLN (Calbiochem) was added to cells at a final concentration of 20 μm as described (Siliciano et al. 1997). The deacetylase inhibitor TSA (WAKO) was added at a final concentration of 5 μm immediately after DNA damage or ALLN treatment. In a preliminary experiment similar to that shown in Figure 7, no signal was observed after DNA damage with either acetylation-specific antibody when TSA was omitted.
Cultures were harvested at the indicated times, washed twice with ice-cold PBS, and lysed on ice in ice-cold lysis buffer (50 mm Tris-HCl at pH 7.5, 5 mm EDTA, 50 mm NaCl, 1% Triton X-100, 50 mm NaF, 10 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 5 μg/ml pepstatin, 0.5 mm PMSF, and 5 μm TSA). Lysates were clarified at 20,000g for 30 min at 4°C. Before immunoprecipitation, each sample (containing 1.3 mg of total cellular protein) was precleared by protein G–Sepharose (GammaBind Plus Sepharose, Pharmacia) for 1 hr; then 2.5 μg of affinity-purified mouse monoclonal anti-p53 antibody (Ab-2, Calbiochem) was added. Immunoprecipitation reactions were incubated on ice for 1 hr, protein G-Sepharose was added, and lysates were further incubated at 4°C for 1 hr, with rotation. The beads were collected by centrifugation and then washed five times with ice-cold lysis buffer. The immunoprecipitates were dissolved in 2× SDS sample buffer (125 mm Tris-HCl at pH 6.8, 20% glycerol, 5.2% SDS, and 0.05% bromophenol blue) and boiled for 3 min. Electrophoresis was on a 10% SDS–polyacrylamide gel. After separation, proteins were electrophoretically transferred to PVDF (polyvinylidene difluoride) membranes. Blocking was performed with a 5% solution of reconstituted dried milk powder for 2 hr at room temperature. Primary antibodies, PAbLys(Ac)320, PAbLys(Ac)382, and affinity-purified mouse monoclonal antibody to p53 (Ab-6, Calbiochem), were incubated with the blot for 1 hr at room temperature. Detection was by the enhanced chemiluminescence method (ECL, Amersham) with the appropriate peroxidase-conjugated secondary antibody.
Acknowledgments
We are indebted to P. Tegtmeyer for gifts of reagents, and to M.E. Anderson, C.A. Pise-Masison, and Y. Nakatani for advice and suggestions. C.W.A. is supported by U. S. Public Health Services grant GM52825 and by the Office of Health and Environmental Research of the U.S. Department of Energy.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cwa@bnl.gov; FAX (516) 344-3407.
References
- Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- Anderson ME, Woelker B, Reed M, Wang P, Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol Cell Biol. 1997;17:6255–6264. doi: 10.1128/mcb.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Banin, S., L. Moyal, S.-Y. Shieh, Y. Taya, C.W. Anderson, L. Chessa, N.I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science (in press). [DOI] [PubMed]
- Canman, C.E., D.-S. Lim, K.A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M.B. Kastan, and J.D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science (in press). [DOI] [PubMed]
- Chernov MV, Ramana CV, Adler VV, Stark GR. Stabilization and activation of p53 are regulated independently by different phosphorylation events. Proc Natl Acad Sci. 1998;95:2284–2289. doi: 10.1073/pnas.95.5.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N, Duthu A, Ehrhart JC, Drougard C, Appella E, Anderson CW, May P, Sarasin A, Daya-Grosjean L. Prolonged p53 protein accumulation in trichothiodystrophy fibroblasts dependent on unrepaired pyrimidine dimers on the transcribed strands of cellular genes. Mol Carcinog. 1997;20:340–347. [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Gu W, Shi X-L, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- Haapajärvi, Pitkänen K, Tsubari M, Laiho M. p53 transactivation and protein accumulation are independently regulated by UV light in different phases of the cell cycle. Mol Cell Biol. 1997;17:3074–3080. doi: 10.1128/mcb.17.6.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- Herrera JE, Bergel M, Yang X-J, Nakatani Y, Bustin M. The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J Biol Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- Herrlich P, Blattner C, Knebel A, Bender K, Rahmsdorf HJ. Nuclear and non-nuclear targets of genotoxic agents in the induction of gene expression. Shared principles in yeast, rodents, man and plants. Biol Chem. 1997;378:1217–1229. doi: 10.1515/bchm.1997.378.11.1217. [DOI] [PubMed] [Google Scholar]
- Huang LC, Clarkin KC, Wahl GM. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc Natl Acad Sci. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Lane DP. Two distinct signaling pathways activate the latent DNA-binding function of p53 in a casein kinase II-independent manner. J Biol Chem. 1995;270:18165–18174. doi: 10.1074/jbc.270.30.18165. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA-binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Imhof A, Yang X-J, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- Juven T, Barak Y, Zauberman A, George DL, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- Kapoor M, Lozano G. Functional activation of p53 via phosphorylation following DNA damage by UV but not γ radiation. Proc Natl Acad Sci. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Lavin MF. Ionizing radiation and UV induction of p53 protein by different pathways in ataxia-telangiectasia cells. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- Meek DW. Multisite phosphorylation and the integration of stress signals at p53. Cell Signal. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- Milne DM, Campbell LE, Campbell DG, Meek DW. p53 is phosphorylated in vitro and in vivo by an ultraviolet radiation-induced protein kinase characteristic of the c-Jun kinase, JNK1. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Lee C-G, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Ouchi T, Monteiro ANA, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p53-dependent gene expression. Proc Natl Acad Sci. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson RM, He C, Selkirk JK, Merrick BA. Human p53 expressed in baculovirus-infected Sf9 cells displays a two-dimensional isoform pattern identical to wild-type p53 from human cells. Arch Biochem Biophys. 1996;330:71–79. doi: 10.1006/abbi.1996.0227. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Chambers KA, Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes & Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- Perry ME, Piette J, Zawadzki JA, Harvey D, Levine AJ. The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc Natl Acad Sci. 1993;90:11623–11627. doi: 10.1073/pnas.90.24.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkänen K, Haapajärvi T, Laiho M. U.V.C.-induction of p53 activation and accumulation is dependent on cell cycle and pathways involving protein synthesis and phosphorylation. Oncogene. 1998;16:459–469. doi: 10.1038/sj.onc.1201528. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Lewis MS, Kodama H, Appella E, Sakaguchi K. Specific sequences from the carboxyl terminus of human p53 gene product form anti-parallel tetramers in solution. Proc Natl Acad Sci. 1994;91:8974–8978. doi: 10.1073/pnas.91.19.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Kodama H, Higashimoto Y, Kondo M, Lewis MS, Anderson CW, Appella E, Sakaguchi K. Chemical synthesis of phosphorylated peptides of the carboxy-terminal domain of human p53 by a segment condensation method. Int J Pept Protein Res. 1996;48:429–442. doi: 10.1111/j.1399-3011.1996.tb00861.x. [DOI] [PubMed] [Google Scholar]
- Scolnick DM, Chehab NH, Stavridi ES, Lien MC, Caruso L, Moran E, Berger SL, Halazonetis TD. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes & Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA-binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reed M, Wang P, Stenger JE, Mayr G, Anderson ME, Schwedes JF, Tegtmeyer P. p53 domains: Identification and characterization of two autonomous DNA-binding regions. Genes & Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes & Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Yang X-J, Ogryzko VV, Nishikawa J-i, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]