Abstract
DNA polymerases can misinsert ribonucleotides that lead to genomic instability. DNA polymerase β discourages ribonucleotide insertion with the backbone carbonyl of Tyr-271; alanine substitution of Tyr-271, but not Phe-272, resulted in a >10-fold loss in discrimination. The Y271A mutant also inserted ribonucleotides more efficiently than wild type on a variety of ribonucleoside (rNMP)-containing DNA substrates. Substituting Mn2+ for Mg2+ decreased sugar discrimination for both wild-type and mutant enzymes primarily by increasing the affinity for rCTP. This facilitated crystallization of ternary substrate complexes of both the wild-type and Y271A mutant enzymes. Crystallographic structures of Y271A- and wild type-substrate complexes indicated that rCTP is well accommodated in the active site but that O2′ of rCTP and the carbonyl oxygen of Tyr-271 or Ala-271 are unusually close (∼2.5 and 2.6 Å, respectively). Structure-based modeling indicates that the local energetic cost of positioning these closely spaced oxygens is ∼2.2 kcal/mol for the wild-type enzyme. Because the side chain of Tyr-271 also hydrogen bonds with the primer terminus, loss of this interaction affects its catalytic positioning. Our results support a model where DNA polymerase β utilizes two strategies, steric and geometric, with a single protein residue to deter ribonucleotide insertion.
Keywords: DNA Polymerase, DNA Repair, DNA-Protein Interaction, Enzyme Kinetics, Enzyme Structure, Ribonucleotide Discrimination
Introduction
DNA polymerases must select the right deoxynucleoside triphosphate (dNTP) from a pool of chemically and structurally similar molecules to preserve genomic sequence and structure. The mechanism by which polymerases discriminate against base substitution errors (i.e. insertion of the wrong dNTP) has been described in kinetic (1) and molecular detail (2). Ribonucleoside triphosphates differ from their deoxynucleotide counterparts by a single atom (oxygen) at C2′ of the sugar and, more significantly, are present at much higher concentrations in the cell (3, 4). Thus, ribonucleotides are predicted to be inserted during DNA replication and repair at much higher frequencies than incorrect deoxynucleosides (5, 6). The presence of a ribose 2′-hydroxyl group stabilizes the glycosyl bond but makes the phosphodiester bond more susceptible to hydrolysis (7), thereby diminishing the chemical stability of the genome. Spontaneous or enzyme-induced strand breaks would be expected to initiate DNA repair and cellular signaling events that would also impact genome stability and cell survival.
In most instances, DNA polymerases discriminate against ribonucleotide insertion by binding them weakly and inserting them more slowly than their natural substrate. Thus, the catalytic efficiency (kcat/Km) by which they are inserted is very low (i.e. ∼10−3–10−4/μm-s). Interestingly, different DNA polymerases insert ribonucleotides with very similar efficiencies so that discrimination is often dictated by the efficiency for insertion of the deoxynucleoside triphosphate (i.e. polymerases that exhibit low ribonucleotide discrimination also insert dNTPs poorly) (5).
Based on sequence homology, the 15 human DNA polymerases are grouped into five families. The general architecture and kinetic mechanism among these polymerases are very similar, binding substrates and magnesium in a comparable manner. However, they have each evolved to perform distinct cellular roles. DNA polymerase (pol)2 β is the smallest polymerase and belongs to a group of DNA polymerases (X-family) involved in short gap-filling during DNA repair. Although X-family DNA polymerases share many structural similarities, they have differing template (i.e. DNA gap) specificities that are imposed by their primary protein sequence (8).
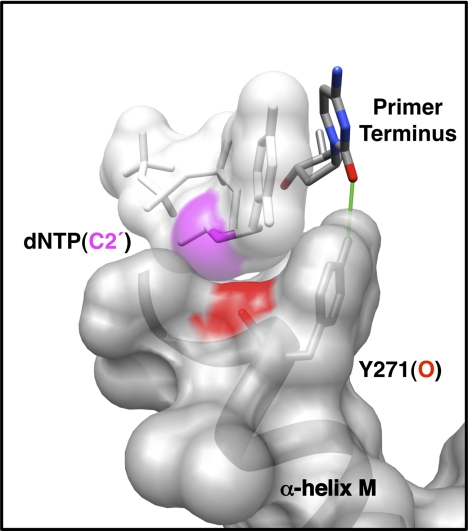
In general, DNA polymerases are in an open inactive conformation in the absence of ligands, but tightly sandwich the nascent base pair between duplex DNA and protein during catalytic activation. This is accompanied by enzyme subdomain motions, side chain repositioning, and substrate conformational adjustments. Crystallographic structures of substrate complexes of DNA polymerases from most families have suggested that a side chain could sterically interfere with binding a ribonucleoside triphosphate (rNTP) (9, 10). This side chain has been referred to as a “steric gate.” In contrast, X-family DNA polymerases have been suggested to deter insertion of a ribonucleotide using the protein backbone near the carboxyl terminus of α-helix M (i.e. pol β Tyr-271; Fig. 1) (5, 11, 12). The carbonyl of this side chain would be predicted to clash with the hydroxyl group on C2′ of the incoming ribonucleotide. Upon binding a nucleotide, α-helix M is repositioned so that Tyr-271 forms a hydrogen bond with the minor groove edge of the primer terminus (13, 14). Consequently, binding a ribonucleotide could perturb interactions at the primer terminus transmitted through altered interactions with α-helix M.
FIGURE 1.
Molecular interactions of Tyr-271 with the incoming nucleotide and primer terminus. Tyr-271 of α-helix M forms part of the dNTP-binding pocket and interacts with the minor groove edge of the DNA substrate. In the closed polymerase conformation, the side chain of Tyr-271 hydrogen bonds with the minor groove edge of the primer terminal base (green line). The backbone carbonyl of Tyr-271 (red) is within 3.5 Å of C2′ (magenta) of the incoming nucleotide. The van der Waals semi-transparent surfaces (gray) of the incoming nucleotide and α-helix M are shown. In the open polymerase conformation, Tyr-271 interacts with the minor groove edge of the templating base (not shown).
EXPERIMENTAL PROCEDURES
Nucleotides
Ultrapure rNTP and dNTP solutions were purchased from Sigma, [γ-32P]ATP from PerkinElmer Life Sciences, ddNTPs from Amersham Biosciences, and araCTP from Jena Biosciences (Germany). All other reagents were purchased in the highest purity available.
Protein Expression and Purification
Wild-type and mutant (Y271A, Y271F, and F272A) human pol β was expressed and purified as described previously (13, 15). Enzyme concentrations were determined from their absorbance at 280 nm (ϵ = 23,380 m−1 cm−1) (16).
DNA Preparation
High pressure liquid chromatography-purified oligonucleotides were purchased from Integrated DNA Technologies, dissolved in 10 mm Tris-HCl, pH 7.4, and 1 mm EDTA, and their concentrations determined by UV absorbance at 260 nm. A single-nucleotide gap was produced by annealing three oligonucleotides (1:1.2:1.2, 15-mer primer/18-mer downstream oligonucleotide/34-mer template) as described previously (5). The downstream oligonucleotide was synthesized with a 5′-phosphate. The sequences are shown in supplemental Fig. S1; changes in the primer terminus or template sugar/base identity are noted in the tables or figures. The upstream primer was 5′-labeled with [γ-32P]ATP using Optikinase (United States Biochemical Corp.), and free radioactive ATP was removed using a Bio-Spin 6 column (Bio-Rad).
