Abstract
During DNA synthesis, DNA polymerases must select against ribonucleotides, present at much higher levels compared with deoxyribonucleotides. Most DNA polymerases are equipped to exclude ribonucleotides from their active site through a bulky side chain residue that can sterically block the 2′-hydroxyl group of the ribose ring. However, many nuclear replicative and repair DNA polymerases incorporate ribonucleotides into DNA, suggesting that the exclusion mechanism is not perfect. In this study, we show that the human mitochondrial DNA polymerase γ discriminates ribonucleotides efficiently but differentially based on the base identity. Whereas UTP is discriminated by 77,000-fold compared with dTTP, the discrimination drops to 1,100-fold for GTP versus dGTP. In addition, the efficiency of the enzyme was reduced 3–14-fold, depending on the identity of the incoming nucleotide, when it extended from a primer containing a 3′-terminal ribonucleotide. DNA polymerase γ is also proficient in performing single-nucleotide reverse transcription reactions from both DNA and RNA primer terminus, although its bypass efficiency is significantly diminished with increasing stretches of ribonucleotides in template DNA. Furthermore, we show that the E895A mutant enzyme is compromised in its ability to discriminate ribonucleotides, mainly due to its defects in deoxyribonucleoside triphosphate binding, and is also a poor reverse transcriptase. The potential biochemical defects of a patient harboring a disease mutation in the same amino acid (E895G) are discussed.
Keywords: DNA Polymerase, DNA Replication, Mitochondrial DNA, Nucleic Acid Enzymology, Nucleoside Nucleotide Analogs, Nucleotide, Reverse Transcription, Ribonucleotide
Introduction
Replicative DNA polymerases must have the ability not only to discriminate between the correct and incorrect nucleotide to be incorporated during DNA synthesis but also to distinguish between the correct and incorrect sugar moiety to maintain its fidelity. This process is crucial because ribonucleotides are more prone to strand cleavage compared with deoxyribonucleotides due to the presence of a reactive hydroxyl group at the 2′-position of the ribose sugar. The discrimination against ribonucleotide incorporation during various DNA metabolic processes has been studied for many nuclear replicative DNA polymerases, including pol2 α, δ, and ϵ and other polymerases involved in DNA repair like pol β, λ, and μ (1–5). Studies on these eukaryotic nuclear replicative and repair enzymes indicate that the levels of ribonucleotide discrimination vary from 2 to 3 orders of magnitude between them, suggesting that the mechanism employed by each polymerase in excluding ribonucleotide from its active site may be different during DNA synthesis. Analysis of the human DNA polymerase β revealed that this polymerase discriminates against ribonucleotide incorporation by nearly 4 orders of magnitude (3).
DNA pol γ, encoded by the nuclear gene POLG, is the sole DNA polymerase in human mitochondria and hence bears the burden of replicating and repairing the entire mitochondrial DNA (6, 7). The holoenzyme of pol γ is a heterotrimer composed of a catalytic subunit (p140) and a homodimeric accessory subunit (p55) (8). The catalytic subunit belongs to family A DNA polymerases and consists of an exonuclease domain at its N terminus and a polymerase domain at its C terminus with a linker region connecting these domains. The p140 enzyme possesses DNA polymerase, 3′ → 5′ exonuclease, and 5′-dRP lyase activities (for reviews, see Refs. 6 and 7). The p55 accessory subunit confers processive DNA synthesis and tight DNA binding to the p140 catalytic subunit (9).
Two mechanisms, the asynchronous strand displacement model and the coupled replication model, have been proposed to replicate the 16,569-bp human mitochondrial genome by pol γ in conjunction with its accessory protein (10). In the asynchronous model, pol γ extends the H-strand DNA from an RNA primer, suggesting that the polymerase can initiate DNA synthesis from a ribonucleotide (11). In the strand-coupled model, the replication intermediates contain RNA/DNA heteroduplexes, and extensive RNA-rich regions have also been identified in the lagging strand, indicating a role for pol γ in incorporating these ribonucleotides during nascent strand synthesis (12). RNase H enzymes can remove the RNA incorporated into DNA, where RNase H1 functions to processively cleave long stretches of RNA/DNA hybrids, and RNase H2 removes singly incorporated ribonucleoside 5′-monophosphate (rNMP) residues in DNA. Although both H1 and H2 enzymes are found in the nucleus, only RNase H1 has been implicated in the mitochondria (13). Thus, it is presently unknown if any mechanism exists to remove singly incorporated rNMP residues in the mtDNA. However, because pol γ possesses reverse transcriptase activity, the single ribonucleotides present in the template DNA strand could be bypassed during replication.
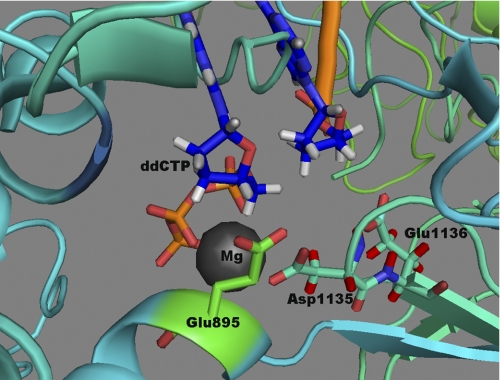
DNA polymerases preferentially incorporate deoxyribonucleotides over ribonucleotides, with as much as a 10,000-fold preference for deoxyribonucleotides, although ribonucleotides are present at much higher concentrations compared with their deoxyribonucleotide counterparts (2, 14). Efficient discrimination of ribonucleotides by these enzymes is controlled by specific amino acid residues, which sterically block the incoming rNMP to the enzyme's active site. However, the identity of these steric gate side chains varies between different polymerases (15–18). Goff and colleagues (14) first identified a phenylalanine residue (Phe-155) in Moloney murine leukemia viral reverse transcriptase that confers selectivity against ribonucleotides. Phe-155 in Moloney murine leukemia viral reverse transcriptase is analogous to Escherichia coli pol I Glu-710 and to T7 DNA polymerase Glu-480. Alteration of Glu-710 in E. coli pol I to Ala results in an 800-fold decrease in overall discrimination against ribonucleoside 5′-triphosphates (rNTPs), and hence Glu-710 is thought to provide discrimination against rNTPs by sterically blocking the 2′-OH group of an incoming rNTP (15). The structure of T7 DNA polymerase indicates that Glu-480 interacts with the ribose ring of the incoming deoxyribonucleoside 5′-triphosphates (dNTPs) as well as through hydrogen bonding to Tyr-530 (19). Because Glu-895 of human DNA pol γ is analogous to E. coli pol I Glu-710 and T7 DNA polymerase Glu-480, it was suggested that changing Glu-895 to Ala or Gly could cause the enzyme to lose discrimination against ribonucleotides (20). Fig. 1 depicts the role of Glu-895 in nucleotide recognition in the active site of the human pol γ (21). Biochemical analysis of the recombinant pol γ harboring the E895A mutation demonstrated severely reduced catalytic activity to ∼0.2% wild type levels (20); however, the role of Glu-895 in ribonucleotide discrimination is still not known.
FIGURE 1.
Involvement of Glu-895 in the discrimination of ribonucleotide. Shown is a molecular model of the human DNA pol γ active site with incoming ddCTP (dark blue), and Mg2+ (gray spheres). The side chain of Glu-895 is shown in green and is in close contact with the ribose ring of the incoming ddCTP. This ddCTP is chelated to Mg2+ ion, which is also held into place by Asp-1135 and Glu-1136 side chains. This image was generated by PyMOL using the pol γ coordinates for the human pol γ, Protein Data Bank entry 3IKM (21).
Previous biochemical studies using purified DNA polymerase γ suggested that the enzyme is an efficient reverse transcriptase and possesses the ability to incorporate and extend ribonucleotides (22, 23). However, a comprehensive analysis of incorporation, discrimination, extension, and bypass of ribonucleotides by DNA pol γ is still lacking. In this study, we employed steady state kinetic analysis to investigate the efficiency of all four rNMP incorporations compared with their cognate deoxyribonucleoside 5′-monophosphate (dNMP), the level of discrimination imposed by pol γ on individual ribonucleotides, the efficiency of the wild type (WT) enzyme to extend primers terminating with a ribonucleotide, and its ability to perform single-nucleotide reverse transcription (RT) and bypass multiple ribonucleotides in a DNA template. The results from these analyses revealed that DNA pol γ differentially discriminates rNTPs with as much as 1,100–77,000-fold preference for dNTPs, depending on the identity of nucleotide. However, the catalytic efficiency of the enzyme decreased during extension from a ribonucleotide and also when it encountered increasing numbers of ribonucleotides in template DNA. Furthermore, analysis with E895A pol γ suggested that the enzyme had impaired ability to discriminate ribonucleotides, but this effect is mainly due to its inability to efficiently incorporate dNMPs. In addition, the RT and ability to extend a ribonucleotide were also compromised in the mutant enzyme.
