Abstract
The bone marrow failure syndrome dyskeratosis congenita (DC) has been considered to be a disorder of telomere maintenance in which disease features arise due to accelerated shortening of telomeres. By screening core components of the telomerase and shelterin complexes in patients with DC and related bone marrow failure syndromes we have identified 24 novel mutations: 11 in the RNA component of telomerase (TERC), 8 in the reverse transcriptase component (TERT), 4 in dyskerin (DKC1) and 1 in TRF1-interacting nuclear factor 2 (TINF2). This has prompted us to review these genetic subtypes in terms of telomere length, telomerase activity and clinical presentation among 194 genetically characterised index cases recruited onto the registry in London. While those with DKC1 and TINF2 mutations present at a younger age and have more disease features than those with TERC or TERT mutations, there is no difference in telomere length between these groups. There is no difference in the age of onset and numbers of disease features seen in those with TERC and TERT mutations despite the fact that the latter show higher levels of telomerase activity in vitro. The incidence of aplastic anaemia is greater in patients with TERC or TINF2 mutations compared to patients with DKC1 mutations, and cancer incidence is highest in patients with TERC mutations. These data are the first to provide robust comparisons between different genetic subtypes of telomerase and shelterin mutations (the “telomereopathies”) and clearly demonstrate that disease severity is not explained by telomere length alone.
Introduction
Telomeres are nucleoprotein structures that protect chromosome ends, distinguishing them from double strand breaks that might occur elsewhere in the genome and need repair [1]. Telomeric DNA consists of a TTAGGG repeat sequence that is bound by a protein complex known as shelterin [2]. Because of the end-replication problem [3] telomeres shorten with each cell division and when they become critically short, a p53-dependent checkpoint is activated that leads to apoptosis or cell senescence [4]–[6]. It has been suggested that this mitotic clock has a tumour-suppressor function in long lived mammals, giving cells a limited lifespan and hence restricting the accumulation of DNA damage [7]. Telomere shortening has therefore been linked to the processes of both aging [8] and cancer [9]. In the germ line and in some stem cells, telomeric DNA can be replenished by telomerase, a ribonucloeprotein complex that involves an RNA template and a reverse transcriptase [10].
Dyskeratosis congenita (DC) is an inherited multi-system disorder classically characterized by a mucocutaneous triad of abnormal skin pigmentation, nail dystrophy and leukoplakia [11]. DC patients frequently develop bone marrow failure and are at a high risk of developing cancer [12] as well as a variety of other features [13], [14]. DC is clinically and genetically heterogeneous. Eight disease genes (DKC1, TERC, TERT, NOP10, NHP2, TINF2, C16orf57 and TCAB1) have been identified to date [15]–[22] and all but one are known to be involved in telomere maintenance. The four most common genetic subtypes of DC are those that involve either the core components of the telomerase complex [23], the RNA (TERC), the reverse transcriptase (TERT) and the accessory protein dyskerin (DKC1), or the shelterin component, the telomeric repeat binding factor 1-interacting nuclear factor 2 (TINF2).
It is not surprising therefore that DC has been considered to represent the clinical manifestation of defective telomere maintenance and indeed, very short telomeres are a hallmark of this disease [24]–[26]. However, there is a broad clinical spectrum associated with these mutations and patients can present with a number of overlapping disorders, characterised by one or more of a constellation of clinical features [7], [27]. In addition to DC, these include the Hoyeraal-Hreidarsson (HH) syndrome [28], which is characterised by a variety of features including bone marrow failure, growth retardation, cerebellar hypoplasia, enteropathy and immunodeficiency. Other patients may present with only aplastic anaemia (AA) [29], [30], myelodysplastic syndrome (MDS) [31], [32], acute myeloid leukaemia (AML) [33] pulmonary fibrosis [34], [35] and liver fibrosis [36], and collectively these can be referred to as the ‘telomereopathies’.
We have been screening telomere-related genes among patients referred, primarily with bone marrow failure, to our registry in London and in this paper we report on 24 novel mutations. The variable clinical presentation among these patients has prompted us to review the clinical features, telomere length and mutation status among 194 genetically characterised index cases. While there are clear differences between the genetic subgroups in terms of clinical presentation, we find that the range of telomere length is similar in each group. We conclude that telomere length alone does not account for variation in disease severity between the different genetic subtypes.
