Abstract
The Ω-loop at the active site of β-lactamases exerts significant impact on the kinetics and substrate profile of these enzymes by forming part of the substrate binding site and posing as steric hindrance toward bulky substrates. Mutating certain residues on the Ω-loop has been a general strategy for molecular evolution of β-lactamases to expand their hydrolytic activity toward extended-spectrum antibiotics through a mechanism believed to involve enhanced structural flexibility of the Ω-loop. Yet no structural information is available that demonstrates such flexibility or its relation to substrate profile and enzyme kinetics. Here we report an engineered β-lactamase that contains an environment-sensitive fluorophore conjugated near its active site to probe the structural dynamics of the Ω-loop and to detect the binding of diverse substrates. Our results show that this engineered β-lactamase has improved binding kinetics and positive fluorescence signal toward oxyimino-cephalosporins, but shows little such effect to non-oxyimino-cephalosporins. Structural studies reveal that the Ω-loop adopts a less stabilized structure, and readily undergoes conformational change to accommodate the binding of bulky oxyimino-cephalosporins while no such change is observed for non-oxyimino-cephalosporins. Mutational studies further confirm that this substrate-induced structural change is directly responsible for the positive fluorescence signal specific to oxyimino-cephalosporins. Our data provide mechanistic evidence to support the long-standing model that the evolutionary strategy of mutating the Ω-loop leads to increased structural flexibility of this region, which in turn facilitates the binding of extended spectrum β-lactam antibiotics. The oxyimino-cephalosporin-specific fluorescence profile of our engineered β-lactamase also demonstrates the possibility of designing substrate-selective biosensing systems.
Keywords: Antibiotics, Enzyme Kinetics, Enzyme Mutation, Fluorescence, Protein Chemical Modification, Protein Conformation, Protein Structure
Introduction
Numerous β-lactamases (over 400) have emerged in the past decades; as a result of rapid molecular evolution under the selective pressure of extensive usage of β-lactam antibiotics (reviewed in Refs. 1–2). These enzymes are divided into four classes (A–D) based on the homology of their amino acid sequence and catalytic mechanism. The catalytic mechanisms of Classes A, C, and D are highly similar, consisting of an acylation step catalyzed by Ser-70 leading to the formation of an acyl-enzyme intermediate with the β-lactam acylated to the side chain Ser-70 (E + S → ES*), and a subsequent deacylation step catalyzed by Glu-166 to release the hydrolyzed antibiotics in the form of “opened” β-lactam ring and restore the enzyme to its free form (ES* → E + P) (residue numbering according to the clinically significant TEM-1 β-lactamase). Class B employs a distinct metal-dependent acylation and deacylation process (1–3).
While sharing a common catalytic mechanism, the various β-lactamases differ significantly in their substrate profile toward the large number of clinically available β-lactam antibiotics (1–2). These enzymes undergo rapid molecular evolution on a time scale of several years by acquiring single or multiple mutations at certain strategic “hot-spots” that result in drastic expansion of their substrate profile toward newer generation antibiotics (1–2). One such hot spot for evolutionary mutation is the Ω-loop, a short stretch of residues on the surface of the β-lactamase structure that folds back to form part of the enzyme active site (1–3). For the narrow-spectrum β-lactamases such as PenP and PenPC used in our study as well as the clinically significant TEM-1 and SHV-1 enzymes the Ω-loop is tightly packed onto the enzyme active site through hydrophobic and electrostatic interactions, posing as steric hindrance for the binding of second or third generation cephalosporins with bulky side chains attached onto the cephem nucleus (1–3). Many mutant strains of TEM- and SHV-like β-lactamases overcome this inefficiency and become extended spectrum β-lactamases (ESBLs)4 by acquiring mutations in the Ω-loop region that disrupt the stabilizing interactions in this region, and potentially render this region more flexible for reconfiguration to accommodate large-sized cephalosporins (1–3). Mutation at one residue of the Ω-loop, i.e. residue Arg-164, is found in over 50% of TEM-1 variants (1–2). For certain Class C β-lactamases that are generally ESBLs, the corresponding region of the Ω-loop is significantly extended as compared with that of Class A β-lactamases, resulting in an enlarged active site that readily binds to and hydrolyzes almost all cephalosporins (1–2).
We are interested in the molecular mechanism of how enhanced structural flexibility in the Ω-loop of a β-lactamase affects its substrate profile. So far among the large number of experimentally determined β-lactamase structures, the Ω-loop appears to adopt a well-defined conformation that is little affected by different substrates. No experimental data are yet available that clearly demonstrate the range of possible dynamic conformations of the Ω-loop and correlate such flexibility to the substrate profile. To address this issue we set out to engineer a β-lactamase that allows us to probe its structural dynamics and substrate binding at the active site. The principle of our design is to conjugate an environment-sensitive fluorophore reporter group to residue 166 on the Ω-loop after E166C mutation. Such an engineered β-lactamase enzyme would lose its critical residue (E166) for the deacylation step of the catalysis and becomes an acylation-competent but deacylation-impaired β-lactam-binding protein that readily forms the stable ES* acyl-enzyme intermediate. The fluorophore molecule, with its proximity to the active site and the Ω-loop, could potentially report the event of substrate binding as well as any conformational change in the Ω-loop induced by such binding, if these events lead to changes in the polarity of the local environment (4–6).
