Abstract
Background
Renal cell carcinoma is a common malignancy that often presents as a metastatic-disease for which there are no effective treatments. To gain insights into the mechanism of renal cell carcinogenesis, a number of genome-wide expression profiling studies have been performed. Surprisingly, there is very poor agreement among these studies as to which genes are differentially regulated. To better understand this lack of agreement we profiled renal cell tumor gene expression using genome-wide microarrays (45,000 probe sets) and compare our analysis to previous microarray studies.
Methods
We hybridized total RNA isolated from renal cell tumors and adjacent normal tissue to Affymetrix U133A and U133B arrays. We removed samples with technical defects and removed probesets that failed to exhibit sequence-specific hybridization in any of the samples. We detected differential gene expression in the resulting dataset with parametric methods and identified keywords that are overrepresented in the differentially expressed genes with the Fisher-exact test.
Results
We identify 1,234 genes that are more than three-fold changed in renal tumors by t-test, 800 of which have not been previously reported to be altered in renal cell tumors. Of the only 37 genes that have been identified as being differentially expressed in three or more of five previous microarray studies of renal tumor gene expression, our analysis finds 33 of these genes (89%). A key to the sensitivity and power of our analysis is filtering out defective samples and genes that are not reliably detected.
Conclusions
The widespread use of sample-wise voting schemes for detecting differential expression that do not control for false positives likely account for the poor overlap among previous studies. Among the many genes we identified using parametric methods that were not previously reported as being differentially expressed in renal cell tumors are several oncogenes and tumor suppressor genes that likely play important roles in renal cell carcinogenesis. This highlights the need for rigorous statistical approaches in microarray studies.
Background
Renal cancer will be diagnosed in 31,900 Americans in 2003 [1], making it the ninth most common malignancy. Clear-cell renal cancer (RCC) is the most frequent type of renal cancer, accounting for 80–85% of adult renal neoplasms. Moreover, many RCC patients (20–30%) present with metastatic disease. Despite substantial progress in the understanding of renal cancer biology, the most effective treatment for RCC remains surgical extirpation. Improved medical therapies are greatly needed for patients who present at an advanced stage, and development of rational approaches will be driven by insights into renal cancer biology.
The von Hippel-Lindau tumor suppressor VHL plays a key role in RCC. Biallelic VHL gene defects are found in 75% of sporadic renal cancers [2]. Moreover, loss of VHL or chromosome 3p is likely an initiating event in renal cancer pathogenesis. Comparative genome hybridization and mathematical modeling support this assertion [3]. In addition, renal tumorigenesis in VHL disease appears to be a multistep process, as development of VHL-deficient renal cysts precedes onset of renal cancers [4]. VHL mutations impair ubiquitination of the hypoxia-inducible HIF alpha transcription factors and thereby promote overexpression of HIF target genes, such as angiogenic factors [5]. Thus, early VHL loss initiates a molecular cascade that facilitates renal oncogenesis. Gene regulatory and genetic events subsequent to VHL loss must therefore be critical to renal cancer pathogenesis.
Renal cancers are highly resistant to chemotherapy and to radiotherapy. Although the multi-drug resistant protein transporter has been implicated in chemo-resistance, no mechanism has been proposed for irradiation resistance. Gene expression may provide insight into the refractory nature of these tumors. Other molecular pathways involving the initiation, progression, and metastasis of RCC await identification.
Genome-wide expression analysis using high-density nucleic acid microarrays is one approach to identifying key molecular events and pathways involved in renal cancer. Several microarray studies of RCC gene expression have already been published but our analysis indicated that these studies either did not use enough samples, complete enough arrays and/or appropriate analytic methods to identify the bulk of genes that are affected by renal cancer. We have analyzed gene expression in RCC tumors and adjacent normal tissue using Affymetrix U133 microarrays and have identified 1,234 that are significantly differentially expressed in RCC by three-fold or more and estimate that this list includes > 95% of all such genes. Among these differentially expressed genes are a number of genes involved in hypoxia, angiogenesis, apoptosis, and metastasis as well as a number of oncogenes and tumor suppressor genes that have not been reported previously as being differentially expressed in other microarray-based RCC expression profiling studies. We discuss possible explanations for the sensitivity of our parametric analysis.
Methods
Sample preparation and RNA isolation
IRB approval and informed consent were obtained for the procurement of tissue at the Cleveland Clinic Foundation. Patient samples were chosen to represent the grading spectrum (Fuhrman 1–3) of kidney clear cell carcinoma. Eighteen frozen tissue samples of RCC tissues and patient-matched normal kidney tissues from 9 patients were mechanically disrupted in TriZol reagent (Life technologies) using a PowerGen 35 tissue homogenizer (Fisher Scientific). Total RNA was isolated from each sample after chloroform extraction and isopropanal precipitation following the manufacturer's procedures (Life technologies).
RNA labeling and hybridization
Using a poly-dT primer incorporating a T7 promoter, double-stranded cDNA was synthesized from 10 μg total RNA using a Superscript cDNA Synthesis Kit (Invitrogen, Carlsbad, CA). Double-stranded cDNA was purified by phenol/chloroform extraction. The aqueous phase was isolated using Phase-Lock Gel Heavy (Brinkmann Instruments, Westbury, NY) and the cDNA ethanol precipitated. Subsequently, biotin-labeled cRNA was generated from the double-stranded cDNA template though in-vitro transcription with T7 polymerase using a BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). The biotinylated cRNA was purified using RNeasy affinity columns (Qiagen, Valencia, CA). Biotinylated cRNA (20 μg)was fragmented in 40 mM Tris-acetate, pH 8.1, 100 mM KOAc, 30 mM MgOAc, for 35 min. at 94°C, to an average size of 35 to 200 bases. Fragmented, biotinylated cRNA (10 μg), along with hybridization controls (Affymetrix, Santa Clara, CA), were hybridized to Affymetrix Human Genome U133A and U133B GeneChip arrays. The arrays were hybridized for 16 hrs. at 45°C and 60 rpm. Following hybridization, arrays were washed and stained according to the standard Antibody Amplification for Eukaryotic Targets protocol (Affymetrix, Santa Clara, CA). The stained GeneChip arrays were scanned at 488 nm using a G2500AGeneArray Scanner (Agilent, Palo Alto, CA) and Microarray Suite 5.0 software (Affymetrix, Santa Clara, CA).
Data Quantification and Normalization
Following data acquisition, the scanned images were quantified using Microarray Suite 5.0 (MAS 5.0) software (Affymetrix, Santa Clara, CA) yielding a signal intensity for each probe on the GeneChip. The signal intensities from the twenty-two probes for each gene were then used to determine an overall expression level and measure of sequence-specific hybridization according to algorithms implemented in MAS 5.0. The arrays were then linearly scaled to an average expression level of 500 units on each chip.
Data Analysis
Most analytic steps were performed using Excel (Microsoft, Redmond, WA) with a combination of built in functions and custom formulae except as follows. Principal Components Analysis was performed with DecisionSite (Spotfire, Sommerville, MA). Spearman Correlation Analysis was performed with an on-line calculator at the Institute for Phonetic Science at the University of Amsterdam [6]. Hierarchical clustering was performed with Cluster and visualized with Treeview [7]. The distribution of matched and unmatched tumor / adjacent-normal-tissue hybridization-intensity ratios was calculated with a PERL script we wrote for this purpose. The Fisher Exact Test was performed with an online calculator that is part of Daan Uitenbroek's Simple Interactive Statistical Analysis (SISA) package [8]. Power calculations were performed using a javascript we customized for this purpose based on the SISA javascript power calculator.
Results
To determine the genome-wide changes in gene expression that accompany clear-cell carcinogenesis, we performed expression analysis using the Affymetrix U133A and U133B GeneChips. Together, these arrays contain approximately 45,000 probesets designed to detect the expression of approximately 39,000 different transcripts derived from 33,000 well-substantiated genes. Total RNA was isolated from clear-cell carcinoma tissue removed from nine patients during radial nephrectomy as well as adjacent normal renal tissue present in the same surgical samples. We refer to the patients by an anonymous ID, and to the carcinoma tissue obtained from each patient by prefixing the patient ID with a "C" and prefixing the patient ID with a "N" when referring to their normal tissue. For three patients, the RNA from either the RCC or adjacent-normal tissue was not of sufficient quality for hybridization (Table 1).
Table 1.
Clinical data on samples for gene-expression analysis
| Sample Code | age | gender | kidney | grade | capsule penetration | sinus invasion | tumor RNA | normal RNA |
| 1 | 51 | M | left | III | - | + | - | + |
| 2 | 67 | M | right | II | - | + | + | + |
| 3 | 50 | M | left | II | - | - | + | + |
| 4 | 65 | M | left | I | + | - | + | + |
| 001 | 58 | M | left | I | + | - | + | + |
| 005 | 64 | M | right | I | - | - | + | + |
| 011 | 55 | M | left | III | + | - | + | - |
| 023 | 72 | F | right | I | + | - | + | + |
| 032 | 65 | F | right | III | + | + | + | -* |
| 035 | 70 | F | right | III | + | - | + | + |
* The RNA from patient 032's normal kidney tissue is described in detail in the text.
