Abstract
Collagens make up the most abundant component of interstitial extracellular matrices and basement membranes. Collagen remodeling is a crucial process in many normal physiological events and in several pathological conditions. Some collagen subtypes contain specific carbohydrate side chains, the function of which is poorly known. The endocytic collagen receptor urokinase plasminogen activator receptor-associated protein (uPARAP)/Endo180 plays an important role in matrix remodeling through its ability to internalize collagen for lysosomal degradation. uPARAP/Endo180 is a member of the mannose receptor protein family. These proteins all include a fibronectin type II domain and a series of C-type lectin-like domains, of which only a minor part possess carbohydrate recognition activity. At least two of the family members, uPARAP/Endo180 and the mannose receptor, interact with collagens. The molecular basis for this interaction is known to involve the fibronectin type II domain but nothing is known about the function of the lectin domains in this respect. In this study, we have investigated a possible role of the single active lectin domain of uPARAP/Endo180 in the interaction with collagens. By expressing truncated recombinant uPARAP/Endo180 proteins and analyzing their interaction with collagens with high and low levels of glycosylation we demonstrated that this lectin domain interacts directly with glycosylated collagens. This interaction is functionally important because it was found to modulate the endocytic efficiency of the receptor toward highly glycosylated collagens such as basement membrane collagen IV. Surprisingly, this property was not shared by the mannose receptor, which internalized glycosylated collagens independently of its lectin function. This role of modulating its uptake efficiency by a specific receptor is a previously unrecognized function of collagen glycosylation.
Keywords: Collagen, Connective Tissue, Endocytosis, Extracellular Matrix Proteins, Glycoprotein, CD206, CD280, MRC1, MRC2, Urokinase Plasminogen Activator Receptor-associated Protein
Introduction
The breakdown and remodeling of the extracellular matrix (ECM)2 including the basement membrane are important steps in embryonic growth, tissue rearrangements in the healthy body, and invasive cancer growth (1–3). The ECM is composed of a range of different structural proteins, including collagens, laminins, fibronectin, and proteoglycans. The collagens make up by far the most abundant component. Collagens are trimeric proteins that form unique triple helices and assemble into large supramolecular structures such as fibers and sheets, enabling them to form the barriers and structures of the ECM (4).
Collagens undergo a range of post-translational modifications, including extensive hydroxylation of prolyl and lysyl residues, N- and O-linked glycosylation, and processing of proforms (5). The hydroxylation of proline and lysine residues plays a role in triple helix stabilization and cross-linking of collagen molecules and the processing of proforms is important for the assembly of collagens into fibrillar structures (6). In contrast, the role of collagen glycosylation is poorly understood. Most is known about the O-linked glycosylation and this type of collagen glycosylation is composed of a single galactose unit or a disaccharide consisting of galactose and glucose attached to hydroxylated lysine residues (7). The level of O-glycosylation varies greatly between different types of collagen and between different tissues. The network-forming collagen type IV of the basement membrane and the fibrillar collagen type V have high levels of glycosylation (8–11), whereas collagen type I in the skin and tendon has very low levels (12, 13). Some studies have demonstrated an importance of glycosylation in collagen function in processes of mouse development, however, the mechanism of action remains unknown (14–16). In particular, it is an open question if collagen glycosylation affects collagen interaction with other proteins and processes like collagen turnover.
Collagen turnover is complex and involves different mechanisms and degradation pathways. The unique structure of collagens makes them resilient to most means of proteolytic attack and only relatively few proteases are known to degrade collagens in their native state. Most of these active collagenases are secreted or membrane-bound members of the matrix metalloproteinase family (17, 18).
However, in addition to extracellular degradation, a pathway for intracellular degradation of collagen has been identified (19). This pathway involves collagen endocytosis mediated by the urokinase plasminogen activator receptor-associated protein (uPARAP) (20), also designated Endo180 (21). This protein, designated uPARAP in the following, is a member of the mannose receptor family of endocytic receptors (22–24). uPARAP is expressed at sites with ongoing tissue remodeling (25–28) and functions in delivering collagens for intracellular degradation in lysosomes (29–35). In some processes the receptor operates together with extracellular collagen degradation mechanisms and it is effective in internalizing the large cleavage products resulting from initial extracellular collagenase digestion (36–38). The importance of this pathway of collagen turnover in vivo was emphasized in a recent study of a devastating hereditary bone defect in cattle, which turned out to be caused by an inactivating frameshift mutation in the uPARAP encoding gene (39).
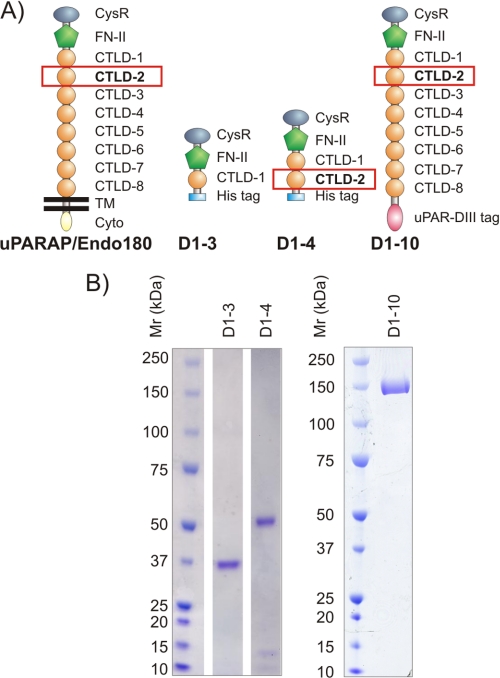
The involvement of uPARAP in extracellular matrix remodeling has underscored the importance of understanding the molecular basis of the ligand interactions of the receptor and several studies during the last decade have provided information in this regard. uPARAP consists of a large extracellular part, a transmembrane domain, and a short cytoplasmic domain (Fig. 1A). The extracellular part of uPARAP includes 10 domains as predicted from the amino acid sequence (21, 40). The collagen binding activity of uPARAP has been ascribed to a fibronectin type II (FN-II) domain in the protein structure, whereas the other domains of the receptor have so far been considered devoid of collagen interaction sites (30, 31). Interestingly, the receptor includes eight C-type lectin-like domains (CTLD1–8) but only one of these, CTLD-2, possesses lectin activity (41). This domain includes an essential calcium-binding site and specifically binds a range of monosaccharides, including mannose, glucose, and N-acetylglucosamine (GlcNAc), in a calcium-dependent reaction (41). However, no biological ligand has so far been associated with the lectin activity of uPARAP.
FIGURE 1.
uPARAP recombinant proteins. A, domain composition of uPARAP and recombinant uPARAP constructs. The constructs D1–3, D1–4, and D1–10 comprise the first 3, 4, and 10 N-terminal domains, respectively, of the complete sequence of mature uPARAP. CysR, cysteine-rich domain; CTLD, C-type lectin-like domain with the indicated number. Note that only CTLD-2 contains an intact Ca2+-binding site and possesses carbohydrate binding activity. His tag and uPAR-DIII tag designate purification tags on the recombinant proteins; see “Experimental Procedures.” TM and Cyto, transmembrane and cytoplasmic regions of native uPARAP. B, SDS-PAGE analysis of purified recombinant uPARAP proteins. The calculated theoretical molecular masses are 38 (D1–3), 55 (D1–4), and 179 kDa (D1–10), respectively. The electrophoretic mobilities of molecular mass marker proteins are indicated to the left.
In the present work, we show that CTLD-2 of uPARAP participates in the binding to specific collagens with a high degree of glycosylation, that CTLD-2 indeed interacts directly with carbohydrate residues on collagens through its lectin function, and that the resulting combined activity of FN-II and CTLD-2 serves to specifically increase the binding of uPARAP to glycosylated collagens. Furthermore, we demonstrate an impact of this lectin activity on the efficiency of uPARAP-dependent collagen internalization in cultured cells. Through this mechanism, the type-specific and dynamic pattern of collagen glycosylation may add to determine the efficiency of collagen turnover through the uPARAP-mediated endocytic degradation route. Surprisingly, we find that this lectin dependence in collagen internalization is limited to uPARAP because the related collagen endocytosis receptor, the mannose receptor (MR) (42, 43), does not depend on lectin activity in enabling the internalization of collagen by macrophages.
