Abstract
Generalized vitiligo is a common autoimmune disease in which acquired patchy depigmentation of skin, hair, and mucous membranes results from loss of melanocytes from involved areas. Previous genetic analyses have focused on vitiligo susceptibility, and have identified a number of genes involved in disease risk. Age of onset of generalized vitiligo also involves a substantial genetic component, but has not previously been studied systematically. In this study, we report a genome-wide association study of vitiligo age of onset in 1,339 generalized vitiligo patients, with replication in an independent cohort of 677 cases. We identified a quantitative trait locus for vitiligo age of onset in the major histocompatibility complex (MHC) class II region, located near c6orf10-BTNL2 (rs7758128; P = 8.14×10−11), a region that is also associated with generalized vitiligo susceptibility. In contrast, there was no association of vitiligo age of onset with any other MHC or non-MHC loci that are associated with vitiligo susceptibility. These findings highlight the differing roles played by genes involved in vitiligo susceptibility versus vitiligo age of onset, and illustrate that genome-wide analyses can be used to identify genes involved in quantitative aspects of disease natural history, as well as disease susceptibility per se.
INTRODUCTION
Generalized vitiligo is a common autoimmune disease in which acquired patchy depigmentation of skin, hair, and mucous membranes results from loss of melanocytes from involved areas (Rezaei et al., 2006). Moreover, generalized vitiligo is epidemiologically associated with increased risk of several other autoimmune diseases, both in patients and their close relatives (Alkhateeb et al., 2003; Laberge et al., 2005), suggesting that these autoimmune diseases involve shared susceptibility genes. Twin and epidemiological studies indicate that generalized vitiligo is a complex disease involving multiple genetic and environmental susceptibility factors, and genetic linkage, candidate gene, and recent genome-wide association studies have implicated a number of genes in vitiligo susceptibility (Spritz, 2008; Jin et al., 2010a, b; Quan et al., 2010; Birlea et al., 2011). Most of these genes encode immunoregulatory proteins, and many have also been implicated in susceptibility to some of the other autoimmune diseases that are epidemiologically associated with generalized vitiligo.
In addition to vitiligo susceptibility per se, variation in vitiligo age of onset also involves a genetic component. Vitiligo age of onset is correlated among affected relatives (Majumder et al., 1993), and mean age of onset is significantly earlier in multiplex families (21.5±15.0 years) than among sporadic cases (24.2±16.2 years) (Laberge et al., 2005). Furthermore, patients with early disease onset tend to have more affected relatives and more extensive disease involvement (Laberge et al., 2005). In this study, using MENDEL 7.0 (Lange et al., 2001), we calculated heritability of vitiligo age of onset as 0.45 in 184 multiplex families.
We previously described a genome-wide association study of generalized vitiligo susceptibility in whites of European descent, in which we identified a number of loci that contribute to disease susceptibility (Jin et al., 2010a, b; Birlea et al., 2011). These included major association signals in the major histocompatibility complex (MHC) class I and class II regions and at least 11 non-MHC loci. Here, we have specifically addressed vitiligo age of onset as a heritable trait distinct from disease susceptibility per se, re-analyzing our previous genome-wide data set by coding patients’ self-reported age of disease onset as a quantitative trait, testing for association genome-wide, and then re-testing significant association signals in an independent replication cohort. These analyses identified a quantitative trait locus in the MHC class II region that contributes significantly to generalized vitiligo age of onset, thus illustrating that genome-wide analyses can be used to identify genes involved in quantitative aspects of disease natural history, as well as disease susceptibility.
