We consider the vertebrate somite segmentation clock as an example of a rhythmic phenomenon that occurs in development. Using mouse genetics and mathematical analyses, we found that the period of the clock in each presomitic cell is sensitive to Notch activity. It may be a system for each cell to adapt to its local environment.
Abstract
The number of vertebrae is defined strictly for a given species and depends on the number of somites, which are the earliest metameric structures that form in development. Somites are formed by sequential segmentation. The periodicity of somite segmentation is orchestrated by the synchronous oscillation of gene expression in the presomitic mesoderm (PSM), termed the “somite segmentation clock,” in which Notch signaling plays a crucial role. Here we show that the clock period is sensitive to Notch activity, which is fine-tuned by its feedback regulator, Notch-regulated ankyrin repeat protein (Nrarp), and that Nrarp is essential for forming the proper number and morphology of axial skeleton components. Null-mutant mice for Nrarp have fewer vertebrae and have defective morphologies. Notch activity is enhanced in the PSM of the Nrarp−/– embryo, where the ∼2-h segmentation period is extended by 5 min, thereby forming fewer somites and their resultant vertebrae. Reduced Notch activity partially rescues the Nrarp−/– phenotype in the number of somites, but not in morphology. Therefore we propose that the period of the somite segmentation clock is sensitive to Notch activity and that Nrarp plays essential roles in the morphology of vertebrae and ribs.
INTRODUCTION
The somite is the earliest discernible metameric structure in vertebrates. It gives rise to vertebrae, ribs, and skeletal muscles, thereby providing a segmental pattern along the anterior–posterior axis. Somites are aligned along both sides of the neural tube. A pair of somites buds off sequentially from the anterior extremity of the presomitic mesoderm (PSM) in a rhythmic manner (Pourquie, 2001). The periodicity of this repetitive process is orchestrated by the synchronous oscillation of gene expression in the PSM, termed the “somite segmentation clock” (Pourquie, 2003). Some of the genes that are cyclically activated and deactivated, including Hes7 and Lunatic fringe (Lfng), are components of Notch signaling (Forsberg et al., 1998; McGrew et al., 1998; Aulehla and Johnson, 1999; Bessho et al., 2001b).
Notch signaling is cell–cell contact–dependent and plays a major role in development (Bolos et al., 2007). After Notch binds to its ligand, Delta, on an adjacent cell, it is activated and undergoes limited proteolysis, which is dependent on γ-secretase activity. Here the Notch intracellular domain (NICD) moves into the nucleus, where it forms a complex with recombinant signal binding protein for immunoglobulin kappa J region (RBPj) to activate target genes, including Hes7 and Lfng (Bessho et al., 2001a; Cole et al., 2002; Morales et al., 2002). In mouse, Hes7 inhibits transcription of its own gene and of Lfng to establish a negative feedback loop that induces oscillatory Hes7 and Lfng expression (Bessho et al., 2003). Hes7 or Lfng knockout mice have severe defects in their somites and resultant vertebrae (Evrard et al., 1998; Zhang and Gridley, 1998; Bessho et al., 2001b). Therefore Notch signaling regulates the segmentation clock, which in turn orchestrates somite patternings and the resultant vertebrae.
In addition to Notch components, the amount of NICD oscillates in synchrony with Hes7 and Lfng expression in the PSM. This periodicity of Notch activity contributes to periodic somite segmentation (Morimoto et al., 2005). NICD activates Lfng transcription, and Lfng inhibits NICD production to form a negative feedback loop (Dale et al., 2003). Because Lfng expression is cyclical in the PSM, this negative feedback loop probably contributes to oscillatory NICD production. The dynamic Notch activity that is produced by the compound feedback loops may play a crucial role in regulating the segmentation clock because it acts upstream of Hes7 and Lfng.
Notch-regulated ankyrin repeat protein (Nrarp) encodes a small 114-amino-acid residue protein with two ankyrin repeat motifs in the carboxy terminus. This protein is highly conserved among vertebrates. Nrarp is induced by Notch signaling (Krebs et al., 2001; Lamar et al., 2001; Topczewska et al., 2003; Pirot et al., 2004), and it promotes NICD loss by forming a ternary complex with NICD and RBPj (Lamar et al., 2001). Thus Nrarp acts as a feedback regulator in Notch signaling. In addition, it is expressed periodically in the PSM (Sewell et al., 2009; Wright et al., 2009). The roles of Nrarp in the segmentation clock and somitogenesis, however, remain largely unclear.
To investigate the roles of Nrarp in these processes, we generated an Nrarp knockout mouse and inspected its segmentation clock and somitogenesis. Here we show that Nrarp is essential for proper axial skeleton formation. Null-mutant Nrarp mice have small but substantial defects in their vertebrae and ribs. In addition, Nrarp mutants lose two vertebrae as a consequence of the extended somite segmentation clock period. The loss of Nrarp increases Notch signaling but does not affect any other signaling, including Wnt signaling in the PSM. Remarkably, an inhibitor of Notch signaling partially rescues the clock period phenotype. It failed, however, to improve the defects in the axial skeleton components. Thus our studies suggested that Nrarp plays indispensable roles in securing the number and morphology of axial skeleton components.
RESULTS
Nrarp−/– mutant mice had defects in axial skeletons and fewer vertebrae
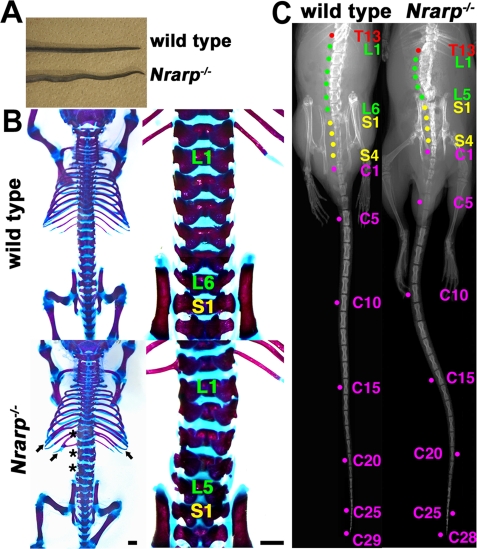
Nrarp is strongly expressed in the PSM in a cyclical manner during mouse development (Sewell et al., 2009; Wright et al., 2009). We expected it to play a role in vertebrae and rib formation, because the PSM is the primordial somite, which gives rise to the vertebrae and ribs. To investigate the role of Nrarp, we disrupted the Nrarp gene (Supplemental Figure S1). Twenty-two of 55 Nrarp-homozygous mice had kinked tails (Figure 1A). Interestingly, the Hes7-heterozygous mutants had similarly kinked tails (Bessho et al., 2001b), although Nrarp-homozygous mutants express normal levels of Hes7 in the PSM (Figures 3D and 4B). According to this phenotype, we anticipated defects to be present in the axial skeleton. Thus we visualized the bones and cartilage of newborn mice, and inspected the shape and numbers of their vertebrae and ribs. In all of the Nrarp−/– mutant mice, a few vertebrae and ribs had small defects, which included partial fusion of the ribs and abnormally shaped vertebrae (Figure 1B). The majority of the vertebrae and ribs were normal. We next examined the skeletal patterns of adult mice by x-ray computed tomography (CT). Similar to the newborns, Nrarp−/– mutant adult mice had a few abnormal vertebrae and ribs (Figure 1C). Therefore Nrarp was essential for minute configuration of the axial skeleton.
