Abstract
Aminoacyl-tRNA synthetases attach specific amino acids to cognate tRNAs. Prolyl-tRNA synthetases are known to mischarge tRNAPro with the smaller amino acid alanine and with cysteine, which is the same size as proline. Quality control in proline codon translation is partly ensured by an editing domain (INS) present in most bacterial prolyl-tRNA synthetases that hydrolyzes smaller Ala-tRNAPro and excludes Pro-tRNAPro. In contrast, Cys-tRNAPro is cleared by a freestanding INS domain homolog, YbaK. Here, we have investigated the molecular mechanism of catalysis and substrate recognition by Hemophilus influenzae YbaK using site-directed mutagenesis, enzymatic assays of isosteric substrates and functional group analogs, and computational modeling. These studies together with mass spectrometric characterization of the YbaK-catalyzed reaction products support a novel substrate-assisted mechanism of Cys-tRNAPro deacylation that prevents nonspecific Pro-tRNAPro hydrolysis. Collectively, we propose that the INS and YbaK domains co-evolved distinct mechanisms involving steric exclusion and thiol-specific chemistry, respectively, to ensure accurate decoding of proline codons.
Keywords: Aminoacyl tRNA Synthetase, Enzyme Mechanisms, Molecular Dynamics, Transfer RNA (tRNA), Translation Control, Editing, Translational Fidelity
Introduction
Aminoacyl-tRNA synthetases (aaRSs)3 play a pivotal role in the decoding of genetic information by catalyzing the esterification of cognate amino acids onto specific transfer RNAs (tRNAs) in a two-step reaction (1). The first step involves amino acid activation with ATP to form an aminoacyl-adenylate intermediate and concomitant pyrophosphate release. The second step involves aminoacyl transfer to the cognate tRNA via a transesterification reaction. Although aaRSs display high substrate specificity, they often misactivate structurally similar amino acids. If left unchecked, these mistakes lead to errors in protein synthesis, and accumulation of such errors can be deleterious to cells (2, 3).
To ensure high fidelity in translation, aaRSs have evolved several proofreading mechanisms (4–7). In the first type of proofreading, misactivated aminoacyl-adenylates are enzymatically hydrolyzed or selectively released from the active site followed by solvent hydrolysis, in a process termed “pretransfer” editing. Another mechanism of proofreading known as “post-transfer” editing is generally believed to function via the so-called “double-sieve” mechanism (8). The model predicted that, whereas one active site could not completely discriminate Ile and Val, two separate active sites with distinct strategies for recognition could significantly enhance fidelity. The aminoacylation active site of the aaRS would act as a “coarse” sieve for adenylate synthesis, activating the cognate amino acid but also allowing, to a lesser extent, activation of isosteric or smaller amino acids that could fit into the amino acid binding pocket. The second “fine” sieve would selectively bind misactivated amino acids for editing while excluding the original cognate amino acid. Thus, substrates synthesized in the first sieve would be further screened by the second sieve to enhance fidelity. Subsequently, biochemical and structural data validated this steric exclusion mechanism for valyl-tRNA synthetase and isoleucyl-tRNA synthetase (9, 10). Indeed, members of both classes of synthetases are now known to possess a second active site spatially separated from the ancient catalytic core to carry out post-transfer editing. Structures of class I isoleucyl-tRNA synthetase (10), leucyl-tRNA synthetase (11), and valyl-tRNA synthetase (12) reveal the highly conserved connective peptide 1 domain that functions in editing. Class II alanyl-tRNA synthetase (AlaRS) (13, 14), threonyl-tRNA synthetase (15), prolyl-tRNA synthetase (ProRS) (16, 17), and phenylalanyl-tRNA synthetase (18, 19) also possess distinct domains for post-transfer editing. A recent study of AlaRS revealed an exception to the size exclusion-based double-sieve model of editing. In this case, a coordinated zinc ion in the editing site of AlaRS may help provide specificity for non-cognate serine, which is larger than cognate alanine (20, 21). Insights into the relative contributions of pre- and post-transfer editing have also begun to emerge in several Class I and Class II synthetase systems (22–26).
Due to its unique side chain with special chemical properties, cysteine should be relatively easy for synthetases to discriminate. Similarly, proline is the only imino acid and would not be predicted to be especially problematic. Paradoxically, ProRSs from all domains of life confuse cysteine for proline (27, 28). ProRSs also misactivate alanine, and the majority of bacterial ProRSs possess an editing domain (INS) that can edit mischarged Ala-tRNAPro but not Cys-tRNAPro (17, 29). We imagine that the smaller size of alanine relative to proline facilitates binding and catalysis by INS, whereas the editing domain rejects cysteine, which is very similar to proline in molecular volume (30). Consistent with this idea, the Methanobacterium thermoautotrophicus (31) and Rhodopseudomonas palustris (16) ProRS structures solved in complex with Cys- and Pro-sulfamoyl-adenylates showed that the aminoacylation active site of ProRS could accommodate both adenylates in a very similar manner. Thus, it is likely that a distinct post-transfer editing mechanism that does not rely on steric exclusion is needed to clear mischarged Cys-tRNAPro. Indeed, the latter is hydrolyzed by a freestanding domain known as YbaK, which is proposed to function in collaboration with ProRS in trans (29, 32, 33).
YbaK belongs to a larger protein superfamily that is widely distributed among all three kingdoms. Members of the YbaK superfamily share significant sequence and structural homology with the INS domain of bacterial ProRS (16, 34–36). Interestingly, in contrast to YbaK, the freestanding PrdX domain within the YbaK superfamily possesses the same substrate specificity for Ala-tRNAPro as the INS domain (37). Freestanding editing domains have also been identified based on homology to the AlaRS and threonyl-tRNA synthetase editing domains. AlaXs generally display the same Ser- and Gly-tRNAAla editing specificity as the AlaRS protein (20, 37–39), and ThrX possesses Ser-tRNAThr specificity similar to threonyl-tRNA synthetase (40). Thus, to date, YbaK is the only known editing domain homolog with distinct substrate specificity relative to the homologous synthetase domain.
Although the phenomenon of post-transfer editing in aaRSs is well established, relatively little is known about the detailed hydrolysis mechanism of freestanding editing proteins like YbaK at the molecular level (6, 41–44). We were especially interested in understanding the basis for the unique Cys-tRNA specificity of YbaK and how discrimination of similar sized Pro-tRNA is achieved. The x-ray crystal structures of several members of the YbaK superfamily (PrdX, ProX, YbaK, and ProRS INS) from a variety of organisms (Protein Data Bank codes 2DXA, 1DBX, 1VJF, 1WDV, 1VKI, 2CX5, 2ZOX, 2ZOK and 2J3L) have been solved (16, 35, 36). However, no structures of these proteins bound to post-transfer editing substrates are available to date. To understand the chemical basis of the distinct substrate specificities of these homologous editing domains, we investigated the mechanism of YbaK hydrolysis. Collectively, our experimental and computational data support a mechanism of catalysis that exploits the special side chain chemistry of cysteine.
EXPERIMENTAL PROCEDURES
Materials
All amino acids and chemicals were purchased from Sigma unless otherwise noted. [3H]Alanine (54 Ci/mmol), [3H]proline (99 Ci/mmol), [3H]serine (33 Ci/mmol), and α-[32P]ATP were from Amersham Biosciences, and [35S]cysteine (1075 Ci/mmol) was from PerkinElmer Life Sciences.
Multiple-sequence Alignments
Multiple-sequence alignments were performed using the ClustalW multiple-sequence realignment program (45).