Sample filtering
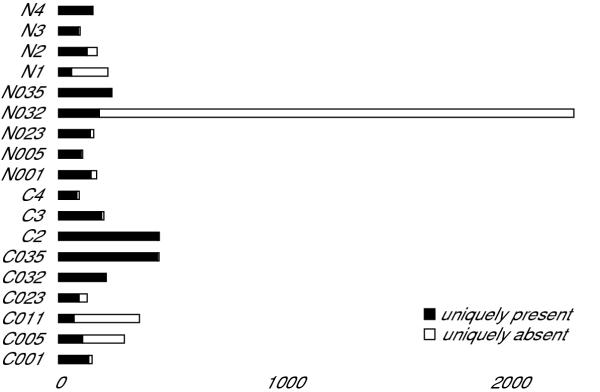
To increase the power of our analysis of differential gene expression, we first performed several tests to identify any defective samples in our dataset while retaining those with interesting biological variability. We used the ratio of sequence-specific to non-specific hybridization summarized in the "absent/present calls" generated by MicroarraySuite 5.0 for each probeset to determine the fraction of probesets in each sample with insignificant sequence-specific hybridization in that sample but significant sequence-specific hybridization for the same probeset in each of the remaining samples (fraction of uniquely absent probesets). For comparison, we also determined the fraction of uniquely present probesets for each sample – the fraction of probesets where significant sequence-specific hybridization is only detected in that one sample. There were 2,939 probesets where sequence-specific hybridization is detected in all but one of the samples and 2,980 where sequence-specific hybridization is detected in only one sample. Figure 1 shows how many of these uniquely absent and uniquely present probesets are contributed by each sample. While the distribution of uniquely present probesets per sample has a roughly normal distribution, the distribution of uniquely absent probesets per sample does not. N032 contributes 2,104 or 71.6% of the uniquely absent calls and is an outlier by the Grubb's Test (mean = 163.28, SD = 491.11, Z = 3.95, p < 0.01). We also found that the scaling factor required to bring the mean intensity of sample N032 hybridized to the U133A array to 500 (the arbitrary mean target value to which we normalized all the arrays) was an extremely high outlier (N032 U133A scaling factor = 21.15, mean = 7.06, SD = 4.05, Z = 3.47, p < 0.01) while the scaling factor of sample N032 hybridized to the U133B, while the highest U133B scaling factor, was not an outlier (N032 U133B scaling factor = 41.47, mean = 20.98, SD = 8.10, Z = 2.53, p > 0.01). Since these data argue that differences in sample N032 are unlikely to result from a biological difference between N032 and the other samples, we have concluded that N032 has a technical defect and have excluded the data obtained from this sample from further analysis. The gene expression data generated by this study is freely available from the NCBI Gene Expression Omnibus and has been submitted under accession GSE781 [9].
Figure 1.
Identification of failed samples. Histogram of uniquely absent (uniquely poor sequence-specific hybridization, open bars) and uniquely present (uniquely significant sequence-specific hybridization, closed bars) contributed by each sample
Gene filtering
Since not every gene that is interrogated by probesets on the U133A and U133B arrays is likely to be expressed in normal or diseased kidney tissue, we sought to eliminate the hybridization intensity data from probesets that detect genes that are not expressed in our samples. We retained all probeset hybridization intensities where significant sequence-specific hybridization is detected in at least one of the samples (as measured by being called present). This cutoff likely retains a large number probesets that are in fact not reliably detected but also allows our dataset to capture the greatest amount of variability in gene expression between samples. Removing probesets where there is no significant sequence-specific hybridization in any of the samples reduces the number of probesets in our dataset from 44,792 to 27,609. We next annotated the probesets with information about the transcripts that they detect using information obtained from the Affymetrix NETAFFX database (version March 1, 2003) [10]. We were able to map all but 492 of these probesets to a unigene cluster using this database. The remaining 492 probesets are designed to detect the expression of EST's that are not assigned to unigene clusters.
Sample relatedness
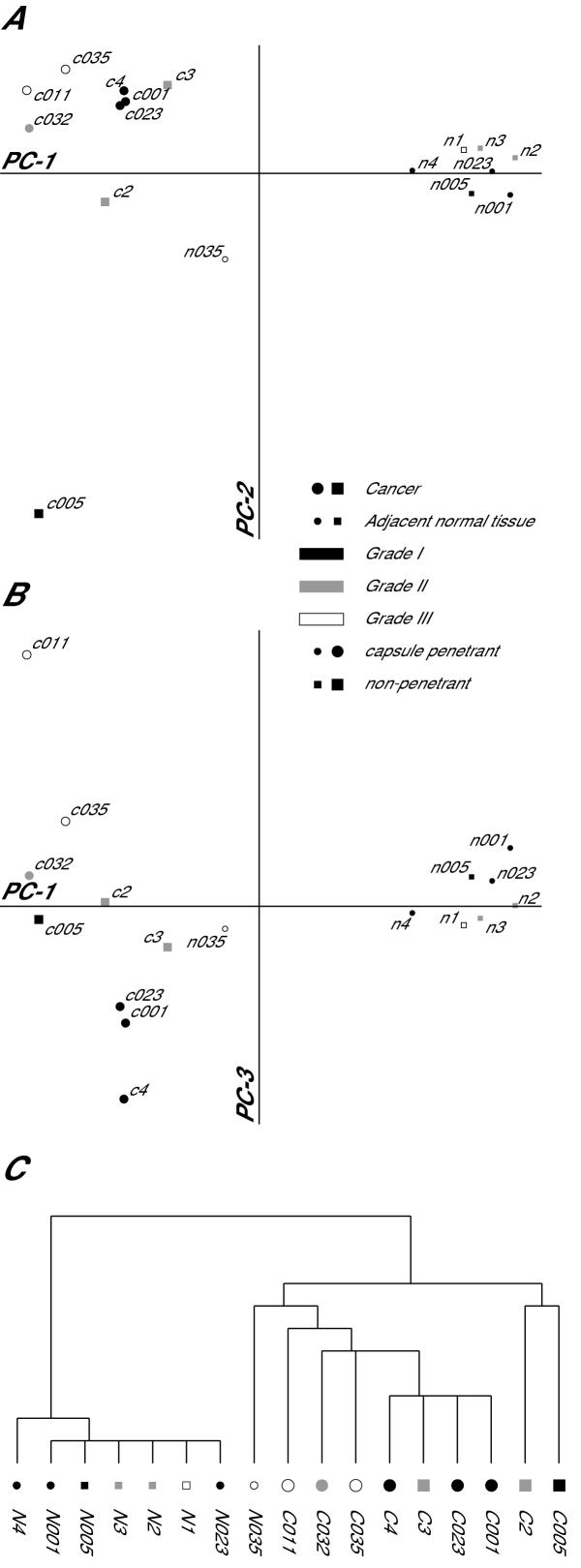
With this filtered dataset we sought to test the hypothesis that clear-cell carcinoma tumors would have significant differences in gene expression when compared with neighboring normal renal tissue. To test this hypothesis we performed a principal-components analysis to identify the primary axes upon which the samples vary and how the samples are distributed along these axes. As expected, the most significant axis of variation in gene expression among the samples accounts for 91.5% of the variation and predominantly distinguishes gene expression in tumors from gene expression in adjacent normal tissue (Figure 2A). While all of the cancer samples cluster at one end of the major axis of variation (PC-1) and most of the normal tissues cluster at the other end, one normal tissue sample (N035) has a pattern of gene expression that is intermediate between tumor and normal. We could not identify any unique clinical parameter in patient 035 which would account for the abnormal pattern of gene expression in the tissue adjacent to her tumor. One possibility that we cannot exclude is that this normal tissue sample contains a mixture of normal and cancerous tissue. The second major axis of variation accounts for 4.1% of the total variation in gene expression and also highlights a heterogeneity in gene expression for which we cannot provide a satisfactory biological explanation: the bulk of the cancer samples cluster at one end of the PC-2 axis while the tumor from patient 005 is alone at the other end of this axis. The third major axis of variation (PC-3, Figure 2B) accounts for 1.5% of the total variation in gene expression and separates the tumor samples roughly according to the Fuhrman grade of the tumor: Fuhrman grade I tumors are at one end of the axis, grade III tumors are at the other, and grade II tumors are largely in between grade I and grade III. The ordered relationship between tumor grade and position along the PC-3 axis is highly significant (Spearman rank-order correlation coefficient rs = 0.88, p = 0.003). Interestingly the three grades of tumors do not fall into obvious tight clusters, but rather spread out in a continuum on the PC-3 axis. This suggests that the patterns of gene expression that underlie the morphologic differences between the different tumor grades may be more continuous than discrete. We compared the ability of principal-components analysis to identify similarities in the patterns of gene expression between samples with hierarchical clustering (Figure 2C) and found that the principal-components analysis was more informative. The strengths of PCA for this analysis are that it does not require the samples to be organized into a binary tree structure and allows for minor sources of variation in gene expression to be visualized independently of major sources.
Figure 2.
Variation in gene expression as a function of sample-type. A & B. Samples plotted as a function of their loading for the primary axes of variation in gene expression as identified by principal components analysis. A. Samples plotted on the primary (PC-1) and secondary (PC-2) axes of variation. PC-1 accounts for 91.5% of the variation in gene expression and PC-2 accounts for 4.1% of the variation. The primary axis of variation in gene expression organizes the samples according to whether they are from tumor or adjacent normal tissue. B. Samples plotted on the primary and tertiary (PC-3) aces of variation. PC-3 accounts for 1.5% of the variation in gene expression. The tertiary axis of variation in gene expression organizes the tumor samples according to their Fuhrman grade. C. Hierarchical clustering of the samples in which the height of the vertical lines are proportional to the degree of dissimilarity between nodes. In all three panels the samples are labeled with the sample ID and the symbols are coded to represent sample type (large symbols represent tumor samples, small symbols represent normal tissue adjacent to the tumor), the Fuhrman grade of the tumor (filled symbols represent grade I, shaded symbols represent grade II, open symbols represent grade III), and whether the tumor had penetrated the renal capsule (round symbols represent penetrant tumors).