Results
Novel mutations in telomerase and shelterin components
Through mutation screening in patients with DC and related bone marrow failure syndromes, we have identified 23 novel mutations in core components of telomerase: 11 in TERC, 8 in TERT and 4 in DKC1 (Table 1, Figures S1 and S2). Two additional TERT mutations were identified during the course of this study which have been reported elsewhere in unrelated families [15], [37]. One mutation in the shelterin component TINF2 has been identified (reported also by Sasa et al [38] while this paper was in preparation) which is in addition to the 8 TINF2 mutations that we have recently reported elsewhere [39]. All TERC, TERT and TINF2 mutations were heterozygous, while the DKC1 mutations are hemizygous (X-linked). None have been reported previously and none are reported among previous screens of normal healthy individuals [15], [16], [20], [30] or on dbSNP or the 1000 genomes database. The clear majority (8/11) of the TERC mutations disrupt base pairing in the pseudoknot region of the molecule and they look like classic disease causing mutations (Figure S1). Of all 45 disease-associated TERC mutations that are known to us, 30 are located in the pseudoknot. Also consistent with previous findings [30], [36], [37], the TERT mutations are spread throughout the molecule and show variable degrees of conservation across species (Figure S2). Polyphen conservation scores support the notion that most of these mutations are disruptive: 8/10 are predicted to be probably damaging (score = 0.949−1.0) and one is possibly damaging (Val56Leu, score = 0.433); the only one predicted to be benign (Arg972His, score = 0.059) is seen in combination with a second substitution, Gly1063Asp, which has been reported in another patient elsewhere [37].
Table 1. Characteristics of patients with novel telomerase and shelterin mutations.
| Age (years) | Disease1 | Additional disease features2 | FH3 | Relatives4 | Nucleotide | Location5 or amino acid | TRAP6 | Δtel7 |
| TERC | ||||||||
| 18 | CAA | dysphagia, skin pigmentation, low NK cells | No | AsM | 36C>T | Ps P1b | 11.3 | |
| 22 | CAA | skin pigmentation | No | 67G>A | Ps P2a.1 | 4.6 | ||
| 24 | AA/MDS | non-cirrhotic portal hypertension with oesophageal varices | No | 83T>G | Ps P2a | 4.3 | −2.52 | |
| 40 | DC | dental & hair loss, pulmonary disease, cancer | No | AsM, AsSo | 95_96delGC | Ps P3 | 0.6 | −2.82 |
| 29 | CAA | microcephaly, aseptic necrosis of femoral head | No | AsF | 107G>T | Ps P3 | 0.9 | −4.30 |
| 28 | NSAA | No | 126A>G | Ps P2a | 4.8 | −6.22 | ||
| 34 | AA | aseptic necrosis of femoral head | No | AsSo | 176A>C | Ps P3 | 0.5 | −3.58 |
| 18 | CAA | nail dystrophy | No | 182G>A | Ps P3 | 1.3 | −3.03 | |
| 5 | HH | microcephaly, cerebellar hypoplasia, hair loss, low NK cells | No | AsM, AsSi | 242C>T | Loop P5 | 46.8 | −0.57 |
| 55 | MDS | mucocutaneous features, lung disease, abnormal LFT | No | 287C>G | CR4-CR5 P6b | 8.1 | −0.57 | |
| 31 | CAA | skin pigmentation, VSD | Yes | S? | 377A>G | H box | ||
| TERT | ||||||||
| 14 | NSAA | easy fracture | No | AsM | 166G>C | Val56Leu | 4.3 | −3.48 |
| 26 | AA/MDS | premature greying, telangiectasia | Yes | SF, SGm | 248G>C | Arg83Pro | ||
| 5 | DC | No | 1142G>C | Arg381Pro | 43.1 | |||
| 12 | CAA | short digit | Yes | AsF, Ssi | 2147C>T | Ala716Val8 | 16.6 | −3.14 |
| 48 | DC | MDS, IPF, hair loss, dental caries | Yes | SB, AsSi | 2152G>A | Asp718Asn | 44.1 | −3.61 |
| 25 | MDS | liver cirrhosis, congenital hearing loss, bilateral nystagmus | No | AsM | 2581G>A | Gly861Arg | 1.0 | |
| 11 | DC | hyperhiderosis | Yes | SB, SB | 2915G>A | Arg972His | ||
| 19 | DC | hypogonadism | No | 3082A>C | Asn1028His | 16.7 | −2.20 | |
| 11 | DC | hyperhiderosis | Yes | SB, SB | 3187G>A | Gly1063Ser8 | 29.0 | |
| 34 | NSAA | abnormal LFT | No | 3388A>G | Ile1130Val | |||
| DKC1 | ||||||||
| 1 | HH | microcephaly, cerebellar hypoplasia, low B cells | Yes | SB | 202C>T | His68Tyr | ||
| 15 | DC | No | 227C>T | Ser76Leu | −2.66 | |||
| 4 | HH | microcephaly, enteropathy, immunodeficiency | No | 1133G>A | Arg378Gln | −0.7 | ||
| 19 | DC | No | 114C>G | Ile38Met | ||||
| TINF2 | ||||||||
| 6 | AA | Intracranial calcification, ocular hemorrhage | No | 811C>T | Gln271X |
AA: aplastic anaemia, CAA: constitutional AA, NSAA: non-severe AA, MDS: myelodysplastic syndrome, HH: Hoyeraal Hreidarrson syndrome.
NK: natural killer cells; VSD: ventricular septal defect; IPF: idiopathic pulmonary fibrosis; LFT: liver function test.
FH: family history.
Relatives are listed where they were available and tested positive for the mutation. As = asymptomatic, S = symptomatic, M = mother, F = father, So = son, Si = sister, B = brother, Gm = grandmother.
