Abstract
The promoter selectivity factor Sp1 often cooperates with other enhancer-binding proteins to activate transcription. To study the molecular underpinnings of these regulatory events, we have reconstituted in vitro the synergy observed in vivo between Sp1 and the sterol-regulated factor SREBP-1a at the low density lipoprotein receptor (LDLR) promoter. Using a highly purified human transcription system, we found that chromatin, TAFs, and a novel SREBP-binding coactivator activity, which includes CBP, are all required to mediate full synergistic activation by Sp1 and SREBP-1a. The SREBP-binding domain of CBP inhibits activation by SREBP-1a and Sp1 in a dominant-negative fashion that is both chromatin- and activator-specific. Whereas recombinant CBP alone is not sufficient to mediate activation, a human cellular fraction containing CBP can support high levels of chromatin-dependent synergistic activation. Purification of this activity to near homogeneity resulted in the identification of a multiprotein coactivator, including CBP, that selectively binds to the SREBP-1a activation domain and is capable of mediating high levels of synergistic activation by SREBP/Sp1 on chromatin templates. The development of a reconstituted chromatin transcription system has allowed us to isolate a novel coactivator that is recruited by the SREBP-1a activation domain and that functions in concert with TFIID to coordinate the action of multiple activators at complex promoters in the context of chromatin.
Keywords: Transcription, Sp1, chromatin, TAFs, coactivators, SREBP1a, CBP, LDLR
The transcriptional control regions of many eukaryotic genes harbor multiple enhancer elements recognized by different sequence-specific factors necessary to achieve high levels of transcription (Tjian and Maniatis 1994; Thanos and Maniatis 1995; Sauer and Tjian 1997). The transcription factor Sp1 is frequently found to be one of the contributing enhancer factors on cognate promoters. Although the molecular mechanisms of Sp1 activation have been extensively studied (Pascal and Tjian 1991; Hoey et al. 1993; Chen et al. 1994), the function of Sp1 working in concert with other enhancer-binding factors to regulate inducible promoters remains largely unexplored. Recent evidence suggests that many of these inducible genes are governed by gene-selective activators that work synergistically with Sp1 to direct transcription (Krey et al. 1995; Look et al. 1995; Merika and Orkin 1995; Sheridan et al. 1995; Pazin et al. 1996). However, the cofactor requirements and molecular interactions that allow combinatorial regulation by multiple transcriptional activators are poorly understood.
The regulation of the low density lipoprotein receptor (LDLR) gene by cholesterol provides a useful model system to explore the molecular mechanisms of synergistic activation by Sp1 together with other enhancer-binding factors (Goldstein and Brown 1990; Sanchez et al. 1995; Brown and Goldstein 1997). The sequences within the LDLR promoter mediating cholesterol regulation consist of a DNA element recognized by the sterol-responsive element-binding proteins (SREBP-1 and 2) flanked by Sp1-binding sites (Südhof et al. 1987; Dawson et al. 1988; Briggs et al. 1993; Hua et al. 1993; Wang et al. 1993, 1994; Yokoyama et al. 1993; Yieh et al. 1995). SREBPs belong to the basic/helix-loop-helix class of transcription factors, with the unusual property of being anchored to the ER membrane until released by cholesterol-regulated proteolysis (Brown and Goldstein 1997). Transient transfection experiments suggested that SREBPs and Sp1 can function synergistically to activate transcription of the LDLR gene (Sanchez et al. 1995; Athanikar et al. 1997). To investigate the molecular requirements for cooperative activation by SREBP and Sp1, we have attempted to recapitulate SREBP/Sp1 synergy in an in vitro transcription system.
Results
Synergistic activation by Sp1 and SREBP-1a in vitro depends on chromatin and TAFs
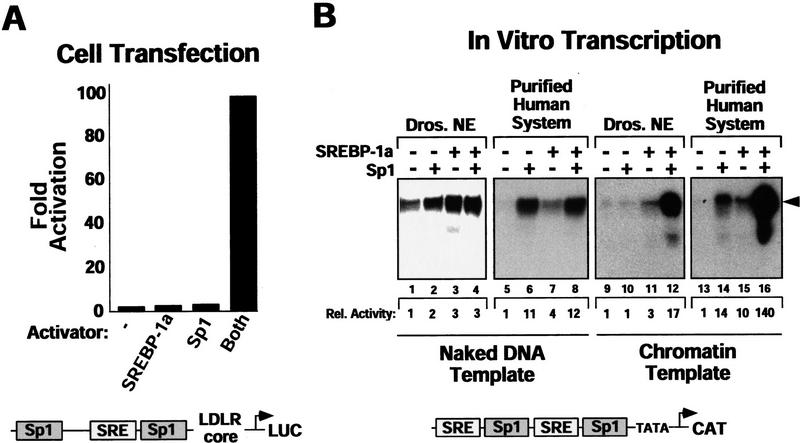
Because we and others (Sanchez et al. 1995; Athanikar et al. 1997) observed synergistic activation by SREBP-1a/Sp1 in a Drosophila cell line (Fig. 1A), we initially evaluated transcriptional activation in a crude Drosophila transcription system (Hansen and Tjian 1995). A DNA template containing the SREBP- and Sp1-responsive elements from the LDLR promoter was used together with purified recombinant human SREBP-1a (amino acids 1–487) corresponding to the active proteolytic cleavage product and recombinant human Sp1 (Fig. 1C). Interestingly, in contrast to the synergy observed in vivo, only additive levels of transcription were found in the presence of both activators under these conditions in vitro (Fig. 1B, lanes 1–4). Next, we tested activation by SREBP-1a and Sp1 on the LDLR-derived template in a highly purified human in vitro transcription reaction consisting of recombinant TFIIA, TFIIB, TFIIE, TFIIF, immuno-affinity purified TFIID, TFIIH purified over five columns, RNA polymerase II (Pol II) purified from HeLa cell nuclear pellets over three columns, and a cofactor purified from HeLa cell nuclear extract over four columns (Goodrich and Tjian 1994; Materials and Methods). Although this system supported high levels of activated transcription, no synergy of Sp1 and SREBP-1a was observed on this naked DNA template (Fig. 1B, lanes 5–8).
Figure 1.
