Abstract
The Mycobacterium tuberculosis membrane protein Rv0899 confers adaptation of the bacterium to acidic environments. Due to strong sequence homology of its C-terminus to bacterial OmpA-like domains, Rv0899 has been proposed to constitute an outer membrane porin of M. tuberculosis. However, OmpA-like domains are widespread in a wide variety of bacterial proteins with different functions. Furthermore, the three-dimensional structure of Rv0899 does not contain a transmembrane β-barrel, and recent evidence demonstrates that it does not have porin activity. Instead, the rv0899 gene is part of an operon (rv0899-rv0901) that is required for fast ammonia secretion, pH neutralization and growth of M. tuberculosis in acidic environments. The mechanism whereby these functions are accomplished is not known. To gain further functional insights, a targeted search of the genomic databases was performed for proteins with sequence similarity beyond the OmpA-like C-terminus. The results presented here, show that Rv0899-like proteins are widespread in bacteria with functions in nitrogen metabolism, adaptation to nutrient poor environments, and/or establishing symbiosis with the host organism, and appear to form a protein family. These findings suggest that M. tuberculosis Rv0899 may also assist similar processes and lend further support to its role in ammonia secretion and M. tuberculosis adaptation to the host environment.
Keywords: mycobacterium, tuberculosis, Rv0899, OmpATb, membrane protein, OmpA-like, porin
INTRODUCTION
The genome of Mycobacterium tuberculosis H37Rv contains several stress response genes, which contribute to pathogenicity.1 Among these, rv0899 encodes a 326-residue membrane protein (Rv0899) that is involved in conferring adaptation of M. tuberculosis to acidic environments.2,3 The rv0899 gene and its two neighbors rv0900 and rv0901, also encoding predicted membrane proteins, are found in pathogenic mycobacteria associated with tuberculosis (M. tuberculosis, M. bovis) and other diseases (M. marinum, M. ulcerans, M. kansasii), but are absent from non-pathogenic mycobacteria, suggesting that they may be important for pathogenicity and, thus, may be attractive candidates for the development of chemotherapeutic agents.
Rv0899 contains three independently structured domains.4 The N-terminus (~ residues 1–80) includes a membrane-anchoring sequence of 20 hydrophobic amino acids (~ residues 28–50) that is required for membrane translocation but is not cleaved.5 The central region (~ residues 81–195) contains two consecutive repeats of the BON (Bacterial OsmY and Nodulation) domain (pfam04972), a conserved, putative lipid binding sequence found in some bacterial osmotic shock protection proteins, secretins, haemolysins, channels, and nodulation specificity proteins.6 Finally, the C-terminus (residues 196–326) contains an OmpA-like domain (pfam00691), a periplasmic peptidoglycan-binding structure found in several types of bacterial membrane proteins, including in the C-terminus of the outer membrane protein OmpA from E. coli.
Owing to its strong homology with E. coli OmpA, Rv0899 was originally annotated as OmpATb in the published genome sequence of M. tuberculosis and was proposed to be an outer membrane porin.2,5,7–9 However, the three-dimensional structure shows that Rv0899 does not form a membrane-spanning β-barrel and, thus, is not compatible with porin function.4,10 Recent studies also show that Rv0899 is not a porin, but rather, that it is encoded by an operon (also known as ammonia release facilitator, arf, operon), which includes rv0899, rv0900 and rv0901, and which is required for fast ammonia secretion, rapid pH neutralization and growth of M. tuberculosis in acidic environments.3 However, the mechanism whereby Rv0899, Rv0900, and Rv0901 contribute to this function is not known.
The C-terminus of Rv0899 adopts the typical α/β structure of peptidoglycan-binding domains in the OmpA-like superfamily, reflecting association with the peptidoglycan layer. However, theα/β fold of the central domain, with three parallel/antiparallel α-helices packed against a six-stranded parallel/antiparallel β-sheet, was unprecedented in the Protein Data Bank (PDB) and, thus, provided limited insights about function.
A simple BLAST (Basic Local Alignment Search Tool) search of the protein databases for sequences similar to Rv0899 also provides little insights about the function of the central domain, because it is dominated by hits with strong homology to the C-terminus, but little homology to the rest of the protein. This reflects the strong sequence conservation and ubiquitous nature of bacterial OmpA-like domains, which are widespread in outer membrane proteins (e.g. OmpA inserts in the outer membrane as a β-barrel), as well as outer membrane lipoproteins (e.g. Pal is bound to the outer membrane through a lipid anchor), and inner membrane proteins (e.g. MotB inserts in the inner membrane through a transmembrane helix).11,12 In contrast, the central BON-containing domain of Rv0899 has only weak similarity to database sequences. Thus, matches to this region are obscured by numerous, much more pronounced matches to the OmpA-like region. Furthermore, performing the BLAST search with individual domains as query (i.e. only BON or only OmpA-like) yields sequences with homology primarily restricted to the specific query region, obscuring sequences that cover the entire length of Rv0899.