Location: domain of TERC: Ps = pseudoknot.
TRAP: telomere repeat amplification protocol, % wild type.
Δtel: age-adjusted telomere length measurement.
There is a broad spectrum of clinical presentation among these patients, ranging from early onset of features resembling the HH syndrome to a patient presenting at the age of 55 years with MDS. It is noteworthy that of the 21 index cases with TERC or TERT mutations only 7 had sufficient clinical features to be classified as DC or HH; 14 out of the 21 were classified as having either idiopathic AA, constitutional AA or MDS. Another striking observation here is that of the patients with TERC mutations, only one reports that a family member had presented with any features related to the disease (Table 1, column ‘FH’). A lack of family history is normal among patients with TINF2 mutations due to the fact that these mutations usually arise de novo. However, this is not the case for the TERC families: when no family history is reported and both parents are available, one of these parents is usually an asymptomatic carrier.
TERC mutations cause lower telomerase activity than TERT mutations
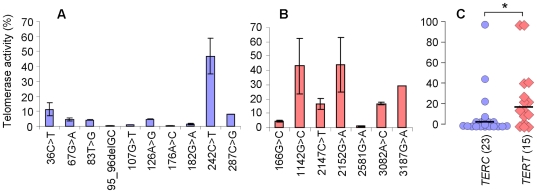
The in vitro telomerase activity of the TERC and TERT mutations, as determined by the telomere repeat amplification protocol (TRAP assay) may act as a guide to the clinical penetrance of these different alleles. It is noticeable that for all but one of the TERC mutations, the TRAP activity is <12% of wild type (Table 1, Figure 1A). The exception is the TERC 242C>T mutation, which gives an activity of 47±11%; this mutation is located in a loop bordering the CR4-CR5 domain of the molecule, away from other disease-causing alleles. It is seen in a patient who has HH, consanguineous parents, and whose asymptomatic mother and sister are also heterozygous; this may therefore not be the pathogenic lesion in this family.
Figure 1. TERC mutations have lower residual telomerase activity than TERT mutations.
Telomerase activity of (A) novel TERC mutations and (B) novel TERT mutations were determined using the TRAP assay and are expressed as a % of wild-type activity. (C) A comparison of all TERC mutations (n = 23, blue circles) and TERT mutations (n = 15, pink diamonds) that we have studied shows that as a group, the TERC mutations give significantly lower residual telomerase activity (* P value = 0.04). Black bars indicate median values.
In contrast, only 2/7 of the TERT mutations give a TRAP activity of <10% wild type, while the others give an activity >15% normal (Table 1, Figure 1B). When we include mutations that we have analysed previously, (14 in TERC and 8 in TERT), we see that as a group, the TERC mutations give significantly lower telomerase activity than the TERT mutations (Figure 1C, Student’s t-test P-value = 0.04).
Disease severity varies between different genetic subtypes
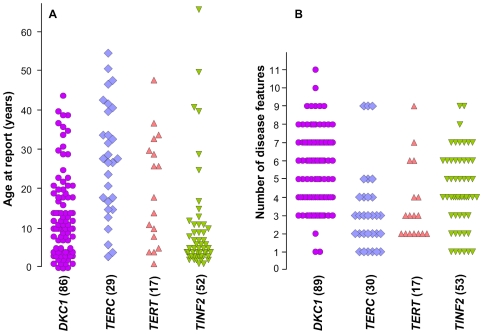
These novel data have prompted us to review the clinical and biological manifestations of different mutations across a large series of patients referred to our registry in London. We have information relating to 194 index cases in which we have defined the genetic basis of the disease: 91 with DKC1, 56 with TINF2, 30 with TERC and 17 with TERT mutations. As indicators of disease severity, we have compared the age of report and the number of presenting disease features between the different subtypes (Figure 2). No difference is seen in either category between patients with TERC and TERT mutations. However, those with DKC1 mutations present at a significantly younger age compared to those with TERC mutations (Student's t-test, P-value <.0001) and have a significantly greater number of disease features than those with both TERC and TERT mutations (P-value <.0001). Those with TINF2 mutations present at a younger age than those with both TERC and TERT mutations (P-value <.0001 and 0.008, respectively) and have more disease features than those with TERC mutations (P-value = 0.027).
Figure 2. Differences in clinical presentation between genetic subgroups of patients.
(A) Age at report of patients with different telomerase or shelterin mutations, as indicated below the panel. (B) The number of disease features observed in the same group of patients. The numbers of subjects (in brackets) differ slightly, reflecting the fact that some information is missing. The disease features that were scored included: abnormal skin pigmentation, nail dystrophy, leucoplakia, AA/cytopenia, immunodeficiency, cancer, oesophageal stenosis/dysphagia, enteropathy, liver disease, splenomegaly, pulmonary disease, epiphora, ear abnormality, deafness, dental decay/loss, hair greying/loss, retinopathy, microcephaly, cerebellar hypoplasia/ataxia, learning disabilities/developmental delay, abnormal facies, intrauterine growth retardation/low birth weight, short stature/growth retardation, aseptic necrosis, osteoporosis, renal disease, gonadal abnormality, phimosis, hyperhiderosis and cardiac disease.