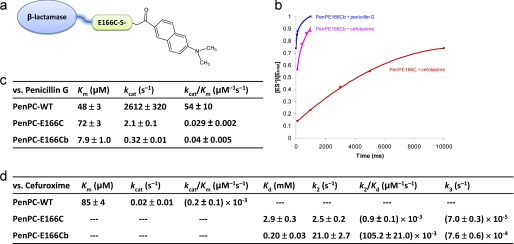
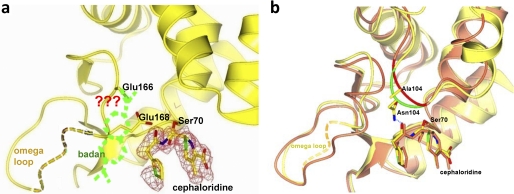
For our design we decided to use the naphthalene-type fluorophore molecules because of their exceptional solvachromatic properties that can reflect the polarity of the local environmental at the molecule level, as well as their relative small size comparable to the amino acid, tryptophan, to allow them to be positioned close to the protein structure (7–13). This fluorophore has been applied to probe the structural dynamics of membrane lipids (11) and ion channels (12), and to detect the binding of peptide ligands to SH2 domains (9) and Class II MHC proteins (13). We examined several fluorophores and report here one such engineered β-lactamase, PenP-E166Cb, which is based on the Class A β-lactamase PenP from Bacillus licheniformis. An environment-sensitive fluorophore BADAN (6-bromoacetyl-2-dimethylaminonaphthalene) is conjugated to the side chain thiol group of residue 166 after E166C mutation (Fig. 1a). The kinetic profile and fluorescence spectra of PenP-E166Cb toward a variety of cephalosporins, as well as structural studies to correlate this profile with the flexibility of the Ω-loop are reported in detail below.
FIGURE 1.
The kinetic profile of BADAN-labeled β-lactamases. a, scheme for BADAN labeling at residue 166 of a β-lactamase enzyme. b, time course of ES* formation as monitored by ESI-MS when PenP-E166Cb was mixed with cefotaxime or penicillin G at a concentration of 2.5 μm (enzyme) versus 5 μm (substrate). Such spectra obtained at different substrate concentrations were used to calculate the kinetic parameters of the labeled enzyme following the procedure as previously reported (4). The solid squares represent the [ES*]/[Etotal] values of the PenP-E166C-antibiotic and PenP-E166Cb-antibiotic complexes respectively as determined by ESI-MS. Buffer: 20 mm ammonium acetate (pH 7.0). Curve fitting of the time profile was performed using single-binding site. c and d, kinetic parameters of the wild-type PenPC β-lactamase, its PenPC-E166C mutant, and the BADAN-labeled PenPC-E166Cb mutant for the hydrolysis of penicillin G (c) and cefuroxime (d). The kinetic parameters of the wild-type PenPC for the hydrolysis of cefuroxime were quoted from our previously published results (4).
EXPERIMENTAL PROCEDURES
Protein Preparation and BADAN Labeling
The expression and purification of PenP-E166C are described under supplemental data. Before BADAN labeling, the purified PenP-E166C protein was mixed with 2 m guanidine HCl (GdHCl) (pH adjusted to 7–7.5 with KOH), and the final concentration of protein was adjusted to 1 mg/ml. The protein solution was incubated at room temperature for 30 min. A 10-fold molar excess of BADAN at the concentration of 20 mm was dissolved in DMF and added to the protein solution drop by drop. The mixture was shaken at 300 rpm at room temperature for 2 h in darkness, and then dialyzed against 1 liter of 50 mm potassium phosphate buffer (pH 7.0) at 4 °C overnight by dialysis tubing with the cut-off size of 12 kDa to remove excess dyes or GdHCl. Buffer exchanges were carried out regularly during dialysis. After three buffer exchanges the protein solution was concentrated by Amicon® Ultra-4 Centrifugal Filter Devices (Millipore NMWL = 10,000) to less than 1 ml and loaded into the SuperdexTM 75 gel filtration column from GE Healthcare. The running buffer contains 20 mm Tris-HCl, 50 mm NaCl with a pH value of 7.5. The flow-rate was 1 ml/min. The eluted protein was pooled and concentrated by Amicon Ultra to 20 mg/ml. Assessment of the labeling efficiency by ESI-MS is described under supplemental data. The expression, purification, and BADAN labeling of PenP-E166C-E168L protein were identical to those of PenP-E166C. The E168L mutation was constructed by site-directed mutagenesis.
Fluorescence Measurement
Measurement of the fluorescence scanning spectra (from 410 to 600 nm) of PenP-E166Cb and PenP-E166Cb-E168L in the presence of six cephalosporins and in the presence of cefotaxime at various concentrations were performed in the same procedure as previously published (6) and are described in detail under supplemental data.
Detection of Covalent Acyl Enzyme-Substrate Complexes (ES*) and Kinetic Measurements by Mass Spectrometry
The binding kinetics of PenP and PenPC, both in wild-type form, the E166C mutant form and the BADAN-labeled E166Cb form in the presence of cefotaxime and penicillin G were measured by ESI-MS following the same procedure as reported in our previous study (4). The enzyme-substrate binding reaction was initiated by mixing 70 μl of 5 μm His6-tagged PenP-E166C and His6-tagged PenP-E166Cb (in 20 mm ammonium acetate buffer, pH 7.0) with 70 μl of 10 μm antibiotic (penicillin G and cefotaxime, in 20 mm ammonium acetate buffer, pH 7.0) at room temperature. At desired time intervals, the reaction was quenched, and the reaction mixture was subjected to ESI-MS characterization. Mass spectra identified two major peaks that corresponded to the free enzyme E and the enzyme-substrate complex ES*, respectively. The relative amount of ES* can be expressed as the ratio of the amount of ES* to the total amount of the enzyme, [ES*]/[Etotal], where [ES*] is the peak intensity of ES* and [Etotal] (= [E] + [ES*]) is the sum of peak intensities of ES* and E. Further technical details can be found under the supplemental data.