To overcome this problem and gain further insights about the potential function of Rv0899, a detailed iterative search of the NCBI (National Center for Biotechnology Information) database was performed to identify other Rv0899-like proteins with homology spanning the entire Rv0899 sequence, including the transmembrane, BON and OmpA-like domains. This analysis uncovers a family of Rv0899-like proteins in bacteria with functions in nitrogen metabolism, adaptation to nutrient poor environments, and/or establishing symbiosis with the host organism.
METHODS
A PSI-BLAST (Position-Specific Iterated BLAST) search of the NCBI (National Center for Biotechnology Information) database was performed using the parameter “Max Matches in a Query Range”, which is useful in cases where many strong matches to one region of a query sequence may prevent BLAST from presenting weaker matches to another region.13 In each PSI-BLAST iteration, the hits were visually inspected to remove false positives by retaining only those having sequence matches across at least 70% of the Rv0899 sequence length. After seven cycles the search converged to yield 39 hits with E values <10−30 (Table 1).
Table 1.
Proteins in the Rv0899 family listed by taxonomic groups.
|
CLASS | |||
|---|---|---|---|
| ORDER | |||
| GENUS | |||
| SPECIES | ORGANISM | GENE | NCBI RefSeq |
|
ACTINOBACTERIA
| |||
| Actinomycetales | |||
| Mycobacteriaceae | |||
| Mycobacterium | Mycobacterium tuberculosis H37Rv | Rv0899 | NP_215414 |
| Mycobacterium ulcerans Agy99 | MUL_0252 | YP_904461 | |
| Mycobacterium marinum M | MMAR_4637 | YP_001852895 | |
| Mycobacterium kansasii ATCC 12478 | MkanA1_010100023014 | ZP_04750863 | |
| Nocardioidaceae | |||
| Kribbella | Kribbella flavida DSM 17836 | Kfla_4948 | YP_003382763 |
| Frankiaceae | |||
| Frankia | Frankia sp. CcI3 | Francci3_1913 | YP_481018 |
| Frankia sp. EAN1pec | Franean1_2246 | YP_001506587 | |
| Frankia sp. EuI1c | FraEuI1c_4798 | YP_004018658 | |
| Frankia alni ACN14a | FRAAL4505 | YP_714692 | |
|
| |||
|
NITROSPIRA
| |||
| Nitrospirales | |||
| Nitrospiraceae | |||
| Nitrospira | Candidatus Nitrospira defluvii | NIDE2575 | YP_003798208 |
|
| |||
|
α-PROTEOBACTERIA
| |||
| Rhizobiales | |||
| Beijerinckiaceae | |||
| Beijerinckia | Beijerinckia indica subsp. indica ATCC 9039 | Bind_1196 | YP_001832327 |
| Bradyrhizobiaceae | |||
| Nitrobacter | Nitrobacter hamburgensis X14 | Nham_0631 | YP_575979 |
| Bradyrhizobium | Bradyrhizobium japonicum USDA 110 | blr1084 | NP_767724 |
| Oligotropha | Oligotropha carboxidovorans OM5 | OCAR_4642 | YP_002287650 |
| Rhodopseudomonas | Rhodopseudomonas palustris CGA009 | RPA4784 | NP_950118 |
| Afipia | Afipia sp. 1NLS2 | AfiDRAFT_2335 | ZP_07027206 |
|
| |||
|
β-PROTEOBACTERIA
| |||
| Rhodocyclales | |||
| Rhodocyclaceae | |||
| Dechloromonas | Dechloromonas aromatica RCB | Daro_2186 | YP_285395 |
| Burkholderiales | |||
| Oxalobacteraceae | |||
| Herbaspirillum | Herbaspirillum seropedicae SmR1 | Hsero_4029 | YP_003777408 |
| Cupriavidus | Cupriavidus taiwanensis | RALTA_B1027 | YP_002007692 |
| Ralstonia solanacearum GMI1000 | RS01947 | NP_522322 | |
| Burkholderiaceae | |||
| Burkholderia (cepacia complex) | Burkholderia multivorans ATCC 17616 | BMULJ_00324 | YP_001944835 |
| Burkholderia vietnamiensis G4 | Bcep1808_0469 | YP_001118316 | |
| Burkholderia sp. 383 | Bcep18194_A3573 | YP_367818 | |
| Burkholderia cenocepacia J2315 | BCAL0349 | YP_002229515 | |
| Burkholderia dolosa AUO158 | BDAG_02659 | ZP_04946714 | |