Kinetic Assays
Steady-state kinetic parameters for single-nucleotide gap-filling reactions were determined as described previously (5). Reactions typically contained 50 mm Tris-HCl, pH 7.4, 100 mm KCl, 5 mm MgCl2, 200 nm single-nucleotide gapped DNA, and varying concentrations of nucleoside triphosphate. Reactions were initiated by the addition of enzyme and incubated at 37 °C. Enzyme concentration and reaction time intervals were chosen so that substrate depletion or product inhibition did not influence initial velocity measurements. In some instances, 5 mm MnCl2 replaced MgCl2. Reactions were quenched with 0.5 m EDTA and an equal (total) volume of 95% formamide solution (bromophenol blue and xylene cyanol). Products were separated on 20% denaturing polyacrylamide gels and quantified using phosphorimagery and ImageQuant software. Steady-state kinetic parameters were determined by fitting rate data to the Michaelis-Menten equation.
Crystallography Conditions
The double-stranded DNA substrate consisting of a 16-mer template (5′-CCGACGCCGCATCAGC), a 9-mer primer (5′-GCTGATGCG), and a 5-mer downstream oligonucleotide (5′-pGTCGG) were dissolved in 20 mm MgCl2 and 100 mm Tris-HCl, pH 7.5, and annealed (1:1:1) using a PCR thermocycler by heating for 10 min at 90 °C and cooling to 4 °C (1 °C/min) creating 1 mm two-nucleotide gapped DNA. This solution was mixed with an equal volume of pol β (15 mg/ml in 20 mm BisTris, pH 7.0) or Y271A mutant enzyme (13.5 mg/ml) at 4 °C, then warmed to 35 °C, and gradually cooled to 4 °C. A 2-fold excess of 2′,3′-dideoxyguanosine 5′-triphosphate was added to obtain a one-nucleotide gapped complex with a dideoxy-primer terminus. Enzyme-DNA complexes were crystallized by sitting drop vapor diffusion by mixing protein 1 to 1 with mother liquor consisting of 16% PEG-3350, 350 mm sodium acetate, and 50 mm imidazole, pH 7.0–7.5, at 18 °C. These trays were streak-seeded after 1 day. Crystals (2–4 days after seeding) were soaked in a solution consisting of 50 mm MnCl2, 90 mm sodium acetate, 10 mm dCTP or rCTP, 20% PEG-3350, and 12% ethylene glycol and flash-frozen in liquid nitrogen. All crystals belong to the space group P21.
Data Collection and Structure Determination
X-ray crystal diffraction data were collected at 100 K on a Saturn 92 CCD detector system mounted on a MicroMax-007HF (Rigaku Corp.) rotating anode generator. Data were integrated and reduced with HKL2000 software (17). Ternary substrate complex structures were modeled by molecular replacement with a previously determined structure of pol β complexed with one-nucleotide gapped DNA and a complementary incoming ddCTP (PDB code 2FMP). The crystal structures have similar lattices and are sufficiently isomorphous to determine the molecular replacement model using PHENIX (18) and manual model building using O (19). The molecular graphic images were prepared in Chimera (20).
Structural Modeling
The interaction between O2′ of the incoming ribonucleotide with the backbone carbonyl of Tyr-271 was examined using molecular dynamics (MD) simulations. The initial simulation structures were based on the crystallographic structure of the ternary substrate complex of wild-type enzyme with an incoming rCTP. All metal ion and crystallographic water positions were preserved along with the additional ribonucleotide bound to pol β at a site distal to the active site. Hydrogen atoms were added to conserve valence. Additional counter-ions were introduced to neutralize the charged system, and the final assembly was solvated in a box of 24,828 water molecules. For comparison of the ribose and deoxyribose interactions of the incoming nucleotide with pol β, an additional system was prepared by converting the ribonucleotide at the incoming nucleotide position to the corresponding deoxynucleotide.
Prior to general structural equilibration, both systems were subjected to several stages of energy minimizations and relaxations under constant volume. As the first step, 100 ps of belly dynamics on the water molecules relaxed their initial positions, whereas the rest of the system was fixed. A subsequent energy minimization step was followed by a low temperature (constant temperature/constant pressure) step with fixed protein, DNA, and nucleotide to obtain a reasonable starting density (∼1 g/ml). After a complete energy minimization (10,000 conjugate gradient steps), and a stepwise heating procedure at constant volume (1 ns), equilibration runs were carried out for 13 ns at 300 K while gradually releasing constraints to ensure that the system was not disrupted due to initialization of unconstrained molecule dynamics. Unconstrained final trajectories were then calculated at 300 K under constant temperature/constant pressure for over 10 ns. Final MD runs were carried out with time steps of 1 fs. The particle mesh Ewald method (21) was used to treat long range electrostatics in all simulations. The PMEMD module of the AMBER10 molecular dynamics package was used for all energy minimizations and MD trajectory calculations. The force field parameters were taken from the ff99SB force field included in the AMBER10 package (22).
Quantum mechanical calculations were carried out with Gaussian 09 (23) using the B3LYP/aug-cc-pvtz method/basis. Atomic charges estimates were made with the CHELPG module. Additional details are outlined in supplemental Methods.
RESULTS
We recently examined the impact of modified sugars of the incoming nucleoside triphosphate on the insertion efficiency of pol β and found that this X-family polymerase discriminated against ribonucleotides (possessing a 2′-OH) while efficiently inserting araCTP (2′-OH nearer cytosine) and dideoxynucleotides (removing 3′-OH) (5). Here, we examined the structural features of pol β that contribute to ribonucleotide discrimination. Based on previously reported crystallographic structures of X-family DNA polymerases, we predict that the backbone of an α-helix would sterically interfere with binding of a ribonucleotide (12, 14). In pol β, the backbone of Tyr-271 is in the vicinity of C2′, whereas the phenol side chain hydrogen bonds with the base of the primer terminus (Fig. 1). We now examine the impact of removing the Tyr-271 side chain on ribonucleotide discrimination.
Removing Tyr-271 Does Not Affect Deoxynucleotide Insertion
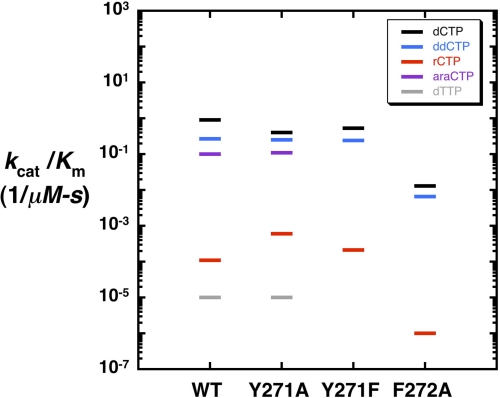
Alanine substitution for Tyr-271 (Y271A) resulted in a mutant that inserted dCTP into single-nucleotide gapped DNA with comparable efficiency as wild-type enzyme. This result is similar to that previously observed for the Y271F and Y271H mutants (13). Y271A inserted dCTP opposite a templating guanine only 2-fold less efficiently than wild type and misinserted dTTP with the same efficiency as wild-type pol β (Table 1 and Fig. 2). These results indicate that removing the tyrosine side chain does not change the ability of the mutant polymerase to insert/discriminate between correct and incorrect deoxynucleotides. Because the catalytic efficiencies in Fig. 2 are plotted on a logarithmic scale, the distance between two points corresponds to substrate discrimination, i.e. the larger the difference, the greater the discrimination (24).
TABLE 1.