Crystallization and Structure Determination
Crystals of PenP-E166Cb were grown by hanging-drop vapor diffusion method. A concentrated solution of PenP-E166Cb protein (∼20 mg/ml) in 20 mm Tris-HCl buffer with 50 mm NaCl at pH 7.5 was mixed with an equal volume of reservoir solution containing 25% (w/v) PEG 4000, 0.1 m Hepes at pH 7.2. Crystals grew to ∼100 μm size after 2 weeks. The presence of the BADAN fluorophore in the crystals was confirmed by their fluorescence signal (supplemental Fig. S6). For data collection, crystals were harvested and cryoprotected in reservoir solution plus 20% ethylene glycol for 1 min before mounted to the Rigaku MicroMaxTM-007HF x-ray machine. Data sets were collected and processed by CrystalClearTM 1.3.5 SP2. The crystals belonged to the monoclinic group P21 with a cell parameter of a = 43.8 Å, b = 91.3 Å, c = 66.5 Å and diffracted to 1.86 Å. There were two molecules per asymmetric unit. Cefotaxime and cephaloridine were soaked into crystals of PenP-E166Cb by incubating crystals in reservoir solution with addition of cefotaxime and cephaloridine respectively at final concentration of 0.01 m for 20 min. A summary of the data collection and soaking conditions is presented in supplemental Table ST1. When crystals were initially mounted to the x-ray machine after quick cryoprotection, the electron density for BADAN was rather poor, while that for the rest of the protein structure was excellent. The electron density for BADAN was greatly improved after the crystals had been subjected to a long period of stabilization (1–3 h) in the cryoprotection step. Finally an electron density map of high quality was obtained for BADAN to allow us to build the BADAN molecule with confidence. For one data set the maps were sufficiently good to allow fitting of the whole BADAN molecule into the electron density for both PenP-E166Cb molecules in the asymmetric unit. Subsequent structure refinement confirmed the coordinates for BADAN molecule were stable after several rounds of refinement. Data processing and structure determination were done using the CCP4i software package. COOT program was used for manual structure rebuilding. The coordinates and structure factors from this study have been deposited into Protein Data Bank (PDB) under accession codes 3KGM (PenP-E166Cb), 3KGN (PenP-E166Cb-cefotaxime), and 3KGO (PenP-E166Cb-cephaloridine).
RESULTS
Mutation of E166C for PenP and subsequent expression and purification of PenP-E166C were carried out following standard protocols. BADAN labeling of PenP-E166Cb was performed by mixing the purified PenP-E166C protein with BADAN solution in excess at a molar ration of 1:10 under mildly denaturing conditions of 2 m GdHCl (Fig. 1a) to achieve best efficiency and the labeled PenP-E166Cb protein was subsequently refolded by dialysis. Labeling efficiency was significantly enhanced by this denaturing step as measured by electrospray ionization mass spectroscopy (ESI-MS) (supplemental Fig. S1). The denaturing and refolding steps in the process of BADAN labeling induced little overall structural change of the enzyme as measured by CD spectrum (supplemental Fig. S2).
Kinetics of Acylation-competent and Deacylation-impaired PenP-E166Cb
To confirm that the engineered PenP-E166Cb was indeed acylation-competent and deacylation-impaired, its enzyme kinetics as well as that of a related β-lactamase with the same BADAN labeling (PenPC-E166Cb), were measured by ESI-MS. The ESI-MS spectra demonstrate that PenP-E166Cb can readily form stable ES* acyl adducts with two representative substrates of penicillin G and cefotaxime (Fig. 1b). The binding profile of PenP-E166Cb toward cefotaxime, a third-generation extended-spectrum cephalosporin and a naturally poor substrate for the wild-type PenP, is significantly improved as compared with that of PenP-E166C (Fig. 1b). For the related enzyme PenPC from Bacillus cereus, the BADAN-labeled PenPC-E166Cb is acylation-competent, with slightly improved acylation kinetics (Km) of ∼6 times as compared with that of the wild-type and unlabeled mutant PenPC-E166C toward its natural substrate penicillin G (Fig. 1c). Yet PenPC-E166Cb is deacylation-impaired with the deacylation kinetics (kcat) drastically reduced by ∼5 orders of magnitude as compared with that of the wild-type (Fig. 1c). PenPC shows a noteworthy improvement in kinetics toward cefuroxime, a cephalosporin highly similar to cefotaxime and a naturally poor substrate for PenPC. The acylation kinetics (k2/Kd) of PenPC-E166Cb is actually ∼2 orders of magnitude faster than that of the unlabeled PenP-E166C mutant. The deacylation kinetics (k3) is also slightly improved by ∼10 times (Fig. 1d), suggesting that BADAN-labeled β-lactamases may have a distinct binding mode for extended-spectrum cephalosporins such as cefotaxime and cefuroxime as compared with that for the naturally good substrates of penicillins.
