Abstract
Members of the TGF-β superfamily influence a broad range of biological activities including stimulation of wound healing and inhibition of cell growth. TGF-β signals through type I and II receptor serine/ threonine kinases and induces transcription of many genes including plasminogen activator inhibitor-1 (PAI-1). To identify proteins that participate in TGF-β-induced gene expression, we developed a novel retrovirus-mediated expression cloning strategy; and using this approach, we established that transcription factor μE3 (TFE3) is involved in TGF-β-induced activation of the PAI-1 promoter. We showed that TFE3 binds to an E-box sequence in PE2, a 56-bp promoter fragment of the PAI-1 promoter, and that mutation of this sequence abolishes both TFE3 binding as well as TGF-β-dependent activation. TFE3 and Smad3 synergistically activate the PE2 promoter and phosphorylated Smad3 and Smad4 bind to a sequence adjacent to the TFE3-binding site in this promoter. Binding of both TFE3 and the Smad proteins to their cognate sequences is indispensable for TGF-β-inducible activation of the PE2 promoter. Hence, TFE3 is an important transcription factor in at least one TGF-β-activated signal transduction pathway.
Keywords: TFE3, Smads, TGF-β, E box, PAI-1
TGF-β has a diverse range of biological activities including inhibition of cell growth, induction of cell differentiation, inhibition of the immune response, and production of extracellular matrix proteins. TGF-β rapidly induces transcription of extracellular matrix proteins such as plasminogen activator inhibitor-1 (PAI-1) and cell-cycle inhibitors such as p15INK4B and p21WAF1/CIP (Attisano et al. 1994; Hannon and Beach 1994; Datto et al. 1995; Li et al. 1995). TGF-β signals through the sequential activation of two homodimeric cell-surface receptors, termed type I and type II (TβRI and TβRII), both of which are serine–threonine protein kinases (Franzen et al. 1993; Lin and Lodish 1993; Wrana et al. 1994; Luo and Lodish 1996; Weis-Garcia and Massagué 1996). Ligand-activated TβRI phosphorylates conserved serines at the carboxyl termini of either Smad2 or Smad3, which promote their binding to one or more molecules of Smad4, a common partner for all phosphorylated Smads involved in signaling by both TGF-β and bone morphogenetic proteins (Heldin et al. 1997; Massagué et al. 1997; Attisano and Wrana 1998). Smad complexes then enter the nucleus and activate transcription of a variety of genes.
Ectopic expression of Smad2 and Smad4 activates transcription of a reporter gene driven by the artificial TGF-β responsive promoter 3TP, and addition of TGF-β further stimulates its expression (Lagna et al. 1996; Macias-Silva et al. 1996). Similarly, overexpression of Smad3 and Smad4 activates transcription from the TGF-β inducible PAI-1 promoter (Zhang et al. 1996; X. Liu et al. 1997). Recent evidence indicates that Smad3 and Smad4 can directly bind to specific DNA sequences in either artificial or natural TGF-β-inducible promoters and thus activate gene transcription (Yingling et al. 1997; Dennler et al. 1998; Vindevoghel et al. 1998; Zawel et al. 1998). However, it is not yet clear whether binding only of Smad proteins to DNA is sufficient to confer maximal TGF-β-induced transcription. Given the diversity of genes that are activated by TGF-β, it seems likely that other transcription factors, some possibly expressed only in certain kinds of cells, partner with phosphorylated Smads to induce the expression of different genes. In Xenopus, activin induces phosphorylation of Smad2, which then forms a complex with Smad4 and the transcription factor FAST-1. This complex then binds to the promoter of the developmentally regulated gene Mix2 and induces its transcription (X. Chen et al. 1996, 1997; F. Liu et al. 1997).
PAI-1 is a component of the extracellular matrix and also plays an important role in regulating blood coagulation. Addition of TGF-β to cultured human Hep G2 hepatoma cells dramatically induces PAI-1 gene expression (Westerhausen et al. 1991). Coexpression of Smad2 or Smad3 with Smad4 also increases the expression of reporter genes driven by the PAI-1 promoter (Y. Chen et al. 1996; Lagna et al. 1996; Zhang et al. 1996), and mutation of Smad4 leads to loss of the TGF-β response (F. Liu et al. 1997; Zawel et al. 1998). Thus, Smad proteins are involved in TGF-β-induced transcription of the PAI-1 gene. AP-1 binding sites as well as an E-box sequence have been implicated in TGF-β-induced transcription of the PAI-1 gene (Keeton et al. 1991; Riccio et al. 1992). However, little is known about how transcription factors in concert with Smad proteins stimulate TGF-β-dependent PAI-1 transcription.
We developed a novel expression cloning system involving an engineered TGF-β responsive cell line and a retroviral cDNA library. Using this system, we cloned a transcription factor, TFE3, which when ectopically expressed activates TGF-β-induced expression of the PAI-1 gene. We identified two E-box DNA sequences in the natural PAI-1 promoter that specifically bind to TFE3. Ectopic expression of TFE3 increases TGF-β-dependent expression of a reporter gene driven by a natural PAI-1 promoter fivefold, whereas mutation of the E-box sequence in a fragment of the PAI-1 promoter completely abolishes TGF-β-inducible transcription. Moreover, we showed that TFE3 and Smad3 and Smad4 synergize in enhancing TGF-β-dependent transcription from this minimal 56-bp promoter fragment, a synergy strictly dependent on the phosphorylation of serine residues at the carboxyl terminus of Smad3. Furthermore, we show that within a 36-bp PAI-1 promoter, a complex of Smad3 and Smad4 bind to a sequence adjacent to the TFE3-binding site. Binding of this DNA by a Smad3–Smad4 complex requires phosphorylation of the carboxyl terminus of Smad3, and binding of both TFE3 and Smad proteins to this promoter is essential for TGF-β-inducible transcription. Together, our data show that TFE3 and Smad proteins synergistically cooperate in transcription of at least one TGF-β-inducible gene.
Results
Isolation of cell clones with constitutive TGF-β signaling on infection with a retroviral cDNA library
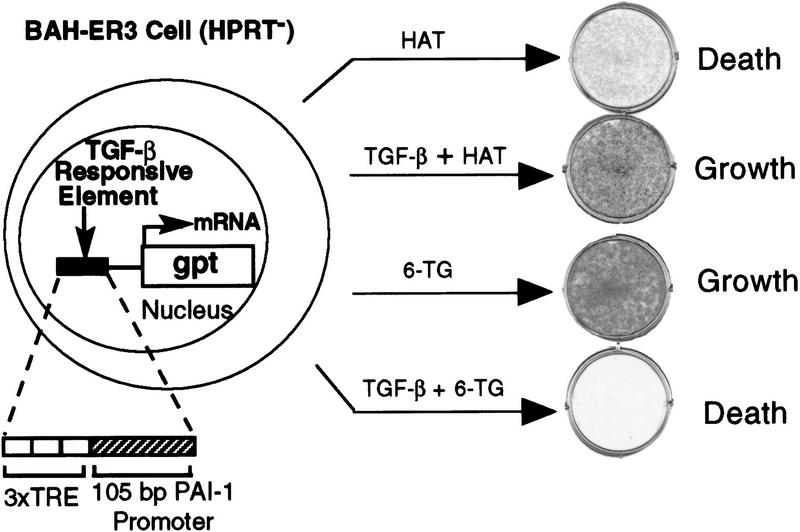
To identify proteins that participate in the TGF-β-induced transcription of the PAI-1 gene, we established an expression cloning system involving an engineered cell line and a high titer retroviral cDNA library. Our expression cloning strategy was based on the finding that overexpression of either Smad2 or Smad3 alone or together with Smad4 induces TGF-β responses in the absence of ligand (Y. Chen et al. 1996; Lagna et al. 1996; Zhang et al. 1996; X. Liu et al. 1997); thus overexpression of other proteins in the TGF-β-signaling pathway might also induce transcription of genes normally activated by TGF-β. To infect the engineered TGF-β-responsive BAH-gpt ( uanosine phosphoribosyl transferase) cells (Hocevar and Howe 1996) with a retroviral cDNA library, we stably introduced into the cells the cDNA encoding the receptor for the ecotropic murine Moloney retrovirus (Baker et al. 1992). Growth of the resulting BAH-ER3 cells, like parental BAH-gpt cells, was not inhibited by TGF-β but transcription of the PAI-1 gene was fully inducible by TGF-β. These cells are deficient in hypoxanthine phosphoribosyl transferase (HPRT), but contain the bacterial gpt gene under the control of 3TP, an artificial TGF-β inducible promoter (Wrana et al. 1992; Hocevar and Howe 1996) (Fig. 1). Thus, BAH-ER3 cells grow in HAT medium only in the presence of TGF-β. In contrast, addition of 6-thioguanine (6-TG) to normal medium kills the cells, but only in the presence of TGF-β, as gpt converts 6-TG to a toxic product (Fig. 1).
uanosine phosphoribosyl transferase) cells (Hocevar and Howe 1996) with a retroviral cDNA library, we stably introduced into the cells the cDNA encoding the receptor for the ecotropic murine Moloney retrovirus (Baker et al. 1992). Growth of the resulting BAH-ER3 cells, like parental BAH-gpt cells, was not inhibited by TGF-β but transcription of the PAI-1 gene was fully inducible by TGF-β. These cells are deficient in hypoxanthine phosphoribosyl transferase (HPRT), but contain the bacterial gpt gene under the control of 3TP, an artificial TGF-β inducible promoter (Wrana et al. 1992; Hocevar and Howe 1996) (Fig. 1). Thus, BAH-ER3 cells grow in HAT medium only in the presence of TGF-β. In contrast, addition of 6-thioguanine (6-TG) to normal medium kills the cells, but only in the presence of TGF-β, as gpt converts 6-TG to a toxic product (Fig. 1).
Figure 1.
TGF-β-regulated growth of BAH-ER3 cells in the presence of drug selection. On day 0, BAH-ER3 cells were seeded at a density of 5 × 104 cells/well in a six-well plate in DMEM containing 10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin. On day 1, the cells were switched to medium with 1× HAT or 6-TG (30 μg/ml) with or without 200 pm TGF-β as indicated. On day 9, the growing cells were stained with crystal violet. (TRE) Phorbol ester TPA response element; (gpt) guanosine phosphoribosyl transferase.
