Abstract
Transglutaminase 2 (TG2) is secreted by a non-classical pathway into the extracellular space, where it has several activities pertinent to fibronectin (FN), including binding to the gelatin-binding domain of FN and acting as an integrin co-receptor. Glutamines in the N-terminal tail of FN are known to be susceptible to transamidation by both TG2 and activated blood coagulation factor XIII (FXIIIa). We used immunoblotting, limited proteolysis, and mass spectrometry to localize glutamines within FN that are subject to TG2-catalyzed incorporation of dansylcadaverine in comparison to residues modified by FXIIIa. Such analysis of plasma FN indicated that Gln-3, Gln-7, and Gln-9 in the N-terminal tail and Gln-246 of the linker between fifth and sixth type I modules (5F1 and 6F1) are transamidated by both enzymes. Only minor incorporation of dansylcadaverine was detected elsewhere. Labeling of C-terminally truncated FN constructs revealed efficient TG2- or FXIIIa-catalyzed dansylcadaverine incorporation into the N-terminal residues of constructs as small as the 29-kDa fragment that includes 1–5F1 and lacks modules from the adjacent gelatin-binding domain. However, when only 1–3F1 were present, dansylcadaverine incorporation into the N-terminal residues of FN was lost and instead was in the enzymes, near the active site of TG2 and terminal domains of FXIIIa. Thus, these results demonstrate that FXIIIa and TG2 act similarly on glutamines at either end of 1–5F1 and transamidation specificity of both enzymes is achieved through interactions with the intact 29K fragment.
Keywords: Collagen, Extracellular Matrix, Fibronectin, Mass Spectrometry (MS), Protein Cross-linking, Factor XIII, Thermolysin, Transglutaminase 2
Introduction
The transglutaminase (TG)2 family of calcium-dependent enzymes is involved in numerous processes, including hemostasis and thrombosis, wound healing, extracellular matrix (ECM) formation, cell signaling, and cell-adhesion (1–4).
TGs are expressed in a cell-specific manner and consist of eight active enzymes, TG1–7 and activated blood coagulation factor XIII (FXIIIa), along with Band 4.2, which is inactive as an enzyme (1–3, 5). TGs catalyze incorporation of amines into proteins and formation of ϵ-(γ-glutamyl)-lysine isopeptide bonds between proteins (1–4). The catalytic FXIII A subunit originates from monocyte/macrophages and platelets and circulates in association with the carrier B subunit made in the liver (1–2). After activation with thrombin, FXIIIa stabilizes the hemostatic plug by cross-linking fibrin and introducing cross-links among fibrin, fibronectin (FN), and α2-antiplasmin (6–8). Deficiency in FXIIIa leads to a distinctive bleeding phenotype (9). TG2 is localized in the cytoplasm, is secreted into the extracellular space by an ill-defined mechanism, and binds to the cell surface; it is abundant in endothelial cells, smooth muscle cells, and other cells (4). TG2−/− mice suffer from impaired wound healing, autoimmunity, susceptibility to develop diabetes, and other pathophysiological conditions (4, 10–12).
FN is a large, multi-modular glycoprotein found in plasma that is produced by hepatocytes and also secreted by many other cell types (13–14). It is a dimer of 230–250-kDa subunits, consisting of 12 type 1 (F1) modules, two type 2 (F2) modules, and 15–17 type 3 (F3) modules per subunit depending on splice variation (Fig. 1) (13–15). FN is cross-linked by FXIIIa to fibrin (16), itself (17), and fibrillar collagens (18). The FN N-terminal region contains a 70-kDa (70K) region comprised primarily of F1 modules (13–14). Limited trypsinization of FN yields a 29-kDa (29K) N-terminal fragment containing modules 1–5F1 (Fig. 1), which interacts with fibrin and other FN-binding proteins (13–14). Gln-3 (numbering starting after processing of pre-propeptide sequences) is the major target of amine incorporation into FN by FXIIIa (19–20). Gln-4 and Gln-16 have been predicted to be additional sites of modification by in vitro mutagenesis and studies of peptide mimics (14, 21–23). These glutamines are in the unstructured N-terminal tail of FN (24). TG2 also incorporates amines into the N-terminal region of FN (25). However, strong binding of TG2 is to the 40–42-kDa (40K) gelatin-binding domain of FN (26–27) adjacent to the 29K N-terminal domain. Reduced TG2 expression in human endothelial cells results in less polymerization of FN into the ECM (28), and evidence suggests that TG2 stabilizes the ECM by enhancing FN matrix formation and cross-linking of extracellular matrix proteins (29–31). Because binding to the gelatin-binding domain does not require that the TG2 be catalytically active (26, 32–33) the relationship of this binding activity to the enzymatic action of TG2 on FN is obscure.
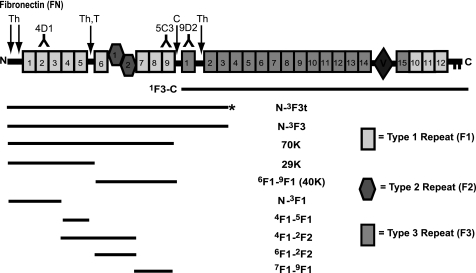
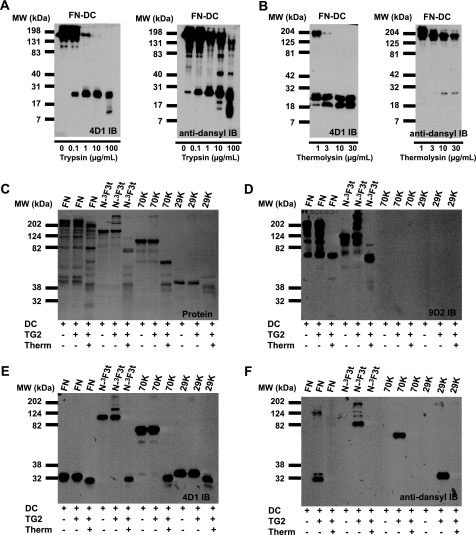
FIGURE 1.
Schematic diagram of FN and proteins used to study FXIIIa- and TG2-catalyzed dansylcadaverine incorporation into FN. FN purified from human plasma was the source of the 70-kDa (70K), 40-kDa (40K), and 29-kDa (29K) proteolytic fragments. Additional proteins were produced recombinantly in the baculovirus expression system. The * on N-3F3t indicates an extra C-terminal tail sequence found in a truncated FN splice variant. Sites of cathepsin D (C), trypsin (T), and thermolysin (Th) N-terminal cleavages and epitopes for 4D1, 5C3, and 9D2 monoclonal antibodies are also indicated. FN and 1F3-C are held together as dimers by disulfides near the C termini.
The comparative reactivities of specific glutamines within FN for TG2 and FXIIIa are not known. The two enzymes have different specificities for fibrinogen and casein (8, 34–37). On the other hand, Gln-2 of Asn-α2-anti-plasmin is the preferred target of both TG2 and FXIIIa (8, 38). This finding is consistent with the conclusion that glutamines in unstructured regions like N termini or flexible loops are reactive to transglutaminases (2). In addition, peptide mimics with Gln-Gln sequences, as is found at the N terminus of FN, have been shown to be substrates with incorporation of amines into both residues (22–23, 35, 39). Positively charged residues two to four residues away from the reactive glutamine substrate promote transamidation as well, whereas flanking positive residues seem to inhibit (1–2). Finally, evidence suggests that binding of distal regions of the substrate protein with the enzyme helps direct transamidation (40). Tight binding of TG2 to the gelatin-binding domain of FN (26–27), therefore, raises the possibility that such binding may direct incorporation of amines to the structured F1 and F2 modules within this region or to other parts of FN.
We have used N-(5-aminopentyl)-5-dimethylaminonaphthalene-1-sulfonamide (dansylcadaverine), a versatile model amine substrate (41–42), to address this and related issues. The goals of our study were to identify the glutamines of FN subject to TG2-catalyzed dansylcadaverine incorporation, compare the substrate specificities of TG2 and FXIIIa, and explore the roles of distal enzyme-FN interactions in directing the enzymes to specific glutamines.