RESULTS
Genome-wide scan
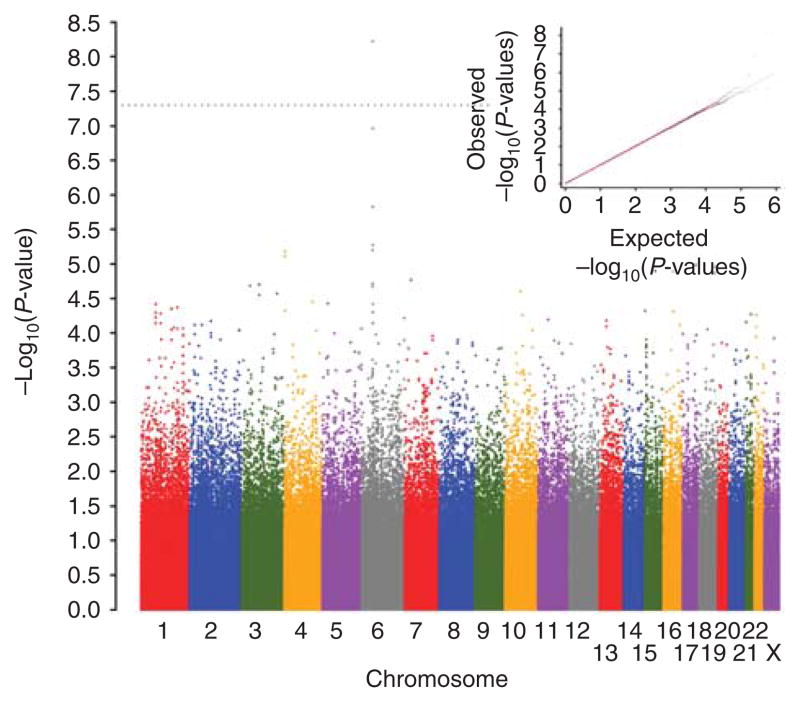
Considering all 2,016 cases in the present study, the mean vitiligo age-of-onset was 24.45 years; 24.84 for males (634 cases) and 24.27 for females (1,382 cases) (Table 1). After excluding some cases and markers by applying stringent quality control filters (Jin et al., 2010a), we tested association of 520,460 autosomal and X-chromosomal single-nucleotide polymorphisms (SNPs) with generalized vitiligo age of onset in 1,339 unrelated cases (Table 1) (Jin et al., 2010a) by linear regression, considering P<5×10−8 as the criterion for genome-wide significance (Ioannidis et al., 2009). A quantile– quantile plot of the genome-wide P-values (Figure 1) generally fit the null expectation, except at the extreme of the tail where observed P-values departed from expectation. The overall genomic inflation factor (Bacanu et al., 2002) was 1.007, indicating minimal inflation of the genome-wide statistics because of population stratification.
Table 1.
Summary description of the patients with generalized vitiligo used in this study
| Study cohort | No. of patients | Generalized vitiligo age of onset | Female (%) | Occurrence of other autoimmune diseases (%)1 | |
|---|---|---|---|---|---|
| Mean | SD | ||||
| Genome-wide scan | 1,339 | 24.06 | 16.40 | 70.20 | 31.29 |
| Replication cohort | 677 | 25.23 | 15.73 | 65.29 | 31.31 |
Includes autoimmune thyroid disease, rheumatoid arthritis, psoriasis, adult-onset autoimmune diabetes, pernicious anemia, Addison’s disease, and systemic lupus erythematosus, which are the principal autoimmune diseases that are epidemiologically associated with generalized vitiligo (Alkhateeb et al., 2003; Laberge et al., 2005).
Figure 1. Genome-wide association results for generalized vitiligo age of onset.
The genome-wide distribution of −log10(P-values) from the unadjusted linear regression analysis for 520,460 polymorphic single-nucleotide polymorphisms (SNPs) that passed quality control (QC) filters in 1,339 unrelated generalized vitiligo patients is shown plotted across the chromosomes. The dotted line indicates the genome-wide significance criterion (P<5×10−8). The inset shows quantile–quantile (Q–Q) plots of the observed versus expected −log10(P-values) for unadjusted linear regression statistics. The plot in red shows P-values for all 520,460 SNPs, whereas the plot in blue shows P-values excluding the 3,400 SNPs located across the extended major histocompatibility complex (MHC; chromosome 6: 25.9–33.4 Mb, genome build GRCh37).
As shown in Figure 1 and Supplementary Table S1 online, the five top-ranked SNPs genome-wide (based on P-values) were all located at chromosome 6p21.3 in the MHC class II region. SNP rs7758128, located near c6orf10-BTNL2, exceeded the criterion for genome-wide significance (P= 5.98×10−9), and three nearby SNPs showed suggestive association: rs28362680 (P= 1.48×10−6), rs28362683 (P= 1.09×10−7), and rs10947262 (P= 1.48×10−6). One additional MHC class II region SNP, rs532098, located >200 kb distal to the others, in the vicinity of HLA-DRB1-DQA1, also showed suggestive evidence of association (P= 2.03×10−6).