EXPERIMENTAL PROCEDURES
Materials
Ultrapure dNTP and NTP solutions were obtained from Amersham Biosciences, and [γ-32P]ATP was from PerkinElmer Life Sciences. Oligonucleotides were synthesized by Integrated DNA Technologies, and T4 polynucleotide kinase was purchased from New England Biolabs.
Expression and Purification
The His6 affinity-tagged recombinant catalytic subunits of WT and E895A human pol γ were overproduced in baculovirus-infected Sf9 cells, and the proteins were purified to homogeneity as described previously (20, 24, 25). The His6 affinity-tagged p55 accessory subunit was expressed in E. coli and purified to homogeneity as described (9, 25, 26). The eluted protein samples were visualized with SDS-PAGE, and enzyme concentrations were determined by a Bradford (Bio-Rad) assay with bovine serum albumin as the standard. After purification, the proteins were frozen in small aliquots in liquid nitrogen and stored at −80 °C.
Substrate Preparation
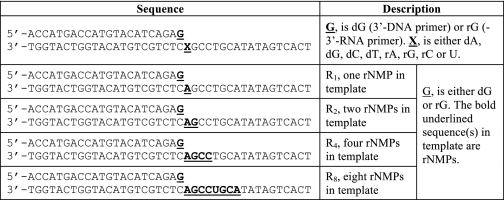
Substrates were prepared by annealing a 22-mer oligonucleotide (primer) radiolabeled at its 5′-end with [γ-32P]ATP and T4 polynucleotide kinase, to 1.2-fold molar excess of a 40-mer oligonucleotide (template) using standard protocols. Table 1 summarizes the list of substrates used in this study.
TABLE 1.
Sequence of DNA substrates used in this study
Kinetic Assays
Steady state kinetic parameters were determined using a polyacrylamide gel-based single-nucleotide extension assay. Because it was previously shown that the affinity of the human DNA pol γ with RNA/DNA substrates is similar to that with DNA/DNA substrates, comparison of steady state kinetic parameters becomes valid (22, 23). Reaction mixtures (10 μl) contained 25 mm HEPES-KOH (pH 7.6), 2 mm 2-mercaptoethanol, 0.1 mm EDTA, 5 mm MgCl2, 50 nm radiolabeled substrate, 10 nm exonuclease-deficient WT or E895A pol γ, and 20 nm WT p55. The reactions were started by the addition of various concentrations of one of the four dNTPs or rNTPs (depending on the substrate and analysis). After incubation at 37 °C for 10 min, reactions were terminated by the addition (10 μl) of 95% deionized formamide and 10 mm EDTA. Samples (3 μl) were boiled for 5 min at 95 °C and resolved by electrophoresis on 12% polyacrylamide gels containing 6 m urea. Gels were dried and exposed to a phospor screen, and radioactive bands were detected with a Typhoon 9400 PhosphorImager (GE Healthcare) and quantified with NIH Image software. Km and Vmax values were determined by fitting the data to the steady state Michaelis-Menten model using KaleidaGraph (version 4.0; Synergy).
Ribonucleotide Bypass Assays
Bypass reactions were performed similar to the single-nucleotide extension assays with the following exceptions: (i) all four dNTPs were added to the reaction mixtures, and (ii) the incubation time was 5 min at 37 °C.
RESULTS
Ribonucleotides Are Strongly and Differentially Discriminated by Human DNA Pol γ
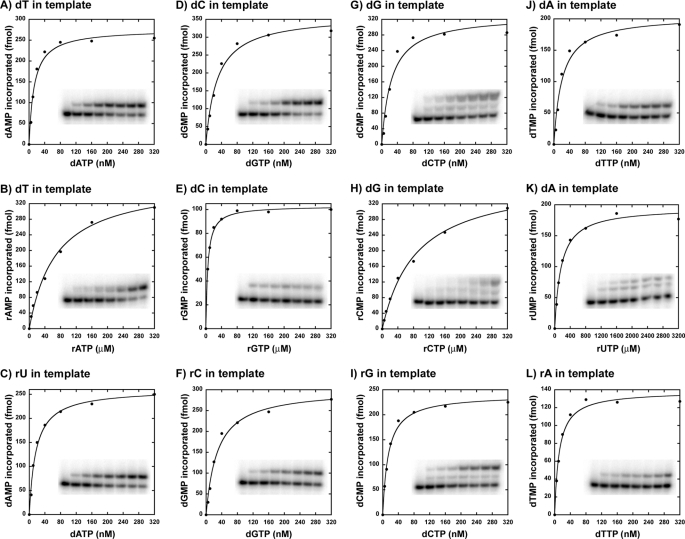
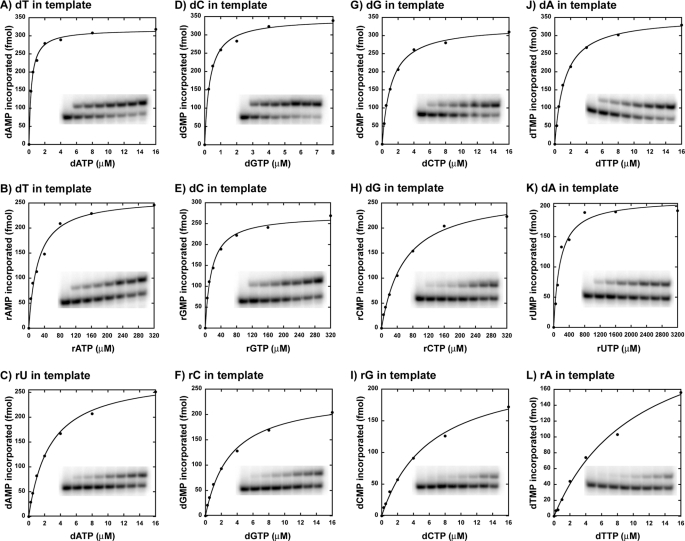
At least 30 ribonucleotides are found scattered in the mitochondrial genome (27), and it has been shown that DNA pol γ can incorporate a single ribonucleotide into a DNA primer (22), suggesting a role for pol γ in this activity. However, a detailed kinetic analysis for incorporation of all four individual ribonucleotides by DNA pol γ is still lacking. Hence, in order to determine how efficiently human DNA polymerase γ can incorporate the four individual ribonucleotides opposite to its cognate deoxyribonucleotides, single-nucleotide incorporation assays were performed on four different substrates containing dT, dC, dG, or dA on the template strand in otherwise the same sequence context (see Table 1 for sequence). Representative saturation curves of WT pol γ to obtain the steady state kinetic values in Table 2 for incorporation of a ribonucleotide opposite to its cognate deoxyribonucleotide are shown in Fig. 2. This and all other analyses in the study were carried out with an exonuclease-deficient DNA pol γ (D198A/E200A) (24), which abolishes the 3′ → 5′ exonuclease activity that can interfere with biochemical assays involving nucleic acids and in the presence of a 2-fold molar excess of p55 accessory subunit. DNA pol γ was able to incorporate all of the four ribonucleotides against its cognate deoxyribonucleotides, albeit with significant reduction in catalytic efficiency (compare kcat/Km values for rNMP versus dNMP incorporation opposite to dNMP in template in Table 2). In addition, the discrimination factors (DF) were highly variable for the individual ribonucleotide incorporation by pol γ (see Table 2 and Fig. 3A) from 1,100-fold (for rGMP versus dGMP incorporation opposite to dC in template) to 77,000-fold (for rUMP versus dTMP incorporation opposite to dA in template). The reason for the ∼2-order of magnitude difference in discrimination among different ribonucleotide incorporations is mainly due to a tighter nucleotide binding exhibited by pol γ for rGTP (Km = 5.13 μm; see Table 2) and reduced catalysis for rUMP incorporation (kcat = 0.038 min−1; see Table 2) compared with other rNTP binding and rNMP incorporations.
TABLE 2.
Steady state kinetic parameters for ribonucleotide incorporation and reverse transcriptase activity of wild type DNA pol γ
The average values of at least two independent experiments are shown with errors expressed as S.D.
| Incoming nucleotide | Template base | Km | kcat | kcat/Km | DF |
|---|---|---|---|---|---|
| μm | min−1 | μm−1min−1 | |||
| dATP | dT | 0.015 ± 0.002 | 0.57 ± 0.02 | 39 ± 4 | |
| rATP | dT | 100 ± 45 | 0.38 ± 0.01 | 0.004 ± 0.002 | 9,300a |
| dATP | rU | 0.018 ± 0.002 | 0.58 ± 0.01 | 32 ± 1 | 1.2b |
| dGTP | dC | 0.032 ± 0.004 | 0.77 ± 0.07 | 25 ± 1 | |
| rGTP | dC | 5.13 ± 0.16 | 0.11 ± 0.01 | 0.022 ± 0.002 | 1,100a |
| dGTP | rC | 0.027 ± 0.003 | 0.56 ± 0.06 | 21 ± 0 | 1.2b |
| dCTP | dG | 0.023 ± 0.002 | 0.64 ± 0.04 | 27 ± 1 | |
| rCTP | dG | 100 ± 19 | 0.41 ± 0.04 | 0.0041 ± 0.0004 | 6,600a |
| dCTP | rG | 0.014 ± 0.001 | 0.50 ± 0.03 | 36 ± 3 | 0.8b |
| dTTP | dA | 0.021 ± 0.001 | 0.47 ± 0.08 | 22 ± 3 | |
| UTP | dA | 140 ± 32 | 0.038 ± 0.001 | 0.0003 ± 0.0001 | 77,000a |
| dTTP | rA | 0.015 ± 0.004 | 0.26 ± 0.02 | 19 ± 7 | 1.2b |
a For ribonucleotide incorporation, the discrimination factor (DF) = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of rNTP incorporation opposite to its cognate dNMP in template.
b For reverse transcriptase activity, DF = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of dNTP incorporation opposite to its cognate rNMP in template.