FIGURE 1:
Skeletal defects of Nrarp-null mice. (A) A kinked tail that was typical of adult homozygous mutants. (B) Dorsal views and high magnifications (right) of skeletal preparations at postnatal day 1. The arrows and the asterisk are used to indicate the location of defective ribs and vertebrae, respectively. L, lumbar vertebra; S, sacral vertebra. Scale bars, 1 mm. (C) x-ray analysis was used to reveal malformation and decreased numbers of vertebra in adult Nrarp−/– mice. T, thoracic vertebra; L, lumbar vertebra; S, sacral vertebra; C, caudal vertebra.
FIGURE 3:
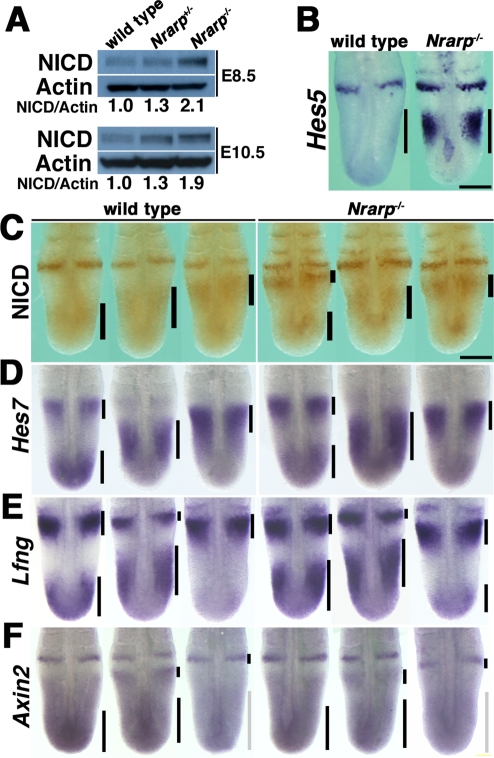
Loss of Nrarp specifically increases Hes5 expression but does not affect cyclical gene expression patterns. (A) Quantification of NICD by immunoblotting (n = 3). The ratios of NICD to β-actin are indicated. (C) Immunoreactivity of NICD in the PSM. The black bars are used to indicate the location of the NICD. The activity in the PSM and the caudal parts of the somites was higher in the Nrarp−/– embryos, although dynamic NICD distribution patterns were maintained. Scale bar, 200 μm. (B, D–F) Whole-mount in situ hybridization for Hes5, Hes7, Lfng, and Axin2, at E 10.5. Scale bar, 200 μm. (B) Hes5 expression increased dramatically in the PSM of the Nrarp−/– embryos, but the pattern was not affected. (D–F) The expression pattern was divided into three phases, Phase I (left), Phase II (middle), and Phase III (right), according to a previous report. The expression patterns of Hes7, Lfng, and Axin2 were not affected in the Nrarp−/– embryos. The bars are used to indicate the expression domain: the black bars indicate strong expression and the gray bars indicate weak expression.
FIGURE 4:
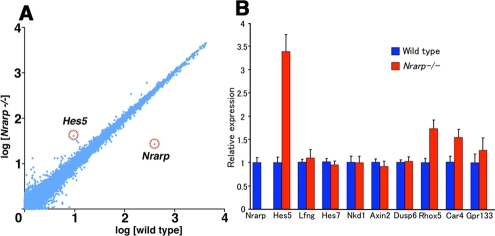
Comparison of gene expression levels between wild-type and Nrarp−/– embryos. (A) Comparison of gene expression levels in the PSM between the Nrarp−/– and the wild-type embryos (n = 3). In this microarray analysis, the average of the normalized signal intensity was plotted. (B) The expression levels of the Notch, Wnt, and FGF signaling downstream genes were compared by using quantitative RT-PCR (n = 3; mean ± SD). Hes5 expression increased dramatically, and that of the Nrarp was depleted in the PSM of Nrarp−/– embryos. There was no difference in the expression of other genes except for Rhox5, Car4, and Gpr133. These data are consistent with the results of the microarray analysis.
We counted the number of vertebrae. The total number of vertebrae was 57.4 ± 0.2 and 59.0 ± 0.0 in the Nrarp−/– mutant mice (n = 9) and in their wild-type littermate mice (n = 7), respectively (Table 1). Most of the Nrarp−/– mice lost a lumber vertebra and a caudal vertebra (Table 1). These results were consistent with the results for the newborn mice, for which all of the Nrarp−/– had five lumbar vertebrae, whereas their wild-type littermate newborn mice had six (Figure 1B and Supplemental Table S1). We did not observe total fusion of the vertebrae or ribs in the Nrarp−/– mice; even some of them had small defects. Therefore it was not likely that the fusion of two adjacent vertebrae reduced the number of vertebrae. Thus Nrarp was essential for securing the number and configuration of axial skeleton components.