Distinct Fluorescence Profile of PenP-E166Cb toward Diverse Cephalosporins
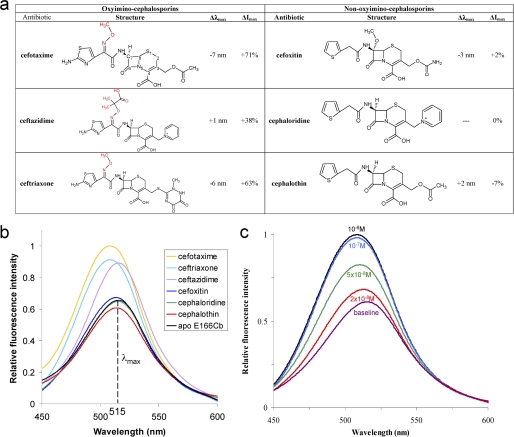
To understand the dynamic properties of the Ω-loop and the distinct binding modes of PenP-E166Cb toward different substrates, the profile of fluorescence emission by PenP-E166Cb in the presence of various cephalosporins was measured by both scanning and time-dependent spectrum at steady state. We mainly focused on cephalosporins because of the improved binding kinetics toward this group of substrates as measured by ESI-MS. Nearly 20 cephalosporins were tested and six of them are reported as representatives of first-, second-, and third-generation cephalosporins containing diverse side chain groups conjugated onto the C3 and C7 positions of the cephem nucleus (Fig. 2a). The R1 group conjugated onto the C3 position is usually named the “leaving” group because this group is quickly rearranged and released when the cephalosporin is acylated to β-lactamase (14). Several R1 groups including the acetoxymethyl group in cefotaxime, the carbamoyloxymethyl group in cefoxitin and the pyridine group in cephaloridine were chosen. For the R2 group at the C7 position two major types were chosen, namely the non-branched thiozoldine-only group as seen in cefoxitin and cephaloridine (non-oxyimino-cephalosporins) and the branched side chains that contains a methoxyimini group in addition to the thiozoldine ring as in cefotaxime (oxyimino-cephalosporins). Ceftazidime represents an oxyimino-cephalosporin with the most complicated R2 group containing two additional methyl groups and one additional carboxylic group at the methoxy moiety.
FIGURE 2.
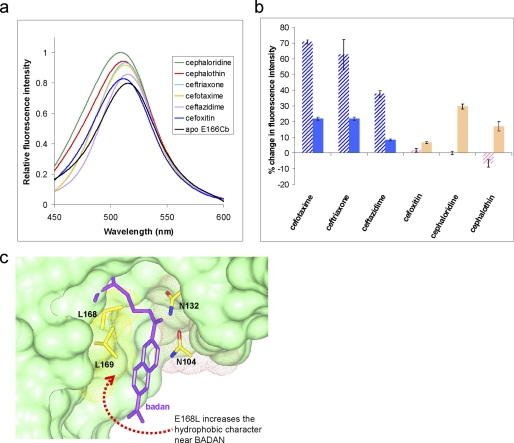
The fluorescence profile of PenP-E166Cb toward diverse cephalosporins. a, fluorescence profile of PenP-E166Cb in the presence of selected cephalosporins at 0.1 mm. Δλmax refers to the shift in wavelength at maximum emission intensity. ΔImax refers to change in emission intensity at λmax. b, fluorescence scanning spectra of PenP-E166Cb (10−7 m) in the presence of the 6 listed cephalosporins (10−4 m), respectively. c, fluorescence scanning spectra of PenP-E166Cb (10−7 m) in the presence of cefotaxime at different concentrations.
PenP-E166Cb demonstrated a selective fluorescence response toward the six representative cephalosporins. Only oxyimino-cephalosporins with a bulky branched R2 group at the C7 position containing the methoxyimini moiety, including cefotaxime, ceftazidime, and ceftriaxone, produced measurable fluorescence change, (Fig. 2, a and b). For the case of cefotaxime, a noticeable increase of ∼20% in the intensity of emitted fluorescence was detected in the scanning fluorescence spectra at near nanomolar concentrations (2 × 10−8 m), while at a concentration of 10−7 m, >70% (Fig. 2c). Non-oxyimino-cephalosporins including cefoxitin, cephaloridine, and cephalothin that contain non-branched R2 groups at the C7 position induced weak (less than 10%) or no measurable fluorescence changes, even at concentration as high as 10−4 m (Fig. 2b). The different R1 groups at the C3 position did not affect the fluorescence signal.
Structure of PenP-E166Cb Reveals a Less-stabilized Ω-loop
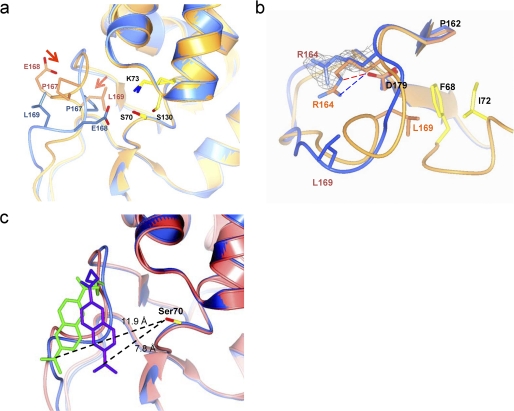
To investigate the structural properties of the Ω-loop, we solved the structure of PenP-E166Cb by molecular replacement using the known structure of PenP (PDB ID 4BLM) as a search model (supplemental Table ST1). The overall folding of the mutant enzyme PenP-E166Cb is largely identical to that of the wild type PenP. The RMSD between these two structures is just ∼1.5 Å for all atoms and only 0.8 Å for the main chain atoms. Key residues lining the catalytic site, including Ser-70, Lys-73, Ser-130, and Lys-234 are virtually in identical positions as that in the wild-type enzyme and the PenP-E166C mutant (Fig. 3a).