Molecular Modeling of CCA-Cys Bound to H. influenzae YbaK
The crystal structure of monomeric H. influenzae YbaK was used as the starting structure (Protein Data Bank entry 1DBX) (35). Missing residues 25–29 (NNQHF) in the flexible loop region were added using the template-based loop structure prediction server ArchPRED (46). The protonation states of the residues were calculated by PropKa (47). To relax the resulting structure and to sample the flexibility of the protein, 15 ns of molecular dynamics (MD) simulation was performed in explicit solvent (TIP3P) (48) using AMBER 9 (49). Twenty-five snapshots of the protein structure from the resulting MD trajectory were extracted at equal time intervals and used for molecular docking. The structure of the 5′-CCA-Cys ligand was generated using the xleap module of AMBER 9. This ligand was docked onto the 25 structures of YbaK using AutoDock 4.0 (50). All of the ligand torsions were kept flexible, whereas the protein torsions were fixed. Each docking simulation involved generation of 200 different conformers, which were then clustered using a root mean square deviation cut-off of 2.0 Å. Resulting clusters were manually inspected, and the five docked structures most consistent with available mutagenesis data were chosen for further analysis. These structures were minimized using the ff99 force field in AMBER 9, followed by 15 ns of MD simulation. The complex structure displaying the most stable trajectory was selected for further analysis. The final complex structure was subjected to 15 ns of MD to obtain time-averaged distance information and identify long lived H-bonds. These analyses were performed using the ptraj module in AMBER 9.
Protein and tRNA Preparation
All of the proteins and tRNA substrates were prepared using published protocols as described in the supplemental Methods.
Aminoacylation Assays
To test the ability of YbaK to prevent mischarging of β-aminoalanine (β-Aa) onto tRNAPro, 1.5 μm editing-defective Escherichia coli K279A ProRS was incubated with 10 μm 3′-32P-labeled tRNAPro and 150 mm β-Aa in 50 mm HEPES (pH 7.5), 4 mm ATP, 20 mm KCl, 50 mm dithiothreitol, 25 mm MgCl2, and 0.1 mg/ml BSA at 37 °C, in the absence or presence of 6 μm YbaK. For each time point, 1.5 μl of reaction mixture was quenched into 4.5 μl of 200 mm NaOAc containing 0.4 unit/μl P1 nuclease at 4 °C. After digestion for 20 min at room temperature, 1 μl of quenched mixture was spotted onto a polyethyleneimine-cellulose TLC plate prerun with water. Separation of β-Aa-AMP and AMP was accomplished by developing the TLC in 0.1 m ammonium acetate and 5% acetic acid. The radioactivity was analyzed by using a Typhoon PhosphorImager, and data were analyzed using Bio-Rad Quantity One software. The fraction of free tRNA was determined from the ratio of AMP formed over the total AMP plus β-Aa-AMP.
Preparation of aminoacyl-tRNAs for use in deacylation assays was carried out as described (29). Briefly, [3H]Ser-tRNAPro was prepared by incubating 2 μm E. coli AlaRS-CQ and 8 μm E. coli G1:C72/U70 tRNAPro in a reaction mixture containing [3H]serine (9.1 μm) and unlabeled serine (0.5 mm) in 50 mm HEPES (pH 7.5), 4 mm ATP, 20 mm KCl, 20 mm β-mercaptoethanol, 25 mm MgCl2, and 0.1 mg/ml BSA. [3H]Ala-tRNAPro was prepared using E. coli WT AlaRS and G1:C72/U70 tRNAPro in a similar manner. [35S]Cys-tRNAPro was prepared by incubating E. coli ProRS (8 μm), E. coli tRNAPro (8 μm) with [35S]cysteine (0.9 μm) and unlabeled cysteine (50 μm) in a reaction buffer containing 20 mm Tris-HCl (pH 7.5), 20 mm KCl, 10 mm MgCl2, 25 mm dithiothreitol, and 2 mm ATP as described previously. [35S]Cys-tRNACys was prepared by incubating E. coli cysteinyl-tRNA synthetase (CysRS) (30 nm) and E. coli tRNACys (8 μm) in the same reaction buffer. 3′-End-modified (2′-dA76 or 3′-dA76) E. coli tRNACys was charged with [35S]cysteine in a similar manner, except a higher concentration of E. coli CysRS (5 μm) was used. Sec-3′-[32P]tRNACys was prepared by incubating E. coli CysRS (2 μm) and E. coli tRNACys (10 μm) with freshly prepared 50 mm selenocysteine in a reaction buffer containing 20 mm Tris-HCl (pH 8.0), 20 mm KCl, 10 mm MgCl2, 0.1 mg/ml BSA, 2 mm ATP, and 50 mm tris-(2-carboxyethyl)phosphine in 0.1 m Tris-HCl buffer (pH 9.0) at 37 °C for 1 h. A fresh solution of 140 mm selenocysteine was prepared immediately prior to use by dissolving seleno-l-cystine (Fluka) (11.7 mg, 0.035 mmol) in ∼40 μl of 0.2 n HCl. The diselenide bond was reduced in situ by the addition of an equal volume of 0.2 m tris-(2-carboxyethyl)phosphine in 0.1 m Tris-HCl (pH 9.0) (51). The final volume (500 μl) was made up with 0.1 m tris-(2-carboxyethyl)phosphine in 0.1 m Tris-HCl (pH 9.0), and the pH of the resulting solution was adjusted to ∼7.5–8.0 with a few drops of 14 n NaOH. During ethanol precipitation of the Sec-tRNA, 50 μl of 0.1 m tris-(2-carboxyethyl)phosphine solution was added to prevent auto-oxidation. 2-aminobutyric acid (2-Abu)-3′-[32P]tRNAPro was prepared by incubating 1.5 μm E. coli K279A ProRS with 10 μm 3′-32P-labeled tRNAPro and 150 mm 2-Abu in 50 mm HEPES (pH 7.5), 4 mm ATP, 20 mm KCl, 50 mm dithiothreitol, 25 mm MgCl2, and 0.1 mg/ml BSA at 37 °C.
Deacylation Assays
Deacylation assays were generally carried out at 37 °C according to published protocols. Reactions contained ∼1.0 μm G1:C72/U70 [3H]Ala-tRNAPro, 1.0 μm G1:C72/U70 [3H]Ser-tRNAPro, 0.5 μm [35S]Cys-tRNAPro, 1.0 μm [3H]Pro-tRNAPro, 0.7 μm [35S]Cys-2′-dA76-tRNACys, and 0.5 μm [35S]Cys-3′-dA76-tRNACys. Reactions were initiated with 0.5–1.0 μm WT or mutant YbaK, quenched, and worked up as described (29). A background reaction was carried out in which buffer (0.15 m KPO4, pH 7.0) was used to initiate the reaction for each substrate. For competition assays, various concentrations of G1:C72/U70 Ser-tRNAPro were mixed with 0.5 μm [35S]Cys-tRNAPro, and reactions were initiated by the addition of 0.1 μm YbaK. In other competition assays, Sec-tRNACys, Cys-2′-dA76-tRNACys, or Cys-tRNACys (∼6–8 μm each) were mixed with 0.5 μm [35S]Cys-tRNACys, and reactions were initiated by the addition of 0.5 μm YbaK. 2-Abu-[32P]tRNAPro, Sec-[32P]tRNACys, and Cys-[32P]tRNACys deacylation assays contained a 1 μm concentration of each tRNA and 0.5–5 μm YbaK. Reactions were quenched by adding 0.4 unit/μl P1 nuclease in 200 mm NaOAc, and the deacylation level was analyzed using polyethyleneimine-cellulose TLC, as described previously (52). The fraction of aminoacyl-tRNA remaining was plotted as a function of time and fitted to a single-exponential equation to obtain kobs. All assays were carried out in triplicate. In some cases, analysis of reaction products was performed by electrospray ionization mass spectrometry (ESI-MS) and LC/MS as described in the supplemental Methods.