| Burkholderia ambifaria | BamMC406_0418 | YP_001807131 | |
| Burkholderia | Burkholderia glumae BGR1 | bglu_1g03970 | YP_002910300 |
| Burkholderia graminis C4D1M | BgramDRAFT_6351 | ZP_02887541 | |
| Burkholderia phytofirmans PsJN | Bphyt_4921 | YP_001888655 | |
| Burkholderia (pseudomallei group) | Burkholderia thailandensis TXDOH | BthaT_010100033018 | ZP_02375888 |
| Burkholderia pseudomallei K96243 | BPSL3099 | YP_109694 | |
| Burkholderia oklahomensis C6786 | BoklC_010100016479 | ZP_02364314 | |
| Burkholderia mallei ATCC 23344 | BMA2828 | YP_104341 | |
|
| |||
|
γ-PROTEOBACTERIA
| |||
| Pseudomonadales | |||
| Moraxellaceae | |||
| Acinetobacter | Acinetobacter radioresistens SK82 | ACIRA0001_2398 | ZP_05359661 |
| Acinetobacter baumannii AYE | ABAYE2406 | YP_001714238 | |
| Xanthomonadales | |||
| Xanthomonadaceae | |||
| Xanthomonas | Xanthomonas oryzae pv. oryzae KACC10331 | XOO3515 | YP_202154 |
| Stenotrophomonas | Stenotrophomonas sp. SKA14 | SSKA14_3746 | ZP_05136662 |
|
| |||
|
ε-PROTEOBACTERIA
| |||
| Campylobacterales | |||
| Campylobacteraceae | Campylobacterales bacterium GD1 | CBGD1_825 | ZP_05070558 |
| Acrobacter | Arcobacter butzleri RM4018 | Abu_0025 | YP_001488979 |
To test the validity of the results, a similar PSI-BLAST search was conducted in revers, starting from individual, randomly selected hits in Table 1 rather than from Rv0899. Indeed, when the search was initiated from hit sequences selected from each of the identified bacterial classes (NCBI RefSeq: YP_481018; YP_003798208; YP_001832327; YP_001944835; YP_001714238), Rv0899 as well as the other hit sequences were readily identified. The results were further validated by performing a BLAST search using the UniProtKB/SwissProt Databases, and manually selecting for hits with matches across at least 70% of the Rv0899 sequence length, to include both central (BON) and C-terminal (OmpA-like) domains. Finally, the hit sequences were analyzed for the presence of conserved domains using the NCBI Conserved Domains Database,14 and transmembrane regions were identified using TMHMM.15
ClustalW and manual editing, performed with the program Jalview, were used to generate the final sequence alignment.16,17 The aligned sequences (Figures 1, S1) were further examined for correct alignment by generating homology-based structural models, using SWISS-MODEL,18–20 with the template coordinates of the BON domain of Rv0899 (PDB: 2KSM). A neighbour-joining phylogeny tree was generated with the aligned Rv0899 family sequences using the neighbor-joining method, with the NCBI BLAST Tree View and Archaeopteryx programs (Figure 2).21,22 Alignments in FASTA format are provided as supporting information (Supporting Data S1–S3).
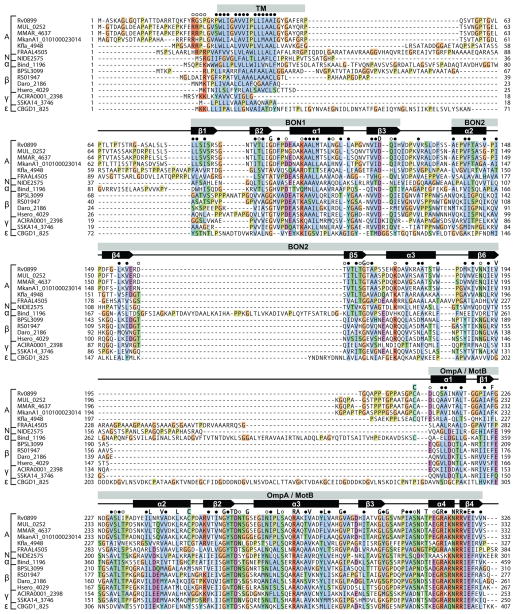
Figure 1. Amino acid sequence alignment for Rv0899 family proteins.
Alignment is shown for representative sequences from each species. Fully conserved residues (single letter), conserved hydrophobic residues (●), and conserved polar residues (○) are marked above the sequences. Secondary structure is derived from the structure of M. tuberculosis Rv0899. Conserved regions include a transmembrane domain (TM), two BON domains (pfam04972) and one OmpA-like domain (pfam00691). Alignments were rendered with ClustalX coloring using Jalview.17 Sequences are grouped by organism class: (A) actinobacteria; (N) nitrospira; (α) α-proteobacteria; (β) β-proteobacteria; (γ) γ-proteobacteria; (ε) ε-proteobacteria.