Kinetic summary for insertion/misinsertion of nucleotides with modified sugars
Assays were performed as described under “Experimental Procedures.” The DNA substrate was single-nucleotide gapped DNA with a templating guanine. The results represent the mean ± S.E. of at least two independent determinations.
| Enzyme | xNTPa | kcat | Km, xNTP | kcat/Km | Discriminationb |
|---|---|---|---|---|---|
| 10−2(s−1) | μm | 10−3(1/s-μm) | |||
| Wild typec | dC | 38.8 (0.6) | 0.42 (0.05) | 900 (100) | 1 |
| ddC | 12 (3) | 0.45 (0.07) | 270 (80) | 3.3 | |
| rC | 4.6 (0.7) | 400 (90) | 0.11 (0.03) | 8,200 | |
| araC | 34 (7) | 3.5 (0.4) | 100 (20) | 9 | |
| dT | 1.3 (0.2) | 1300 (300) | 0.010 (0.003) | 90,000 | |
| Y271A | dC | 39 (6) | 1.0 (0.2) | 400 (100) | 1 |
| ddC | 13 (2) | 0.51 (0.05) | 250 (50) | 1.6 | |
| rC | 3.4 (0.6) | 60 (10) | 0.6 (0.1) | 670 | |
| araC | 40 (10) | 3.4 (0.4) | 110 (30) | 3.6 | |
| dT | 1.1 (0.8) | 1100 (200) | 0.010 (0.007) | 40,000 | |
| Y271F | dC | 40 (4) | 0.75 (0.08) | 530 (80) | 1 |
| ddC | 10 (2) | 0.41 (0.08) | 240 (70) | 2.2 | |
| rC | 3.8 (0.2) | 180 (30) | 0.21 (0.04) | 2,500 | |
| F272A | dC | 4.2 (0.7) | 3.2 (0.2) | 13 (2) | 1 |
| ddC | 1.30 (0.06) | 1.9 (0.8) | 6.6 (0.8) | 2.0 | |
| rC | 0.041 (0.005) | 400 (90) | 0.0010 (0.0003) | 13,000 |
a For xNTP, x denotes the sugar and N denotes the base for each nucleotide listed (i.e. rC denotes rCTP).
b Discrimination is defined as (kcat/Km)dCTP/(kcat/Km)xNTP.
c Wild type data from Ref. (5).
FIGURE 2.
Substrate discrimination by human DNA polymerase β mutants. A discrimination plot where the catalytic efficiencies (−) for nucleotide insertion (Table 1) are plotted on a log ordinate scale. Nucleotide insertion was measured on a single-nucleotide gapped substrate with a templating “dG” as described under “Experimental Procedures.” Discrimination is proportional to the distance between a colored bar and a black bar (corresponding to correct insertion of dCTP) for each enzyme. Thus, the greater the distance between catalytic efficiencies, the greater the discrimination. The catalytic efficiencies for ddCTP (blue), araCTP (purple), rCTP (red), and dTTP (gray) are plotted for wild-type pol β and Y271A, Y271F, and F272A mutants.
Y271A Exhibits Lower Discrimination for Ribonucleotides
The ability of Y271A to insert nucleotides with modified sugars is summarized in Table 1 and Fig. 2. Y271A inserted dideoxycytidine (ddCTP) and 1-β-d-arabinofuranosylcytidine (cytarabine or araCTP) opposite guanine only 1.6- and 3.6-fold less efficiently than dCTP, respectively. The insertion efficiency of ddCTP and araCTP was similar for both wild-type and Y271A enzymes. However, the mutant exhibited a slightly lower discrimination for araCTP because of the small loss of dCTP insertion efficiency (Table 1). These results indicate that replacing Tyr-271 with alanine had little effect on discrimination of nucleotides with arabinose or dideoxynucleotide sugars as long as the incoming base satisfied Watson-Crick base pairing.
The Y271A variant displayed a significant effect on the discrimination of ribose sugars. Y271A inserted rCTP opposite template dG 670-fold less efficiently than dCTP (12-fold lower in ribonucleotide discrimination than wild type; Table 1 and Fig. 2). This resulted primarily from an increase in binding affinity for the ribonucleotide rather than a change in the rate of insertion.3 Thus, the greatest impact of the tyrosine to alanine mutation was more efficient insertion of ribonucleotides compared with wild type, although still strongly discriminating against misinsertion of dTTP and exhibiting insertion efficiencies for other modified sugars and dCTP similar to wild-type pol β (Table 1 and Fig. 2). A similar result was observed when the reciprocal insertion (rGTP-dC) of a ribonucleotide was measured.4
F272A Does Not Affect Ribonucleotide Discrimination
There is an adjacent aromatic side chain (Phe-272) to Tyr-271 that is also part of α-helix M and very close to the ribose ring of the incoming nucleotide. We explored the possibility that an alanine mutant for this residue may also exhibit a similar effect on ribonucleotide discrimination. As tabulated in Table 1, removing the phenyl side chain decreased correct dCTP insertion efficiency 69-fold compared with wild type. F272A inserted ddCTP and rCTP 41- and 110-fold less efficiently than wild-type enzyme, respectively. However, the relative ratio of the catalytic efficiencies (i.e. discrimination) for right (dCTP) and wrong sugar (rCTP) insertion for F272A was similar to wild-type pol β (Table 1). Therefore, ribonucleotide discrimination for pol β was associated specifically with Tyr-271 rather than its aromatic neighbor.
Because removing the tyrosine side chain decreased ribonucleotide discrimination, we determined whether the loss of discrimination might be restored by the addition of an aromatic side chain. An alternate mutant, Y271F, incorporated dCTP and ddCTP opposite dG with nearly the same efficiency as wild-type enzyme (Table 1 and Fig. 2), consistent with previously reported results (13). In terms of ribonucleotide discrimination, however, Y271F inserted rCTP opposite dG 3.7-fold less efficiently than Y271A and 3.3-fold more efficiently than wild type, indicating that adding a phenyl ring at this position only partially recovered ribonucleotide discrimination (Table 1 and Fig. 2).
Y271A Efficiently Extends Ribonucleotide-terminated Primers
Because Y271A could insert a ribonucleotide more efficiently than wild type, we examined how well the mutant could extend a ribonucleotide at the primer terminus, i.e. addition of the correct dNTP. Wild-type and Y271A enzymes inserted the next correct nucleotide, dCTP, with comparable efficiencies whereas Y271A misinserted dTTP 3-fold more efficiently than wild-type enzyme (Table 1). Importantly, Y271A extended a primer terminus with a ribonucleotide with a second ribonucleotide, rCTP, >12-fold more efficiently than wild-type enzyme, yet again indicating the relaxed ribonucleotide discrimination by the mutant. The discrimination factor for the second rCTP insertion by Y271A was only 260 compared with 670 for the first insertion, suggesting that insertion of a ribonucleotide facilitated the addition of a second ribonucleotide.
Y271A Inserts rNTPs More Efficiently than Wild Type on All rNMP-containing DNA Substrates
We examined the effect of placing a ribonucleotide at different positions of the DNA template-primer by substituting the upstream template base, altering the base pair at the primer terminus to a ribonucleotide pair, or substituting a ribonucleotide at the template coding position and comparing wild-type and mutant activities on these modified hybrid gapped substrates (supplemental Table S1). In all cases, only the sugar was modified, although the identity of the base was maintained to directly compare with the original DNA substrate. Wild-type pol β and Y271A exhibited similar catalytic efficiencies for insertion of dCTP and misinsertion of dTTP on all of the gapped substrates, whereas Y271A incorporated rCTP with higher catalytic efficiency than wild type (10–32-fold depending on the position of the ribonucleotide in the DNA substrate). These results demonstrate that the Y271A mutation specifically affects ribonucleotide discrimination at the insertion step.