FIGURE 2.
Single-nucleotide incorporation catalyzed by WT DNA pol γ. Representative nucleotide incorporation saturation curves for WT pol γ on a 3′-DNA primer-template substrate are shown with the corresponding polyacrylamide gels as insets. A, D, G, and J show incorporation of dAMP, dGMP, dCMP, and dTMP opposite to dTMP, dCMP, dGMP, and dAMP in template, respectively. B, E, H, and K show incorporation of rAMP, rGMP, rCMP, and rUMP opposite to dTMP, dCMP, dGMP, and dAMP in template, respectively. C, F, I, and L show incorporation of dAMP, dGMP, dCMP, and dTMP opposite to rUMP, rCMP, rGMP, and rAMP in template, respectively. The fraction of nucleotide incorporated (in fmol) is plotted against the concentration of incoming nucleotide (in nm or μm). The data were fitted to the steady state Michaelis-Menten model using KaleidaGraph to determine the Km and Vmax values. Each incorporation analysis was performed at least twice, and the steady state kinetic parameters with S.D. values are tabulated in Table 2.
FIGURE 3.
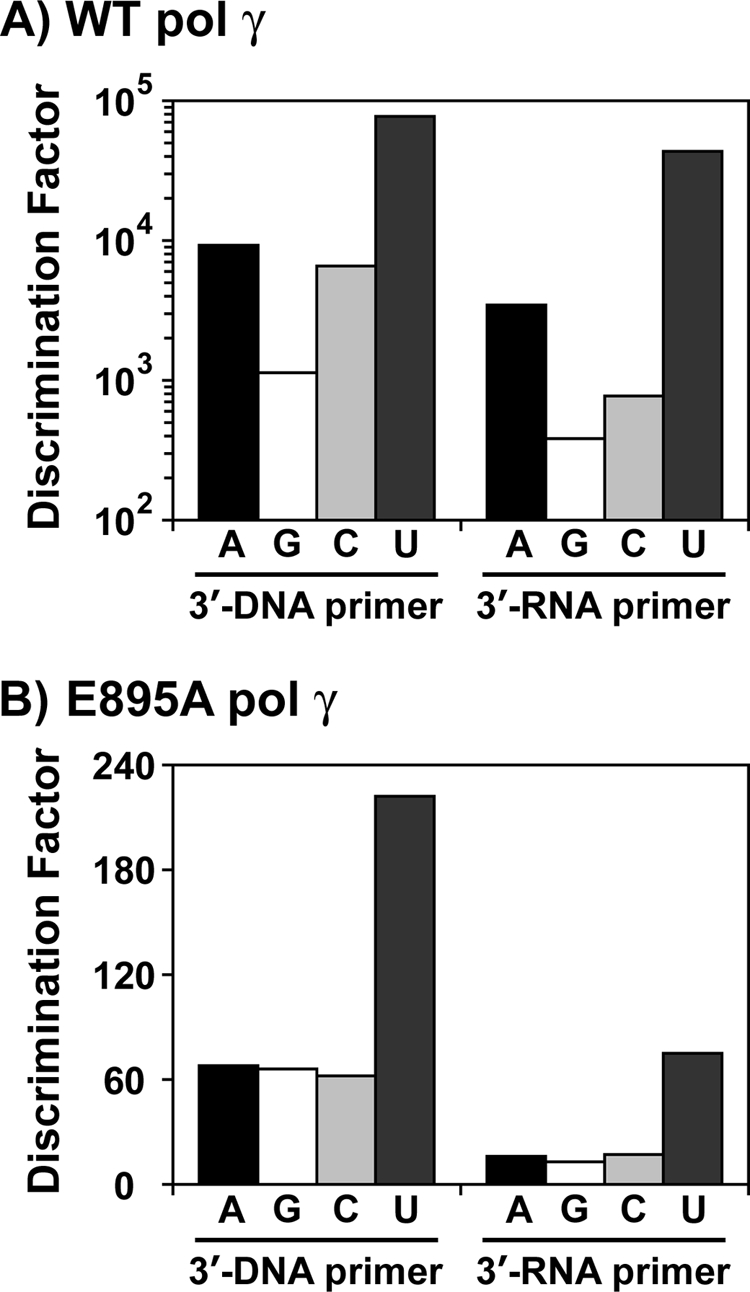
Ribonucleotide discrimination by human DNA pol γ. Discrimination factor for ribonucleotide incorporation for WT pol γ (from Tables 2 and 3) is expressed in logarithmic scale (A), and that for E895A pol γ (from Tables 5 and 6) is expressed in linear scale (B). A (black filled bars), G (white filled bars), C (light gray filled bars), and U (dark gray filled bars), discrimination of incorporation for rA, rG, rC, and U against dA, dG, dC, and dT, respectively. The 3′-DNA primer contains dGMP, and the 3′-RNA primer contains rGMP at the 3′ terminus (see Table 1 for oligonucleotide sequences).
Because the kcat for rUMP incorporation is an order of magnitude lower compared with the other kcat values in Table 2, it is likely that the kcat value of rUMP incorporation reflects only the chemistry step during incorporation, whereas the kcat for other nucleotide incorporations might also include the dissociation of polymerase after incorporation of a nucleotide. In order to gain further insight into this mechanism, we performed an experiment to compare the rate of dTMP and rUMP incorporations opposite to dA in the template at various time points (see supplemental Fig. S1 for details). Analysis of the product formed over time revealed that the y intercept was greater than zero for dTMP incorporation and less than zero for rUMP incorporation opposite to template dA (supplemental Fig. S1). This suggests that the observed kcat for rUMP incorporation is mainly due to the chemistry step as hypothesized, a result of the slower reaction rate. However, the kcat values in Table 2 for other nucleotide incorporations reflect dissociation due to a faster product formation as a result of an initial kinetic burst.
Catalytic Activity of WT DNA Pol γ Is Reduced during Extension from a Ribonucleotide
For the incorporated ribonucleotides to remain in the mtDNA, it has to be extended by DNA pol γ either with a deoxyribonucleotide or ribonucleotide. Therefore, a comprehensive analysis was performed to determine the kinetic parameters of all four dNMP and rNMP incorporations from a 3′-rG-terminated primer (3′-RNA primer). In this experiment, four different templates containing either a dT, dC, dG, or dA as the first template base that pol γ will encounter were annealed to the 3′-rG terminal primer and used as substrates (see Table 1 for sequence). The results from the analysis revealed that the overall catalytic efficiency of pol γ to incorporate dNMPs was slightly compromised when the 3′-end of the primer terminus was a ribonucleotide instead of a deoxyribonucleotide (compare kcat/Km values for individual dNMP incorporations on substrates containing a 3′-RNA primer with DNA template in Table 3 versus the dNMP incorporations on substrates containing a 3′-DNA primer with DNA template in Table 2). Furthermore, this decrease in catalytic efficiency for dNMP incorporations in the 3′-RNA primer substrate also depended on the identity of the nucleotide because only a 3-fold reduction was observed for dGMP incorporation opposite to dC in template (kcat/Km = 9.2 μm−1 min−1 for 3′-RNA primer (Table 3) versus 25 μm−1 min−1 for 3′-DNA primer (Table 2)), but 14-fold reduction was observed for dCMP incorporation opposite to dG in template (kcat/Km = 2.03 μm−1 min−1 for 3′-RNA primer (Table 3) versus 27 μm−1 min−1 for 3′-DNA primer (Table 2)). Fig. 4A summarizes the catalytic efficiencies (kcat/Km) of pol γ for dNMP incorporations from a 3′-dG primer terminus and from a 3′-rG primer terminus. However, the catalytic efficiencies were similar for rNMP incorporations opposite to its cognate dNMP in template from both 3′-DNA primer and 3′-RNA primer (compare kcat/Km values of incoming rNTP in Tables 2 and 3), suggesting that ribonucleotide incorporation by DNA pol γ is not modulated by the sugar identity of the primer terminus. Hence, the reduction in the discrimination factors (3–14-fold) for ribonucleotide incorporations from the 3′-rG primer terminus (Table 3) compared with the 3′-dG primer terminus (Table 2) is mainly due to a decrease in the kcat/Km values of dNMP incorporations by pol γ from the 3′-rG primer. In addition, as previously shown in supplemental Fig. S1 for 3′-dG primer, time course experiments suggested that the kcat values for dTMP and rUMP incorporations opposite to template dA in Table 3 for 3′-RNA primer reflect dissociation and the chemistry step, respectively (data not shown).