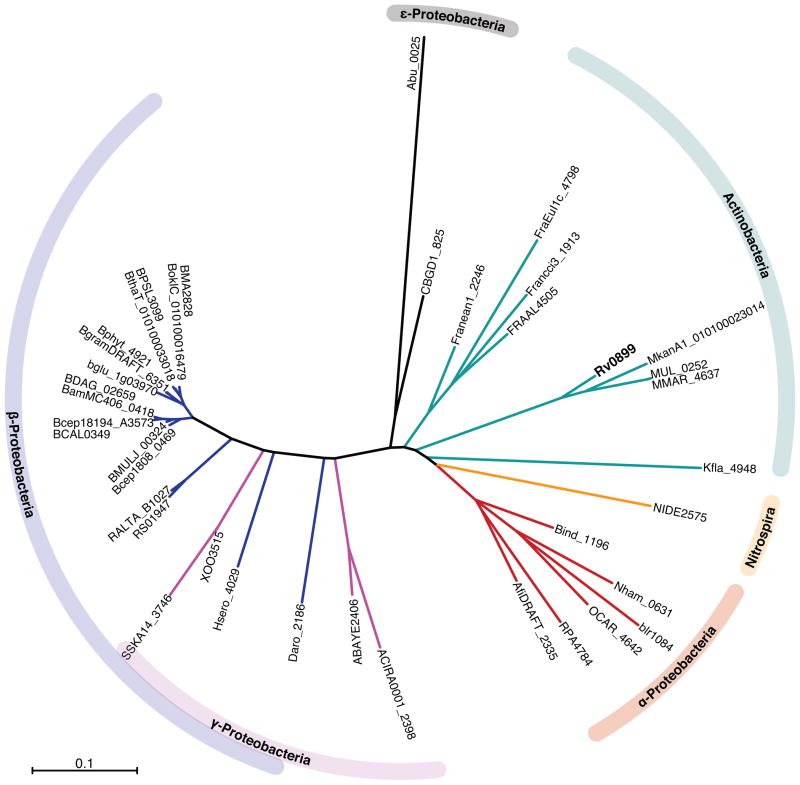
Figure 2. Neighbour-joining phylogeny tree for the aligned Rv0899 family proteins.
The tree was generated with the ClustalW aligned sequences using the neighbor-joining method,21 using the NCBI BLAST Tree View and Archaeopteryx22 programs. The scale bar represents mean residue difference.
RESULTS AND DISCUSSION
Identification of the Rv0899 protein family
The resulting hits share homology across the single transmembrane domain, the central BON domain, as well as the C-terminal OmpA-like domain (Figures 1, S1). They span a variety of bacterial species in GC-rich Gram-positive actinobacteria as well as Gram-negative bacteria and proteobacteria (Figure 2). Phylogenetic analysis suggests that they may have descended from a common ancestor and, therefore, that they can be viewed as orthologous members of the same protein family.
Although Rv0899 is predicted to be an outer membrane protein,3,5,7,9,23,24 its topology with respect to the mycobacterial cell envelope is not known. Within each bacterial genus, the hydrophobic amino acid sequence of the transmembrane (TM) region is highly conserved. For all species, the average pairwise identity of the sequences to M. tuberculosis Rv0899 is 32%. In all sequences, with the exception of Stenotrophomonas, the transmembrane region is preceded by up to three positively charged Arg residues, which could facilitate insertion across the outer bacterial membrane (as in the Wza translocon25), or across the inner membrane with the N-terminus exposed to the cytoplasm, according to the “positive inside rule”.26 The transmembrane region is followed by a 30- to 80-residue sequence, rich in Gly and Pro, with similarity that is limited to within each genus.
Sequence conservation in the central region of the proteins begins at the start of β1 (L81 of M. tuberculosis Rv0899), the first β-strand in the ββαβαββαβ structure of the central domain. In this structure, the first BON domain (BON1) spans β1-β2-α1-β3 while the second (BON2) spansα2-β4-β5-α3-β6. BON1 and BON2 share both a similar ββαβ topology (except for an additional N-terminal α-helix in BON2) as well as significant amino acid sequence homology, with other previously described BON domain sequences.6 Overall, in this region, the average pairwise identity of the sequences to M. tuberculosis Rv0899 is 28%. The most prominent difference is that the loop connecting β4 to β5 is much longer (~50-residue) in the Rv0899-like proteins from α-proteobacteria compared to all other proteins, where it is only a short hairpin.
The BON domain is characterized by a conserved glycine and several conserved hydrophobic residues.6 In all of the Rv0899 family proteins, the characteristic Gly is fully conserved in both BON1 (G95 at the end of β2) and BON2 (G164 at the end of β5), with the exception of sequences from Kribbella and Frankia where the BON1 Gly is replaced by Ala, a conservative difference that preserves the small size of the side-chain at this position. Sequence alignment further indicates that acidic residues preceding α1 and α3 and following β3 and β4 are conserved, as are basic residues situated at the start of α1 and α3, and several hydrophobic residues throughout the sequences.