Structures of Ternary Substrate (rCTP, dCTP) Complexes
The low binding affinity of rNTPs has hindered structural studies attempting to assemble a ternary substrate complex. However, the Y271A mutant binds rCTP 6-fold more tightly than wild-type enzyme in the presence of Mg2+ (Table 1).3 Furthermore, we exploited the observation that substituting Mg2+ with Mn2+ dramatically increases the binding affinity for incorrect nucleotides, as demonstrated previously by substrate complex structures of pol β with active site mismatches (16, 25). Manganese (Mn2+) has a similar ionic radius and coordination geometry as Mg2+ making it an excellent substitute.
Binary complex crystals of wild-type pol β and the Y271A mutant were prepared using a one-nucleotide gapped DNA substrate containing a dideoxy-terminated primer. rCTP or dCTP was soaked into the crystal as described under “Experimental Procedures.” Ternary substrate complex crystals with an incoming rCTP diffracted to 1.92 and 2.05 Å for wild-type and mutant enzymes, respectively. In addition, a ternary complex crystal structure of the Y271A mutant with an incoming dCTP was obtained for comparison. The crystallographic data for these structures are summarized in Table 2.
TABLE 2.
Crystallographic data and refinement statistics
| Enzyme incoming nucleotide | Wild type, rCTP | Y271A |
|
|---|---|---|---|
| dCTP | rCTP | ||
| PDB code | 3RH4 | 3RH5 | 3RH6 |
| Data collection | |||
| a | 51.17 Å | 51.22 Å | 51.06 Å |
| b | 79.79 Å | 79.92 Å | 79.81 Å |
| c | 55.00 Å | 55.09 Å | 55.06 Å |
| β | 107.95° | 107.09° | 107.68° |
| dmin | 1.92 Å | 2.10 Å | 2.05 Å |
| Rmergea | 5.7% (42.1)b | 11.1% (56.7) | 11.1% (52.1) |
| Completeness | 94.1% (67.1) | 98.1% (97.4) | 97.2% (95.8) |
| I/σI | 22.2 (2.1) | 10.0 (2.3) | 11.3 (2.3) |
| No. of observed reflections | 101,462 | 85,107 | 92,422 |
| No. of unique reflections | 30,404 (2,164) | 24,357 (2,390) | 25,790 (2,513) |
| Refinement | |||
| Root mean square deviations | |||
| Bond lengths | 0.007 Å | 0.007 Å | 0.007 Å |
| Bond angles | 1.164° | 1.202° | 1.180° |
| Rworkc | 17.5% | 20.8% | 19.5% |
| Rfreed | 24.0% | 28.1% | 26.6% |
| Average B factor | |||
| Protein | 26.9 Å2 | 31.8 Å2 | 32.6 Å2 |
| DNA | 30.9 Å2 | 33.8 Å2 | 35.3 Å2 |
| rCTP/dCTP | 16.5 Å2 | 22.7 Å2 | 22.3 Å2 |
| Ramachandran analysise | |||
| Favored | 97.6% | 96.0% | 98.5% |
| Allowed | 100% | 100% | 100% |
a Rmerge = 100 × ΣhΣi|Ih,i − Ih| ΣhΣiIh,i, where Ih is the mean intensity of symmetry-related reflections Ih,i.
b Numbers in parentheses refer to the highest resolution shell of data (10%).
c Rwork = 100 × Σ‖Fobs|−|Fcalc‖/Σ|Fobs|.
d Rfree for a 5% subset of reflections withheld from refinement.
e Data were as determined by MolProbity (47).
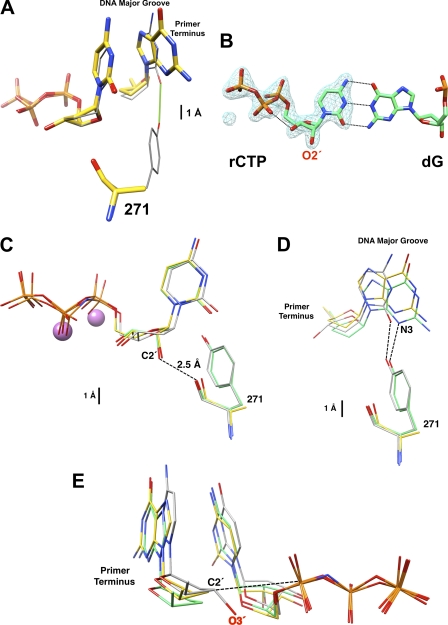
The Tyr-271 hydroxyl group hydrogen bonds with the minor groove edge of the base of the primer terminus nucleotide in the closed ternary substrate complex (Fig. 1). Comparing the structure of wild-type enzyme (PDB code 2FMP) with Y271A with an incoming dCTP indicates that the enzyme is in a closed conformation, but the loss of the hydrogen bond with the Y271A mutant results in a modest shift (difference in O2 and N3 positions ∼0.7 Å) of the primer terminus toward the major groove (Fig. 3A). In contrast, the incoming nucleotide position is not significantly altered, and the overall structure is identical to that of wild-type enzyme (Fig. 3A) (26).
FIGURE 3.
Structural characterization of dCTP/rCTP in the confines of wild-type and Y271A DNA polymerase β. A, active site comparison between superimposed ternary complex structures with a correct active site base pair for wild-type (PDB ID 2FMP; gray carbons) (26) and Y271A mutant (yellow carbons) of pol β. The incoming nucleotide (ddCTP, wild type; dCTP, Y271A) is shown along with the dideoxy-terminated primer terminus and residue 271. Loss of the hydrogen bond (green) with the minor groove edge of the base of the primer terminus in the Y271A structure results in a modest displacement into the DNA major groove. B, Fo − Fc simulated annealing electron density omit map (blue) contoured at 5.0σ showing electron density corresponding to rCTP bound to wild-type pol β. The O2′ of the ribose sugar is clearly observed and the base forms Watson-Crick base pairs with the templating deoxyguanine (dG). C, comparison of the incoming nucleotides with superimposed structures of wild-type enzyme with an incoming nucleotide analog of dTTP (PDB code 2FMS, gray) (26) and wild-type (green) and Y271A mutant (yellow) pol β with an incoming rCTP. The structures were superimposed using all Cαs. The root mean square deviations are given in the text. Residue 271 (tyrosine or alanine) is also illustrated. The O2′ of the ribose sugar of rCTP is unusually close (2.54 Å) to the carbonyl backbone of residue 271 in the wild-type structure. The two purple spheres represent the active site Mn2+ ions from the wild-type structure with an incoming rCTP. D, from the position of the primer terminus in the superimposed structures from C, the loss of the hydrogen bond between the side chain of Tyr-271 and the minor groove edge of the base of the primer terminus results in a similar displacement into the major groove as observed with a correct incoming nucleotide. This is most easily seen by looking at the position of N3 of the guanine base in the structures of wild-type and mutant enzymes with an incoming rCTP. E, to compare the relative position of C3′/O3′ of the primer terminus relative to Pα of the incoming nucleotide, the triphosphate portion of wild-type and mutant enzymes was superimposed. The structures with an incoming rCTP do not include O3′ of the primer terminus (i.e. dideoxy-terminated), whereas the wild-type enzyme with an inert dTTP analog includes O3′ of the primer terminus and represents the reference structure for ideal active site geometry. The relative position of the primer termini, as judged by the position of C2′, suggests that the Y271A mutant might achieve a modestly better active site geometry with an incoming rCTP than wild-type enzyme.