EXPERIMENTAL PROCEDURES
Reagents and Cultured Cells
The following proteins and other reagents were purchased from commercial sources as indicated: endoglucosidase H (EndoH) from Streptomyces plicatus (New England Biolabs, Ipswich, MA), native, trypsin-resistant collagen type I from rat tail (BD Biosciences), native collagen type IV and V isolated from human placenta, holotransferrin, and cysteine protease inhibitor E64d (Merck Biosciences, Darmstadt, Germany), mannose-BSA (Dextra Laboratories, Reading, United Kingdom), d-mannose, N- acetyl-d-glucosamine (GlcNAc), α-methyl-d-galactopyranoside (α-Me-Gal), bovine serum albumin (BSA), granulocyte macrophage-colony stimulating factor, and trifluoromethanesulfonic acid (TFMS) deglycosylation kit (Sigma), 125I for protein labeling (PerkinElmer Life Sciences), Oregon Green-conjugated gelatin and Alexa Fluor 488 microscale protein labeling kit (Invitrogen), goat polyclonal antibody against human MR (pAb a-MR) (R&D Systems, Minneapolis, MN), and Cy3-conjugated goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). The monoclonal mouse anti-uPARAP antibodies 5f4 and 2h9 have been described previously (37, 44). The cell surface marker, monoclonal antibody mR3 against murine urokinase receptor was produced as described (45).
Fibroblasts from the skin of newborn homozygous uPARAP-deficient mice and littermate wild type mice were isolated and cultured as described (29). Human osteosarcoma cells (MG63 cell line; ATCC number CRL-1427) were cultured in minimum Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Human macrophages were generated as previously described (43, 46) with minor modifications. In brief, mononuclear cells were isolated from whole blood from normal, healthy volunteers, using density gradient centrifugation (Lymphoprep, Axis Shield, Dundee UK). After this monocytes were isolated using an automated cell isolator (RoboSep) and EasySep Human Buffy Coat CD14 Selection Kit (Stemcell Technologies, Grenoble, France). Following isolation, monocytes were cultured for 8 days in AIM-V medium with l-glutamine, streptomycin, gentamycin, and human serum albumin (Invitrogen) and supplemented with 10% FCS and 5 ng/ml of granulocyte macrophage-colony stimulating factor for differentiation into macrophages. The medium was replenished on days 4 and 7.
Recombinant uPARAP Variants
Three recombinant truncated uPARAP variant proteins designated D1–3, D1–4, and D1–10 were generated (Fig. 1). These proteins comprise the first three N-terminal domains (Gly31–Ala365), the first four N-terminal domains (Gly31–Leu510), and all of the 10 extracellular domains (Gly31–Ser1402) in the human uPARAP sequence, respectively (21, 40). D1–3 and D1–4 were produced in a Pichia pastoris-based expression system (Invitrogen). In brief, the DNA sequences encoding D1–3 and D1–4 were amplified by PCR from a uPARAP expression vector (37) and fused with a sequence encoding a C-terminal His6 tag, a stop codon, and appropriate restriction sites for subsequent cloning into the P. pastoris expression vector pPICZα as previously described (47). The following synthetic oligonucleotide primers were used in the PCR: 5′-TCTCTCGAGAAAAGAGGCGCCCCTGGGGACGCCGC-3′ (for amplification of both D1–3 and D1–4), 5′-GCTCTAGATTAATGATGATGATGATGATGGGCGTTGGGCTTCTTCTTGC-3′ (for D1–3), and 5′-GCTCTAGATTAATGATGATGATGATGATGCAGCTGGCCTGCCTTCTTGC-3′ (for D1–4). Restriction enzyme recognition sites are in bold. The DNA encoding the specified sequences were inserted at a position leading to N-terminal fusion with a P. pastoris signal sequence to allow protein export into the growth medium and processing mediated by the host cell. The recombinant proteins were purified from filtered supernatant from overnight P. pastoris cultures using Ni2+-chelating chromatography on an ÄKTA purifier system (GE Healthcare) and elution with a linear imidazole gradient ranging from 0.005 to 1 m. Homogenous protein preparations were obtained by enzymatic deglycosylation, treating the elution fractions (protein concentration 0.1–0.5 mg/ml) with EndoH (2000 units/ml) in 50 mm sodium citrate buffer, pH 5.5, for 1 h at room temperature. A second round of Ni2+-chelating chromatography was then performed to remove EndoH enzyme. The D1–10 recombinant uPARAP protein construct was fused to a purification tag (the third domain of the urokinase receptor), produced in Drosophila Schneider S2 cells, and purified by affinity chromatography as previously described (37, 48).
Gel Electrophoresis Analysis of Recombinant uPARAP
SDS-PAGE analysis of purified D1–3 and D1–4 was performed under reducing conditions with ∼3 μg of protein loaded onto the gel. SDS-PAGE analysis of D1–10 was performed under nonreducing conditions and 5 μg of this protein was loaded. Following SDS-PAGE proteins were stained with Coomassie Brilliant Blue.
Surface Plasmon Resonance Analysis
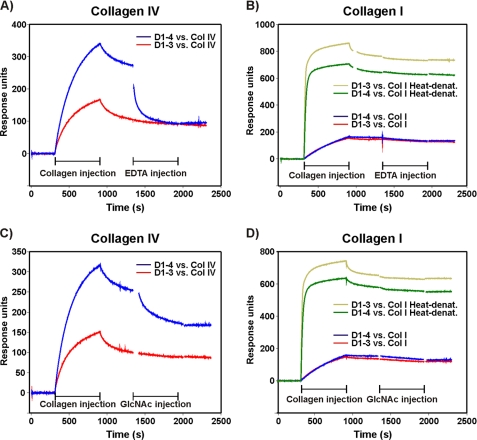
Dynamic analysis of collagen-uPARAP interactions was performed using a BIAcore2000 instrument (GE Healthcare), using conditions slightly modified from the method previously described (37). The anti-uPARAP catching antibody (mAb 2h9) was coupled to the surface of a BIAcore CM5 sensor chip, after which D1–3 and D1–4 recombinant proteins were immobilized by injection into individual flow channels. Due to the binding characteristics of mAb 2h9, this leads to a near irreversible immobilization of the uPARAP proteins (37). To allow comparison, approximately equal molar amounts of D1–4 and D1–3 were immobilized by this procedure (molar ratios D1–4/D1–3 of 1:0.87 and 1:1.18 for the experiments shown in Fig. 2, A and C and Fig. 2, B and D, respectively). Collagens type I and IV were preincubated in assay buffer (10 mm HEPES, 150 mm NaCl, 1 mm CaCl2, 0.005% surfactant P20, pH 7.4) and were then injected into the flow cells with immobilized uPARAP variants at a flow rate of 10 μl/min for 10 min. In experiments with denatured collagen, collagen I was incubated at 65 °C for 20 min prior to injection. Following injection, dissociation was recorded in two phases: an initial 4-min phase performed in assay buffer alone followed by a 10-min phase performed in assay buffer supplemented with interaction inhibitors (EDTA (10 mm) or GlcNAc (50 mm)). Binding and dissociation were recorded at 20 °C. After each round of collagen injection and dissociation, the chip was regenerated by two pulses of 1 min each with 10 mm glycine/HCl, pH 2.0. A reference flow channel without coupled protein or with mAb 2h9 alone (no recombinant uPARAP) allowed for buffer bulk subtraction throughout each sensorgram. No binding of collagen was observed in the reference channels.
FIGURE 2.
SPR analysis of uPARAP interactions with solubilized collagens. Approximately equal amounts of D1–4 and D1–3 were immobilized in parallel flow channels of a BIAcore sensor chip, using near irreversible capture on mAb 2h9. Solubilized collagens (10 μg/ml) were then injected into parallel flow channels with captured D1–4 or D1–3. Collagen IV (A and C) was injected in the native state, whereas collagen I (B and D) was injected both in the form of native and heat-enatured protein. Blue and red curves depict the binding of native collagens to D1–4 and D1–3, respectively. Dark and light green curves represent the binding of heat-denatured collagen I to the same two uPARAP constructs. After the indicated 600-s phase of collagen injection, dissociation was allowed to proceed in running buffer for 240 s, followed by a 600-s injection phase with the indicated inhibitory reagents. Finally, the flow was shifted back to running buffer. The sensorgrams are shown after subtraction of buffer bulk effect (parallel flow cell without uPARAP) and a blank run with injection of running buffer instead of collagen material.