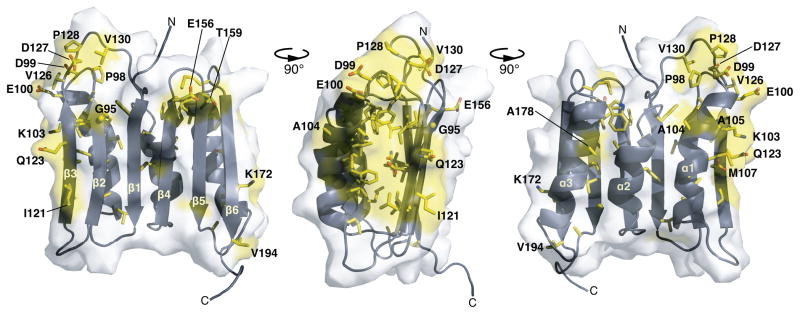
Mapping these conserved residues (or conserved residue types) onto the structure of Rv0899 (Figure 3) shows that most hydrophobic side-chains and a few polar side-chains (e.g. D122; N190) are buried in the core of the protein where the three α-helices contact the β-sheet. In contrast, several polar or charged side chains are exposed on the molecular surface, suggesting that they could play a functional role beyond maintaining the protein’s structural integrity. For example, the conserved P98-D99-E100 triplet and D127-P128 pair are situated in two spatially proximal, loops, and give rise to a surface-exposed acidic patch. Furthermore, I121, E156, T159, Q123, protrude from the β-sheet and have surface exposed side-chains, as do A104, A105, and A178 on helices α1 and α3. Finally, K103 and M107 in α1, and K172 in α3, protrude from each of the sides of the structure and are surface-exposed. In all of the identified Rv0899 family sequences, the central BON-containing domain is connected to the OmpA-like C-terminus by a stretch of about 10 to 80 amino acids that are only conserved within each bacterial order.
Figure 3. Amino acid conservation in the three-dimensional structure of the BON domain region of M. tuberculosis Rv0899.
Residues that are fully conserved in all Rv0899 family proteins are colored red; residues with surface-exposed side chains are labeled. The molecular structure was rendered with PyMol.40 Amino acid numbering is according to the sequence of M. tuberculosis Rv0899.
Amino acid conservation in the OmpA-like domain is very strong across all identified sequences, with an average pairwise identity of each sequence to M. tuberculosis Rv0899 of 40%. The sequence conservation and the three-dimensional structures of several OmpA-like domains from different organisms have been described.27–29 They all share a common, basic, α/β fold with a four-stranded β-sheet, three core α-helices, and one or two additional α-helices. The OmpA-like domain of M. tuberculosis Rv0899 has the basic αβαβαβαβ topology of Pal,28 and the high sequence similarity (with little or no alignment gaps) of all Rv0899-family proteins in this region indicates that they all adopt the same fold. The structure of Rv0899 is stabilized by a disulfide bond (C208-C250) linking the N-terminus of α1 to the C-terminus of α2. This disulfide bond appears to be conserved in the sequences from mycobacteria, from Kribbella, and from α-proteobacteria, which all have two Cys residues at similar positions. Furthermore, several amino acids have been shown to play an important role in mediating the association of OmpA-like domains with peptidoglycan.28 These residues are highly conserved in the sequences of Rv0899 from M. tuberculosis (F225, D228, T261, D262, R277, R319) and from all other bacteria.
Identification of Rv0900- and Rv0901-like proteins in other bacteria
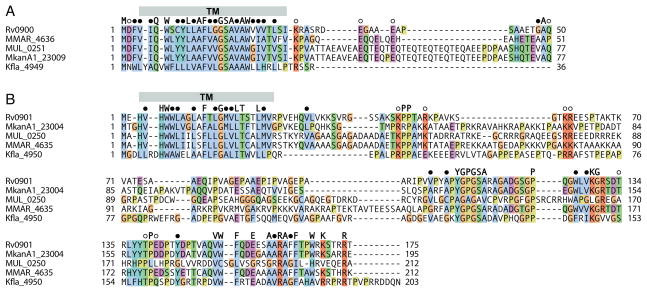
The M. tuberculosis genes rv0899-rv0901 were recently identified as components of an operon that is required for facilitating ammonia release when the bacterium encounters an acidic environment.3 Proteins encoded by genes in the same configuration are also found in the other pathogenic mycobacteria, and the Kribbella gene (Kfla4948) encoding the Rv0899-like protein is adjacent to two genes encoding Rv0900-like (Kfla4949) and Rv0901-like (Kfla4950) proteins (Table 2; Figure 4). Interestingly, similar genes organized in a similar configuration, neighbouring their rv0899-like counterpart, appear to be also present in α-proteobacteria (Table 2; Additional Files 2 and 3) suggesting that this operon is also conserved in other species.
Table 2.
Rv0900 and Rv0901 orthologs. Gene names and NCBI RefSeq identifiers are provided.