Soaking crystals of DNA binary complexes of wild-type or mutant enzymes with rCTP and Mn2+ resulted in closed ternary complexes that superimpose well (root mean square deviations = 0.21 Å for all 326 Cα; supplemental Fig. S2). Comparing the wild-type/mutant enzyme structures with dCTP or rCTP indicates that rCTP binding is well tolerated (root mean square deviation = 0.29 Å for 324 Cα and 0.17 Å for 326 Cα, respectively; see supplemental Fig. S2). Although X-family DNA polymerases sterically exclude insertion of ribonucleotides through backbone interactions near the carboxyl terminus of α-helix M (Fig. 1), few structural adjustments are required to bind rCTP. An omit map illustrates that the density for rCTP bound to wild-type enzyme is clearly observed (Fig. 3B). rCTP binds in a similar conformation to dCTP so that the conformation of the nascent base pair is preserved. The structure of wild-type enzyme with an incoming rCTP positions O2′ within 2.54 Å of the carbonyl of Tyr-271. For the mutant enzyme, the carbonyl rotates away from O2′ an additional ∼0.1 Å compared with the wild type (Fig. 3C).
As with an incoming dCTP, the primer terminus of the mutant enzyme has made minor adjustments in response to rCTP binding (Fig. 3D). The loss of the hydrogen bond by removing the tyrosine side chain results in a 0.8-Å shift in the primer terminus toward the major groove. The sugar of the primer terminus appears to be in a more advantageous position (distance) to attack Pα of the incoming ribonucleotide (Fig. 3E). Because the primer terminus lacks O3′, the precise distance and geometry cannot be assessed from these structures. Additionally, the coordination of the catalytic metal does not exhibit octahedral geometry because of the lack of O3′.
Manganese Increases Efficiency for rCTP Insertion
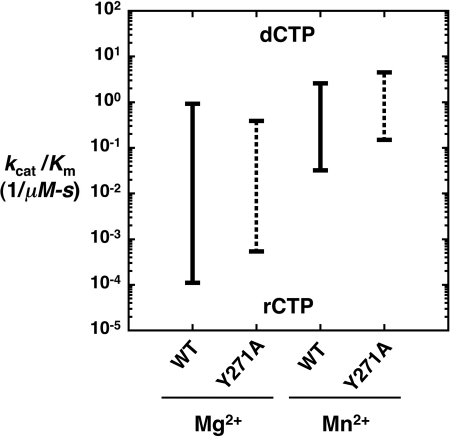
Substituting Mn2+ for Mg2+ enhances nucleotide binding without affecting the rate of correct dNTP insertion (25). For rCTP insertion, replacing Mg2+ with Mn2+ increased catalytic efficiency 290-fold for wild-type enzyme while altering dCTP insertion very little (<3-fold) (Fig. 4). For the mutant enzyme, rCTP and dCTP insertions are increased 280- and 12-fold, respectively, by replacing Mg2+ with Mn2+. Thus, as with dNTP misinsertions, the catalytic efficiency is increased to a much greater extent for a poor nucleotide substrate (wrong sugar or base) than for the correct substrate (right sugar and base) in the presence of Mn2+. This reduces sugar discrimination to 80 and 30 for wild-type and mutant enzymes, respectively; significantly lower than in the presence of Mg2+ (Table 1).
FIGURE 4.
Effect on Mn2+ on ribonucleotide discrimination. A discrimination plot comparing catalytic efficiencies (−) for wild-type and Y271A pol β in the presence of Mg2+ and Mn2+. Nucleotide (dCTP or rCTP) insertion was measured on a single-nucleotide gapped substrate with a templating dG as described under “Experimental Procedures.” Discrimination is proportional to the distance between the two catalytic efficiencies for wild type (solid line) and Y271A (dotted line).
Molecular Modeling
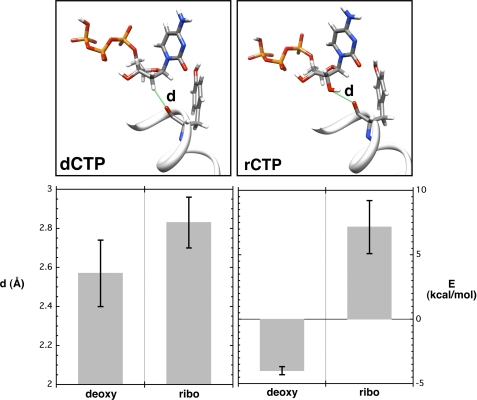
Even in the presence of Mn2+, rCTP binds to pol β more weakly than dCTP. Molecular dynamics simulations were employed to study the interaction of the backbone carbonyl oxygen of Tyr-271 with O2′ of the incoming nucleotide. In the crystallographic structure, the distance between the two oxygens was found to be 2.54 Å. This observation gives rise to the possibility of a significant hydrogen bond between these atoms. However, during the MD simulation, the average distance was found to be significantly larger (∼2.83 Å) than that found in the crystallographic structure (Fig. 3C and Fig. 5). For comparison, we have also illustrated the nearest proton distance to the carbonyl oxygen of Tyr-271 when the incoming nucleotide is the natural deoxynucleotide.
FIGURE 5.
Interaction of the carbonyl of Tyr-271 with an incoming deoxy- or ribonucleotide. MD simulations were performed as described under “Experimental Procedures,” and the final unconstrained runs (10 ns) were carried out with time steps of 1 fs. The upper illustrations highlight the distance (d, green line) of the interacting atoms that are monitored for dCTP (left panel) and rCTP (right panel). A portion of α-helix M with Tyr-271 is also illustrated. The average dCTP(H2′)/Tyr-271(O) and rCTP(O2′)/Tyr-271(O) distances and local interaction energies (E) are plotted below.
The only change in the chemical environment of the carbonyl of Tyr-271 when accommodating a ribonucleotide is the presence of a hydroxyl group in place of a proton bonded to C2′. Although the comparison is somewhat indirect, one can estimate the change in average interaction energies of the carbonyl of Tyr-271 with the constituents at C2′ positions for ribose and deoxyribose environments. To model this local interaction energy, we selected the carbon at 2′ and the two protons attached at this position for the incoming deoxynucleotide. For the incoming ribonucleotide, the C2′ group is bonded to a proton and a hydroxyl group. The sum of the averaged Coulomb and van der Waals interactions (ff99SB force field parameters/AMBER10) between the carbonyl Tyr-271 and the proton at C2′ in the deoxynucleotide is about −3.99 ± 0.32 kcal/mol, and therefore, this simple model indicates that the interaction of the natural incoming deoxynucleotide is energetically favorable. However, for an incoming ribonucleotide, the average local interaction energy between the carbonyl of Tyr-271 and the hydroxyl at C2′ evaluated from a 10-ns simulation is found to be unfavorable (7.16 ± 2.06 kcal/mol). For comparison, a pair of optimally configured water molecules can give rise to −5 kcal/mol (average oxygen distance ∼2.9 Å) (27).
The high resolution substrate structures of pol β indicate that rNTP or dNTP can stably bind to the active site. The MD simulations described above suggest that the dNTP is more stable in the region of C2′/Tyr-271(O) than the rNTP. However, the observed 2.54 Å separation between rCTP(O2′) and Tyr-271(O) suggests that one of the interacting oxygens could be negatively charged. Indeed, increasing the negative charge on the carbonyl oxygen during the MD simulation decreased the distance between rCTP(O2′) and Tyr-271(O) (see supplemental “Results” and supplemental Fig. S3). Thus, if the carbonyl of Tyr-271 is more negative, the hydrogen bond decreases in length in the direction of the experimental observation.
To analyze the direction and degree of charge flow in the ribonucleotide system, two small complexes were constructed to test quantum mechanically (supplemental “Results” and supplemental Fig. S4). In crystallographic structures, Gly-268 and Tyr-271 are near the carboxyl terminus of a short α-helix where the helix dipole points at the carbonyl of Tyr-271. A calculation of atomic charge on the carbonyl oxygen for the two and three body systems shows a small increase (∼0.020e) in the negative charge for the three body system (includes an additional molecule that mimics the Gly-268 backbone interaction) relative to the two body system. Although the right direction, this is not large enough to account for the short rCTP(O2′)/Tyr-271(O) distance observed experimentally.