EXPERIMENTAL PROCEDURES
Plasma FN, FXIII, and TG2
FN was purified from a concentrate of fibrinogen, FN, and FXIII obtained by fractionation of plasma obtained from a community blood bank or from a side-fraction of commercial Factor VIII production (16, 43). The mixture of FN, fibrinogen, and FXIII was heated at 56 °C for 3 min to precipitate fibrinogen, and FXIII and FN in the supernatant were separated from one another on DEAE-cellulose. TG2, purified from human red cells (26, 44), was a kind gift from Dr. Prasanna Murthy (Northwestern University, Chicago, IL). Concentrations of FN, FXIII, TG2, and other proteins were estimated based on extinction coefficients at 280 nm predicted by amino acid content (ExPASy Proteomics Server).
Proteolytic 29K, 40K, and 70K N-terminal FN Fragments
70K (see Fig. 1) was purified by affinity chromatography on insolubilized gelatin after cathepsin D cleavage of FN; 70K was then subjected to limited cleavage with trypsin to yield 29K and 40K (45). Alternatively, 29K was immunopurified after limited trypsinization of FN on a column with monoclonal antibody 4D1 to module 2F1 (46) coupled to CNBr-activated Sepharose 4B (Sigma). Separation of 29K from trace amounts of uncleaved FN was accomplished by fast performance liquid chromatography separation on a Superose 12 HR 10/300 gel filtration column (GE Healthcare).
Expression of FN Modules Using pAcGP67.coco (COCO)
A schematic of FN constructs is shown in Fig. 1. Constructs are named according to their modular content, and residues are numbered after the 31 amino acid pre-prosequences are released, i.e. residue one is the first glutamine in sequence QAQQMVQPQSPVAVSQ…. Recombinant N-9F1 (residues 1–577), N-3F1 (residues 1–152), 4FN1-5FN1 (residues 153–259), 6FN1-9FN1 (residues 260–577), 7FN1-9FN1 (residues 437–577), N-3F3 (residues 1–873), and N-3F3t (residues 1–873 plus 17 residues encoded in the adjacent intron, GNFFKKTLPMLSYQDCS; this is the human homolog to a splice variant found in zebrafish (47)) were expressed as secreted His-tagged proteins using the baculovirus vector pAcGP67.coco as described previously (48–49). Segments of DNA encoding for the constructs 4F1-2F2 (residues 153–436) and 6F1-2F2 (residues 260–436) were amplified by PCR, cloned into pAcGP67.coco and expressed in the same manner as the other recombinant proteins. 1F3-C was expressed as in previous literature (50). As a consequence of the cloning and purification strategy, the recombinant proteins contained the sequence ADPG at the N terminus and a polyhistidine tag at the C terminus. The polyhistidine-tagged proteins were purified by Ni2+-chelate chromatography.
Labeling of Transglutaminase-reactive Glutamines by Amine Incorporation
TG2 from human erythrocytes was received frozen in 20 mm imidazole-HCl, pH 6.5, 10% glycerol, 1 mm EDTA, 4 mm GDP, 4 mm MgCl2 as previously described (26, 44), thawed, and diluted to 1 mg/ml with 20 mm imidazole-HCl buffer, pH 6.5 containing 10% glycerol, and stored in smaller portions at −80 °C. Prior to experiments, TG2 was diluted further to 0.1 mg/ml in Tris-buffered saline (TBS; 10 mm trizma base, 150 mm NaCl, pH 7.4), and incubated with 1 mm dithiothreitol in the presence of 2 mm CaCl2 for 30 min at 37 °C. This was added to a final concentration of 5 μg/ml (62 nm) to potential substrates at concentrations of 0.4 μm along with 0.5 mm dansylcadaverine and incubated at 37 °C for 3 h, after which reaction mixtures were prepared for immunoblotting and mass spectrometry as indicated below. Typically, the volume of the incubations was 50 μl. FXIII was activated with 1 units/ml thrombin and 2 mm CaCl2 at 37 °C for 30 min. FXIIIa was added at a final concentration of 10 μg/ml, (62 nm AB dimer), and reactions were carried out as with TG2. For cross-linking of FN and type I collagen, the proteins were added to give final concentrations of 200 μg/ml each.
SDS-PAGE and Immunoblotting
Samples were mixed with 0.2 volumes of sodium dodecyl sulfate (SDS) loading buffer (TBS, pH 6.8 including 8 m urea, 4% SDS, 1 mm EDTA, 1 mm EGTA, with or without 1% 2-mercaptoethanol), heated at 95 °C for 5 min, and subjected to discontinuous polyacrylamide gel electrophoresis with a 3.3% stacking gel and 8 or 10% separating gel. Kaleidoscope Prestained Molecular Weight Markers (Bio-Rad) were used as size determinants. Immunoblots were blocked with 5% nonfat dried milk and 1% BSA in TBS with 0.05% Tween-20 (TBS-T). Immunoblots were washed four times (10 min each) in TBS-T between primary and secondary antibody application. Rabbit anti-dansyl IgG (Molecular Probes, Invitrogen) was used at 1:2500 in block solution diluted 50-fold in TBS-T. Peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratory) was used at 1:20,000. Mouse monoclonal antibody 4D1 to 2F1 or 5C3 to 9F1 (46) or 9D2 to 1F3 (51) and the peroxidase-conjugated goat anti-mouse IgG were diluted similarly. To develop immunoblots, Enhanced Chemiluminescence Substrate (ECL) reagent (PerkinElmer) was used followed by exposure on Kodak Biomax XAR Film.
Pulse Proteolysis of FN and FN Constructs
Trypsin and thermolysin were used, respectively, at concentrations of 0.1–100 μg/ml and 0.1–30 μg/ml for 5 min at 37 °C, as described in the figure legends and results. Thermolysin was inhibited with reducing SDS loading buffer and heating at 95 °C prior to SDS-PAGE or by acidification and zip-tipping for matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-MS).
MALDI-MS Analysis
A Bruker Reflex II mass spectrometer (UW-Madison Chemistry MS Facility) was used for MALDI-MS on thermolysin-treated protein samples. Digested samples were subject to C4 zip-tip desalting and concentration (see protocol, Michrom Biosciences) before applying to a MALDI plate. Elution buffer for the zip-tips was 3–5 μl of saturated α-cyano-4-hydroxycinnammic acid in 70% acetonitrile/30% water for spotting onto the MALDI plate. Laser power of the MALDI-TOF mass spectrometer was varied to give maximum peak resolution. The machine was operated under positive reflectron mode. Peak masses were compared against known terminal amino acid sequences of intact FN and proteolytic fragments. Calibrants used were adenosine diphosphate (ADP, 427.2-Da), a calcium/calmodulin-dependent kinase peptide (CCDK, 1450.6-Da), and bovine insulin (5734.6-Da).
Linear Trap Quadrupole (LTQ)-MS/MS and LTQ-Fourier Transform Ion Cyclotron Resonance MS (LTQ-FT ICR MS) Analysis
Reaction mixtures (see above), were reduced in 10 mm dithiothreitol at 56 °C for 1 h, cooled to 22 °C, and alkylated with 20 mm iodoacetamide at 22 °C for 30 min in the dark. Extensive trypsinization was performed at a 20:1 substrate:protease ratio using sequence grade modified trypsin (Promega, Cat. #V511A) for 16–20 h at 37 °C. An electrospray ThermoScientific LTQ mass spectrometer (UW-Madison Human Proteomics Facility) coupled to an Eksigent nano-liquid chromatography (LC) system fitted with a Michrom Bioresources C4 column (5 μm beads and 300 Å pore) was used for tandem MS. Results were analyzed and compared against the human NCBI database using Xcalibur and Bioworks/Sequest software. Possible alykylation (+57-Da) and dansylcadaverine (318.46-Da; 335.47-Da minus 17-Da for amine loss) modifications were queried for all peptides. Search results were exported to Excel, and spectral counting comparing specific modified and unmodified glutamine-containing peptides was performed using modified/total ×100 to yield percent modified for at least three replicates of each sample separated by the same 5–80% acetonitrile gradient over 60 min, then averaged. Bioworks mass spectrometric data were filtered using a mass tolerance window of 2-Da, p ≤ 5e−02, and Xcorr (z = +1, +2, +3) ≥ 1.5, 2.0, 2.5. MS/MS ion spectra were also examined to confirm that matches were correct for modified peptides. Coverage of glutamine-containing tryptic peptides was 86% for 29K and 73% for 70K.