Two million BAH-ER3 cells were infected with a retroviral cDNA library prepared from HPRT-deficient HT1080 cells, and grown in HAT medium in the absence of TGF-β. After 2 weeks, we isolated 12 HAT-resistant clones. We reasoned that if overexpression of a particular cDNA caused the HAT-resistant phenotype, then we could rescue the cDNA sequence by packaging of the retroviral RNA by the Gag, Pol, and Env proteins derived from superinfecting wild-type Moloney retroviruses (Rasheed 1995). The rescued retrovirus transducing the cDNA should allow transfer of the HAT-resistant phenotype to fresh BAH-ER3 cells. Retroviruses produced by 3 of the 12 lines, including HATR4 and HATR7 cells, caused BAH-ER3 cells to acquire the ability to grow in HAT medium in the absence of TGF-β and to be killed in normal medium in the presence of 6-TG (Fig. 2A).
Figure 2.
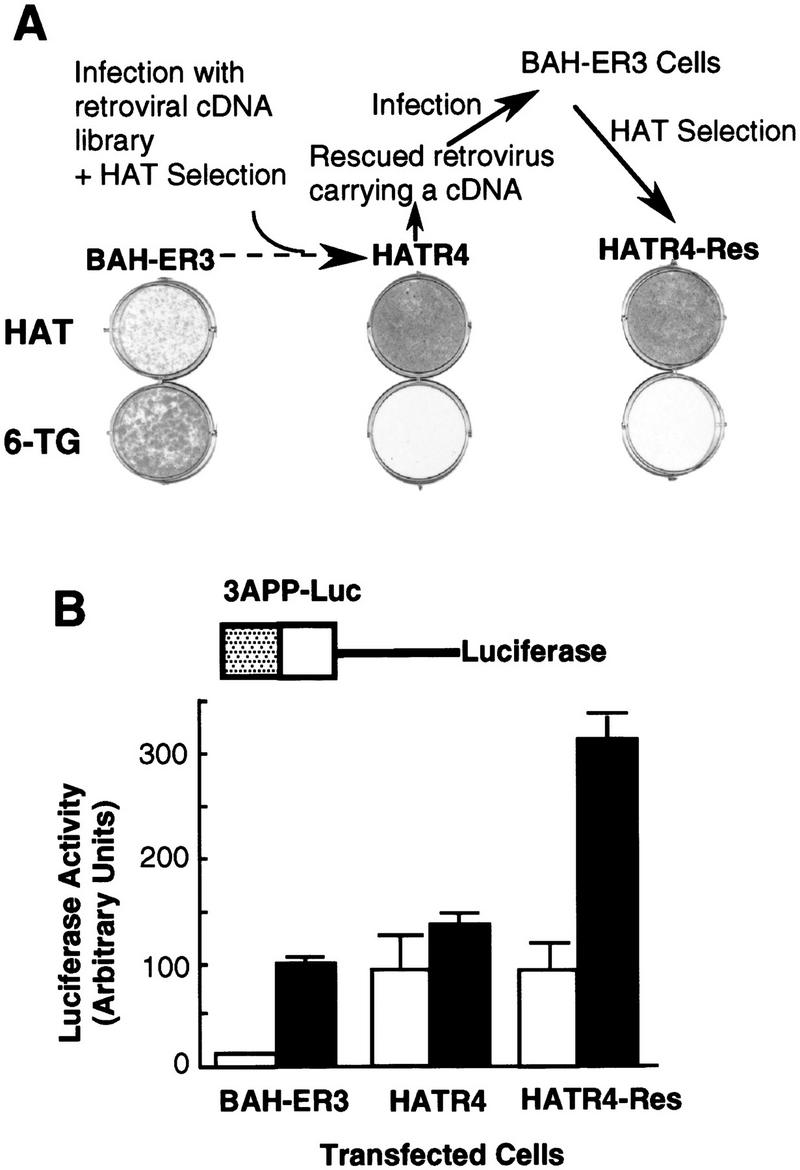
Isolation of a HAT-resistant cell clone that activates the TGF-β-inducible 3TP promoter in the absence of TGF-β. (A) After infection of BAH-ER3 cells with a retroviral cDNA library, the cells were grown in HAT medium for 2 weeks. A HAT-resistant clone, HATR4, was isolated, and then infected with wild-type Moloney retrovirus; the supernatant containing the rescued retrovirus was used to infect normal BAH-ER3 cells as described in Materials and Methods. Infected cells were also subjected to HAT selection and the resulting HAT-resistant cells, HATR4–Res cells, were isolated. BAH-ER3, HATR4, and HATR4–Res cells were plated at 5 × 104 cells/well in a 6-well plate; HAT medium or 6-TG medium was added as indicated, and cells were incubated for 10 days in the absence of TGF-β. Growing cells were stained with crystal violet. (B) On day 0, 105 cells were plated in each well of a 12-well plate. On day 2, the cells in each well were transfected with 2 μg of 3APP–Luc DNA and 0.5 μg of pSVβ. After overnight culture the cells were switched to serum-free medium with (█) or without (□) 200 pm TGF-β as indicated, and then incubated for 20 hr before being harvested for luciferase and β-galactosidase assays as described in Materials and Methods. Luciferase activities, plotted in arbitrary units, have been normalized to β-galactosidase activity. Each value represents an average of duplicate samples, and the error bar denotes the standard deviation of the duplicates.
The HAT-resistant HATR4-Res cells were derived from the BAH-ER3 cells infected with the retrovirus rescued from HATR4 cells (Fig. 2A). If constitutive expression of the gpt gene is caused by constitutive activation of the TGF-β-inducible promoter upstream of the gpt gene, then we expect that a similar TGF-β-inducible promoter in a luciferase reporter construct should drive expression of luciferase even in the absence of TGF-β. To test this hypothesis, we transfected a TGF-β-inducible luciferase reporter construct, 3APP–Luc, into parental BAH-ER3 cells, HATR4 cells, and the rescued HATR4–Res cells (Fig. 2B). In BAH-ER3 cells, expression of the TGF-β-inducible 3APP–Luc is low in the absence of TGF-β and induced sevenfold by TGF-β. In contrast, in the absence of TGF-β expression of 3APP–Luc is much higher in both HATR4 and HATR4–Res cells than that in BAH-ER3 cells. Addition of TGF-β has little effect on expression of the reporter gene in HATR4 cells. In HATR4–Res cells, which may contain multiple copies of the retroviral genome, TGF-β stimulates reporter gene expression threefold (Fig. 2B). These results suggest that a retrovirus-introduced cDNA is responsible for HAT resistance as well as constitutive expression of the normally TGF-β-inducible reporter gene.
Cloning of TFE3 from the cell clone with constitutive TGF-β signaling
To clone the cDNA responsible for the HAT-resistant phenotype of HATR4 cells by use of PCR, we amplified genomic DNA with a pair of oligonucleotides flanking the multiple cloning site in the retroviral vector. A single 2.7-kb DNA fragment was amplified from the genomic DNA of HATR4 cells, but not from control BAH-ER3 cells (data not shown). Sequencing of this DNA fragment indicates that it encodes the full-length transcription factor μE3 (TFE3), a ubiquitously expressed basic helix–loop–helix transcription factor originally isolated as a factor binding to the E-box sequence (CACGTG) in the enhancer of an immunoglobulin gene (Beckmann et al. 1990; Zhao et al. 1993). A 1.9-kb cDNA encoding the full-length TFE3 was also cloned from HATR7 cells (data not shown).
TFE3 enhances TGF-β-dependent activation of the PAI-1 promoter
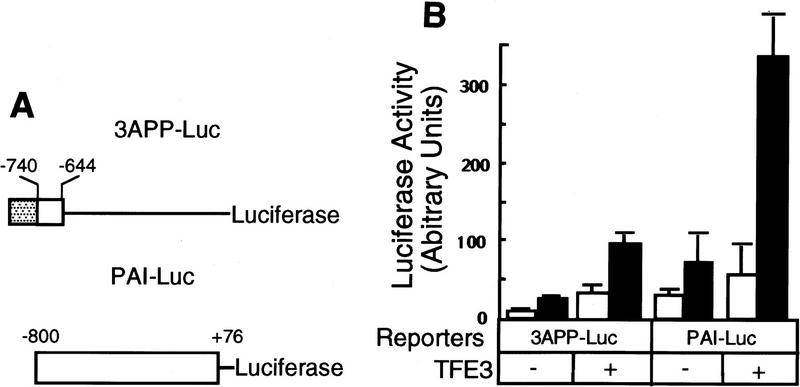
To determine whether TFE3 activates the expression of the luciferase reporter gene driven by the natural PAI-1 promoter, which is well induced by TGF-β (Keeton et al. 1991; Westerhausen et al. 1991; Riccio et al. 1992), we transfected PAI-Luc into BAH-ER3 cells. Coexpression of TFE3 enhanced TGF-β-independent expression of PAI–Luc less than twofold (Fig. 3B). Importantly, cotransfection of TFE3 enhanced PAI-1 promoter activity fivefold in the presence of TGF-β. This suggests that TFE3 is involved in TGF-β-induced transcription of the PAI-1 gene.
Figure 3.
TFE3 activates the PAI-1 promoter in a TGF-β-dependent fashion. (A) A diagram of the luciferase reporter genes driven by various promoters. The three tandem repeats of a 7-bp AP1-binding site [(stippled box) TGA(G/C)TCA] separated by an XbaI site were inserted upstream of −740 to −644 fragment of the PAI-1 promoter (open box) in the 3APP–Luc construct. Hence, the short AP1-binding sequence was used to replace the 32-bp fragment containing an AP1-binding sequence in the 3TP promoter (Wrana et al. 1992) to eliminate the potential influence of sequences other than the AP1-binding site. (B) Activation of PAI–Luc expression by TFE3 is dependent on TGF-β. BAH-ER3 cells were transfected as described in Fig. 2B. Each well received 1.5 μg of reporter gene, 0.5 μg of pSVβ, and also 1.0 μg of plasmid encoding TFE3 as indicated. The total amount of DNA per well was adjusted to 3.0 μg. Transfected cells were treated with (█) or without (□) TGF-β, and processed for both luciferase and β-galactosidase assays as described in Fig. 2B.