TABLE 3.
Steady-state kinetic parameters of wild-type DNA pol γ for single nucleotide extension and reverse transcription from a DNA primer terminating with a ribonucleotide at its 3′-end
The average values of at least two independent experiments are shown with errors expressed as S.D.
| Incoming nucleotide | Template base | Km | kcat | kcat/Km | DF |
|---|---|---|---|---|---|
| μm | min−1 | μm−1min−1 | |||
| dATP | dT | 0.023 ± 0.003 | 0.31 ± 0.01 | 13.2 ± 1.1 | |
| rATP | dT | 62.2 ± 13.7 | 0.23 ± 0.01 | 0.004 ± 0.001 | 3,500a |
| dATP | rU | 0.026 ± 0.002 | 0.31 ± 0.01 | 11.8 ± 1.4 | 1.1b |
| dGTP | dC | 0.045 ± 0.004 | 0.42 ± 0.02 | 9.2 ± 0.3 | |
| rGTP | dC | 4.94 ± 0.44 | 0.12 ± 0.01 | 0.024 ± 0.001 | 380a |
| dGTP | rC | 0.042 ± 0.001 | 0.33 ± 0.01 | 7.9 ± 0.4 | 1.2b |
| dCTP | dG | 0.13 ± 0.02 | 0.26 ± 0.05 | 2.03 ± 0.06 | |
| rCTP | dG | 97.2 ± 39.2 | 0.23 ± 0.01 | 0.003 ± 0.001 | 770a |
| dCTP | rG | 0.14 ± 0.09 | 0.18 ± 0.06 | 1.5 ± 0.6 | 1.4b |
| dTTP | dA | 0.033 ± 0.008 | 0.19 ± 0.01 | 5.7 ± 1.0 | |
| UTP | dA | 231 ± 57 | 0.029 ± 0.004 | 0.0001 ± 0.0001 | 43,000a |
| dTTP | rA | 0.13 ± 0.01 | 0.204 ± 0.003 | 1.5 ± 0.1 | 3.8b |
a For ribonucleotide incorporation, DF = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of rNTP incorporation opposite to its cognate dNMP in template.
b For reverse transcriptase activity, DF = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of dNTP incorporation opposite to its cognate rNMP in template.
FIGURE 4.
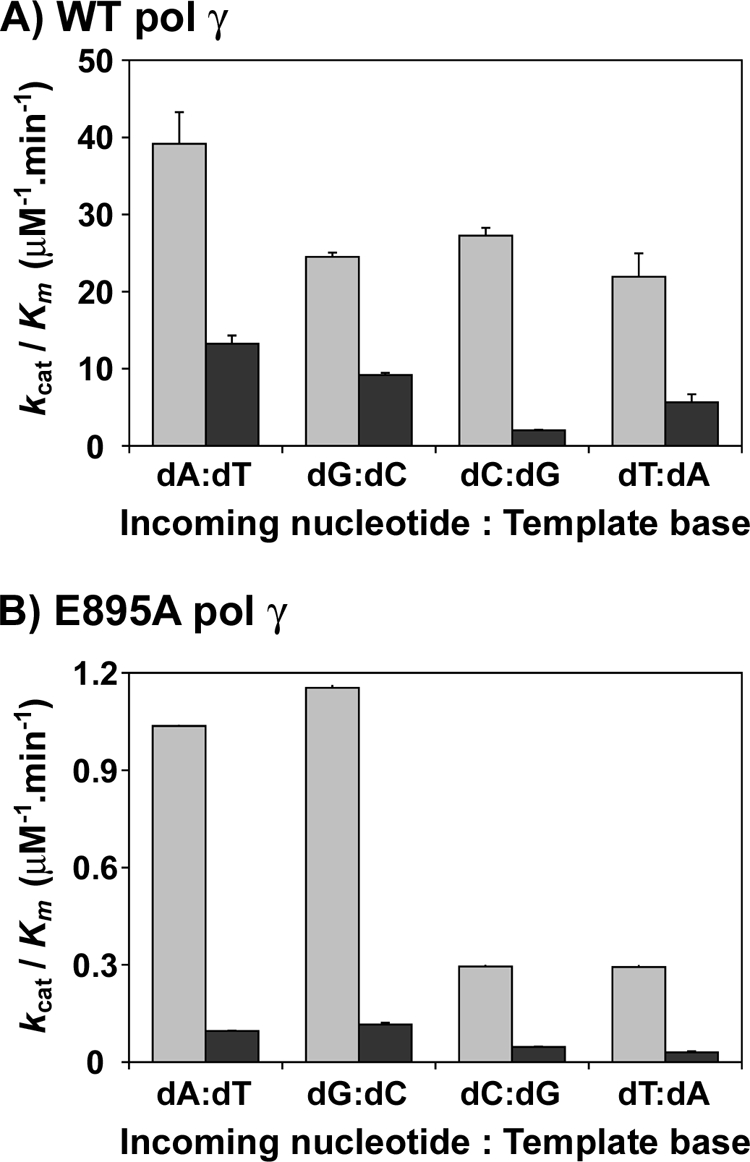
Differential incorporation of deoxyribonucleotides by DNA pol γ. Shown is catalytic efficiency (kcat/Km) of dA, dG, dC, and dT incorporation opposite to their cognate bases dT, dG, dC, and dA in template for WT pol γ (A) and E895A pol γ (B). Light gray filled bars, substrate containing 3′-dG primer terminus; dark gray filled bars, substrate containing 3′-rG primer terminus. The catalytic efficiencies are tabulated in Tables 2, 3, 5, and 6. The average result of at least two experiments is shown with S.D. values (error bars).
Single-nucleotide Reverse Transcription Is Efficiently Catalyzed by DNA Pol γ
Because DNA pol γ can incorporate ribonucleotides and extend them, these rNMPs could be present on the template during the next round of replication and might potentially deplete the catalytic activity of the enzyme. Hence, bypass efficiency of pol γ on DNA templates containing a single ribonucleotide was determined using steady state kinetic analysis. As before, four different templates, each containing a different ribonucleotide in otherwise the same sequence, were annealed to the complementary primer such that pol γ will encounter a ribonucleotide after binding to the 3′-end of the primer terminus, which could be either a dG or rG (see Table 1 for sequence). Results of the single-nucleotide incorporation assays for each of the four dNMPs opposite to their cognate rNMPs in template are summarized in Table 2 (for 3′-DNA primer) and Table 3 (for 3′-RNA primer). Representative saturation curves to obtain the steady state kinetic values in Table 2 for this analysis are shown in Fig. 2. The data suggest that the incorporation efficiency of pol γ is very similar compared with the control substrate (contains only DNA in template). For instance, the catalytic efficiencies (kcat/Km) of dAMP incorporation opposite to dT and rU in the template are 39 and 32 μm−1 min−1, respectively; for dGMP incorporation opposite to dC and rC in template, the values are 25 and 21 μm−1 min−1 respectively, and so on (see Table 2). Similar results were obtained with primer terminating with a ribonucleotide. The catalytic efficiencies for dAMP incorporation opposite to dT and rU in template are 13.2 and 11.8 μm−1 min−1, respectively; for dGMP incorporation opposite to dC and rC in template, the values are 9.2 and 7.9 μm−1 min−1, respectively, and so on (see Table 3). Therefore, pol γ does not discriminate dNMP incorporation opposite to a single deoxyribonucleotide or a ribonucleotide in template, suggesting that single-nucleotide reverse transcription activity of pol γ is robust.