Analysis of differential gene expression
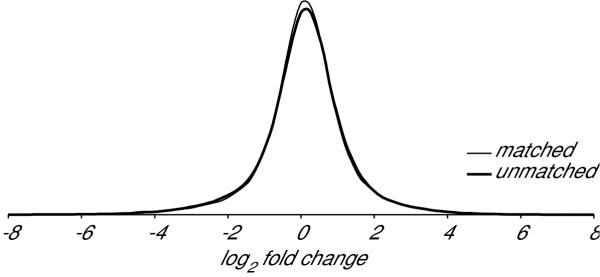
We next sought to determine if variation in gene expression between patients that is independent of disease state would affect the dispersal of gene-expression ratios. If renal gene expression varies strongly among individuals we would need to perform a paired t-test and discard any samples for which we did not have matched tumor and normal samples from the same patient. However if renal gene expression is relatively constant among individuals, we could perform an unpaired test and include all of the samples for which we had either normal or tumor hybridization intensities. To determine if renal gene expression is relatively constant between individuals we first determined the distribution of the hybridization intensity ratios for every probeset, comparing each patient's tumor with their adjacent normal tissue. We next determined the distribution of ratios when we compared each patient's tumor with the normal tissue sample from each of the other patients. The distribution of these ratios is presented in Figure 3 which indicates that a patient's tumor has a pattern of gene expression slightly more similar to their own adjacent normal tissue than to the normal tissue of other individuals. These patient-specific differences in renal gene expression could be the result of environmental or genetic factors.
Figure 3.
Patient-specific variation in renal gene expression. Hybridization intensities are slightly more similar between a tumor and the tissue adjacent to that tumor than they are to tissue adjacent to the tumors of other patients. The frequency (y-axis) of varying degrees of differential expression (x-axis) when comparing the hybridization intensity for every tumor-sample probeset with the corresponding measurement in adjacent tissue is shown with the thin line. The frequency of varying degrees of differential expression observed when comparing the same tumor hybridization intensities with the corresponding measurements from tissues adjacent to the tumors of other patients is shown with the thick line.
We next determined if these inter-patient differences in gene expression were strong enough to make a paired-sample t-test more powerful than an unpaired t-test for detecting gene expression differences between tumor and normal renal tissue. The paired-sample design would only use samples from seven patients and would result in an error measurement with six-degrees of freedom. An unpaired design would use nine tumor samples and eight normal tissue samples and would result in an error measurement with sixteen-degrees of freedom. As a result of the reduced degrees of freedom in the paired-sample design, we would therefore only expect the paired-sample design to be more powerful at detecting differential expression if the inter-patient variability in gene expression were stronger than the variability between tumor and normal renal tissue. To determine if this is the case, we compared the results of both types of analysis.
We first identified probesets that exhibit differential hybridization intensity in clear-cell carcinoma using a two-tailed Student's t-test in which we assigned the tumor samples to one group and the normal kidney tissue to a second group. We performed t-tests sequentially on each probeset in our data set and established a dual threshold of maximum p-value and minimum fold-change to identify the subset of probesets that we consider to have significantly and meaningfully changed expression levels between the clear-cell carcinoma tissue and normal renal tissue. We chose to focus on those probesets with a t-test p-value < 0.03. We chose this threshold because if there is no difference between the two types of tissue, a false-positive threshold of p < 0.03 would on average identify 828 changed genes in our dataset of 27,609 probesets while we observed 7,685 probesets that showed differential expression at this false-positive threshold: giving us an estimated false-discovery rate of 11% [11]. We chose to focus on those probesets amongst these 7,685 that are also either induced or repressed greater than 3-fold as determined by dividing the geometric mean of the hybridization intensities obtained from the tumor samples by the geometric mean of the adjacent normal tissue intensities. We chose a three-fold-changed threshold because we felt that the differential expression of less-changed genes would be more difficult to interpret. We estimate our false-negative rate at the chip-wide mean hybridization intensity to be less than five percent with these false-positive and fold-change thresholds. Combining the 3-fold change threshold with the p < 0.03 threshold leaves 1,706 probesets (approximately 6.2% of the probesets in our filtered dataset) that are differentially expressed in clear-cell renal-cell carcinoma.
We next compared the results of this unpaired analysis to an analysis of the seven patient-matched tumor/normal pairs using a paired-sample t-test. With this analysis there are 12 probesets that meet the dual 3-fold-change and p < 0.03 threshold that are not identified as being differentially expressed in the unpaired analysis. In contrast, there are 622 probesets that we identify as being differentially expressed in the unpaired analysis that are not identified in the paired-sample analysis. These results suggest that while there may be some genes that vary strongly both between patients and between tumor and normal renal tissue, the majority of genes that vary strongly between tumor and normal renal tissue show relatively little inter-patient variability and that as a result the unpaired t-test is more powerful for detecting these differences. The 12 probesets identified as being > 3-fold differentially expressed only by the paired-sample t-test are listed in Additional file 1.
As there are multiple probesets for some of the genes interrogated by the Affymetrix U133 arrays, we next sought to reduce our dataset from the probeset level to the gene level. We found that the 27,609 probesets for which we had detected sequence-specific hybridization correspond to 19,700 unique unigene clusters and 492 probesets for which no unigene information is currently available. The 1,706 probesets that we identified as being differentially expressed in the tumor tissue samples correspond to 1,448 unigene clusters and 23 probesets for which no unigene information is available. 353 of the 1,448 unigene clusters for which we had identified a differentially expressed probeset have at least one additional probeset that did not meet our criteria for differential expression.
There are several possible explanations for heterogeneous differential expression among multiple probesets that map to a single unigene cluster including alternative splicing, false positives, incorrect unigene cluster assignment and probeset-specific differences in hybridization variability. Upon finding little evidence for alternative splicing among several unigene clusters with multiple probesets that we picked at random, and with our primary goal being the identification of differentially expressed genes, we decided to focus on the other possible causes of probeset heterogeneity. While the most conservative approach for minimizing false positives would be to exclude any unigene cluster for which at least one probeset failed to provide evidence of differential expression, we felt that this approach is overly conservative. As an alternative, for all the unigene clusters that are interrogated by multiple probesets we calculated the geometric means of the p-value and the fold-change of the individual probesets to arrive at a summary value. The average values of 237 of the 353 heterogeneous probeset clusters were now below the critical thresholds we had established for differential expression and were eliminated. This results in 1,211 unigene clusters and 23 unannotated probesets that show differential expression in the renal-cell carcinoma samples. A complete list of the 1,706 differentially expressed probesets can be found in Additional file 2. This table includes columns which indicate if multiple probesets map to the same unigene cluster, whether one or more of these additional probesets failed to meet our criteria for differential expression, and whether the corresponding unigene cluster was retained on our list of differentially expressed unigene clusters. A complete list of the 929 probesets that map to a unigene cluster where at least one probeset provides evidence for differential gene expression and at least one probeset does not show evidence of differential expression can be found in Additional file 3.
Comparison with other studies of RCC gene expression
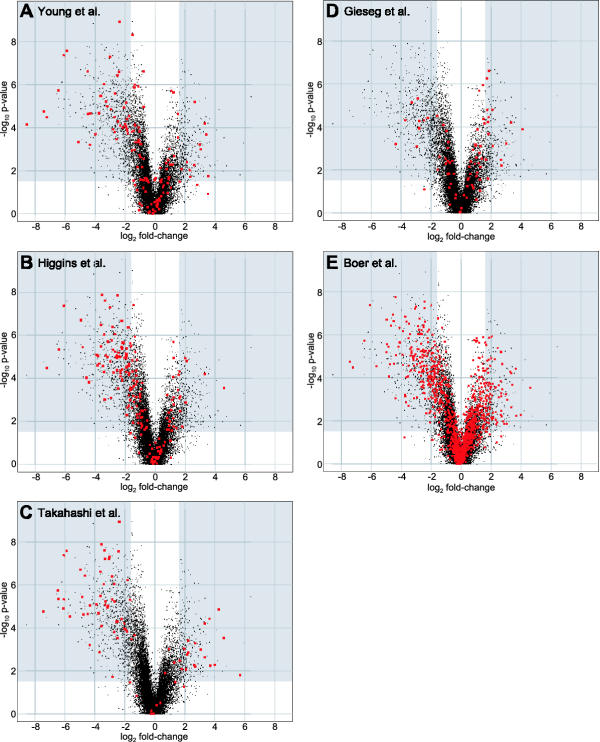
We are aware of seven previous studies that use large-scale expression analysis to identify differences in gene expression between renal tumors and normal kidney tissue. Young et al. hybridized RNA from seven tumors and seven patient-matched normal kidney tissue samples to a 7,075 element cDNA microarray [12]. Their panel of tumors contained four clear-cell carcinomas (two Fuhrman grade II, one grade III and one grade IV), two onocytomas, and one chromophobe carcinoma. They identified 189 genes that showed noteworthy differential expression using a criterion of being either induced or repressed more than two-fold in two or more of the tumor samples. We were able to map 137 of these 189 genes to unigene clusters represented on the U133A and B arrays that had significant gene-specific hybridization in at least one of our samples. We did this by translating the GenBank accession number reported by Young et al into the corresponding unigene ID and then searching our dataset annotation for this ID. We observed differential expression for 50 of these 137 genes (Figure 4A) or 36.5%.
Figure 4.
Our observations of genes identified as being differentially expressed in other studies of RCC gene expression. The 20,000 genes for which we measured significant sequence-specific hybridization are plotted in each scatter plot as a function of the fold change between the tumor and adjacent normal tissue (log2, on the x-axis) and the statistical significance of the change (-log10, on the y-axis). In each panel, our observations of genes identified as being differentially expressed in the indicated study are highlighted in red.