FIGURE 3.
The structure of PenP-E166Cb. a, structure of BADAN-labeled PenP-E166Cb reveals an altered conformation of the Ω-loop. Structures of PenP-E166Cb and wild-type PenP (PDB ID 4BLM) are shown in blue and orange, respectively. Residues on Ω-loop (E166, E166Cb, and P167) and key catalytic residues for acylation (Lys-73, Ser-70, and Ser-130) are displayed in cpk color scheme. The BADAN fluorophore is omitted for clarity purpose. The arrows indicate the induced conformational change of certain residues. b, altered conformation of the Ω-loop is less-stabilized. The Ω-loop of PenP-E166Cb (shown in blue) is aligned with the corresponding region in the wild-type PenP structure (shown in orange). The salt bridge and the hydrogen bond between residues Arg-164 and Asp-179 in the wild-type structure are indicated by dashed lines. The residues forming favorable hydrophobic interaction with Leu-169 (Phe-68 and Ile-72) in the wild-type structure are colored in cpk scheme in the wild-type structure but colored blue in the PenP-E166Cb structure. The fo-fc density for residue Arg-164 in PenP-E166Cb structure is shown in mesh format and contoured at 2.0 σ. c, superposition of the two PenP-E166Cb molecules in the asymmetric unit by aligning all the protein atoms. Ser-70 is shown in cpk. The two BADAN molecules are colored purple and green, respectively. The two superimposed PenP-E166Cb molecules are colored pink and blue, respectively. The distances between the dimethyl group of the BADAN molecule and the -OH of Ser-70 are shown.
The only region that has undergone distinct reconfiguration in the E166Cb structure is the Ω-loop region consisting of residues 166–172. In the wild-type PenP structure, this segment is a short 2-turn helix closely packed toward the active site of the enzyme. In the PenP-E166Cb structure this helical element is unwound and adopts a new extended loop-like conformation to accommodate the bulky BADAN fluorophore linked to Cys-166 (Fig. 3a). All residues within this segment are displaced accordingly by ∼5 Å. Most notably the side chain of Glu-168, a highly conserved residue in Class A β-lactamases, is flipped nearly 180° from the original solvent-exposed orientation to point into the interior of the PenP active site, occupying a position approximate to Asn-170 in the wild-type structure (Fig. 3a). In this new conformation the carboxylate of Glu-168 is largely shielded from bulk solvent with a Solvent Accessible Area (SAA) of ∼20 Å2, exerting a strong electrostatic effect on the local environment.
The structure of the Ω-loop of PenP-E166Cb is largely less stabilized than that of the wild type. Several distinct features that contribute to the structural stabilization of the Ω-loop, including the salt bridge and the hydrogen bonds between residue Arg-164 and Asp-179, and the hydrophobic packing of Leu-169 against the PenP enzyme, are all disrupted in the PenP-E166Cb structure (Fig. 3b).
The BADAN fluorophore is clearly visible in the fo-fc map (supplemental Fig. S7). The fluorophore is located on the surface of the protein at the outer edge of the antibiotic binding site. The naphthalene plane is lodged into a surface groove formed by residues 166–170 of the Ω-loop on one side and by residues 103–104 of a loop segment on the other side (Fig. 3c). No specific interactions were found between BADAN and the protein atoms of PenP-E166Cb. The local environment of BADAN is highly polar. Atoms located within 4.5 Å from BADAN molecule include the charged carboxylate side chain of Glu-168, the polar side chains of Asn-132 and Asn-104, and 5 water molecules. Two symmetry-related BADAN molecules in the asymmetric unit adopt largely identical orientation, with one BADAN molecule nudged a little closer to the active site by crystallographic contact and both molecules are in close proximity to the active site, being ∼10.0 Å away from the -OH of Ser-70 (Fig. 3c). The crystallographic B-factor of the BADAN molecule is noticeably higher than the rest of the protein, suggesting high thermodynamic mobility.
Structure of PenP-E166Cb with Cefotaxime (Oxyimino-cephalosporin) Acylated to Its Active Site Reveals an Induced Conformational Change of the Ω-Loop
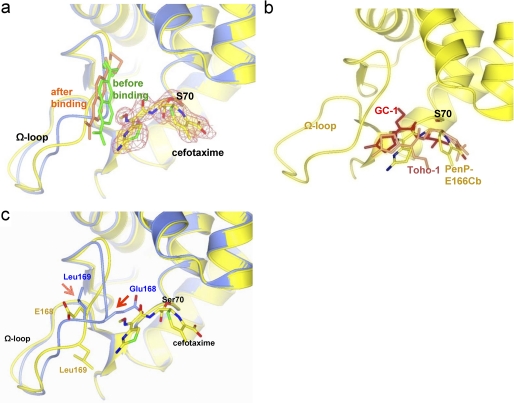
In the PenP-E166Cb-cefotaxime structure both cefotaxime and BADAN are clearly visible on the fo-fc map (Fig. 4a). The binding and acylation of cefotaxime exerts little effect on the position of the BADAN molecule (Fig. 4a). No specific interaction was observed between BADAN and cefotaxime. Thus the BADAN molecule is not directly involved with the acylation kinetics of E166Cb, but serves solely as a probe of the local environment instead.