TABLE 1:
X-ray analyses of adult mice vertebrae.
| Genotype (Nrarp) | Cervical | Thoracic | Lumbar | Sacral | Caudal | Total |
|---|---|---|---|---|---|---|
| +/+ | 7 | 13 | 6 | 4 | 29 | 59 |
| +/+ | 7 | 13 | 6 | 4 | 29 | 59 |
| +/+ | 7 | 13 | 6 | 4 | 29 | 59 |
| +/+ | 7 | 13 | 6 | 4 | 29 | 59 |
| +/+ | 7 | 13 | 6 | 4 | 29 | 59 |
| +/+ | 7 | 13 | 6 | 4 | 29 | 59 |
| +/+ | 7 | 13 | 6 | 4 | 29 | 59 |
| +/− | 7 | 13 | 5 | 5 | 29 | 59 |
| +/− | 7 | 13 | 6 | 4 | 29 | 59 |
| +/− | 7 | 13 | 6 | 4 | 29 | 59 |
| +/− | 7 | 13 | 6 | 4 | 29 | 59 |
| +/− | 7 | 13 | 6 | 4 | 29 | 59 |
| +/− | 7 | 13 | 6 | 4 | 30 | 60 |
| −/− | 7 | 13 | 5 | 5 | 28 | 58 |
| −/− | 7 | 13 | 5 | 5 | 27 | 57 |
| −/− | 7 | 13 | 5 | 4 | 29 | 58 |
| −/− | 7 | 13 | 5 | 4 | 29 | 58 |
| −/− | 7 | 13 | 5 | 5 | 28 | 58 |
| −/− | 7 | 13 | 5 | 4 | 28 | 57 |
| −/− | 7 | 13 | 6 | 4 | 28 | 58 |
| −/− | 7 | 13 | 5 | 4 | 28 | 57 |
| −/− | 7 | 13 | 5 | 4 | 27 | 56 |
We counted vertebrae by using x-ray CT imaging. Although there were some fluctuations in the number of lumbar and sacral vertebrae, the Nrarp−/− mice had fewer vertebrae than their wild-type littermates did.
Nrarp−/– mutant mice had fewer somites
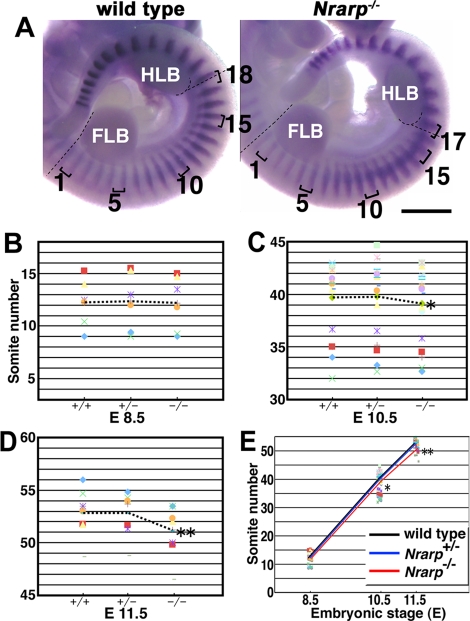
Somites give rise to vertebrae in a one-to-one ratio; the caudal half of a pair of somites and the rostral half of the next pair merge into a vertebra (Gossler and Hrabe de Angelis, 1998). Thus the number of vertebrae corresponds to the number of somites. Somites are transient structures in development and differentiate subsequently into bones, muscle, and skin. Therefore it was difficult to count the total number of somites. We compared the number of somites located between the fore- and hind-limb buds (FLBs and HLBs, respectively) at embryonic day (E) 10.5. The Nrarp−/– embryos had 17 somites between the limb buds (n = 13), whereas their wild-type littermate embryos had 18 somites (n = 34) (Figure 2A). This result was consistent with the skeletal phenotype, and also suggested that the positions of the limb buds were not affected in the absence of Nrarp.
FIGURE 2:
Nrarp-null mice have smaller numbers of somites. (A) Comparison of the numbers of somites between the FLB and the HLB. The dashed lines are used to indicate the rostral borders of the FLB and the HLB. Uncx4.1 expression was detected in the posterior half of each somite. The brackets are used to indicate single somites. Scale bar, 1 mm. (B–E) The average number of somites for each indicated genotype was plotted, and the same symbol was used to indicate the littermate. The averages of the total numbers of somites within littermates were compared using the paired t test. The dashed lines are used to indicate the average number of somites for each genotype at each stage (mean ± SEM). (B) At E 8.5, no significant difference was detected between the wild type (12.24 ± 0.93) and the Nrarp+/− (12.36 ± 1.14) or Nrarp−/– (12.20 ± 1.08) embryos (n = 6 pregnant females). (C) At E 10.5, the number of somites in the Nrarp−/– embryos (39.16 ± 0.84) was significantly lower than in the wild-type embryos (39.69 ± 0.83) (n = 18; *p < 0.02). The average of the difference was 0.5. (D) At E 11.5, the number of somites in Nrarp−/– embryos (51.11 ± 0.95) was significantly lower than in the wild-type embryos (52.85 ± 0.78) (n = 8; **p < 0.01). The average of the difference was 1.7. (E) The average number of somites of the wild-type embryos (black) and of the Nrarp+/− embryos (blue) and the Nrarp−/– embryos (red) were compared at three different stages. There were no significant differences between the wild-type embryos and the Nrarp+/− embryos at any stage.
Judging from the expression of Uncx4.1, which was exclusively restricted to the posterior compartment of each somite, at E 8.5, E 10.5, and E 11.5 (Figure 2A and Supplemental Figure S2), the morphology of the somites was almost normal. We did not find any defects in their shape, so we ruled out the possibility that the fusion of two adjacent somites caused a reduction in the number of somites. In addition, because the Nrarp−/– embryos were not distinguishable from the Nrarp+/− or wild-type embryos, and because they showed similar expression pattern of Fgf8 (Supplemental Figure S3), we ruled out the possibility of a small delay in the general development of the Nrarp−/– embryos.
The segmentation clock period was longer in Nrarp−/– mutant mice
We showed that Nrarp−/– mice had fewer numbers of somites and vertebrae, respectively. This result led us to two possibilities: The somite formation period was longer and/or the total duration of somite formation was shorter in the Nrarp−/– embryos. Thus we counted the number of somites at various stages to measure the somite formation period. Somite formation starts between E 7.5 and E 8.0. We failed to detect a significant difference in the number between the genotypes throughout the initial stage of somite formation at the early stage, E 8.5 (approximately 12; Figure 2B). Thus we presumed that the start of somite formation was not affected. At E 10.5 and E 11.5, the number of somites in the Nrarp−/– embryos was significantly lower than that of their wild-type littermate embryos (Figure 2, C and D). Numerically speaking, the 95% confidence interval (CI) of mean decrease in the somite number of Nrarp−/– from that of wild type was (–2.65, –1.10) at E 11.5. Because the average number of somites formed during the 72 h from E 8.5 to E 11.5 was 40.6 in the wild-type embryos, on average somites of (37.75, 39.30) were produced (95% CI) during the 72 h in the Nrarp−/– embryos, to correspond to the above CI of the mean decrease. This difference in number of somites between the Nrarp−/– embryos and their wild-type littermate embryos was consistent with the difference in the total number of vertebrae (59.0 vs. 57.4). Using this result, we estimated that the period of somite formation was ∼106 min and 111 min in the wild-type embryos and the Nrarp−/– embryos, respectively (Figure 2E). Therefore the loss of Nrarp extends the segmentation clock period by ∼5 min, and this extension resulted in fewer numbers of somites and vertebrae.