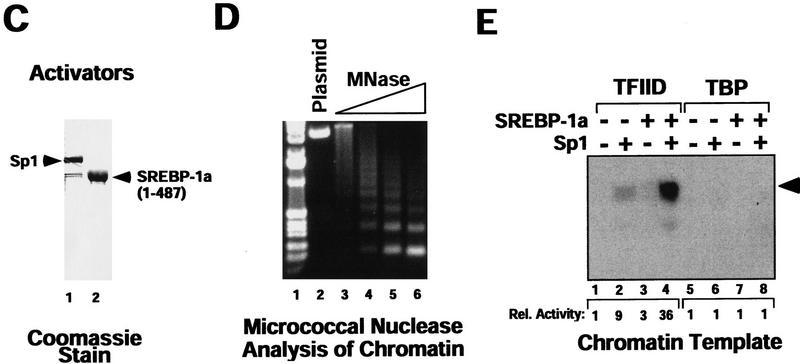
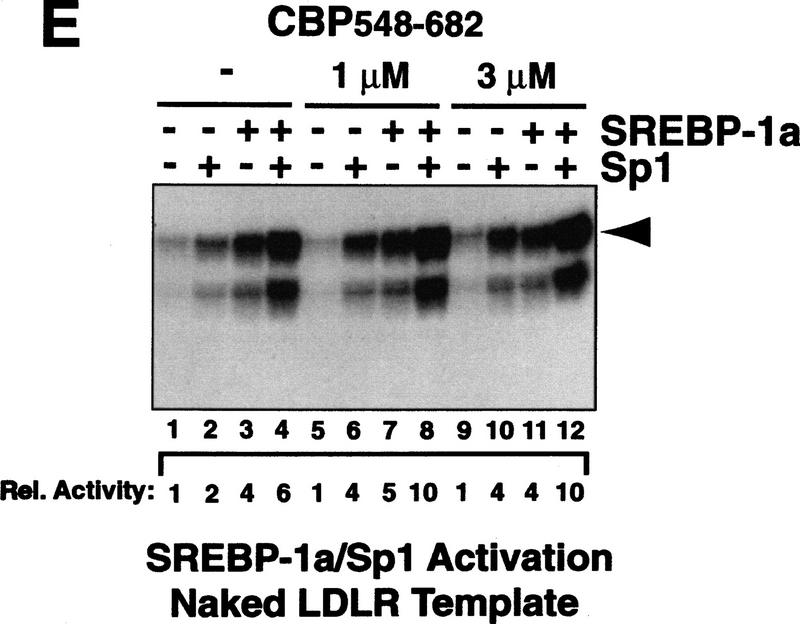
Sp1/SREBP-1a-dependent transcriptional synergy in a purified in vitro transcription system requires chromatin templates and TAFs. (A) Synergistic activation of transcription by Sp1 and SREBP-1a on the LDL receptor promoter in vivo. Transient transfection of Drosophila Schneider cells with vectors directing expression of human Sp1 and SREBP (SREBP-1a amino acids 1–487) or vector alone (none) together with a reporter construct containing the human LDLR promoter and enhancer sequences fused to the luciferase reporter gene. The organization of the LDLR promoter/enhancer is schematically depicted below the bar graph. (B) Sp1 and SREBP-1a require chromatin for synergistic activation of the LDLR-derived template. PhosphorImager (Fuji) scan and autoradiographs of the in vitro transcription results with no activator added (lanes 1,5,9,13), Sp1 (lanes 2,6,10,14), SREBP-1a (lanes 3,7,11,15), or both activators added (lanes 4,8,12,16). The activity relative to basal transcription (lane 1, arbitrarily set to 1) is shown below the transcription panels (Rel. Activity). In vitro transcription reactions were assembled by purified activators, a crude Drosophila transcription system (lanes 1–4,9–12) or a highly purified human transcription system (lanes 5–8,13–16), and a LDLR promoter-derived reporter plasmid known to support SREBP-1a/Sp1 synergy in vivo. Activators were tested for activation with the crude Drosophila transcription system (lanes 9–12) or the purified human transcription system (lanes 13–16) by use of naked (lanes 1–8) or chromatin (lanes 6–16) templates. Transcription products were analyzed by primer extension and denaturing PAGE. (C) Purified activators used in transcription reactions. Human recombinant Sp1 (lane 1) was expessed by vaccinia virus infection of HeLa cells and purified from nuclear extract by wheatgerm- and DNA-affinity chromatography. Human His-tagged SREBP-1a (amino acids 1–487, corresponding to the nuclear proteolytic product) (lane 2) was generated by baculovirus infection of Sf9 insect cells and purified over Ni-NTA and DNA affinity columns. (D) Analysis of the LDLR chromatin template by micrococcal nuclease digestion. The DNA template was assembled into chromatin by use of a Drosophila chromatin assembly system and the resulting nucleosomal template was analyzed by digestion with increasing concentrations of micrococcal nuclease. (E) Activation by Sp1/SREBP-1a on the LDLR chromatin template requires TAFs. This panel shows the results of in vitro transcription reactions by use of the purified transcription system and chromatin templates containing either no activators (lanes 1,5), Sp1 (lanes 2,6), SREBP-1a (lanes 3,7), or both activators (lanes 4,8) in the presence of either immuno-purified human TFIID (lanes 1–4) or recombinant human TBP (lanes 5–8).
One difference between the in vitro and in vivo systems that could influence the coordinate action of multiple activators is the packaging of the in vivo template into chromatin (Pazin et al. 1996). To investigate this possibility, we assembled the LDLR template into chromatin in vitro using a Drosophila chromatin assembly system (Kamakaka et al. 1993) (Fig. 1D) and tested it with SREBP-1a and Sp1 in the Drosophila nuclear extract-based transcription system. In contrast to the results obtained with naked DNA templates, high levels of transactivation were observed only when both activators were added together, whereas little transcription was seen with either activator alone (Fig. 1B, lanes 9–12). When analyzing SREBP-1a/Sp1 activation on chromatin with the purified human transcription system, we also observed a high degree of synergy (Fig. 1B, lanes 13–16). The absolute levels of Sp1/SREBP-1a-dependent transcription on chromatin templates were typically 20%–50% of that observed with naked DNA templates. These results suggest that it is possible to reconstitute transcription reactions in vitro by use of a chromatin template and purified factors that recapitulate synergistic activation by SREBP-1a and Sp1 on the LDLR-derived template. The purified transcription system also provided the opportunity to investigate various cofactor requirements for Sp1/SREBP-1a activation not possible to address with cruder transcription systems on chromatin templates.
Previous studies identified the TBP-associated factors (TAFs) as potential coactivators for mediating activation in Drosophila and human reconstituted transcription reactions on naked DNA templates (Goodrich et al. 1993; Hoey et al. 1993; Chen et al. 1994; Sauer et al. 1995; Thut et al. 1995). Therefore, we tested whether the TAF subunits of TFIID are also required for activation on chromatin templates. Substituting recombinant human TBP for antibody-affinity purified human TFIID resulted in the loss of activation observed with Sp1 alone, as well as synergistic activation by Sp1 and SREBP-1a (Fig. 1E). This finding suggests that TAFs serve important functions in directing transcriptional responses to activators on chromatin templates. In addition to TFIID, high levels of activation with both naked DNA and chromatin templates also require a fraction containing a novel human cofactor important for Sp1 activation (CRSP, S. Ryu and R. Tjian, unpubl.). However, because Sp1/SREBP-1a-dependent synergy only occurs on nucleosomal LDLR templates, additional chromatin-specific cofactors may also be required to observe cooperative activation on chromatin templates.
The coactivator CBP is implicated in SREBP-1a/Sp1-dependent chromatin-specific synergy
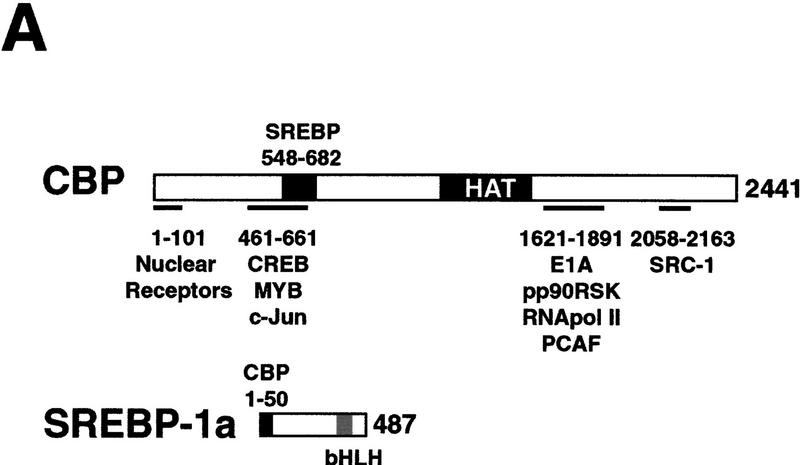
Our previous investigation of SREBP transcriptional function revealed that the coactivator CBP interacts with the SREBP activation domain (Oliner et al. 1996). The reconstitution of SREBP-1a and Sp1 synergy on the LDLR template in a defined in vitro transcription system allowed us to address the potential involvement of CBP in chromatin-dependent synergy in vitro. To examine this, we first used a combination of molecular deletion analysis and proteolytic digestion of SREBP-1a and CBP to identify the sequences responsible for the interaction between SREBP and CBP. Our studies of the activation domain of SREBP-1a revealed that the amino-terminal 50 amino acids were sufficient for interaction with CBP (Fig. 2A,B). This region is the most highly conserved within the activation domains of SREBPs. Additionally, a natural splicing variant of SREBP-1 that lacks the first 29 amino acids (SREBP-1c) is severely compromised for transcriptional activation, indicating the functional importance of this region in activation by SREBPs (Shimano et al. 1997), and truncation of the first 51 amino acids inactivated SREBP-1a in transfection studies (Sato et al. 1994).