RESULTS
Sequence Analysis of the YbaK Superfamily
Freestanding domains with homology to the ProRS editing domain have been identified across a variety of species and can be grouped together to constitute a “YbaK superfamily.” In addition to the INS domain of bacterial ProRS, phylogenetic analysis using available sequences in the Protein Data Bank indicates that members of this superfamily include at least five distinct freestanding protein domains designated YbaK, YeaK, ProX, PrdX, and PA2301. Despite their relatively low sequence identity, these proteins possess high structural homology (16, 34, 35). Interestingly, members of the YbaK superfamily investigated to date show distinct substrate specificities. Whereas the INS domain specifically hydrolyzes Ala-tRNAPro, bacterial YbaK possesses post-transfer editing activity against Cys-tRNA in vitro and in vivo (29, 32). In contrast, Clostridium sticklandii PrdX has been reported to display Ala-tRNAPro deacylation specificity (37). The chemical basis for the altered substrate specificity within the YbaK superfamily is not understood.
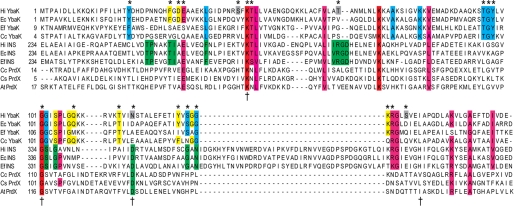
To begin to address this question, a multiple-sequence alignment of YbaK superfamily members with known function, including YbaK, the INS domain of ProRS, and PrdX, was performed using the ClustalW multiple-sequence realignment program (Fig. 1). A separate alignment of the 105-member YbaK subfamily was also carried out (supplemental Fig. 1). As noted previously (35), a Lys residue (Lys-46 in H. influenzae YbaK) and a GXXXP motif are almost universally conserved in the YbaK superfamily (Fig. 1). The GXXXP motif, which resembles a common sequence element in serine proteases (35) and d-Tyr-tRNATyr deacylases (53), forms part of a flexible loop in the x-ray crystal structures of Enterococcus faecalis ProRS (16) and H. influenzae YbaK (35). In addition, several other highly conserved residues in the YbaK subfamily include surface-exposed lysines and glycines in the flexible random coil regions of YbaK.
FIGURE 1.
Multiple-sequence alignment of YbaK superfamily. Multiple-sequence alignment of members of the YbaK superfamily performed using the ClustalW multiple-sequence realignment program (45). The alignment of YbaK, INS domain of prokaryotic-like ProRS, and PrdX is shown. Functionally important residues in E. coli ProRS for editing are indicated by a dagger (42). The residues in H. influenzae YbaK investigated by mutagenesis in this study are indicated by an asterisk. Residues are colored as follows. Red, strictly conserved in YbaK, INS, and PrdX; pink, highly conserved in YbaK/INS; green, highly conserved in INS; blue, highly conserved in YbaK; yellow, partially conserved in YbaK; gray, not conserved but selected for mutagenesis. Ec, E. coli; Hi, H. influenzae; Ef, E. faecalis V583; Cc, Caulobacter crescentus CB15; At, Agrobacterium tumefaciens.
Computational Modeling of CCA-Cys Bound to H. influenzae YbaK
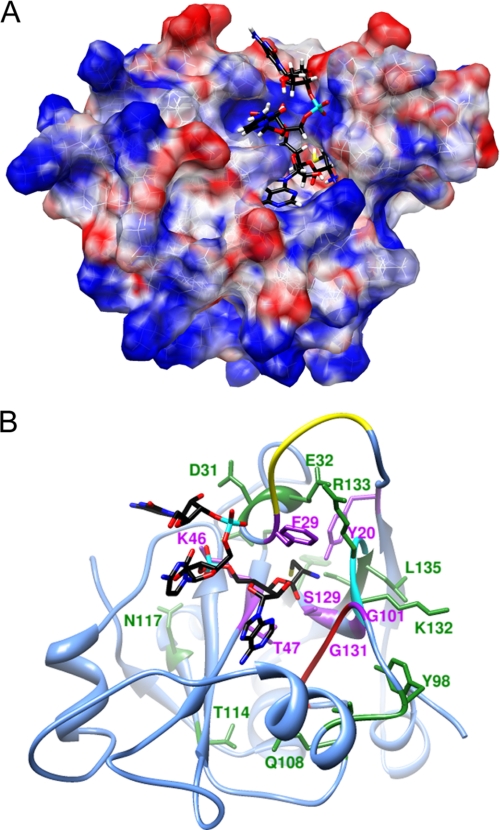
There are presently no available structures of post-transfer editing substrates or substrate analogs bound to the active site of H. influenzae YbaK or its homologs. To begin to probe the chemical basis of substrate recognition and the mechanism of Cys-tRNAPro hydrolysis by YbaK, we used computational approaches to generate a docking model of a 5′-CCA-Cys substrate mimic in the H. influenzae YbaK active site. The model of the YbaK·CCA-Cys complex was generated using a judicious combination of molecular docking (AUTODOCK 4) and MD simulations (AMBER 9), as described under “Experimental Procedures.” Stability of the complex, distances between important functional groups, and the occupancy (lifetimes) of the H-bonds were assessed by analyzing the MD trajectory of the complex structure.
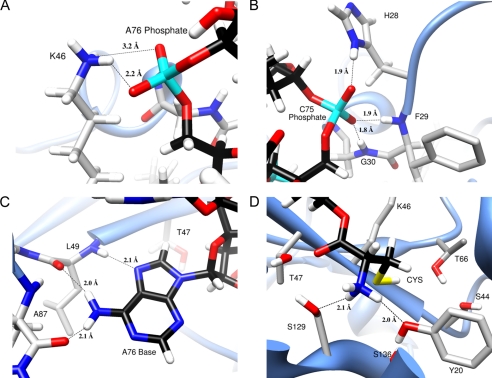
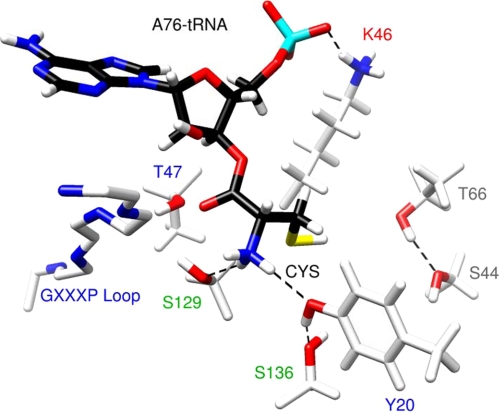
The modeled complex shows an extended active site binding pocket that accommodates the three nucleotides of the ligand (Fig. 2A). The cysteine moiety binds in a deep cleft and interacts directly with several functionally important residues identified by the mutagenesis studies described below (Fig. 2B, purple residues). The Phe-29 of the NNQHF loop (Fig. 2B, yellow), which is missing in the x-ray structure of H. influenzae YbaK and was therefore built into the model, is positioned on top of the bound ligand and presumably stabilizes the complex. The O1P and O2P oxygen atoms of the A76 phosphate form H-bonds with the Lys-46 amine with 96.8 and 65.6% occupancy, respectively (Fig. 3A). These interactions are consistent with the critical nature of this lysine residue for activity (29) as well as with the x-ray structure of apo-E. faecalis ProRS, where the equivalent lysine is bound to a solvent SO42− moiety (16). Thus, the highly conserved lysine in the YbaK superfamily appears to play a general role in stabilizing the bound conformation of the tRNA in the active site. The C75 phosphate also forms strong H-bonds (greater than 90% occupancy) with the protein. The O1P oxygen forms an H-bond with the side chain NδH of His-28, whereas the O2P oxygen forms two H-bonds with the backbone NH groups of Phe-29 and Gly-30 (Fig. 3B).
FIGURE 2.
Computational model of H. influenzae YbaK·CCA-Cys complex. A. The electrostatic potential surface of YbaK was generated by APBS software (65). The cysteine moiety of the substrate (yellow) is located in a deep cleft in the active site. B, ribbon representation of YbaK structure with CCA-Cys substrate shown primarily in black. The NNQHF loop, GXXXP loop, and KRG loop are highlighted in yellow, red, and cyan, respectively. Residues changed in this study are highlighted in purple (deacylation rate decreased >5-fold upon mutagenesis) or green (deacylation rate affected <3-fold upon mutagenesis).