At this point, the energetic cost of “forcing” rCTP(O2′)/Tyr-271(O) to the experimentally observed distance was estimated. It is possible that favorable interactions elsewhere in the enzyme-substrate complex (e.g. divalent metal and acidic side chains) may force these atoms near one another creating a locally repulsive environment. To quantify the energetic cost, the distance between the two oxygens in our two body model system (methyl alcohol, amide bond blocked with methyl groups) that mimics rCTP(O2′)/Tyr-271(O) was reduced stepwise (supplemental Methods and supplemental Fig. S4) while optimizing all atoms not in this “bond.” When the 2.54-Å distance (the experimental rCTP(O2′)/Tyr-271(O) distance) was reached, the energy cost to force the repulsion was found to be ∼2.2 kcal/mol. This is a relatively small cost to collapse this interaction almost 0.3 Å.
DISCUSSION
Sugar Discrimination
Substrate discrimination represents the ability of the enzyme to choose between alternative substrates and is calculated from the ratio of catalytic efficiencies (right/wrong). Accordingly, changes in discrimination as the result of modifying the conditions of the assay (e.g. metal substitution) or the enzyme (e.g. site-directed mutagenesis) occur through a change in efficiency for one or the other substrate. Thus, a change in discrimination is the result of a differential effect of some factor on two catalytic efficiencies, and both need to be considered in the interpretation. The plots illustrated in Figs. 2 and 4 provide a straightforward graphical approach that reveals the source of a change in discrimination (24). When base substitution discrimination among various low and high fidelity DNA polymerases is considered, fidelity is nearly completely determined by the efficiency of correct nucleotide insertion, i.e. incorrect insertion efficiency is independent of the identity of the polymerase (28). Likewise, ribonucleotide sugar discrimination is also primarily determined by how efficiently the polymerase utilizes deoxynucleotide (correct) insertion because ribonucleotide insertion efficiency is only weakly dependent on the identity of the DNA polymerase (5).
Ribonucleotide Discrimination by DNA Polymerase β
Wild-type pol β inserts ribonucleotides with an efficiency comparable with other polymerases that have been examined (5). We have demonstrated how ribonucleotide discrimination by pol β is diminished through alanine substitution for Tyr-271 (Fig. 2) and by substituting Mn2+ for Mg2+ (Fig. 4). Ribonucleotide discrimination is specific to Tyr-271 because phenylalanine substitution (i.e. Y271F) only partially recovers sugar fidelity, and alanine substitution of the neighboring residue (i.e. F272A, which is also within proximity of the incoming nucleotide sugar) has little effect on discrimination (Table 1).
The ternary substrate complexes of wild-type and Y271A pol β with an incoming rCTP indicated that the incoming nucleotide is well accommodated in the nascent base pair binding pocket of pol β. Similarly, Kirouac et al. (29) reported that rADP and dADP bound similarly in the Y12A mutant of Dpo4 DNA polymerase (Y-family). Attempts to soak rCTP into crystals of pol β binary-DNA complexes failed to form ternary complex structures in the presence of Mg2+. Substituting Mn2+ for Mg2+ increased the binding affinity for rCTP over 200-fold (Kd ∼400 and 1.7 μm for Mg2+ and Mn2+, respectively). Like incorrect deoxynucleotide insertion (25), Mn2+ increases ribonucleotide insertion to a greater degree than correct insertion thereby decreasing sugar discrimination (Fig. 4).
Comparing the crystallographic structure of wild-type enzyme with bound ddCTP (PDB code 2FMP) or rCTP and that of the Y271A mutant with rCTP, Tyr-271 appears to play two significant roles. The backbone carbonyl at Tyr-271 gets unfavorably close to 2′-OH of the ribose in the wild-type structure, and the structure of the mutant indicates that this carbonyl attempts to move further from O2′ of the incoming ribonucleotide. However, because the carbonyl of residue-271 is situated in an α-helix and hydrogen bonds with Gly-274(N) and Asn-279(ND2), it can only make minor adjustments even though the large aromatic side chain has been replaced with a small methyl group. Likewise, other residues of α-helix M, including Phe-272, would be expected to preserve the overall backbone architecture because the backbone hydrogen bonding pattern would not be affected by alanine substitution for Tyr-271. More importantly, the loss of the side chain at Tyr-271 removes a hydrogen bond between the tyrosine hydroxyl group and the minor groove edge of the terminal primer base in the closed ternary substrate conformation. In the wild-type structure, this hydrogen bond may modulate active site geometry thereby deterring insertion of substrates with the wrong sugar and/or wrong base. Thus, ribonucleotide discrimination by pol β is attributed to the loss of a contact with the primer terminus as well as a steric clash with the protein backbone carbonyl.
The polymerase is in a closed conformation like that observed for correct and incorrect nucleotide insertion (14, 25, 26). However, in contrast to the structures with an incorrect incoming nucleotide where the templating base evacuated its coding position (25), the cytosine base of the ribonucleotide forms Watson-Crick hydrogen bonds with the templating deoxyguanine. The sugar pucker of the incoming nucleotide is C3′-endo, like that observed for the natural incoming nucleotide (25). Surprisingly, O2′ of the incoming ribonucleotide is within 2.54 Å of the Tyr-271 carbonyl (Fig. 3C), and replacing the tyrosine side chain with alanine only results in ∼0.1-Å increase in this distance.
Structure-based MD simulations indicate that the distance between O2′ of the incoming ribonucleotide and the backbone carbonyl of Tyr-271 would be greater than 2.8 Å, which is significantly larger than the observed (2.54 Å; Figs. 3C and 5). The estimated interaction energies from a qualitative perspective are what might be expected; the binding of deoxynucleotides is favorable although binding ribonucleotides is unfavorable. In an attempt to probe the molecular origin for the close juxtaposition of O2′ of the incoming ribonucleotide and the carbonyl of Tyr-271, the charge on the carbonyl was increased to −1.0e (supplemental “Results”). With this enhanced charge, MD simulation decreased the distance between these oxygens to 2.58 Å, more in line with experimental observations, but an enhanced interaction energy (−8.69 ± 1.92 kcal/mol) is not consistent with the lower binding affinity of a ribonucleotide (Table 1).
To follow the degree and direction of charge flow in our system, we performed quantum calculations. The results indicate that although Tyr-271 is near the carboxyl terminus of α-helix M, the helix dipole would only decrease the charge (i.e. make more negative) on the carbonyl and the distances between the two oxygens to a small extent, not enough to account for the experimentally observed distance. In an attempt to probe the local energetic cost of bringing the carbonyl of Tyr-271 within 2.54 Å of O2′ of an incoming nucleotide, a two body system (methyl alcohol and amide bond blocked with two methyls) was used to calculate the repulsive cost as the distance between the two oxygens of interest was reduced to the experimentally observed distance. Surprisingly, the cost was only ∼2.2 kcal/mol, which is similar to the observed loss in binding affinity when wild-type enzyme binds ribonucleotides in the presence of Mg2+ (i.e. Kd, rCTP/Kd, dCTP = 40). Thus, it appears that favorable interactions in one region of an overall stable system can reasonably force smaller repulsive interactions in adjacent regions. In this view, the ribose position as seen in the x-ray crystal structure is not unreasonable.