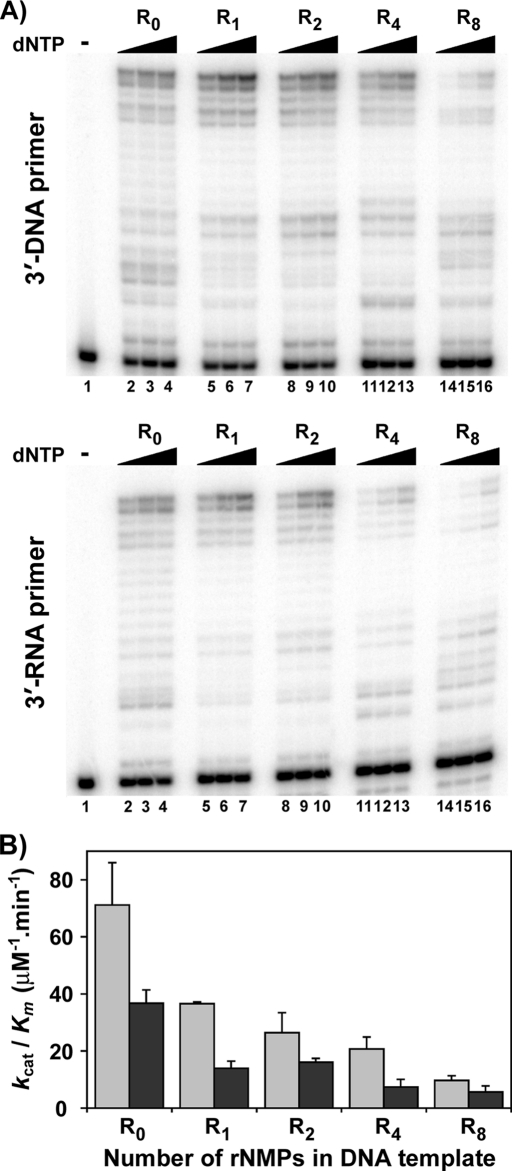
Increasing Stretches of Ribonucleotides in the DNA Template Reduces the Bypass Efficiency of Pol γ
DNA polymerase γ was initially identified by its ability to efficiently perform reverse transcription of DNA from homopolymeric RNA templates (28). In order to perform a processive RT reaction, the enzyme must not stall when bypassing extensive lengths of ribonucleotides on a template. To determine this, bypass experiments were performed on substrates containing various stretches of ribonucleotides (1, 2, 4, or 8 rNMPs; see Table 1 for sequence) on a DNA template. The results from this experiment indicated that DNA pol γ could bypass ribonucleotides and extend until the end of the DNA template; however, the efficiency of extension decreased with increasing lengths of ribonucleotides (Fig. 5A, compare lanes 2–4 with lanes 5–7, 8–10, 11–13, and 14–16). Furthermore, the sugar identity at the 3′-primer terminus did have a moderate effect on the enzyme's catalysis (Fig. 5A, compare top panel (primer terminating with a dG) with bottom panel (primer terminating with an rG)). Steady state kinetic analysis of bypass and extension revealed that the catalytic efficiency of WT pol γ decreased as the number of ribonucleotides in the template multiplied, independent of the nature of sugar at the primer terminus (Table 4, compare kcat/Km for substrate containing no rNMP (R0) with kcat/Km for substrates containing different lengths of ribonucleotides (R1, R2, R4, and R8) for both 3′-DNA primer and 3′-RNA primer). In addition, a 2-fold difference in the efficiency of the enzyme was also observed on control substrates containing primers terminating with an rG compared with dG (Table 4, kcat/Km = 71 and 37 μm−1 min−1 for R0 substrates with 3′-DNA primer and 3′-RNA primer, respectively). The catalytic efficiencies of DNA pol γ on substrates containing varying stretches of ribonucleotides are summarized in Fig. 5B.
FIGURE 5.
Bypass of ribonucleotides in DNA template by WT pol γ. A, representative gels of bypass reactions. The assay was performed as described under “Experimental Procedures.” Top, the 3′-primer terminus is dGMP; bottom, the 3′-primer terminus is rGMP. In both panels, lane 1 contains no dNTP; lanes 2, 5, 8, 11, and 13 contain 0.25 μm dNTPs; lanes 3, 6, 9, 12, and 15 contain 0.5 μm dNTPs; lanes 4, 7, 10, 13, and 16 contain 1 μm dNTPs. R0, DNA template with no rNMP; R1, DNA template with one rNMP; R2, DNA template with two rNMPs; R4, DNA template with four rNMPs; R8, DNA template with eight rNMPs. B, catalytic efficiency (kcat/Km) of bypass of ribonucleotides of varying lengths (R0–R8) in a DNA template. Light gray filled bars, extension from a 3′-dGMP primer terminus; dark gray filled bars, extension from a 3′-rGMP primer terminus (see Table 1 for oligonucleotide sequences). The average result of at least two experiments is shown with S.D. values (error bars).
TABLE 4.
Steady-state kinetics for bypass of rNMPs in DNA templates by DNA pol γ
The average values of at least two independent experiments are shown with errors expressed as S.D.
| Substrate | Km | kcat | kcat/Km | Relative efficiencya |
|---|---|---|---|---|
| μm | min−1 | μm−1min−1 | ||
| 3′-DNA primer | ||||
| R0 | 0.017 ± 0.003 | 1.19 ± 0.02 | 71 ± 15 | 1 |
| R1 | 0.032 ± 0.003 | 1.17 ± 0.13 | 37 ± 1 | 0.51 |
| R2 | 0.042 ± 0.006 | 1.08 ± 0.13 | 26 ± 7 | 0.37 |
| R4 | 0.049 ± 0.003 | 1.01 ± 0.13 | 21 ± 4 | 0.29 |
| R8 | 0.085 ± 0.013 | 0.81 ± 0.02 | 10 ± 2 | 0.14 |
| 3′-RNA primer | ||||
| R0 | 0.031 ± 0.002 | 1.12 ± 0.07 | 37 ± 5 | 1 |
| R1 | 0.081 ± 0.006 | 1.12 ± 0.13 | 14 ± 3 | 0.38 |
| R2 | 0.069 ± 0.009 | 1.10 ± 0.05 | 16 ± 1 | 0.44 |
| R4 | 0.119 ± 0.047 | 0.82 ± 0.03 | 7 ± 3 | 0.20 |
| R8 | 0.167 ± 0.067 | 0.88 ± 0.04 | 6 ± 2 | 0.15 |
a Relative efficiency is defined as (kcat/Km) of primer extension opposite to DNA templates containing various lengths of rNMPs (R1, R2, R4, or R8)/(kcat/Km) of primer extension opposite to DNA template containing no rNMP (R0).
Loss of Ribonucleotide Discrimination in E895A Pol γ Is Primarily Due to Defects in dNTP Incorporation
Human DNA pol γ efficiently discriminates deoxyribonucleotides over ribonucleotides, with as much as a 1,100–77,000-fold preference for dNTPs, depending on the nucleotide (Table 2). A strong discrimination is also observed across DNA polymerases from different families (1–3), and amino acid side chains from many DNA polymerases have been identified that control the discrimination of ribonucleotide incorporation (14, 15). The side chain that confers this discrimination to DNA pol γ is thought to be Glu-895 (Fig. 1) based on sequence alignments with E. coli pol I (Glu-710) and T7 DNA polymerase (Glu-480). In order to elucidate the efficiency of ribonucleotide discrimination imparted by the E895A mutation on DNA pol γ, steady state kinetic analyses were carried out with the mutant enzyme on the same substrates used for characterization of ribonucleotide incorporation by WT pol γ (see Table 1). Single-ribonucleotide incorporation assays revealed that the discrimination for ribonucleotide decreased 17-fold (from 1,100- to 66-fold, for rGMP incorporation for WT versus E895A pol γ; see Tables 2 and 5) to 347-fold (from 77,000- to 222-fold for rUMP incorporation by WT versus E895A pol γ; see Tables 2 and 5) with mutant DNA pol γ compared with the WT enzyme, depending on the identity of the incoming ribonucleotide (compare DF in Table 2 with Table 5 for rNMP incorporation). This 1–2-order of magnitude decrease in discrimination factor for ribonucleotide incorporation (summarized in Fig. 3, A and B) exhibited by the mutant enzyme in comparison with WT pol γ is not due to an enhanced ability to incorporate ribonucleotides by the E895A enzyme (compare kcat/Km for rNMP incorporation opposite to its cognate dNMP in template by WT and E895A pol γ from Tables 2 and 5). The decrease in ribonucleotide discrimination by the mutant enzyme is in fact primarily due to its poor dNMP insertion efficiency (compare kcat/Km for dNMP incorporation opposite to its cognate deoxyribonucleotide in template by WT and E895A enzyme from Tables 2 and 5 and Fig. 4, A and B). The E895A pol γ incorporated dNMPs with 22-fold (kcat/Km = 39 μm−1 min−1 for WT pol γ from Table 2 versus kcat/Km = 1.04 μm−1 min−1 for E895A pol γ from Table 5, for dGMP incorporation opposite to dC in the template) to 92-fold (kcat/Km = 27 μm−1 min−1 for WT enzyme from Table 2 versus kcat/Km = 0.295 μm−1 min−1 for E895A enzyme from Table 5, for dCMP incorporation opposite to dG in template) lower efficiency compared with the WT enzyme, depending on the incoming nucleotide. Fig. 6 shows representative saturation curves of E895A pol γ to obtain the steady state kinetic values summarized in Table 5 for ribonucleotide incorporation.
TABLE 5.
