Abstract
Oxidative protein folding in the endoplasmic reticulum is supported by efficient electron relays driven by enzymatic reactions centering on the ERO1-protein-disulfide isomerase (PDI) pathway. A controlled in vitro oxygen consumption assay was carried out to analyze the ERO1-PDI reaction. The results showed the pH-dependent oxidation of PDI by ERO1α. Among several possible disulfide bonds regulating ERO1α activity, Cys94–Cys131 and Cys99–Cys104 disulfide bonds are dominant regulators by excluding the involvement of the Cys85–Cys391 disulfide in the regulation. The fine-tuned species specificity of the ERO1-PDI pathway was demonstrated by functional in vitro complementation assays using yeast and mammalian oxidoreductases. Finally, the results provide experimental evidence for the intramolecular electron transfer from the a domain to the a′ domain within PDI during its oxidation by ERO1α.
Keywords: Disulfide, Electron Transfer, Endoplasmic Reticulum, Oxidase, Protein Isomerase
Introduction
Endoplasmic reticulum (ER)2 is the primary intracellular compartment of the secretory pathway (1, 2). Most secretory and membrane proteins cotranslationally enter the ER and are subjected to post-translational modifications, including glycosylation and disulfide bond formation. Disulfide bonds are vital for the correct folding and maintenance of the conformation of secretory proteins (3). The oxidative environment of the ER is suitable for efficient disulfide bond formation, and it harbors a number of oxidoreductases that participate in the oxidation, isomerization, and reduction of disulfide bonds (4).
Protein disulfide isomerase (PDI) is a representative ER-resident oxidoreductase that catalyzes disulfide formation and acts as a molecular chaperone (5, 6). PDI has two catalytically active domains (a and a′), each of which possesses a catalytic Cys-X-X-Cys motif and two non-catalytic b and b′ domains (see Fig. 4A). The hydrophobic region of the b′ domain is known to serve as a substrate-binding domain. ERO1 (Ero1p in yeast), which encodes a flavoprotein oxidase, was identified as the primary oxidative source in the ER (7). In conjunction with the oxidation of the substrate proteins, ERO1 consumes molecular oxygen as the terminal electron acceptor and produces hydrogen peroxide (H2O2). ERO1 does not transfer oxidizing equivalents directly to substrate proteins. Instead, ERO1 oxidizes PDI, which oxidizes substrate proteins by the formation of disulfide bonds (Fig. 1A).
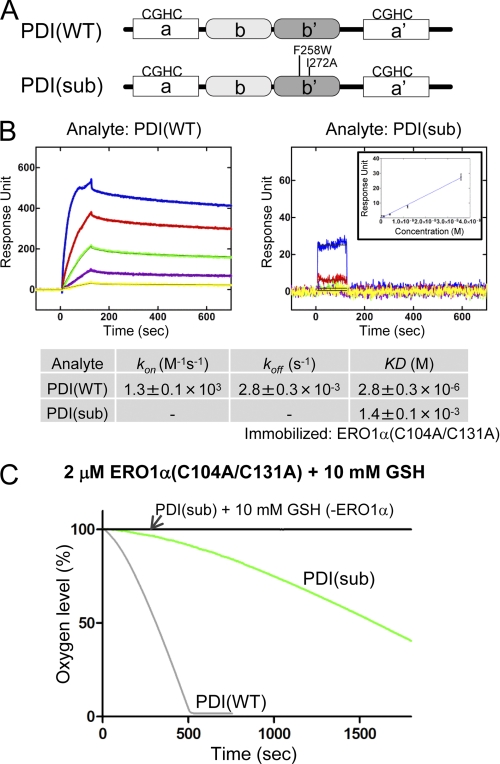
FIGURE 4.
Effect of point mutations of the PDI b′ domain on the binding affinity for ERO1α and the rate of oxygen consumption. A, schematic representation of human PDI protein and the point mutations introduced into the b′ domain of PDI, PDI(sub). B, affinity measurements between wild-type PDI and constitutively active ERO1α, or PDI(sub) and constitutively active ERO1α by SPR spectroscopy. Blue, red, green, purple, and yellow curves correspond to 36, 12, 4, 1.33, and 0.44 μm analyte, respectively. The inset in the right panel shows equilibrium analysis based on a set of response units of the steady state (y-axis) at each analyte concentration (x-axis). The lower column shows the calculated kinetic parameters. C, kinetics of oxygen consumption by 2 μm constitutively active ERO1α during the reaction with 5 μm PDI(WT) or PDI(sub) in the presence of GSH (10 mm).