Steric Gates
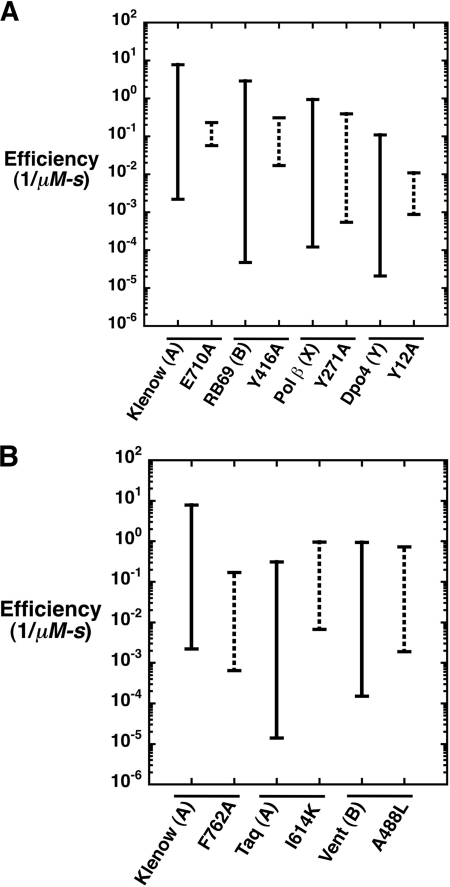
Ribonucleotide discrimination has been examined for several DNA polymerases across families by removing a side chain that would be predicted to sterically interfere with 2′-OH of the incoming rNTP. The residues that contribute these side chains have been referred to as “steric gates” (9, 10). For A-family polymerases, this residue is a glutamate, although for B-, Y-, and RT-families, a tyrosine or phenylalanine residue provides the steric clash with O2′ of an incoming nucleotide. A discrimination plot illustrating sugar discrimination for wild-type and mutant polymerases indicates that the loss of discrimination is due to an increase in the insertion efficiency of the ribonucleotide and often due to a loss in the insertion efficiency for the deoxynucleotide (Fig. 6A). Thus, the mutation is affecting both pathways, correct and incorrect sugar selection.
FIGURE 6.
Comparing ribonucleotide discrimination for wild-type and mutant DNA polymerases from different families. Ribonucleotide discrimination has been typically attributed to a steric gate residue in the active site (10). Discrimination plots compare catalytic efficiencies (−) for dCTP and rCTP insertion by wild-type (solid line) and mutant DNA polymerases (dotted line) from A-, B-, X-, and Y-family DNA polymerases (5, 33, 44–46). Ribonucleotide discrimination is proportional to the distance between the catalytic efficiencies for dCTP and rCTP. A, ribonucleotide discrimination of wild-type and mutant DNA polymerases when the steric gate or fence residue is removed. B, ribonucleotide discrimination of wild-type and mutant DNA polymerases when a nonsteric gate residue is altered.
Not surprisingly, removing the steric gate residue with the A-, B-, and Y-family polymerases decreases ribonucleotide discrimination (2–3 orders of magnitude). This is significantly greater than observed with the Y271A mutant of pol β. An analogous change in pol λ, Y505A, likewise resulted in a modest decrease in ribonucleotide discrimination (11). In these two cases, insertion of the correct deoxynucleotide was hardly affected by mutation; however, mutation in A- and B-family polymerases affected dNTP insertion (Fig. 6A). As suggested recently (9) and demonstrated here, the backbone carbonyl of Tyr-271 of pol β, rather than the aromatic side chain, deters ribonucleotide insertion. Thus, removing the Tyr-271 side chain only permits a small adjustment in the protein backbone. In this situation, Tyr-271 behaves more as a steric “fence” than a “gate.” In contrast, pol μ and terminal deoxynucleotidyltransferase, also X-family members, have a glycine at this position (8). Interestingly, the sugar specificities for the pairs of enzymes correlate to their amino acid composition, i.e. pol β and pol λ strongly discriminate against ribonucleotides (Table 1) (11), whereas pol μ and terminal deoxynucleotidyltransferase insert dNTPs and rNTPs with similar efficiencies (30, 31). Because the efficiency of ribonucleotide insertion is similar for all four enzymes, the loss in sugar discrimination is due to the poor dNTP insertion efficiency exhibited by enzymes that possess a glycine, rather than tyrosine, at this position. This was clearly demonstrated by the Y505G mutant of pol λ, which exhibited higher sugar discrimination than the Y505A mutant and only modestly decreased dNTP insertion (11). Taken together, these observations indicate that the contribution of each active site residue to overall dNTP or rNTP insertion efficiency is unique to each polymerase and is determined by more than a single amino acid side chain.
DNA polymerase sugar discrimination can also be modulated by residues that would not be predicted to sterically exclude O2′ of an incoming nucleotide (Fig. 6B). For example, random mutagenesis of Thermus aquaticus DNA polymerase I identified a single amino acid change (I614K) that resulted in a 160-fold decrease in dCTP/rCTP discrimination (32). As illustrated in Fig. 6B, the loss in discrimination is entirely due to an increase in the efficiency of ribonucleotide insertion. The backbone of Ile-614 forms a hydrogen bond with a nonbridging oxygen on Pβ of the incoming nucleotide. In this case, the mutant enzyme (I614K) inserts rCTP 80-fold more efficiently than wild-type enzyme. Similarly, the F762A mutant of Klenow fragment also exhibited a loss of sugar discrimination (33). In this case, however, it is entirely due to a loss of efficiency for insertion of dCTP (Fig. 6B). Insertion efficiency of rCTP for the mutant enzyme is lower than that for wild-type enzyme. From the ternary complex structure of the related Bacillus polymerase I (34), this aromatic residue is believed to stack with the base of the incoming nucleotide.
RNA polymerases appear to use an alternate mechanism to discourage dNTP insertion. Crystallographic structures of RNA polymerase from prokaryotes and eukaryotes suggest that sugar selection is, in part, due to a hydrogen bond that could form between O2′ of the incoming nucleotide and a polymerase side chain (35, 36). Significantly, DNA and RNA polymerases bind ribonucleotides with comparable affinities (Kd) (37), which is probably a reflection of the higher concentration of rNTPs in the cell. Thus, DNA polymerases sterically deter misinsertion of ribonucleotides, whereas RNA polymerases hasten insertion of ribonucleotides by providing favorable interactions that would promote binding. Both approaches lead to decreased binding and insertion of the nucleotide with the wrong sugar. As observed with DNA polymerases, conformational changes occurring in the vicinity of the incoming ribonucleotide of yeast RNA polymerase II have also been suggested to play a role in sugar selection (38).
Biological Implications
When the cellular imbalance in rNTP and dNTP concentrations is considered, replicative and DNA repair polymerases are predicted to insert a large numbers of ribonucleotides (5, 6). The dNTP binding affinity for many DNA polymerases is similar to the concentration found in cells (3, 39). In contrast, the concentration of ribonucleotides is ∼40–350-fold higher in cycling cells (4). Not surprisingly, however, the apparent binding affinity of RNA polymerase for rNTPs is considerably higher than the binding affinities of DNA polymerases for dNTPs (37). Critically however, the low binding affinity that DNA polymerases exhibit for ribonucleotides is still in a range that would be predicted to significantly compete with their deoxy-counterparts. Although they may not be incorporated, they would impact the rate of polymerization. Because dNTPs bind weakly to RNA polymerase and their cellular concentration is low, they would exert less of an impact on RNA polymerization.
Overall, polymerase sugar specificity is determined by its intrinsic sugar specificity as determined by the ratio of specificity constants, (right + wrong)/wrong, as well as the relative concentration of competing substrates (ribonucleotides and deoxynucleotides). For wild-type pol β, its intrinsic specificity is ∼8,400, but the overall specificity is reduced to 51 when the cellular concentration of dCTP and rCTP in nondividing cells is considered (3). When this is calculated for the Y271A mutant utilizing Mn2+, the overall specificity is <2 indicating that rCTP would be preferentially inserted relative to dCTP. Significantly, X-family DNA polymerases that exhibit low ribonucleotide discrimination (e.g. pol μ and terminal deoxynucleotidyltransferase) would be predicted to insert more ribonucleotides than deoxynucleotides when the dNTP/rNTP imbalance is taken into consideration.