Notch activity was up-regulated in the Nrarp−/– PSM
Nrarp reduces Notch activity by enhancing NICD degradation (Lamar et al., 2001). Thus we predicted that Notch activity was increased in the PSM cells of the Nrarp−/– embryos. We collected the PSM of several embryos and determined the quantity of NICD to measure the Notch activity in the PSM. The amount of NICD increased 1.9-fold in the PSM of the Nrarp−/– embryos at E 10.5 (Figure 3A) over that in their wild-type littermate embryos. However, the cyclical pattern of NICD immunoreactivity was not affected in the mutant PSM (Figure 3C). Hes5 expression, another target gene of Notch, was up-regulated (Figure 3B). Similar to the NICD immunoreactivity, Hes5, Hes7, and Lfng expression was cyclical in the Nrarp−/– PSM in the same way as in wild-type PSM (Figure 3, D and E). Therefore the loss of Nrarp enhanced Notch activity but did not affect its cyclical pattern of activity.
Because Nrarp has been reported to up-regulate Wnt signaling (Ishitani et al., 2005; Phng et al., 2009), we checked the expression of Axin2, which acts downstream of Wnt signaling. We detected the same level of Axin2 mRNA in the Nrarp−/– PSM as in the wild-type PSM, and the cyclical pattern was not affected (Figure 3F). We next carried out a microarray analysis to compare comprehensively the gene expression in the Nrarp−/– PSM to that in their wild-type littermate PSM. We detected an increase in Hes5 expression as well as a dramatic decrease in Nrarp expression (Figure 4A). The expression of the Wnt target genes (including Axin2, Lef1, and mesogenin) was not altered, however, in the Nrarp−/– PSM (Figure 4B and Supplemental Table S2). In addition, the expression of target genes including dusp and sprouty of fibroblast growth factor (FGF) signaling was not altered (Figure 4B and Supplemental Table S2), although FGF signaling plays an important role in somitogenesis. Thus we found that a loss of Nrarp leads to a remarkable increase in Notch activity in the PSM.
Reduced Notch activity shortened the period of the segmentation clock
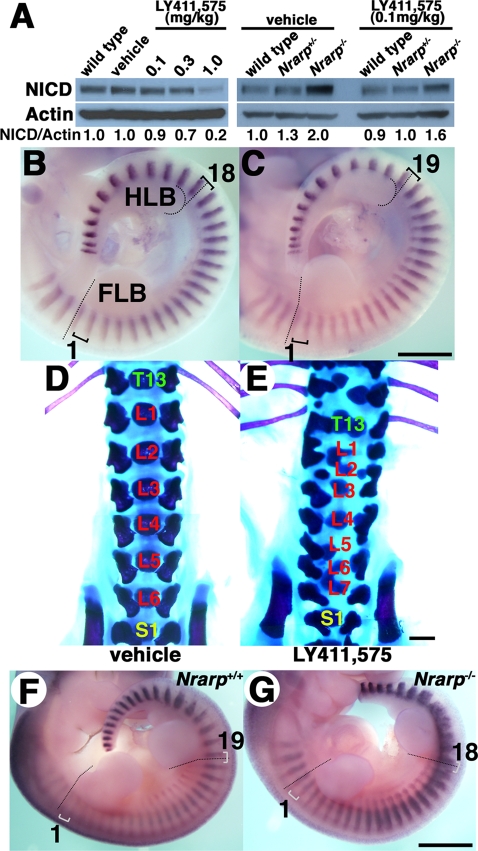
We found that the period of the somite segmentation clock was longer in Nrarp−/– mutant cells and that Notch activity in the PSM cells was enhanced. We assumed that the period of the somite segmentation clock was sensitive to Notch activity; higher Notch activity leads to a longer clock period, and lower Notch activity leads to a shorter clock period. To test this assumption, we decreased Notch activity in the embryo PSM by administering a Notch inhibitor to pregnant female mice. We administered the γ-secretase inhibitor LY411,575 (Wong et al., 2004) to wild-type pregnant mice three times every 24 h beginning on E 7.5 and then determined the amount of NICD in the PSM and examined the number of somites on E 10.5. Administration of 0.1, 0.3, and 1.0 mg/kg of LY411,575 reduced Notch activity in the PSM by 10%, 30%, and 80%, respectively (Figure 5A). A 10% reduction in Notch activity resulted in an extra somite forming: 19 somites between the limb buds at E 10.5 (n = 4/7), in comparison with 18 in the control (Figure 5, B and C). A larger reduction in Notch activity disrupted the somite pattern, however, and we could not count the number of somites (Supplemental Figure S4). The mice treated with 0.1 mg/kg LY411,575 had consistently seven lumbar vertebrae (n = 4/11) compared with six in the control, and they had some malformation in axial skeletons (Figure 5, D and E, and Supplemental Table S3). Thus these results suggested that a mild reduction in Notch activity in the PSM led to a shorter clock period. The possibility that the Notch inhibitor affects other phenomena could not be excluded, however, because even lower dose administration of the Notch inhibitor resulted in malformation in axial skeletons (Figure 5E).
FIGURE 5:
A γ-secretase inhibitor reduces Notch activity and decreases the somite segmentation clock period. (A) NICD quantification by using immunoblotting (n = 3). The ratios of NICD to β-actin are indicated. Administering 0.1 mg/kg LY411,575 reduced Notch activity by 10% over 24 h. Whole-mount in situ hybridization for Uncx4.1 of embryos that were treated with the vehicle (B) or 0.1 mg/kg LY411,575 (C). The dashed lines are used to indicate the rostral border of the FLB and HLB, and the brackets are used to indicate single somites. Skeletal preparations at postnatal day 1 for mice treated with the vehicle (D) or 0.1 mg/kg LY411,575 (E). T, thoracic vertebra; L, lumbar vertebra; S, sacral vertebra. Whole-mount in situ hybridization for Uncx4.1 in wild-type embryos (F) or Nrarp−/– embryos (G) that were treated with the 0.1 mg/kg LY411,575. Scale bars, 1 mm.