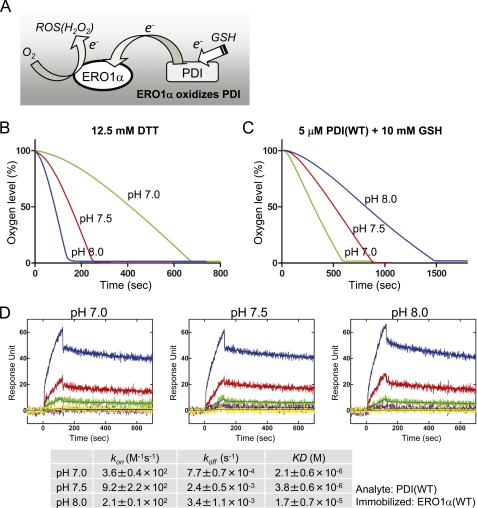
FIGURE 1.
pH-dependent oxygen consumption of ERO1α(WT). A, a schematic model of Ero1α-driven electron transfer. During the oxidation of PDI by ERO1α, ERO1α transfers electrons from PDI to O2, which in turn becomes a reactive oxygen species (ROS(H2O2)). GSH supplies electrons to PDI. Oxygen consumption by 2 μm wild-type ERO1α oxidation of 12.5 mm DTT (B), or 5 μm human PDI in the presence of 10 mm GSH (C) under different pH conditions (pH 7.0, 7.5, and 8.0). D, affinity of human PDI and wild-type ERO1α as measured by SPR spectroscopy under different pH conditions. ERO1α was immobilized on a biosensor chip, and PDI was injected as an analyte. Blue, red, green, purple, and yellow curves correspond to 36, 12, 4, 1.33, and 0.44 μm analyte, respectively. The lower column shows the calculated kinetic parameters.
Molecular functions and regulatory mechanisms of the yeast and mammalian ERO1 family members have been clarified (8–11). To activate the yeast Ero1p oxidase, its Cys150–Cys295 disulfide bond must be reduced (see Fig. 3A) (12). The C150A/C295A Ero1p mutant exhibited a deregulated increase in its oxidase activity and disturbed redox homeostasis by hyperoxidizing the yeast ER (8, 13). Thus, one or more regulatory intra-disulfide bonds have an important role in optimizing the ER redox environment.
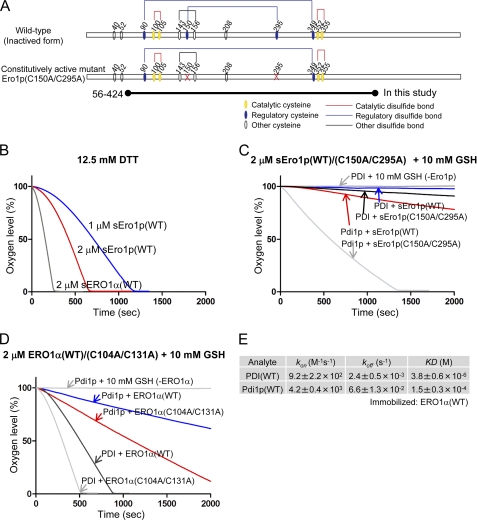
FIGURE 3.
In vitro complementation assay between the human PDI-ERO1α and yeast Pdi1p-Ero1p pathway. A, wild-type and constitutively active mutant of yeast Ero1p. Cysteine residues are shown with circles. Structural, regulatory, and catalytic disulfides are represented by black, blue, and red lines, respectively (7). The short form of yeast Ero1p, spanning amino acid residues 56–424, was examined; it is named sEro1p (12, 21). B, oxygen consumption by yeast sEro1p or human ERO1α oxidation of 12.5 mm DTT. Kinetics of oxygen consumption by 2 μm yeast wild-type or constitutively active sEro1p (C), or 2 μm human wild-type or constitutively active ERO1α (D), during the reaction with 5 μm yeast Pdi1p or human PDI in the presence of GSH (10 mm). E, calculated kinetic parameters for affinity measurements between human PDI and ERO1α(WT) or yeast Pdi1p and ERO1α(WT) by SPR spectroscopy. Sensorgrams are shown in supplemental Fig. S2.