LTQ-FT ICR MS analysis (UW-Madison Human Proteomics Facility) was performed on immunopurified 29K FN looking for post-translational modifications, including the percent of conversion of the N-terminal Gln to pyroglutamic acid. 29K was dialyzed into 0.1% acetic acid, and diluted to 200 μg/ml in 1% acetic acid and 30% acetonitrile, of which 10 μl were injected. Approximately 200 scans were collected in FT mode. Signals due to modified and unmodified protein were summed, and percent of modification was determined as modified/total ×100 for a given charge state.
RESULTS
TG2-catalyzed Dansylcadaverine Incorporation into FN and N-terminal FN Constructs
Previous studies of radiolabeled putrescine and full-length FN demonstrated TG2-catalyzed incorporation of four moles of amine per FN monomer as compared with two when FXIIIa was tested (25). Higher putrescine incorporation was detected if FN was fragmented first and then treated with the TG2 (25). Building on this study, we incubated plasma FN, N-3F3t, N-3F3, 70K, or 1F3-C with activated TG2 and dansylcadaverine. All proteins incorporated dansylcadaverine as assessed by immunoblotting with anti-dansyl mAb (Figs. 2 and 3). No formation of multimers was noted except with N-3F3t and N-3F3. Incubation of these proteins with TG2 in the absence and presence of dansylcadaverine demonstrated inhibition of multimer formation as monitored by immunoblotting with 4D1.7 anti-FN and by protein staining (Fig. 3), suggesting that the multimers are held together by ϵ-(γ-glutamyl)-lysine isopeptide bonds introduced by TG2.
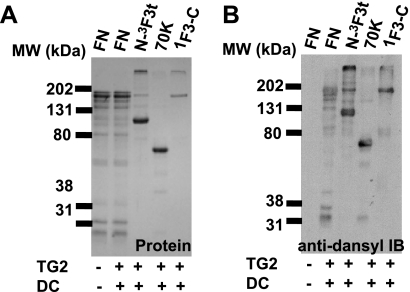
FIGURE 2.
TG2-catalyzed DC incorporation into FN, N-3F3t, 70K, and 3F1-C. The FN sample had some degradation, yielding a 29K fragment. A, protein staining and B, anti-dansyl immunoblotting of TG2-catalyzed dansylcadaverine incorporation.
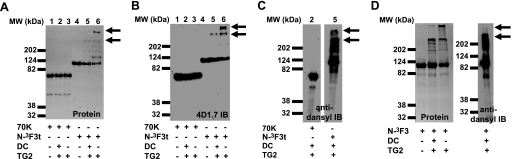
FIGURE 3.
Effects of TG2-catalyzed DC incorporation into N-3F3t, 70K, and N-3F3. A, protein staining of TG2-catalyzed dansylcadaverine incorporation and aggregate formation of N-3F3t and 70K. B, 4D1 immunoblot (IB) of samples in A. Numbers (1–6) on top of the gel indicate similar lanes in A–C. Note aggregate formation of N-3F3t, and inhibition with DC. C, anti-dansyl immunoblotting of indicated lanes from A and B. D, TG2-catalyzed cross-linking and dansylcadaverine incorporation into N-3F3 FN visualized by protein staining and anti-dansyl immunoblotting. N-3F3 acts similar to N-3F3t, indicating the C-terminal tail in N-3F3t is not responsible for aggregate formation. The top arrow in the figures indicate the top of the stacking gel, and the bottom arrow indicates the top of the separating gel.
LTQ-MS/MS Analysis of TG2- and FXIIIa-catalyzed Dansylcadaverine Incorporation into FN
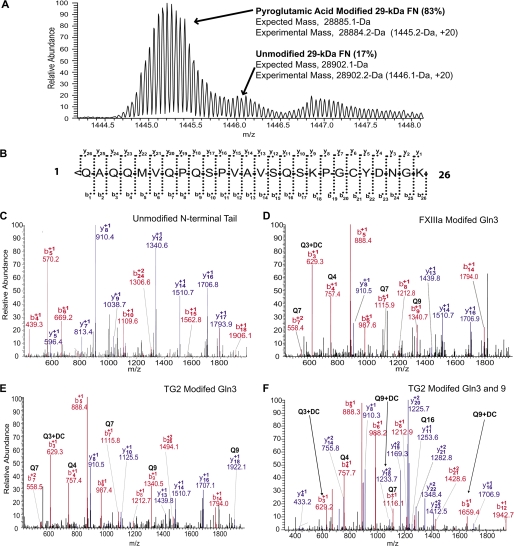
Immunoblotting indicated that TG2 acts predominantly on glutamines near the N terminus of FN. Gln1 of FN is known to exist as pyroglutamic acid (52). To search for this and other prior post-translational modifications, proteolytic 29K immunopurified as described in methods was subject to LTQ-FT ICR MS. Such analysis indicated the modification to pyroglutamate (−17 Da) occurs in ∼85% of molecules (Fig. 4A). This modification was included in the search algorithm for peptides containing dansylcadaverine. No evidence of other modifications was found.
FIGURE 4.
MS analysis of 29K and FXIIIa- and TG2-catalyzed dansylcadaverine modification of the FN N-terminal tail peptide. A, LTQ-FT ICR MS analysis of proteolytic 29K determining percentage of pyroglutamic acid modification (% = (modified/total) x 100). B, potential b- and y-ions for MS/MS of the FN N-terminal tail tryptic peptide (residues 1–26, < denotes pyroglutamic acid modification). C, LTQ LC-MS/MS b- and y-ion spectrum for the unmodified N-terminal tail tryptic peptide. LTQ LC-MS/MS b- and y-ion spectra for D, FXIIIa-catalyzed dansylcadaverine modification of Gln-3 from the FN N-terminal tail tryptic peptide, E, TG2-catalyzed dansylcadaverine modification of Gln-3 from the FN N-terminal tail tryptic peptide, and F, TG2-catalyzed dansycadaverine modification of Gln-3 and Gln-9 from the same peptide.
TG2-catalyzed dansylcadaverine incorporation into FN N-terminal constructs was compared with incorporation catalyzed by FXIIIa, which is known to modify Gln-3 (19) and may also utilize Gln-4, and Gln-16 as predicted by mutagenesis and studies of peptides mimicking the FN N-terminal sequence (21–23). After dansylcadaverine incorporation into N-3F3t, 70K, or 29K, the proteins were subjected to reduction, alkylation, extensive trypsinization, chromatographic separation of tryptic peptides, and tandem MS analysis to map modified glutaminyl residues of transamidation. FXIIIa- and TG2-catalyzed dansylcadaverine incorporation demonstrated major overlap (Tables 1, 2, and 3). Spectral counting of FXIIIa-catalyzed dansylcadaverine incorporation into the N-terminal peptide, containing Gln-3, Gln-4, Gln-7, Gln-9, and Gln-16, indicated 81% N-terminal modification for 29K and 94% for 70K (Fig. 4C and Table 1). TG2-catalyzed dansylcadaverine incorporation into the N-terminal peptide was 95% for 29K and ∼100% for 70K (Table 2). FN N-terminal peptides primarily contained the single dansylcadaverine modification at Gln-3 after FXIIIa-catalyzed reactions, with double modification of Gln-3 and Gln-7 or Gln-9 being found at a lesser frequency (Fig. 4D and Table 1). The N-terminal peptides in TG2-treated samples consisted of a mix of a single Gln-3 modification and of Gln-3 modified along with Gln-4, Gln-7, Gln-9, and/or Gln-16 (Fig. 4, E and F, and Table 2). Earlier mutagenesis studies on the FN N terminus indicated that Gln-4 and Gln16 are subject to transamidation by FXIIIa when Gln-3 was mutated (21), and studies of peptide mimics demonstrated major incorporation of dansylcadaverine into both Gln-3 and Gln-4 (22–23). However, in our tandem MS analyses of various FN constructs, modification of Gln-4 or Gln-16 with dansylcadaverine was detected minimally (Tables 1 and 2). The fifth residue in the sequence ADPGQAQQ… of recombinant N-3F3t, corresponding to Gln-1 in FN, was modified by both enzymes in conjunction with modification of the seventh residue corresponding to Gln-3 (Table 3). Thus, Gln-3 is the major target of both FXIIIa and TG2, and additional glutamines in the immediate vicinity of Gln-3 are secondary targets.