ELISA Interaction Analysis
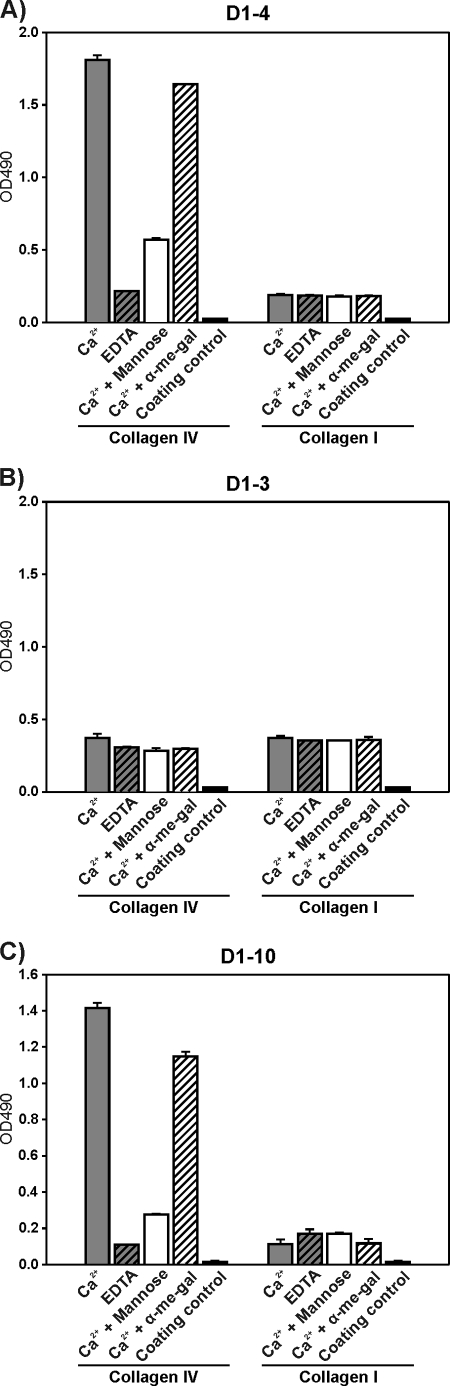
The interaction of uPARAP with immobilized collagens was analyzed using an ELISA setup with heat-denatured collagens type I, IV, and V. Collagens were diluted to 10 μg/ml in coating buffer (0.1 m Na2CO3, pH 9.8) and incubated for 20 min at 65 °C before being coated onto the surface of wells in 96-well Nunc Maxisorp plates (Thermo Fisher Scientific, Waltham, MA) at 4 °C overnight. Subsequently, blocking of nonspecific binding sites was performed with either 2.5% ELISA blocking reagent (Roche Diagnostics, used for D1–3 and D1–4 experiments) or 2% skimmed milk powder (Irma, Copenhagen, Denmark, used for D1–10) for 30 min at room temperature. Solutions of 27 nm D1–3 or D1–4, or 55 nm D1–10 were then added to the wells in binding buffer (50 mm HEPES, 20 mm KCl, 100 mm NaCl, 10 mg/ml of BSA, 0.1% Tween 20, pH 7.4), either supplemented with CaCl2 (1 mm), or with interaction inhibitors (EDTA (3 mm) or mannose (50 mm)). Ligand binding was performed at 4 °C overnight. Binding was detected with primary mouse anti-uPARAP antibody (mAb 2h9, 10 μg/ml) followed by horseradish peroxidase (HRP)-coupled rabbit anti-mouse antibody ((Dako, Glostrup, Denmark) diluted 1:2000) and finally ∼20 min incubation with HRP substrate (0.0025% H2O2 and 0.33 mg/ml of o-phenylenediamine) (Dako). The reaction was stopped by addition of 1.3 m H2SO4, after which the reaction product was quantified by absorbance measurements at 490 nm with absorbance at 540 nm used for background subtraction. In between each incubation step, ELISA plates were washed three times in PBS with 1.7 mm Mg2+, 1.3 mm Ca2+, and 0.1% Tween 20. After the incubation step with secondary antibody three additional washes were performed with Milli-Q water.
Treatment of Collagens with Trifluoromethanesulfonic Acid (TFMS)
For deglycosylation studies, collagens type I, IV, and V were treated with TFMS (49), using a commercial protein deglycosylation kit (Sigma). TFMS treatment was performed according to the manufacturer's instructions and 200 μg of each collagen was treated. Immediately after treatment, collagens were diluted to 10 μg/ml in ELISA coating buffer (0.1 m Na2CO3, pH 9.8). In the mock treated samples, collagens were lyophilized and then redissolved in 10 mm acetic acid before being diluted to 10 μg/ml in ELISA coating buffer.
Endocytosis Assays
Internalization assays with 125I-labeled ligands in mouse skin fibroblasts isolated from newborn uPARAP-deficient or wild type mice and MG63 cells were performed as previously described (29). In brief, samples of 1 × 105 cells were seeded in 24-well tissue culture plates and cultured overnight. In experiments with function-blocking antibody (mAb 5f4 against uPARAP), 10 μg/ml of the antibody was added to MG63 cells 5 h after seeding. The next day, culture medium was removed and cells were washed with serum-free assay buffer (low glucose DMEM with sodium pyruvate for fibroblasts and minimum Eagle's medium for MG63 cells; both from Invitrogen) supplemented with 20 mm HEPES and 15 mg/ml of BSA. Subsequently, fresh assay medium was added, with the inclusion of either no inhibitor, 50 mm mannose, 50 mm GlcNAc, 50 mm α-Me-Gal, or 10 μg/ml of mAb 5f4. In addition, the cysteine protease inhibitor E64d (10 μm) was added to all samples to optimize lysosomal accumulation of internalized proteins (32). After a 30-min incubation, 133 ng/ml of 125I-labeled ligand (collagens type I, IV, V, holotransferrin, or anti-uPARAP mAb 2h9) was added to each well. Cultures were then incubated for 4–5 h at 37 °C before the intracellular fraction was isolated as described (29).
For endocytosis assays with human macrophages, 2 × 105 cells were seeded in each well of a 24-well tissue culture plate and assayed on day 8 according to the isolation and differentiation procedure described above. Internalization assays with 125I-labeled ligands were performed essentially as described for fibroblasts and MG63 cells. In experiments including mAb 5f4, 10 μg/ml of the antibody was added to the macrophage cultures on day 7. On day 8 assay buffer (AIM-V supplemented with 20 mm HEPES and 15 mg/ml of BSA, 10 μm E64d) was added to the cells with the inclusion of either no inhibitor, 50 mm mannose, 10 μg/ml of pAb a-MR, or 10 μg/ml of mAb 5f4.
Internalization assays with fluorescent collagen or gelatin were performed essentially as described (32). In brief, samples of 2 × 104 uPARAP-deficient or wild type fibroblasts were seeded overnight, after which ligands for internalization (25 μg/ml of Oregon Green-gelatin or 10 μg/ml of Alexa 488-labeled collagen IV) were added in assay buffer (low glucose DMEM with sodium pyruvate, supplemented with 20 mm HEPES 15 mg/ml of BSA, 1% penicillin/streptomycin, and 10 μm E64d). Internalization was then allowed to proceed overnight in the presence or absence of 50 mm mannose. Following internalization, cells were treated with trypsin-EDTA for 5 min and re-seeded on polylysine-coated coverslips. Cells were allowed to attach for 4 h before being fixed with 4% paraformaldehyde and surface stained with anti-uPAR antibody (10 μg/ml) and secondary Cy3-conjugated goat anti-mouse antibody (diluted 1:200) (Jackson ImmunoResearch). Cell nuclei were stained with DAPI. Fluorescence microscopy was performed as previously described (32), except that a Zeiss LSM 780 microscope was used.
RESULTS
Recombinant uPARAP Variants
To address a possible function of the calcium-dependent lectin domain, CTLD-2, in the uPARAP-collagen interaction we designed two recombinant uPARAP proteins. Both recombinant proteins comprised the first three domains in the uPARAP sequence and in addition one of them contained the fourth domain, CTLD-2. The recombinant proteins were designated D1–3 and D1–4, respectively (Fig. 1A). These recombinant proteins were expressed in P. pastoris and both were purified to yield homogenous products (Fig. 1B, left panel). By comparing D1–3 and D1–4 constructs with respect to the binding of various collagen subtypes, the importance of CTLD-2 could be directly assessed.
Differential Binding of uPARAP to Different Collagen Subtypes
The interactions between collagens and the D1–3 and D1–4 constructs were initially analyzed by surface plasmon resonance (SPR) using an established setup where recombinant uPARAP is immobilized on a supporting, high affinity monoclonal antibody (mAb 2h9) (37). Because this setup includes a divalent capture of the receptor on a catching IgG, it precludes the derivation of quantitative kinetic parameters of monomeric uPARAP. However, it does enable the qualitative analysis of separate collagen association and dissociation phases.
First, we studied the interaction between the recombinant uPARAP constructs and collagen IV, a collagen with a high degree of glycosylation (9). When injected in a calcium containing running buffer, collagen IV clearly bound more rapidly to D1–4 than to the equivalent molar amount of D1–3 (Fig. 2A). To study whether this difference in binding was dependent on calcium, the collagen injection pulse was followed by a short dissociation phase, after which an EDTA-containing buffer was injected. Strikingly, the chelation of calcium strongly affected the dissociation of collagen IV from D1–4 and brought the binding signal to a level similar to that obtained with D1–3. The binding between collagen IV and D1–3 itself was unaffected by EDTA.