| Rv0899-like | Rv0900-like | Rv0901-like | Organism | |||
|---|---|---|---|---|---|---|
| GENE | NCBI RefSeq | GENE | NCBI RefSeq | GENE | NCBI RefSeq | |
| Rv0899 | NP_215414 | Rv0900 | NP_215415 | Rv0901 | NP_215416 | M. tuberculosis H37Rv |
| MUL_0252 | YP_904461 | MUL_0251 | YP_904460 | * MUL_0250 | * | M. ulcerans Agy99 |
| MMAR_4637 | YP_001852895 | MMAR_4636 | YP_00185289 | MMAR_4635 | YP_001852893 | M. marinum M |
| MkanA1_010100023014 | ZP_04750863 | MkanA1_23009 | ZP_04750862 | MkanA1_23004 | ZP_04750861 | M. kansasii ATCC 12478 |
| Kfla_4948 | YP_003382763 | Kfla_4949 | YP_003382764 | Kfla_4950 | YP_003382765 | K. flavida DSM 17836 |
| Bind_1196 | YP_001832327 | Bind_1197 | YP_001832328 | B. indica subsp. indica ATCC 9039 | ||
| Nham_0631 | YP_575979 | Nham_0632 | YP_575980 | Nham_0633 | YP_575980 | N. hamburgensis X14 |
| blr1084 | NP_767724 | blr1085 | NP_767725 | blr1088 | NP_767728 | B. japonicum USDA 110 |
| OCAR_4642 | YP_002287650 | OCAR_4643 | YP_002287651 | OCAR_4644 | YP_002287652 | O. carboxidovorans OM5 |
| RPA4784 | NP_950118 | RPA4783 | NP_950117 | RPA4782 | NP_9501186 | R. palustris CGA009 |
| AfiDRAFT_2335 | ZP_07027206 | AfiDRAFT_2336 | ZP_07027207 | AfiDRAFT_2337 | ZP_07027208 | Afipia sp. 1NLS2 |
The amino acid sequence of MUL_0250 is not available in the databank and was deduced from the nucleotide sequence of the gene.
Figure 4. Amino acid sequence alignment of Rv0900 and Rv0901 orthologs from Mycobacterium and Kribbella species.
Alignments are shown for (a) Rv0900 and (b) Rv0901 sequences. Fully conserved residues (single letter), conserved hydrophobic residues (●), and conserved polar residues (○), are marked above the sequences. Secondary structures are derived from the structure of M. tuberculosis Rv0899. Alignments were rendered with ClustalX coloring using Jalview.17
Rv0899 family members in actinobacteria
In actinobacteria, Rv0899 orthologs are found in four pathogenic mycobacteria, as well as in Frankia, a nitrogen-fixing symbiont, and Kribbella, a soil bacterium. Actinobacteria, including mycobacteria, are widely distributed in both aquatic and terrestrial environments, especially in soil, where they play an important role in recycling biomaterials by decomposition and humus formation (recently reviewed30). Notably, while M. tuberculosis has never been isolated from other environments than humans, M. ulcerans, which causes a devastating necrotic disease of the skin, has also been isolated from soil, and has been proposed to exist in symbiosis with certain tropical plants.31–33 Frankia alni establishes nitrogen-fixing symbiosis with certain non-leguminous plants enabling them to grow in soils where nitrogen is the limiting factor (forest clearings, mine wastes, sand dunes, glacial moraines).
Rv0899 family members in gram-negative bacteria and proteobacteria
Rv0899-like proteins are also found in gram-negative α-proteobacteria that are plant-associated (Bradyrhizobium), animal-associated (Afipia), or free-living (Nitrobacter hamburgensis; Beijerinckia indica; Oligotropha carboxidovorans; Rhodopseudomonas). These bacteria display very high metabolic versatility and are capable of using light, CO2, CO, organic (including aromatics) or inorganic compounds as energy sources. Among them, Bradyrhizobium japonicum is an agriculturally important N2-fixing legume symbiont, which colonizes root nodules resembling those induced by Frankia Alni. Furthermore, Nitrobacter hamburgensis and the closely related bacterium C. Nitrospira defluvii are nitrite-oxidizing organisms that play an important role in the global nitrogen cycle. They are found in marine, freshwater, and terrestrial habitats, often in association with ammonia-oxidizing bacteria and are also important for the removal of nitrogen in wastewater treatment plants. The nitrite oxidoreductase enzyme, involved in nitrite oxidation, can also reduce nitrate to nitrite in the absence of oxygen, allowing Nitrobacter sp. to grow anaerobically.
A large number of Rv0899-like proteins are found in β-proteobacteria, including in Dechloromonas aromatica, a bacterium used for bioremediation that can oxidize aromatic hydrocarbon compounds in the absence of oxygen, in the N2-fixing, root-nodule-forming, plant symbionts: Ralstonia solanacearum, Cupriavidus taiwanensis, and Herbaspirillum seropedicae, and in several Burkholderia, a diverse and important species of bacteria, which includes: human and animal pathogens (e.g. members of the B. pseudomallei group and of the B. cepacia complex), plant pathogens (e.g. B. glumae causes seedling rot and panicle blight of rice), as well as plant growth-promoting species of biotechnological interest (e.g. B. phytofirmans). These bacteria are extremely adaptable to diverse environments, and are capable of degrading water and soil pollutants as well as fixing atmospheric nitrogen.