Finally, we tried to rescue the Nrarp−/– phenotypes by reducing Notch activity by administering LY411,575. Administration of 0.1 mg/kg LY411,575 reduced Notch activity by 20% in the Nrarp−/– PSM (Figure 5A). Some Nrarp−/– embryos that were treated with LY411,575 had 18 somites between both limb buds (n = 6/11), whereas the Nrarp−/– embryos without treatment had 17 somites between both limb buds (n = 13/13; Figures 2A and 5F). We propose that the clock period was sensitive to Notch activity. Interestingly, inhibitor-treated Nrarp−/– neonates had more severe defective axial skeleton configurations than did the untreated Nrarp−/– neonates or inhibitor-treated wild-type neonates. Thus, because reduced Notch activity did not correct the defective axial skeleton morphology of Nrarp−/–, Nrarp itself was likely essential for proper morphogenesis. Furthermore, the skeletal morphology of the Nrarp−/– neonates was more severe than that of the wild-type neonates for both conditions of Notch inhibitor administration and no treatment. Therefore Nrarp may have contributed to maintaining proper skeletal morphology.
The segmentation clock period was sensitive to Notch activity
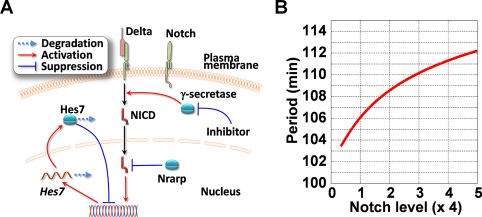
We found that the segmentation clock period is sensitive to Notch activity from our genetic and pharmacological experiments. The segmentation clock is regulated by synchronous gene oscillation in the PSM. In PSM cells, Notch signaling activates Hes7 transcription, and Hes7 inhibits transcription of its own gene and provokes the oscillatory gene expression over a 2-h cycle in the mouse, where the Hes7 negative feedback loop provides a core mechanism for the clock to run on (Figure 6A). A mathematical model was constructed based on this negative feedback loop, and was used to successfully reproduce the oscillatory gene expression (Lewis, 2003; Hirata et al., 2004). Model analysis revealed that the oscillation amplitude becomes small when Notch activity is low (Lewis, 2003). We carried out further analyses of the model and found that the oscillation period also decreases with low Notch activity, because a smaller amplitude means that less time is required for a cycle to pass (see Supplemental Note 1). Strikingly, we found from a simulation based on the model that the oscillation period is sensitive to the average level of Notch activity in the model: Higher Notch activity extends the period and gradually increases it with extremely large activity, whereas lower Notch activity shortens it (Figure 6B). This result supports the experimental results that the clock period was sensitive to Notch activity. In addition, by using the model analysis we predicted that the Hes7 negative feedback loop provides a molecular mechanism for clock period sensitivity to Notch activity.
FIGURE 6:
Mathematical analyses of the effect of Notch activity on the oscillation period. (A) The molecular mechanism of generation of oscillatory gene expression in the mouse PSM cell. Activated Notch receives limited proteolysis, which is dependent on γ-secretase activity, and then NICD activates Hes7 transcription. The accumulated Hes7 protein inhibits its own transcription. This negative feedback loop serves as a core mechanism of gene oscillation in the mouse PSM. Nrarp enhances the degradation of NICD, thereby inhibiting Notch activity, and the γ-secretase inhibitor reduces the production of NICD to inhibit Notch activity. In this research, we increased Notch activity by disrupting Nrarp and decreased it by administration of the γ-secretase inhibitor. (B) Dependence of the oscillation period on Notch activity. The mathematical model based on the negative feedback loop of Hes7 shown in (A) was used for the mathematical simulation. The oscillation period has a positive relationship with the average Notch activity, which is described as the maximum rate of Hes7 mRNA production, k (see Supplemental Note 1). Model parameters are listed in Supplemental Note 1.
DISCUSSION
We examined the roles of Nrarp in mouse in this report. Nrarp was essential for generating the proper number and morphology of axial skeleton components. Notch activity in the PSM was enhanced in the absence of Nrarp, and this activity extends over somite segmentation period, thereby decreasing the number of somites and their resultant vertebrae. In contrast, reduced Notch activity caused by a Notch inhibitor shortened the period of segmentation and increased the number of somites and their resultant vertebrae. We propose that the oscillation period for gene expression was sensitive to Notch activity and that they were positively proportional: Higher Notch activity led to a longer oscillation period, and lower Notch activity led to a shorter oscillation period.
Likewise, the number of Nrarp−/– embryo somites was lower than the number of wild-type somites at E 10.5 and E 11.5, whereas we did not observe the fusion of two adjacent somites or a difference in size of each somite. Thus vertebrae or somite fusion was not likely the cause of fewer numbers of vertebrae or somites. We failed to detect differences in somite numbers between the Nrarp−/– embryos and the wild-type embryos at E 8.5. Therefore the loss of Nrarp may not have affected the timing of the initiation of somite formation, because E 8.5 is quite an early stage of somitogenesis (Gossler and Hrabe de Angelis, 1998). In addition, the numbers of somites increased proportionally in both genotypes, and the number of somites in the Nrarp−/– embryo was significantly lower than that in the wild-type embryos at E 10.5 and E 11.5, respectively. Therefore the period of somite formation was longer in the absence of Nrarp. We could not rule out the possibility that the duration of somite formation in the Nrarp−/– was shorter than in the wild-type embryos, so the numbers of somites and vertebrae were lower in the Nrarp−/– embryos because it was not possible to detect the end of somite formation. This possibility did not likely happen, however, because the loss of two vertebrae (59.0 vs. 57.4, wild-type and Nrarp−/–, respectively) is consistent with the number of somites that formed between E 8.5 and E 11.5 (40.6 vs. 38.9). Thus we conclude that extending the period of somite formation was the major cause of the lower number of vertebrae in the Nrarp−/– embryos.
We showed that the clock period was sensitive to Notch activity. It was reported that other signaling, Wnt or retinoic acid, affects the pace of the segmentation clock (Kawakami et al., 2005; Vermot et al., 2005; Vermot and Pourquie, 2005; Gibb et al., 2009). In addition, the period of the somite segmentation clock is reported to be sensitive to the surrounding temperature in zebrafish development (Schroter et al., 2008). To our kowledge our report here, however, is the first for which the correlation between a fluctuating segmentation period and the intensity of molecular signaling was analyzed quantitatively. In addition, we interpreted the correlation by using mathematical analyses. In the mouse, the Hes7 negative feedback loop, which acts downstream of Notch, is essential for oscillatory gene expression in the PSM (Bessho et al., 2003). The results of our mathematical simulation led us to speculate that increasing Notch activity leads to extending the oscillation period. This speculation is in agreement with our experimental results.