Human ERO1α has similar regulatory disulfide bonds; however, the regulatory mechanism is different from that of yeast Ero1p. Regulatory (non-catalytic) cysteines of ERO1α form disulfide bonds with the catalytic cysteines (10, 11). Specifically, when ERO1α is under inactivated condition, Cys94, which is known to be involved in receiving electrons from PDI, forms a disulfide bond with Cys131. The other cysteine of the outer active site (Cys99) forms a disulfide bond with Cys104. When both disulfide bonds are reduced/isomerized, the Cys94–Cys99 shuttle disulfide is formed and ERO1α is fully activated (see Fig. 2A) (9, 10, 14).
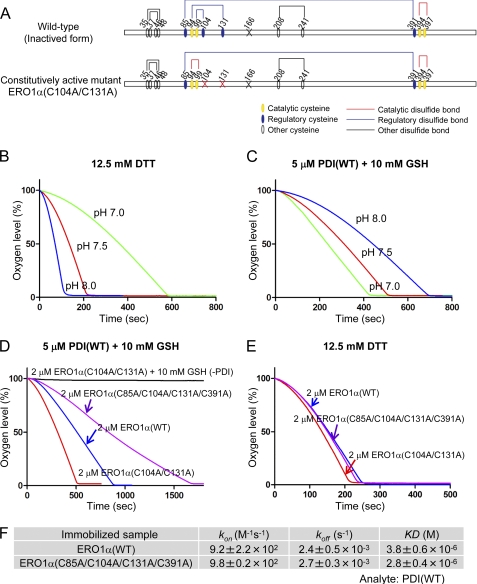
FIGURE 2.
Evaluation of regulatory disulfide bonds of ERO1α. A, wild-type and constitutively active (deregulated or hyperactive) mutant of human ERO1α. Cysteine residues are shown with circles, and the cysteine to alanine mutation sites in ERO1 are represented by an X. Structural, regulatory, and catalytic disulfides are represented by black, blue, and red lines, respectively (11). Free cysteine Cys166 of ERO1 was mutated to alanine to purify homogeneous recombinant proteins (11). Oxygen consumption by the constitutively active ERO1α oxidation of 12.5 mm DTT (B), or 5 μm human PDI in the presence of 10 mm GSH (C), under different pH conditions (pH 7.0, 7.5, and 8.0). D, kinetics of oxygen consumption by ERO1 (2 μm) wild-type or mutants, as depicted in the figure, during a reaction with 5 μm PDI in the presence of GSH (10 mm). E, oxygen consumption by 2 μm ERO1α wild type or mutants in the presence of 12.5 mm DTT. F, calculated kinetic parameters for affinity measurements between human PDI and ERO1α(WT) or ERO1α(C85A/C104A/C131A/C391A) by SPR spectroscopy.
In the present study, a detailed in vitro analysis of the regulatory disulfides in mammalian ERO1α was performed using an oxygen consumption assay that had been optimized for pH, temperature, and ionic strength. The functional differences between yeast and mammalian ERO1 proteins were compared in terms of their specificities for the substrates, PDI/Pdi1p proteins. Finally, the study yielded experimental evidence that ERO1α catalyzes the PDI oxidation by way of intramolecular electron transfer from its a domain to its a′ domain.
EXPERIMENTAL PROCEDURES
Plasmids
Human PDI and ERO1α cDNAs without the signal sequence were amplified by PCR from a Matchmaker Pretransformed Human HeLa library (Clontech) and subcloned into the modified pETDuet-1 vector (Novagen) modified with an N-terminal His6 tag and TEV (tobacco etch virus) protease recognition site called the pETDuet-TEV vector. The C166A mutation was introduced into all ERO1α constructs to avoid formation of disulfide-linked ERO1α dimers or oligomers as mentioned previously, except in supplemental Fig. S4 (11). Yeast Ero1p and Pdi1p were amplified from Yeast ORF Collections (ID, YML130C; Ero1p and YCL043C; Pdi1p) and subcloned into the pETDuet-TEV vector. All mutations for PDI, ERO1α, and Ero1p were created using the QuikChange site-directed mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing.