Among Burkholderia containing Rv0899-like proteins, at least three are N2-fixing, root-nodule- forming, plant symbionts (B. phytofirmans; B. glumae; B. graminis). Members of the B. pseudomallei group include B. mallei, the etiologic agent of glanders, a painful and incapacitating disease, where rapid-onset pneumonia, bacteremia (spread of the organism through the blood), pustules, and death are common outcomes. Because B. mallei is highly infectious as an aerosol, it is regarded as a potential biological weapon.34,35 B. mallei is an obligate mammalian parasite; in contrast, B. pseudomallei and B. thailandensis are human and animal pathogens as well as environmental soil inhabitants. Members of the B. cepacia complex are commonly found in soil, but are all opportunistic pathogens, especially in cystic fibrosis patients where they colonize the major airways, leading to debilitating pulmonary infection and death.36
In γ-proteobacteria, Rv0899 family members are found in two closely related members of Acinetobacter, in Stenotrophomonas and in Xanthomonas. Acinetobacter radioresistens and Acinetobacter baumannii are aquatic bacteria commonly isolated from hospital environments and hospitalized patients. Although they have low virulence, they can cause infection in the blood or in organs with a high fluid content, such as the lungs or urinary tract. Stenotrophomonas are found in varied environmental settings, particularly in close association with plants, and play major roles in the nitrogen as well as sulfur cycles. The Rv0899-containing species, Stenotrophomonas sp. SKA14, is a nitrogen-fixing bacterium from the Baltic Sea. Xanthomonas oryzae is a major pathogen of rice plants that enters rice leaves through water pores or wounds, and causes bacterial blight.37
Finally, more distant orthologs of Rv0899 are found in the gram-negative ε-proteobacteria, Campylobacterales bacterium and Arcobacter butzleri. The first is an anaerobic chemolithotrophic bacterium that plays an important role in dark, anaerobic CO2 fixation and nitrate reduction at marine oxic-anoxic transition zones. The second is a close relative of established human pathogens, such as Helicobacter pylori, that are found primarily in livestock and marine environments and can cause gastroenteritis and bacteremia in humans.
CONCLUSIONS
The identification of Rv0899-like proteins in organisms with related life-styles suggests the existence of a protein family defined by Rv0899. Protein families typically share common functionality and structure, thus, functional and structural analysis of the Rv0899-like proteins is needed to confirm whether they constitute a family. Nevertheless, several observations indicate that this is indeed the case. The central domain of Rv0899 adopts a fold that was previously unprecedented in the database and homology modelling indicates that this fold is shared by the other family members. Furthermore, at least some of the rv0899-like genes are found next to rv0900- and rv0901-like genes in the same operon. The presence of similar sequence, structure and genetic context, strongly suggests the existence of a common family.
The most striking finding of this analysis is that Rv0899-like proteins are present predominantly in bacteria that specialize in nitrogen fixation or metabolism, adaptation to nutrient poor environments, and/or establishing symbiosis with the host organism, suggesting that Rv0899 of M. tuberculosis may also assist similar processes. Free-living bacteria generally induce nitrogen fixation only under nitrogen stress, while symbiotic bacteria convert atmospheric N2 to ammonia to satisfy the needs of the host.38 The bacteria penetrate plant root cells, and induce the formation of nodules, which differentiate into spherical, thick-walled vesicles, formed by the plant’s plasma membrane, that provide a protective oxygen-free environment for the organism where reductive N2 fixation takes place. These vesicles act as a physical barrier between the symbiotic partners and it is tempting to draw analogy to the granulomas observed in M. tuberculosis infection.
Interestingly, M. tuberculosis is known to generate substantial quantities of ammonia, which inhibits phagosome fusion in infected macrophages,39 and expression of the Rv0899-Rv0901 proteins was shown to significantly accelerate ammonia secretion conferring adaptation of M. tuberculosis to acidic environments.3 The discovery of Rv0899 family proteins, and of at least some Rv0900- and Rv0901-like proteins, in bacteria active in nitrogen fixation or nitrogen metabolism lends further support to the hypothesis that Rv0899 an its operon are involved in promoting resistance of M. tuberculosis to the host environment by facilitating the release of ammonia. Finally the strong signal of the OmpA-like domain shared by all Rv0899-like proteins underscores the importance of examining the association of Rv0899 with peptidoglycan in the mycobacterial cell envelope. Most antibiotics developed for M. tuberculosis target the cell envelope, therefore identifying its components and understanding how they interact to provide a mechanically strong, highly impermeable barrier, that also maintains communication with the outside world, is important for identifying new drug targets.
Supplementary Material
Acknowledgments
We thank Kutbuddin Doctor, Michael Niederweis and Yong Yao for helpful discussions. This research was supported by a grant from the National Institutes of Health (AI074805).