FIGURE 3.
Views of Docked YbaK·CCA-Cys complex showing predicted H-bonds (dashed lines) between YbaK and the substrate. A, H-bonds between Lys-46 and A76 phosphate; B, H-bonding interactions of C75 phosphate; C, H-bonding interactions of A76 adenine; D, H-bonds between the amino group of substrate cysteine and Tyr-20 and Ser-129 hydroxyl groups. Colors for atoms used are as follows. Black, carbons of CCA-Cys substrate; cyan, phosphorus; red, oxygen; white, hydrogen; gray, carbons of YbaK residues; yellow, sulfur.
One of the most stable binding interactions between YbaK and CCA-Cys involves the adenine base A76. Our analysis showed three long lived H-bonds, one between the N7 of A76 and the Leu-49 backbone NH and two between the N6 hydrogens of A76 and the backbone carbonyls of Leu-49 and Ala-87 (Fig. 3C). These interactions appear to play a critical role in properly orienting the substrate in the active site.
The orientation of the cysteine was highly stable over the course of the MD simulation. Two H-bonds form between the amine of the substrate cysteine moiety and the side chain oxygen atoms of Tyr-20 and Ser-129 (Fig. 3D). The substrate amine group also forms H-bonds with backbone carbonyls of the KRG loop (Lys-132, Arg-133, and Gly-134; Fig. 2B, cyan). The carbonyl oxygen of the cysteine moiety points to the center of a cluster of hydroxyl groups belonging to A76 (2′-OH), Thr-47, and Ser-129, with average distances (to the oxygen of the hydroxyl groups) of 3.3, 2.9, and 3.6 Å, respectively (Fig. 3D). Although the modeling results support close interactions between the carbonyl and amine groups of the substrate amino acid with the YbaK active site, the position of the side chain thiol, which lies in a rather large active site pocket, is notable for its lack of direct interaction with protein residues. Three polar residues, Ser-44, Thr-66, and Ser-136, in this active site pocket surround the thiol group, with their side chain hydroxyl oxygen atoms located 4.1, 5.3, and 4.3 Å away from the thiol proton, respectively.
Cys-tRNAPro Deacylation by WT and Mutant H. influenzae YbaK
To probe the role of conserved positions in the YbaK subfamily, as well as residues in the substrate binding pocket based on the computational modeling, 22 residues were individually mutated to alanine. These were chosen based on the sequence alignments described above. Residues in four categories were chosen for alanine-scanning mutagenesis: 1) residues highly conserved within the entire YbaK superfamily (Lys-46 and Gly-101; numbering based on H. influenzae YbaK); 2) residues highly conserved in the YbaK subfamily only (Thr-47, Thr-96, Gly-97, Tyr-98, Ser-129, Gly-131, and Gly-134); 3) residues only partially conserved in the YbaK subfamily (Tyr-20, Phe-29, Gln-108, Thr-114, Asn-117, Tyr-127, Lys-132, and Arg-133); and 4) residues not conserved but close to the predicted substrate-binding pocket based on modeling studies (Asp-31, Glu-32, Ser-44, Thr-66, and Ser-136).
Initial assays were performed using 1 μm WT or mutant H. influenzae YbaK and 1 μm E. coli Cys-tRNAPro, and relative deacylation activity was calculated based on the level of deacylation at 30 min (supplemental Fig. 2). Most of the mutants tested in these assays (D31A, E32A, S44A, T66V, T96A, G97A, Q108A, T114A, N117A, Y127A, K132A, R133A, G134A, and S136A) displayed robust deacylation activity (≥50% of WT YbaK). For the mutants that showed lower than 50% activity, a more detailed kinetic analysis was performed, wherein the rate of deacylation was measured as a function of time (kobs) and compared with that of WT YbaK (Table 1). In addition, some positions close to the substrate binding pocket were probed in more detail, and in a few cases, mutants to non-alanine residues were made and tested.
TABLE 1.
Observed rate constants (kobs) for Cys-tRNAPro deacylation by WT and mutant H. influenzae YbaK
| Mutant | kobs × 102a | Decrease |
|---|---|---|
| min−1 | -fold | |
| WT | 31.6 ± 0.74 | |
| Y20A | 0.70 ± 0.02 | 45 |
| F29A | 1.09 ± 0.03 | 29 |
| S44A | 24.74 ± 2.66 | 1.3 |
| K46A | 0.48 ± 0.08 | 66 |
| K46R | 0.71 ± 0.16 | 44 |
| K46I | 0.83 ± 0.07 | 38 |
| T47A | 1.53 ± 0.03 | 21 |
| T66V | 15.30 ± 1.23 | 2.1 |
| G101A | 0.89 ± 0.08 | 36 |
| S129A | 5.68 ± 0.13 | 5.6 |
| G131A | 1.14 ± 0.03 | 28 |
| G134A | 4.29 ± 0.08 | 7.4 |
| S136A | 8.67 ± 0.12 | 3.6 |
| S136H | 1.93 ± 0.03 | 16 |
a The kobs (min−1) values were calculated by fitting the percentage of Cys-tRNAPro remaining as a function of time to a first order exponential decay equation, y = Ae−kobs× t + B. Values represent the average of three trials with the S.D. indicated.
Previously, we showed that Lys-46 of H. influenzae YbaK is critical for Cys-tRNAPro deacylation activity (29). The kinetic analysis performed here showed a 66-fold decrease in deacylation efficiency for the K46A variant relative to WT YbaK (Table 1). To gain further insights into the role of this key residue, K46I and K46R variants were tested. Both the hydrophobic mutation (K46I) and the conservative mutation (K48R) significantly reduced activity (38- and 44-fold, respectively). Due to its critical nature, the Lys-46 residue was earlier suggested to play a catalytic role in Cys-tRNAPro deacylation (29). In contrast, our modeling suggests that Lys-46 is involved in binding tRNAPro and positioning the 3′-end into the substrate binding pocket (Fig. 3A). This second hypothesis is supported by the mutagenesis data showing sensitivity to even a conservative arginine substitution. A similar role for the corresponding Lys-279 residue has been postulated for Ala-tRNAPro hydrolysis by the INS domain of E. coli ProRS (16). We also probed Gly-101, which is a part of the GXXXP motif proposed to be analogous to the “oxyanion hole” found in serine proteases and d-Tyr-tRNATyr deacylases (35, 53). The G101A mutation results in a 36-fold reduction in deacylation activity, consistent with a possible role in stabilizing the oxyanionic tetrahedral intermediate generated during ester bond hydrolysis.
In addition to Lys-46 and Gly-101, which are almost universally conserved in the YbaK superfamily (Fig. 1 and supplemental Fig. 1), several other highly conserved residues in the YbaK subfamily were probed in more detail. Alanine substitutions of Thr-47, Ser-129, Gly-131, and Gly-134 resulted in 6–28-fold decreases in deacylation efficiency. Upon mutation to alanine, semiconserved aromatic residues in the YbaK subfamily, Tyr-20 and Phe-29, displayed 45- and 29-fold decreases in activity, respectively. Although Ser-44, Thr-66, and Ser-136 are not conserved in the YbaK subfamily, these residues were selected for mutation due to their location proximal to the substrate-binding pocket. Whereas a S136A substitution resulted in 4-fold reduced activity, S136H displayed a 16-fold reduction. The greater effect of histidine substitution implies that YbaK catalysis is sensitive to the substrate binding pocket size or polarity. Alanine and valine substitutions of hydrophilic Ser-44 and Thr-66 side chains, respectively, resulted in small (≤2.1-fold) reductions in activity compared with WT YbaK (Table 1). In summary, the mutagenesis study provided experimental support for the computationally derived docking model, identifying functionally important interactions in and around the substrate binding pocket (Fig. 2B).