To more accurately assess the influence of these five SNPs on vitiligo age of onset, we applied Cox proportional hazards models (Cox, 1972) to test the multiplicative effect of each SNP on the hazard. As shown in Supplementary Table S1 online, Cox models yielded results very similar to the genome-wide linear regression results, with P-values of 6.24×10−9, 1.44×10−6, 9.82×10−8, 1.44×10−6, and 5.12×10−6 for SNPs rs7758128, rs28362680, rs28362683, rs10947262, and rs532098, respectively.
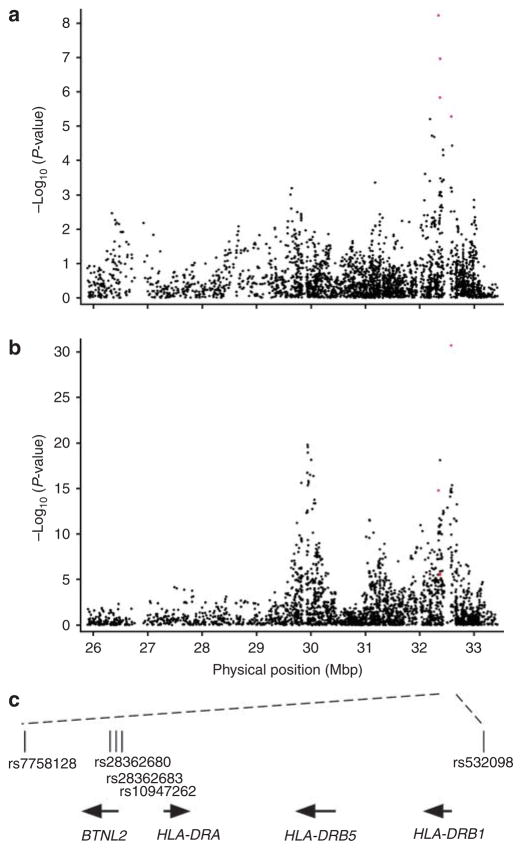
As shown in Figure 2a, these five SNPs derive from a prominent association peak for generalized vitiligo age of onset in the MHC class II region, which corresponds closely to a highly significant association peak for generalized vitiligo susceptibility (Figure 2b, and Jin et al., 2010a). In contrast, there was no association signal for vitiligo age of onset in the MHC class I region that is similarly strongly associated with vitiligo susceptibility (Jin et al., 2010a), indicating that the MHC class II region has a specific influence on vitiligo age of onset, whereas the class I region does not.
Figure 2. Major histocompatibility complex association results from genome-wide analyses of generalized vitiligo age of onset and susceptibility.
(a) Results of association analyses of the 3,400 (black dots) MHC region (chromosome 6: 25.9–33.4 Mb, genome build GRCh37) single-nucleotide polymorphisms (SNPs) with generalized vitiligo age of onset in 1,339 unrelated patients. The five top-ranked SNPs genome-wide are represented by red dots (dots representing rs28362680 and rs10947262 are too close to resolve). (b) Results of association analyses of the 3,400 (black dots) MHC region SNPs with generalized vitiligo susceptibility in 1,392 unrelated patients versus 2,629 unrelated controls (Jin et al., 2010a) using Cochran–Armitage trend tests implemented in PLINK (Purcell et al., 2007). The five top-ranked SNPs from the age of onset analysis are represented by red dots (dots representing rs28362680, rs28362683, and rs10947262 are too close to resolve). (c) A genomic map of the five top-ranked age-of-onsetassociated SNPs genome-wide. The arrows indicate gene orientations.
The SNPs rs7758128, rs28362680, rs28362683, and rs10947262 span 37.8 kb within one block of strong linkage disequilibrium, whereas rs532098, over 200 kb distal, is not in linkage disequilibrium with the other four SNPs (Supplementary Figure S1 online). To determine which of these five SNPs exert independent effects on the age of onset, and to test whether family history of vitiligo, co-occurrence of other concomitant autoimmune disorders, and gender might also contribute to vitiligo age of onset, we included these eight variables in a Cox model, utilizing L1 penalized (lasso) estimation and a small L2 penalty (Goeman, 2010) to identify variables with the strongest effects on the age of onset. Under the optimal value of λ1= 5.04 and λ2= 0.01, three variables (SNP rs7758128, SNP rs532098, and family history) showed non-zero coefficients. Accordingly, we re-tested association of rs7758128 and rs532098 with the age of onset in both linear regression analysis and in a Cox model adjusted for family history. As shown in Table 2, incorporating family history as a covariate slightly improved the linear regression and the Cox model P-values for rs7758128, P= 3.22×10−9 and P= 4.92×10−9, respectively, whereas for rs532098 the P-values were slightly worse, P= 1.56×10−5 and P= 1.30× 10−5, respectively. Together, these results indicate that SNPs rs7758128 and rs532098 may exert independent effects on vitiligo age of onset, and may partially explain the previously observed association between vitiligo age of onset and positive family history (Laberge et al., 2005).