Next, we analyzed the interaction between the recombinant uPARAP proteins and collagen I, a collagen with a low degree of glycosylation (13). This ligand bound with similar efficiency to D1–4 and D1–3, leading to nearly identical association and dissociation phases, and EDTA did not appear to affect the dissociation of collagen I from either construct (Fig. 2B, lower curves).
However, although the complexes formed between collagen I and the uPARAP constructs were highly stable as indicated by the low dissociation rate, the slow binding process with this ligand led to relatively low binding levels under the conditions used. Therefore, it was difficult to rule out the possibility that EDTA could have a small effect on the stability of the complexes formed with collagen I. To exclude this, an additional experiment was set up in which collagen I was denatured prior to injection. We have previously shown that collagen cleaved by specific collagenases binds more rapidly to uPARAP than native collagen and that collagen cleavage can be mimicked in this respect by heat-induced denaturation while retaining binding specificity (37). Indeed, denaturation of collagen I resulted in a faster association reaction and, consequently, a higher binding level than that obtained with native collagen I, allowing for a detailed evaluation of the dissociation curves (Fig. 2B, upper curves). Also under these conditions, EDTA was clearly without effect on the dissociation of collagen I from D1–4 and D1–3.
The Active Lectin Domain of uPARAP Participates in the Collagen Interaction
To further investigate if the observed calcium-dependent effects were related to the lectin activity of CTLD-2, the effect of monosaccharide on collagen interaction was analyzed using the same SPR setup. The injection of GlcNAc, which has previously been identified as a ligand for CTLD-2 (41), efficiently accelerated the dissociation of collagen IV from D1–4 (Fig. 2C), although the effect obtained was slightly weaker than that found with EDTA. The binding between collagen IV and D1–3 was unaffected by GlcNAc injection. Furthermore, GlcNAc did not affect the interaction between native or denaturated collagen I and D1–4 or D1–3 (Fig. 2D). A similar result as obtained here with GlcNAc could also be obtained with mannose, another positive monosaccharide ligand for CTLD-2 (41) (result not shown). The effect of α-Me-Gal, a monosaccharide found to be a very poor ligand for CTLD-2 (41), was also analyzed. This monosaccharide did not influence the interaction between collagen IV and D1–4 or D1–3 (result not shown, but see Fig. 3, below). Together these experiments suggested a role of the lectin activity of CTLD-2 in the interaction between uPARAP and collagen IV and a lack of importance of this domain in the interaction with collagen I.
FIGURE 3.
ELISA analysis of uPARAP interactions with immobilized collagens. Heat-denatured collagen types IV and I were immobilized in ELISA wells as indicated below each panel. D1–4 (27 nm, A), D1–3 (27 nm, B), or D1–10 (55 nm, C) was added to each well. The binding was analyzed in assay buffer with 1 mm CaCl2 and no further additions (gray columns), buffer with 3 mm EDTA (hatched gray columns), buffer with 1 mm CaCl2 and 50 mm mannose (white columns), and buffer with 1 mm CaCl2 and 50 mm α-Me-Gal (hatched white columns). Binding of the recombinant uPARAP proteins was detected with mAb 2h9 against uPARAP, followed by a secondary HRP-coupled rabbit anti-mouse antibody. Controls comprised wells without immobilized collagen (coating control, black columns) and wells without recombinant uPARAP (negative in all cases). Error bars represent S.D. of duplicate samples.
To examine this mechanism in an independent system, an ELISA setup was developed that allowed us to study binding between recombinant uPARAP in solution and collagens immobilized in microtiter wells. Initial experiments revealed that in this setup, uPARAP binding levels well suited for quantitative analysis could be obtained with immobilized denatured collagens, whereas the signals obtained with native collagens were too low for a detailed comparison of binding conditions. Consequently, denatured collagens with high and low levels of glycosylation were coated onto the ELISA well surface, after which D1–4 or D1–3 were added in solution. In the presence of calcium, D1–4 bound much more efficiently to collagen IV than to collagen I (Fig. 3A). Calcium chelation by EDTA or competition with mannose efficiently reduced the binding of D1–4 to collagen IV, whereas α-Me-Gal had no effect. The binding of D1–4 to collagen I was unaffected by EDTA, mannose, or α-Me-Gal. Importantly, these differences were not observed in the case of D1–3 (Fig. 3B). D1–3 bound with similar efficiency to collagen IV and I, and neither EDTA, mannose, nor α-Me-Gal had an effect on the binding. Thus, the ELISA experiments were in accordance with the SPR analyses and confirmed that CTLD-2 is engaged in the interaction of uPARAP with collagen IV and that it contributes to the binding through its calcium-dependent lectin activity.
To exclude that the observed effects were caused by artifacts related to the expression of short truncated recombinant proteins in a P. pastoris-based expression system, a previously characterized recombinant protein, D1–10, was included in the study. This purified protein includes all of the extracellular domains of uPARAP and was expressed in insect cells (37) (Fig. 1, A and B, right panels). In the ELISA setup the binding of D1–10 to collagen IV was sensitive to calcium chelation by EDTA and competition by mannose, whereas α-Me-Gal only had very limited effect. The binding to collagen I was not inhibited by any of the tested reagents (Fig. 3C). Therefore, D1–10 showed binding characteristics toward collagen IV and I very similar to those observed for D1–4.
The Active Lectin Domain of uPARAP Interacts Directly with Carbohydrates on Collagen
A likely hypothesis to explain the results above was that CTLD-2 strengthens the receptor-collagen binding by interacting directly with carbohydrates on collagen.
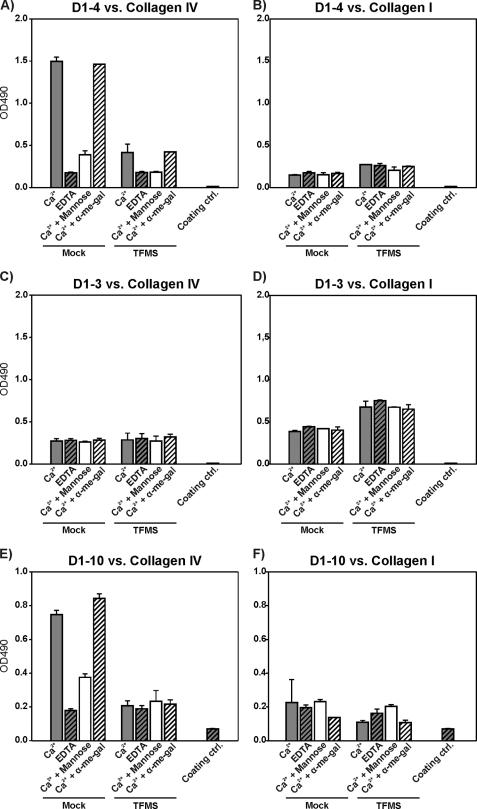
To test this hypothesis, collagen IV and I were treated with TFMS to remove covalently attached carbohydrates (49). Following TFMS treatment, interaction between the deglycosylated collagens and D1–4, D1–3, or D1–10 was compared in the same ELISA system as used above (Fig. 4). In the presence of calcium, the binding of D1–4 to TFMS-treated collagen IV proved strongly reduced as compared with the binding to the same type of collagen after mock treatment (Fig. 4A). In fact, in the absence of inhibitors, this binding was nearly reduced to the same level as that obtained with mock treated collagen IV in the presence of EDTA. Furthermore, after TFMS treatment of collagen IV, the inhibitory effects of EDTA and mannose on D1–4 binding were markedly reduced, leading to a residual binding close to that obtained with mock treated collagen IV under the same conditions. In contrast, the binding of D1–4 to collagen I was unaffected by TFMS treatment (Fig. 4B). TFMS treatment of collagen IV or I did not change the pattern of D1–3 binding and so binding remained equal in the presence or absence of competitors (Fig. 4, C and D). The longer construct, D1–10, showed binding characteristics toward mock and TFMS-treated collagen IV and I very similar to those observed for D1–4 (Fig. 4, E and F), as was also the case in the situation described above (Fig. 3).
FIGURE 4.
Collagen deglycosylation affects the interaction with uPARAP. Collagen types IV (A, C, and E) and I (B, D, and F) were mock treated or deglycosylated with TFMS under anhydrous conditions. Following treatment, collagens were immobilized on ELISA plates, after which D1–4 (A and B), D1–3 (C and D), or D1–10 (E and F) were added. Analysis of binding and representation of results were performed as described in the legend to Fig. 3.
Finally, because collagen I and IV belong to different structural classes (fibrillar and sheet-forming collagens, respectively (4)), we wanted to exclude that the differential binding to CTLD-2 of uPARAP could be related to this structural difference. Therefore, we included experiments with collagen V, a fibrillar collagen carrying high levels of glycosylation (10) (supplemental Fig. S1). D1–4 and D1–10 bound strongly to mock treated collagen V and binding was inhibited by EDTA and mannose. The binding to TFMS-treated collagen V was markedly reduced along with the inhibitory effects of EDTA and mannose when compared with mock treated collagen V. D1–3 binding to mock or TFMS-treated collagen V was unaffected by the inhibitors. Therefore the binding profiles of D1–4, D1–3, and D1–10 to mock and TFMS-treated collagen V was very similar to the one for collagen IV.