The relationship between the somite formation period and Notch activity in zebrafish was reported recently: Attenuation of Notch activity led to a longer somite formation period (Herrgen et al., 2010), and this observation is opposite to our findings. In zebrafish, Notch signaling plays a major role in the synchronization of the oscillatory gene expression between each PSM cell (Jiang et al., 2000; Horikawa et al., 2006). The authors of the recent study focused on this Notch-dependent synchronization between the PSM cells, and, by using mathematical analyses, they accounted for the delay in the somite formation period by the attenuation of Notch activity. Their model assumes, however, that the oscillation period is not affected by the change in Notch activity in each PSM cell. By contrast, we conducted mathematical analyses of the oscillatory gene expression in each cell, although we did not consider the synchronization between cells because the role of Notch signaling in the synchronization remains to be elucidated in the mouse. Further study will be essential to gaining a comprehensive understanding of the mechanism of tuning of the somite formation period in vertebrates.
The small defects in the morphologies of the vertebrae and ribs in the Nrarp−/– mice were not repaired by using the Notch inhibitor. Therefore they were not caused by higher Notch activity, but rather Nrarp was essential for proper axial skeleton morphogenesis. Feedback regulation of Notch activity via Nrarp may have contributed to axial skeleton morphogenesis. Nrarp mediates Wnt-signaling–dependent neural-crest-cell development by stabilizing LEF1, which is a downstream effector of Wnt signaling (Ishitani et al., 2005). Nrarp also coordinates Notch and Wnt signaling in zebrafish and mouse endothelial cells, thereby controlling angiogenesis (Phng et al., 2009). In contrast, we detected enhanced Notch activity but failed to detect a change in Wnt signaling. We could have inferred this from the expression of Wnt downstream genes in our knockout mice. Consistent with this observation, the extended period of somite segmentation was restored by using the Notch inhibitor. Thus Nrarp seemed to regulate Notch signaling but not Wnt signaling in somitogenesis, in which Nrarp maintained the proper period of somite segmentation and the proper numbers of somites and vertebrae.
We failed to detect abnormalities in the morphology of the somites, although we found defects in the vertebrae and ribs in the Nrarp−/– embryos. It is possible that there were minute discrepancies in the size of the somites or in their gene-expression pattern. Further investigation is necessary to clarify this. We found that administering the Notch inhibitor caused vertebra and rib malformation. This result is consistent with the results of a report in which cyclical Notch activity was crucial for somite formation (Morimoto et al., 2005). The Notch inhibitor may have perturbed the cyclical Notch activity in PSM so that the morphology of the vertebrae and ribs was affected. We could not, however, rule out the other possibility that the Notch inhibitor may affect the ossification or some other process of skeletal morphogenesis but the somite segmentation clock, because lower dose administration of the Notch inhibitor affected the morphology of the axial skeleton but not the somite morphology. Strikingly, the Notch inhibitor disturbed the configuration of the axial skeleton more severely in the absence of Nrarp than in the wild-type condition. Thus we speculate that Nrarp may contribute to the robustness against environmental perturbation of somite formation or oscillatory gene expression. Further study is needed to elucidate the role of Nrarp in somite formation.
MATERIALS AND METHODS
Generation of Nrarp-knockout mice and genotyping
Nrarp-deficient mice were generated by homologous recombination with a targeting vector in which the Nrarp coding region was replaced with IRES-LacZ and PGK-neo (Supplemental Figure S2a). TT2 ES cell lines (Yagi et al., 1993) that were missing Nrarp were identified by using Southern blot analysis with 5′ and 3′ probes (Supplemental Figure S2b). Chimeric mice were generated as described previously (Bessho et al., 2001b). Nrarp-deficient mice were back-crossed with CD1 more than six times. We crossed pairs of 8- to 50-wk-old Nrarp+/– mice and collected their embryos at a certain embryonic stage to compare Nrarp mutants with their wild-type littermates. Chimera, F1, and F2 mice were initially genotyped by Southern blot analysis (Supplemental Figure S2b) and allele-specific PCR of somatic cell (skin) DNA. Embryos were genotyped by PCR of yolk-sac DNA by using the primers Nrarp-F CTCATACTATTGCTGAATGAGTGGAAGGGCTGC (which binds to the 5′UTR region of Nrarp) and Nrarp-R CAGCAGCACTTCTACGAAGGGGAAACCTCA (which binds to the 5′UTR region and downstream of Nrarp-F), which detect a 320-base-pair fragment indicative of the wild-type allele, and primers Neo-F AGCCCAGAAAGCGAAGGAGCAAAGCTGCTAT and Nrarp-R, which detect a 520-base-pair fragment indicative of the mutated allele.
Immunoblotting
PSMs were dissected from the embryos at E 8.5 or E10.5 and lysed in a buffer that contained 1% NP-40, 50 mM Tris-HCl (pH7.5), 150 mM NaCl, and 5 mM EDTA. The tissue lysates were fractionated by using SDS–PAGE and then blotted onto Hybond-P membranes (Amersham, Piscataway, NJ). Next they were probed with anti-cleaved Notch1 monoclonal antibody (Cell Signaling Technology, Danvers, MA) or anti-β-actin monoclonal antibody (Sigma-Aldrich, St. Louis, MO), and signals were then detected with a chemiluminescence detection system (Amersham). Quantification was performed using the public domain National Institutes of Health (NIH) image program, ImageJ. Notch signaling activity was evaluated by measuring the amount of NICD that was normalized to the amount of β-actin, which was used as an internal control.
Microarray analysis and quantitative RT-PCR
PSMs were dissected from the E 10.5 embryos of 8- to 12-mo-old Nrarp+/− female mice that were crossed with Nrarp+/− male mice. The part of the neural tube that was attached to the PSM was removed. The PSMs were then stored immediately in liquid nitrogen for several days. Total RNA was extracted using the SV Total RNA Isolation System (Promega, Madison, WI). We prepared three independent biological replicates. Microarray analysis was performed as described previously (Watanabe et al., 2009). The microarray data set that we analyzed (GSE18419) is available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus. Reverse transcription was performed by using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). Quantitative PCR reactions were performed by using KAPA SYBR FAST Universal qPCR (Kapa Biosystems, Woburn, MA). Each reaction was carried out in triplicate using gene-specific primers. The expression level of each gene was first normalized to that of G3PDH. The primers for the mouse genes that we used in the quantitative PCRs were as follows.