We have also investigated the relative incidence of several clinically significant disease features: the presence of 2 or more of the classical mucocutaneous features, AA, cancer, pulmonary disease, cerebellar hypoplasia and short stature (Table 2). Again, no difference is seen in any category between patients with TERC and TERT mutations. However, mucocutaneous features are far less common in patients with either TERC or TERT mutations compared to DKC1 (Pearson's chi-squared test, P-value <0.0001). Patients with TINF2 mutations are also less likely to have mucocutaneous features than the DKC1 patients (P-value = 0.006), but are more likely to have them than in either TERC or TERT patients (P-value = 0.014 and 0.019, respectively). The incidence of AA is less among patients with DKC1 mutations than either TERC or TINF2 patients (P-value = 0.007 and 0.006, respectively); this is despite the fact that the TINF2 patient group is younger than the DKC1 group, while the TERC group is older than the DKC1 group. Cancer incidence is higher among patients with TERC and TERT mutations, significantly so compared to the TINF2 group (P-value = 0.003 and 0.011, respectively); this may well reflect the fact that they are generally an older patient group.
Table 2. Number of index cases with different disease features amongst different genetic subtypes.
| Gene | Number of cases | Aplastic anaemia (%) | 2 or more mucocutaneous features (%) | Cancer (%) | Pulmonary disease (%) | Cerebellar hypoplasia (%) | Short stature (%) |
| DKC1 | 91 | 44 (48.4) | 60 (65.6) | 7 (7.7) | 6 (6.6) | 15 (16.5) | 19 (20.8) |
| TINF2 | 56 | 40 (71.4) | 24 (42.9) | 1 (1.8) | 3 (5.5) | 6 (10.7) | 12 (21.4) |
| TERC | 30 | 23 (76.7) | 5 (16.7) | 6 (20.0) | 4 (13.3) | 1 (3.3) | 3 (10) |
| TERT * | 17 | 12 (70.6) | 2 (11.8) | 3 (17.6) | 3 (17.6) | 1 (5.8) | 2 (11.8) |
| All | 194 | 119 (61.3) | 91 (46.9) | 17 (8.8) | 16 (8.3) | 23 (11.9) | 36 (18.6) |
*Homozygotes excluded.
Telomere lengths do not explain differences in disease severity between genetic subtypes
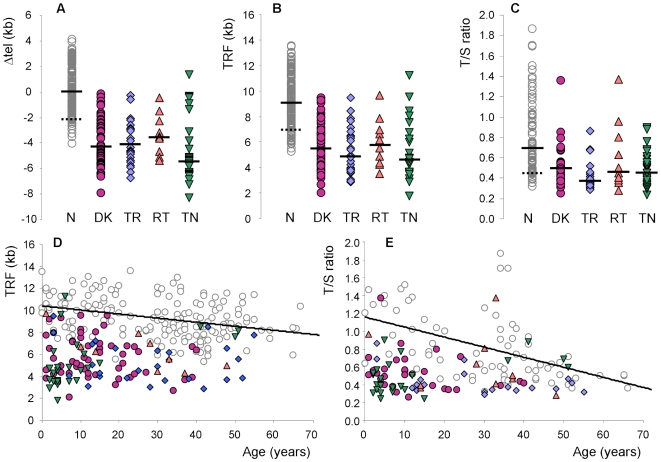
Despite the fact that we see significant differences in clinical severity between the different genetic subtypes of patients, both in terms of age of presentation and the number of disease features, we do not see any difference in telomere lengths between these groups of index cases. This is true whether we compare age adjusted (Figure 3A) or absolute (Figure 3B) telomere lengths as measured by Southern blot analysis, or when we compare telomere lengths determined by quantitative PCR (Figure 3C). Plots of telomere length against age (Figure 3D and 3E) are consistent with the idea that disease presents when telomeres shorten below a certain length, independent of age.
Figure 3. Telomere length measurements in patients with telomerase and shelterin mutations.
Different genetic subgroups of patients with telomerase and shelterin mutations, shown as purple circles (DKC1 mutations), blue diamonds (TERC mutations), pink triangles (TERT mutations) and inverted green triangles (TINF2 mutations) are compared to healthy controls (open grey circles). Solid bars indicate median values and the dotted bars indicate the 10th centiles of the healthy control measurements. (A) Telomere lengths were measured by Southern blot analysis and adjusted for age to give a Δtel value (54). N = normal healthy controls (n = 176), DK = patients with DKC1 mutations (n = 56), TR = patients with TERC mutations (n = 26), RT = patients with TERT mutations (n = 10), TN = patients with TINF2 mutations (n = 26). (B) Absolute telomere lengths, measured as terminal restriction fragments for the same group of patients as shown in panel A. (C) Telomere lengths measured as a T/S ratio, determined by monochrome multiplex quantitative PCR for a subset of patients shown in panel A (73 healthy controls and 29, 19, 10 and 23 patients with DKC1, TERC, TERT and TINF2 mutations, respectively). (D) Absolute telomere lengths (terminal restriction fragments) plotted against age in years, for the patients and health controls shown in panel A. A line of best fit is drawn for the healthy controls. (E) Telomere lengths measured by monochrome multiplex quantitative PCR analysis plotted against age in years for all patients in panel C. A line of best fit is drawn for the healthy controls.