Higgins et al hybridized RNA from 41 tumor and 3 normal tissue samples to a 22,648 element cDNA microarray using a common reference RNA for normalization [13]. 23 of the tumor samples were from clear cell renal carcinomas. We downloaded the Higgins et al dataset and removed data points where the pixel to pixel correlation coefficient between the experimental RNA and the reference RNA channel is less than 0.6. We next excluded those genes that had fewer than two data points in either the clear cell carcinoma group or the normal kidney group. Due to the small number of adjacent normal tissue samples in the Higgins et al dataset, this resulted in a dataset with measurements for only 7,943 genes – of which we were able to map 6,285 to one or more genes in our dataset using the unigene cluster annotation. We performed t-tests on the filtered Higgins et al. dataset to compare the expression of these genes in clear-cell carcinoma and normal tissue and found 1,177 cDNA's that showed evidence of differential expression (p < 0.03). We used the same p-value threshold as the one we used in analyzing our dataset despite the fact that this gives a false-discovery rate of approximately 20% (as compared with the 11% false-discovery rate in our dataset). Of these 1,177 cDNA's, 217 were also induced or repressed by more than 3-fold. 182 of these changed cDNA's were among those that we were able to map to our dataset. We observed differential expression for 86 of these 182 genes (Figure 4B) or 47.3%.
Takahashi et al hybridized RNA from 29 clear-cell carcinomas to a 21,632 element cDNA microarray [14]. They identified 77 genes that are down-regulated by three-fold or more in 75% or more of the tumor samples compared to adjacent normal tissue and 32 genes that were up-regulated by three-fold or more by similar criteria. We were able to map 82 of these 109 genes to our dataset by mapping the Genbank accession numbers they reported to Unigene identifiers and/or gene symbols. We observe differential expression for 68 of these 82 genes (Figure 4C) or 82.9%.
Gieseg et al hybridized RNA from 13 renal cell carcinoma samples (9 clear cell, 2 chromaphobe, 1 urothelial carcinoma, and 1 adenoma) and 9 adjacent normal tissue samples [15] to the Affymetrix HuGeneFL array that contains probes for approximately 5600 genes. They identified 456 genes that were changed greater than 2-fold and called as significantly changed by the Affymetrix Data Mining Tool software package. Of these genes, 235 were changed only in the chromaphobe samples while 221 were changed in the chromaphobe and clear-cell or the clear-cell samples alone. The authors report the gene symbols for 85 of these 221 genes and we were able to map 79 of these 85 to genes in our dataset. We observed differential expression for 25 of these 79 genes (Figure 4D) or 31.6%.
Boer et al. hybridized RNA from clear-cell renal-cell carcinoma tissue samples and adjacent normal tissue from 37 patients to a 31,500 element cDNA microarray [16]. They identified 1,581 cDNA's that are differentially expressed in the tumor tissue and we were able to map the IMAGE clone ID these cDNAs to 1,139 genes in our dataset via either the Unigene cluster ID or gene symbol. We were able to translate each IMAGE Clone ID to Unigene ID's and/or gene symbols using the SOURCE program from Stanford [17]. We observed differential expression for 282 of these 1,139 genes (Figure 4E) or 24.8%
We were unable to make a useful comparison between our results and the remaining two RCC microarray studies. The study by Moch et al. reports the observation of 89 genes that are differentially expressed in the tumor cell line CRL-1933 compared with normal kidney tissue [18]. They identified these genes by hybridizing radioactively labeled RNA to the 5,184 element Research Genetics Human Gene Filters, Release 1 array. However they only report the overexpression of vimentin specifically. We see vimentin significantly upregulated in our dataset (2.3 fold, p < 10-4) but have not included it on our list of changed genes because the gene is less than three-fold induced. Skubitz and Skubitz hybridized RNA obtained from 8 renal cell carcinoma, 11 normal kidney, and 8 diseased non-malignant kidney tissue samples to the Affymetrix U95 array set [19]. They report the gene titles for 69 genes that are induced 4.9-fold or more and 29 genes that are repressed 10-fold or more in the RCC samples compared with normal kidney. However since gene title descriptors are not standardized, we were unable to map these gene descriptors to genes in our dataset. As a result, we have not determined how many of the genes that they report are also significantly changed in our dataset.
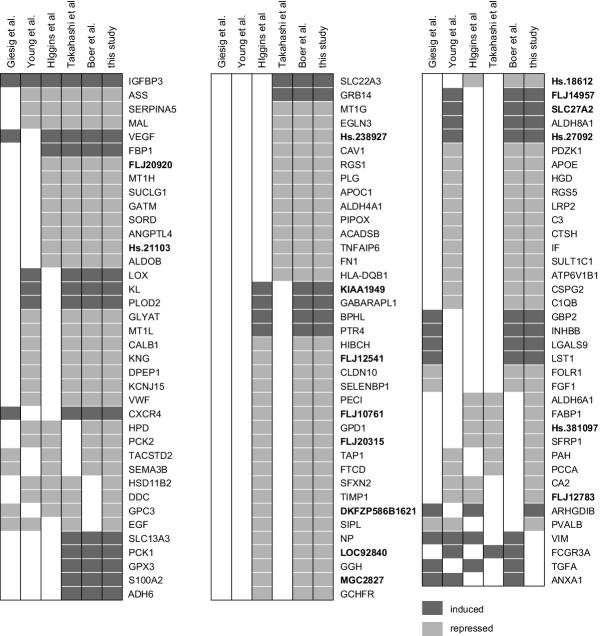
Having performed these basic comparisons between our dataset and other RCC-expression profiling experiments, we next sought to identify genes that have been identified as differentially regulated in multiple studies. Given the different microarray platforms, analytic techniques, and reporting strategies employed in the different studies negative results from this comparison should be interpreted cautiously (the failure of a gene to be reported as being differentially expressed in multiple studies does not indicate that the gene is not in fact differentially expressed in RCC), but agreement across multiple papers may be significant – especially given the differences between the studies. We chose to focus on those genes that have been identified as having noteworthy differential gene expression in three or more studies. We identified 113 genes for which three or more studies (including this study) identified a noteworthy change in RCC gene expression (Figure 5). In Figure 5 we report the gene symbols used in the unigene database as reported through NETAFFX.
Figure 5.
Genes identified as being differentially expressed in three or more studies of RCC gene expression. Each column represents a different microarray study of RCC gene expression. In each column, filled boxes indicate that the gene was reported as being differentially expressed in RCC except in the case of the Higgins et al. column where filled boxes indicate that the gene is differentially expressed based on our re-analysis of their data. Dark grey boxes indicate that a gene was reported as induced in RCC compared to normal kidney tissue while light grey boxes indicate that a gene is repressed. The gene identifier reported is either the gene symbols associated with a sequence in the unigene database as reported by NETAFFX or the unigene cluster ID. The gene identifiers in bold type refer to currently unannotated sequences.
Only four of the genes reported to be differentially expressed in three or more microarray studies of renal carcinoma gene expression are not on our list of differentially expressed genes. In all four cases we detected a significant change in the expression of these genes but the magnitude of the observed change was less than our three-fold change threshold (Table 2). Takahashi et al. use a three-fold change threshold similar to ours and also do not report Vimentin, TGF-alpha and Annexin A1 among their list of noteworthy changed genes. However, Takahashi et al. do report the differential expression of FCGR3A suggesting that the small fold-change we observe for this gene could be erroneous despite observing similar differential expression across two probesets for this gene.
Table 2.
Genes previously identified as differentially expressed in RCC not identified in our study.
| Gene | -log10 p-value | log2 fold-change |
| VIM | 5.66 | 1.22 |
| FCGR3A | 3.04 | 0.94 |
| TGFA | 2.78 | 1.23 |
| ANXA1 | 2.30 | 1.17 |
Keyword analysis of differentially expressed genes
We looked for classes of genes that are over represented in our list of differentially expressed genes relative to the complete list of genes we assayed. To do this, we picked keywords that are associated with processes known to be important in renal cell carcinogenesis in particular as well as additional keywords that describe processes involved generally in carcinogenesis. We searched the BioKnowledge Library (Incyte Genomics, Palo Alto, CA) [20] for genes that had these keywords anywhere in their description to come up with a list of gene symbols associated with each keyword. We next used these lists of keyword-associated genes to query the entire list of genes for which we had detected sequence-specific gene expression and tabulated the number of genes we were able to match as well as those matching genes that we had identified as being either differentially induced or repressed. To determine if the genes associated with each of the thirteen keywords are over or under represented among the differentially expressed genes, we used a two-tailed Fisher exact test to determine whether the fraction of induced or repressed keyword-associated genes is significantly different from the overall fraction of genes that are either induced or repressed. Since this process involves testing twenty-six hypotheses, we adjusted the threshold p-value for evidence of significant over- or under-representation to 1.92 × 10-3 to maintain an approximate overall 5% probability of incorrectly identifying any of the keywords as being over- or under-represented.
We did not find evidence of significant under-representation of genes associated with any of the thirteen keywords in our lists of differentially expressed genes. There is however strong evidence for over-representation of genes associated with the keywords hypoxia, angiogenesis, tumor-necrosis factor, apoptosis, interferon, drug resistance and metastasis among the genes that are up regulated in renal cell tumors as well as strong evidence for over-representation of genes associated with the keyword kidney among the genes that are down regulated in the tumors (Table 3). 190 genes, (about 15%) of the significantly differentially regulated genes, are associated with one or more of these statistically significant keywords (Additional file 4.)
Table 3.