Steady-state kinetic parameters for ribonucleotide incorporation and reverse transcriptase activity of E895A DNA pol γ
The average values of at least two independent experiments are shown with errors expressed as S.D.
| Incoming nucleotide | Template base | Km | kcat | kcat/Km | DF |
|---|---|---|---|---|---|
| μm | min−1 | μm−1min−1 | |||
| dATP | dT | 0.31 ± 0.01 | 0.316 ± 0.002 | 1.04 ± 0.01 | |
| rATP | dT | 19.6 ± 5.5 | 0.28 ± 0.03 | 0.015 ± 0.006 | 68a |
| dATP | rU | 2.33 ± 0.48 | 0.28 ± 0.01 | 0.12 ± 0.02 | 8b |
| dGTP | dC | 0.30 ± 0.04 | 0.34 ± 0.01 | 1.15 ± 0.11 | |
| rGTP | dC | 16.8 ± 1.1 | 0.30 ± 0.04 | 0.018 ± 0.001 | 66a |
| dGTP | rC | 3.33 ± 0.25 | 0.26 ± 0.03 | 0.077 ± 0.002 | 15b |
| dCTP | dG | 1.14 ± 0.03 | 0.34 ± 0.01 | 0.295 ± 0.004 | |
| rCTP | dG | 59.7 ± 2.3 | 0.29 ± 0.02 | 0.0048 ± 0.0002 | 62a |
| dCTP | rG | 3.63 ± 3.61 | 0.16 ± 0.11 | 0.06 ± 0.02 | 5b |
| dTTP | dA | 1.18 ± 0.08 | 0.35 ± 0.01 | 0.29 ± 0.01 | |
| UTP | dA | 181 ± 24 | 0.24 ± 0.04 | 0.0013 ± 0.0001 | 222a |
| dTTP | rA | 17.9 ± 10.4 | 0.32 ± 0.10 | 0.019 ± 0.006 | 15b |
a For ribonucleotide incorporation, DF = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of rNTP incorporation opposite to its cognate dNMP in template.
b For reverse transcriptase activity, DF = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of dNTP incorporation opposite to its cognate rNMP in template.
FIGURE 6.
Single-nucleotide incorporation catalyzed by E895A DNA pol γ. Representative nucleotide incorporation saturation curves for E895A pol γ on a 3′-DNA primer template substrate are shown with the corresponding polyacrylamide gels as insets. A, D, G, and J, incorporation of dAMP, dGMP, dCMP, and dTMP opposite to dTMP, dCMP, dGMP, and dAMP in template, respectively. B, E, H, and K, incorporation of rAMP, rGMP, rCMP, and rUMP opposite to dTMP, dCMP, dGMP, and dAMP in template, respectively. C, F, I, and L, incorporation of dAMP, dGMP, dCMP, and dTMP opposite to rUMP, rCMP, rGMP, and rAMP in template, respectively. The fraction of nucleotide incorporated (in fmol) is plotted against the concentration of incoming nucleotide (in μm). The data were fitted to the steady state Michaelis-Menten model using KaleidaGraph to determine the Km and Vmax values. Each incorporation analysis was performed at least twice, and the steady state kinetic parameters with S.D. values are tabulated in Table 5.
Reverse Transcription and Extension from a Ribonucleotide Are Poorly Catalyzed by the E895A Pol γ
The reduced catalytic efficiency displayed by the E895A mutant enzyme in incorporating dNMPs suggested that RT activity might also be affected in this mutant. Steady state kinetic analysis for single-nucleotide incorporation performed with E895A pol γ on substrates containing a single ribonucleotide in template revealed that the catalytic efficiency for reverse transcriptase activity decreased by 5-fold (for dCTP incorporation opposite to rG in template; see DF in Table 5) to 15-fold (for dTMP incorporation opposite to rA in template; see DF in Table 5) in comparison with its DNA polymerase activity, depending on the incoming nucleotide (see Fig. 6 for representative saturation curves for E895A mutant enzyme's RT activity). However, the WT enzyme showed no significant discrimination between DNA polymerase and reverse transcriptase activities in single-nucleotide incorporation assays (see DF values in Table 2), suggesting a possible role for Glu-895 in the enzyme's reverse transcriptase activity.
To probe whether the E895A mutant displayed a reduction in catalyzing single-nucleotide extension from a ribonucleotide similar to the WT enzyme, steady state kinetic analyses were performed using substrates containing a 3′-rG primer terminus (see Table 1 for sequence). The analysis revealed that the catalytic efficiency was reduced by 6-fold (compare kcat/Km for dCTP incorporation opposite to dG in template in Tables 5 and 6) to 11-fold (compare kcat/Km for dATP incorporation opposite to dT in template in Tables 5 and 6), depending on the dNMP incorporated. This result is similar to the WT enzyme extending a 3′-rG primer, where it exhibited a 3–11-fold reduction in efficiency (compare kcat/Km for incorporation of all four dNMPs opposite to its cognate deoxyribonucleotide in the template in Tables 2 and 3). Furthermore, this reduction in catalytic efficiency for dNMP incorporations in E895A from a 3′-RNA primer also lowered the ribonucleotide discrimination factor by 3–4-fold in comparison with the 3′-DNA primer (compare DF for ribonucleotide incorporation in Tables 5 and 6).
TABLE 6.
Steady-state kinetic parameters of E895A DNA pol γ for single nucleotide extension and reverse transcription from a DNA primer terminating with a ribonucleotide at its 3′-end
The average values of at least two independent experiments are shown with errors expressed as S.D.
| Incoming nucleotide | Template base | Km | kcat | kcat/Km | DF |
|---|---|---|---|---|---|
| μm | min−1 | μm−1min−1 | |||
| dATP | dT | 1.61 ± 0.14 | 0.16 ± 0.01 | 0.0965 ± 0.0005 | |
| rATP | dT | 25.7 ± 1.5 | 0.151 ± 0.004 | 0.0059 ± 0.0002 | 16a |
| dATP | rU | 4.75 ± 0.12 | 0.11 ± 0.01 | 0.023 ± 0.002 | 4b |
| dGTP | dC | 1.51 ± 0.14 | 0.18 ± 0.01 | 0.12 ± 0.01 | |
| rGTP | dC | 18.6 ± 1.3 | 0.169 ± 0.001 | 0.009 ± 0.001 | 13a |
| dGTP | rC | 3.88 ± 0.76 | 0.13 ± 0.02 | 0.033 ± 0.001 | 4b |
| dCTP | dG | 3.36 ± 0.29 | 0.16 ± 0.01 | 0.047 ± 0.002 | |
| rCTP | dG | 50.1 ± 6.1 | 0.141 ± 0.005 | 0.0028 ± 0.0002 | 17a |
| dCTP | rG | 15.8 ± 2.2 | 0.14 ± 0.03 | 0.0088 ± 0.0004 | 5b |
| dTTP | dA | 5.28 ± 0.98 | 0.16 ± 0.01 | 0.031 ± 0.003 | |
| UTP | dA | 236 ± 60 | 0.094 ± 0.007 | 0.0004 ± 0.0001 | 75a |
| dTTP | rA | 11.5 ± 4.2 | 0.068 ± 0.003 | 0.006 ± 0.003 | 5b |
a For ribonucleotide incorporation, DF = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of rNTP incorporation opposite to its cognate dNMP in template.
b For reverse transcriptase activity, DF = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/(kcat/Km) of dNTP incorporation opposite to its cognate rNMP in template.
DISCUSSION
Human DNA Pol γ Differentially Discriminates during the Incorporation of a Single rNMP into DNA
Preliminary analysis revealed that pol γ possesses the ability to incorporate a ribonucleotide into DNA (20, 22). We have extended this initial observation through a steady state kinetic analysis in order to determine the discrimination against each of the four ribonucleoside triphosphates. We found that pol γ provides the least degree of discrimination against rGTP (1,100-fold; Table 2) and the greatest degree of discrimination against rUMP insertion (77,000-fold; Table 2). rUTP and dTTP differ not only by the presence of a 2′-OH, but thymidine also contains a C5-methyl group, both moieties that could account for the higher discrimination. In 1988, Mosbaugh (29) demonstrated that the porcine liver DNA pol γ could incorporate dUMP but with a 3-fold lower efficiency compared with dTMP due to a 3-fold lower Km for dTTP. This suggests that the absence of the C5-methyl group in dUTP decreases the affinity for the polymerase by only 3-fold, whereas the majority of the discrimination against the rUTP is due to the 2′-OH group. Unlike pol ϵ and α, pol δ also shows an enhanced discrimination against rUTP compared with dTTP relative to the other three ribonucleotides (2).