Altogether, the removal of carbohydrate from glycosylated collagens by TFMS treatment strongly reduced the interaction with D1–4 or D1–10 but had no effect on the binding to D1–3. Therefore, we can conclude that the binding contribution of CTLD-2 is directed to the carbohydrates on glycosylated collagens.
The Lectin Function of uPARAP Is Active in the Cellular Uptake of Glycosylated Collagen
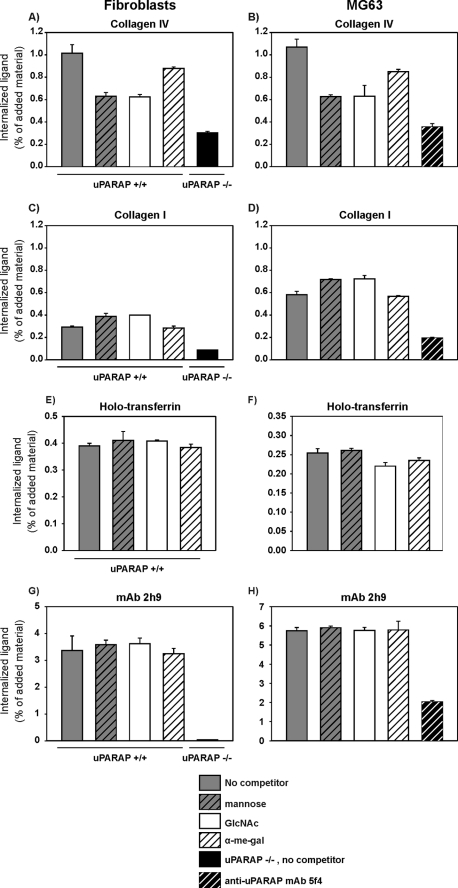
The results presented above were so far limited to a system with purified proteins. Therefore, we wanted to investigate if the binding contribution of CTLD-2 would also be important for the cellular endocytosis of collagen for lysosomal degradation. To address this question, we utilized cultures of wild type mouse skin fibroblasts (uPARAP+/+ fibroblasts) and human MG63 cells, which both degrade collagen through uPARAP-mediated collagen endocytosis (29, 43). Collagen type IV was labeled with 125I and added to these cells and the effect of monosaccharide competitors on collagen internalization was examined.
In this system, both of the cell types exhibited a marked reduction of collagen IV internalization in the presence of mannose or GlcNAc (∼40% reduction) and only a very small reduction in the presence of α-Me-Gal (∼15% reduction) (Fig. 5, A and B). As a control of the uPARAP dependence in the endocytic process, cultures with littermate mouse skin fibroblasts from uPARAP-deficient mice (uPARAP−/− fibroblasts) and MG63 cells treated with the uPARAP blocking mAb 5f4 (43) were included in the experiments. As expected these cells only internalized very low levels of collagen IV.
FIGURE 5.
Carbohydrate dependence of uPARAP-mediated collagen endocytosis. 125I-Labeled collagens types IV (A and B) or I (C and D), holotransferrin (E and F),or mAb 2h9 (G and H) were added to uPARAP+/+ and uPARAP−/− fibroblasts, as indicated (A, C, E, and G). In a parallel experiment, the same ligands were added to MG63 cells (B, D, F, and H). The labeled proteins (133 ng/ml) were added in endocytosis assay buffer alone (no competitor), in buffer including 50 mm of the indicated monosaccharides, or in buffer including 10 μg/ml of mAb 5f4. After 4 h at 37 °C the cells were harvested, the intracellular fraction of cell samples was isolated, and the amount of internalized labeled protein was determined. Data are presented as radioactivity in the intracellular fraction of cells, relative to the total amount of radioactivity added. Error bars represent the S.D. of triplicate samples.
When 125I-labeled collagen I was used instead of collagen IV, the level of internalization was not reduced in the presence of any of the monosaccharides (Fig. 5, C and D). Finally, we examined the effect of GlcNAc, mannose, and α-Me-Gal on the internalization of collagen V. In accordance with the high degree of glycosylation of this subtype, collagen V displayed an internalization pattern and carbohydrate dependence very similar to that of collagen IV (supplemental Fig. S2).
To exclude the possibility that the monosaccharides could also influence the clathrin-dependent endocytic machinery in a less specific manner, two control experiments were performed. First, we examined the uptake of holotransferrin, a ligand endocytosed through a clathrin-dependent mechanism similar to the collagen-uPARAP system, but mediated by the transferrin receptor (50). When 125I-labeled holotransferrin was added to the same cell types as used above, no effect of the monosaccharides was noted (Fig. 5, E and F), demonstrating that these reagents had no general effect on clathrin-dependent endocytosis. Second, to address any effect of the monosaccharides specifically against the cellular endocytosis of uPARAP, we exploited the fact that uPARAP is capable of internalizing mAbs directed against it (51). When mAb 2h9 against uPARAP was labeled with 125I and added to uPARAP+/+ fibroblasts or MG63 cells, none of the monosaccharides had any effect on internalization (Fig. 5, G and H). In uPARAP−/− fibroblasts and MG63 cells treated with the blocking mAb 5f4, the internalization of mAb 2h9 was very low, documenting that the endocytosis was uPARAP dependent. Therefore, carbohydrate sensitivity of the internalization experiments above was indeed limited to the uPARAP-mediated uptake of glycosylated collagens.
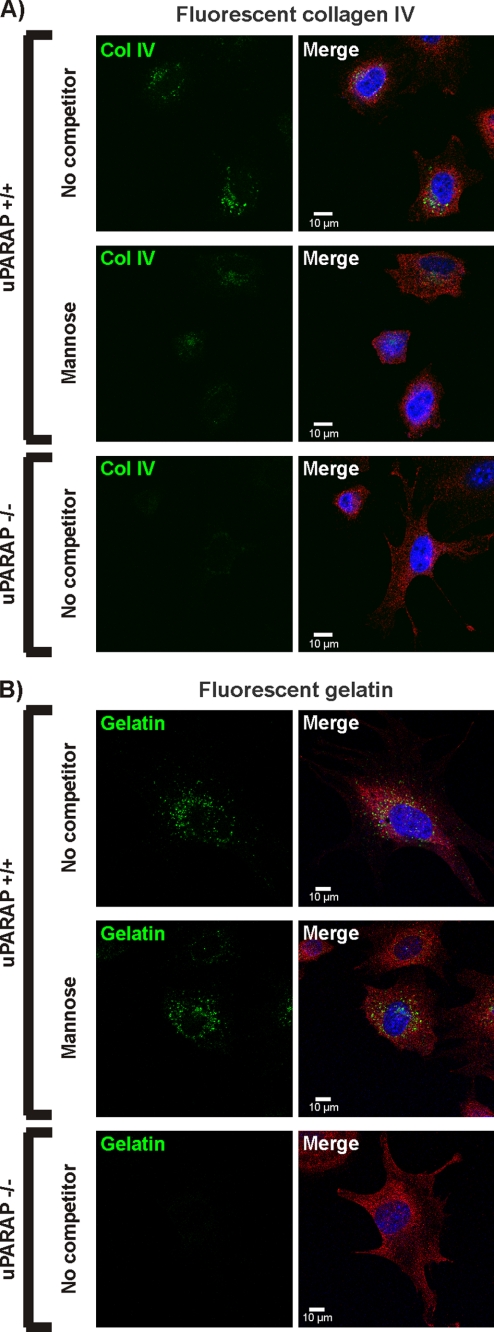
Finally, to directly visualize the uPARAP-dependent uptake and the lysosomal accumulation of collagens, we employed an experimental setup with fluorescently labeled collagens, which allows the demonstration of the internalized ligand in endosomes and lysosomes by fluorescence microscopy (32, 37). Fluorescence-labeled collagen IV and gelatin (denatured collagen I) were added to uPARAP+/+ fibroblasts in the absence or presence of mannose. When fluorescence-labeled collagen IV was used, a strong reduction in the vesicular accumulation of fluorescence was detected in the presence of mannose, compared with cells where no competitor was added (Fig. 6A, upper and center panels). In contrast, no effect of mannose was observed in the case of the gelatin ligand (Fig. 6B, upper and center panels). uPARAP−/− fibroblasts were used as a specificity control and as expected failed to accumulate intracellular collagen IV as well as gelatin (Fig. 6, A and B, lower panels). Altogether, these experiments were in complete accordance with results from the protein-protein interaction analyses and demonstrated a specific inhibitory effect of CTLD-2 monosaccharide ligands on the uPARAP-mediated endocytosis and lysosomal routing of highly glycosylated collagens.