FIGURE 4.
The structure of PenP-E166Cb with cefotaxime acylated at its active site. a, cefotaxime is acylated to Ser-70 at the active site of PenP-E166Cb. The PenP-E166Cb-cefotaxime complex structure (in yellow with its BADAN molecule in gold) is aligned with that of PenP-E166Cb (blue with its BADAN molecule in green). The cefotaxime molecule is depicted in cpk color scheme. The fo-fc map of cefotaxime is contoured at 2.0 σ. b, comparison of the binding mode of cefotaxime in PenP-E166Cb (cpk color) with that in Toho-1 (coral) and GC1 (red). The protein structure of PenP-E166Cb is in yellow. c, induced conformational change of the Ω-loop upon acylation of cefotaxime. The PenP-E166Cb-cefotaxime complex structure (yellow) is aligned with that of PenP-E166Cb (blue). Residues on the Ω-loop (E166, E166Cb, and P167) and key catalytic residues for acylation (K73, S70, and S130) are displayed in cpk color scheme. The BADAN fluorophore is omitted for clarity purpose. The arrows indicate the induced conformational change of certain residues.
Cefotaxime is covalently linked to the Oγ atom of Ser-70 through C8 of its β-lactam ring. The bulky and branched side chain group containing the methoxyimino moiety is inserted into the PenP active site, occupying a location that overlaps with the position of Glu-168 in the apo structure (Fig. 4b). This binding mode of cefotaxime shows distinct difference as compared with that of Toho-1, an extended spectrum Class A β-lactamase with a tightly packed Ω-loop that poses steric hindrance (15). Instead it resembles the binding mode seen in GC1, a Class C extended-spectrum β-lactamase with a loosely packed Ω-loop that does not pose any steric hindrance (16) (Fig. 4b). In the Toho-1 structure the methoxyimino side chain points away from the active site and is solvent-exposed. Such an orientation packs the methoxyimino side chain tightly against the thiozolyl ring, leading to a distorted configuration of the cephem nucleus that is catalytically incompetent for deacylation (15). In the GC1 structure the transition analog of cefotaxime binds to GC1 in a fully extended conformation, with the oxyimino group inserted to the active site and extended away from the thiozolyl ring. This conformation is regarded as catalytically competent to facilitate deacylation because the distortion on the cephem nucleus is released (16). Thus PenP-E166Cb preferentially interacts with cefotaxime in a manner that resembles an ESBL. To confirm the new binding mode of oxyimino-cephalosporins observed here we also solved the structure of PenP-E166Cb in acyl adduct complex with cefuroxime. The binding mode for cefuroxime is essentially the same as cefotaxime, with the oxyimino group inserted into the protein body of PenP-E166Cb (supplemental Fig. S8).
The new binding mode of cefotaxime with its methoxyimino group inserted into the PenP active site leads to significant conformational change for a segment of the Ω-loop (residues 168–172). To avoid steric clash with the inserted methoxyimino group the side chain carboxylate of residue Glu-168 is flipped 180° from its solvent-shielded position adjacent to the antibiotic binding site as seen in the PenP-E166Cb structure to become completely solvent exposed in juxtaposition to the BADAN fluorophore (Fig. 4c). The rest of the Ω-loop region becomes highly dynamic and we were not able to build the side chain conformation for a few residues on the Ω-loop because of the relatively poor electron density in this region. The polarity of the local environment for BADAN is likely significantly decreased when the charged carboxylic side chain of Glu-168 relocates from the interior of the protein molecule to become fully solvent exposed because the electrostatic effect Glu-168 exerts on local environment is weakened when its side chain is fully solvated.
Structure of PenP-E166Cb with Cephaloridine (Non-oxyimino-cephalosporins) Acylated at Its Active Site Reveals a Highly Flexible Ω-loop
In the PenP-E166Cb-cephaloridine structure the electron density unambiguously reveals a covalent acyl-enzyme adduct connected at C7 of cephem nucleus with the C-3 leaving group lost (Fig. 5a). The interaction mode of cephaloridine with PenP-E166Cb is nearly identical to that observed in another Class A β-lactamase PC1 (Fig. 5b) (17–18). The acylamide of cephaloridine forms one extra hydrogen bond with the polar side chain of Asn-104, a bond that is absent in PC-1 structure due to a slightly different conformation of the 103–105 loop region (Fig. 5b).
FIGURE 5.
The structure of PenP-E166Cb with cephaloridine acylated at its active site. a, the fo-fc density map of cephaloridine in the complex structure of PenP-E166Cb-cephaloridine is shown in mesh format in gray and contoured at 2.0 σ. The disordered BADAN molecule is illustrated in dashed lines. The yellow dotted line represents the poorly ordered segment of residues L169 to V174 on the Ω-loop. b, overlapping of PenP-E166Cb (lemon) and PC1 (coral) with cephaloridine. PenP-E166Cb and PC-1 are shown in green and red, respectively. Residues S70, N104, and cephaloridine are displayed in a cpk cylinder model. The blue dotted line represents the H-bond between side chains of N104 and acylamide of cephaloridine.