Ribonucleotide Pools Differentially Affect Incorporation
Ribonucleotide pools are known to exceed deoxyribonucleotide pools in the cell, which provides an opportunity to effectively compete for its incorporation into genomic DNA by DNA polymerases. Ribonucleotide pools have been reported in E. coli and Salmonella to be 7–16-fold higher than dNTP pools (30). In eukaryotic cells, this difference appears to be much greater because a recent study in yeast reported dNTP pools in the range of 12–30 μm and rNTP pools in the range of 500–3000 μm (2). This large difference in pool size between rNTPs and dNTPs has been used to investigate whether the yeast replicative polymerases can incorporate appreciable amounts of rNMPs into genomic DNA. Nick McElhinny et al. (2) found that yeast DNA polymerase ϵ can incorporate one rNMP for every 1,250 dNMPs, whereas pol δ and α incorporate one rNMP for every 5,000 and 625 dNMPs, respectively. Further analysis by this group revealed in vivo incorporation with a mutant pol ϵ and in an RNase H2-deleted strain (1). This incorporation was accompanied by elevated deletion mutagenesis of 2–5 base pairs.
A recent study by Wheeler and Mathews (31) revealed that rat tissue mitochondria rNTP/dNTP levels varied depending on the nucleotide and tissue, but in general, the ATP/dATP ratio is ∼1,000, 9–73 for UTP/dTTP, 6–12 for CTP/dCTP, and 2–26 for GTP/dGTP in these rat tissues. In our analysis of the discrimination by the human pol γ against ribonucleotides, the binding affinity (Km) appeared to be the greater determinant in the kinetic analysis. Thus, consideration of the higher ribonucleotide concentration in mitochondria will lower the discrimination. For example, because the Km determinant is the major discrimination factor with rAMP incorporation (∼6,700-fold; Table 2), a ∼1,000-fold higher level of ATP should lower this value to ∼6.7-fold, and hence pol γ may have the potential to incorporate 1 rAMP for every 6–7 dAMP nucleotides into mtDNA. This may account for the high degree of ribonucleotides observed in mammalian mtDNA (27). However, because one source of mitochondrial dNTP pools appears to involve the reduction of ribonucleotides by ribonucleotide reductase (32, 33), the up-regulation of ribonucleotide reductase offers a possible means to alter this balance by reducing rNTP pools and increasing dNTP pools. This concept has been tested in yeast, where up-regulation of ribonucleotide reductase in the presence of certain disease mutations of POLG reduces the petite frequency and lower mutagenesis (34).
Extension from a Ribonucleotide Primer
Extensive evidence suggests that mtDNA replication is primed by the transcription machinery with the mtRNA polymerase (11, 35–37). Thus, pol γ must be able to extend from a ribonucleotide primer or DNA primer with an RNA at the 3′-OH. We investigated the efficiency of pol γ to extend a ribonucleotide located at the 3′-primer terminus during normal DNA synthesis, during rNMP incorporation, and during reverse transcription events. The efficiency of extension from a 3′-rG-terminated primer during DNA synthesis and reverse transcription was 3–14-fold and 3–24-fold lower, respectively, relative to the 3′-dG-terminated primer, mainly due to decreased nucleotide binding and catalysis (see Tables 2 and 3). The difference in activity could not be attributed to the affinity of pol γ to 3′-dG- or 3′-rG-terminated primers because previous studies suggested that the enzyme had similar affinity for DNA/DNA and DNA/RNA substrates (22).
The Glu-895 Residue Is Responsible for rNTP Discrimination, and a Disease Mutation at This Site Is Deleterious to Function
The steric gate that imparts a large degree of discrimination against ribonucleotides is afforded by Glu-710 in Klenow (38). Alteration of Glu-710 to Ala in Klenow lowers the kcat by 30–40-fold, increases the Km(dNTP) by 5-fold, and reduces the affinity for DNA by 5-fold (15, 39). A similarly large decrease in kcat and increase in Km were observed for the analogous E895A mutation in human pol γ (20). Additional analysis of the E. coli mutant polymerase demonstrated that alteration of this Glu to Ala decreased fidelity at the nucleotide insertion step, including a notable increase in A·dCTP mispairing (40). The additional space provided by Ala substitution may allow the accommodation of aberrant base pairs in the active site, thus increasing the likelihood of mispairing events. Alteration of Glu-895 in pol γ to Ala increased the discrimination against D4T-TP and ddNTPs by 500- and 17-fold, respectively (20). The decreased intrinsic activity of the E895A mutant derivative prevented the detection of incorporation with the other analogs. The E710A mutation in Klenow also increased the discrimination against ddNTPs as compared with the F762Y Klenow polymerase (15). Thus, Glu-895 in human pol γ, like Glu-710 in E. coli pol I, appears to enhance discrimination by interacting with the ribose ring (20) (Fig. 1).
The gene for the catalytic subunit of the human pol γ, POLG, is one of several nuclear genes that is associated with mitochondrial DNA depletion or deletion disorders (33). To date, more than 150 disease mutations have been identified in the POLG, and these mutations are distributed over the entire length of the protein (see the Human DNA Polymerase Gamma Mutation Database at the NIEHS, National Institutes of Health, Web site) (41). Disorders associated with POLG mutations have been defined as either mitochondrial depletion syndromes, which include myocerebrohepatopathy spectrum and Alpers syndrome, or mitochondrial disease due to mtDNA deletions, which include ataxia neuropathy spectrum, myoclonus epilepsy myopathy sensory ataxia, and autosomal recessive and dominant forms of progressive external ophthalmoplegia (42).
The Glu-895 codon of POLG was recently found mutated to a Gly codon in a newborn boy with severe lactic acidosis and generalized hypotonia and marked hyporeflexia (43). The infant died 36 h after birth. Post-mortem analysis revealed only 20% mtDNA levels in muscle tissue and drastically reduced respiratory chain activity (43). This E895G mutation was found as a heterozygous mutation without any other detected POLG mutation, and due the absence of paternal DNA analysis, the dominance versus recessive modes of inheritance or de novo acquired mutation could not be determined. Analysis of the E895A mutant derivative of pol γ in ribonucleotide insertion indicated that the E895A pol γ had reduced discrimination of ribonucleotide incorporation relative to dNMP insertion. This discrimination dropped from 1,100- to 66-fold for rATP and from 77,000- to 222-fold for rUTP between the WT and E895A mutant pol γ. However, the decrease in ribonucleotide discrimination is two-part and mostly due to a decrease in overall catalytic efficiency of dNMP incorporation. For example, as in the case for ATP/dATP, the apparent decrease in ribonucleotide discrimination is mostly due to a large decrease in dAMP insertion efficiency by the mutant protein (a 38-fold decrease in kcat/Km) as well as a 4-fold increase in kcat/Km for rAMP insertion as compared with the wild type enzyme (Tables 2 and 5). This difference is similar for the other three rNMPs during insertion. In addition, both the RT and extension by the E895A mutant derivative were also inefficient, most likely due to its intrinsic defect in catalysis. Thus, the most detrimental affect of eliminating the Glu-895 side chain is the large decrease in overall catalytic efficiency, which is more likely the scenario for POLG defect in the observed patient (43) versus enhanced ribonucleotide insertion.
Reverse Transcriptase Activity of Pol γ Is Inefficient with Increasing Ribonucleotides in the Template
Because mitochondria appear to lack a known RNase H2 activity, it is likely that singly incorporated rNMPs in DNA, regardless of the source, are not removed and persist in mitochondria. The absence of RNase H2 could very well explain the observed ribonucleotides found in mammalian mtDNA (27). These ribonucleotides present a challenge to DNA polymerases that cannot incorporate opposite RNA. DNA pol γ was originally discovered in mammalian cells as a possible reverse transcriptase during a screen for cellular reverse transcriptase activity (28). However, the standard reverse transcriptase assay uses homopolymer RNA primed by DNA oligonucleotides in an Mn2+-catalyzed reaction. Mn2+ accelerates the catalysis of DNA polymerases at the expense of fidelity. On random DNA substrates, Mn2+ causes substantial misincorporation, which stalls replication due to the inefficiency of the DNA polymerase to extend a mismatch (44). However, on homopolymer substrates, such as with oligo(dT)12–18/poly(rA) with only dTTP, the catalytic rate appears higher as compared with natural DNA. In vivo, DNA polymerases use Mg2+ as the metal co-activator, which promotes high fidelity polymerization.
Lee and Johnson (23) explored the physiological consequence of RT activity by human pol γ opposite a single rNMP and found that pol γ stalls after incorporating a single dNMP. We found that the efficiency of bypass of one rNMP was 51% compared with a dNMP template base (Fig. 5B). However, longer stretches of RNA in the template caused a diminishing return because the bypass efficiency dropped to only 29% with four consecutive ribonucleotides and 14% with eight ribonucleotides. Thus, this sudden drop in catalytic activity indicates that pol γ will stall while bypassing long stretches of ribonucleotides in DNA, most likely due to the A-form conformation of a RNA-DNA heteroduplex, which is less tolerated by most replicative DNA polymerases. The efficiency was slightly lower when the 3′-primer terminus was rG (compare 0.38 versus 0.51 in Table 4). This difference is diminished as the ribonucleotide stretch gets longer, or as the distance from the DNA primer end lengthens.