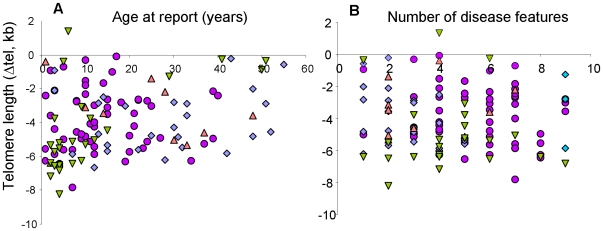
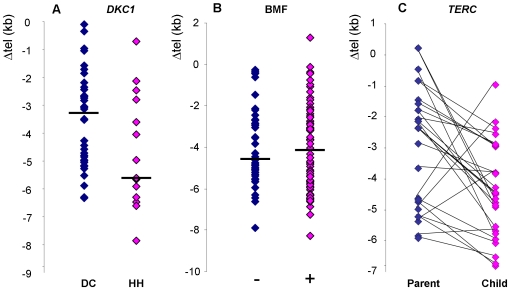
Taking all index cases together, there is no correlation between telomere length and either age of report (Figure 4A) or number of disease features (Figure 4B). The same is true if each subtype is analysed separately (Figures S3 and S4), although there is a trend toward earlier presentation with shorter telomeres among patients with TINF2 mutations (R2 = 0.424). However, we do see that among patients with DKC1 mutations, those that present with the severe HH syndrome have shorter telomeres than those that present with more classical DC (P-value = 0.03, Figure 5A). Somewhat surprisingly, when we compare all patients who did and did not present with bone marrow failure, we do not see any difference in telomere length (Figure 5B).
Figure 4. No correlation between telomere length and clinical presentation.
Among all index cases (n = 118) there is no correlation seen between telomere length and either (A) the age at report (R2 = 0.062) or (B) the number of disease features observed (R2 = 0.007). Symbols for the different disease subgroups are as described for Figure 3.
Figure 5. Comparison of telomere lengths in specific patient subgroups.
(A) Amongst patients with DKC1 mutations, telomere lengths are longer in those that have the more classical disease, DC (n = 42, indigo diamonds), compared to those that have the more severe HH phenotype (n = 14, pink diamonds). (B) No difference in telomere length is seen between patients who presented with bone marrow failure (n = 66, pink diamonds) and those that did not have bone marrow failure (n = 46, indigo diamonds). (C) In 27 parent-child combinations of subjects with heterozygous TERC mutations, the children (with 4 exceptions) have shorter telomeres than their parents.
Another factor that can be taken into consideration is the number of asymptomatic relatives (both younger and older) that exist in each of the subtypes: while we know of very few asymptomatic relatives with TINF2 or DKC1 mutations, this is a common feature of families with TERC and TERT mutations. It has been proposed that is at least in part due to disease anticipation in families with both TERC and TERT mutations, and that this is associated with progressive telomere shortening through the generations. We now know of 27 parent-child combinations from 15 different families in which both are TERC heterozygotes. In all but 4 of these combinations, the affected child has a shorter age-adjusted telomere length than their affected parent (Figure 5C). As a group, the children have significantly shorter telomeres than the parents (P value = 0.0026) with a median difference in Δtel between parent and child (Δtel, child-Δtel, parent) of −1.53 kb.
Discussion
This is the first study to systematically review the relationship between telomere length and disease severity in a large cohort of unrelated patients with mutations in different telomere-related genes. While clear phenotypic differences exist between the genetic subtypes, there is no difference in the range of telomere lengths observed. This conclusion may seem somewhat surprising as several previous studies have indicated that disease severity may be related to telomere length. These include the phenomenon of disease anticipation associated with telomere shortening in families with TERC and TERT mutations [17], [40], [41], shorter telomeres among patients with TINF2 mutations [42], and patients with severe disease caused by DKC1 mutations having shorter telomeres than those with mild disease [43].
There are several differences between previous studies and the one we present here. Firstly, we are now looking at a considerably larger number of patients with telomere defects, representing the largest series published to date. Secondly, we have focused on the index cases only to avoid the issues relating to asymptomatic individuals with telomerase/shelterin mutations, particularly in the context of disease anticipation. Thirdly, we are comparing different genetic subtypes of the disease, rather than looking within a single subtype.