TABLE 1.
LTQ-MS/MS and spectral counting of dansylcadaverine-modified (+318.46-Da) tryptic peptides after FXIIIa-catalyzed incorporation into 29K, 70K, or N-3F3t
| Residues (Module) | Modified Gln | Est. mass | Exp. mass | FXIIIa % modified |
||
|---|---|---|---|---|---|---|
| 29K | 70K | N-3F3t | ||||
| Da | Da | |||||
| 1–26 (N-terminal tail) | None | 2815.13 | 2814.75 | 19% | 6% | a |
| 3 | 3133.80 | 3132.99 | 81% | 82% | a | |
| 3,7 | 3452.26 | 3451.74 | NDb | 6% | a | |
| 3,9 | 3452.26 | 3451.55 | ND | 6% | a | |
| 27–36 (1F1) | None | 1401.67 | 1401.70 | 79% | 90% | 76% |
| 29 | 1720.13 | 1720.51 | ND | 2% | 3% | |
| 32 | 1720.13 | 1720.38 | 18% | 8% | 17% | |
| 33 | 1720.13 | 1719.54 | 3% | ND | 3% | |
| 242–259 (Linker) | None | 1863.88 | 1864.21 | 83% | 97% | 75% |
| 246 | 2182.34 | 2183.35 | 17% | 3% | 25% | |
| 349–366 (6F1-1F2) | None | 2196.93 | 2197.11 | NAc | 98% | 89% |
| 365 | 2515.39 | 2514.76 | NA | 2% | 11% | |
| 367–380 (1F2) | None | 1726.82 | 1726.77 | NA | 100% | 99% |
| 378 | 2045.28 | 2045.61 | NA | ND | 1% | |
| 473–484 (7F1-8F1) | None | 1483.70 | 1483.81 | NA | 100% | 98% |
| 482 | 1802.16 | 1802.55 | NA | ND | 2% | |
| 485–502 (8F1) | None | 2196.99 | 2195.97 | NA | ND | 86% |
| 486 | 2515.45 | 2514.96 | NA | ND | 14% | |
| 639–663 (3F3) | None | 2799.48 | 2798.62 | NA | NA | 98% |
| 659 | 3117.94 | 3117.38 | NA | NA | 2% | |
| 800–811 (3F3) | None | 1431.75 | 1431.62 | NA | NA | 99% |
| 804 | 1750.21 | 1749.59 | NA | NA | 1% | |
a See Table III.
b ND indicates not detected.
c NA indicates not applicable.
TABLE 2.
LTQ-MS/MS and spectral counting of dansylcadaverine-modified (+318.46-Da) tryptic peptides after TG2-catalyzed incorporation into 29K, 70K, or N-3F3t
| Residues (Module) | Modified Gln | Est. Mass | Exp. Mass | TG2% modified |
||
|---|---|---|---|---|---|---|
| 29K | 70K | N-3F3t | ||||
| Da | Da | |||||
| 1–26 (N-terminal Tail) | None | 2815.13 | 2815.81 | 5% | NDa | b |
| 3 | 3133.80 | 3132.99 | 45% | 21% | b | |
| 3,7 | 3452.26 | 3451.74 | ND | 5% | b | |
| 3,9 | 3452.26 | 3451.55 | 23% | 62% | b | |
| 3,16 | 3452.26 | 3452.67 | 9% | 4% | b | |
| 3,4,9 | 3770.72 | 3770.55 | 18% | 8% | b | |
| 27–36 (1F1) | None | 1401.67 | 1401.70 | 48% | 100% | 87% |
| 29 | 1720.13 | 1720.70 | 23% | ND | 5% | |
| 32 | 1720.13 | 1720.62 | 7% | ND | 4% | |
| 33 | 1720.13 | 1719.61 | 22% | ND | 4% | |
| 242–259 (Linker) | None | 1863.88 | 1864.21 | 20% | 59% | 44% |
| 246 | 2182.34 | 2181.92 | 80% | 41% | 56% | |
| 367–380 (1F2) | None | 1726.82 | 1726.93 | NAc | 100% | 99% |
| 378 | 2045.28 | 2044.71 | NA | ND | 1% | |
| 473–484 (7F1-8F1) | None | 1483.70 | 1484.09 | NA | 100% | 95% |
| 482 | 1802.16 | 1802.53 | NA | ND | 5% | |
| 524–547 (8F1-9F1) | None | 2865.16 | 2865.21 | NA | 94% | 100% |
| 529 | 3183.62 | 3182.50 | NA | 6% | ND | |
| 800–811 (3F3) | None | 1431.75 | 1431.81 | NA | NA | 65% |
| 804 | 1750.21 | 1750.62 | NA | NA | 35% | |
a ND, not detected.
b See Table III.
c NA, not applicable.
TABLE 3.
LTQ-MS/MS analysis and spectral counting of the tryptic N-terminal tail (ADPG plus residues 1–26) generated after FXIIIa- or TG2-catalyzed dansylcadaverine incorporation (+318.46-Da) into recombinant N-3F3t
| Residues (Module) | Modified Gln | Est. Mass | Exp. Mass (% FXIIIa Modified) | Exp. Mass (% TG2 Modified) |
|---|---|---|---|---|
| Da | Da | Da | ||
| ADPG+1–26 (N-terminal tail) | None | 3172.47 | NDa | 3171.53 (33%) |
| 3 | 3490.93 | 3489.56 (6%) | ND | |
| 1,3 | 3809.40 | 3808.28 (61%) | ND | |
| 1,4 | 3809.40 | 3808.28 (6%) | ND | |
| 3,4 | 3809.40 | 3809.08 (8%) | ND | |
| 3,9 | 3452.26 | ND | 3451.74 (23%) | |
| 1,3,4 | 4127.86 | 4126.86 (4%) | ND | |
| 1,3,9 | 4127.86 | 4127.15 (15%) | 4127.95 (44%) |
a ND, not detected.
N-3F3t, 70K, or 29K were modified by TG2 or FXIIIa at Gln-246 in the N-terminal half of the sequence linking repeats 5F1 and 6F1 (Tables 1 and 2). As quantified by spectral counting, dansylcadaverine was found in 41–80% and 3–25% of peptide 242–259 after reaction with TG2 and FXIIIa, respectively (Tables 1 and 2). Such degrees of Gln-246 modification approached the level that was detected of N-terminal tail glutamines after incubation with TG2 but was less with FXIIIa.
Additional glutamines within the modules of N-3F3t, 70K, and 29K were found to be modified by dansylcadaverine, but the modifications were minor for both TG2 and FXIIIa as determined by spectral counting of unmodified and modified peptides detected by tandem MS analysis (Tables 1 and 2). In addition to the residues described in the tables, the Gln226-containing tryptic peptide from the 5F1 module and the Gln-419-containing tryptic peptide from the 2F2 module were found to be modified by either FXIIIa or TG2, but unmodified peptide was not detected consistently in control samples not treated with TGs, making it impossible to perform meaningful spectral counting on this peptide.