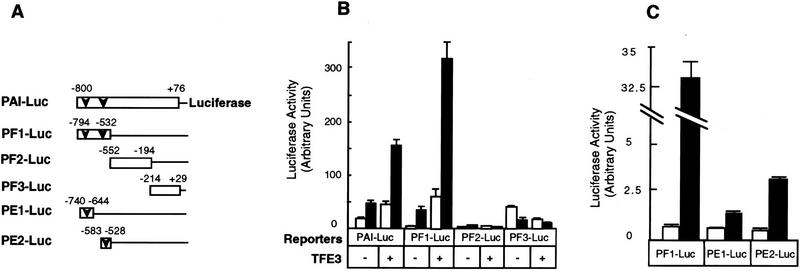
To identify the minimal element(s) in the PAI-1 promoter that are responsive to both TFE3 and TGF-β, we tested the activity of three fragments of the full-length PAI-1 promoter. Figure 4B shows that activity of fragment PF1 (bases −794 to −532) of the PAI-1 promoter, which contains two perfect TFE3-binding E-box sequences (CACGTG), is stimulated fivefold by TGF-β. Notably, coexpression of TFE3 enhances PF-1 promoter activity dramatically in the presence of TGF-β but only slightly in the absence of TGF-β. In contrast, segments PF2 and PF3, containing bases −552 to −194 and −214 to +29, respectively, are not responsive to TGF-β and are unaffected by TFE3 overexpression (Fig. 4B). Subdivision of the PF1 promoter into smaller pieces showed that at least two subfragments, PE1 and PE2, each of which contains one E box, are responsive to TGF-β; this experiment (Fig. 4C) was done by use of Hep G2 cells because we found that the expression of these luciferase reporter genes were more regulatable by TGF-β in this cell line. The higher level of expression of PF1–Luc compared with PE1–Luc and PE2–Luc is probably the result of the effect of tandem repeats of PE1 and PE2 in the PF1 promoter fragment.
Figure 4.
Identification of small subfragments, PE1 and PE2, of the PAI-1 promoter that are activated by TFE3 and TGF-β. (A) A diagram of the reporter constructs. (Open bars) PAI-1 promoter; (arrowhead) E-box sequence (CACGTG). BAH-ER3 cells (B) and Hep G2 cells (C) were transfected and the luciferase and β-galactosidase assays were carried out as detailed in Fig. 3B. (□) Without TGF-β; (█) with TGF-β.
E box, the TFE3-binding sequence, is essential for TGF-β-induced activation of the PE2 promoter
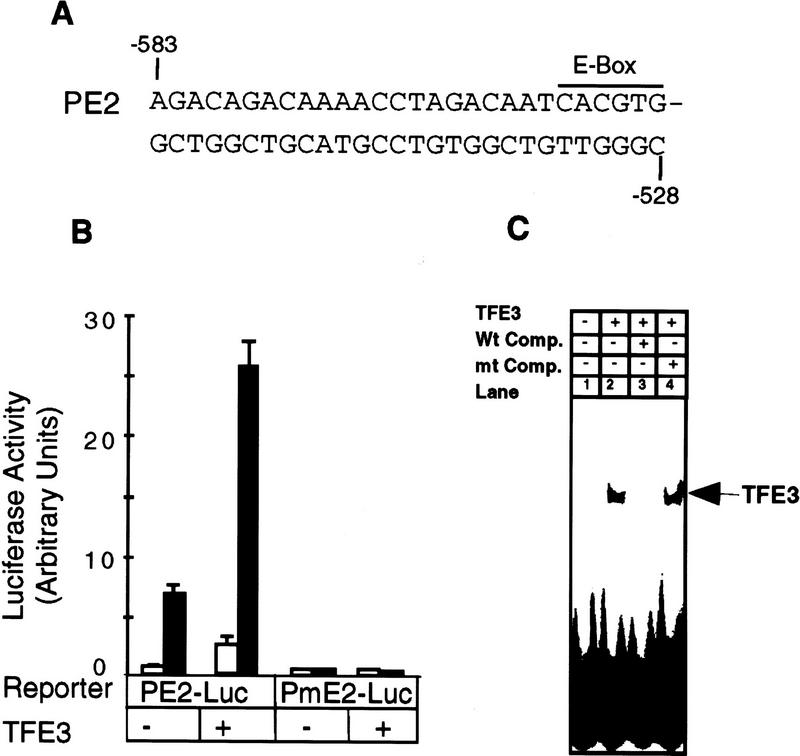
Each of the two TGF-β-responsive elements, PE1 and PE2, contains a perfect E-box sequence. We tested the importance of the E box in the PE2 promoter by transfecting luciferase reporter genes driven by the wild-type PE2 promoter (PE2–Luc) or a promoter with the mutant E box (PmE2-Luc, CACGTG → acCGac) (Fig. 5A). All activity of the PE2 promoter was dependent on the presence of a functional E box, because the mutant was inactive. In contrast, activity of the wild-type PE2 promoter was stimulated fourfold by TFE3 in the presence of TGF-β (Fig. 5B). This result is consistent with a previous report showing that the E-box sequence in the PE2 fragment of the PAI-1 promoter is critical for TGF-β-induced transcription of the PAI-1 gene (Riccio et al. 1992).
Figure 5.
The E-box sequence is essential for TFE3-mediated and TGF-β-dependent activation of the PE2 fragment of the PAI-1 promoter. (A) The sequence of the PE2 promoter. (B) Hep G2 cells were transfected with 1 μg of PE2–Luc or PmE2–Luc DNA (CACGTG → acCGac), 1 μg of pSVβ, and also 1.0 μg of plasmid encoding TFE3 as indicated. Transfected cells were treated with (█) or without (□) TGF-β, processed, and assayed as described in Fig. 3B. (C) TFE3 was synthesized in vitro from pET–TFE3 by the TNT T7 Coupled Reticulocyte Lysate System (Promega). Gel-shift reactions contained 3 μl of the in vitro TFE3 translation reaction and 1 μl of (4 × 103 cpm) of 32P-labeled PE2 DNA probe. Reactions in lanes 3 and 4 contained a 50-fold excess of wild type or mutant PE2 oligonucleotides, respectively.
The gel-shift assay in Figure 5C shows that TFE3 synthesized in vitro binds to a 32P-labeled PE2 DNA probe (Fig. 5, lane 2). Binding was competed completely by unlabeled wild-type PE2 oligonucleotides (Fig. 5, lane 3) but not by oligonucleotides bearing a scrambled mutation in the E-box sequence (CACGTG → acCGac; Fig. 5, lane 4). Together, these data suggest that the E-box sequence in the PE2 promoter is essential for the binding of TFE3 to the promoter as well as for TFE3- and TGF-β-dependent activation of the promoter.
TFE3 and Smad3–Smad4 synergize in TGF-β-dependent transcription
The data in Figure 6A show that TFE3 synergizes with Smad3 in enhancing TGF-β-dependent activation of the PE2 PAI-1 promoter. We transfected Hep G2 cells with the PE2-Luc reporter and various Smad constructs. Transfection of either Smad3 or Smad4 or both together had little effect on the PE2 promoter activity, either in the absence or presence of TGF-β. Transfection of a small amount of TFE3 plasmid DNA alone slightly stimulated the PE2 promoter activity in the presence of TGF-β. Importantly, cotransfection of TFE3 and Smad3, or TFE3, Smad3, and Smad4 together, markedly stimulated the PE2 promoter activity in the presence of TGF-β. In contrast, only a slight stimulation was observed in the absence of TGF-β. The reporter construct PmE2–Luc, containing a mutant E box, was inactive even after cotransfection of TFE3, Smad3, and Smad4 and stimulation with TGF-β.
Figure 6.
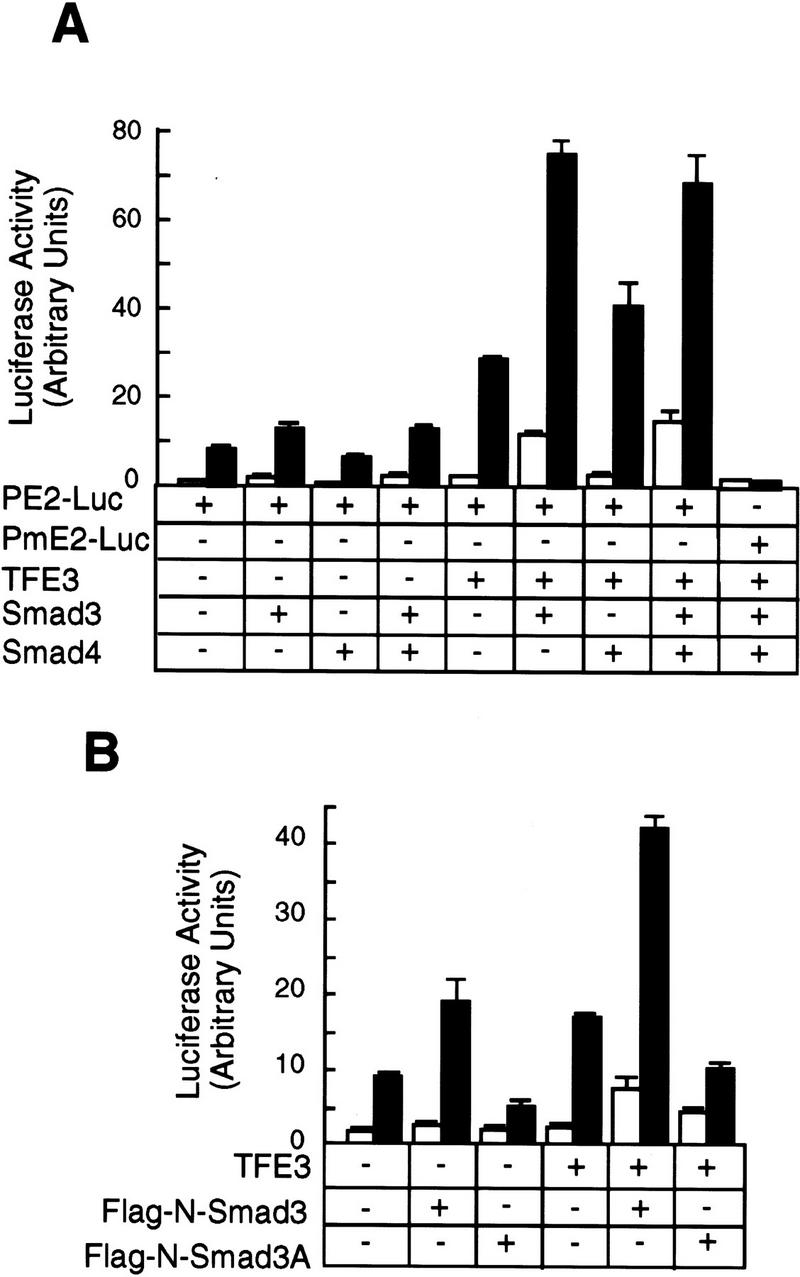
Synergy between TFE3 and Smad3 in the activation of the PE2 promoter. (A) Hep G2 cells were transfected as described in the legend to Fig. 5A. The following plasmids were used in transfection as indicated: 0.5 μg of plasmid encoding TFE3, 1 μg of plasmid encoding Smad3, and 1 μg of plasmid encoding Smad4; every well received 1 μg of PE2–Luc and 0.2 μg of pCMV–β-gal. The total amount of DNA was adjusted to 3.7 μg per well with pcDNA3. (B) Hep G2 cells were transfected with the following plasmids: 0.5 μg of TFE3, 1 μg of Flag-N–Smad3 or Flag-N–Smad3A; every well received 1 μg of PE2–Luc and 0.2 μg of pCMV-β. The total amount of DNA per well was adjusted to 3.7 μg with a control plasmid, pEXL–GFP. The cells were transfected, treated with (█) or without (□) TGF-β, and assayed as described for panel A.