Substrate Specificity and Role of Substrate Functional Groups in YbaK Catalysis
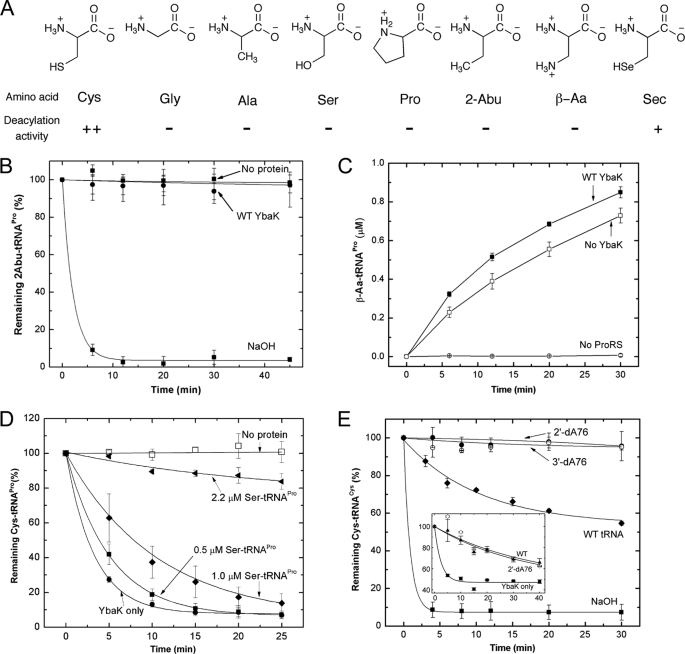
YbaK catalyzes hydrolysis of Cys-tRNA and does not show activity against isosteric substrates Ala-tRNAPro, Gly-tRNAAla, and Ser-tRNAAla (Fig. 4A) using 1 μm protein (29). At very high enzyme concentration (21 μm), weak hydrolysis of these substrates was observed (29). To determine whether any of the YbaK mutants display altered substrate specificity, we tested the ability of the majority of the YbaK variants to hydrolyze nonspecific substrates Ala-, Ser-, and Pro-tRNAPro. In all cases, the substrate specificity of the mutants was similar to that of WT YbaK (i.e. none of the mutants displayed significantly enhanced ability to hydrolyze the nonspecific substrates even in the presence of very high concentrations of mutant proteins (21 μm) (supplemental Table 1)).
FIGURE 4.
Substrate specificity of H. influenzae YbaK. A, structures of amino acids tested as substrates for post-transfer editing by H. influenzae YbaK. ++, hydrolyzed; +, hydrolyzed (deacylation activity is ∼10-fold reduced relative to Cys-tRNA); −, not hydrolyzed (deacylation activity >100-fold reduced relative to Cys-tRNA). B, deacylation of 2-Abu-tRNAPro by H. influenzae YbaK (0.5 μm). Hydrolysis in the absence of protein or in the presence of 2 m NaOH is also shown. C, mischarging of β-Aa onto 10 μm tRNAPro by 1.5 μm E. coli K279A ProRS in the absence or presence of WT H. influenzae YbaK (2 μm). D, YbaK (0.1 μm) deacylation of 0.5 μm Cys-tRNAPro in the absence (YbaK only) and presence of increasing concentrations of Ser-tRNAPro. E, deacylation of 2′-dA76 or 3′-dA76 Cys-tRNACys by WT H. influenzae YbaK (1 μm). Hydrolysis of WT Cys-tRNACys by YbaK or 2 m NaOH is also shown. Inset, deacylation of [35S]Cys-tRNACys by E. coli YbaK (0.5 μm) in the absence (YbaK only) and presence of Cys-tRNACys (WT) and Cys-2′-dA76-tRNACys (2′dA76) at 30 °C. Axis labels are the same as in the main panel. Error bars, S.D.
We also tested two non-natural amino acids, 2-Abu and β-Aa (Fig. 4A), for hydrolysis by WT YbaK. These substrates are isosteres of cysteine where the thiol group is replaced by either a methyl or amine group, respectively. As shown in Fig. 4B, 2-Abu-tRNAPro was not a substrate for deacylation by YbaK (0.5 μm). In the case of β-Aa, sufficient quantities of mischarged tRNA could not be readily generated for deacylation assays; however, a mischarging assay conducted in the presence and absence of YbaK is consistent with a lack of hydrolysis activity (Fig. 4C). Taken together, our data suggest that the substrate cysteine thiol side chain is required for catalysis by YbaK.
Although Ser-tRNA is not a substrate for YbaK deacylation, we wanted to test whether Ser-tRNA binds to the same active site as Cys-tRNA. To address this question, Cys-tRNAPro deacylation assays were performed in the presence of various concentrations of Ser-tRNAPro (Fig. 4D). These studies showed that YbaK deacylation of Cys-tRNAPro was inhibited in the presence of Ser-tRNAPro, with ∼65% inhibition observed at 1 μm Ser-tRNAPro. These results support the conclusion that cysteine and serine occupy the same active site, but only Cys-tRNA is a substrate for YbaK deacylation, suggesting a critical role for the cysteine thiol group in catalysis.
We next wanted to establish whether the specificity for cysteine over serine was conferred by the presence of a zinc ion in the active site. A similar role for zinc has been demonstrated in the case of CysRS (54). Atomic absorption spectroscopic analysis revealed that purified H. influenzae YbaK did not contain bound zinc ions, as expected from the available H. influenzae YbaK crystal structure (35). Nevertheless, we tested the effect of zinc on YbaK deacylation by supplementing the reaction buffer with 5 mm ZnCl2. The results showed that the zinc addition had no effect on deacylation activity (supplemental Fig. 3). Furthermore, EDTA treatment to chelate any residual zinc following protein purification also failed to alter catalysis by YbaK (supplemental Fig. 3).
Previous studies have shown that the cis-hydroxyl groups of the terminal A76 of tRNA are critical for deacylation by aaRSs (44, 55–57). To examine the role of the 3′-terminal hydroxyl groups in YbaK deacylation, Cys-tRNA analogs lacking either the 2′- or 3′-hydroxyl group were prepared. Due to poor levels of cysteine charging onto dA76 tRNAPro variants by ProRS, dA76 tRNACys variants were used for this study. CysRS has previously been shown to aminoacylate cysteine on 2′-dA76- or 3′-dA76-tRNACys (58). As shown in Fig. 4E, neither the 2′- nor the 3′-dA76 Cys-tRNACys variant was hydrolyzed by YbaK, whereas WT Cys-tRNACys was readily deacylated. Based on our level of detection, we estimate that the single deoxy substitutions result in at least a 100-fold decrease in deacylation efficiency. We showed that Cys-2′-dA76-tRNACys serves as a competitive inhibitor of Cys-tRNACys deacylation by YbaK (Fig. 4E, inset), indicating that binding is not significantly affected by the deoxy substitution. These data suggest that the hydroxyl group proximal to the site of esterification plays a critical role in catalysis by YbaK.
Role of Cysteine Thiol Group in Hydrolysis by YbaK
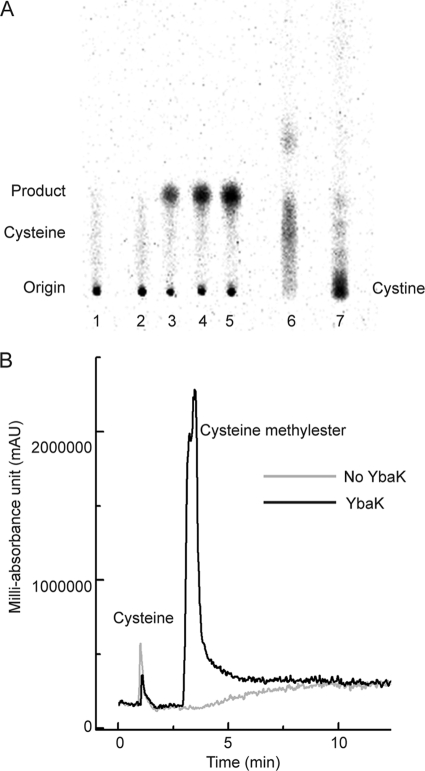
A previous study demonstrated that E. coli CysRS displays weak hydrolysis of Cys-tRNACys in the aminoacylation site and suggested that this may occur via cysteine-thiolactone formation (59). To test this as a possible mechanism for YbaK deacylation, we analyzed the YbaK-catalyzed [35S]Cys-tRNAPro deacylation products using cellulose TLC. A major species (Fig. 5A, lane 3–5, Product) is observed with an Rf value that is distinct from that of free cysteine (Fig. 5A, lane 6). In the absence of YbaK, this compound was not detected by TLC (Fig. 5A, lanes 1 and 2), suggesting that this compound is generated during YbaK catalysis. We also showed that the major YbaK product does not co-migrate with cystine (Fig. 5A, lane 7).