The four common genetic subtypes of DC (caused by mutations in the DKC1, TINF2, TERC, and TERT genes) can be divided into three groups: (i) the inherited autosomal dominant mutations in the telomerase-specific components, TERC and TERT; (ii) the X-linked recessive mutations in DKC1, encoding a protein that acts as a pseudouridine synthase in H/ACA small nuclear ribonucleoprotein (snoRNP) complexes [44], predominantly involved in ribosomal RNA processing [45], as well as a core component of telomerase; and (iii) the predominantly de novo mutations in TINF2, encoding a core component of the telomere-binding protein complex, shelterin [46], [47]. Mutations in any of these genes can clearly impact dramatically on the ability to maintain telomere length and result in a multisystem disease. However, the fact that three different pathways are affected in the different genetic subtypes may explain why telomere length alone does not correlate with disease severity. This suggests other biological defects are likely to be important in the disease pathology and the resulting overall clinical phenotype. Although there is no clear evidence of a defect in pseudouridylation in patients with DKC1 mutations [48], [49], several studies have indicated that in mice they can have a significant impact beyond telomere maintenance, affecting snoRNA accumulation [50], the DNA damage response [51] and IRES-mediated translation [52].
No significant differences were observed in clinical presentation of patients with TERC and TERT mutations in our cohort. This is despite the fact that as a group, the TERC mutations gave lower residual telomerase activity than TERT mutations in the in vitro TRAP assay. No equivalent comparison has been made previously although a trend toward higher residual TRAP activity of TERT mutations has been apparent in other studies [26], [30], [32]. In the largest series of TERT mutations published to date [37], the mean age of 134 carriers was 51 years; 50% of these heterozygotes had pulmonary disease but only 16% had haematological abnormalities. This highlights the variable penetrance as well as the diverse and heterogeneous nature of the disease caused by TERT mutations. A similar spectrum of disease severity can be observed in families with TERC mutations, although these appear more likely to present with haematological abnormalities than pulmonary disease. Nevertheless, the disease seen in patients with both TERC and TERT mutations clearly contrasts dramatically with the severe early onset of DC, the HH syndrome, and/or AA that can be seen in patients with mutations in either the shelterin component, TINF2 or the snoRNP component, DKC1.
It is interesting to consider how the rate of telomere shortening differs between the genetic subtypes, even though this is very difficult to measure empirically in individual patients. However, in a typical patient with a TINF2 mutation (e.g. the recurrent Arg282His substitution), this will have arisen de novo and presentation, with very short telomeres occurs in the early years of life. A similar rapid shortening of telomeres will have occurred in approximately 1/3 of patients with DKC1 mutations (e.g. the recurrent Ala353Val substitution) in which the mutation will also have occurred de novo; in patients that inherit a DKC1 mutation, they do so from an asymptomatic heterozygous mother, who will show a highly skewed pattern of X-inactivation in peripheral blood. Both situations are completely different to the typical presentation of TERC and TERT mutations which, although dominant, are usually present in the family for at least one asymptomatic generation. The rate of telomere shortening here appears to be much slower, with critically short telomeres manifesting in disease at an older age and after transmission through the germ line.
One confounding factor in the comparison of different mutations within and between genetic subtypes is the diversity of the mutations themselves. Some mutations are clearly and recognizably pathogenic because they recur (such as Ala353Val in DKC1 and Arg282His in TINF2) or because they abolish telomerase activity (as seen in TERC and TERT). However with private missense mutations in DKC1 and TINF2 or with mutations that only partially reduce telomerase activity in TERC and TERT, it is more difficult to be sure that they are disease causing, rather than bystander mutations.
The significant clinical differences observed between the genetic subgroups (such as the early onset of disease and greater number of disease features in DKC1 and TINF2 patients compared to TERC and TERT patients and the higher incidence of cancer in patients with TERC mutations) will facilitate more informed decisions in the clinic. Equally these findings serve to highlight that whilst these genetic subtypes are unified by having a defect in telomere biology (the “telomereopathies”) the clear differences they exhibit suggest other factors (inherited and acquired) contribute to the overall clinical phenotype. Finally, as a major subgroup of the novel mutations were observed in patients who did not fulfil the criteria to be classified as DC, this study highlights the importance of screening the telomerase and shelterin genes in patients with a spectrum of clinical phenotypes in addition to DC.
Methods
Subjects
All individuals included in this study have given written informed consent in accordance with the Declaration of Helsinki and our study design, approved by the East London and The City Research Ethics Committee. The novel mutations reported in this study were identified among 732 patients referred to our registry over a period of 69 months. They comprised 318 patients with AA, 99 patients who had AA in combination with a family history and/or other somatic disease features (constitutional AA), 81 patients with MDS, 123 patients who had features overlapping those of DC, but insufficient for a clinical diagnosis to be made and 111 who were classified as having DC on clinical grounds. The latter were defined as having either (i) the diagnostic triad of mucocutaneous features, or (ii) 1 or more of these features combined with a hypoplastic bone marrow and at least 2 other features known to occur in DC, or (iii) at least 4 of the 6 features commonly associated with the HH syndrome (intrauterine growth retardation, developmental delay, microcephaly, cerebellar hypoplasia, immunodeficiency and bone marrow failure). From this same group of patients we have previously reported on 4 families with MDS/AML and mutations in TERC or TERT [53], as well as 33 families with mutations in the TINF2 gene [39], [42] and have also identified 14 families with recurrent DKC1 mutations and 3 families with recurrent TERC mutations.