TGF-β induces phosphorylation of the serine residues at the carboxyl terminus of Smad3. Phosphorylation is essential for signaling because overexpression of the mutant Smad3A, in which the three carboxy-terminal serines are changed to alanines, blocks the ability of TGF-β to inhibit cell division and stimulate the PAI-1 promoter (X. Liu et al. 1997). Consistent with these observations, Smad3A had little effect on the PE2 promoter activity, either in the absence or presence of TFE3 or TGF-β (Fig. 6B). Taken together, these results show a functional synergy between TFE3 and phosphorylated Smad3 in activation of the PE2 promoter.
Smad 4 and phosphorylated but not unphosphorylated Smad3 together bind to the PE2.1 element of the PAI-1 promoter
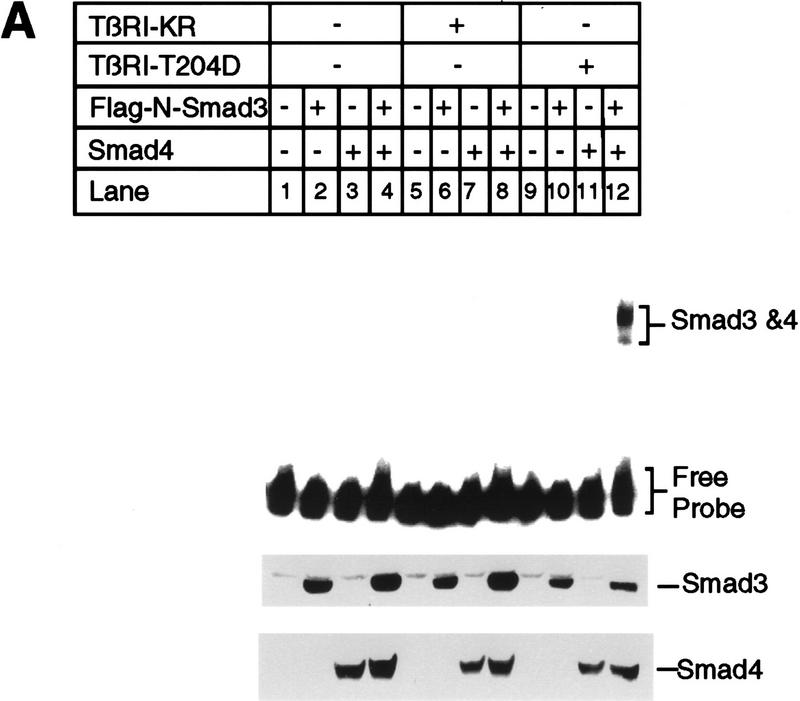
The PE2 fragment of the PAI-1 promoter, bases −583 to −528, contains a perfect E box at −561 to −556. As detailed below, we surmised that a Smad3–Smad4 complex binds to nucleotides within −583 to −528; thus, we tested the PE2.1 probe, containing two tandem segments of DNA spanning bases −586 to −551 of the PAI-1 promoter. The gel-shift experiment in Figure 7A shows that a complex of Smad4 and phosphorylated Smad3 binds to this DNA fragment. In this study we transfected Bosc23 cells with plasmids encoding Smad4 and/or Flag-tagged Smad3, together with the constitutively active type I TGF-β receptor, TβRI–T204D. Lysates from transfected cells were then incubated with the 32P-labeled PE2.1 probe and analyzed on a native polyacrylamide gel (Fig. 7).
Figure 7.
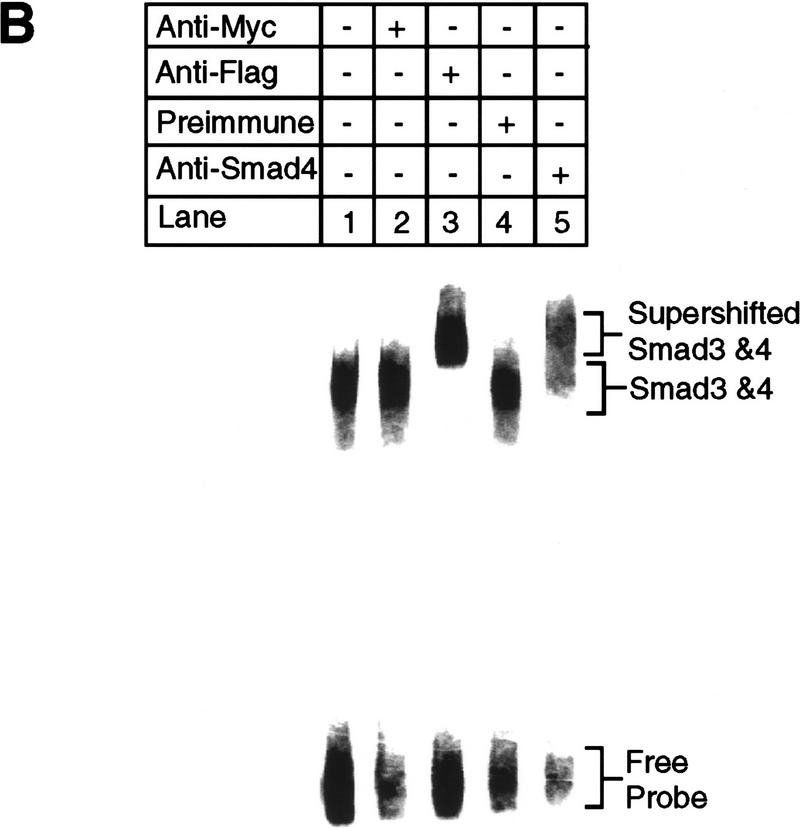
Smad3 and Smad4 together bind to the PE2.1 element of the PAI-1 promoter. (A) Bosc23 cells were transfected as described in Materials and Methods; cells in each dish were transfected with 2 μg of plasmid encoding Flag-N-Smad3 or Smad4 (pEXL–Smad4), and 1 μg of plasmid encoding TβRI-KR (pCMV5–TβRI–KR) or TβRI-T204D (pCMV5-TβRI-T204D) as indicated. The total amount of DNA for each dish was adjusted to 5 μg with pEXL–GFP. The gel-shift assay at top was carried out with the 32P-labeled probe and 1 μl of cell lysate as described in Materials and Methods, and exposed to a Fuji PhosphorImager plate. The minor lower band in lane 12 at top probably represents Smad3 and Smad4 protein binding to only one of the two tandem repeats of the PE2.1 element. (Middle, bottom) Immunoblots with 5 μl (150 μg of proteins) of cell lysates in each lane that were blotted with an anti-Flag M2 antibody, recognizing the Flag epitope-tagged Smad3, and with an anti-Smad4 antibody, respectively, as described in Materials and Methods. As indicated, the levels of expression of Smad4 were the same in all cases (lanes 3,4,7,8,11,12) as were those of the Flag-tagged Smad3 (lanes 2,4,6,8,10,12). (B) Cell lysates containing both Smad4 and the Flag-tagged Smad3 were incubated with 32P-labeled PE2.1 DNA, followed by addition of 1 μl of an irrelevant control antibody or preimmune serum (lanes 2,4) or the anti-Flag antibody (anti-Smad3) (lane 3) and the anti-Smad4 antibody (lane 5), followed by gel electrophoresis as described in Materials and Methods.
Only lysates from cells expressing Smad3, Smad 4, and the active TβRI–T204D bound this PE2.1 element (Fig. 7A, lane 12); lysates from cells transfected with either Smad3 or Smad4 failed to bind (Fig. 7A, lanes 10,11), indicating that a complex of Smad 3 and Smad4 is binding to this probe. This gel-shifted complex can be supershifted by either an anti-Flag antibody, recognizing the epitope-tagged Smad3, or by an anti-Smad4 antibody (Fig. 7B, lanes 3,5), but not by control antibodies (Fig. 7B, lanes 2,4), confirming the presence of both Smad3 and Smad4 in the complex. Figure 7A, lanes 4–8 provide additional controls, showing that cells transfected with a kinase-deficient type I receptor fail to generate a functional DNA-binding complex. Furthermore, cotransfection of cells with TβRI–T204D, Smad4, and the mutant Smad3A did not yield a complex capable of binding the PE2.1 probe (Fig. 8, lane 6). Dennler et al. (1998) reported that GST fusion proteins of both full-length Smad4 and the amino-terminal half of Smad3 independently and directly bind to multiple CAGA sequences derived from the PAI-1 promoter. There is one CAGA sequence in the PE2.1 fragment. In contrast, our experiments show that binding of Smad3 and Smad4 to the 36-bp PE2.1 promoter fragment depends on the presence of the constitutively active type I receptor, and that the complex contains Smad4 and phosphorylated Smad3.
Figure 8.