Table 2.
Loci with strongest association with generalized vitiligo age of onset
| Chr. | Locus region1 | SNP | Location (nt) | Genome-wide association analysis
|
Replication analysis
|
Combined analysis
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Early-onset allele | Risk allele frequency | Linear regression P-value (two tailed) | Cox model P-value (two tailed) | Hazard ratio2 | Early-onset allele | Linear regression P-value (one tailed) | Cox model P-value (one tailed) | Hazard ratio2 | Early-onset allele | Linear regression P-value (two tailed) | Cox model P-value (two tailed) | Hazard ratio2 | ||||
| 6p21.3 | C6orf10/BTNL2 | rs7758128 | 32345283 | A | 0.06 | 3.22 × 10−9 | 4.92 × 10−9 | 1.61 | A | 2.05 × 10−3 | 0.014 | 1.30 | A | 8.14 × 10−11 | 1.36 × 10−9 | 1.50 |
|
| ||||||||||||||||
| 6p21.3 | HLA-DRB1-DQA1 | rs532098 | 32578052 | T | 0.60 | 1.56 × 10−5 | 1.30 × 10−5 | 1.19 | T | 0.081 | 0.044 | 1.09 | T | 1.83 × 10−5 | 4.23 × 10−6 | 1.15 |
Abbreviations: Chr., chromosome; SNP, single-nucleotide polymorphism.
Genes in close proximity to the designated SNP.
Effect sizes are measured as multiplicative effects, corresponding to the average change in phenotype when one later-onset allele is replaced with one early-onset-associated allele.
For each SNP, the P-values were calculated with family history as a covariate in a linear regression model or in a Cox model.
SNP nucleotide positions are from genome build GRCh37.
Replication analysis
To test the replication of association of rs7758128 and rs532098 with vitiligo age of onset, we genotyped these two SNPs in an independent cohort of 677 European-derived white generalized vitiligo patients (Table 1) (Jin et al., 2010a). As shown in Table 2, for rs7758128 we confirmed association with vitiligo age of onset in the replication cohort (linear regression P= 2.05×10−3; Cox model P= 0.014). Furthermore, the meta-analysis of rs7758128 in the genome-wide and replication data sets yielded highly significant P-values (linear regression P= 8.14×10−11; Cox model P= 1.36× 10−9), with a hazard ratio of 1.50 per A allele. For rs532098, we observed the same effect direction in the replication cohort, although the P-values were only marginal (linear regression P= 0.081; Cox model P= 0.044) and the combined P-values did not achieve genome-wide significance (linear regression P= 1.83×10−5; Cox model P= 4.23×10−6).
DISCUSSION
Our findings show that variation in the MHC class II region, best represented by rs7758128 in the vicinity of BTNL2, is strongly associated with generalized vitiligo age of onset. In contrast, other loci that are associated with generalized vitiligo susceptibility (Jin et al., 2010a, b; Birlea et al., 2011), including variation in the MHC class I region, are not associated with disease age of onset (Supplementary Table S2 online). These findings thus indicate that different loci have differing roles in vitiligo susceptibility versus vitiligo age of onset.
Penalized regression analysis suggested that MHC class II SNPs rs7758128 and rs532098 may contribute independently to vitiligo age of onset, although replication of rs532098 association with vitiligo age of onset was only marginally significant. Linear regression analyses indicated that rs7758128 accounts for 2.5% of the variance of vitiligo age of onset, rs532098 accounts for 1.5%, and both SNPs together account for 3.4% of the variance of vitiligo age of onset. The early age-of-onset-associated alleles of these two SNPs are also associated with increased disease susceptibility (Jin et al., 2010a), and the associated allele of rs532098 is in strong linkage disequilibrium with HLA-DR4, which we previously showed is associated with both vitiligo susceptibility and early disease onset (Fain et al., 2005).