Keywords associated with RCC-differentially-expressed genes
| keyword | # of genes | % up | % down | p up | p down |
| all genes | 20,199 | 1.8 | 4.3 | ||
| hypoxia | 42 | 28.6 | 2.4 | < 10-4 | 1.0000 |
| angiogenesis | 115 | 10.4 | 7.8 | < 10-4 | 0.1044 |
| tnf | 140 | 8.6 | 1.4 | < 10-4 | 0.1329 |
| apoptosis | 562 | 5.2 | 3.4 | < 10-4 | 0.3936 |
| interferon | 90 | 10.0 | 0.0 | 0.0001 | 0.0539 |
| resistance | 40 | 12.5 | 2.5 | 0.0013 | 1.0000 |
| metastasis | 142 | 6.3 | 1.4 | 0.0019 | 0.1340 |
| kidney | 403 | 4.0 | 15.1 | 0.0055 | < 10-4 |
| nfκb | 34 | 11.8 | 0.0 | 0.0049 | 0.4024 |
| tumor suppressor | 138 | 3.6 | 5.8 | 0.1916 | 0.3971 |
| chromatin | 93 | 1.1 | 1.1 | 1.0000 | 0.1886 |
| oncogene | 110 | 0.9 | 2.7 | 0.7261 | 0.6324 |
| telomerase | 27 | 0.0 | 0.0 | 1.0000 | 0.6269 |
Significant associations are bolded.
Discussion
We have identified 1,233 genes that are significantly differentially expressed by 3-fold or more in clear cell tumors relative to adjacent normal tissue isolated from the same surgical samples. Our ability to identify so many differentially expressed genes by measuring mRNA hybridization intensities from just ten surgical samples was strongly effected by our data analysis strategy.
Perhaps most importantly, we found that an unpaired t-test strategy was more powerful than a paired-sample t-test for identifying differentially expressed genes. This suggests that the variability between patients might be less than our hybridization-intensity measurement error. We propose that in two-channel hybridization experiments (like many of the previous renal cell tumor expression profiling experiments) it may also be beneficial to average hybridization intensities across many samples rather than average the ratio of hybridization intensities – despite the added inter-array normalization requirements – to arrive at more accurate estimates of the difference in expression between groups.
Another key to the sensitivity of our analysis was removing samples that showed subtle technical defects. We did this by looking for samples with an unusually large number of intensity measurements in which the ratio of sequence-specific to non-specific hybridization is low, and identified one sample, N032, that was a clear outlier in this regard. Had we included this sample in our analysis we would have only identified about 6,800 probesets that display evidence of significant differential expression (p < 0.03) compared with the nearly 7,700 probesets we identified when not including this sample. In light of the dramatic effect of excluding this sample, we also examined the consequences of having included other samples that might be atypical for technical or non-technical reasons.
One such sample is the adjacent normal tissue from patient 035. This sample shows a pattern of gene expression that by principal-components analysis appears to be a mixture of the patterns seen in the normal tissue and clear cell tumor samples. Since we have no compelling reason to think that the pattern of gene expression we observe in sample N035 results from something other than normal biological variability, we have retained this sample in our analysis. Had we excluded N035, the number of probesets with evidence of significant differential expression would have increased to almost 8,100.
The other atypical sample that we have retained in our analysis is the tumor sample from patient 005. This sample has a pattern of gene expression that is the largest source of variation among the patterns of gene expression seen in the tumor samples as measured by principal components analysis. Excluding this sample would have increased the number of differentially expressed probesets by 89. Taken together, the effect of removing N035 and C005 suggests that our estimate of being able to identify greater than 95% of the genes with average signal intensity that are changed three-fold or more needs to interpreted somewhat cautiously as our data is unlikely to be uniformly normal and our power to detect differential expression might therefore be lower.
We validated our approach of using the number of uniquely poor sequence-specific probeset hybridizations to eliminate sample N032 with a more generalizable three-step approach of first identifying probesets that contain an intensity measurement that significantly deviates from the mean; next counting how many of these outlier values are contributed by each sample; and then determining if the fraction contributed by any one of the samples is significantly different from the other samples. This analysis also identifies sample N032 as a significant outlier (data not shown). But the probability of N032 being an outlier is lower with this analysis than with our analysis of hybridization sequence-specificity and is more complex. The scaling factor N032 hybridized to the U133A array is also exceptionally large suggesting that the defect we detected in our "uniquely absent" analysis might also be reflected in the scaling factor.
Another aspect of our data analysis strategy involved minimizing the number of false positives – genes that appear to be differentially expressed in our particular samples but are in fact expressed similarly in tumor and normal tissue. One approach we have taken to reduce the absolute number of false positives has been to reduce the number of hypotheses of differential expression we have tested. To do this, we chose not to test the hypothesis that any individual probeset is differentially expressed in the absence of strong evidence of sequence-specific hybridization to this probset in at least one of the tumor or normal samples. By including only those probesets where there is significant sequence-specific hybridization in one or more sample, we reduced the number of probesets we tested for differential expression from about 45,000 to just over 27,600. Our rationale for requiring significant sequence-specific hybridization is that it would be otherwise impossible to interpret the meaning of a differentially expressed probeset if that differential expression couldn't then be associated with a particular sequence and thereby a particular transcript. We set our threshold for evidence of significant differential expression such that the number of probesets that we would expect to exceed this threshold by chance alone is about 11% of the total number of probesets that we observed to exceed this threshold. It is quite likely that after applying our secondary threshold – of only considering those probesets that are differentially expressed by three-fold or more – that the fraction of false-positives is less than 11% but this cannot be determined directly.
By further characterization of the multiple probesets that interrogate some of the genes represented on the Affymetrix arrays, it may be possible to further reduce the number of hypotheses tested by testing for evidence of differential gene-level rather than probset-level hybridization. We found that the 27,600 probesets we analyzed for differential expression only interrogate 20,000 distinct unigene clusters. Of the 12,500 probesets that map to unigene clusters that are interrogated by multiple probesets, 5.8% are significantly differentially expressed by more than 3-fold, compared to 6.6% of the 14,600 probesets that map to a unique unigene cluster (chi-square test, p = 5 × 10-4). This suggests either that a subset of the multiple probesets that interrogate a particular gene have a reduced ability to detect differential expression or that it is more difficult to reliably detect the expression of those genes that are interrogated by multiple probesets. With a large enough collection of array experiments it might be possible to extend the probeset filtering protocol to determine if any of the multiple probesets that detect a single gene have a reduced ability to detect differential expression using one portion of the dataset, and eliminate these probeset measurements from the remaining data which would be used for the analysis of differential expression. Another strategy would be to filter out the probesets using the same data for filtering and analysis in a post-hoc scheme, but this type of approach could easily result in over-filtering. We took a conservative approach to the issue of multiple probesets by averaging both the significance and the fold-change across the probesets and this led us to discard about 16% of the three-fold changed genes that we would have retained using the post-hoc filtering strategy. An important issue associated with all three of these approaches for handling multiple probesets for any individual gene is that if heterogeneous evidence for differential expression across multiple probesets is the result of alternative splicing or incorrect unigene cluster assignment, any conclusion about the expression of that gene as a single entity will be invalid. Thus, while the most conservative approach would be to eliminate genes for which there is heterogeneous evidence of differential expression across multiple probesets, we chose the slightly less conservative approach of averaging the probesets since many of the probsets for a single unigene cluster had similar values.
Given the differences in gene-expression measurement platforms, data-analysis strategies, and reporting techniques, we were pleased to find that over a hundred of the twelve-hundred genes that we identified as being differentially expressed by three-fold or more in renal cell carcinoma had also been identified in two or more of the five previous studies of RCC gene expression with which we compared our results. We failed to identify only four genes that had been identified in three or more previous studies. For all four genes, we found that each had statistically significant evidence of a change in expression but that our requirement for a three-fold or larger change prevented us from scoring these genes as being differentially expressed. The fact that we identified so many of the genes that had been identified in multiple other studies suggests that our list contains an extensive catalog of three-fold differentially expressed genes. Of the twelve-hundred genes that we identify as being differentially expressed in RCC, 870 have not been previously reported.
Several factors related to the reporting of the data may have reduced our ability to identify additional shared conclusions about genes that are differentially expressed between the studies. First, Gieseg et al. only report 38% of the genes they identified as being differentially expressed. Second, for four of the datasets, we were only able to match 72 – 79% of the reported gene identifiers with genes in our dataset, and there might be additional agreements among those genes that we were unable to compare directly. Third, most previous studies of RCC gene expression have identified many fewer differentially expressed genes than we report here and this decreases the probability of finding agreements between our study and at least two additional studies.
We saw the highest level of agreement (83%) between our data set and the differentially expressed genes reported by Takahashi et al. Several factors likely contribute to our having identified differential expression for such a large fraction of the genes identified in this study. First, Takahashi et al. use a three-fold-change threshold similar to our analysis. Second, Takahashi et al. use a large number of patient samples in their analysis and this increases their power to detect differentially expressed genes. Third, while Takahashi et al. do not use an analysis strategy that calculates a probability of differential expression, they do use a very stringent sample-wise voting strategy (requiring greater than three-fold differential expression in more than 75% of the patient to normal comparisons) that may in fact be more stringent than the parametric statistical methods we used. It appears from highlighting the Takahashi et al. genes on the "volcano plot" of our dataset (genes plotted as a function of fold-change and statistical significance) that of the fourteen genes that are differentially expressed in the Takashi et al. dataset that are not differentially expressed in our dataset, there appear to be two classes of genes: six that we observe to be unchanged and eight that are changed but where the differences do not meet our criteria for differential expression. We suggest that the six Takahashi et al. genes that are unchanged in our dataset may reflect differences between the probes used for detecting the expression of these genes in the two distinct microarray systems.