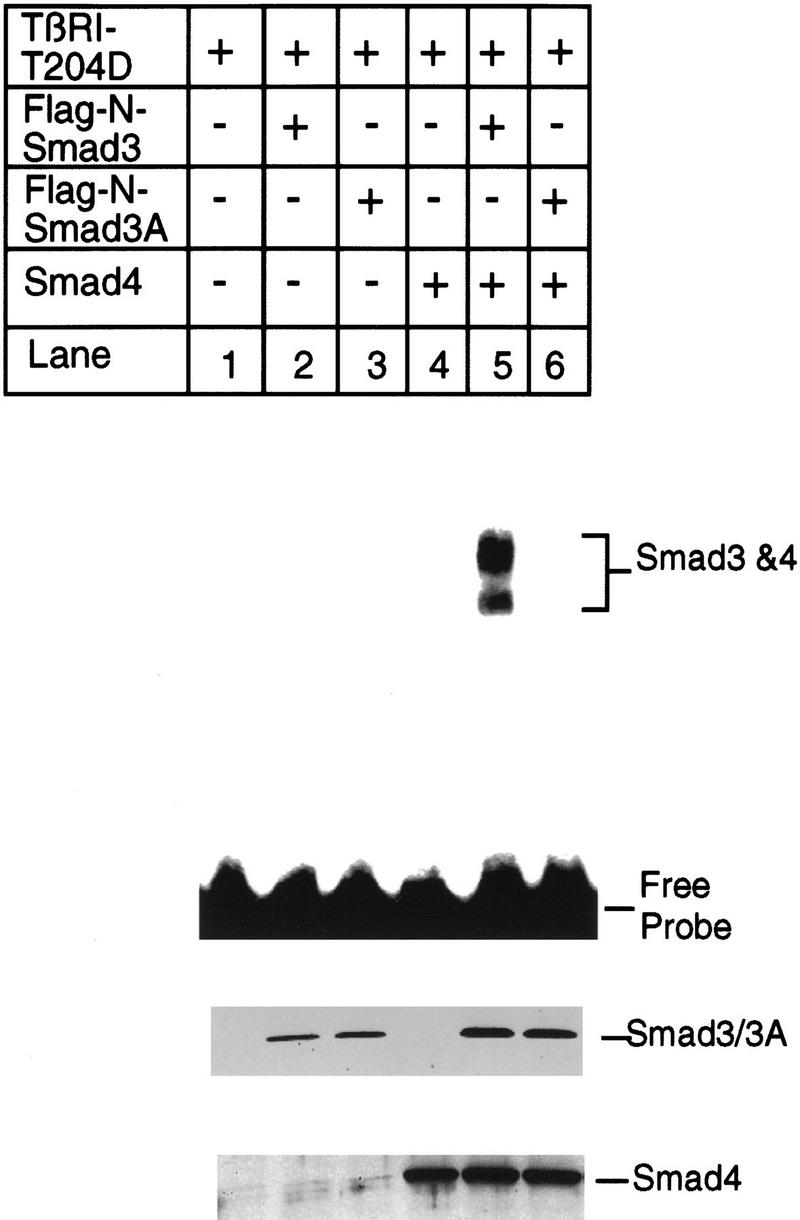
Phosphorylation of Smad3 at the carboxyl terminus is essential for binding of Smad3 and Smad4 to the PE2.1 element of the PAI-1 promoter. Bosc23 cells were transfected with the constitutively active TβRI–T204D, Smad4, and wild-type Smad3 or mutant Smad3A as described in Fig. 7. (Top) Results of a gel shift assay. (Middle, bottom) Immunoblots with 5 μl (150 μg of protein) of cell lysates in each lane that were blotted with an anti-Flag M2 antibody, recognizing the Flag epitope-tagged Smad3, and with an anti-Smad4 antibody, respectively, as described in the legend to Fig 7A.
Binding of TFE3 and Smad3–Smad4 to adjacent sites in the 36 bp of the PE2.1 element induces TGF-β-dependent transcription
Figure 9 shows that TFE3 and the complex of Smad4 and phosphorylated Smad3 bind to adjacent sequence of the PE2.1 promoter, and that both binding sites are essential for TGF-β-induced promoter activity. This experiment compares the activity of wild-type PE2.1 DNA (−586 to −551) with that of its two mutant versions, namely a mutation in the potential Smad3 and Smad4 binding site, PE2.1Sm, and a mutation in the E box, PE2.1Em (Fig. 9A). Lysates from cells overexpressing Smad4 and phosphorylated Smad3 form a gel-shifted band with radiolabeled PE2.1 DNA (Fig. 9B, lane 2) that is blocked by an excess of unlabeled PE2.1 DNA (Fig. 9B, lane 3), but not by unlabeled PE2.1Sm DNA (Fig. 9B, lane 4). This indicates that at least part of the sequence 5′-CCTAGAC-3′, located 3 nucleotides in front of the E-box sequence, is bound by the complex of Smad4 and activated Smad3.
Figure 9.
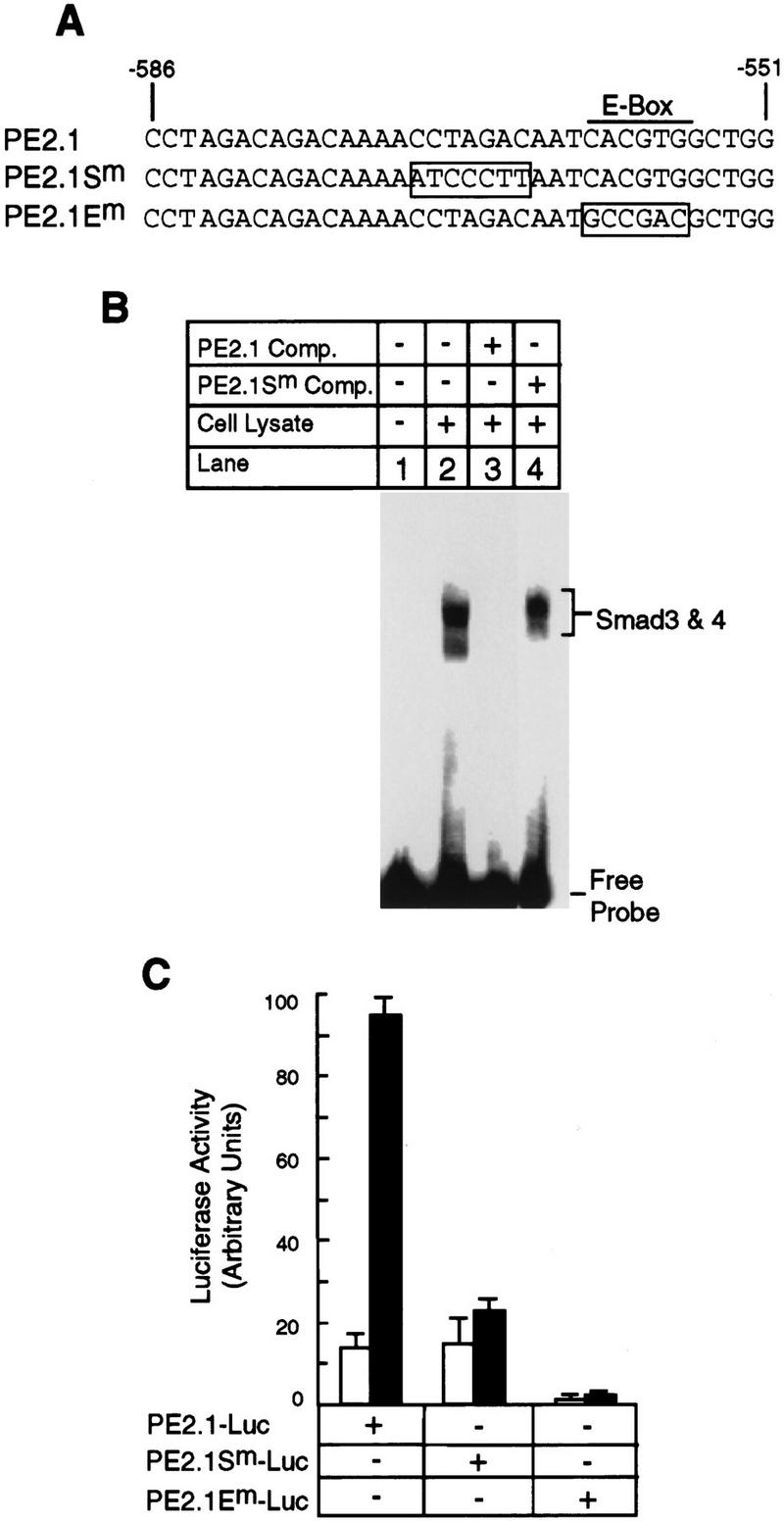
Both the Smad3–Smad4 binding site and the TFE3 binding site are essential for TGF-β-induced activation of the 36 nucleotide PE2.1 element of the PAI-1 promoter. (A) The sequences of one of the two identical repeats of the wild-type and two mutant PE2.1 elements. (B) Lysate from cells overexpressing TβRI-T204D, Smad3, and Smad4 as described in Fig. 7 were incubated with radiolabeled PE2.1 DNA alone (lane 2) or in the presence of unlabeled PE2.1 (lane 3) or mutant PE2.1Sm oligonucleotides (lane 4), followed by gel electrophoresis. Both the radiolabeled DNA and the competing DNA contained two tandem repeats of the corresponding sequence. (C) Luciferase reporter genes driven by two tandem repeats of either the PE2.1, PE2.1Sm, or PE2.1 Em elements were transfected into Hep G2 cells, and luciferase assays were carried out as described in Materials and Methods. (█) With TGF-β; (□) without TGF-β.
Figure 9C shows that two tandem repeats of the PE2.1 sequence, inserted upstream of a luciferase reporter gene, forms a functional TGF-β-inducible promoter. Mutation of the E boxes in this sequence inactivated promoter activity, both in the presence and absence of TGF-β. In contrast, mutation of the Smad binding site, PE2.1Sm, had no effect on basal (TGF-β-independent) promoter activity, but abolished the ability of TGF-β to stimulate transcription.
Together, these experiments indicate that both the Smad3–Smad4-binding site and the TFE3-binding site in PE2.1, the 36-bp TGF-β-inducible PAI-1 promoter, are essential for TGF-β-induced gene activation, suggesting that TFE3 synergizes with the Smad3–Smad4 protein complex in TGF-β signaling by binding to adjacent sites in the promoter. The gel-shift experiment in Figure 10 supports this notion by showing that a complex of TFE3 and Smad3–Smad4 bind to the same PE2.1 element. TFE3 was generated by in vitro translation, and the complex of Smad4 and phosphorylated Smad 3 was produced in transfected cells. TFE3 (Fig. 10, lane 2) generates a single-shifted band, whereas the activated Smad3–Smad4 (Fig. 10, lane 4) generate two shifted bands. A distinct, slower-migrating band was detected in the sample containing both TFE3 and the activated Smad3–Smad4 complex (Fig. 10, lane 3). These results show that both TFE3 and the activated Smad3–Smad4 complex bind to the adjacent sequences of the same 36 nucleotide segment of the PE2.1 promoter.
Figure 10.