FIGURE 5.
Analysis of deacylation reaction by TLC and LC/MS. A, reaction products of [35S]Cys-tRNAPro deacylation by H. influenzae YbaK detected by cellulose TLC. Eluent and carrier solvents were n-butanol/acetic acid/water (4:1:1, v/v/v) and methanol, respectively. A unique product band (Rf = 0.21, lanes 3–5, Product) distinct from free cysteine (Rf = 0.15, lane 6) is observed. Lane 1, deacylation reaction in the absence of YbaK; lanes 2–5, deacylation reaction in the presence of YbaK at 0, 10, 20, and 30 min; lane 6, authentic [35S]cysteine; lane 7, [35S]cystine. B, LC traces of crude reaction mixtures following H. influenzae YbaK-catalyzed Cys-tRNAPro deacylation (black) or a control reaction in the absence of YbaK (gray). Free cysteine eluted at 1 min, and cysteine methyl ester eluted at 3.5 min.
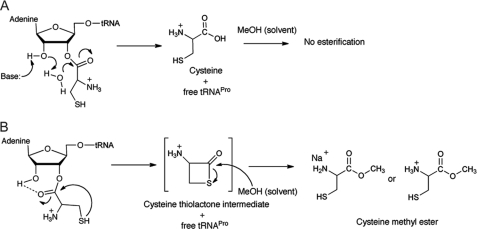
To identify the product formed upon YbaK catalysis, we analyzed the crude reaction mixture using high resolution ESI-MS with methanol as a carrier solvent (supplemental Fig. 4). The major species in the spectrum was observed at 158.0246 atomic mass units and was assigned as the Na+ adduct of cysteine methyl ester (theoretical m/z = 158.0246). This assignment was further validated with an authentic sample of cysteine methyl ester. We propose that the cysteine methyl ester is produced by methanol-activated transesterification of a thiolactone intermediate formed upon YbaK-catalyzed hydrolysis of Cys-tRNA (Fig. 6B). A similar esterification reaction between free cysteine and methanol is kinetically less feasible and thus not observed (Fig. 6A). Reaction mixtures were also analyzed by tandem LC/MS. In the presence of YbaK, a distinct peak was observed at 3.5 min that is well separated from free cysteine at 1 min (Fig. 5B), and this major peak was confirmed to be the cysteine methyl ester by MS/MS analysis (M + H = 136). YbaK-independent transesterification of Cys-tRNAPro by methanol can be ruled out because only free cysteine is observed in the absence of YbaK (Fig. 5B). In a control reaction, free cysteine was analyzed in methanol solution, and no cysteine methyl ester peak was detected. These data support the conclusion that YbaK-catalyzed Cys-tRNAPro hydrolysis proceeds via a cysteine-thiolactone intermediate.
FIGURE 6.
Possible Cys-tRNAPro hydrolysis mechanisms. A, water hydrolysis pathway. The free product cysteine formed does not react with methanol (carrier solvent in ESI-MS). B, hydrolysis by cyclization of cysteine. The cysteine thiolactone intermediate reacts with methanol to form cysteine methyl ester.
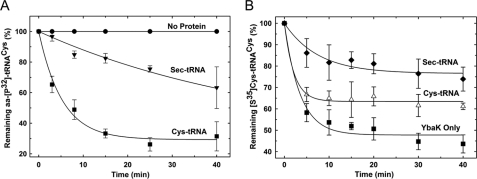
Based on the chemical similarity between cysteine and selenocysteine, we hypothesized that substitution of the side chain sulfur for a selenium atom would support deacylation by YbaK. To test this hypothesis, we misacylated tRNACys with selenocysteine using CysRS and measured the rates of deacylation by YbaK. YbaK deacylates Sec-tRNACys, albeit with an ∼10-fold reduced rate relative to Cys-tRNACys (Fig. 7A). Nevertheless, these results further support the mechanism shown in Fig. 6B.
FIGURE 7.
Deacylation of aminoacyl-tRNAs by E. coli YbaK. A, deacylation of ∼1 μm Cys-3′-[32P]tRNACys and Sec-3′-[32P]tRNACys by 5 μm WT E. coli YbaK at 25 °C. A representative buffer (0.15 m KPO4, pH 7.0) hydrolysis is also shown. B, deacylation of [35S]Cys-tRNACys by E. coli YbaK (0.5 μm) in the absence (YbaK only) and presence of Cys-tRNACys and Sec-tRNACys at 25 °C.
DISCUSSION
Several biochemical, computational, and x-ray crystallographic studies examining post-transfer editing mechanisms of aaRSs have proposed water-mediated cleavage of the aminoacyl-tRNA ester bond (Fig. 6A) (43, 44, 55–57). Class I leucyl-tRNA synthetase and isoleucyl-tRNA synthetase and class II threonyl-tRNA synthetase and phenylalanyl-tRNA synthetase activate water molecules via interaction with conserved residues in their editing active sites. In addition, editing often occurs via substrate-assisted catalysis involving the hydroxyl groups of the A76 ribose on tRNA (44, 56, 57). In all of these systems, a double-sieve mechanism is used, involving steric exclusion of the cognate aminoacyl-tRNA from the editing domain. In contrast, our data are consistent with an alternate mechanism that exploits the unique chemistry of the thiol side chain.
The thiol group of free cysteine is a strong nucleophile with a pKa value of 8.3. Furthermore, the pKa of cysteine in the active site of a protein may be suppressed by as much as 2 units when close to polar or charged residues (60). In our model of H. influenzae YbaK, the substrate cysteine thiol is located close to several non-conserved polar residues (Ser-44, Thr-66, and Ser-136) (Fig. 8). In E. coli YbaK, Ser-136 is replaced by a charged aspartate residue. As expected, mutation of these non-conserved residues is not critical for hydrolysis. Nevertheless, the environment of the active site may lower the cysteine pKa, resulting in thiolate attack of the carbonyl center and formation of a four-membered thiolactone intermediate (Fig. 6B). Interestingly, the average side chain dihedral χ1 of cysteine in our model is 54.2°, which brings the thiol sulfur in close proximity to the carbonyl carbon (3.3 Å) at an angle of 105°, making it facile for nucleophilic attack. Cyclization would be less favorable for a serine substrate because of the higher pKa (∼13) of its hydroxyl side chain. In addition, a four-membered lactone would be highly constrained due to the shorter C–O bond length. On the other hand, selenocysteine was found to be a substrate for YbaK. Given the lower pKa of the selenol group (∼5.2), the apparent 10-fold reduction in rate for the Sec-tRNACys substrate is somewhat surprising. Computational studies showed that the selenolactone ring strain is actually lower than that of the thiolactone (supplemental Table 3), supporting the prediction that selenocysteine should be a better substrate than cysteine. In addition, using a competition assay, we showed that ∼6 μm Cys-tRNACys or Sec-tRNACys both competitively inhibit YbaK deacylation of Cys-tRNACys (Fig. 7B), suggesting that there is not a significant tRNA binding defect. Thus, although global tRNA binding is not affected, we hypothesize that the larger selenocysteine group may perturb the active site leading to slight destabilization of the transition state and decrease the rate of catalysis.