When highlighting the differentially expressed genes from our re-analysis of the Higgins et al. dataset on the volcano plot of our dataset, we observe a similar apparent bifurcation of our observations into a group of genes that show very little change and another group of genes that is more highly changed. Despite using a similar analysis strategy for analyzing our dataset and our reanalysis of the Higgins et al. dataset we only find evidence of differential expression for 47% of the genes identified in the re-analysis of the Higgins et al. dataset. If we are correct that the genes that show very little change result from probe differences, it is possible that the rate of shared conclusions is influenced by a large number of probe-specific platform differences as well as the false-discovery rates of each analysis and/or an overestimation of our power to detect three-fold changed genes (especially for genes with low hybridization intensity which tend to have higher measurement error).
It is interesting that our observations of the differentially expressed genes identified in the remaining three studies do not show the same degree of apparent bifurcation between genes that are unchanged and genes that are changed but which did not meet our criteria for differential expression. These studies all use some sort of sample-voting analysis strategy though it appears that the fold-change and/or percent-vote thresholds are less stringent than those used by Takahashi et al.
Despite the differences in analysis strategies, the degree of shared conclusions about specific genes that are differentially expressed in RCC tumors is much higher than would be expected by chance alone. We were particularly interested to find that among the genes identified as differentially expressed in three or more studies, sixteen have no known function or informative homology to other genes with known function. This argues strongly for the ability of microarray-based expression profiling to identify genes that are differentially expressed in a manner that is not informed by our current understanding of underlying biological processes.
To begin the process of understanding what differential gene expression can tell us about the biological processes underlying renal cell carcinogenesis, we have taken a directed approach of asking whether specific keywords related to biological processes known to be important in RCC and cancer are associated with a larger number of the genes we have identified as being either induced or repressed in RCC than would be expected by chance alone. We found that genes associated with the keywords hypoxia, angiogenesis, tumor-necrosis factor, apoptosis, interferon, drug- and radiation-resistance, and metastasis are enriched among our list of induced genes. Genes associated with either the keyword kidney or renal are enriched among our list of repressed genes.
Among the eleven differentially expressed genes related to the keyword metastasis we were intrigued to find that some are connected directly or indirectly to MAP kinase and TNF-alpha. Expression of TIMP1 (induced 3.2-fold in RCC) and TNFAIP6 (induced 15.7 fold) are both induced by treatment with TNF-alpha [21,22]. GPR54 (induced 8.2-fold) is the receptor for metastin and is upstream of MAPK [23] as is endothelin1 (EDN1, induced 3.2 fold) [24]. Endothelin1 and VEGF (both induced 3.2 fold) promote the migration of endothelial cells [25] and angiogenesis [26].
ITGA5, MCAM, FXYD5, FUT3 and CHI3L1 – all associated with metastasis – are involved in cell adhesion like TIMP1. A number of other genes involved in cell adhesion, cell migration, and/or cytoskeletal organization – but not specifically known to be involved in metastasis – are also changed in RCC.
A number of the genes associated with metastasis that are induced in RCC also regulate or are regulated through HIF1 in response to hypoxia. Transcription of VEGF and endothelin1 are both stimulated by HIF1 [27] and endothelin1 promotes the stabilization and accumulation of HIF1-alpha [28]. EGLN3 a homolog of C. elegans EGL9 which is a prolyl hydroxylase that targets HIF1-alpha for destruction [29] is induced 12.5-fold in RCC. HIF1-alpha induces expression of carbonic anhydrase IX (induced 17 fold) [30] which is the renal cell carcinoma-associated antigen G250 [31] and is induced in many cancer types. HIF1-alpha is also responsible for the hypoxia-induced expression of angiopoietin-like 4 (induced 24.2-fold), insulin-like growth factor binding protein 3 (induced 10.0-fold), RTP801 (induced 4.2-fold) and PLOD2 (induced 3.5-fold) [30]. These changes in gene expression are consistent with the model that HIF1-alpha accumulation is a hallmark of renal cell carcinogenesis. The increase in HIF1-alpha-dependent gene expression despite the strong induction of EGLN3 suggests that HIF1-alpha is resistant to degradation perhaps as a result of defects in VHL-mediated proteolysis.
We were also intrigued to see the induction of the apoptosis-inhibitor BIRC3 (3.3-fold) which is induced by hypoxia through a HIF1-independent mechanism [32] as well as the induction of the chemokine receptor CCL5 (3.5-fold) which is repressed under hypoxic conditions [33]. HIG2 (induced 7.4-fold) and ADORA3 (induced 3.4-fold) are also known to be induced in response to hypoxia [34,35] though it is unclear if this is a HIF1-dependent process.
We found a number of genes with oncogenic potential that are induced in RCC that have not been noted in other microarray studies of RCC gene expression. One such gene is Axl (induced 3.5-fold), a receptor tyrosine kinase that causes transformation when overexpressed in NIH 3T3 cells [36]. APOBEC3G (induced 4.0-fold) is highly similar to the catalytic subunit of the RNA editing enzyme APOBEC1 – overexpression of which causes elevated rates of carcinoma in transgenic mice [37]. Both APOBEC3G and APOBEC1 have a potent DNA-mutator phenotype when expressed in E. coli [38]. We observed that IMUP, a possible transcription factor that is up-regulated in SV40-transformed cells [39], is induced 5.1-fold.
Additional genes identified as upregulated in renal cancer are interesting from a tissue perspective. Several groups of genes suggest increases in tumor vasculature and inflammation. Endothelial cell specific genes found at increased abundance include von Willebrand factor (8.2-fold increased) and endothelial cell specific molecule-1 (ESM1) (3.0-fold increased) [40]. Platelet/endothelial cell adhesion molecule-1 (PECAM1 or CD31) (4.1-fold increased) is expressed highly on endothelium and in leukocytes. Consistent with an overall inflammatory response, several T cell expressed genes are increased, which likely reflect T cell invasion. Moreover, classic major histocompatibility complex, class II DQ beta 1 (4.2-fold increased) and DP beta 1 (3.0 fold increased) are also increased. Perhaps consistent with tumor necrosis, several toll-like receptors are upregulated as well, including TLR7 (4.7-fold increased), TLR3 (4.5-fold increased) and TLR2 (3.4-fold increased).
A suprising number of G protein coupled receptors (GPR) and G protein signaling molecules are upregulated. These include GPR4 (3.4-fold increased), GPR54 (8.2-fold increased), GPR92 (3.4-fold increased), Rho GTPase activating protein 9 (3.3-fold increased), and particularly regulators of G protein signaling RGS1 (12.8-fold increased) and RGS5 (7.8-fold increased). Although activity of these pathways might be expected to increase with cellular transformation, their increased expression suggests an additional higher level of control of these pathways.
The majority of differentially expressed genes were downregulated. Moreover, many of the genes expressed at lower levels in the renal cancers are ones normally expressed at high levels in differentiated kidney tissue, such as renal epithelial transporters. In addition, some lower expressed genes are cell type-restricted and are therefore excluded from the cancer, such as glomerular podocyte-specific genes encoding the Wilms' tumor suppressor WT1, nephrin and podocin. The notion that primarily "kidney" genes are expressed at lower levels is also supported by the keyword results.
Nevertheless, a small number of the downregulated genes may be important in renal cancer biology due to their tumor suppressor phenotypes. For example, the FHIT tumor suppressor gene is frequently lost in renal cancer. Reduced FHIT expression noted here might also reflect this phenomenon. Surprisingly, the sequence-specific hybridization to the VHL tumor suppressor gene probeset was not detected in normal kidney tissue on the Affymetrix U133B array, which may indicate a problem with the VHL probeset. Thus, the group of downregulated genes might still include some additional important growth suppressors. For example, DLEC1, a gene deleted or mutated in a variety of cancers which inhibits cell growth when reintroduced into DLEC1-defective tumor cell lines [41], is repressed 6.9-fold. GAS1, overexpression of which causes growth arrest in p53-positive cell lines [42,43], is repressed 5.4-fold. SSAT2, repressed 4.8-fold, is highly similar to murine SAT which catalyzes the rate-limiting step in polyamine biosynthesis. Overexpression of SAT causes growth arrest in human breast carcinoma cells [44]. CALML3 which is induced in terminally differentiated epithelial cells [45] and which may have reduced expression in lung carcinoma [46] is repressed 4.0-fold. GADD45A, a p53-dependent DNA-damage inducible gene that inhibits mitotic CDK activity [47] and is required for DNA-damage induced growth arrest, is repressed 3.9-fold. AUH, which is involved in degrading AU-rich mRNA's – including many proto oncogenes [48] – is repressed 3.9-fold. HRASLS2 (down 3.9-fold) is highly similar to HRASL3 which is an anti-apoptotic tumor suppressor [49]. ARHI (repressed 3.3-fold) is a member of the Ras homolog gene family. Expression of ARHI is lost in many ovarian and breast cancers [50]. We also found three tumor suppressor genes to be upregulated in RCC: the BRCA1-binding protein BARD1, the CDK4 inhibitor CDKN2B, and Cystatin A.
Several genes involved in DNA synthesis (MCM5, TOP2A), G2 (BUB2, HEC) and mitosis (DOCK2, PRC1) are also induced in RCC. This could be the result of the higher mitotic index of the tumor cells or could reflect tumor-specific alterations to the cell-cycle machinery.
Among the genes that have not been previously noted to be differentially regulated in RCC, we found that aldeyhyde oxidase and sulfite oxidase but not xanthine dehydrogenase (the three human enzymes that use a molybdenum cofactor [51]), are down regulated. We also found that MOCS1 and molybdopterin synthase [52] as well as gephyrin [53], three of the four enzymes involved in molybdenum cofactor biosynthesis, are also down regulated. It is unclear if or how the differential expression of these molybdenum-related genes relates to RCC disease.