Pulse Proteolysis of FN and FN Constructs
Previous studies (25) indicated that incorporation of amines into FN fragments may be different than incorporation into intact FN. We could not perform consistent high coverage LC-tandem MS on the peptides resulting from exhaustive trypsin digestion of plasma FN or of 1F3-C that labeled with dansylcadavereine (Fig. 2) because of the complexity of the peptide mixtures. Therefore, we used thermolysin, which is known to cleave efficiently at hydrophobic residues near the terminal regions of proteins (53), to examine whether the major sites of dansylcadaverine incorporation identified in the N-terminal constructs are the major sites found in intact plasma FN or additional sites are present in the part of the protein represented by 1F3-C. Upon digestion of 29K with thermolysin, peptides arising from cleavages in the N-terminal tail and the linker region proximal to 5F1 were detected (Table 4). When intact plasma FN was labeled with dansylcadaverine using FXIIIa, digested with increasing amounts of trypsin, separated, and immunoblotted, the 29K N-terminal fragment was detected in the limited trypsin digest by both 4D1 and anti-dansyl (Fig. 5A). Limited thermolysin digest, in contrast, yielded a ∼25-kDa doublet recognized by 4D1 but not by anti-dansyl (Fig. 5B). Residual staining for dansylcadaverine was present in high molecular fragments although the staining was much diminished compared with the no-thermolysin control. Further comparisons of trypsin versus thermolysin digests of plasma FN, N-3F3t, 70K, or 29K that had been subjected to TG2-catalyzed dansylcadaverine incorporation and after digestion analyzed by SDS-PAGE, protein staining (Fig. 5C), and immunoblotting with anti-1F3 antibody 9D2 (Fig. 5D), anti-2F1 antibody 4D1 (Fig. 5E), and anti-dansyl (Fig. 5F) yielded similar results. Dansylcadaverine incorporated into FN, N-3F3t, 70K, or 29K was lost upon thermolysin digestion as assessed by anti-dansyl immunoblotting (Fig. 5F). These results indicate that most FXIIIa- and TG2-catalyzed dansylcadaverine incorporation is in glutamines N- and C-terminal to the 1F1-5F1 tandem modules that were detected in analyses of 29K, verifying spectral counting results.
TABLE 4.
MALDI-MS analysis of peptides generated by limited thermolysin cleavage of 29K (residues 1–259)
| Residues | Sequence | Est. mass | Exp. mass |
|---|---|---|---|
| Da | Da | ||
| 1–5 | QAQQM | 604.26 | 603.53 |
| 1–5 | <QAQQMa | 587.24 | 586.50 |
| 1–6 | <QAQQMV | 686.31 | 686.10 |
| 1–11 | QAQQMVQPQSP | 1240.59 | 1238.40 |
| 1–11 | <QAQQMVQPQSP | 1223.56 | 1221.64 |
| 1–12 | QAQQMVQPQSPV | 1339.66 | 1340.29 |
| 1–12 | <QAQQMVQPQSPV | 1322.63 | 1323.26 |
| 245–259 | VQTTSSGSGPFTDVR | 1537.74 | 1538.78 |
a <, modification with pyroglutamic acid.
FIGURE 5.
Characterization of FXIIIa- and TG2-catalyzed DC incorporation into plasma FN, N-3F3t, 70K, and 29K via limited proteolytic treatment of varying concentrations for 5 min at 37 °C. A, digestion with variable trypsin concentrations after FXIIIa-catalyzed dansylcadaverine incorporation into FN, followed by 4D1 and anti-dansyl immunoblotting. B, digestion with variable trypsin concentrations after FXIIIa-catalyzed dansylcadaverine incorporation into FN, followed by 4D1 and anti-dansyl immunoblotting. Note the loss of dansylcadaverine incorporation after digestion with thermolysin in B but not with trypsin in A. C–F, staining for protein (C) and 9D2 (D), 4D1 (E), and α-dansyl (F) immunoblotting after TG2-catalyzed dansylcadaverine incorporation in FN, N-3F3t 70K, or 29K followed by digestion with 30 μg/ml thermolysin. In F, dansylcadaverine incorporation is seen in all proteins and lost with digestion, indicating that most of the amine is in the peptides described in Table 4. Note that a portion of the plasma FN degraded during the 3 h of incubation at 37 °C dansylcadaverine incorporation; this preparation showed the same sensitivity to thermolysin as in panel B.
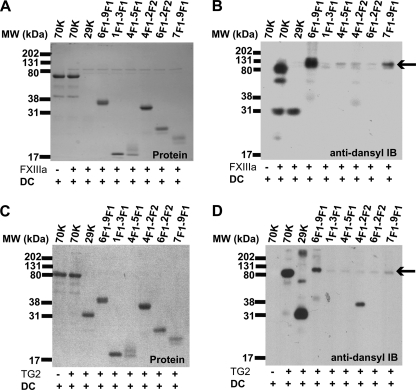
FN Modules Required for Efficient FXIIIa- and TG2-catalyzed Transamidation
Proteolytic 70K and 29K or recombinant 6F1-9F1, N-3F1, 4F1-5F1, 4F1-2F2, 6F1-2F2, or 7F1-9F1 at equal molar concentrations were subjected to dansylcadaverine labeling by FXIIIa or TG2. Dansylcadaverine incorporation was detected in 70K or 29K with both enzymes (Fig. 6), as well as incorporation into 4F1-2F2 with TG2. The 29K multimers found after incubation with TG2 but not with FXIIIa were most likely due to residual dithiothreitol from TG2 activation reducing and causing aggregation of F1 modules as described previously (54). There was no detectable dansylcadaverine incorporation in N-3F1, 4F1-5F1, 6F1-9F1 (40K), 6F1-2F2, or 7F1-9F1 (Fig. 6). The findings of dansylcadaverine incorporation into 29K and into 4F1-2F2 containing the linker between modules 5F1 and 6F1, but not into N-3F1 or 4F1-5F1 individually, indicate that transamidation of the N-terminal glutamines by either FXIIIa or TG2 requires an intact 29K N terminus.
FIGURE 6.
FXIIIa- and TG2-catalyzed DC incoporation into FN constructs. A and B, protein staining and anti-dansyl immunoblot (IB) of FXIIIa-catalyzed dansylcadaverine incorporation. C and D, protein staining and anti-dansyl immunoblotting of TG2-catalyzed dansylcadaverine incorporation. Arrows indicate positions of FXIIIa and TG2 as determined by immunoblotting (data not shown).
The reaction mixtures of 6F1-9F1 and FXIIIa or TG2 contained ∼80-kDa bands recognized by anti-dansyl immunoblotting (Fig. 6). Protein staining (Fig. 6, A and C) and 5C3 immunoblotting of the same reaction mixtures (data not shown) indicated 6F1-9F1 FN was at the expected ∼40-kDa position. Because FXIIIa and TG2 are 80-kDa proteins, we suspected self-transamidation. Immunoblotting of the reaction mixtures with anti-TG2 or anti-FXIIIa yielded bands that co-migrated with anti-dansyl labeling (data not shown). Tandem MS analysis and spectral counting revealed FXIIIa-catalyzed self-incorporation of dansylcadaverine into peptides containing Gln-43, Gln-488, and Gln-725. Similar analysis revealed that TG2-catalyzed self-incorporation occurred at Gln-234 and Gln-307.
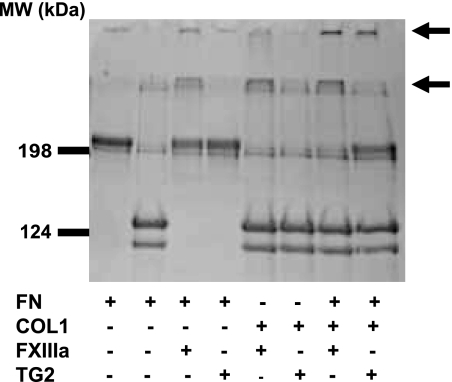
FN and Collagen Cross-linking
Given the similarities in the sites of TG2- and FXIIIa-catalyzed transamidation near the FN N terminus seen in this study, we asked if there were similarities between the enzymes in relation to cross-linking of FN and collagen, as has been shown previously for FXIIIa (18). This is an important question because binding of TG2 to the gelatin-binding region of FN could prevent FN-collagen complex formation (55). FN and collagen, both at 200 μg/ml, were incubated alone or together with active TG2 or FXIIIa. Only in reactions containing both proteins plus TG2 or FXIIIa were large cross-linked aggregates observed at the top of the stacking gel (Fig. 7), in accordance with cross-linked aggregates described previously (18). These results indicate overlapping activities of TG2 and FXIIIa in relation to FN and collagen cross-linking.
FIGURE 7.
FXIIIa-and TG2-catalyzed FN and Type I collagen (COL1) cross-link aggregate formation. Both enzymes caused appearance of protein staining bands at the top of the separating and stacking gels (arrows) in comparison to FN or collagen alone.