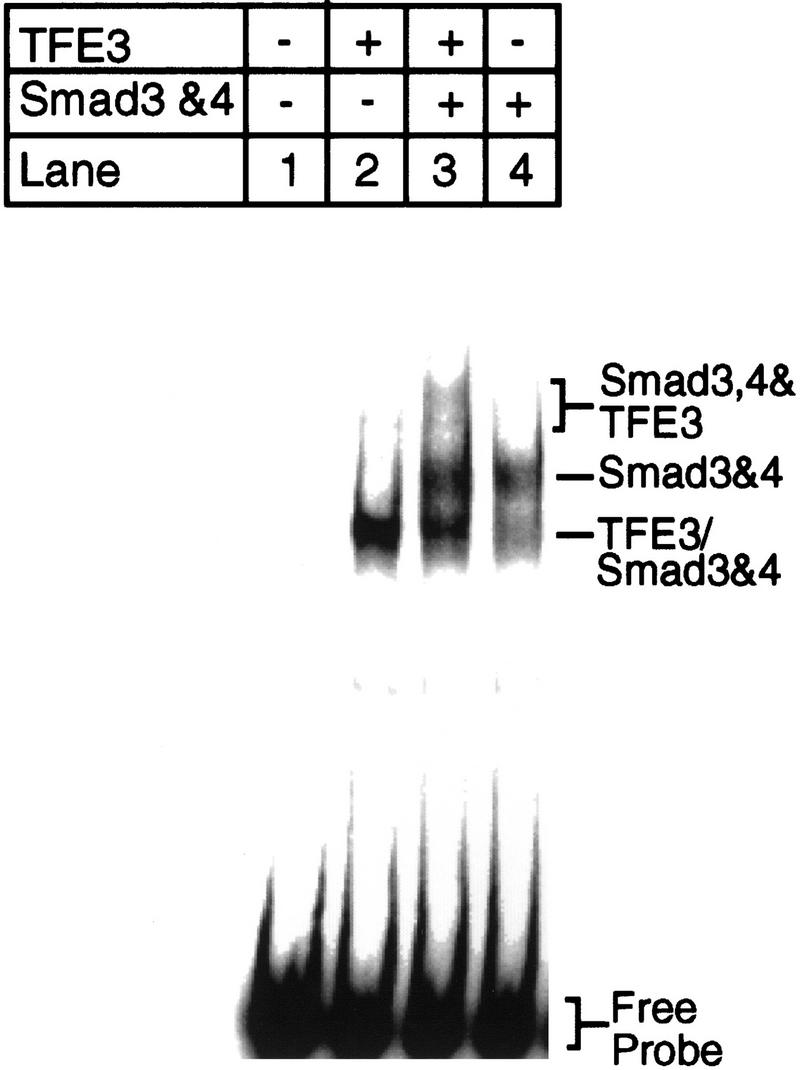
Both TFE3 and Smad3–Smad4 bind to the PE2.1 promoter fragment of the PAI-1 promoter. TFE3 was synthesized in vitro with a Promega TNT kit as instructed by the manufacturer. In vitro synthesized TFE3 (2 μl; lanes 2,3), and/or lysates from cells overexpressing TβRI-T204D, Smad3, and Smad4 (1 μl; lanes 3,4) were incubated with radiolabeled PE2.1 DNA before gel electrophoresis. (Lanes 1,4) Contained 2 μl of control reticulocyte lysate; (lanes 1,2) contained 1 μl of lysate from mock-transfected cells. The lower band in lane 3 (TFE3/Smad3 and Smad4) probably contains TFE3 and/or a Smad3–Smad4 complex that is bound to one of the two identical tandem repeats of the PE2.1 promoter fragment present in the probe.
Discussion
TFE3 activates TGF-β-induced transcription by binding to the E-box sequence in the PAI-1 promoter
To identify proteins that mediate TGF-β induction of the PAI-1 promoter, we developed an expression cloning strategy utilizing an engineered TGF-β-responsive cell line and a retroviral cDNA library. Our strategy was based on the demonstration that the engineered TGF-β-responsive cell line, BAH–gpt, constructed by Hocevar and Howe (1996), grows in HAT medium only in the presence of TGF-β. In this sense, TGF-β was converted from a growth-inhibitory factor, its normal function, into a growth-promoting hormone. Our strategy also made use of the observation that retroviruses deliver recombinant DNA sequences into the genome of recipient cells at a very high efficiency (Kitamura et al. 1995), and that the ecotropic retrovirus receptor is essential and sufficient for infection of cells by murine retroviruses (Baker et al. 1992).
Using this approach, we cloned the transcription factor, TFE3, which slightly activates transcription of the PAI-1 gene in the absence of TGF-β but strongly potentiates the ability of TGF-β to induce transcription (Figs. 3,4). TFE3 has not been implicated previously in TGF-β signaling. It was isolated previously by screening a phage expression library with a 32P-labeled E box-containing sequence from the immunoglobulin heavy chain gene, and is ubiquitously expressed (Beckmann et al. 1990; Zhao et al. 1993).
Several lines of evidence suggest that TFE3 plays a critical role in activating TGF-β-dependent transcription of the PAI-1 gene. First, cotransfection of TFE3 and a reporter gene containing ∼800 bp of the natural PAI-1 promoter enhances expression of the reporter gene fivefold in the presence of TGF-β (Fig. 3B). Second, serial truncation of the PAI-1 promoter identified 36- to 56-bp segments that are responsive to both TGF-β addition and overexpression of TFE3 (Figs. 4, 5, 9C), and these elements contain an E-box sequence. Moreover, mutation of the E-box sequence in either the 56-bp PE2 promoter or the 36-bp PE2.1 promoter abolished TGF-β-induced transcription as well as its binding to TFE3 (Figs. 5 and 9C). Third, Smad3, a critical signal transducer in TGF-β signaling, synergizes with TFE3 in TGF-β-induced transcription (Fig. 6A) and mutation (Smad3A) of the TGF-β-inducible phosphorylation sites in Smad3 abolished its ability to activate transcription (Fig. 6B). Fourth, USF1, a basic helix–loop–helix transcription factor that also binds the E-box sequence (Beckman et al. 1992), activated transcription of a luciferase gene driven by the PE2.1 promoter in BAH-ER3 cells, but transcription of the reporter gene was no longer regulated by TGF-β (data not shown).
Phosphorylation of Smad3 triggers binding of a Smad3–Smad4 complex to a sequence in the PAI-1 promoter adjacent to the TFE3-binding site
Smad3 and Smad4 together, but neither alone, bind to the 36-bp PE2.1 promoter. A prerequisite for formation of this DNA-binding complex is that the cells express a constitutively active type I receptor TβRI–T204D (Fig. 7). The constitutively active TβRI phosphorylates Smad2 and Smad3, which are normally phosphorylated by the wild-type I receptor only on addition of TGF-β (Macias-Silva et al. 1996; Abdollah et al. 1997; Souchenlnytskyi et al. 1997). Nevertheless, mutation of the TGF-β-inducible phosphorylation sites in Smad3 abolished the formation of a complex of Smad3 and Smad4 capable of binding to the PE2.1 sequence (Fig. 8).
Binding of the PE2.1 promoter by Smad3 and Smad4 was unaffected by mutation of the E box (data not shown), but was abrogated by mutation of the 7-bp sequence (5′-CCTAGAC-3′) located 3 bp upstream of the E box (Fig. 9). This suggests that at least part of the 5′-CCTAGAC-3′ sequence contains the Smad binding site. Dennler et al (1988) reported GST fusion proteins of both full-length Smad4 and the amino-terminal half of Smad3 directly bind to the CAGA sequence 5′ to the CCTAGAC sequence in the PE2.1 promoter. In contrast, our evidence suggests that phosphorylation of Smad3 not only triggers its association with Smad4, as reported previously (Nakao et al. 1997) but also is indispensable for binding to the PE2.1 element and subsequent activation of gene transcription (Figs. 6B and 9C). We have not yet precisely mapped the Smad binding site in this promoter segment. Phosphorylation may induce exposure of the DNA-binding domain in Smad3, or a multimer of Smad4 and phosphorylated Smad3 may have higher affinity for the PE2.1 sequence than does an unphosphorylated Smad3 monomer.
A number of recent reports show direct binding of Smad3 and Smad4 to specific DNA sequences, but these reports disagree on the consensus binding sequences (Yingling et al. 1997; Dennler et al. 1998; Vindevoghel et al. 1998; Zawel et al. 1998). As an example, Drosophila Mad binds to the consensus sequence GCCGnCGc (Kim et al. 1997); whereas human Smad 3 and Smad4 was reported to preferentially bind to GACACC (Yingling et al. 1997), GTCTAGAC (Zawel et al. 1998), or AG(C/A)CAGACA (Dennler et al. 1998); the latter sequence is also present in the PE2.1 element in the PAI-1 promoter. Hence, Smad3 and Smad4 appear to bind to DNA with a relative but not absolute specificity. Multiple tandem repeats of a Smad-binding sequence are required for TGF-β-induced transcription of a reporter gene, and even two tandem repeats of the GTCTAGAC sequence cannot support TGF-β-induced expression of a luciferase reporter gene (Zawel et al. 1998). These observations raise the possibility that a complex of multiple Smad proteins, together with other transcription factors such as TFE3, are required for maximal TGF-β-inducible transcription.
Synergism of TFE3 and Smad proteins in TGF-β-induced gene transcription by binding to adjacent sites in the PAI-1 promoter
TFE3 and a complex of Smad3 and Smad4 bind to adjacent sites in the 36-bp PE2.1 promoter (Figs. 9 and 10), and both binding sites are required for maximal TGF-β-induced gene transcription (Fig. 9C). This synergy requires TGF-β-induced phosphorylation of the carboxyl terminus of Smad3, as mutant Smad3A, lacking the TGF-β-induced phosphorylation sites, cannot synergize with TFE3 to activate TGF-β-dependent transcription from the PE2.1 promoter (Fig. 6B).
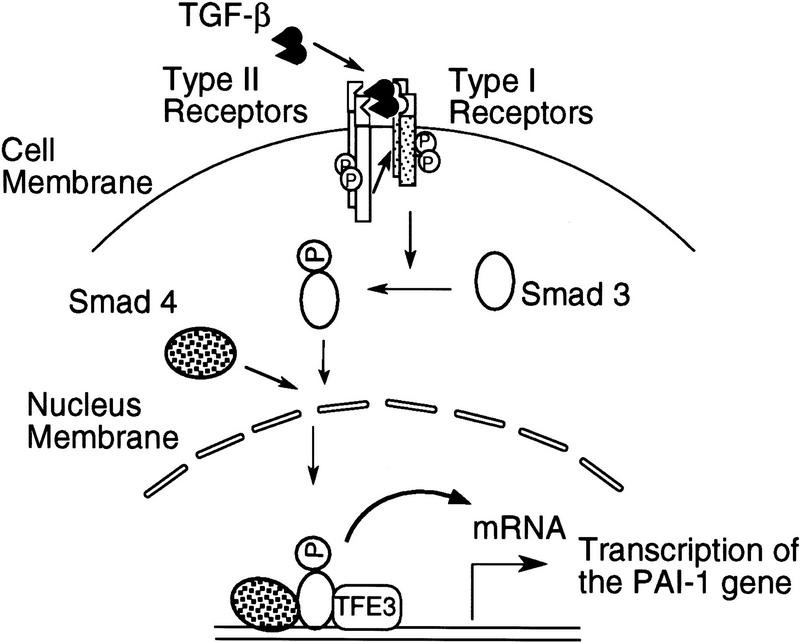
The model for TGF-β-induced transcription of the PAI-1 gene in Figure 11 summarizes our results. A TGF-β-activated type I receptor phosphorylates Smad3, which then associates with Smad4. The complex of Smad3 and Smad4 then enters the nucleus and binds to a sequence upstream of the E box, which is already occupied by TFE3. Binding of both the Smad3–Smad4 complex and TFE3 within the 36-bp PE2.1 element is essential for maximal transcription of the PAI-1 gene.