To determine which HLA class II haplotypes include the SNP alleles that affect vitiligo age of onset, we analyzed class II alleles and SNP data from 74 previously reported vitiligo trios (Fain et al., 2005) and 37 CEPH (Centre d’Etude du Polymorphisme Humain) trios (Bugawan et al., 2000). This analysis showed that the A allele of rs7758128 that is associated with earlier vitiligo age of onset is exclusively located on the HLA class II haplotype DRB1*1301-DQA1*0103-DQB1*0603, although the majority of this haplotype (0.61) carry the C allele.
As shown in Figure 2c, the 27-kb rs7758128/rs28362680/ rs28362683/rs10947262 SNP cluster that is associated with vitiligo age of onset is located in close proximity to only one known gene, BTNL2, which encodes an immunoglobulin superfamily membrane protein implicated in T-cell activation, and one predicted gene of unknown function, c6orf10. The c6orf10/BTNL2 gene region has been associated with susceptibility to many other autoimmune diseases, including type 1 diabetes (Orozco et al., 2005; He et al., 2009), rheumatoid arthritis (Orozco et al., 2005; Cui et al., 2009), systemic lupus erythematosus (Orozco et al., 2005), ulcerative colitis (Pathan et al., 2009), psoriasis (Feng et al., 2009), and Graves’ disease (Simmonds et al., 2006), as well as with sarcoidosis (Valentonyte et al., 2005).
Biological evidence indicates that variation in the HLA class I region (specifically, HLA-A*0201) may contribute to disease susceptibility by mediating ongoing immune surveillance against malignant melanomas (and thus recognition of melanocytes) by the immune system (Spritz, 2010; Jin et al., 2010a). In contrast, association of the MHC class II region with both disease susceptibility and disease age of onset suggests that variation in this region might mediate response to environmental triggers encountered over the course of the life of genetically susceptible individuals.
SUBJECTS AND METHODS
Subjects
The discovery cohort comprised 1,339 unrelated generalized vitiligo patients and the replication cohort 677 additional unrelated patients, all of European white origin (Jin et al., 2010a). The age of onset was by self-report. All study participants provided written informed consent, and the study was approved by each institutional review board and was conducted according to the Declaration of Helsinki Principles.
Genotyping and quality control
DNA preparation, genome-wide genotyping using the Illumina 610-Quad BeadChip (Illumina, San Diego, CA) and replication genotyping using the Sequnom MassArray iPLEX system (Sequenom, San Diego, CA), and quality control filtering of the data have been described previously (Jin et al., 2010a).
Statistical analyses
For both the genome-wide and replication analyses, linear regression analyses were performed using PLINK (Purcell et al., 2007), considering the age of onset as a quantitative trait. To control for population stratification, we performed principal component analysis in the 1,339 discovery phase cases using EIGENSOFT (Price et al., 2006), using a subset of 21,642 independent SNPs, and included the three significant principal components (Tracy–Widom P<0.1) detected as continuous covariates in the linear regression analyses. We applied a Cox proportional hazards model (Cox, 1972) to the five top-ranked SNPs from the genome-wide scan and the two SNPs in the replication analysis using STATA 10 (www.stata.com). To obtain combined linear regression P-values, we performed meta-analysis using the inverse variance-weighted method (de Bakker et al., 2008). To obtain combined Cox model P-values, we specified genome-wide scan and replication data as different strata in the Cox model. We used L1 penalized (lasso) estimation with a small L2 penalty in a Cox proporational hazards model to determine those variables exhibiting the strongest effects on age-of-onset, using cross-validation to calculate the global optimal value of the tuning parameter λ (Goeman, 2010).
PHASE (Stephens et al., 2001), version 2.1.1, was used to determine which HLA class II haplotypes include the SNP alleles that affect vitiligo age of onset. Linkage disequilibrium was calculated using Haploview (Barrett et al., 2005) version 4.1.
Supplementary Material
Acknowledgments
This study was supported by grants AR45584 and AR056292 from the National Institutes of Health. We thank the University of Colorado Denver Microarray Core and the Washington University St Louis Genotyping Core, Janelle Noble, and Henry Erlich for CEPH HLA genotype data, and the memberships of Vitiligo Support International, the Vitiligo Society, the National Vitiligo Foundation, the American Vitiligo Research Foundation, and Associazione Ricerca Informazione per la Vitiligine for their participation.