In the present study, we investigated the ribonucleotide discrimination and reverse transcription activities of human DNA pol γ using steady state kinetic analysis. Our results indicate that pol γ incorporates ribonucleotides and that the base identity plays a role in discrimination. Next, although pol γ was efficient in performing single-nucleotide RT, the bypass efficiency of the enzyme decreases with increasing sequences of rNMPs in a DNA template, indicating that ribonucleotides in DNA could slow down the rate of DNA synthesis by pol γ. Furthermore, the polymerase was also capable of extending from a 3′-terminal ribonucleotide, albeit with slightly lower efficiency, suggesting that rNMPs incorporated in the mtDNA may become substrates for future rounds of replication. Finally, the E895A mutant enzyme displayed poor ribonucleotide discrimination mainly due to its catalytic defect for DNA synthesis and does not have enhanced rNMP incorporation. This study represents the first in-depth analysis on the role of DNA pol γ in ribonucleotide incorporation, extension, bypass, and reverse transcription and the function of Glu-895 in ribonucleotide discrimination.
Supplementary Material
Acknowledgments
We thank Drs. Nisha A. Cavanaugh and Danielle L. Watt for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program, NIEHS, Grants ES 065078 and ES 065080.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- pol
- polymerase
- mtDNA
- mitochondrial DNA
- dNTP
- deoxyribonucleoside 5′-triphosphate
- dNMP
- deoxyribonucleoside 5′-monophosphate
- rNTP
- ribonucleoside 5′-triphosphate
- rNMP
- ribonucleoside 5′-monophosphate
- RT
- reverse transcription
- DF
- discrimination factor(s).
REFERENCES
- 1. Nick McElhinny S. A., Kumar D., Clark A. B., Watt D. L., Watts B. E., Lundström E. B., Johansson E., Chabes A., Kunkel T. A. (2010) Nat. Chem. Biol. 6, 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavanaugh N. A., Beard W. A., Wilson S. H. (2010) J. Biol. Chem. 285, 24457–24465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nick McElhinny S. A., Ramsden D. A. (2003) Mol. Cell Biol. 23, 2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown J. A., Fiala K. A., Fowler J. D., Sherrer S. M., Newmister S. A., Duym W. W., Suo Z. (2010) J. Mol. Biol. 395, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graziewicz M. A., Longley M. J., Copeland W. C. (2006) Chem. Rev. 106, 383–405 [DOI] [PubMed] [Google Scholar]
- 7. Kaguni L. S. (2004) Annu. Rev. Biochem. 73, 293–320 [DOI] [PubMed] [Google Scholar]
- 8. Yakubovskaya E., Chen Z., Carrodeguas J. A., Kisker C., Bogenhagen D. F. (2006) J. Biol. Chem. 281, 374–382 [DOI] [PubMed] [Google Scholar]
- 9. Lim S. E., Longley M. J., Copeland W. C. (1999) J. Biol. Chem. 274, 38197–38203 [DOI] [PubMed] [Google Scholar]
- 10. Brown T. A., Cecconi C., Tkachuk A. N., Bustamante C., Clayton D. A. (2005) Genes Dev. 19, 2466–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shadel G. S., Clayton D. A. (1997) Annu. Rev. Biochem. 66, 409–435 [DOI] [PubMed] [Google Scholar]
- 12. Bowmaker M., Yang M. Y., Yasukawa T., Reyes A., Jacobs H. T., Huberman J. A., Holt I. J. (2003) J. Biol. Chem. 278, 50961–50969 [DOI] [PubMed] [Google Scholar]
- 13. Cerritelli S. M., Frolova E. G., Feng C., Grinberg A., Love P. E., Crouch R. J. (2003) Mol. Cell 11, 807–815 [DOI] [PubMed] [Google Scholar]
- 14. Gao G., Orlova M., Georgiadis M. M., Hendrickson W. A., Goff S. P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Astatke M., Ng K., Grindley N. D., Joyce C. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3402–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang G., Franklin M., Li J., Lin T. C., Konigsberg W. (2002) Biochemistry 41, 10256–10261 [DOI] [PubMed] [Google Scholar]
- 17. Bonnin A., Lázaro J. M., Blanco L., Salas M. (1999) J. Mol. Biol. 290, 241–251 [DOI] [PubMed] [Google Scholar]
- 18. Boyer P. L., Sarafianos S. G., Arnold E., Hughes S. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. (1998) Nature 391, 251–258 [DOI] [PubMed] [Google Scholar]
- 20. Lim S. E., Ponamarev M. V., Longley M. J., Copeland W. C. (2003) J. Mol. Biol. 329, 45–57 [DOI] [PubMed] [Google Scholar]
- 21. Lee Y. S., Kennedy W. D., Yin Y. W. (2009) Cell 139, 312–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murakami E., Feng J. Y., Lee H., Hanes J., Johnson K. A., Anderson K. S. (2003) J. Biol. Chem. 278, 36403–36409 [DOI] [PubMed] [Google Scholar]
- 23. Lee H. R., Johnson K. A. (2007) J. Biol. Chem. 282, 31982–31989 [DOI] [PubMed] [Google Scholar]
- 24. Longley M. J., Ropp P. A., Lim S. E., Copeland W. C. (1998) Biochemistry 37, 10529–10539 [DOI] [PubMed] [Google Scholar]
- 25. Kasiviswanathan R., Longley M. J., Young M. J., Copeland W. C. (2010) Methods 51, 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Longley M. J., Clark S., Yu Wai Man C., Hudson G., Durham S. E., Taylor R. W., Nightingale S., Turnbull D. M., Copeland W. C., Chinnery P. F. (2006) Am. J. Hum. Genet. 78, 1026–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang M. Y., Bowmaker M., Reyes A., Vergani L., Angeli P., Gringeri E., Jacobs H. T., Holt I. J. (2002) Cell 111, 495–505 [DOI] [PubMed] [Google Scholar]
- 28. Gallo R. C., Yang S. S., Ting R. C. (1970) Nature 228, 927–929 [DOI] [PubMed] [Google Scholar]
- 29. Mosbaugh D. W. (1988) Nucleic Acids Res. 16, 5645–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kornberg A., Baker T. A. (1992) DNA Replication, 2nd Ed., p. 54, W.H. Freeman and Co., New York [Google Scholar]
- 31. Wheeler L. J., Mathews C. K. (2011) J. Biol. Chem. 286, 16992–16996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bourdon A., Minai L., Serre V., Jais J. P., Sarzi E., Aubert S., Chrétien D., de Lonlay P., Paquis-Flucklinger V., Arakawa H., Nakamura Y., Munnich A., Rötig A. (2007) Nat. Genet. 39, 776–780 [DOI] [PubMed] [Google Scholar]
- 33. Copeland W. C. (2008) Annu. Rev. Med. 59, 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stumpf J. D., Bailey C. M., Spell D., Stillwagon M., Anderson K. S., Copeland W. C. (2010) Hum. Mol. Genet. 19, 2123–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clayton D. A. (1982) Cell 28, 693–705 [DOI] [PubMed] [Google Scholar]
- 36. Wanrooij S., Fusté J. M., Farge G., Shi Y., Gustafsson C. M., Falkenberg M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11122–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fusté J. M., Wanrooij S., Jemt E., Granycome C. E., Cluett T. J., Shi Y., Atanassova N., Holt I. J., Gustafsson C. M., Falkenberg M. (2010) Mol. Cell 37, 67–78 [DOI] [PubMed] [Google Scholar]
- 38. Astatke M., Grindley N. D., Joyce C. M. (1998) J. Mol. Biol. 278, 147–165 [DOI] [PubMed] [Google Scholar]
- 39. Polesky A. H., Dahlberg M. E., Benkovic S. J., Grindley N. D., Joyce C. M. (1992) J. Biol. Chem. 267, 8417–8428 [PubMed] [Google Scholar]
- 40. Minnick D. T., Bebenek K., Osheroff W. P., Turner R. M., Jr., Astatke M., Liu L., Kunkel T. A., Joyce C. M. (1999) J. Biol. Chem. 274, 3067–3075 [DOI] [PubMed] [Google Scholar]
- 41. Stumpf J. D., Copeland W. C. (2011) Cell Mol. Life Sci. 68, 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wong L. J., Naviaux R. K., Brunetti-Pierri N., Zhang Q., Schmitt E. S., Truong C., Milone M., Cohen B. H., Wical B., Ganesh J., Basinger A. A., Burton B. K., Swoboda K., Gilbert D. L., Vanderver A., Saneto R. P., Maranda B., Arnold G., Abdenur J. E., Waters P. J., Copeland W. C. (2008) Hum. Mutat. 29, E150–E172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spinazzola A., Invernizzi F., Carrara F., Lamantea E., Donati A., Dirocco M., Giordano I., Meznaric-Petrusa M., Baruffini E., Ferrero I., Zeviani M. (2009) J. Inherit. Metab. Dis. 32, 143–158 [DOI] [PubMed] [Google Scholar]
- 44. Copeland W. C., Lam N. K., Wang T. S. (1993) J. Biol. Chem. 268, 11041–11049 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.