Figure 2.
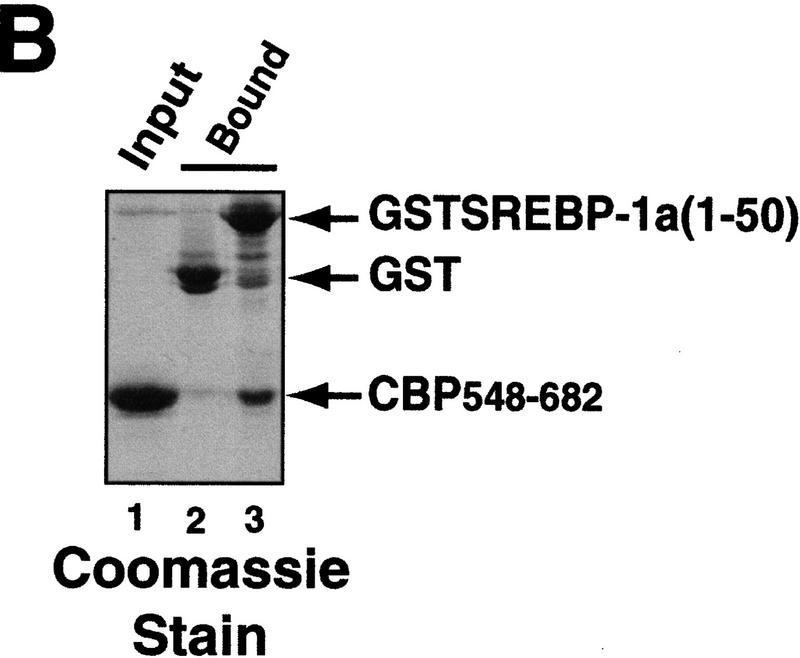
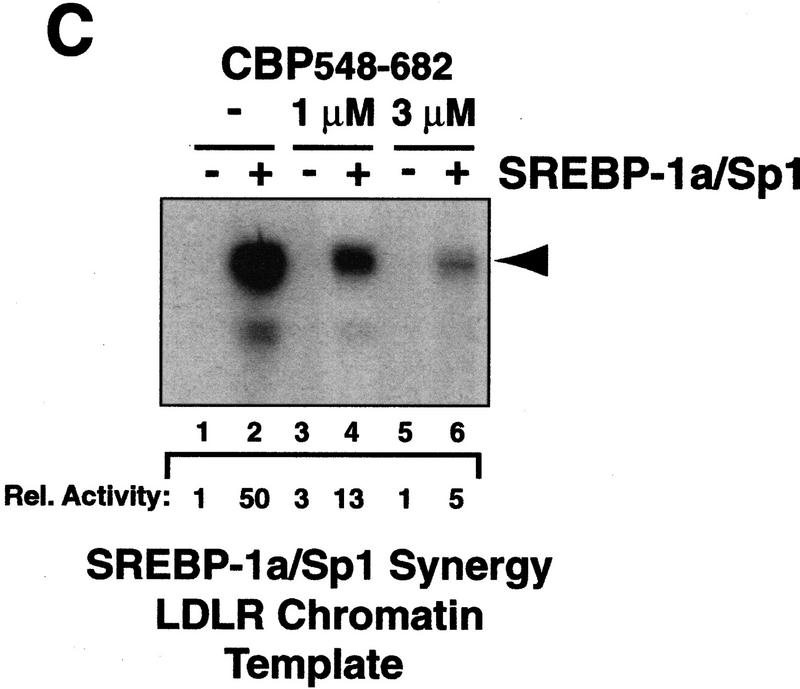
Interaction between CBP and SREBP-1a is implicated in chromatin-specific transcriptional synergy. (A) Schematic depiction of the domains of CBP that are involved in protein–protein interactions and HAT activity. The CBP domain interacting with the amino-terminal 50 amino acids of SREBP-1a was determined by partial V8 protease digestion. The regions that are involved in interaction between CBP and SREBP-1a are highlighted by solid boxes in both proteins. (B) Binding assay with CBP548–682 and GST–SREBP-1a (amino acids 1–50). Purified CBP548–682 was incubated with glutathione-agarose beads prebound to either GST (lane 2) or GST–SREBP-1a (amino acids 1–50) (lane 3). Lane 1 shows the input CBP548–682 (50%). The binding of CBP548–682 to the beads was analyzed by SDS-PAGE and visualized by Coomassie staining. (C) Effect of adding CBP548–682 protein (lanes 3–6) to a highly purified transcription system using the LDLR-promoter chromatin template and SREBP-1a/Sp1. (D) Effect of adding CBP548–682 protein (lanes 3–6) to a highly purified transcription system using a HIV–LTR chromatin template and NF-κB/Sp1. (E) The same CBP548–682 protein fragment was added (lanes 5–12) to a highly purified transcription system using a naked LDLR-promoter template and SREBP-1a/Sp1.
In a previous study, we found that the first 682 amino acids of CBP bind strongly to SREBP-1a (Oliner et al. 1996). Partial Staphylococcal V8 protease digestion of this region resulted in the isolation of a stable 135-amino-acid fragment spanning amino acids 548–682 that was selectively retained on a SREBP affinity column. As expected, a recombinant CBP548–682 polypeptide also interacted strongly with the SREBP-1a activation domain (Fig. 2B) and was predicted to compete with endogenous CBP for interaction with SREBP-1a and potentially disrupt SREBP–1a/Sp1 cooperativity in a dominant-negative fashion. To test this possibility, we added increasing concentrations of the CBP peptide to in vitro transcription reactions reconstituted with the LDLR chromatin template. The SREBP–1a/Sp1-mediated transcriptional activation was strongly inhibited by the CBP peptide in a dose-dependent manner (Fig. 2C). The chromatin-dependent synergistic activation by NF-κB and Sp1 on the HIV–LTR was used as a control for the specificity of the inhibition. Because NF-κB interacts with a different portion of CBP (P. Beaurang, D. Avizonis, and R. Tjian, unpubl.), NF-κB/Sp1 synergy should be refractory to inhibition by the SREBP-specific CBP peptide. Addition of CBP548–682 did not significantly affect NF-κB/Sp1 synergy on the HIV–LTR chromatin template, attesting to the specificity of the peptide (Fig. 2D).
As an additional control, we also carried out experiments with increasing amounts of the CBP peptide added to transcription reactions with naked DNA templates. The CBP peptide failed to significantly inhibit either basal or activated transcription on the naked DNA template (Fig. 2E). These results taken together suggest that an activity, possibly related to CBP, present in our chromatin-based transcription system is a functionally important target for the SREBP-1a activation domain specifically in the context of the chromatin-dependent synergy with Sp1 on the LDLR promoter template.