FIGURE 6.
Influence of carbohydrate on lysosomal collagen accumulation. uPARAP+/+ and uPARAP−/− fibroblasts were incubated with Alexa 488-labeled collagen IV (A) or Oregon Green-labeled gelatin (B) in the absence (no competitor) or presence of 50 mm mannose. Endocytosis was allowed to proceed during incubation overnight. Subsequently, cells were released, re-seeded on coverslips, fixed, and stained with a cell surface marker (anti-uPAR antibody followed by Cy3-conjugated secondary antibody; red fluorescence). Cell nuclei were stained with DAPI (blue). Cells were then examined by confocal microscopy. The left panels show the channel with green fluorescence alone (fluorescent collagen IV/gelatin), whereas the right panels show the merged images with green, red, and blue fluorescence. Note the vesicular accumulation of internalized collagen/gelatin.
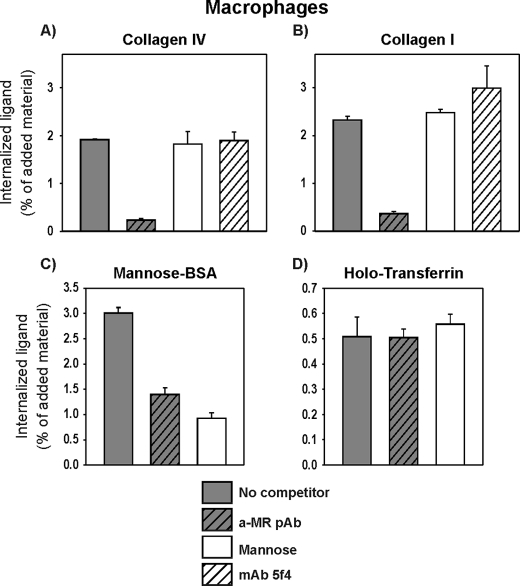
Mannose Receptor-mediated Collagen Uptake Is Independent of Lectin Activity
The demonstration of an active lectin domain being important for uPARAP binding and internalization of the glycosylated collagens opened the possibility that a similar mechanism could be employed by the MR. This receptor is another collagen binding member of the same protein family, having the same domain composition as uPARAP (23) and has been shown to be active in collagen internalization in macrophages (42, 43).
To study the cellular collagen uptake through MR, 125I-labeled collagen IV and I were added to human macrophages. For both ligands, in accordance with studies published previously (43), this uptake process was governed by MR because a polyclonal antibody against this receptor (a-MR pAb) efficiently blocked the internalization of both collagens (Fig. 7, A and B). Strikingly, however, mannose did not have any effect on the internalization of either collagen IV or I in these cells. In contrast, both mannose and the a-MR pAb strongly inhibited the ability of the macrophage to internalize mannose-BSA (Fig. 7C), a well established ligand for the active CTLDs of MR (52), thus documenting that mannose is indeed a competitor of CTLD-dependent internalization of MR-ligands.
FIGURE 7.
Collagen internalization in macrophages. Cultured macrophages were incubated with 125I-labeled collagens type IV (A) or type I (B), mannose-BSA (C), or holotransferrin (D). Incubation was performed in the absence (no competitor) or presence of the following reagents: mannose (50 mm), a-MR pAb (10 μg/ml), or mAb 5f4 against uPARAP (5f4; 10 μg/ml). Following incubation for 4 h at 37 °C, the internalized fraction of labeled protein was determined. Data are presented as described in the legend to Fig. 5.
In a control experiment, neither mannose nor the a-MR pAb had any effect on the internalization of holotransferrin by macrophages (Fig. 7D) and furthermore, the a-MR pAb did not reduce the uPARAP-dependent internalization of collagens in MG63 cells (supplemental Fig. S3). Altogether, the cell-based experiments demonstrate an MR-dependent, but lectin-independent, internalization of glycosylated as well as unglycosylated collagens by cultured macrophages and point to a clear difference in lectin functions between uPARAP and MR.
DISCUSSION
In this study, we have examined the importance of collagen glycosylation for interaction with the collagen endocytosis receptor uPARAP. We showed that CTLD-2, the single active lectin domain of uPARAP (41), contributes strongly to the interaction between uPARAP and glycosylated collagens. Competition experiments with monosaccharides and collagen deglycosylation studies pointed to this being the result of a direct binding between CTLD-2 and protein carbohydrate on collagen ligands. We first demonstrated in two independent purified systems that CTLD-2 strengthens the interaction between uPARAP and glycosylated collagens in a calcium- and carbohydrate-dependent manner and that this contribution could be blocked by monosaccharide ligands of CTLD-2. Second, we showed that the uptake and lysosomal routing of these collagens by mouse skin fibroblasts and human osteosarcoma cells is increased because of this binding contribution. To our knowledge this is the first time that a specific receptor interaction with biologically occurring collagen glycosylation has been identified and it is the first time that a biological ligand has been identified for CTLD-2 in uPARAP.
Previous studies with recombinant, truncated uPARAP constructs have convincingly shown that the FN-II domain plays a central role in collagen binding (30), but did not identify a role of CTLD-2 in this respect. However, these studies did not focus on differential glycosylation among different types of collagen and the findings were mostly based on a positive collagen binding capability of a recombinant protein comprising just the first three domains of uPARAP. This is in complete accordance with our findings that the D1–3 construct binds to collagens and does so independently of carbohydrate. Relative to this binding, we found an increased binding of the D1–4 construct exclusively to glycosylated collagens. In this connection, it is noteworthy that the three-domain construct in the previous work was also found to bind less efficiently to collagen IV than a longer construct (30), although the background for this finding was not identified.
Whereas our findings thus show that CTLD-2 contributes to the binding of glycosylated collagens through its lectin activity, this interaction is clearly not responsible for binding alone. Although both monosaccharides and EDTA inhibition led to a pronounced reduction in binding, neither of these reagents abolished binding completely. In line with these observations, previous studies have demonstrated that CTLD-2 is not sufficient for collagen binding in the absence of the N-terminal part of the receptor because a truncated uPARAP variant, including CTLD2–8 but lacking the FN-II domain, is unable to bind collagen IV (31).
The relative positioning of the FN-II domains and the active lectin domains differs within the MR protein family (see Ref. 23). This may be important for the lectin contribution to collagen binding because we found that in contrast to uPARAP, MR does not depend on lectin activity in the uptake of collagens. This is noteworthy because MR includes active lectin domains (52) with several known carbohydrate ligands (for a review, see Ref. 23) but it does support previously published results on the MR-collagen interaction in a purified system, which suggest that MR binds collagen IV independently of the lectin activity (42, 53). Therefore, strong evidence now supports distinct functions of lectin domains found in the closely related receptors uPARAP and MR.
Although the molecular details of the collagen carbohydrate that binds to uPARAP are not known, it is quite likely that the interaction involves the unique type of collagen O-glycosylation (7). This type of glycosylation is elevated in collagen types IV and V, found in this study to be CTLD-2 ligands, versus the non-ligand collagen I. Furthermore, it includes terminal glucose residues (7), in accordance with glucose being an established monosaccharide ligand of CTLD-2 (41). Finally, this type of glycosylation is scattered across the helical regions of the collagen α-chains and therefore is positioned close to regions containing the presumed binding sites for the FN-II domain of uPARAP (37, 54).
The demonstration of an interaction between collagen carbohydrate and CTLD-2 opens new questions concerning the function of collagen glycosylation as well as uPARAP. The importance of collagen post-translational modifications is well recognized and mutations in responsible genes have been implicated in various diseases in both mice and humans (55). However, the role of collagen glycosylation is generally not well defined in molecular terms, although the importance of collagen O-glycosylation has recently become more apparent. In mice it has been demonstrated that O-glycosylation is essential for the proper structural assembly of collagens, especially collagen IV, and that genetic deletion of the enzyme catalyzing this glycosylation process leads to incomplete formation of basement membranes and, as a consequence, to embryonic lethality (14–16). In humans, mutations in the corresponding gene, which result in decreased collagen glycosylation activity, have now been shown to cause severe connective tissue disorders resembling disorders caused by mutations in collagen-encoding genes (56, 57). The structural importance of collagen O-glycosylation is further demonstrated by the finding that increased levels of O-glycosylation decrease the diameter of fibers formed by fibrillar collagens II and V in vitro (58, 59), and it has been suggested that an increase in collagen I glycosylation in vivo can lead to the altered collagen architecture seen in sclerotic skin (60).