We also found that a component of the variation in gene expression among RCC tumors is significantly correlated with the Fuhrman grade of the tumor, and that this variation is more continuous than distinct between the Fuhrman grades. Since our study was designed to identify genes that are differentially regulated in RCC, we did not analyze enough samples of each of the different tumor grades to identify genes that vary as a function of Fuhrman grade with a rigorous false-positive threshold. Analysis of additional tumors from the different Fuhrman grades should allow us to identify these genes and may provide additional insight into the biological processes underlying RCC progression.
Conclusions
Filtering of microarray-derived gene expression data to remove defective samples and undetected genes accompanied by parametric analysis resulted in the identification of 1,234 genes that are differentially expressed by more than three-fold in renal cell carcinoma which we estimate account for > 95% of all such genes. 800 of these genes had not been reported as being differentially expressed in any of five previous studies of RCC gene expression. Many of the previous studies of RCC gene expression use analytic strategies that identify differential expression by the fraction of tumor / normal comparisons that show a greater than some threshold fold-change (a strategy we refer to as sample-wise voting) that do not control for false-positives or allow for direct estimations of power. Of the genes previously identified as being differentially expressed in RCC with these types of sample-wise voting strategies, we found that we also identified these genes by a parametric method only when stringent majority and fold-change thresholds had been used. We also identified more genes as being differentially expressed with a more stringent fold-change threshold than most of the previous studies had identified. These types of sample-wise voting analyses are therefore likely to make false-positive and false-negative classification errors and this likely accounts for some of the failure of multiple previous microarray studies to identify the same genes as being differentially expressed. However a similar parametric analysis of data from a distinct microarray platform also results in the identification of a set of differentially expressed genes that dissimilar to a degree that is beyond what would be expected from false-positive errors alone. This could be due to differences in probe efficiency between the microarray platforms or may indicate that our power estimation is incorrect. Among the genes we are the first to identify as being differentially expressed in RCC are several oncogenes and tumor suppressor genes that likely play important roles in renal cell carcinogenesis. This highlights the need for rigorous statistical analysis in microarray studies.
Competing Interests
None declared.
Authors' Contributions
MEL designed and carried out the analysis, participated in the biological interpretation and drafted the manuscript. LSL participated in the conception of the study, prepared the tissue samples, isolated the RNA, and participated in the biological interpretation. NPG participated in the sample preparation. GMF performed the array hybridizations and participated in the analysis. HTC participated in the biological interpretation. MFC conceived the study, participated in its design, coordination and the biological interpretation as well as helped draft the manuscript.
All authors read and approved the final manuscript
Abbreviations
RCC, renal cell carcinoma; IRB, institutional review board; M, male; F, female; RNA, ribonucleic acid; DNA, deoxyribonucleic acid; cDNA, complementary deoxyribonucleic acid; cRNA, complementary ribonucleic acid; mRNA, messenger ribonucleic acid; MAS, microarray suite; PCA, principal-components analysis; PERL, practical extraction and report language; ID, identifier; SD, standard deviation; EST, expressed-sequence tag; PC principal component; p, probability.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Tab-delimited text file (1 KB) listing the genes >3.0 fold changed in RCC that are identified by the paired-sample t-test with p < 0.03 and are not also identified by the unpaired t-test. Columns include: the Affymetrix probeset ID, the gene symbol, a descriptive title, the genbank accession of the sequence which was used in the design of the probeset, the -log10 p-value of the paired-sample t-test, and the log2 of the ratio of the geometric mean tumor intensity to the geometric mean adjacent normal tissue intensity.
Tab-delimited text file (345 KB) listing all the probesets > 3.0 fold changed in RCC that are identified by the unpaired t-test with p < 0.03. Columns include: the Affymetrix probeset ID, the genbank accession of the sequence which was used in the design of the probeset, the gene symbol, a descriptive title, the cytologic map position of the gene, the unigene cluster ID, the gene ontology biological process ID, the gene ontology cellular component ID, the gene ontology molecular function ID, the -log10 p-value of the paired-sample t-test, the log2 of the ratio of the geometric mean tumor intensity to the geometric mean adjacent normal tissue intensity, whether the probeset had a paired t-test p < 0.03, whether there are multiple probesets with sequence-specific hybridization for this unigene cluster, whether any additional probesets are < 3.0 fold changed or have a t-test p > 0.03 (column labeled "heterogeneous"), and whether the average fold-change and t-test p-value meet our criteria for differential expression (column labeled "kept in final list").
Tab-delimited text file (196 KB) listing all the probesets that map to a unigene cluster for which there is at least one probeset with a > 3.0 fold change in RCC and an unpaired t-test with p < 0.03 and at least one additional probeset with a < 3.0 fold change or t-test p > 0.03. Columns include: the Affymetrix probeset ID, a descriptive title, the gene symbol, the cytologic map position of the gene, the genbank accession of the sequence which was used in the design of the probeset, the unigene cluster ID, the gene ontology biological process ID, the gene ontology cellular component ID, the gene ontology molecular function ID, the -log10 p-value of the paired-sample t-test, the log2 of the ratio of the geometric mean tumor intensity to the geometric mean adjacent normal tissue intensity, whether the probeset has a > 3.0 fold change in RCC and an unpaired t-test with p < 0.03.
Adobe portable document file (PDF, 36 KB) listing the subset of genes differentially expressed in RCC that are associated with the keywords hypoxia, angiogenesis, tnf, apoptosis, interferon, resistance, metastasis, and kidney.
Acknowledgments
Acknowledgments
We are grateful to members of the Boston University Microarray Resource for comments on the manuscript. This work was supported by a grant from the NIH to MFC.
Contributor Information
Marc E Lenburg, Email: mlenburg@bu.edu.
Louis S Liou, Email: lioul@ccf.org.
Norman P Gerry, Email: npgerry@bu.edu.
Garrett M Frampton, Email: gmframpt@bu.edu.
Herbert T Cohen, Email: htcohen@bu.edu.
Michael F Christman, Email: mfc@bu.edu.
References
- Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Lerman MI, Zbar B, Linehan WM. Genetics of renal-cell carcinoma and evidence for a critical role for von Hippel-Lindau in renal tumorigenesis. Semin Oncol. 1995;22:3–8. [PubMed] [Google Scholar]
- Jiang F, Desper R, Papadimitriou CH, Schaffer AA, Kallioniemi OP, Richter J, Schraml P, Sauter G, Mihatsch MJ, Moch H. Construction of evolutionary tree models for renal cell carcinoma from comparative genomic hybridization data. Cancer Res. 2000;60:6503–6509. [PubMed] [Google Scholar]
- Lubensky IA, Gnarra JR, Bertheau P, Walther MM, Linehan WM, Zhuang Z. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel-Lindau disease patients. Am J Pathol. 1996;149:2089–2094. [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Institute of Phonetic Science, University of Amsterdam http://www.fon.hum.uvo.ni/service/statistics.html
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simple Interactive Statistical Analysis http://home.clara.net/sisa
- NCBI Gene Expression Omnibus Accession Display GSE781 http://www.ncbi.nih.gov/geo/query/?acc=gse781
- Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639–1651. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Rhodes DR, Furge KA, Kanayama H, Kagawa S, Haab BB, Teh BT. Gene expression profiling of clear cell renal cell carcinoma: gene identification and prognostic classification. Proc Natl Acad Sci U S A. 2001;98:9754–9759. doi: 10.1073/pnas.171209998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseg MA, Cody T, Man MZ, Madore SJ, Rubin MA, Kaldjian EP. Expression profiling of human renal carcinomas with functional taxonomic analysis. BMC Bioinformatics. 2002;3:26. doi: 10.1186/1471-2105-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer JM, Huber WK, Sultmann H, Wilmer F, von Heydebreck A, Haas S, Korn B, Gunawan B, Vente A, Fuzesi L, Vingron M, Poustka A. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 2001;11:1861–1870. doi: 10.1101/gr.184501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, Alizadeh AA. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moch H, Schraml P, Bubendorf L, Mirlacher M, Kononen J, Gasser T, Mihatsch MJ, Kallioniemi OP, Sauter G. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol. 1999;154:981–986. doi: 10.1016/S0002-9440(10)65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubitz KM, Skubitz AP. Differential gene expression in renal-cell cancer. J Lab Clin Med. 2002;140:52–64. doi: 10.1067/mlc.2002.125213. [DOI] [PubMed] [Google Scholar]
- Hodges PE, Carrico PM, Hogan JD, O'Neill KE, Owen JJ, Mangan M, Davis BP, Brooks JE, Garrels JI. Annotating the human proteome: the Human Proteome Survey Database (HumanPSD) and an in-depth target database for G protein-coupled receptors (GPCR-PD) from Incyte Genomics. Nucleic Acids Res. 2002;30:137–141. doi: 10.1093/nar/30.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler W, Friedrichs U, Thies A, Tratz C, Wiedemann P. Modulation of matrix metalloproteinase and TIMP-1 expression by cytokines in human RPE cells. Invest Ophthalmol Vis Sci. 2002;43:2767–2773. [PubMed] [Google Scholar]