Reconstitution of SREBP-1a/Sp1 activation on chromatin requires a CBP-containing activity
To identify and characterize the chromatin-dependent cofactor activity, we next attempted to modify the transcription reaction so that it gained responsiveness to the addition of exogenous cofactor/CBP activity. Because the dominant-negative inhibition by the CBP peptide indicated that our chromatin transcription reactions contain an activity that may be related to CBP, and immunoblotting results showed that the purified transcription system is devoid of CBP (data not shown), we suspected that a CBP-like activity is present in the S-190 chromatin assembly extract. To limit this potential cofactor contribution, we reconstituted transcription reactions with low levels of S-190 extract. Decreasing the amount of S-190 extract used to assemble chromatin by 50% did not significantly affect chromatin assembly; however, transcriptional activation by SREBP-1a/Sp1 was substantially reduced (Fig. 3A, cf. lanes 1 and 2 and 5 and 6). Additionally, purification of the chromatin template over a sucrose gradient revealed a requirement for the S-190 extract to observe synergistic activation (data not shown). These results suggest that the S-190 extract provides one or more cofactor(s) important for SREBP-1a/Sp1 activation. To determine whether addition of a human CBP-containing cofactor could substitute for the limiting cofactor(s) in the S-190 extract, we partially purified CBP from HeLa cell nuclear extract by phosphocellulose chromatography and assayed for stimulatory activity in the cofactor-dependent chromatin transcription system. Whereas the CBP-containing cofactor fraction [phosphocellulose fraction eluting at 0.5 m salt (PC 0.5 m) stimulated transcription modestly when added to reactions with high levels of S-190 extract, addition of the PC 0.5 m fraction to reactions reconstituted with low amounts of S-190 extract resulted in greatly increased transcriptional response (Fig. 3A, cf. lanes 1–4 with lanes 5–8). Because RNA Pol II and the general transcription factors TFIIB, TFIIE, TFIIF, and TFIIH are not limiting in our transcription reactions (data not shown), this result suggests that the CBP-containing fraction can at least partially replace the S-190 activity.
Figure 3.
A CBP-containing fraction is required for full transactivation by SREBP-1a/Sp1 on chromatin templates and cannot be replaced by recombinant CBP. (A) A human cofactor fraction stimulates transcriptional activation by SREBP-1a/Sp1 on chromatin template. The transcriptional response to a CBP-containing fraction (PC 0.5 m) derived from HeLa cell nuclear extract was tested with chromatin templates assembled by use of high (lanes 1–4) or low (lanes 5–8) amounts of S-190 extract in the absence (lanes 1,3,5,7) or presence of SREBP-1a/Sp1 (lanes 2,4,6,8). (B) Analysis of purified recombinant CBP. Recombinant Flag epitope-tagged CBP (rCBP) was expressed in Sf9 insect cells by baculovirus infection and affinity purified from nuclear extracts by use of M2 Flag beads. The peptide-eluted rCBP was analyzed on a 7% SDS–polyacrylamide gel followed by protein staining with Coomassie brilliant blue. (Arrowhead) Migration of full-length rCBP; (asterisk) position of a CBP proteolytic product. (C) Stimulation of Sp1/SREBP-1a-dependent activation on chromatin by a CBP-containing fraction, but not by rCBP. The effect of adding a CBP-containing fraction from HeLa cell nuclear extract (PC 0.5 m) (lanes 3,4) to S-190-limited chromatin transcription reactions performed in the absence (lanes 1,3) or presence (lanes 2,4) of SREBP-1a/Sp1 (1–2 nm) is shown. Recombinant CBP (lanes 5–10) were added in the indicated nanogram amounts to chromatin transcription reactions performed in the absence (lanes 5,7,9) or presence (lanes 6,8,10) of activators (1–2 nm). (Arrowhead) Migration of transcription/primer extension products. (D) Depletion of CBP from the PC 0.5 m fraction by the activation domain of SREBP-1a. The CBP-containing cofactor fraction (PC 0.5 m) (lane 1) was passed over glutathione–Sepharose loaded either with GST or GST–SREBP-1a (amino acids 1–50). The bound (lanes 2,3) and flowthrough (FT; lanes 4,5) fractions were analyzed by Western blotting with anti-CBP antiserum (Santa Cruz Biotechnology). (E) A coactivator activity is depleted from the PC 0.5 m fraction by the SREBP-1a activation domain. The GST and GST–SREBP-1a (amino acids 1–50)-flowthrough fractions were tested for stimulatory activity in the in vitro transcription reactions on chromatin template. Depletion by GST did not significantly diminish cofactor activity (lanes 3,4), whereas the cofactor activity in the GST–SREBP-1a (amino acids 1–50)-depleted fraction was dramatically reduced (lanes 5,6).
Next, we wished to test whether CBP alone was sufficient to reconstitute this cofactor activity. When increasing concentrations of recombinant CBP (rCBP) (Fig. 3B) were added to transcription reactions with the S-190-limited chromatin template, no stimulation of transcriptional activation by SREBP-1a/Sp1 was observed (Fig. 3C, cf. lanes 1 and 2 and 5–10). At higher concentrations of rCBP, we observed apparent squelching of activated transcription. Similarly, recombinant P/CAF, which has been shown to interact with CBP, either alone or in combination with rCBP failed to enhance activation (data not shown). This is in contrast to the strong stimulatory activity observed with the PC 0.5 m fraction (Fig. 3C, lanes 3,4). The recombinant CBP interacts strongly with the activation domain of SREBP-1a and mediates histone acetyl transferase (HAT) activity (Bannister and Kouzarides 1996; Ogryzko et al. 1996) (data not shown), suggesting that recruitment of HAT activity is not sufficient for full transcriptional activation by SREBP-1a/Sp1 on chromatin templates. Additionally, recombinant p300, demonstrated previously to enhance activation by the estrogen receptor in a crude chromatin transcription system (Kraus and Kadonaga 1998), was not capable of replacing the CBP fraction in our purified chromatin transcription system (data not shown). Taken together, these experiments suggest that CBP/p300 alone or in combination with P/CAF are not sufficient to mediate transcriptional activation by SREBP-1a/Sp1 on chromatin templates, and indicate that other activities present in the CBP-containing fraction may provide important functions.
Because the activation domain of SREBP-1a binds to CBP and perhaps other activities, we attempted to use it as an affinity resin to deplete the CBP-containing cofactor fraction of stimulatory activity. After passing the cofactor fraction over the SREBP-affinity resin or a GST control resin, we analyzed the bound and flowthrough fractions by CBP immunoblotting (Fig. 3D). We found that >95% of CBP present in the cofactor fraction input was depleted (Fig. 3D, lane 5) and remained bound to the activation domain of SREBP-1a even after extensive washing of the resin (Fig. 3D, lane 3). In contrast, the GST control resin did not bind any CBP (Fig. 3D, lanes 2,4). When testing the two flowthrough fractions for stimulation of transcription in the SREBP-1a/Sp1-dependent chromatin transcription system, the GST flowthrough fraction contained cofactor activity, whereas the SREBP activation-domain flowthrough was significantly depleted of cofactor activity (Fig. 3E). These results suggest that an activity present in the cofactor fraction binds to and is depleted by the SREBP-1a activation domain. Although we have obtained multiple lines of evidence suggesting that the coactivator CBP may be an important mediator of chromatin-dependent synergy by SREBP-1a/Sp1, our results could also be consistent with the notion that other factors present in the CBP-containing fraction contribute to the coactivator activity.