DISCUSSION
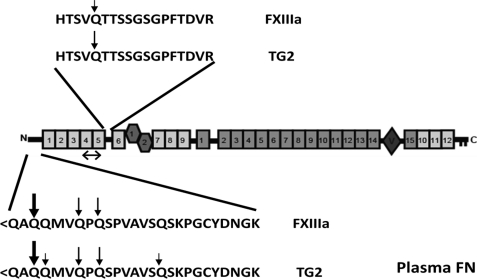
The goals of the present study were to identify the glutamines of FN subject to TG2-catalyzed dansylcadaverine incorporation, compare the substrate specificities of TG2 and FXIIIa, and explore features of FN that direct the enzymes to specific glutamines. To achieve these goals, we subjected a number of FN constructs to a single set of conditions with an enzyme:substrate (M:M) ratio of ∼0.15, dansylcadaverine concentration of 0.5 mm, and a 3-h incubation at 37 °C. Immunoblotting and tandem MS were used as independent, yet supporting, read-outs for dansylcadaverine incorporation. Several findings emerged from these studies, some of which are summarized in Fig. 8. 1) The same glutamines are targets for both enzymes. 2) Gln3 in the flexible N-terminal tail of FN (24) is the preferred target for both enzymes, and surrounding glutamines in the tail are secondary targets. 3) Gln246, which is at the opposite end of the N-terminal tandem 1F1-5F1 modules from Gln3, is a target for both enzymes but a better substrate for TG2 than FXIIIa. Gln-246 is in a sequence that can be cleaved readily by a number of proteases (13) and presumably is flexible. 4) Modification of the N-terminal glutamines occurs efficiently when 4–5F1 is present. While 29K exhibited dansylcadaverine incorporation, N-3F1 and 4F1-5F1 subconstructs alone had no detectible incorporated dansylcadaverine. Importantly, TG2 incorporated dansylcadaverine into 29K, suggesting that the 29K region binds to an exosite on active TG2 that directs the enzyme to the reactive glutamines in a way that is independent of its binding site in the gelatin-binding domain. 5) In the absence of suitable substrate glutamines, TG2 and FXIIIa catalyze dansylcadaverine incorporation into themselves. 6) N-3F3t and N-3F3 treated with TG2 underwent intermolecular cross-linking that was inhibited by the presence of dansylcadaverine (Figs. 2 and 3), suggesting the presence of exposed lysines suitable for cross-linking in 1F3-3F3 modules of the truncated constructs that are positioned differently in intact FN. Modules 1F3 and 3F3 associate with the 29K N-terminal region when these parts of FN are studied as fragments (56–58). 7) Both TG2 and FXIIIa crosslink FN and collagen.
FIGURE 8.
Summary of FXIIIa- and TG2-catalyzed transamidation of the plasma FN N-terminal region. Primary glutamines of modification for each enzyme are indicated by the boldest arrows with descending arrow size indicating decreasing modification. The double arrow indicates a site of potential transglutaminase exosite binding in 29K FN for directing transamidation.
Previously, TG2 was found to incorporate four moles of radioactive putrescine into FN in conditions which FXIIIa incorporated two (25). When FN fragments were tested, the majority of incorporation was in N-terminal fragments. In our studies, there was a tendency toward increasing modification by TG2 compared with FXIIIa, but we found no all-or-none differences in individual residues that would account for a doubling in the incorporation of amine. Thermolysin, which cleaves N- and C-terminally to hydrophobic residues (53), was used to relate the tandem MS studies of truncated or fragmented pieces of FN to immunoblotting of intact FN for incorporated dansylcadaverine. Thermolysin is known to cleave between modules 5F1 and 6F1 and between modules 1F3 and 2F3 (13, 59–60). Our MALDI-MS analysis revealed cleavages that release peptides containing Gln-1, Gln-3, Gln-4, Gln-7, Gln-9, and Gln-246, leaving 1F1-5F1 without tails. Concomitant loss of dansylcadaverine as assessed by immunoblotting indicated that glutamines in the N-terminal tail and flexible linker sequence between 5F1 and 6F1 are the major sites for TG2- and FXIIIa-catalyzed amine incorporation (Fig. 5). Thus, minor and variable incorporation of dansylcadaverine into glutamines within the 1F1-5F1 core detected by tandem MS (Tables 1 and 2) was not enough to detect by immunoblotting when the major glutamine substrates were lost.
The above results are consistent with the finding that TG2 and FXIIIa both preferentially modify Gln2 of Asn-α2-anti-plasmin, the major serpin-type plasmin inhibitor of blood plasma (8, 38, 61). α2-Anti-plasmin also circulates as Met-α2-anti-plasmin, i.e. 12 residues at its N terminus are removed by anti-plasmin-converting enzyme to yield Asn-α2-anti-plasmin (8, 38). Gln-14 of Met-α2-anti-plasmin (the same as Gln-2 of Asn-α2-anti-plasmin) remains the major glutamine modified by FXIIIa (38). Similarly, Gln-7 of recombinant N-3F3t (the same as Gln-3 of FN) was the preferred target of FXIII and TG2 despite the extra 4 residues at the N terminus (Table 3). In the case of FN, interaction of the enzymes with 4–5F1, which one may assume is some distance from Gln3 based on NMR structures of tandem F1 modules in 29K (62–63), seems to direct FXIIIa or TG2 to N-terminal glutamines. Modification of Gln-3 and Gln-4 of osteonectin by TG2 is greater when the adjacent osteonectin acidic and α-helical N-domain are present (64–65), suggesting that osteonectin is like FN in having a remote binding site that determines activity. The structural motifs of osteonectin and FN outside of the N-terminal tails are quite different. Thus, it is difficult to predict whether TG2 recognizes a common feature on FN and osteonectin that directs enzymatic activity to the N-terminal glutamines. In the case of Asn-α2-anti-plasmin, the N-terminal tail seems to be sufficient to determine specificity of FXIIIa based on the finding that a fusion protein in which the Asn-α2-anti-plasmin tail is joined to albumin exhibits only minimal loss of amine incorporation by FXIIIa compared with Asn-α2-anti-plasmin itself (8, 66). Our observations taken together with observations in the literature indicate that specificities of FXIIIa and TG2 for FN are determined by properties of the N-terminal tail that are suboptimal compared with the tail of Asn-α2-anti-plasmin but compensated for by an exosite in 4–5F1.
Finding that the TG2-catalyzed transamidation of the proteolytic 29K N-terminal fragment is independent of the 40K gelatin-binding domain of FN squares with the finding that mutant TG2 without transamidation activity (67) or the compact, enzymatically inactive, calcium-depleted form of non-mutant TG2 binds to the gelatin-binding region of FN. The results raise the questions of whether FXIIIa is like TG2 in binding to the 40K gelatin-binding region and whether the presumed exosites in the two enzymes for 29K can be demonstrated by binding assays. To address these questions, we did a set of ELISAs to test abilities of the soluble FN fragments or constructs, all assayed at a single invariant concentration, to compete for binding of the transglutaminases to substrate-adsorbed FN (supplemental Fig. S1). The ELISA is based on the published assay that demonstrated interaction of TG2 in EDTA with soluble 40K (26). In the absence of potential competitor, FXIIIa and both inactive TG2 (in EDTA) and active TG2 (in calcium) bound to substrate-adsorbed FN. However, a higher concentration of soluble FXIIIa than TG2 was needed to obtain a good signal and more soluble FN was required to compete for binding of FXIIIa than to compete for binding of TG2 (data not shown). We surveyed 29K, N-3F1, 4F1-5F1, and 70K or N-3F3 in addition to 40K as soluble competitors for binding of inactive or active TG2 (supplemental Fig. 1A) or FXIIIa (supplemental Fig. 1B). Soluble 40K inhibited binding of inactive and active TG2 but not binding of FXIIIa. Soluble 29K, in contrast, inhibited binding of active TG2 and FXIIIa but not binding of inactive TG2. Subconstructs of 29K, N-3F1 and 4–5F1, were ineffective in inhibiting binding of either transglutaminase. These experiments indicate that binding of TG2 to 40K is an attribute that is not shared with FXIIIa and active TG2 and FXIIIa both bind to 29K, although we could not map their binding sites further to 4–5F1.