G3pdh: G3pdh-S, 5′-ACCACAGTCCATGCCATCAC-3′, and G3pdh-AS, 5′-TCCACCACCCTGTTGCTGTA-3′; Nrarp: Nrarp-S, 5′-CTCGCACTTAGGAAGGGAAG-3′, and Nrarp-AS, 5′-ACCACGCACAATATTTCCAA-3′; Hes5: Hes5-S, 5′-GCAGCATAGAGCAGCTGAAG-3′, and Hes5-AS, 5′-GAAGGCTTTGCTGTGTTTCA-3′; Car4: Car4-S, 5′-GTCAAATGGGAATGACAACG-3′, and Car4-AS, 5′-TTGTCCTTCGAGTCCTCCTT-3′; Gpr133: Gpr133-S, 5′-TCATTACTGGCCATTGGAAA-3′, and Gpr133-AS, 5′-GAGAGGCACAGTGAGGTTGA-3′; Rhox5: Rhox5-S, 5′-CAGGTATGGAAGCTGAGGGT-3′, and Rhox5-AS, 5′-GCTGTTCTTCCGAGTCTTCC-3′; Hes7: Hes7-S, 5′-TAGAAGAGCTGAGGCTGCTG-3′, and Hes7-AS, 5′-CTTTCTCCAGCTTCGGGTT-3′; Lfng: Lfng-S, 5′-TGTTTGAGAACAAGCGGAAC-3′, and Lfng-AS, 5′-CAGGGTGTGTCTGGGTACAG-3′; Dusp6: Dusp6-S, 5′-GGAATGAGAACACTGGTGGA-3′, and Dusp6-AS, 5′-GAAGCCACCTTCCAGGTAGA-3′; Spry2: Spry2-S, 5′-GAAGAGGATTCAAGGGAGAGG-3′, and Spry2-AS, 5′-GTCTTGGCAGTGTGTTCACC-3′; Axin2: Axin2-S, 5′-CTGGCTCCAGAAGATCACAA-3′, and Axin2-AS, 5′-TCAGCATCCTCCTGTATGGA-3′; Nkd1: Nkd1-S, 5′-CGTGGCTGGGAGAAGAAGC-3′, and Nkd1-AS, 5′-CAGGTCTAGGTAGTGGTTTCTCC-3′.
Skeletal preparation, x-ray CT, and whole-mount in situ hybridization
The cartilage and bones of newborn mice were stained with alcian blue and alizarin red, respectively, after being fixed in 95% ethanol (Bessho et al., 2001b). The skeletal structures of 6- to 12-wk-old mice were examined by using a LaTheta LCT-100 Series x-ray CT (Aloka, Wallingford, CT). Whole-mount in situ hybridizations were carried out (Bessho et al., 2001a) with minor modifications.
Immunostaining
Whole-mount immunostaining was carried out as described previously (Bessho et al., 2003) with minor modifications. E 10.5 embryos were fixed with 4% paraformaldehyde at 4°C for 1 h and then treated with 6% H2O2 for 10 min and 2% Triton X-100 for 10 min. Next, we incubated the embryos with anti-cleaved Notch1 monoclonal antibody (Cell Signaling Technology) overnight at 4°C, and then with peroxidase-conjugated anti–rabbit immunoglobulin G antibody for 3 h. We then visualized the peroxidase activity as deposits of 3,3′-diaminobenzidine tetrahydrochloride (Sigma).
Counting somites and vertebrae and statistical analysis
We detected the expression of Uncx4.1 mRNA by using in situ hybridization to count the number of somites. The stain localizes to the caudal domain in somites; therefore it was used as a landmark of the somite caudal border. We conducted experiments that compared wild type, Nrarp+/ and Nrarp−/– using littermates and counted the somites for each embryo before genotyping. The bud positions were determined as described previously (Chan et al., 2004/2005) for counting somites that were located between the FLBs and HLBs. For accuracy, we laterally and dorsally observed embryos by using high-magnification fields. Following this, we counted the number of somites of each embryo and calculated the average number of somites for each genotype for individual pregnant females. This process allowed us to determine the changes in the numbers of somites for Nrarp−/– or Nrarp+/− mice in comparison to their wild-type littermates. Then, we determined the average number of somites for each genotype of the embryos that we collected from multiple pregnant females and compared the number with the one that was determined for the wild-type embryos by using the paired t test. We performed this comparison at E 8.5 (n = 6 pregnant mothers), E 10.5 (n = 18), and E 11.5 (n = 8). We also estimated a 95% CI of the segment clock extension in Nrarp−/– embryos relative to the wild type. The CI was estimated by applying maximum likelihood estimation to a hierarchical generative model of the somite number (see Supplemental Note 2 for details). Although mice treated with 0.1 mg/ml LY411,575 had substantial malformation of the axial skeleton, we did not observe a split vertebral body. Thus we counted their vertebral bodies. For small or malformed vertebral bodies, we checked the vertebral arches, which should be on either side of each vertebral body.
Animals and γ-secretase inhibitor administration
We then administered LY411,575 to pregnant female mice per os following a 12-h fasting period. The LY411,575 was formulated as a 30 mg/ml solution in dimethyl sulfoxide and was diluted in 5% ethanol in sunflower seed oil. It was administered every 24 h from E 7.5 to E 9.5. The amount of NICD in the PSM was quantified 24 h after administering the LY411,575 or the vehicle. Our experiments were approved by the Animal Care Committee of Nara Institute of Science and Technology. They were conducted in accordance with guidelines that were established by the Science Council of Japan.
Supplementary Material
Acknowledgments
We thank Shigeru Kondo, Naoyuki Inagaki, Takashi Kondo, Kentaro Hirata, Miguel Maroto, Kim Dale, Alexander Aulehla, and Siripong Thitamadee for discussion; David Ish-Horowicz, Achim Gossler, Olivier Pourquie, Thomas N. Sato, Yukie Takabatake, Jan Moren, and Ian Smith for critically reading the manuscript; and Michiko Saitou for generating the knockout mice.
This work was supported by a Kakenhi (Grant-in-Aid for Scientific Research) for the priority area “Systems Genomics”; Kakenhi B and Wakate Initiatives A and B from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; and by the Uehara Memorial Foundation. This work was also supported in part by the Global COE Program in NAIST (Frontier Biosciences: strategies for survival and adaptation in a changing global environment), MEXT, Japan.