Mutation analysis
Genes were scanned for mutation as previously described [43]. Briefly, fragments encompassing all coding exons and flanking intronic sequences of each gene were amplified from genomic DNA by PCR. These fragments were then subjected to denaturing HPLC analysis (in the presence of equal amounts of wild-type DNA fragments in the case of the X-linked DKC1 gene). Where abnormal patterns of elution were identified, the fragments were re-amplified and sequenced directly using the BigDye™ chain termination method and a 3130xl genetic analyzer (Applied Biosystems).
Telomere length measurement
Telomere terminal restriction fragment lengths were determined by Southern blot analysis using the sub-telomeric probe pTelBam8 in DNA extracted from total peripheral blood white cells, as previously described [16]. Age adjusted lengths (Δtel) for any one individual were determined by subtracting the expected telomere length, obtained from a regression line drawn from 176 healthy controls, from the observed telomere length as described [54].
We have also used a monochrome multiplex quantitative PCR method to measure telomere length [55] in which the amount of telomeric DNA (T) and the amount of a single copy gene (S) are quantified using standard curves established by titration of a reference genomic DNA sample. All samples were measured in triplicate. T/S ratios, which are proportional to the telomere length [55], were then normalised against a second reference DNA sample that was run on every plate. We have adapted the method for use on a LightCycler 480 real time thermocycler (Roche) as described previously [21].
Telomerase repeat amplification protocol (TRAP) assay
The TRAP assay was performed as previously described [53]. Briefly, mutations identified in the TERC or TERT genes were introduced into plasmids harbouring the wild-type gene using a QuikChange Site-Directed Mutagenesis kit (Stratagene). Plasmids were then transfected into WI-38 VA13 cells and after 48 hours the telomerase activity was measured using the TRAPeze RT Telomerase Detection Kit (Millipore/Chemicon).
Statistical methods
Student's t-tests were performed in ExCel and the Pearson's chi-squred test was performed using VassarStats (http://faculty.vassar.edu/lowry/VassarStats.html).
Supporting Information
Sequence traces and location of novel TERC mutations. Arrows indicate the heterozygous base change named beneath each panel. Their location is shown on a sketch of the TERC molecule.
(PPT)
Sequence traces and conservation of novel TERT mutations. Arrows indicate the heterozygous base change named beneath each panel. Alignment of the human, mouse, chicken, frog, yeast and plant TERT protein sequences (generated by MUSCLE at the NCBI) indicates the degree of conservation of the affected amino acid, shown in bold font.
(PPT)
No correlation between age at report and telomere length. Age at report versus telomere length of index cases in different genetic subtypes, as indicated on each panel.
(PPT)
No correlation between number of disease features and telomere length. Number of disease features versus telomere length in index cases with different genetic subtypes, as indicated on each panel.
(PPT)
Acknowledgments
We thank all patients and family members who have given the samples that made this study possible. We also thank the doctors who referred samples to our registry, in particular Drs. F. Bonifazi, J. Cavenagh, E. A. Chalmers, N. Chassing, V. Chowdhury, N. Daguindau, G. Davies, M. Fuhrer, E. C. Gordon-Smith, D. Luk, M. Mohamed, J. Nathilde, C. O'Connell, C. Oudot, A. Schwarer, R. Skinner, J. Soulier, C. Steward and J. Zonana.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors are grateful to The Wellcome Trust (http://www.wellcome.ac.uk/) for financial support, through project grant number 085937/Z/08/Z. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 3.Olovnikov AM. Telomeres, telomerase, and aging: origin of the theory. Exp Gerontol. 1996;31:443–448. doi: 10.1016/0531-5565(96)00005-8. [DOI] [PubMed] [Google Scholar]
- 4.Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, et al. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 6.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 7.Lansdorp PM. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 11.Dokal I. Dyskeratosis congeinta in all its forms. Br J Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 12.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113:6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessler M, Wilson DB, Mason PJ. Dyskeratosis congenita. FEBS Lett. 2010;584:3831–3838. doi: 10.1016/j.febslet.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savage SA, Dokal I, Armanios M, Aubert G, Cowen EW, et al. Dyskeratosis congenita: the first NIH clinical research workshop. Pediatr Blood Cancer. 2009;53:520–523. doi: 10.1002/pbc.22061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heiss NS, Knight SW, Vulliamy TJ, Klauk SM, Wiemann S, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genetics. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 16.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 17.Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci USA. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walne AJ, Vulliamy T, Beswick R, Marrone A, Al-Qurashi F, et al. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase associated protein NOP10. Hum Mol Genet. 2007;16:1619–1629. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vulliamy T, Beswick R, Kirwan M, Marrone M, Digweed M, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci USA. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, et al. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. Mutations in C16orf57 and normal length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and Rothmund-Thomson syndrome. Hum Mol Genet. 