Together, these studies suggest that collagen glycosylation must be tightly regulated to ensure proper deposition and structuring of ECM compartments, including the basement membrane. In addition to this function, our findings now suggest a role of collagen glycosylation in matrix turnover through the link to uPARAP-mediated endocytosis, with collagen carbohydrates modulating turnover efficiency.
uPARAP has been found to be important for numerous processes involving degradation of the major types of collagen, notably including both fibrillar and network-forming collagens. In vivo, uPARAP-dependent collagen turnover has been shown to be central in the development of bone in cattle (39) and mice (25, 36), a tissue rich in fibrillar collagen. In ECM turnover in breast cancer and gliomas (61–63), uPARAP is likely to be important for the degradation of both fibrillar and network forming collagens. Particularly, in mouse mammary cancer a pronounced accumulation of collagen I and collagen IV has been demonstrated inside tumors as a result of uPARAP deficiency (61). Furthermore, the receptor has been found to be expressed in the stroma of several types of cancer in humans (44, 64–66) as well as in wound healing (27).
In vitro, the importance of uPARAP-mediated endocytosis of glycosylated collagen has been underscored by experiments demonstrating that the invasiveness of glioma cell lines in collagen IV-rich matrices is almost completely suppressed by RNAi depletion of uPARAP (66), and by experiments demonstrating that uPARAP mediates endocytosis of collagen IV by stromal cells of mammary tumors explanted from mice (61). Together, these experiments point to a regulatory role of the newly discovered uPARAP lectin function in invasive processes.
The demonstration of an interplay between collagen carbohydrates and uPARAP-mediated endocytosis opens the possibility that uPARAP plays a role in other physiological and pathological processes. Thus, some of the deleterious effects resulting from changes in collagen glycosylation may include an altered cellular uptake through uPARAP and, as a result, an imbalance in collagen turnover.
In rheumatoid arthritis, which involves excessive degradation of ECM in joint cartilage and bone, elevated levels of carbohydrate derivatives on collagen degradation products in the urine of patients have been correlated with tissue destruction (67, 68). Indeed, in mice uPARAP has been found to be expressed by chondrocytes in cartilage during development (26, 36) and it is an intriguing hypothesis that uPARAP may participate in the excessive collagen degradation observed in patients with rheumatoid arthritis. This increase could be influenced by elevated levels of collagen glycosylation.
In diabetes, alterations in extracellular matrices connected with the microvasculature are crucially involved in severe complications of the disease. Diabetes is accompanied by elevated concentrations of glucose in tissues and thickening of basement membranes, particularly in the microvascular system (69, 70). Our results indicate that the interaction between uPARAP and collagen IV is sensitive to high carbohydrate concentrations, suggesting that strongly hyperglycemic conditions could affect the local turnover rate of this basement membrane component. Furthermore, diabetes is characterized by the increased non-enzymatic addition of carbohydrates (glycation) to collagens in the ECM. Collagen glycation includes the addition of glucose to hydroxylysine residues (71), creating structures resembling normal collagen O-glycosylation. These changes in collagen carbohydrate content result in deleterious changes in the functional properties of collagen that may include an altered collagen turnover mediated by uPARAP.
Altogether, our present work on the lectin activity of uPARAP points to a function of collagen glycosylation in the cellular uptake process. This provides a novel regulatory level in the dynamic balance between collagen deposition and turnover with several implications in healthy and pathological processes.
Supplementary Material
Acknowledgments
The excellent technical assistance of Suzanne K. Møller and Katharina H. Stegmann is gratefully acknowledged. We thank the CFIM (Department of Biomedical Sciences, Copenhagen University) for microscope assistance.
This work was supported by the Danish Cancer Society, Danish Medical Research Council, Danish Cancer Research Foundation, Lundbeck Foundation, Danish National Research Foundation (Danish-Chinese Center for Proteases and Cancer), European Community's Seventh Framework Programme FP7/2007–2011 under Grant agreement 201279 (to N. B.), grants from the “Grosserer Alfred Nielsen og Hustrus” foundation (to L. H. E.), the Copenhagen University Hospital (to D. H. M. and H. J. J.), and the University of Copenhagen, Faculty of Science (to S. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ECM
- extracellular matrix
- uPARAP
- urokinase plasminogen activator receptor-associated protein
- FN-II
- fibronectin type II
- CTLD
- C-type lectin-like domain
- MR
- mannose receptor
- EndoH
- endoglucosidase H
- α-Me-Gal
- α-methyl-galactopyranoside
- TFMS
- trifluoromethanesulfonic acid
- pAb a-MR
- polyclonal antibody against MR
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Marastoni S., Ligresti G., Lorenzon E., Colombatti A., Mongiat M. (2008) Connect. Tissue Res. 49, 203–206 [DOI] [PubMed] [Google Scholar]
- 2. Rowe R. G., Weiss S. J. (2008) Trends Cell Biol. 18, 560–574 [DOI] [PubMed] [Google Scholar]
- 3. Holmbeck K., Szabova L. (2006) Birth Defects Res. C Embryo. Today 78, 11–23 [DOI] [PubMed] [Google Scholar]
- 4. Khoshnoodi J., Cartailler J. P., Alvares K., Veis A., Hudson B. G. (2006) J. Biol. Chem. 281, 38117–38121 [DOI] [PubMed] [Google Scholar]
- 5. Prockop D. J., Kivirikko K. I. (1995) Annu. Rev. Biochem. 64, 403–434 [DOI] [PubMed] [Google Scholar]
- 6. Gelse K., Pöschl E., Aigner T. (2003) Adv. Drug Deliv. Rev. 55, 1531–1546 [DOI] [PubMed] [Google Scholar]
- 7. Spiro R. G. (1967) J. Biol. Chem. 242, 4813–4823 [PubMed] [Google Scholar]
- 8. Kresina T. F., Miller E. J. (1979) Biochemistry 18, 3089–3097 [DOI] [PubMed] [Google Scholar]
- 9. Sage H., Woodbury R. G., Bornstein P. (1979) J. Biol. Chem. 254, 9893–9900 [PubMed] [Google Scholar]
- 10. Sage H., Bornstein P. (1979) Biochemistry 18, 3815–3822 [DOI] [PubMed] [Google Scholar]
- 11. Hong B. S., Davison P. F., Cannon D. J. (1979) Biochemistry 18, 4278–4282 [DOI] [PubMed] [Google Scholar]
- 12. Aguilar J. H., Jacobs H. G., Butler W. T., Cunningham L. W. (1973) J. Biol. Chem. 248, 5106–5113 [PubMed] [Google Scholar]
- 13. Spiro R. G. (1969) J. Biol. Chem. 244, 602–612 [PubMed] [Google Scholar]
- 14. Rautavuoma K., Takaluoma K., Sormunen R., Myllyharju J., Kivirikko K. I., Soininen R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14120–14125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruotsalainen H., Sipilä L., Vapola M., Sormunen R., Salo A. M., Uitto L., Mercer D. K., Robins S. P., Risteli M., Aszodi A., Fässler R., Myllylä R. (2006) J. Cell Sci. 119, 625–635 [DOI] [PubMed] [Google Scholar]
- 16. Sipilä L., Ruotsalainen H., Sormunen R., Baker N. L., Lamandé S. R., Vapola M., Wang C., Sado Y., Aszodi A., Myllylä R. (2007) J. Biol. Chem. 282, 33381–33388 [DOI] [PubMed] [Google Scholar]
- 17. Lauer-Fields J. L., Juska D., Fields G. B. (2002) Biopolymers 66, 19–32 [DOI] [PubMed] [Google Scholar]
- 18. Song F., Wisithphrom K., Zhou J., Windsor L. J. (2006) Front. Biosci. 11, 3100–3120 [DOI] [PubMed] [Google Scholar]
- 19. Mohamed M. M., Sloane B. F. (2006) Nat. Rev. Cancer 6, 764–775 [DOI] [PubMed] [Google Scholar]
- 20. Behrendt N. (2004) Biol. Chem. 385, 103–136 [DOI] [PubMed] [Google Scholar]