- Lee TH, Klampfer L, Shows TB, Vilcek J. Transcriptional regulation of TSG6, a tumor necrosis factor- and interleukin-1-inducible primary response gene coding for a secreted hyaluronan-binding protein. J Biol Chem. 1993;268:6154–6160. [PubMed] [Google Scholar]
- Ringel MD, Hardy E, Bernet VJ, Burch HB, Schuppert F, Burman KD, Saji M. Metastin receptor is overexpressed in papillary thyroid cancer and activates MAP kinase in thyroid cancer cells. J Clin Endocrinol Metab. 2002;87:2399. doi: 10.1210/jc.87.5.2399. [DOI] [PubMed] [Google Scholar]
- Wang Y, Simonson MS, Pouyssegur J, Dunn MJ. Endothelin rapidly stimulates mitogen-activated protein kinase activity in rat mesangial cells. Biochem J. 1992;287 ( Pt 2):589–594. doi: 10.1042/bj2870589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salani D, Di Castro V, Nicotra MR, Rosano L, Tecce R, Venuti A, Natali PG, Bagnato A. Role of endothelin-1 in neovascularization of ovarian carcinoma. Am J Pathol. 2000;157:1537–1547. doi: 10.1016/S0002-9440(10)64791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salani D, Taraboletti G, Rosano L, Di Castro V, Borsotti P, Giavazzi R, Bagnato A. Endothelin-1 induces an angiogenic phenotype in cultured endothelial cells and stimulates neovascularization in vivo. Am J Pathol. 2000;157:1703–1711. doi: 10.1016/S0002-9440(10)64807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Spinella F, Rosano L, Di Castro V, Natali PG, Bagnato A. Endothelin-1 induces vascular endothelial growth factor by increasing hypoxia-inducible factor-1alpha in ovarian carcinoma cells. J Biol Chem. 2002;277:27850–27855. doi: 10.1074/jbc.M202421200. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Lal A, Peters H, St Croix B, Haroon ZA, Dewhirst MW, Strausberg RL, Kaanders JH, van der Kogel AJ, Riggins GJ. Transcriptional response to hypoxia in human tumors. J Natl Cancer Inst. 2001;93:1337–1343. doi: 10.1093/jnci/93.17.1337. [DOI] [PubMed] [Google Scholar]
- Grabmaier K, Vissers JL, De Weijert MC, Oosterwijk-Wakka JC, Van Bokhoven A, Brakenhoff RH, Noessner E, Mulders PA, Merkx G, Figdor CG, Adema GJ, Oosterwijk E. Molecular cloning and immunogenicity of renal cell carcinoma-associated antigen G250. Int J Cancer. 2000;85:865–870. doi: 10.1002/(SICI)1097-0215(20000315)85:6<865::AID-IJC21>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Dong Z, Venkatachalam MA, Wang J, Patel Y, Saikumar P, Semenza GL, Force T, Nishiyama J. Up-regulation of apoptosis inhibitory protein IAP-2 by hypoxia. Hif-1-independent mechanisms. J Biol Chem. 2001;276:18702–18709. doi: 10.1074/jbc.M011774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirani N, Antonicelli F, Strieter RM, Wiesener MS, Ratcliffe PJ, Haslett C, Donnelly SC. The regulation of interleukin-8 by hypoxia in human macrophages--a potential role in the pathogenesis of the acute respiratory distress syndrome (ARDS) Mol Med. 2001;7:685–697. [PMC free article] [PubMed] [Google Scholar]
- Denko N, Schindler C, Koong A, Laderoute K, Green C, Giaccia A. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin Cancer Res. 2000;6:480–487. [PubMed] [Google Scholar]
- Dougherty C, Barucha J, Schofield PR, Jacobson KA, Liang BT. Cardiac myocytes rendered ischemia resistant by expressing the human adenosine A1 or A3 receptor. Faseb J. 1998;12:1785–1792. doi: 10.1096/fasebj.12.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bryan JP, Frye RA, Cogswell PC, Neubauer A, Kitch B, Prokop C, Espinosa R., 3rd, Le Beau MM, Earp HS, Liu ET. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11:5016–5031. doi: 10.1128/mcb.11.10.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- Kim JK, Ryll R, Ishizuka Y, Kato S. Identification of cDNAs encoding two novel nuclear proteins, IMUP-1 and IMUP-2, upregulated in SV40-immortalized human fibroblasts. Gene. 2000;257:327–334. doi: 10.1016/S0378-1119(00)00414-5. [DOI] [PubMed] [Google Scholar]
- Lassalle P, Molet S, Janin A, Heyden JV, Tavernier J, Fiers W, Devos R, Tonnel AB. ESM-1 is a novel human endothelial cell-specific molecule expressed in lung and regulated by cytokines. J Biol Chem. 1996;271:20458–20464. doi: 10.1074/jbc.271.34.20458. [DOI] [PubMed] [Google Scholar]
- Daigo Y, Nishiwaki T, Kawasoe T, Tamari M, Tsuchiya E, Nakamura Y. Molecular cloning of a candidate tumor suppressor gene, DLC1, from chromosome 3p21.3. Cancer Res. 1999;59:1966–1972. [PubMed] [Google Scholar]
- Del Sal G, Collavin L, Ruaro ME, Edomi P, Saccone S, Valle GD, Schneider C. Structure, function, and chromosome mapping of the growth-suppressing human homologue of the murine gas1 gene. Proc Natl Acad Sci U S A. 1994;91:1848–1852. doi: 10.1073/pnas.91.5.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokiou A, Cowled PA. Tumor-suppressive activity of the growth arrest-specific gene GAS1 in human tumor cell lines. Int J Cancer. 1998;75:568–577. doi: 10.1002/(SICI)1097-0215(19980209)75:4<568::AID-IJC13>3.3.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Vujcic S, Halmekyto M, Diegelman P, Gan G, Kramer DL, Janne J, Porter CW. Effects of conditional overexpression of spermidine/spermine N1-acetyltransferase on polyamine pool dynamics, cell growth, and sensitivity to polyamine analogs. J Biol Chem. 2000;275:38319–38328. doi: 10.1074/jbc.M003270200. [DOI] [PubMed] [Google Scholar]
- Rogers MS, Kobayashi T, Pittelkow MR, Strehler EE. Human calmodulin-like protein is an epithelial-specific protein regulated during keratinocyte differentiation. Exp Cell Res. 2001;267:216–224. doi: 10.1006/excr.2001.5254. [DOI] [PubMed] [Google Scholar]
- Schraml P, Shipman R, Colombi M, Ludwig CU. Identification of genes differentially expressed in normal lung and non-small cell lung carcinoma tissue. Cancer Res. 1994;54:5236–5240. [PubMed] [Google Scholar]
- Zhan Q, Antinore MJ, Wang XW, Carrier F, Smith ML, Harris CC, Fornace A. J., Jr. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892–2900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jeno P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci U S A. 1995;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sers C, Husmann K, Nazarenko I, Reich S, Wiechen K, Zhumabayeva B, Adhikari P, Schroder K, Gontarewicz A, Schafer R. The class II tumour suppressor gene H-REV107-1 is a target of interferon-regulatory factor-1 and is involved in IFNgamma-induced cell death in human ovarian carcinoma cells. Oncogene. 2002;21:2829–2839. doi: 10.1038/sj.onc.1205377. [DOI] [PubMed] [Google Scholar]
- Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y, Cuevas B, Kuo WL, Gray JW, Siciliano M, Mills GB, Bast R. C., Jr. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci U S A. 1999;96:214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss J. Genetics of molybdenum cofactor deficiency. Hum Genet. 2000;106:157–163. doi: 10.1007/s004390051023. [DOI] [PubMed] [Google Scholar]
- Reiss J, Cohen N, Dorche C, Mandel H, Mendel RR, Stallmeyer B, Zabot MT, Dierks T. Mutations in a polycistronic nuclear gene associated with molybdenum cofactor deficiency. Nat Genet. 1998;20:51–53. doi: 10.1038/1706. [DOI] [PubMed] [Google Scholar]
- Feng G, Tintrup H, Kirsch J, Nichol MC, Kuhse J, Betz H, Sanes JR. Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science. 1998;282:1321–1324. doi: 10.1126/science.282.5392.1321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab-delimited text file (1 KB) listing the genes >3.0 fold changed in RCC that are identified by the paired-sample t-test with p < 0.03 and are not also identified by the unpaired t-test. Columns include: the Affymetrix probeset ID, the gene symbol, a descriptive title, the genbank accession of the sequence which was used in the design of the probeset, the -log10 p-value of the paired-sample t-test, and the log2 of the ratio of the geometric mean tumor intensity to the geometric mean adjacent normal tissue intensity.
Tab-delimited text file (345 KB) listing all the probesets > 3.0 fold changed in RCC that are identified by the unpaired t-test with p < 0.03. Columns include: the Affymetrix probeset ID, the genbank accession of the sequence which was used in the design of the probeset, the gene symbol, a descriptive title, the cytologic map position of the gene, the unigene cluster ID, the gene ontology biological process ID, the gene ontology cellular component ID, the gene ontology molecular function ID, the -log10 p-value of the paired-sample t-test, the log2 of the ratio of the geometric mean tumor intensity to the geometric mean adjacent normal tissue intensity, whether the probeset had a paired t-test p < 0.03, whether there are multiple probesets with sequence-specific hybridization for this unigene cluster, whether any additional probesets are < 3.0 fold changed or have a t-test p > 0.03 (column labeled "heterogeneous"), and whether the average fold-change and t-test p-value meet our criteria for differential expression (column labeled "kept in final list").
Tab-delimited text file (196 KB) listing all the probesets that map to a unigene cluster for which there is at least one probeset with a > 3.0 fold change in RCC and an unpaired t-test with p < 0.03 and at least one additional probeset with a < 3.0 fold change or t-test p > 0.03. Columns include: the Affymetrix probeset ID, a descriptive title, the gene symbol, the cytologic map position of the gene, the genbank accession of the sequence which was used in the design of the probeset, the unigene cluster ID, the gene ontology biological process ID, the gene ontology cellular component ID, the gene ontology molecular function ID, the -log10 p-value of the paired-sample t-test, the log2 of the ratio of the geometric mean tumor intensity to the geometric mean adjacent normal tissue intensity, whether the probeset has a > 3.0 fold change in RCC and an unpaired t-test with p < 0.03.
Adobe portable document file (PDF, 36 KB) listing the subset of genes differentially expressed in RCC that are associated with the keywords hypoxia, angiogenesis, tnf, apoptosis, interferon, resistance, metastasis, and kidney.