The complex of active TG2 and 40K may lack catalytic activity for certain macromolecular substrates, as demonstrated in previous studies (68) and in Fig. 6. In contrast, the interaction of active TG2 and 29K results in transamidation of 29K. Sequences responsible for binding of TG2 with the gelatin-binding domain of FN have been mapped to the N-terminal region of TG2 (69–71). The data do not allow distinction between an additional exosite in TG2 for binding to the 29K N-terminal modules of FN by the catalytic form of TG2 or alternative use of the same exosite that recognizes the gelatin-binding modules. Thus, what determines interaction of extended catalytic TG2 with 29K modules versus 40K modules could depend on enzyme conformation and exosite availability or be random, with the enzyme partitioning between the two sites in FN. Further, none of our data rule out the possibility that TG2 can bind to both sites on FN simultaneously.
Immunoblotting and mass spectrometry demonstrated self-labeling of the enzymes in the absence of exogenous substrate that was enhanced when the 40K gelatin-binding domain of FN was present. Mapping the self-labeled sites on crystal structures of TG2 (67, 72) and FXIIIA dimer (73) indicates that Gln-234 and Gln-307 of TG2 are in the catalytic core, whereas Gln-43, Gln-488, and Gln-725 of FXIIIa are, respectively, at the beginning of the N-terminal β-sandwich, in the catalytic core, and in C-terminal β-barrel 2. Interestingly, the FXIIIa-modified glutamines are not conserved in TG2, and the TG2-modified glutamines are not conserved in FXIIIa. Because Gln-43 and Gln-725 of FXIIIa are at the N and C termini and Gln-488 is on the opposite side of the enzyme away from the active site cysteine, FXIIIa may label adjacent FXIIIa. In contrast, Gln-234 and Gln-307 of TG2 are in flexible regions near the active site cysteine, suggesting possible intramolecular self-labeling.
Results presented in this study indicate similar actions of TG2- and FXIIIa-catalyzed actions on glutamines near the FN N terminus and in FN-collagen cross-linking. Despite these similarities, FXIIIa and TG2 likely act on FN in different contexts. Plasma FXIII is activated by thrombin during blood coagulation and crosslinks fibrin to itself and to FN and α2-anti-plasmin in the hemostatic plug and may also function during wound remodeling (4–6). Exactly when the transamidation action of TG2 is functionally relevant is an area of great interest and likely encompasses physiological and pathophysiological processes in which cells are stressed, including thrombosis and wound healing but perhaps later in the time course of these processes (4, 31, 74–75).
Supplementary Material
Acknowledgments
We thank Dr. Prasanna Murthy for TG2, and the support and suggestions of Dr. Laszlo Lorand; both are at Northwestern University. The MS studies were performed on instruments of the University of Wisconsin-Madison Human Proteomics Program facility funded by the Wisconsin Partnership Fund for a Healthy Future and the Department of Chemistry Mass Spectrometry facility funded in part by National Institutes of Health NCRR 1S10RR024601-1. We gratefully acknowledge the advice and support of the facility directors, Drs. Ying Ge and Martha Vestling.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL021644 (to D. F. M.) and T32-HL007899 (to B. R. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TG
- transglutaminase
- FN
- fibronectin
- FXIIIa
- activated Factor XIII
- TG2
- transglutaminase 2
- ECM
- extracellular matrix
- 29K
- 29-kDa FN N terminus
- 40K
- 40-kDa FN gelatin-binding domain
- 70K
- 70-kDa FN N terminus
- F1
- FN type 1 module
- F2
- FN type 2 module
- F3
- FN type 3 module
- LC
- liquid chromatography
- LTQ
- linear trap quadrapole
- MS
- mass spectrometry
- FT ICR
- fourier transform ion cyclotron resonance
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- DC
- dansylcadaverine.
REFERENCES
- 1. Lorand L., Graham R. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 2. Esposito C., Caputo I. (2005) FEBS J. 272, 615–631 [DOI] [PubMed] [Google Scholar]
- 3. Griffin M., Casadio R., Bergamini C. M. (2002) Biochem. J. 368, 377–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iismaa S. E., Mearns B. M., Lorand L., Graham R. M. (2009) Physiol. Rev. 89, 991–1023 [DOI] [PubMed] [Google Scholar]
- 5. Lorand L. (2007) FASEB J. 21, 1627–1632 [DOI] [PubMed] [Google Scholar]
- 6. Cho J., Mosher D. F. (2006) Blood 107, 3555–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsuka Y. V., Migliorini M. M., Ingham K. C. (1997) J. Protein Chem. 16, 739–745 [DOI] [PubMed] [Google Scholar]
- 8. Lee K. N., Lee C. S., Tae W. C., Jackson K. W., Christiansen V. J., McKee P. A. (2000) J. Biol. Chem. 275, 37382–37389 [DOI] [PubMed] [Google Scholar]
- 9. Karimi M., Bereczky Z., Cohan N., Muszbek L. (2009) Semin. Thromb. Hemost. 35, 426–438 [DOI] [PubMed] [Google Scholar]
- 10. Fesus L., Piacentini M. (2002) Trends Biochem. Sci. 27, 534–539 [DOI] [PubMed] [Google Scholar]
- 11. Sarang Z., Tóth B., Balajthy Z., Köröskényi K., Garabuczi E., Fésüs L., Szondy Z. (2009) Amino Acids 36, 625–631 [DOI] [PubMed] [Google Scholar]
- 12. Sane D. C., Kontos J. L., Greenberg C. S. (2007) Front Biosci. 12, 2530–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pankov R., Yamada K. M. (2002) J. Cell Sci. 115, 3861–3863 [DOI] [PubMed] [Google Scholar]
- 14. Magnusson M. K., Mosher D. F. (1998) Arterioscler. Thromb. Vasc. Biol. 18, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 15. Mao Y., Schwarzbauer J. E. (2005) Matrix Biol. 24, 389–399 [DOI] [PubMed] [Google Scholar]
- 16. Mosher D. F. (1975) J. Biol. Chem. 250, 6614–6621 [PubMed] [Google Scholar]
- 17. Keski-Oja J., Mosher D. F., Vaheri A. (1976) Cell 9, 29–35 [DOI] [PubMed] [Google Scholar]
- 18. Mosher D. F., Schad P. E., Kleinman H. K. (1979) J. Clin. Invest. 64, 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDonagh R. P., McDonagh J., Petersen T. E., Thøgersen H. C., Skorstengaard K., Sottrup-Jensen L., Magnusson S., Dell A., Morris H. R. (1981) FEBS Lett. 127, 174–178 [DOI] [PubMed] [Google Scholar]
- 20. McDonagh J. (1987) in Hemostasis and Thrombsosis (Colman R. W., Hirsh J., Marder V. J., Salzman E. W. eds), pp. 289–300, J.B. Lipincott Co., Philadelphia [Google Scholar]
- 21. Corbett S. A., Lee L., Wilson C. L., Schwarzbauer J. E. (1997) J. Biol. Chem. 272, 24999–25005 [DOI] [PubMed] [Google Scholar]
- 22. Parameswaran K. N., Velasco P. T., Wilson J., Lorand L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8472–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sato H., Yamada N., Shimba N., Takahara Y. (2000) Anal. Biochem. 281, 68–76 [DOI] [PubMed] [Google Scholar]
- 24. Potts J. R., Phan I., Williams M. J., Campbell I. D. (1995) Nature Struct. Biol. 2, 946–950 [DOI] [PubMed] [Google Scholar]