Expression and Purification
The overexpression and purification of ERO1α and its mutants that lacked the non-functional cysteine Cys166 was performed according to the method of Inaba et al. (11). Briefly, ERO1α was overexpressed in Escherichia coli strain BL21(DE3). Cells were grown at 37 °C in Luria-Bertani (LB) medium containing 100 μg/ml ampicillin and 10 μm FAD to an optical density (600 nm) of 0.6, and the expression of the recombinant proteins was induced at 24 °C for 12–16 h by adding 200 μm isopropyl-β-d-thiogalactoside. Cells were collected, and protein purification was performed at 4 °C as follows. Harvested cells were sonicated in 20 mm HEPES-NaOH (pH 7.4) containing 150 mm NaCl, 20 mm imidazole, and protease inhibitor mixture without EDTA (Sigma). The supernatant was loaded onto a HisTrap column (GE Healthcare) and eluted with 0.5 m imidazole after washing the column with the lysis buffer. The eluted ERO1α sample was oxidized with the addition of potassium ferricyanide at a final concentration of 20 mm for 20 min on ice, which was followed by gel-permeation chromatography (HiLoad 16/60 Superdex 200-pg column, GE Healthcare) pre-equilibrated with 20 mm HEPES-NaOH (pH 7.4) containing 150 mm NaCl, 10% glycerol, and no imidazole. The fractions containing monomeric ERO1α were applied onto the Resource Q column (GE Healthcare) pre-equilibrated with 20 mm HEPES-NaOH (pH 8). The sample was eluted with a linear NaCl gradient ranging from 0 to 500 mm. Yeast Ero1p and mutant protein were overexpressed in E. coli strain Origami B (DE3), and the other procedures were the same as those for ERO1α, but without the potassium ferricyanide treatment. The overexpression and purification of PDI/Pdi1p and its variants were performed essentially as described in Ushioda et al. (15).
Oxygen Consumption Assay
Oxygen consumption was measured using a Clark-type oxygen electrode (YSI 5331). Experiments were performed in 50 mm HEPES (pH 7.5), 150 mm NaCl, and 2 mm EDTA at 25 °C using a constant temperature incubator in air-saturated buffer, except for the experiments shown in Figs. 1B, 1C, 2B, and 2C and supplemental Fig. S1A and S1B as described in each figure. Catalytic oxygen consumption was initiated by the addition of 2 μm ERO1α, Ero1p, or their variants in a reaction mixture containing 12.5 mm DTT, or PDI, Pdi1p or their variants and 10 mm GSH, as depicted in each figure. The oxidoreductase concentrations used are also given in each figure.
SPR Measurements
Association or dissociation rate constants (kon or koff) for the direct binding of PDI/Pdi1p or their variants to immobilized ERO1α(WT) or its variants were determined by surface plasmon resonance (SPR) measurements on a ProteOn XPR36 Protein Interaction Array System (Bio-Rad). ERO1 was coupled to the GLC sensor chip (Bio-Rad) using amine coupling chemistry. As a control, one channel was coupled with BSA to exclude background binding. Sensorgrams were recorded simultaneously for five concentrations of analytes ranging from 0.44 μm to 36 μm in triplicate increments at 25 °C for a 2-min association phase, followed by a 10-min dissociation phase by using 20 mm HEPES-NaOH (pH 7.5; except Fig. 1D), 150 mm NaCl, 0.001% Tween 20 and 2 mm EDTA as running and sample buffer. Sensorgrams were analyzed by non-linear regression analysis according to a two-state model using ProteOn Manager Version 2.0.1 software (Bio-Rad), except when PDI(sub) was analyzed. PDI(sub) reached the steady state promptly, and the equilibrium parameter was extracted by analyzing a set of sensorgrams of the steady state. Experiments were replicated at least three times.