Purification and functional characterization of a CBP-containing coactivator
The depletion of coactivator activity from the partially purified CBP fraction by GST–SREBP-1a (amino acids 1–50) indicated the feasibility of using the SREBP-affinity column to purify this coactivator. HeLa cell nuclear extract was initially applied to the GST–SREBP (amino acids 1–50) affinity resin or a control resin consisting of the glutamine-rich activation domain A of Sp1 fused to GST. After extensive washing, the bound fractions were analyzed by SDS-PAGE followed by silver staining. The results showed that a series of polypeptides (280–30 kD), including CBP (immunoblot not shown), interacted strongly with the GST–SREBP (amino acids 1–50) affinity resin, whereas no specific binding to the GST–Sp1A control beads was detected (Fig. 4A). Similar results were obtained when using the PC 0.5 m fraction in place of the HeLa cell nuclear extract (data not shown). Preliminary immunoblotting results indicated that trace amounts of P/CAF, SRC-1/NCoA1, GRIP-1/NCoA2, and RNA Pol II were detected in some GST–SREBP (amino acids 1–50) pulldowns (data not shown), however, these proteins do not appear to be stoichiometric components of this activity. RNA Pol II is not limiting in our transcription reactions, thus this source of RNA Pol II is unlikely to contribute significantly to transcriptional activation in this system. No binding of SWI/SNF proteins or RNA helicase A was detected (data not shown). These results suggest that this CBP-containing coactivator is distinct from previously identified CBP complexes and holo-RNA Pol II activities (Chakravarti et al. 1996; Kamei et al. 1996; Kee et al. 1996; Yang et al. 1996; Zhang et al. 1996; Heery et al. 1997; Nakajima et al. 1997a,b; Perkins et al. 1997; Torchia et al. 1997; Korzus et al. 1998; Kurokawa et al. 1998).
Figure 4.
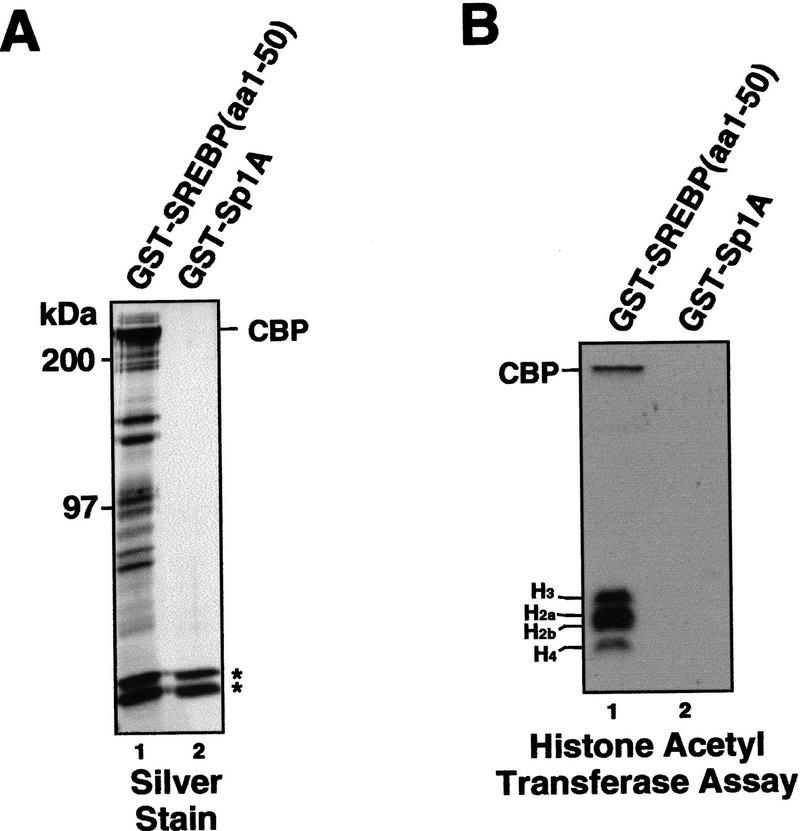
A CBP-containing multiprotein coactivator associates selectively with the activation domain of SREBP-1a and mediates HAT activity. (A) A series of proteins in addition to CBP associate selectively with the SREBP-1a activation domain. HeLa cell nuclear extract was incubated with affinity resins containing the activation domains of SREBP-1a (GST–SREBP-1a amino acids 1–50) (lane 1) or Sp1 (GST–Sp1A amino acids 83–262) (lane 2) and after extensive washing, the bound fractions were analyzed by SDS-PAGE followed by silver staining (shown) and immunoblotting. The migration of CBP is indicated. The two asterisks mark the positions of nonspecific proteins binding to the GST portion of the fusion proteins. (B) The SREBP activation domain recruits HAT activity. Pulldowns from HeLa nuclear extract with GST–SREBP (amino acids 1–50) (lane 1) and GST–Sp1A (amino acids 83–262) (lane 2) were tested for HAT activity by incubating the washed beads with purified Drosophila core histones, 3H-labeled acetyl CoA, and Na Butyrate. The reactions were analyzed by separation on a 15% SDS polyacrylamide gel followed by autoradiography. The migration of core histones and CBP is indicated at left
Because CBP has been reported to harbor HAT activity (Bannister and Kouzarides 1996; Ogryzko et al. 1996), we tested whether the SREBP-1a activation domain could recruit HAT activity. Strong HAT activity was detected only with the SREBP-associated fraction, whereas the Sp1-activation domain did not recruit any HAT activity (Fig. 4B). Because SREBP-1a appears to recruit HAT activity, we tested the effect of adding acetyl CoA and the histone deacetylase inhibitor trichostatin A (TSA) to SREBP-1a/Sp1-driven chromatin transcription reactions. Addition of TSA resulted in a small (less than twofold) dose-dependent increase in levels of activated transcription, whereas addition of acetyl CoA had little influence on transcription (data not shown), suggesting that histone acetylation is unlikely to significantly contribute to the cofactor activity.
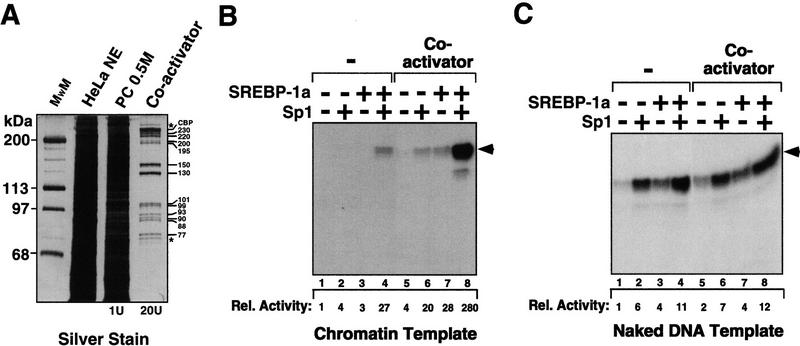
Next, we examined whether the affinity-purified SREBP-binding factors could substitute for the crude PC 0.5 m cofactor fraction when added to transcription reactions. The CBP-containing coactivator was purified from the PC 0.5 m fraction by the SREBP activation domain affinity resin and eluted by use of 0.1% deoxycholate. These two chromatographic steps resulted in purification of the coactivator to near homogeneity (increase in specific activity ∼5000-fold). We estimate that the SREBP affinity-purified coactivator is at least 1000-fold purified relative to the PC 0.5 m cofactor fraction (Fig. 5A). When the highly purified SREBP-binding proteins were added to the S-190-limited chromatin transcription system, strong stimulation of SREBP-1a/Sp1 synergistic activation was observed (Fig. 5B, lanes 1–4 and 5–8), comparable with the enhancement observed with the crude PC 0.5 m cofactor fraction (Fig. 3A, lanes 5–8). In contrast, addition of the eluted SREBP-binding proteins to transcription reactions with naked DNA templates resulted in little effect on basal or activated transcription (Fig. 5C). These findings suggest that a multiprotein coactivator capable of selective binding to the activation domain of SREBP-1a participates in mediating SREBP-1a/Sp1 activation in the context of chromatin.
Figure 5.