- 21. Sheikh H., Yarwood H., Ashworth A., Isacke C. M. (2000) J. Cell Sci. 113, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 22. Wu K., Yuan J., Lasky L. A. (1996) J. Biol. Chem. 271, 21323–21330 [DOI] [PubMed] [Google Scholar]
- 23. East L., Isacke C. M. (2002) Biochim. Biophys. Acta 1572, 364–386 [DOI] [PubMed] [Google Scholar]
- 24. Engelholm L. H., Ingvarsen S., Jürgensen H. J., Hillig T., Madsen D. H., Nielsen B. S., Behrendt N. (2009) Front. Biosci. 14, 2103–2114 [DOI] [PubMed] [Google Scholar]
- 25. Engelholm L. H., Nielsen B. S., Netzel-Arnett S., Solberg H., Chen X. D., Lopez Garcia J. M., Lopez-Otin C., Young M. F., Birkedal-Hansen H., Danø K., Lund L. R., Behrendt N., Bugge T. H. (2001) Lab. Invest. 81, 1403–1414 [DOI] [PubMed] [Google Scholar]
- 26. Howard M. J., Chambers M. G., Mason R. M., Isacke C. M. (2004) Osteoarthritis Cartilage 12, 74–82 [DOI] [PubMed] [Google Scholar]
- 27. Honardoust H. A., Jiang G., Koivisto L., Wienke D., Isacke C. M., Larjava H., Häkkinen L. (2006) Histopathology 49, 634–648 [DOI] [PubMed] [Google Scholar]
- 28. Smith L., Wagner T. E., Huizar I., Schnapp L. M. (2008) Gene Expr. Patterns 8, 486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Engelholm L. H., List K., Netzel-Arnett S., Cukierman E., Mitola D. J., Aaronson H., Kjøller L., Larsen J. K., Yamada K. M., Strickland D. K., Holmbeck K., Danø K., Birkedal-Hansen H., Behrendt N., Bugge T. H. (2003) J. Cell Biol. 160, 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wienke D., MacFadyen J. R., Isacke C. M. (2003) Mol. Biol. Cell 14, 3592–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. East L., McCarthy A., Wienke D., Sturge J., Ashworth A., Isacke C. M. (2003) EMBO Rep. 4, 710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kjøller L., Engelholm L. H., Høyer-Hansen M., Danø K., Bugge T. H., Behrendt N. (2004) Exp. Cell Res. 293, 106–116 [DOI] [PubMed] [Google Scholar]
- 33. Howard M. J., Isacke C. M. (2002) J. Biol. Chem. 277, 32320–32331 [DOI] [PubMed] [Google Scholar]
- 34. Shi F., Harman J., Fujiwara K., Sottile J. (2010) Am. J. Physiol. Cell Physiol. 298, C1265-C1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mousavi S. A., Fønhus M. S., Berg T. (2009) BMC Cell Biol. 10, 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wagenaar-Miller R. A., Engelholm L. H., Gavard J., Yamada S. S., Gutkind J. S., Behrendt N., Bugge T. H., Holmbeck K. (2007) Mol. Cell. Biol. 27, 6309–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Madsen D. H., Engelholm L. H., Ingvarsen S., Hillig T., Wagenaar-Miller R. A., Kjøller L., Gårdsvoll H., Høyer-Hansen G., Holmbeck K., Bugge T. H., Behrendt N. (2007) J. Biol. Chem. 282, 27037–27045 [DOI] [PubMed] [Google Scholar]
- 38. Messaritou G., East L., Roghi C., Isacke C. M., Yarwood H. (2009) J. Cell Sci. 122, 4042–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fasquelle C., Sartelet A., Li W., Dive M., Tamma N., Michaux C., Druet T., Huijbers I. J., Isacke C. M., Coppieters W., Georges M., Charlier C. (2009) PLoS Genet. 5, e1000666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Behrendt N., Jensen O. N., Engelholm L. H., Mørtz E., Mann M., Danø K. (2000) J. Biol. Chem. 275, 1993–2002 [DOI] [PubMed] [Google Scholar]
- 41. East L., Rushton S., Taylor M. E., Isacke C. M. (2002) J. Biol. Chem. 277, 50469–50475 [DOI] [PubMed] [Google Scholar]
- 42. Martinez-Pomares L., Wienke D., Stillion R., McKenzie E. J., Arnold J. N., Harris J., McGreal E., Sim R. B., Isacke C. M., Gordon S. (2006) Eur. J. Immunol. 36, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 43. Madsen D. H., Ingvarsen S., Jürgensen H. J., Melander M. C., Kjøller L., Moyer A., Honoré C., Madsen C. A., Garred P., Burgdorf S., Bugge T. H., Behrendt N., Engelholm L. H. (2011) J. Biol. Chem. 286, 26996–27010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sulek J., Wagenaar-Miller R. A., Shireman J., Molinolo A., Madsen D. H., Engelholm L. H., Behrendt N., Bugge T. H. (2007) J. Histochem. Cytochem. 55, 347–353 [DOI] [PubMed] [Google Scholar]
- 45. Rasch M. G., Pass J., Illemann M., Høyer-Hansen G., Lund I. K. (2008) J. Immunol. Methods 339, 55–65 [DOI] [PubMed] [Google Scholar]
- 46. Honoré C., Rørvig S., Munthe-Fog L., Hummelshøj T., Madsen H. O., Borregaard N., Garred P. (2008) Mol. Immunol. 45, 2782–2789 [DOI] [PubMed] [Google Scholar]
- 47. Kjaergaard M., Gårdsvoll H., Hirschberg D., Nielbo S., Mayasundari A., Peterson C. B., Jansson A., Jørgensen T. J., Poulsen F. M., Ploug M. (2007) Protein Sci. 16, 1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gårdsvoll H., Hansen L. V., Jørgensen T. J., Ploug M. (2007) Protein Expr. Purif. 52, 384–394 [DOI] [PubMed] [Google Scholar]
- 49. Edge A. S. (2003) Biochem. J. 376, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Watts C., Marsh M. (1992) J. Cell Sci. 103, 1–8 [DOI] [PubMed] [Google Scholar]
- 51. Isacke C. M., van der Geer P., Hunter T., Trowbridge I. S. (1990) Mol. Cell Biol. 10, 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taylor M. E., Bezouska K., Drickamer K. (1992) J. Biol. Chem. 267, 1719–1726 [PubMed] [Google Scholar]
- 53. Napper C. E., Drickamer K., Taylor M. E. (2006) Biochem. J. 395, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thomas E. K., Nakamura M., Wienke D., Isacke C. M., Pozzi A., Liang P. (2005) J. Biol. Chem. 280, 22596–22605 [DOI] [PubMed] [Google Scholar]
- 55. Myllyharju J., Kivirikko K. I. (2004) Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 56. Salo A. M., Cox H., Farndon P., Moss C., Grindulis H., Risteli M., Robins S. P., Myllylä R. (2008) Am. J. Hum. Genet. 83, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Risteli M., Ruotsalainen H., Salo A. M., Sormunen R., Sipilä L., Baker N. L., Lamandé S. R., Vimpari-Kauppinen L., Myllylä R. (2009) J. Biol. Chem. 284, 28204–28211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Notbohm H., Nokelainen M., Myllyharju J., Fietzek P. P., Müller P. K., Kivirikko K. I. (1999) J. Biol. Chem. 274, 8988–8992 [DOI] [PubMed] [Google Scholar]
- 59. Mizuno K., Adachi E., Imamura Y., Katsumata O., Hayashi T. (2001) Micron. 32, 317–323 [DOI] [PubMed] [Google Scholar]
- 60. Brinckmann J., Notbohm H., Tronnier M., Açil Y., Fietzek P. P., Schmeller W., Müller P. K., Bätge B. (1999) J. Invest. Dermatol. 113, 617–621 [DOI] [PubMed] [Google Scholar]
- 61. Curino A. C., Engelholm L. H., Yamada S. S., Holmbeck K., Lund L. R., Molinolo A. A., Behrendt N., Nielsen B. S., Bugge T. H. (2005) J. Cell Biol. 169, 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wienke D., Davies G. C., Johnson D. A., Sturge J., Lambros M. B., Savage K., Elsheikh S. E., Green A. R., Ellis I. O., Robertson D., Reis-Filho J. S., Isacke C. M. (2007) Cancer Res. 67, 10230–10240 [DOI] [PubMed] [Google Scholar]
- 63. Huijbers I. J., Iravani M., Popov S., Robertson D., Al-Sarraj S., Jones C., Isacke C. M. (2010) PLoS One 5, e9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schnack Nielsen B., Rank F., Engelholm L. H., Holm A., Danø K., Behrendt N. (2002) Int. J. Cancer 98, 656–664 [DOI] [PubMed] [Google Scholar]
- 65. Kogianni G., Walker M. M., Waxman J., Sturge J. (2009) Eur. J. Cancer 45, 685–693 [DOI] [PubMed] [Google Scholar]
- 66. Takahashi S., Yamada-Okabe H., Hamada K., Ohta S., Kawase T., Yoshida K., Toda M. (2011) J. Neurooncol. 103, 267–276 [DOI] [PubMed] [Google Scholar]
- 67. Gineyts E., Garnero P., Delmas P. D. (2001) Rheumatology 40, 315–323 [DOI] [PubMed] [Google Scholar]
- 68. Garnero P., Gineyts E., Christgau S., Finck B., Delmas P. D. (2002) Arthritis Rheum. 46, 21–30 [DOI] [PubMed] [Google Scholar]
- 69. Tsilibary E. C. (2003) J. Pathol. 200, 537–546 [DOI] [PubMed] [Google Scholar]
- 70. Hayden M. R., Sowers J. R., Tyagi S. C. (2005) Cardiovasc. Diabetol. 4, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Avery N. C., Bailey A. J. (2006) Pathol. Biol. 54, 387–395 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.