The Ω-loop in the PenP-E166Cb-cephaloridine complex retains a conformation largely similar to that seen in the PenP-E166Cb structure (Fig. 5b). Residue E168 is not displaced by the acylated cephaloridine at the active site because cephaloridine lacks the branched oxyimino group to extend deep into the PenP active site. However the quality of electron density for residues beyond Glu-168 on the Ω-loop was poor, rendering it impossible to build a reliable structural model. Furthermore there was no continuous electron density visible beyond the thiol group of the mutated residue Cys-166 to account for the BADAN fluorophore despite our extensive efforts in crystal optimization and structural refinement. Yet electron density for the rest of the PenP-E166Cb-cephaloridine structure is of excellent quality typical of a structure determined at high resolution. The fact that the poor electron density is localized to BADAN and the residues of the Ω-loop surrounding the fluorophore likely suggests that the BADAN fluorophore and the segment of peptide it attaches to become highly dynamic upon acylation of cephaloridine to the active site.
E168L Mutation Correlates the Structural Flexibility of the Ω-loop with the Substrate-specific Fluorescence Profile
To confirm our structural findings that the Ω-loop in the engineered PenP-E166Cb enzyme readily undergoes conformational changes involving displacement of residue Glu-168 and change of local polarity, we mutated Glu-168 to leucine and measured the fluorescence profile of PenP-E166Cb-E168L. We reasoned that by mutating the charged carboxylic side chain of Glu-168 to the hydrophobic side chain of leucine the local polarity for BADAN is likely to be reduced; furthermore the displacement of mutated L168 by the methoxyimino moiety of oxyimino-cephalosporins from the interior solvent-excluded position to the exterior solvent-exposed position should not affect the local polarity as much as Glu-168 did.
Indeed the fluorescence profile for PenP-E166Cb-E168L against the oxyimino-cephalosporins showed a trend of reduced enhancement (Fig. 6, a and b). For cefotaxime the PenP-E166Cb-E168L displayed an enhancement of ∼10% as compared with the ∼70% enhancement by PenP-E166Cb at concentrations between 2 × 10−8 m and 10−6 m. For non-cephalosporins with a non-branched R2 group, the fluorescence response from PenP-E166Cb-E168L was slightly improved as compared with PenP-E166Cb. For cephaloridine the enhancement for PenP-E166Cb-E168L mutant was ∼21% as compared with the ∼3% in PenP-E166Cb.
FIGURE 6.
The fluorescence profile of PenP-E166Cb with E168L mutation. a, fluorescence scanning spectra of PenP-E166Cb-E168L in the presence of the 6 listed cephalosporins at 0.1 mm concentration respectively. b, histogram presentation of the fluorescence profile of PenP-E166Cb-E168L (solid pattern) in comparison to that of PenP-E166Cb (hashed pattern). c, the modeled docking groove of BADAN molecule on the surface of PenP-E166Cb-E168L structure. BADAN and the residues lining up the groove are presented in stick model. The surface profile of the residues is presented as mesh model and colored according to their polarity (yellow: hydrophobic; red: acidic; pink: polar).
DISCUSSION
A Flexible Ω-loop Distinctively Affects the Binding of Oxyimino-cephalosporin
It has long been perceived that the well-ordered Ω-loop of various β-lactamases can become structurally flexible upon acquiring certain destabilizing mutations, and such enhanced flexibility affords the enzyme expanded substrate profile. Yet no experimental data that demonstrates a flexible Ω-loop and confirms its impact on enzyme kinetics and substrate profile are available. Our studies on PenP-E166Cb aim to provide such vital information.
We observed a distinct conformation of the Ω-loop in each of the three crystal structures, including the BADAN-labeled PenP β-lactamase, its acyl adduct complex with cefotaxime (oxyimino-cephalosporin), and another acyl adduct complex with cephaloridine (non-oxyimino-cephalosporin). In all three structures the Ω-loop has lost the stabilizing interactions that would pack the Ω-loop close to the active site to pose as steric hindrance as seen in wild type enzyme. As a result, the Ω-loop becomes highly flexible and readily adopts new conformations to accommodate the acylated cephalosporin substrates. Particularly the oxyimino-cephalosporins can now bind in a catalytically competent form to facilitate their hydrolysis. Furthermore kinetic studies reveal that PenP-E166Cb, with its structurally flexible Ω-loop, possesses improved binding kinetics toward extended-spectrum oxyimino-cephalosporins. Thus our data confirms that the Ω-loop is indeed capable of adopting highly flexible conformations and such dynamic properties correlate with extended substrate profile. The multiple conformations of the Ω-loop seen in the structures of PenP-E166Cb and its acyl adduct with cefotaxime and cephaloridine could serve as prototypes to investigate the various mutant β-lactamases with evolutionary substitutions in the Ω-loop region.
Substrate-specific Biosensing Process in PenP-E166Cb
Fluorescence-based biosensing has become a powerful and popular tool to probe complex and dynamic biological and chemical events at both in vitro and in vivo levels (19–21). Environment-sensitive fluorophores strategically placed near the functional sites of proteins or small molecules have been employed to report a wide range of biological interactions (12, 22–23). An intriguing question particularly related to β-lactam antibiotics is whether a fluorophore conjugated β-lactamase enzyme can give substrate-differentiated signals toward the wide-spectrum of β-lactam antibiotics. Such a design, if successful, would add a new dimension to fluorescence-based biosensing, where the engineered reporter cannot only detect the event of substrate binding, but also indicate the biochemical properties of the bound substrate.
Our engineered PenP-E166Cb can indeed yield differentiated fluorescence signals between oxyimino-cephalosporins and non-oxyimino-cephalosporins. The structural flexibility of the Ω-loop plays an essential role in this substrate-specific biosensing process. The Ω-loop adopts distinctively different conformations depending on the type of the substrate bound, inducing different environmental polarity characteristics near the BADAN fluorophore to influence its fluorescence emission. Our engineered β-lactamase thus provides a new possibility of designing substrate-selective biosensing tools to glean structural and functional information from biological systems that engage a large number of diverse ligands.