An affinity-purified coactivator mediates chromatin-specific activation by SREBP-1a/Sp1. (A) Purification of the coactivator to near homogeneity by phosphocellulose and SREBP-1a-activation domain-affinity chromatography. HeLa cell nuclear extract (lane 2) was first passed over a phosphocellulose column and the main peak of cofactor activity was found in the 0.5 m fraction (PC 0.5 m, lane 3). The cofactor fraction (PC 0.5 m) was then passed over the GST–SREBP-1a (amino acids 1–50) resin, and specifically bound proteins were eluted by buffer containing 0.1% deoxycholate (lane 4). Similar results were observed when using the CBP548–682 peptide or glutathione for elution. The eluate was analyzed on a 7% SDS–polyacrylamide gel followed by silver staining. The relative amounts of coactivator units (as judged by titration of coactivator in the SREBP-1a/Sp1-dependent chromatin transcription system with chromatin assembled by use of low S-190 extract) loaded onto the gel are shown below lanes 3 and 4. Molecular weight markers (lane 1) are indicated at left. The migration of CBP (as judged by immunoblotting) and other specific SREBP-binding proteins is indicated at right. Non-specific proteins that do not consistently copurify with coactivator activity are indicated by asterisks above CBP and below 77 markers. (B) The coactivator is required for full activation by SREBP-1a/Sp1 in S-190-limited chromatin transcription reactions. Shown is the effect of adding the affinity-purified coactivator (lanes 5–8) to S-190 limited chromatin transcription reactions in the absence of activators (lanes 1,5), presence of Sp1 (lanes 2,4,6,8) or presence of SREBP-1a (lanes 3,4,7,8). (Arrowhead) Migration of transcription/primer extension products. (C) The coactivator does not affect transcription from naked DNA templates. Transcription reactions performed with naked DNA templates in the absence (lnes 1–4) or presence (lanes 5–8) of affinity-purified coactivator. SREBP-1a (lanes 3,4,7,8) and Sp1 (lanes 2,4,6,8) were added as indicated. (Arrowhead) Migration of transcription/primer extension products.
Discussion
We have reconstituted Sp1/SREBP-1a -dependent transcriptional synergy at the LDLR promoter in vitro. Our results indicate that the coordinate activation by Sp1 and SREBP-1a requires the LDLR template to be assembled into chromatin. Using a purified human transcription system, we found that multiple distinct coactivators are necessary to mediate chromatin-dependent synergy by SREBP-1a/Sp1, including TAFs, and a highly purified CBP-containing multiprotein coactivator that directly binds to the activation domain of SREBP-1a (Fig. 6). The TAF requirement for high levels of activation by Sp1 and SREBP-1a may be explained at least partly by the well-documented interactions between Sp1 and hTAFII130 in recruiting TFIID to the promoter (Chen et al. 1994; Gill et al. 1994). Because SREBP-1a was not found to contact TFIID, or other components of the core transcriptional machinery (data not shown), cooperativity cannot be explained by direct targeting of multiple general transcription factors (GTFs) (Sauer et al. 1995). SREBP-1a/Sp1 cooperativity may be a function of recruitment of both a CBP-containing coactivator and TFIID to the promoter (Fig. 6). This dual requirement of a CBP-containing coactivator and TAFs for activation of transcription by SREBP-1a and Sp1 is reminiscent of the cofactor interactions utilized by the cAMP/PKA-activated transcription factor CREB. Both recruitment of TFIID and CBP are required for CREB activation, thus it appears that CREB embodies functions of both Sp1 and SREBP-1a (Chrivia et al. 1993; Ferreri et al. 1994; Nakajima et al. 1997b). Although CREB appears to activate transcription efficiently on naked DNA templates and interacts with a CBP–RNA Pol II complex (Kee et al. 1996; Nakajima et al. 1997a), our results suggest that the CBP-containing coactivator recruited by SREBP-1a performs a chromatin-dependent function required for SREBP-1a/Sp1 synergy.
Figure 6.

Model for cofactor requirements necessary for synergistic activation by SREBP-1a and Sp1 on the LDLR chromatin template. Chromatin-mediated repression of transcription could be alleviated by the recruitment of a CBP-containing coactivator by SREBP, which may facilitate access of the transcriptional machinery to a nucleosomal template. Also depicted is the well-characterized recruitment of TFIID by Sp1 via TAFII130 to nucleate formation of the preinitiation complex or aid in reinitiation of transcription. The HAT activity of CBP may participate in remodeling of the nucleosomal structure, whereas the function of the other components of the SREBP-binding coactivator remains to be established.
Chromatin-mediated regulation of transcription may be a strategy utilized by several classes of transcription factors responding to various signaling pathways. Nuclear receptors, STATs, and Jun/Fos, among others have been shown recently to functionally interact with CBP and may require histone-modifying activities to regulate their target genes (Arias et al. 1994; Kwok et al. 1994; Bannister and Kouzarides 1995; Bannister et al. 1995; Chakravarti et al. 1996; Kamei et al. 1996; Zhang et al. 1996; Heery et al. 1997; Torchia et al. 1997; Korzus et al. 1998; Kraus and Kadonaga 1998; Kurokawa et al. 1998). Additionally, NF-κB was found to interact with p300/CBP (Perkins et al. 1997) and was recently shown to synergize with Sp1 on the HIV–LTR in a chromatin-dependent manner (Pazin et al. 1996; Sheridan et al. 1997). Interestingly, p300/CBP recruited by NF-κB is associated with a cyclin-dependent kinase activity (Perkins et al. 1997), suggesting that multiple biochemical activities may be contained within p300/CBP complexes. We are currently investigating whether the functional requirements for NF-κB and SREBP-1a synergy with Sp1 are similar.
In vitro DNase I footprinting showed strong cooperativity in binding of Sp1 and SREBP-1a to the nucleosomal LDLR template, and in vivo DMS footprinting studies also indicated a sterol-regulated cooperative binding of Sp1 and SREBPs to the LDLR promoter in vivo (data not shown). These footprinting results together with the multiple cofactor requirements suggest that cooperative recruitment of factors to the promoter may contribute to the mechanism of transcription activation in a chromatin-regulated environment. Preliminary footprinting experiments suggest that the SREBP binding coactivator exert little effect on DNA binding by SREBP-1a and Sp1 (data not shown). Similarily, little influence on the chromatin structure in the promoter was seen by the SREBP coactivator (data not shown). Chromatin-specific factors distinct from the CBP-containing coactivator, including ATP-dependent nucleosomal remodeling activities such as SWI/SNFs, NURF, CHRAC, ACF, and RSC (Laurent and Carlson 1992; Yoshinaga et al. 1992; Imbalzano et al. 1994; Kwon et al. 1994; Peterson et al. 1994; Tsukiyama and Wu 1995; Cairns et al. 1996; Ito et al. 1997; Varga-Weisz et al. 1997), may also play important roles in activation by Sp1/SREBP-1a by serving in a more general capacity to facilitate binding of activators and the core transcriptional machinery to chromatin templates. We suspect that one or more of these remodeling activities is present in the S-190 chromatin assembly extract and may contribute to the requirement observed for the S-190 extract to activate transcription on chromatin templates. Future studies will be aimed at elucidating the role of ATP-dependent remodeling factors in the synergistic activation by SREBP-1a/Sp1 on the LDLR chromatin template.
The CBP-containing coactivator harbors HAT activity and appears to be necessary for full activation by SREBP-1a/Sp1 on chromatin templates, while having little effect when using naked DNA templates. It is tempting to speculate that acetylation of histones by CBP may facilitate access of SREBP-1a and Sp1 to nucleosomal templates and aid in the recruitment of the preinitiation complex to the core promoter (Fig. 6). However, at present, we cannot conclude that the HAT activity in the CBP-containing coactivator is required for SREBP-1a/Sp1 synergy on chromatin templates. Addition of acetyl CoA and/or the histone deacetylase inhibitor TSA to the transcription reactions resulted in less than twofold effects, suggesting that our transcription system may already contain acetyl CoA and may be limited in histone deacetylase activity, or the histones could already be acetylated. It is also conceivable that HAT activity is not critical for the synergistic activation by SREBP-1a/Sp1 on chromatin templates. Further studies of the role of HAT activity in the chromatin-dependent synergistic activation by SREBP-1a/Sp1 will likely require the use of purified chromatin templates, purified or recombinant histone deacetylases and the development of HAT specific inhibitors. Other enzymatic activities, such as kinases, could also be associated with the CBP-containing coactivator and might modify histones or components of the transcriptional apparatus.