Abbreviations
- MHC
major histocompatibility complex
- SNP
single-nucleotide polymorphism
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
References
- Alkhateeb A, Fain PR, Thody A, et al. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–14. doi: 10.1034/j.1600-0749.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Roeder K. Association studies for quantitative traits in structured populations. Genet Epidemiol. 2002;22:78–93. doi: 10.1002/gepi.1045. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Birlea SA, Jin Y, Bennett DC, et al. Comprehensive association analysis of candidate genes for generalized vitiligo supports XBP1, FOXP3, and TSLP. J Invest Dermatol. 2011;131:371–81. doi: 10.1038/jid.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugawan TL, Klitz W, Blair A, et al. High-resolution HLA class I typing in the CEPH families: analysis of linkage disequilibrium among HLA loci. Tissue Antigens. 2000;56:392–404. doi: 10.1034/j.1399-0039.2000.560502.x. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression models and life tables. J Roy Stat Soc Ser B. 1972;34:187–220. [Google Scholar]
- Cui J, Taylor KE, Destefano AL, et al. Genome-wide association study of determinants of anti-cyclic citrullinated peptide antibody titer in adults with rheumatoid arthritis. Mol Med. 2009;15:136–43. doi: 10.2119/molmed.2009.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain PR, Babu SR, Bennett DC, et al. HLA class II haplotype DRB1*04-DQB1*0301 contributes to risk of familial generalized vitiligo and early disease onset. Pigment Cell Res. 2005;19:51–7. doi: 10.1111/j.1600-0749.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- Feng BJ, Sun LD, Soltani-Arabshahi R, et al. Multiple loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 2009;5:e1000606. doi: 10.1371/journal.pgen.1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biometrical J. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- He C, Hamon S, Li D, et al. MHC fine mapping of human type 1 diabetes using the T1DGC data. Diabetes Obes Metab. 2009;11(Suppl 1):53–9. doi: 10.1111/j.1463-1326.2008.01003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JPA, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10:318–29. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. New Engl J Med. 2010a;362:1686–97. doi: 10.1056/NEJMoa0908547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Birlea SA, Fain PR, et al. Common variants in FOXP1 are associated with generalized vitiligo. Nat Genet. 2010b;42:576–8. doi: 10.1038/ng.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge G, Mailloux CM, Gowan K, et al. Early disease onset and increased risk of other autoimmune diseases in familial generalized vitiligo. Pigment Cell Res. 2005;18:300–5. doi: 10.1111/j.1600-0749.2005.00242.x. [DOI] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, et al. MENDEL version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69(Suppl):A1886. [Google Scholar]
- Majumder PP, Nordlund JJ, Nath SK. Pattern of familial aggregation of vitiligo. Arch Dermatol. 1993;129:994–8. [PubMed] [Google Scholar]
- Orozco G, Eerligh P, Sánchez E, et al. Analysis of a functional BTNL2 polymorphism in type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Hum Immunol. 2005;66:1235–41. doi: 10.1016/j.humimm.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Pathan S, Gowdy RE, Cooney R, et al. Confirmation of the novel association at the BTNL2 locus with ulcerative colitis. Tissue Antigens. 2009;74:322–9. doi: 10.1111/j.1399-0039.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan C, Ren YQ, Xiang LH, et al. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat Genet. 2010;42:614–8. doi: 10.1038/ng.603. [DOI] [PubMed] [Google Scholar]
- Rezaei N, Gavalas NG, Weetman AP, et al. Autoimmunity as an aetiological factor in vitiligo. J Eur Acad Dermatol Venereol. 2006;21:865–76. doi: 10.1111/j.1468-3083.2007.02228.x. [DOI] [PubMed] [Google Scholar]
- Simmonds MJ, Heward JM, Barrett JC, et al. Association of the BTNL2 rs2076530 single nucleotide polymorphism with Graves’ disease appears to be secondary to DRB1 exon 2 position beta74. Clin Endocrinol (Oxf) 2006;65:429–32. doi: 10.1111/j.1365-2265.2006.02586.x. [DOI] [PubMed] [Google Scholar]
- Spritz RA. The genetics of generalized vitiligo. Curr Dir Autoimmun. 2008;10:244–57. doi: 10.1159/000131501. [DOI] [PubMed] [Google Scholar]
- Spritz RA. The genetics of generalized vitiligo: autoimmune pathways and an inverse relationship with malignant melanoma. Genome Med. 2010;2:78. doi: 10.1186/gm199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentonyte R, Hampe J, Huse K, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–64. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.