2010;19:4453–4461. doi: 10.1093/hmg/ddq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, et al. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 24.Vulliamy TJ, Knight SW, Mason PJ, Dokal I. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol Dis. 2001;27:353–357. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- 25.Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, et al. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du HY, Pumbo E, Ivanovich J, An P, Maziarz RT, et al. TERC and TERT gene mutations in patients with bone marrow failure and the significance of telomere length measurements. Blood. 2009;113:309–316. doi: 10.1182/blood-2008-07-166421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armanios M. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight SW, Heiss NS, Vulliamy TJ, Aalfs CM, Hennekman RCM, et al. Unexplained aplastic anaemia, immunodeficiency and cerebellar hypoplasia (Hoyeraal-Hreidarsson syndrome) is caused by mutation in the DKC1 gene. Brit J Haematol. 1999;107:335–339. doi: 10.1046/j.1365-2141.1999.01690.x. [DOI] [PubMed] [Google Scholar]
- 29.Vulliamy TJ, Marrone A, Dokal I, Mason PJ. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359:2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi H, Baerlocher GM, Lansdorp PM, Chanock SJ, Nunez O, et al. Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood. 2003;102:916–918. doi: 10.1182/blood-2003-01-0335. [DOI] [PubMed] [Google Scholar]
- 32.Xin ZT, Beauchamp AD, Calado RT, Bradford JW, Regal JA, et al. Functional characterization of natural telomerase mutations found in patients with hematologic disorders. Blood. 2007;109:524–532. doi: 10.1182/blood-2006-07-035089. [DOI] [PubMed] [Google Scholar]
- 33.Calado RT, Regal JA, Hills M, Yewdell WT, Dalmazzo LF, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106:1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 35.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci U S A 2007. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calado RT, Regal JA, Kleiner DE, Schrump DS, Peterson NR, et al. A spectrum of severe familial liver disorders associate with telomerase mutations. PLoS One. 2009;4:e7926. doi: 10.1371/journal.pone.0007926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diaz de Leon A, Cronkhite JT, Katzenstein AL, Godwin JD, Raghu G, et al. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS One. 2010;5:e10680. doi: 10.1371/journal.pone.0010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasa G, Ribes-Zamora A, Nelson N, Bertuch A. Clin Genet. in press; 2011. Three novel truncating TINF2 mutations causing severe dyskeratosis congenita in early childhood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vulliamy T, Beswick R, Kirwan M, Hossain U, Walne A, et al. Clin Genet in press; 2011. Telomere length measurement can distinguish pathogenic from non-pathogenic variants in the shelterin component, TIN2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, et al. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nature Genetics. 2004;36:447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 41.Goldman F, Bouarich R, Kulkarni S, Freeman S, Du HY, et al. The effect of TERC haploinsufficiency on the inheritance of telomere length. Proc Natl Acad Sci USA. 2005;102:17119–17124. doi: 10.1073/pnas.0505318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vulliamy TJ, Marrone M, Knight S, Walne A, Mason PJ, et al. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107:2680–2685. doi: 10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 44.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 46.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye JZ, de Lange T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet. 2004;36:618–623. doi: 10.1038/ng1360. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 49.Wong JM, Kyasa MJ, Hutchins L, Collins K. Telomerase RNA deficiency in peripheral blood mononuclear cells in X-linked dyskeratosis congenita. Hum Genet. 2004;115:448–455. doi: 10.1007/s00439-004-1178-7. [DOI] [PubMed] [Google Scholar]
- 50.Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc Natl Acad Sci U S A. 2004;101:10756–10766. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu BW, Bessler M, Mason PJ. A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc Natl Acad Sci U S A. 2008;105:10173–10178. doi: 10.1073/pnas.0803559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 53.Kirwan M, Vulliamy T, Marrone A, Walne AJ, Beswick R, et al. Defining the pathogenic role of telomerase mutations in myelodysplastic syndrome and acute myeloid leukaemia. Hum Mutation. 2009;30:1567–1573. doi: 10.1002/humu.21115. [DOI] [PubMed] [Google Scholar]
- 54.Brümmendorf TH, Maciejewski JP, Mak J, Young NS, Lansdorp PM. Telomere length in leukocyte subpopulations of patients with aplastic anemia. Blood. 2001;97:895–900. doi: 10.1182/blood.v97.4.895. [DOI] [PubMed] [Google Scholar]
- 55.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence traces and location of novel TERC mutations. Arrows indicate the heterozygous base change named beneath each panel. Their location is shown on a sketch of the TERC molecule.
(PPT)
Sequence traces and conservation of novel TERT mutations. Arrows indicate the heterozygous base change named beneath each panel. Alignment of the human, mouse, chicken, frog, yeast and plant TERT protein sequences (generated by MUSCLE at the NCBI) indicates the degree of conservation of the affected amino acid, shown in bold font.
(PPT)
No correlation between age at report and telomere length. Age at report versus telomere length of index cases in different genetic subtypes, as indicated on each panel.
(PPT)
No correlation between number of disease features and telomere length. Number of disease features versus telomere length in index cases with different genetic subtypes, as indicated on each panel.
(PPT)