We are currently cloning and characterizing the genes encoding the polypeptides in the SREBP-1a activation domain-binding coactivator to elucidate their functional contribution in mediating SREBP-1a/Sp1 synergistic activation on chromatin templates. The active coactivator fraction contains CBP and at least 10 other stoichiometric polypeptides ranging from 220 to 30 kD. Preliminary peptide sequencing revealed that several of these proteins are novel human gene products. The eventual development of a purified chromatin transcription system should provide us with valuable tools for future detailed molecular studies of chromatin-dependent mechanisms of transcription regulation of natural genes by multiple activators.
Materials and methods
Chromatin assembly
The LDLR-derived template plasmid was assembled into chromatin as described (Kamakaka et al. 1993). Briefly, Drosophila embryo cytosolic high-speed supernatant (S-190) extract, Drosophila core histones purified from embryos by use of hydroxylapatite, Mg/ATP, and an ATP-regenerating system were added to supercoiled DNA template and assembly was performed for 4.5 hr at 27°C. The resulting chromatin was analyzed with 0.31, 0.93, 2.8, and 8.3 units of micrococcal nuclease (MNase, Sigma) digestion for 10 min at 20°C. A typical nucleosomal ladder was observed when separated on agarose-TBE gels and visualized with EtBr staining.
In vitro transcription
Transcription reactions were performed basically as described in (Kamakaka et al. 1993). After 4 hr and 30 min of chromatin template assembly, SREBP-1a (1–5 nm) or NF-κB p65/p50 heterodimers (4–5 nm) and Sp1 (2–5 nm) were added and allowed to incubate with the template for 30 min. For control transcription on naked DNA templates, the activators were added to identical amounts of template (0.6 nm) in the presence or absence of S-190 extract (30 μg) and Mg/ATP for 30 min at 27°C. HeLa nuclear extract, Drosophila H 0.4 m fraction (Hansen and Tjian 1995) or purified GTFs (Goodrich and Tjian 1994) were then added and allowed to incubate for 10 min at 27°C. Transcription was initiated by addition of NTPs (0.5 mm final concentration) in a final volume of 50 μl and allowed to proceed for 30 min at 27°C. The products were analyzed by primer extension (Lillie and Green 1989). For studies of the effect of adding acetyl CoA and TSA to SREBP-1a/Sp1-driven transcription reactions with the LDLR chromatin template, acetyl CoA (1, 10, or 100 μm) or TSA (0.01, 0.1, or 1 μm) were added alone or in combination.
Expression and purification of GTFs
The human recombinant/highly purified transcription system is composed of recombinant human TFIIA (5 ng), TFIIB (5 ng), TFIIE56 (2.5 ng), TFIIE34 (10 ng), TFIIF (7.5 ng), and RNA Pol II (24 ng) purified to the phenyl column stage from HeLa nuclear pellets as described (Goodrich and Tjian 1994), TFIIH (15 ng) purified from either HeLa nuclear or cytoplasmic extracts as described (Goodrich and Tjian 1994), immunopurified TFIID (25 ng) and purified CRSP-containing cofactor fraction (100 ng). Recombinant human TFIIA was generated as described (Ozer et al. 1994). Recombinant human TFIIB, TFIIE56, TFIIE34, and TFIIF were expressed and purified as described (Goodrich and Tjian 1994). Human TFIID was immunoprecipitated with mAb 3A6, raised against hTAFII130, from either PC 1.0 m or from a DE-52 1.0 m KCl-eluted fraction from PC 1.0 m. The complex was washed in 0.8 m KCl HEG [20 mm HEPES (pH 7.6), 0.1 mm EDTA, 10% glycerol] and eluted with an epitope-containing peptide in 0.15 m KCl HEGN (HEG with 0.1% NP-40). The CRSP-containing cofactor fraction was purified from PC 1.0 m, followed by DE-52 FT at 0.1 m, POROS-heparin gradient, and finally over POROS-HS. The activity eluted between 0.38 m and 0.5 m KCl HEG and dialyzed to 0.1 m KCl HEG. Purification, cloning, and definition of CRSP will be published elsewhere (S. Ryu and R. Tjian, in prep.).
Generation of SREBP-1a and Sp1 activation domain fusions to GST
The first 50 amino acids of human SREBP-1a and the activation domain A of Sp1 (amino acids 83–262) were amplified by PCR and ligated to pGEX-2TK (Pharmacia). The resulting GST fusion was expressed in Escherichia coli (BL21) and purified over glutathione–Sepharose beads.
Identification of the SREBP-binding domain of CBP
His-tagged recombinant CBP1-682 was expressed in E. coli (BL21). Cells were lysed by French press in 100 mm NaCl, 25 mm Imidazole, 20 mm Tris HCl (pH 7.8), 1 mm PMSF, 1 mm DTT. The recombinant protein was then purified by nickel affinity chromatography followed by POROS-HS ion exchange chromatography. The protein was dialyzed against 100 mm NaCl, 20 mm Tris HCl (pH 7.8). Purified CBP1-682 was then partially digested by endoproteinase Glu-C (Staphlococcus aureus V8 protease; Promega) at 0.01 μg of enzyme per 100 μg of CBP1-682 for 2.5 hr at 37°C resulting in fragments ranging from 5 kD–60 daltons. These fragments were then tested for binding to GST–SREBP1a (amino acids 1–50). The smallest fragment that specifically bound the SREBP fusion was concentrated and partially purified by POROS-HS ion exchange chromatography followed by reverse-phase HPLC. Its exact sequence was determined by amino-terminal sequencing and electro-spray mass spectroscopy to be amino acids 548–682. The DNA sequence corresponding to CBP548–682 was amplified by PCR and subcloned into the pAED4 plasmid.
Expression and purification of CBP548–682
Recombinant CBP548–682 was expressed in E. coli (BL21 pLysS) which were lysed by French press in 25 mm NaCl, 20 mm MES (pH 6.0), 2.5 mm EDTA, 1 mm DTT, 1 mm AEBSF. DNA was partially removed by 0.2% poly(ethylenimine) precipitation. The supernatant was first passed through a fastflow Q-sepharose column then loaded onto a FPLC POROS-HS column. Bound CBP548–682 was eluted by salt gradient 20–500 mm NaCl in buffer [20 mm MES (pH 6.0), 2.5 mm EDTA, 1 mm DTT]. Recombinant CBP548–682 (5 μg) was added to GST- or GST–SREBP-1a (amino acids 1–50) beads and incubated with gentle mixing for 30 min at 4°C. The beads were washed extensively with 0.1 m KCl HEGN and analyzed by SDS-PAGE and Coomassie Brilliant blue staining. For peptide inhibition of transcription, the various activators (1–2 nm) were incubated with purified recombinant CBP548–682 in 0.1 m KCl HEG for 20 min at room temperature prior to addition to the transcription reactions.
Generation of recominant CBP
The cDNA encoding mouse CBP was inserted into a baculovirus transfer vector (pAcSG2) in frame with two amino-terminal FLAG-epitope (Kodak) sequences. Baculovirus was produced, screened for CBP expression, and amplified, according to protocol supplied by the manufacturer (Invitrogen). One liter of Sf9 cells was infected with CBP baculovirus (5 MOI) for 48 hr. Nuclear extract was prepared from pelleted cells (Dignam et al. 1983) and incubated with 500 μl of FLAG-antibody beads (Kodak) for 4 hr at 4°C. After extensive washing, recombinant CBP was eluted using 0.5 mg/ml FLAG peptide in 0.1 m KCl HEG with 0.01% NP-40.