Abbreviations used:
- CI
confidence interval
- CT
computed tomography
- E
embryonic day
- FGF
fibroblast growth factor
- FLB
fore-limb bud
- HLB
hind-limb bud
- Lfng
Lunatic fringe
- NICD
Notch intracellular domain
- Nrarp
Notch-regulated ankyrin repeat protein
- PSM
presomitic mesoderm
- RBPj
recombinant signal binding protein for immunoglobulin kappa J region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0139) on July 27, 2011.
REFERENCES
- Aulehla A, Johnson RL. Dynamic expression of lunatic fringe suggests a link between Notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev Biol. 1999;207:49–61. doi: 10.1006/dbio.1998.9164. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Hirata H, Masamizu Y, Kageyama R. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 2003;17:1451–1456. doi: 10.1101/gad.1092303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Miyoshi G, Sakata R, Kageyama R. Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells. 2001a;6:175–185. doi: 10.1046/j.1365-2443.2001.00409.x. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001b;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Chan AOK, Dong M, Wang L, Chan WY. Somite as a morphological reference for staging and axial levels of developing structures in mouse embryos. Neuroembryol Aging. 2004/2005;3:102–110. [Google Scholar]
- Cole SE, Levorse JM, Tilghman SM, Vogt TF. Clock regulatory elements control cyclic expression of lunatic fringe during somitogenesis. Dev Cell. 2002;3:75–84. doi: 10.1016/s1534-5807(02)00212-5. [DOI] [PubMed] [Google Scholar]
- Dale JK, Maroto M, Dequeant ML, Malapert P, McGrew M, Pourquie O. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature. 2003;421:275–278. doi: 10.1038/nature01244. [DOI] [PubMed] [Google Scholar]
- Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA. Waves of mouse lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr Biol. 1998;8:1027–1030. doi: 10.1016/s0960-9822(07)00424-1. [DOI] [PubMed] [Google Scholar]
- Gibb S, Zagorska A, Melton K, Tenin G, Vacca I, Trainor P, Maroto M, Dale JK. Interfering with Wnt signalling alters the periodicity of the segmentation clock. Dev Biol. 2009;330:21–31. doi: 10.1016/j.ydbio.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler A, Hrabe de Angelis M. Somitogenesis. Curr Top Dev Biol. 1998;38:225–287. [PubMed] [Google Scholar]
- Herrgen L, Ares S, Morelli LG, Schroter C, Julicher F, Oates AC. Intercellular coupling regulates the period of the segmentation clock. Curr Biol. 2010;20:1244–1253. doi: 10.1016/j.cub.2010.06.034. [DOI] [PubMed] [Google Scholar]
- Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H. Noise-resistant and synchronized oscillation of the segmentation clock. Nature. 2006;441:719–723. doi: 10.1038/nature04861. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Matsumoto K, Chitnis AB, Itoh M. Nrarp functions to modulate neural-crest-cell differentiation by regulating LEF1 protein stability. Nat Cell Biol. 2005;7:1106–1112. doi: 10.1038/ncb1311. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Aerne BL, Smithers L, Haddon C, Ish-Horowicz D, Lewis J. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Deftos ML, Bevan MJ, Gridley T. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the Notch signaling pathway. Dev Biol. 2001;238:110–119. doi: 10.1006/dbio.2001.0408. [DOI] [PubMed] [Google Scholar]
- Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, Niehrs C, Kintner C. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 2001;15:1885–1899. doi: 10.1101/gad.908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- McGrew MJ, Dale JK, Fraboulet S, Pourquie O. The lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- Morales AV, Yasuda Y, Ish-Horowicz D. Periodic lunatic fringe expression is controlled during segmentation by a cyclic transcriptional enhancer responsive to notch signaling. Dev Cell. 2002;3:63–74. doi: 10.1016/s1534-5807(02)00211-3. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Takahashi Y, Endo M, Saga Y. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature. 2005;435:354–359. doi: 10.1038/nature03591. [DOI] [PubMed] [Google Scholar]
- Phng LK, et al. Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev Cell. 2009;16:70–82. doi: 10.1016/j.devcel.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirot P, van Grunsven LA, Marine JC, Huylebroeck D, Bellefroid EJ. Direct regulation of the Nrarp gene promoter by the Notch signaling pathway. Biochem Biophys Res Commun. 2004;322:526–534. doi: 10.1016/j.bbrc.2004.07.157. [DOI] [PubMed] [Google Scholar]
- Pourquie O. Vertebrate somitogenesis. Annu Rev Cell Dev Biol. 2001;17:311–350. doi: 10.1146/annurev.cellbio.17.1.311. [DOI] [PubMed] [Google Scholar]
- Pourquie O. The segmentation clock: converting embryonic time into spatial pattern. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- Schroter C, Herrgen L, Cardona A, Brouhard GJ, Feldman B, Oates AC. Dynamics of zebrafish somitogenesis. Dev Dyn. 2008;237:545–553. doi: 10.1002/dvdy.21458. [DOI] [PubMed] [Google Scholar]
- Sewell W, Sparrow DB, Smith AJ, Gonzalez DM, Rappaport EF, Dunwoodie SL, Kusumi K. Cyclical expression of the Notch/Wnt regulator Nrarp requires modulation by Dll3 in somitogenesis. Dev Biol. 2009;329:400–409. doi: 10.1016/j.ydbio.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topczewska JM, Topczewski J, Szostak A, Solnica-Krezel L, Hogan BL. Developmentally regulated expression of two members of the Nrarp family in zebrafish. Gene Expr Patterns. 2003;3:169–171. doi: 10.1016/s1567-133x(03)00009-7. [DOI] [PubMed] [Google Scholar]
- Vermot J, Gallego Llamas J, Fraulob V, Niederreither K, Chambon P, Dolle P. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005;308:563–566. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- Vermot J, Pourquie O. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature. 2005;435:215–220. doi: 10.1038/nature03488. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, et al. Fezf1 is required for penetration of the basal lamina by olfactory axons to promote olfactory development. J Comp Neurol. 2009;515:565–584. doi: 10.1002/cne.22074. [DOI] [PubMed] [Google Scholar]
- Wong GT, et al. Chronic treatment with the gamma-secretase inhibitor LY-411,575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J Biol Chem. 2004;279:12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- Wright D, et al. Cyclic Nrarp mRNA expression is regulated by the somitic oscillator but Nrarp protein levels do not oscillate. Dev Dyn. 2009;238:3043–3055. doi: 10.1002/dvdy.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- Zhang N, Gridley T. Defects in somite formation in lunatic fringe-deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.