Figure 11.
A model for cooperation of TFE3, Smad3, and Smad4 in TGF-β-induced activation of the PAI-1 promoter. A complex of Smad4 and phosphorylated Smad3 bind to the DNA sequence 5′ to the E-box sequence, which is occupied by TFE3. Binding of the Smads potentiates the activity of TFE3, leading to TGF-β-induced transcription of PAI-1 gene. Although indicated as monomers, we do not yet know the oligomeric state of the Smad or TFE3 proteins when they bind to the PAI-1 promoter.
Our model differs somewhat from that proposed for activin-induced activation of the Xenopus transcription factor FAST-1. On addition of activin, a complex of Smad4 and phosphorylated Smad2 forms in the cytosol, translocates into the nucleus, binds FAST1, and then binds to a segment in the promoter of the developmentally regulated Mix2 gene (X. Chen et al. 1996, 1997). In contrast, we have been unable to detect an interaction of TFE3 and the Smad3–Smad4 complex in the absence of DNA. It is possible that the Smad3–Smad4 complex does not directly bind to TFE3, even though they bind to adjacent sites of the PE2.1 promoter. Alternatively, TFE3 may form a complex with another, as yet unidentified, transcription factor, and that only this complex associates with the Smad3–Smad4 complex that is formed after TGF-β stimulation.
TGF-β activates a diverse range of genes in different cell types. Although Smad3–Smad4 complexes may bind specific DNA sequences, our work suggests that to induce expression of specific genes, these complexes need to cooperate with one or more transcription factors. We showed that TFE3 is one such factor essential for induction of the PE2.1 promoter. Because other genes activated by TGF-β do not have E boxes in the promoter sequenced to date, it is likely that other transcription factors interact with Smad3–Smad4 and Smad2–Smad4 complexes to induce transcription of other genes such as p15INK4B and p21WAF1/CIP (Datto et al. 1995; Li et al. 1995). The TFE3-binding sequence E box is essential for both basal and TGF-β-induced transcription of the PE2.1 promoter. We have no evidence to suggest that TFE3 itself is modified or activated following TGF-β addition; consistent with this notion, in vitro translated TFE3 binds to the E box in the PE2 promoter. We speculate that in unstimulated cells, TFE3 is bound to the two E boxes in the PAI-1 promoter and supports a low level of transcription. Binding of an activated Smad3–Smad4 complex leads to a several-fold increase in gene expression.
Such synergistic cooperation of Smad3–Smad4 complexes with specific transcription factors offers the organism a distinct advantage—the same Smad3–Smad4 complex will activate different genes in different cells depending on the sequence of a promoter and the set of cooperative transcription factors that are expressed.
Materials and methods
Plasmid construction
Standard molecular biology techniques were used as described (Sambrook et al. 1989). Oligonucleotides were synthesized by GIBCO–BRL. To construct 3APP–Luc, the TATA box sequence, 5′-AGGGTATATAAT-3′, was inserted into the PstI–BglII site of the pGL3–basic vector (Promega); a pair of oligonucleotides corresponding to −740/−644 of the PAI-1 promoter was inserted upstream of the TATA box sequence; finally a pair of oligonucleotides containing three tandem repeats of the AP1-binding site [TGA(G/C)TCA] separated by an XbaI site was inserted upstream of the PAI-1 promoter sequence. PAI–Luc was constructed by inserting the 0.8-kb HinddIII fragment of the PAI-1 promoter (Westerhausen et al. 1991) into the HindIII site of pGL3–basic. Various DNA fragments PCR-amplified from the PAI-1 promoter were cloned into the KpnI–PstI sites of 3APP–Luc to generate PF1–Luc, PF2-Luc, and PF3-Luc in place of the 3APP promoter. These DNA fragments corresponded to nucleotides −794 to −532, −552 to −194, and −214 to +29 of the PAI-1 promoter (Keeton et al. 1991). Pairs of oligonucleotides including the E-box sequence in the PAI-1 promoter were cloned into the KpnI–Pst I sites of 3APP-Luc to generate PE1-Luc and PE2–Luc. These oligonucleotides corresponded to −740 to −644 and −583 to −528, respectively. To construct PE2.1–Luc, PE2.1Sm-Luc, and PE2.1Em-Luc reporter genes, two tandem repeats of the wild-type or mutant oligonucleotides corresponding to −586 to −551 of the PAI-1 promoter were inserted into KpnI–PstI sites of p3APP–Luc to replace its promoter (see Fig. 9A). All the constructs were sequenced to confirm the cloning junctions and more than one independent clone of each construct were tested in transfection for luciferase assays to confirm the results.
To generate a plasmid encoding Smad3, human Smad3 cDNA was amplified by PCR and cloned into the BamHI–XbaI sites of a modified pcDNA3, resulting in a Smad3 fusion protein with two tandem repeats of a Flag epitope tag at its NH2-terminus. A plasmid encoding Smad4 was generated by insertion of the human Smad4 cDNA into pMX–IRES–GFP (X. Liu et al. 1997). Human Smad4 cDNA was also cloned into the BamHI–NotI sites of the vector pEXL, a derivative of pEGFP-N1 (Clontech) described previously (X. Liu et al. 1997) to generate pEXL–Smad4. Plasmids Flag-N–Smad3 and Flag-N–Smad3A were described previously (X. Liu et al. 1997). PCR-amplified TFE3 was cloned into the BamHI–XhoI sites of pET28a (Novagen) to generate pET–TFE3 for in vitro transcription and translation. The kinase-defective mutant plasmid of the human TGF-β type I receptor, pCMV5–TβRI–KR, and the constitutively active TGF-β type I receptor, pCMV5–TβRI–T204D, were described previously (Wieser et al. 1995).
Tissue culture
BAH–gpt cells and HPRT-deficient HT1080 cells were kindly provided by P. Howe at the Cleveland Clinic Research Foundation (Hocevar and Howe 1996). HepG2 cells were purchased from ATCC. All of these cells were cultured in DMEM containing 10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin, in 5% CO2 at 37°C unless otherwise stated. To treat cells with HAT or 6-TG, 1× HAT medium (GIBCO–BRL) or 30 μg/ml 6-TG (Sigma) was added to the normal medium. TGF-β1 was provided by R&D Systems, Inc., and a concentration of 200 pm was added to cell cultures as indicated.
Construction of a retroviral cDNA library and infection of cells by retroviruses
Poly(A)+ RNA was isolated from HPRT-deficient HT1080 cells. cDNAs were synthesized from the poly(A)+ RNA by use of the Superscript Plasmid System for cDNA Synthesis and Plasmid Cloning (GIBCO BRL) as described previously (Hua et al. 1996), and then cloned into the EcoRI–NotI sites of the retroviral vector pMX (Onishi et al. 1996). The resulting cDNA library was amplified in transformed bacteria and then introduced into a packaging cell line to obtain a high titer retroviral cDNA library. Briefly, Bosc23 cells, a cell line expressing the Gag, Pol, and Env proteins of Moloney Leukemia Virus (Pear et al. 1993), were seeded at a density of 2 × 106 cells per 60-mm dish in DMEM containing 10% fetal calf serum. On day 1, cells in each dish were transfected with 5 μg of the retroviral cDNA library in the presence of chloroquine (25 μm) to increase the virus titer. The transfected cells were switched to fresh medium 9 hr after transfection, and the supernatant containing the recombinant retroviruses was collected 48 hr after transfection.
Supernatant containing the retroviruses was incubated with the target cells for 6–9 hr in normal medium containing 4 μg/ml Polybrene (Sigma). To measure the titer of the library, pMX–LacZ-derived retroviruses were produced in parallel with the retroviral cDNA library and were used to infect BAH–ER3 cells or NIH 3T3 cells; infected cells were stained with X-gal for β-galactosidase expression. The titer of the retroviral cDNA library was deduced from that of the pMX–LacZ retroviruses produced in parallel.
To rescue recombinant retroviruses from infected BAH-ER3 cells, we first transfected Bosc23 cells with the plasmid pZAP (from D. Baltimore’s laboratory, MIT, Cambridge, MA), which carries the entire cDNA sequence of the murine Moloney leukemia retrovirus genome. The supernatant containing the wild-type virus was collected 48 hr after transfection, and 1 ml of a 1:2 dilution of the supernatant was used to superinfect infected BAH-ER3 cells in six-well plates.
Transfection, luciferase assay, and preparation of cell lysates
Cells were transfected by the calcium phosphate precipitation method (Sambrook et al. 1989). For luciferase assays, cells were also transfected with 0.5 μg/well pSV-β or 0.2 μg/well of pCMV-β encoding the lacZ gene (Clontech) as an internal control to normalize the luciferase activity.
To transfect BAH-ER3 cells and Hep G2 cells, cells were seeded at a density of 50,000 cells/well in 12-well plates unless otherwise stated. On day 1, the cells were switched to fresh medium and then transfected by the calcium phosphate precipitation method. After overnight incubation, the cells were switched to normal medium and incubated for 6–8 hr. Afterward, serum-free medium with or without 200 pm TGF-β was added to the transfected cells; cells were harvested 20 hr after incubation with TGF-β for luciferase and β-galactosidase assays. The cells in each well of 12-well plates were lysed with 250 μl of 1× lysis buffer (Promega), and luciferase assays were carried out with 20 μl of cell lysates by use of the Luciferase Assay System (Promega) as detailed by the manufacturer. To normalize the luciferase activity, 20 μl of cell lysate was incubated with 100 μl of reaction buffer from the Luminescent B-galactosidase Detection Kit II (Clontech Laboratories, Inc.) as instructed by the manufacturer. Both the luciferase and β-galactosidase activities were measured by an AutoLumat LB953 luminometer (EG & G Berthold). All luciferase activities were normalized by the β-galactosidase activities and presented as an average from duplicate samples.