Purification and identification of proteins binding to the SREBP-1a activation domain
HeLa nuclear extract was loaded onto P11 phosphocellulose in 0.1 m KCl HEG. The column was eluted with 0.2 m, 0.5 m, and 1.0 m KCl HEG steps. Cofactor activity and CBP immunoreactivity was found to copurify in the PC 0.5 m fraction, which was dialyzed to 0.1 m KCl HEG. One milliliter of HeLa nuclear extract or 500 μl PC 0.5 m fraction was applied to 20 μl of either GST, GST–Sp1 (amino acids 83–262), or GST–SREBP-1a (amino acids 1–50) beads. Gentle mixing was performed at 4°C for 2 hr, the flowthrough fraction collected and the beads washed extensively with 0.3 and 0.5 m KCl HEGN (HEG with 0.1% NP-40) and analyzed by SDS-PAGE followed by silver staining and immunoblotting with affinity-purified anti-CBP, anti-RNA Pol II, anti-N-CoA1 and 2, anti-P/CAF, anti-RHA, and anti-SWI/SNFs. For the depletion experiments, 500 μl of PC 0.5 m was mixed with 100 μl of GST or GST–SREBP-1a (amino acids 1–50) beads for 2 hr, then the supernatant was transferred to 100 μl of fresh beads and mixed for another 2 hr at 4°C. The double-depleted PC 0.5 m was then tested for cofactor activity in chromatin transcription reactions and for presence of CBP by immunoblotting. For elution of the GST–SREBP-1a (amino acids 1–50) associated proteins, 2 vol of 0.1 m KCl HEGN with 0.1% deoxycholate or 250 μm CBP548–682 peptide was added, and nutated at 4°C for 4 hr. The supernatant was collected and dialyzed against 0.1 m KCl HEG for 4 hr.
Evaluation of HAT activity in the GST pull-downs
One milliliter of HeLa nuclear extract was incubated with 20 μl of GST–SREBP (amino acids 1–50) and GST–Sp1A (amino acids 83–262) beads followed by extensive washing as described above. HAT activity was assayed by incubating the beads with 1–2 μg of purified Drosophila core histones, 1 μCi 3H-labeled acetyl CoA, and 10 mm Na butyrate in buffer A [50 mm Tris-HCl (pH 8.0), 50 mm KCl, 0.5 mm EDTA, 10% glycerol, 1 mm DTT, 1 mm PMSF] for 30 min at 27°C. The reactions were analyzed by separation on a 15% SDS–polyacrylamide gel followed by Coomassie staining and autoradiography for 5 days.
Acknowledgments
We thank D. Reinberg, P. Lieberman, and R. Goodman for providing plasmids encoding hTFIIAαβ, hTFIIAγ, and mouse CBP, respectively. The pAED4 plasmid was a gift of D.S. Doering. The NF-κB subunits were kindly supplied by Z. Cao. We thank Y. Nakatani for the kind gift of P/CAF antiserum. The antisera directed toward N-CoA1 and 2 were generously provided by M.G. Rosenfeld. The RHA antiserum was a kind gift of M. Montminy. Antisera directed towards hSWI/SNF subunits were kindly provided by G. Crabtree. We thank T. Osborne for providing the SREBP-1a expression plasmid and LDLR reporter construct used in the transfection studies. We thank L. Kraus for reagents, protocols, and helpful suggestions. We thank D. Rio, P. Kaufman, B. Lemon, I. Haviv, M. Rabenstein, T. O’Brien, and G. Cutler for critical reading of the manuscript. A.M.N. and J.D.O. are fellows with the Damon Runyon-Walther Winchell Cancer Research Foundation, J.Z. and S.S. receive support from the Deutsche Forschungsgemeinschaft, D.A. is a Merck Fellow with the Life Sciences Research Foundation, and K.R. receives a fellowship from the Medical Research Council of Canada. This work was funded in part by National Institutes of Health grant GM46995 to J.T.K. and grants CA25417 and HL20948.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jmlim@uclink4.berkeley.edu; FAX (510) 643-9547.
References
- Arias J, Alberts AS, Brindle P, Claret FX, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- Athanikar JN, Sanchez HB, Osborne TF. Promoter selective transcriptional synergy mediated by sterol regulatory element binding protein and Sp1: A critical role for the Btd domain of Sp1. Mol Cell Biol. 1997;17:5193–5200. doi: 10.1128/mcb.17.9.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- Briggs MR, Yokoyama C, Wang X, Brown MS, Goldstein JL. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Dawson PA, Hofmann SL, van der Westhuyzen DR, Sudhof TC, Brown MS, Goldstein JL. Sterol-dependent repression of low density lipoprotein receptor promoter mediated by 16-base pair sequence adjacent to binding site for transcription factor Sp1. J Biol Chem. 1988;263:3372–3379. [PubMed] [Google Scholar]
- Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Hansen SK, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hoey T, Weinzierl RO, Gill G, Chen JL, Dynlacht BD, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Bulger M, Kadonaga JT. Potentiation of RNA polymerase II transcription by Gal4–VP16 during but not after DNA replication and chromatin assembly. Genes & Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kee BL, Arias J, Montminy MR. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes & Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey G, Mahfoudi A, Wahli W. Functional interactions of peroxisome proliferator-activated receptor, retinoid-X receptor, and Sp1 in the transcriptional regulation of the acyl-coenzyme-A oxidase promoter. Mol Endocrinol. 1995;9:219–231. doi: 10.1210/mend.9.2.7776972. [DOI] [PubMed] [Google Scholar]
- Kurokawa R, Kalafus D, Ogliastro MH, Kioussi C, Xu L, Torchia J, Rosenfeld MG, Glass CK. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Laurent BC, Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes & Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- Lillie JW, Green MR. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Look DC, Pelletier MR, Tidwell RM, Roswit WT, Holtzman MJ. Stat1 depends on transcriptional synergy with Sp1. J Biol Chem. 1995;270:30264–30267. doi: 10.1074/jbc.270.51.30264. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997a;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Parvin JD, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes & Dev. 1997b;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Andresen JM, Hansen SK, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes & Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- Ozer J, Moore PA, Bolden AH, Lee A, Rosen CA, Lieberman PM. Molecular cloning of the small (α) subunit of human TFIIA reveals functions critical for activated transcription. Genes & Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- Pascal E, Tjian R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes & Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, Jones KA. NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes & Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez HB, Yieh L, Osborne TF. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J Biol Chem. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- Sato R, Yang J, Wang X, Evans MJ, Ho YK, Goldstein JL, Brown MS. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1) J Biol Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- Sauer F, Tjian R. Mechanisms of transcriptional activation: Differences and similarities between yeast, Drosophila, and man. Curr Opin Genet Dev. 1997;7:176–181. doi: 10.1016/s0959-437x(97)80126-8. [DOI] [PubMed] [Google Scholar]
- Sauer F, Hansen SK, Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, Jones KA. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes & Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- Sheridan PL, Mayall TP, Verdin E, Jones KA. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes & Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC, Van der Westhuyzen DR, Goldstein JL, Brown MS, Russell DW. Three direct repeats and a TATA-like sequence are required for regulated expression of the human low density lipoprotein receptor gene. J Biol Chem. 1987;262:10773–10779. [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- Tjian R, Maniatis T. Transcriptional activation: A complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional coactivator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–560. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Wang X, Briggs MR, Hua X, Yokoyama C, Goldstein JL, Brown MS. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J Biol Chem. 1993;268:14497–14504. [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Yieh L, Sanchez HB, Osborne TF. Domains of transcription factor Sp1 required for synergistic activation with sterol regulatory element binding protein 1 of low density lipoprotein receptor promoter. Proc Natl Acad Sci. 1995;92:6102–6106. doi: 10.1073/pnas.92.13.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]