- 25. Fesus L., Metsis M. L., Muszbek L., Koteliansky V. E. (1986) Eur J. Biochem. 154, 371–374 [DOI] [PubMed] [Google Scholar]
- 26. Radek J. T., Jeong J. M., Murthy S. N., Ingham K. C., Lorand L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3152–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akimov S. S., Belkin A. M. (2001) J. Cell Sci. 114, 2989–3000 [DOI] [PubMed] [Google Scholar]
- 28. Jones R. A., Nicholas B., Mian S., Davies P. J., Griffin M. (1997) J. Cell Sci. 110, 2461–2472 [DOI] [PubMed] [Google Scholar]
- 29. Aeschlimann D., Thomazy V. (2000) Connect Tissue Res. 41, 1–27 [DOI] [PubMed] [Google Scholar]
- 30. Forsprecher J., Wang Z., Nelea V., Kaartinen M. T. (2009) Amino Acids 36, 747–753 [DOI] [PubMed] [Google Scholar]
- 31. Haroon Z. A., Hettasch J. M., Lai T. S., Dewhirst M. W., Greenberg C. S. (1999) Faseb J. 13, 1787–1795 [DOI] [PubMed] [Google Scholar]
- 32. Akimov S. S., Krylov D., Fleischman L. F., Belkin A. M. (2000) J. Cell Biol. 148, 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akimov S. S., Belkin A. M. (2001) Blood 98, 1567–1576 [DOI] [PubMed] [Google Scholar]
- 34. Gorman J. J., Folk J. E. (1980) J. Biol. Chem. 255, 1175–1180 [PubMed] [Google Scholar]
- 35. Gorman J. J., Folk J. E. (1981) J. Biol. Chem. 256, 2712–2715 [PubMed] [Google Scholar]
- 36. Shainoff J. R., Urbanic D. A., DiBello P. M. (1991) J. Biol. Chem. 266, 6429–6437 [PubMed] [Google Scholar]
- 37. Murthy S. N., Wilson J., Guy S. L., Lorand L. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10601–10604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hevessy Z., Patthy A., Kárpáti L., Muszbek L. (2000) Thromb Res. 99, 399–406 [DOI] [PubMed] [Google Scholar]
- 39. Pastor M. T., Diez A., Pérez-Payá E., Abad C. (1999) FEBS Lett. 451, 231–234 [DOI] [PubMed] [Google Scholar]
- 40. Mitkevich O. V., Shainoff J. R., DiBello P. M., Yee V. C., Teller D. C., Smejkal G. B., Bishop P. D., Kolotushkina I. S., Fickenscher K., Samokhin G. P. (1998) J. Biol. Chem. 273, 14387–14391 [DOI] [PubMed] [Google Scholar]
- 41. Stenberg P., Curtis C. G., Wing D., Tong Y. S., Credo R. B., Gray A., Lorand L. (1975) Biochem. J. 147, 155–163 [PMC free article] [PubMed] [Google Scholar]
- 42. Lorand L., Murthy S. N., Velasco P. T., Karush F. (1986) Biochem. Biophys. Res. Commun. 134, 685–689 [DOI] [PubMed] [Google Scholar]
- 43. Mosher D. F., Johnson R. B. (1983) J. Biol. Chem. 258, 6595–6601 [PubMed] [Google Scholar]
- 44. Murthy S. N., Velasco P. T., Lorand L. (1998) Exp Eye Res. 67, 273–281 [DOI] [PubMed] [Google Scholar]
- 45. McKeown-Longo P. J., Mosher D. F. (1985) J. Cell Biol. 100, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maurer L. M., Tomasini-Johansson B. R., Ma W., Annis D. S., Eickstaedt N. L., Ensenberger M. G., Satyshur K. A., Mosher D. F. (2010) J. Biol. Chem. 285, 41087–41099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu X., Zhao Q., Collodi P. (2003) Matrix Biol. 22, 393–396 [DOI] [PubMed] [Google Scholar]
- 48. Mosher D. F., Huwiler K. G., Misenheimer T. M., Annis D. S. (2002) Methods Cell Biol. 69, 69–81 [DOI] [PubMed] [Google Scholar]
- 49. Sabatier L., Chen D., Fagotto-Kaufmann C., Hubmacher D., McKee M. D., Annis D. S., Mosher D. F., Reinhardt D. P. (2009) Mol. Biol. Cell 20, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu J., Bae E., Zhang Q., Annis D. S., Erickson H. P., Mosher D. F. (2009) PLoS ONE 4, e4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chernousov M. A., Fogerty F. J., Koteliansky V. E., Mosher D. F. (1991) J. Biol. Chem. 266, 10851–10858 [PubMed] [Google Scholar]
- 52. Mosesson M. W., Umfleet R. A. (1970) J. Biol. Chem. 245, 5728–5736 [PubMed] [Google Scholar]
- 53. Bark S. J., Muster N., Yates J. R., 3rd, Siuzdak G. (2001) J. Am. Chem. Soc. 123, 1774–1775 [DOI] [PubMed] [Google Scholar]
- 54. Williams E. C., Janmey P. A., Johnson R. B., Mosher D. F. (1983) J. Biol. Chem. 258, 5911–5914 [PubMed] [Google Scholar]
- 55. Atkin K. E., Brentnall A. S., Harris G., Bingham R. J., Erat M. C., Millard C. J., Schwarz-Linek U., Staunton D., Vakonakis I., Campbell I. D., Potts J. R. (2010) J. Biol. Chem. 285, 36977–36983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hocking D. C., Sottile J., McKeown-Longo P. J. (1994) J. Biol. Chem. 269, 19183–19187 [PubMed] [Google Scholar]
- 57. Morla A., Ruoslahti E. (1992) J. Cell Biol. 118, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vakonakis I., Staunton D., Ellis I. R., Sarkies P., Flanagan A., Schor A. M., Schor S. L., Campbell I. D. (2009) J. Biol. Chem. 284, 15668–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sekiguchi K., Hakomori S. (1983) Biochemistry 22, 1415–1422 [DOI] [PubMed] [Google Scholar]
- 60. Berry H., Pauthe E., Gallet O., Larreta-Garde V. (1998) Ann. N.Y. Acad. Sci. 864, 198–202 [DOI] [PubMed] [Google Scholar]
- 61. Lee K. N., Lee C. S., Tae W. C., Jackson K. W., Christiansen V. J., McKee P. A. (2001) Ann. N.Y. Acad. Sci. 936, 335–339 [DOI] [PubMed] [Google Scholar]
- 62. Staunton D., Millard C. J., Aricescu A. R., Campbell I. D. (2009) Methods Mol. Biol. 522, 73–99 [DOI] [PubMed] [Google Scholar]
- 63. Pickford A. R., Campbell I. D. (2004) Chem Rev 104, 3557–3566 [DOI] [PubMed] [Google Scholar]
- 64. Hohenadl C., Mann K., Mayer U., Timpl R., Paulsson M., Aeschlimann D. (1995) J. Biol. Chem. 270, 23415–23420 [DOI] [PubMed] [Google Scholar]
- 65. Aeschlimann D., Kaupp O., Paulsson M. (1995) J. Cell Biol. 129, 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cleary D. B., Maurer M. C. (2006) Biochim. Biophys. Acta 1764, 1207–1217 [DOI] [PubMed] [Google Scholar]
- 67. Pinkas D. M., Strop P., Brunger A. T., Khosla C. (2007) PLoS Biol. 5, e327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. LeMosy E. K., Erickson H. P., Beyer W. F., Jr., Radek J. T., Jeong J. M., Murthy S. N., Lorand L. (1992) J. Biol. Chem. 267, 7880–7885 [PubMed] [Google Scholar]
- 69. Jeong J. M., Murthy S. N., Radek J. T., Lorand L. (1995) J. Biol. Chem. 270, 5654–5658 [DOI] [PubMed] [Google Scholar]
- 70. Gaudry C. A., Verderio E., Aeschlimann D., Cox A., Smith C., Griffin M. (1999) J. Biol. Chem. 274, 30707–30714 [DOI] [PubMed] [Google Scholar]
- 71. Hang J., Zemskov E. A., Lorand L., Belkin A. M. (2005) J. Biol. Chem. 280, 23675–23683 [DOI] [PubMed] [Google Scholar]
- 72. Liu S., Cerione R. A., Clardy J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2743–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fox B. A., Yee V. C., Pedersen L. C., Le Trong I., Bishop P. D., Stenkamp R. E., Teller D. C. (1999) J. Biol. Chem. 274, 4917–4923 [DOI] [PubMed] [Google Scholar]
- 74. Siegel M., Strnad P., Watts R. E., Choi K., Jabri B., Omary M. B., Khosla C. (2008) PLoS ONE 3, e1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jobe S. M., Leo L., Eastvold J. S., Dickneite G., Ratliff T. L., Lentz S. R., Di Paola J. (2005) Blood 106, 4146–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.