References
- 1.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Raynaud C, Papavinasasundaram KG, Speight RA, Springer B, Sander P, Bottger EC, Colston MJ, Draper P. The functions of OmpATb, a pore-forming protein of Mycobacterium tuberculosis. Mol Microbiol. 2002;46:191–201. doi: 10.1046/j.1365-2958.2002.03152.x. [DOI] [PubMed] [Google Scholar]
- 3.Song H, Huff J, Janik K, Walter K, Keller C, Ehlers S, Bossmann SH, Niederweis M. Expression of the ompATb operon accelerates ammonia secretion and adaptation of Mycobacterium tuberculosis to acidic environments. Mol Microbiol. 2011;80:900–918. doi: 10.1111/j.1365-2958.2011.07619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teriete P, Yao Y, Kolodzik A, Yu J, Song H, Niederweis M, Marassi FM. Mycobacterium tuberculosis Rv0899 adopts a mixed alpha/beta-structure and does not form a transmembrane beta-barrel. Biochemistry. 2010;49:2768–2777. doi: 10.1021/bi100158s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alahari A, Saint N, Campagna S, Molle V, Molle G, Kremer L. The N-Terminal Domain of OmpATb Is Required for Membrane Translocation and Pore-Forming Activity in Mycobacteria. J Bacteriol. 2007;189:6351–6358. doi: 10.1128/JB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeats C, Bateman A. The BON domain: a putative membrane-binding domain. Trends Biochem Sci. 2003;28:352–355. doi: 10.1016/S0968-0004(03)00115-4. [DOI] [PubMed] [Google Scholar]
- 7.Senaratne RH, Mobasheri H, Papavinasasundaram KG, Jenner P, Lea EJ, Draper P. Expression of a gene for a porin-like protein of the OmpA family from Mycobacterium tuberculosis H37Rv. J Bacteriol. 1998;180:3541–3547. doi: 10.1128/jb.180.14.3541-3547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molle V, Saint N, Campagna S, Kremer L, Lea E, Draper P, Molle G. pH-dependent pore-forming activity of OmpATb from Mycobacterium tuberculosis and characterization of the channel by peptidic dissection. Mol Microbiol. 2006;61:826–837. doi: 10.1111/j.1365-2958.2006.05277.x. [DOI] [PubMed] [Google Scholar]
- 9.Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2008;88:526–544. doi: 10.1016/j.tube.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Auguin D, Delbecq S, Dumas E, Molle G, Molle V, Roumestand C, Saint N. Structure of the Mycobacterium tuberculosis OmpATb protein: a model of an oligomeric channel in the mycobacterial cell wall. Proteins. 2011;79:645–661. doi: 10.1002/prot.22912. [DOI] [PubMed] [Google Scholar]
- 11.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 12.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman P, Zhang Z, Wolf YI, Koonin EV, Miller W. Winnowing sequences from a database search. J Comput Biol. 2000;7:293–302. doi: 10.1089/10665270050081531. [DOI] [PubMed] [Google Scholar]
- 14.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 18.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 19.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rezwan M, Laneelle MA, Sander P, Daffe M. Breaking down the wall: fractionation of mycobacteria. J Microbiol Methods. 2007;68:32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffe M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong C, Beis K, Nesper J, Brunkan-Lamontagne AL, Clarke BR, Whitfield C, Naismith JH. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 27.Grizot S, Buchanan SK. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol Microbiol. 2004;51:1027–1037. doi: 10.1111/j.1365-2958.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 28.Parsons LM, Lin F, Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–2128. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- 29.Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc Natl Acad Sci U S A. 2008;105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barker DJ. Epidemiology of Mycobacterium ulcerans infection. Trans R Soc Trop Med Hyg. 1973;67:43–50. doi: 10.1016/0035-9203(73)90317-9. [DOI] [PubMed] [Google Scholar]
- 32.Merritt RW, Walker ED, Small PL, Wallace JR, Johnson PD, Benbow ME, Boakye DA. Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS Negl Trop Dis. 2010;4:e911. doi: 10.1371/journal.pntd.0000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayman J. Postulated epidemiology of Mycobacterium ulcerans infection. Int J Epidemiol. 1991;20:1093–1098. doi: 10.1093/ije/20.4.1093. [DOI] [PubMed] [Google Scholar]
- 34.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, Davidsen TD, Deboy RT, Dimitrov G, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Khouri H, Kolonay JF, Madupu R, Mohammoud Y, Nelson WC, Radune D, Romero CM, Sarria S, Selengut J, Shamblin C, Sullivan SA, White O, Yu Y, Zafar N, Zhou L, Fraser CM. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci U S A. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, Madupu R, Puiu D, Radune D, Shumway M, Trapnell C, Aparna G, Jha G, Pandey A, Patil PB, Ishihara H, Meyer DF, Szurek B, Verdier V, Koebnik R, Dow JM, Ryan RP, Hirata H, Tsuyumu S, Won Lee S, Seo YS, Sriariyanum M, Ronald PC, Sonti RV, Van Sluys MA, Leach JE, White FF, Bogdanove AJ. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204. doi: 10.1186/1471-2164-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patriarca EJ, Tate R, Iaccarino M. Key role of bacterial NH(4)(+) metabolism in Rhizobium-plant symbiosis. Microbiol Mol Biol Rev. 2002;66:203–222. doi: 10.1128/MMBR.66.2.203-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon AH, Hart PD, Young MR. Ammonia inhibits phagosome-lysosome fusion in macrophages. Nature. 1980;286:79–80. doi: 10.1038/286079a0. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. PyMol. 2005 wwwpymolorg.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.