To obtain cell lysates for gel-shift assays, Bosc23 cells were transfected with desired plasmids and the total amount of DNA per 60 mm dish was adjusted to 7.5 μg by use of the plasmid pEXL–GFP. After overnight transfection, the cells were switched to normal medium, and harvested 24 hr later. Cells from each 60-mm dish were lysed in 150 μl of buffer containing the following components: 50 mm Tris at pH 8.0, 500 mm NaCl, 1% NP-40, 25 mm β-glycerophosphate, and 1× protease inhibitor cocktail Complete (Boehringer Mannheim). The lysed cells were rotated at 4°C at 60 rpm for 2 hr, and the supernatant was collected by centrifugation for use in gel-shift assays.
Gel-shift assay and immunoblotting
Gel-shift reactions were carried out in a total volume of 30 μl at room temperature. The components of the reaction buffer are as follows: 20 mm Tris at pH 8.0, 60 mm KCl, 0.7 mg/ml bovine serum albumin, 1 mm EDTA, 1.6 mm dithiothreitol, 1.6 mm MgCl2, 0.3 % NP-40, 66 μg/ml poly(dI–dC)/poly(dI–dC) (Pharmacia), and 12% glycerol. Radiolabeled probes were made either by end labeling the annealed oligonucleotides with [γ-32P]ATP or by PCR amplification in the presence of [α-32P]dCTP. Briefly, a pair of oligonucleotides corresponding to the PE2 fragment (−583 to −528) of the PAI-1 promoter was end-labeled with [γ-32P]ATP; alternatively, the two tandem repeats of the PE2.1 element (5′-CCTAGACAGACAAAACCTAGACAATCACGTGGCTGG-3′), which comprise base pairs −586 to −551 of the human PAI-1 promoter, were amplified by PCR from the reporter construct PE2.1–Luc in the presence of [α-32P]dCTP.
The amplified probe was isolated on a native polyacrylamide gel as described previously (Wang et al. 1993), and 4 × 103 cpm was added to each reaction that had received the cell lysates 15 min earlier. One microliter of cell lysate (∼30 μg of protein) was used in each reaction unless otherwise stated. For competition with wild-type or mutant oligonucleotides, a 50-fold molar excess of unlabeled oligonucleotides was added to the reaction buffer containing the cell lysate 15 min prior to addition of the 32P-labeled probe. Twenty minutes after addition of the probe, the reaction was loaded onto a 4% polyacrylamide gel in 0.5× TBE buffer (Sambrook et al. 1989), and electrophoresis was carried out at 20 mA for 70 min. To supershift the DNA-binding activity with antibodies, 1 μl of the indicated antibody was added to each reaction; the reaction was loaded onto the gel after 15 min of incubation. All signals were detected on a Fujix BAS2000 PhosphorImager.
To detect the expression of Smad3 and Smad4 proteins in transfected Bosc23 cells, cell lysates prepared from the transfected Bosc23 cells were separated on 6%–18% gradient polyacrylamide gels and then transferred to Nitrocellulose blotting filters. The filters were blotted with 1 μg/ml anti-Flag (M2) antibody (Eastman Kodak) for detection of the Flag epitope-tagged Smad3, or with a 1:2000 dilution of an anti-Smad4 rabbit polyclonal antibody (Nakao et al. 1997). Bound primary antibodies were detected with horseradish peroxidase-labeled anti-mouse or anti-rabbit secondary antibodies, respectively, and developed with enhanced chemiluminescence reagents purchased from Pierce.
Acknowledgments
We thank Dr. P. Howe for kindly providing HPRT-deficient HT1080 cells and BAH–gpt cells. The cDNAs encoding human Smad2, Smad3, and Smad4 and TβRI were kind gifts from Dr. J. Massagué and Dr. R. Derynck. We thank Drs. C. Heldin and P. ten Dijke for providing the polyclonal antibody against human Smad4, and Dr. M. Tal for reagents for gel shift assays. TGF-β1 was a kind gift from R&D Systems, Inc. We also thank Drs. B. Schiemann and A. Sirotkin for reading the manuscript, and other members of the Lodish group for stimulating discussions. This work was supported by National Institutes of Health (NIH) grant CA63260 to H.F.L. X.H. was supported by a Damon Runyon–Walter Winchell Cancer Research Fund postdoctoral fellowship (DRG 1429) and X.L. was supported by a postdoctoral fellowship from the NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lodish@wi.mit.edu; FAX (617) 258-6768.
References
- Abdollah S, Macias-Silva M, Tzukazaki T, Hayashi H, Attisano L, Wrana J. TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2–Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana J. Mads and Smads in TGFβ signaling. Curr Opin Cell Biol. 1998;10:188–194. doi: 10.1016/s0955-0674(98)80141-5. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL, Lopez-Casillas F, Massagué J. TGF-β receptors and actions. Biochimica et Biophysica Acta. 1994;1222:71–80. doi: 10.1016/0167-4889(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Baker BW, Boettiger D, Spooncer E, Norton JD. Efficient retroviral-mediated gene transfer into human B lymphoblastoid cells expressing mouse ecotropic viral receptor. Nucleic Acids Res. 1992;20:5234. doi: 10.1093/nar/20.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H, Su L-K, Kadesch T. TFE3: A helix-loop-helix protein that activates transcription through the immunoglobulin enhancer μE3 motif. Genes & Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher F, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lebrun J-J, Vale W. Regulation of transforming growth factor β- and activin-induced transcription by mammalian Mad Proteins. Proc Natl Acad Sci. 1996;93:12992–12997. doi: 10.1073/pnas.93.23.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto MB, Yu Y, Wang X-F. Functional analysis of the transforming growth factor β responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J. Direct binding of Smad3 and Smad4 to critical TGF-β-inducible elements in the promoter of human plasminogen activator inhibitor-type I gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen P, ten Dijke P, Ichijo H, Yamashita H, Schulz P, Heldin C, Miyazono K. Cloning of a TGF-β type I receptor that forms a heteromeric complex with the TGF-β type II receptor. Cell. 1993;75:681–692. doi: 10.1016/0092-8674(93)90489-d. [DOI] [PubMed] [Google Scholar]
- Hannon G, Beach D. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hocevar B, Howe P. Isolation and characterization of mutant cell lines defective in transforming growth factor β signalling. Proc Natl Acad Sci. 1996;93:7655–7600. doi: 10.1073/pnas.93.15.7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Keeton MR, Curriden SA, Zonneveld A-JV, Loskutoff DJ. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforning growth factor-β. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Onishi M, Kinoshita S, Shibuya A, Miyajima A, Nolan GP. Efficient screening of retroviral cDNA expression libraries. Proc Natl Acad Sci. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Li J-M, Nichols MA, Chandrasekharan S, Xiong Y, Wang X-F. Transforming growth factor β activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J Biol Chem. 1995;270:26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- Lin HY, Lodish HF. Receptors for the TGF-β superfamily: Multiple polypeptides and serine/threonine kinases. Trends Cell Biol. 1993;3:14–19. doi: 10.1016/0962-8924(93)90195-7. [DOI] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGF-β-inducible transcriptional complexes. Genes & Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Lodish HF. Signalling by chimeric erythropoietin-TGF-β receptors: Homodimerization of the cytoplasmic domain of the type I TGF-β receptor and heterodimerization with the type II receptor are both required for intracellular signal transduction. EMBO J. 1996;15:4485–4496. [PMC free article] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGF-β receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Massague J, Hata A, Liu F. TGF-β signalling through the Smad pathway. Trends Cell Biol. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- Nakao A, Imamura T, Souchelnystkyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C-H, Miyazono K, ten Dijke P, et al. TGF-β receptor-mediated signalling through Smad2, Smad3, and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier LL, Gorman DM, Nolan GP, Miyajima A, Kitamura T. Applications of retrovirus-mediated expression cloning. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper free retroviruses by transient transfection. Proc Natl Acad Sci. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed S. Retroviruses and oncogenes. In: Levy JA, editor. The retroviride. New York, NY: Plenum Press; 1995. pp. 293–306. [Google Scholar]
- Riccio A, Pedone PV, Lund LR, Olesen T, Olsen HS, Andreasen PA. Transforming growth factor β1-responsive element: Closely associated binding sites for USF and CCAAT-binding transcription factor-nuclear factor I in the type 1 plasminogen activator inhibitor gene. Mol Cell Biol. 1992;12:1846–1855. doi: 10.1128/mcb.12.4.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Souchenlnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin C-H. Phosphorylation of Ser465 and Ser467 in the C terminus of Smads mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J BiolChem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- Vindevoghel L, Kon A, Lechleider R, Uitto J, Roberts A, Mauviel A. Smad-dependent transcriptional activation of human type VII collagen gene (COLL7A1) promoter by transforming growth factor-β. J Biol Chem. 1998;273:13053–13057. doi: 10.1074/jbc.273.21.13053. [DOI] [PubMed] [Google Scholar]
- Wang X, Briggs MR, Hua X, Yokoyama C, Goldstein JL, Brown MS. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter II. J Biol Chem. 1993;286:14497–14504. [PubMed] [Google Scholar]
- Weis-Garcia F, Massagué J. Complementation between kinase-defective and activation-defective TGF-β receptors reveals a novel form of receptor cooperativity essential for signaling. EMBO J. 1996;15:276–289. [PMC free article] [PubMed] [Google Scholar]
- Westerhausen DR, Jr, Hopkins WE, Billadello JJ. Multiple transforming growth factor-β-inducible elements regulate expression of the plasmigogen activator inhibitor type-1 gene in Hep G2 cells. J Biol Chem. 1991;266:1092–1100. [PubMed] [Google Scholar]
- Wieser R, Wrana J, Massague J. GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X-F, Massagué J. TGFβ signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Francesc V, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Yingling J, Datto M, Wong C, Frederick J, Liberati N, Wang X. Tumor suppressor Smad4 is a transforming growth factor β-inducible DNA binding protein. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Dai J, Buckhaults P, Zhou S, Kinzler K, Vogelstein B, Kern S. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng X-H, Wu R-Y, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- Zhao G-Q, Zhao Q, Zhou X, Mattei M-G, Combrugghe B DE. TFEC, a basic helix-loop-helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol Cell Biol. 1993;13:4505–4512. doi: 10.1128/mcb.13.8.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]