The impact of ribonucleotides in the genome is uncertain. It is recognized that genomic ribonucleotides will lead to instability in the nucleic acid backbone (7) and that in some instances a single ribonucleotide in DNA could lead to a B- to A-form conformational transition (40, 41). Because site-specific nucleic acid-binding proteins rely on specific conformations, ribonucleotide contamination of the genome would be expected to have far reaching consequences. Recent evidence suggests that failure to remove ribonucleotides inserted during replication can lead to replication stress and genome instability (42).
Supplementary Material
Acknowledgments
We thank Drs. R. E. London and B. D. Freudenthal for critical reading of this manuscript and E. W. Hou for purification of the F272A mutant. Molecular graphics images were produced using the Chimera package (20) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
This work was supported, in whole or in part, by National Institutes of Health Research Project Grants Z01-ES0500158 and Z01-ES050161 (to S. H. W.) from the Intramural Research Program, NIEHS, in association with Grant 1U19CA105010.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, “Results,” Tables S1 and S2, Figs. S1–S4, and an additional reference.
The atomic coordinates and structure factors (codes 3RH4, 3RH5, and 3RH6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
When the rate of nucleotide insertion is slow (i.e. rate limiting), then Km,N is equivalent to Kd,N, where N refers to nucleotide, rNTP, or dNTP (43).
The insertion efficiencies for rGTP were 4.3 × 10−5 and 45 × 10−5 s−1 μm−1 for wild-type and Y271A enzymes, respectively. This corresponds to discrimination factors (dGTP/rGTP) of 30,000 and 1,900, respectively, which is an ∼15-fold loss in discrimination with the Y271A mutant.
- pol
- DNA polymerase
- rNTP
- ribonucleoside triphosphate
- araCTP
- arabinofuranosylcytosine triphosphate
- MD
- molecular dynamics
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank.
REFERENCES
- 1. Tsai Y. C., Johnson K. A. (2006) Biochemistry 45, 9675–9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Batra V. K., Beard W. A., Hou E. W., Pedersen L. C., Prasad R., Wilson S. H. (2010) Nat. Struct. Mol. Biol. 17, 889–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V. (2010) Nucleic Acids Res. 38, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Traut T. W. (1994) Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 5. Cavanaugh N. A., Beard W. A., Wilson S. H. (2010) J. Biol. Chem. 285, 24457–24465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 8. Moon A. F., Garcia-Diaz M., Batra V. K., Beard W. A., Bebenek K., Kunkel T. A., Wilson S. H., Pedersen L. C. (2007) DNA Repair 6, 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown J. A., Suo Z. (2011) Biochemistry 50, 1135–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joyce C. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1619–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown J. A., Fiala K. A., Fowler J. D., Sherrer S. M., Newmister S. A., Duym W. W., Suo Z. (2010) J. Mol. Biol. 395, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. (1994) Science 264, 1891–1903 [PubMed] [Google Scholar]
- 13. Beard W. A., Osheroff W. P., Prasad R., Sawaya M. R., Jaju M., Wood T. G., Kraut J., Kunkel T. A., Wilson S. H. (1996) J. Biol. Chem. 271, 12141–12144 [DOI] [PubMed] [Google Scholar]
- 14. Sawaya M. R., Prasad R., Wilson S. H., Kraut J., Pelletier H. (1997) Biochemistry 36, 11205–11215 [DOI] [PubMed] [Google Scholar]
- 15. Beard W. A., Wilson S. H. (1995) Methods Enzymol. 262, 98–107 [DOI] [PubMed] [Google Scholar]
- 16. Beard W. A., Shock D. D., Batra V. K., Pedersen L. C., Wilson S. H. (2009) J. Biol. Chem. 284, 31680–31689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 20. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 21. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 22. Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Crowley M. F., Walker R. C., Zhang W., Merz K. M., Wang B., Hayik S., Roitberg G., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wu X., Brozell S., Steinbrecher T., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Mathews D. H., Seetin M. G., Sagui C., Babin V., Kollman P. A. (2008) AMBER 10, University of California, San Francisco [Google Scholar]
- 23. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A., Jr., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam N. J., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J. (2009) Gaussian 09, Revision A.1, Gaussian, Inc., Wallingford, CT [Google Scholar]
- 24. Beard W. A., Batra V. K., Wilson S. H. (2010) Mutat. Res. 703, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. (2008) Mol. Cell 30, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., Wilson S. H. (2006) Structure 14, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schütz M., Brdarski S., Widmark P. O., Lindh R., Karlstrom G. (1997) J. Chem. Phys. 107, 4597–4605 [Google Scholar]
- 28. Beard W. A., Shock D. D., Yang X. P., DeLauder S. F., Wilson S. H. (2002) J. Biol. Chem. 277, 8235–8242 [DOI] [PubMed] [Google Scholar]
- 29. Kirouac K. N., Suo Z., Ling H. (2011) J. Mol. Biol. 407, 382–390 [DOI] [PubMed] [Google Scholar]
- 30. Nick McElhinny S. A., Ramsden D. A. (2003) Mol. Cell. Biol. 23, 2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruiz J. F., Juárez R., García-Díaz M., Terrados G., Picher A. J., González-Barrera S., Fernández de Henestrosa A. R., Blanco L. (2003) Nucleic Acids Res. 31, 4441–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patel P. H., Loeb L. A. (2000) J. Biol. Chem. 275, 40266–40272 [DOI] [PubMed] [Google Scholar]
- 33. Astatke M., Ng K., Grindley N. D., Joyce C. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3402–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson S. J., Taylor J. S., Beese L. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vassylyev D. G., Vassylyeva M. N., Zhang J., Palangat M., Artsimovitch I., Landick R. (2007) Nature 448, 163–168 [DOI] [PubMed] [Google Scholar]
- 36. Wang D., Bushnell D. A., Westover K. D., Kaplan C. D., Kornberg R. D. (2006) Cell 127, 941–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C. E. (2009) Nat. Struct. Mol. Biol. 16, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaplan C. D., Larsson K. M., Kornberg R. D. (2008) Mol. Cell 30, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beard W. A., Wilson S. H. (2003) Structure 11, 489–496 [DOI] [PubMed] [Google Scholar]
- 40. Ban C., Ramakrishnan B., Sundaralingam M. (1994) J. Mol. Biol. 236, 275–285 [DOI] [PubMed] [Google Scholar]
- 41. Wahl M. C., Sundaralingam M. (2000) Nucleic Acids Res. 28, 4356–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nick McElhinny S. A., Kumar D., Clark A. B., Watt D. L., Watts B. E., Lundström E. B., Johansson E., Chabes A., Kunkel T. A. (2010) Nat. Chem. Biol. 6, 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beard W. A., Shock D. D., Wilson S. H. (2004) J. Biol. Chem. 279, 31921–31929 [DOI] [PubMed] [Google Scholar]
- 44. Gardner A. F., Joyce C. M., Jack W. E. (2004) J. Biol. Chem. 279, 11834–11842 [DOI] [PubMed] [Google Scholar]
- 45. Sherrer S. M., Beyer D. C., Xia C. X., Fowler J. D., Suo Z. (2010) Biochemistry 49, 10179–10186 [DOI] [PubMed] [Google Scholar]
- 46. Yang G., Franklin M., Li J., Lin T. C., Konigsberg W. (2002) Biochemistry 41, 10256–10261 [DOI] [PubMed] [Google Scholar]
- 47. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.