FIGURE 8.
Summary of interactions in the substrate binding pocket of H. influenzae YbaK based on computational and experimental results. A polar environment formed by residues Thr-66, Ser-44, and Ser-136 may lower the pKa of the cysteine thiol. The thiolate thus generated attacks the carbonyl center, leading to the cleavage of the ester bond and formation of a four-membered cyclic thiolactone intermediate. The tetrahedral oxyanionic transition state is stabilized by Thr-47, Ser-129, and the 2′-OH of A76 of the tRNA. Tyr-20 is involved in a hydrogen bonding network with the substrate amine group and the hydroxyl group of Ser-136. The universally conserved Lys-46 residue stabilizes the proper substrate orientation via interaction with the backbone A76 phosphate. The most critical residues or functional groups are labeled in red type. Alanine substitution resulted in deacylation efficiency decreases of ≥66-fold (red), 21–45-fold (blue), 4–6-fold (green), and ≤2-fold (gray).
The formation of cyclized adducts in the active sites of aaRSs is not unprecedented. Studies of aaRSs that utilize amino acids featuring nucleophilic side chains as substrates have demonstrated that in addition to Cys-tRNACys hydrolysis by CysRS (59), adenylates of homocysteine and ornithine can be hydrolyzed in the activation sites of MetRS, and LysRS, respectively (59, 61, 62). The latter reactions have been proposed to occur via cyclization mechanisms that result in the formation of homocysteine thiolactone and ornithine lactam.
In addition to the substrate thiol, the 2′-hydroxyl group of A76 is also crucial for YbaK-catalyzed deacylation of Cys-tRNA. This supports our proposed mechanism that YbaK functions by binding the substrate in an orientation that facilitates cyclization of the substrate cysteine while excluding water molecules from the active site. It is noteworthy that all of the functionally important conserved residues in the YbaK subfamily examined here appear to either play a structural role (Gly-131 and Gly-134) or are involved in the stabilization of the aminoacyl-tRNA (Lys-46 and Ser-129) or the oxyanionic reaction intermediate (Thr-47 and Gly-101) (Fig. 8). Although none of these residues provide specificity for cysteine per se, this is consistent with the proposed catalytic role of substrate cysteine. Our proposed mechanism is also consistent with the observation that none of the active site mutations result in a complete loss in enzymatic activity. The most deleterious mutation occurs at a residue (Lys-46) that is present in all members of the YbaK superfamily (Fig. 1) and that has been proposed to play a general role in positioning the catalytically competent orientation of the substrate in the active site of the enzyme (Fig. 8).
Editing defects in AlaRS are toxic to bacteria and linked to neurodegeneration in mice that harbor a gene encoding the mutant synthetase (2, 13). Although AlaRSs edit both Gly- and Ser-tRNAAla, freestanding AlaXps co-evolved as a second checkpoint to ensure editing of Ser-tRNAAla, which poses a greater challenge than glycine (38, 63). Although even mild editing defects have been linked to serious pathology in the mouse (2), the presence of ∼10% mismade protein is not detrimental to E. coli (64). Thus, different organisms and cellular environments display different requirements with respect to error rate tolerance.
The widespread occurrence of a Cys-tRNA deacylase in bacteria (Fig. 1) is consistent with the importance of proline and cysteine residues for protein structure and function. Proline to cysteine mutations can potentially result in drastic consequences as a result of protein misfolding or disruption of regulatory mechanisms in the cell. The distinct mechanism of YbaK catalysis offers insights into the evolutionary pressure to evolve and maintain a separate trans-editing domain to clear mistakes made by ProRS. During aminoacylation, ProRS cannot effectively discriminate between proline (90 Å3) and cysteine (86 Å3) due to their similar Van der Waals volumes (30). This is also evident from the overlapping orientations of the corresponding aminoacyl-adenylates bound in the amino acid activation site of ProRS (16, 31). The INS domain of ProRS (and the homologous PrdX domain (32, 37); see Fig. 1) selectively clears Ala-tRNAPro based on the double-sieve mechanism of editing that functions via steric exclusion of proline. A model of the E. faecalis INS·CCA-Ala complex (supplemental Fig. 5) generated by homology modeling using our final YbaK·CCA-Cys complex model as a starting structure predicts that the methyl side chain of substrate alanine binds in a small hydrophobic pocket formed by conserved residues Ile-263 and Val-266. This pocket is too small to accommodate the larger amino acids proline and cysteine without disrupting active site geometry.4 Had the INS domain evolved a larger binding pocket to accommodate and edit cysteine in addition to alanine, it would have been more challenging to discriminate against cognate proline.
In addition, computational studies showed that CCA-Pro can bind YbaK with an orientation similar to that of CCA-Cys, albeit with somewhat lower affinity (supplemental Results and Table 2). Therefore, if YbaK had evolved to clear cysteine via a general water-mediated mechanism rather than via thiol-specific chemistry, cognate proline would also be hydrolyzed. We propose that the INS domain and YbaK co-evolved distinct mechanisms involving the use of steric exclusion and thiol-specific chemistry, respectively, to prevent nonspecific hydrolysis of correctly charged Pro-tRNAPro. In this manner, the fine sieve of the ProRS INS domain and the chemical sieve of YbaK collaborate to ensure accurate decoding of proline codons.
Supplementary Material
Acknowledgments
We thank Dr. M. Ignatov (Ohio State University) for help with sequence alignments and Drs. T. R. Hoye, J. Jeon, and A. E. May (University of Minnesota) for assistance in ESI-MS and LC/MS analyses. We thank Dr. Michael Ibba (Ohio State University) for helpful discussion and critical reading of the manuscript. The plasmid encoding E. coli tRNA Nucleotidyl transferase was provided by Dr. P. Schimmel (Scripps Research Institute). Generous computational resources were provided by the Ohio Supercomputer Center.
This work was supported, in whole or in part, by National Institutes of Health Grant GM049928.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Results, Tables 1–4, and Figs. 1–5.
S. Kumar, M. Das, C. Hadad, and K. Musier-Forsyth, unpublished data.
- aaRS
- aminoacyl-tRNA synthetase
- ProRS
- prolyl-tRNA synthetase
- AlaRS
- alanyl-tRNA synthetase
- CysRS
- cysteinyl-tRNA synthetase
- INS
- editing domain of bacterial ProRS
- tRNA
- transfer RNA
- MD
- molecular dynamics
- 2-Abu
- 2-aminobutyric acid
- β-Aa
- β-aminoalanine
- Sec
- selenocysteine
- H-bond
- hydrogen bond
- ESI
- electrospray ionization.