Supplemental Information
Experimental details for expression and purification of PenPE166C mutant, protein labeling and purification, fluorescence measurements, mass spectral, and circular dichroism data are available free of charge via the Internet (see supplemental data).
Supplementary Material
Acknowledgments
We thank M. J. Zhang (HKUST) for critically reviewing the manuscript. C. H. Cheng (HK PolyU) is acknowledged for technical assistance with the x-ray crystallography facility. We thank Shanghai Synchrotron Radiation Facility (SSRF) for access to the beam line.
This work was supported by the Research Grants Council (PolyU 5463/05M, PolyU 5380/04M, PolyU 5017/06P, PolyU 5641/08M, and PolyU 5639/09M), the Area of Excellence Fund of the University Grants Committee (AoE/P-10/01), and the Research Committee of the Hong Kong Polytechnic University.
The atomic coordinates and structure factors (codes 3KGM, 3KGN, and 3KGO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table ST1.
- ESBL
- extended spectrum β-lactamase
- BADAN
- 6-bromoacetyl-2-dimethylaminonaphthalene
- GdHCl
- guanidine HCl.
REFERENCES
- 1. Drawz S. M., Bonomo R. A. (2010) Clin. Microbiol. Rev. 23, 160–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fisher J. F., Mobashery S. (2009) Curr. Protein Pept. Sci. 10, 401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caselli E., Powers R. A., Blasczcak L. C., Wu C. Y., Prati F., Shoichet B. K. (2001) Chem. Biol. 8, 17–31 [DOI] [PubMed] [Google Scholar]
- 4. Chan P. H., So P. K., Ma D. L., Zhao Y., Lai T. S., Chung W. H., Chan K. C., Yiu K. F., Chan H. W., Siu F. M., Tsang C. W., Leung Y. C., Wong K. Y. (2008) J. Am. Chem. Soc. 130, 6351–6361 [DOI] [PubMed] [Google Scholar]
- 5. Chan P. H., Chan K. C., Liu H. B., Chung W. H., Leung Y. C., Wong K. Y. (2005) Anal. Chem. 77, 5268–5276 [DOI] [PubMed] [Google Scholar]
- 6. Chan P. H., Liu H. B., Chen Y. W., Chan K. C., Tsang C. W., Leung Y. C., Wong K. Y. (2004) J. Am. Chem. Soc. 126, 4074–4075 [DOI] [PubMed] [Google Scholar]
- 7. Loving G., Imperiali B. (2008) J. Am. Chem. Soc. 130, 13630–13638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loving G., Imperiali B. (2009) Bioconjug. Chem. 20, 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vázquez M. E., Blanco J. B., Imperiali B. (2005) J. Am. Chem. Soc. 127, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 10. Koehorst R. B., Spruijt R. B., Hemminga M. A. (2008) Biophys. J. 94, 3945–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koehorst R. B., Laptenok S., van Oort B., van Hoek A., Spruijt R. B., van Stokkum I. H., van Amerongen H., Hemminga M. A. (2010) Eur. Biophys. J. 39, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen B. E., Pralle A., Yao X., Swaminath G., Gandhi C. S., Jan Y. N., Kobilka B. K., Isacoff E. Y., Jan L. Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 965–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Venkatraman P., Nguyen T. T., Sainlos M., Bilsel O., Chitta S., Imperiali B., Stern L. J. (2007) Nat. Chem. Biol. 3, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strynadka N. C., Jensen S. E., Johns K., Blanchard H., Page M., Matagne A., Frère J. M., James M. N. (1994) Nature 368, 657–660 [DOI] [PubMed] [Google Scholar]
- 15. Shimamura T., Ibuka A., Fushinobu S., Wakagi T., Ishiguro M., Ishii Y., Matsuzawa H. (2002) J. Biol. Chem. 277, 46601–46608 [DOI] [PubMed] [Google Scholar]
- 16. Crichlow G. V., Nukaga M., Doppalapudi V. R., Buynak J. D., Knox J. R. (2001) Biochemistry 40, 6233–6239 [DOI] [PubMed] [Google Scholar]
- 17. Powers R. A., Caselli E., Focia P. J., Prati F., Shoichet B. K. (2001) Biochemistry 40, 9207–9214 [DOI] [PubMed] [Google Scholar]
- 18. Chen C. C., Herzberg O. (2001) Biochemistry 40, 2351–2358 [DOI] [PubMed] [Google Scholar]
- 19. Weiss S. (2000) Nat. Struct. Biol. 7, 724–729 [DOI] [PubMed] [Google Scholar]
- 20. Michalet X., Weiss S., Jäger M. (2006) Chem. Rev. 106, 1785–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weiss S. (1999) Science 283, 1676–1683 [DOI] [PubMed] [Google Scholar]
- 22. Cohen B. E., McAnaney T. B., Park E. S., Jan Y. N., Boxer S. G., Jan L. Y. (2002) Science 296, 1700–1703 [DOI] [PubMed] [Google Scholar]
- 23. de Lorimier R. M., Smith J. J., Dwyer M. A., Looger L. L., Sali K. M., Paavola C. D., Rizk S. S., Sadigov S., Conrad D. W., Loew L., Hellinga H. W. (2002) Protein Sci. 11, 2655–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.