REFERENCES
- 1. Ibba M., Söll D. (2000) Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 2. Lee J. W., Beebe K., Nangle L. A., Jang J., Longo-Guess C. M., Cook S. A., Davisson M. T., Sundberg J. P., Schimmel P., Ackerman S. L. (2006) Nature 443, 50–55 [DOI] [PubMed] [Google Scholar]
- 3. Nangle L. A., Motta C. M., Schimmel P. (2006) Chem. Biol. 13, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 4. Jakubowski H., Goldman E. (1992) Microbiol. Rev. 56, 412–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hendrickson T. L., de Crécy-Lagard V., Schimmel P. (2004) Annu. Rev. Biochem. 73, 147–176 [DOI] [PubMed] [Google Scholar]
- 6. Mascarenhas A. P., Martinis S. A., An S., Rosen A. E., Musier-Forsyth K. (2008) in Protein Engineering (RajBhandary U. L., Koehrer C. eds) pp. 153–200, Springer-Verlag, New York [Google Scholar]
- 7. Ling J., Reynolds N., Ibba M. (2009) Annu. Rev. Microbiol. 63, 61–78 [DOI] [PubMed] [Google Scholar]
- 8. Fersht A. R. (1977) Enzyme Structure and Mechanism, Freeman, San Francisco [Google Scholar]
- 9. Fersht A. R., Dingwall C. (1979) Biochemistry 18, 2627–2631 [DOI] [PubMed] [Google Scholar]
- 10. Nureki O., Vassylyev D. G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T. L., Schimmel P., Yokoyama S. (1998) Science 280, 578–582 [DOI] [PubMed] [Google Scholar]
- 11. Lincecum T. L., Jr., Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., Van Den Eynde W., Link A., Van Calenbergh S., Grøtli M., Martinis S. A., Cusack S. (2003) Mol. Cell 11, 951–963 [DOI] [PubMed] [Google Scholar]
- 12. Fukai S., Nureki O., Sekine S., Shimada A., Tao J., Vassylyev D. G., Yokoyama S. (2000) Cell 103, 793–803 [DOI] [PubMed] [Google Scholar]
- 13. Beebe K., Ribas De Pouplana L., Schimmel P. (2003) EMBO J. 22, 668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naganuma M., Sekine S., Fukunaga R., Yokoyama S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8489–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dock-Bregeon A., Sankaranarayanan R., Romby P., Caillet J., Springer M., Rees B., Francklyn C. S., Ehresmann C., Moras D. (2000) Cell 103, 877–884 [DOI] [PubMed] [Google Scholar]
- 16. Crepin T., Yaremchuk A., Tukalo M., Cusack S. (2006) Structure 14, 1511–1525 [DOI] [PubMed] [Google Scholar]
- 17. Beuning P. J., Musier-Forsyth K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8916–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kotik-Kogan O., Moor N., Tworowski D., Safro M. (2005) Structure 13, 1799–1807 [DOI] [PubMed] [Google Scholar]
- 19. Roy H., Ling J., Irnov M., Ibba M. (2004) EMBO J. 23, 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sokabe M., Okada A., Yao M., Nakashima T., Tanaka I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11669–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sokabe M., Ose T., Nakamura A., Tokunaga K., Nureki O., Yao M., Tanaka I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11028–11033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boniecki M. T., Vu M. T., Betha A. K., Martinis S. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19223–19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dulic M., Cvetesic N., Perona J. J., Gruic-Sovulj I. (2010) J. Biol. Chem. 285, 23799–23809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Splan K. E., Ignatov M. E., Musier-Forsyth K. (2008) J. Biol. Chem. 283, 7128–7134 [DOI] [PubMed] [Google Scholar]
- 25. Yadavalli S. S., Musier-Forsyth K., Ibba M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19031–19032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minajigi A., Francklyn C. S. (2010) J. Biol. Chem. 285, 23810–23817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beuning P. J., Musier-Forsyth K. (2001) J. Biol. Chem. 276, 30779–30785 [DOI] [PubMed] [Google Scholar]
- 28. Ahel I., Stathopoulos C., Ambrogelly A., Sauerwald A., Toogood H., Hartsch T., Söll D. (2002) J. Biol. Chem. 277, 34743–34748 [DOI] [PubMed] [Google Scholar]
- 29. An S., Musier-Forsyth K. (2004) J. Biol. Chem. 279, 42359–42362 [DOI] [PubMed] [Google Scholar]
- 30. Creighton T. E. (1996) Proteins: Structure and Molecular Properties, Freeman, New York [Google Scholar]
- 31. Kamtekar S., Kennedy W. D., Wang J., Stathopoulos C., Söll D., Steitz T. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1673–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruan B., Söll D. (2005) J. Biol. Chem. 280, 25887–25891 [DOI] [PubMed] [Google Scholar]
- 33. An S., Musier-Forsyth K. (2005) J. Biol. Chem. 280, 34465–34472 [DOI] [PubMed] [Google Scholar]
- 34. Wolf Y. I., Aravind L., Grishin N. V., Koonin E. V. (1999) Genome Res. 9, 689–710 [PubMed] [Google Scholar]
- 35. Zhang H., Huang K., Li Z., Banerjei L., Fisher K. E., Grishin N. V., Eisenstein E., Herzberg O. (2000) Proteins Struct. Funct. Bioinform. 40, 86–97 [DOI] [PubMed] [Google Scholar]
- 36. Murayama K., Kato-Murayama M., Katsura K., Uchikubo-Kamo T., Yamaguchi-Hirafuji M., Kawazoe M., Akasaka R., Hanawa-Suetsugu K., Hori-Takemoto C., Terada T., Shirouzu M., Yokoyama S. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 26–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahel I., Korencic D., Ibba M., Söll D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15422–15427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beebe K., Mock M., Merriman E., Schimmel P. (2008) Nature 451, 90–93 [DOI] [PubMed] [Google Scholar]
- 39. Guo M., Chong Y. E., Shapiro R., Beebe K., Yang X. L., Schimmel P. (2009) Nature 462, 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Korencic D., Ahel I., Schelert J., Sacher M., Ruan B., Stathopoulos C., Blum P., Ibba M., Söll D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10260–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hendrickson T. L., Nomanbhoy T. K., de Crécy-Lagard V., Fukai S., Nureki O., Yokoyama S., Schimmel P. (2002) Mol. Cell 9, 353–362 [DOI] [PubMed] [Google Scholar]
- 42. Wong F. C., Beuning P. J., Nagan M., Shiba K., Musier-Forsyth K. (2002) Biochemistry 41, 7108–7115 [DOI] [PubMed] [Google Scholar]
- 43. Dock-Bregeon A. C., Rees B., Torres-Larios A., Bey G., Caillet J., Moras D. (2004) Mol. Cell 16, 375–386 [DOI] [PubMed] [Google Scholar]
- 44. Ling J., Roy H., Ibba M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fernandez-Fuentes N., Zhai J., Fiser A. (2006) Nucleic Acids Res. 34, W173–W176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jensen J. H., Li H., Robertson A. D., Molina P. A. (2005) J. Phys. Chem. A 109, 6634–6643 [DOI] [PubMed] [Google Scholar]
- 48. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 49. Case D. A., Darden T. A., Cheatham T. E., 3rd, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., Kollman P. A. (2006) AMBER 9, University of California, San Francisco [Google Scholar]
- 50. Huey R., Morris G. M., Olson A. J., Goodsell D. S. (2007) J. Comput. Chem. 28, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 51. Hondal R. J., Nilsson B. L., Raines R. T. (2001) J. Am. Chem. Soc. 123, 5140–5141 [DOI] [PubMed] [Google Scholar]
- 52. Wolfson A. D., Uhlenbeck O. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5965–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lim K., Tempczyk A., Bonander N., Toedt J., Howard A., Eisenstein E., Herzberg O. (2003) J. Biol. Chem. 278, 13496–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Newberry K. J., Hou Y. M., Perona J. J. (2002) EMBO J. 21, 2778–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Igloi G. L., von der Haar F., Cramer F. (1977) Biochemistry 16, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 56. Nordin B. E., Schimmel P. (2002) J. Biol. Chem. 277, 20510–20517 [DOI] [PubMed] [Google Scholar]
- 57. Hagiwara Y., Nureki O., Tateno M. (2009) FEBS Lett. 583, 1901–1908 [DOI] [PubMed] [Google Scholar]
- 58. Shitivelband S., Hou Y. M. (2005) J. Mol. Biol. 348, 513–521 [DOI] [PubMed] [Google Scholar]
- 59. Jakubowski H. (1994) Nucleic Acids Res. 22, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Witt A. C., Lakshminarasimhan M., Remington B. C., Hasim S., Pozharski E., Wilson M. A. (2008) Biochemistry 47, 7430–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jakubowski H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4504–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jakubowski H. (1999) Biochemistry 38, 8088–8093 [DOI] [PubMed] [Google Scholar]
- 63. Chong Y. E., Yang X. L., Schimmel P. (2008) J. Biol. Chem. 283, 30073–30078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ruan B., Palioura S., Sabina J., Marvin-Guy L., Kochhar S., Larossa R. A., Söll D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16502–16507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.