RESULTS
pH-dependent ERO1α Activity Using the Controlled Oxygen Consumption Assay
The oxygen consumption assay using the Clark-type oxygen sensor is a useful technique to analyze the oxidase activity of the ERO1 protein family (Fig. 1A) (16). The high sensitivity of this assay system causes the results to be affected by various experimental conditions, including temperature, ionic strength, pH, and stirring rate (17). However, these factors have not been adequately examined. The amount of dissolved oxygen is influenced by changes in temperature and ionic strength of the solvent. As the NaCl concentration and the temperature increase, the amount of dissolved oxygen decreases, which artificially affects the ERO1α oxygen consumption response curve (supplemental Figs. S1A and S1B).
Alteration of pH affects the protonated or deprotonated states of the cysteine thiols within proteins depending on their pKa and directly affects the activity of oxidoreductases (5). When DTT was used as the electron donor, the higher the pH value of the reaction buffer, the faster the oxygen consumption rate (Fig. 1B). This effect may be caused by the increased efficiency of electron transfers between DTT and ERO1 at higher pH values, which provided an apparent higher specific activity of ERO1α at higher pH values. However, when GSH was used as an electron donor in the presence of PDI, which will be oxidized by ERO1α (Fig. 1A), the oxygen consumption rate decreased as the pH value increased (Fig. 1C). This pH dependence in the oxidation reaction could be explained in part by differences in the binding affinity of PDI with ERO1α at different pH values. ERO1α binding affinity to PDI was strong at pH 7 but decreased to the extent of one-tenth at pH 8 (Fig. 1D). Previously, it was reported that oxidase and isomerase activities of PDI are pH-dependent and show maximum activities near pH 7 (18, 19). Therefore, to obtain accurate results for the oxidoreductase activity of ERO1α in the oxygen consumption assay, it is necessary to maintain optimal and fixed conditions for the pH, temperature, and ionic strength of the solvent. Thus, physiologically relevant conditions for ERO1α and PDI were chosen, i.e. pH 7.5, 150 mm NaCl, with constant temperature and stirring speed in the following study.
Regulatory Disulfide Bonds of ERO1α
Isomerization of regulatory disulfide bonds is required for the activation of ERO1α activity (Fig. 2A). Mutations of both cysteine 104 and 131 to alanine produce a constitutively active mutant of ERO1α, ERO1α(C104A/C131A) (11, 20). We confirmed its increased enzymatic activity and observed a pH dependence similar to that of ERO1α(WT) (Fig. 2, B and C).
Some reports suggest the existence of an additional potential regulatory disulfide between cysteine 85 and 391 (10, 14, 20). The effect of this disulfide bond Cys85–Cys391 on the activity of ERO1α was examined by constructing alanine mutations of these cysteines (C85A and C391A) in combination with the C104A and C131A mutations. C85A or C391A single mutants of ERO1α produced a poor yield and were intrinsically unstable, as reported previously (10).3 On the other hand, ERO1α(C85A/C391A), a double cysteine mutant, was significantly more stable compared with the C85A and C391A single mutants, suggesting that the presence of a free counterpart cysteine (Cys391 in the C85A mutant and Cys85 in the C391A mutant) might disturb proper disulfide bond formation, resulting in structural instability. Oxygen consumption analysis confirmed that these mutations did not increase the oxidase activity of ERO1α (supplemental Fig. S2). In fact, ERO1α(C85A/C104A/C131A/C391A) decreased the accelerated activity of ERO1α(C104A/C131A) (Fig. 2D). These results suggest that its structure would not be disturbed by these mutations, because its oxygen consumption rate in the presence of DTT and its binding affinity for PDI were identical to those of ERO1α(WT) (Fig. 2, E and F). These results demonstrate that Cys94–Cys104 and Cys99–Cys131 disulfide bonds serve a major negative regulatory function and that Cys85–Cys391 disulfide bond formation would be necessary for proper oxidative activity of ERO1α. Thereafter, in experiments described below, ERO1α(C104A/C131A) was used as a constitutively active mutant.
In Vitro Complementation of Human PDI-ERO1α and Yeast Pdi1p-Ero1p Pathways
Human PDI-ERO1α and yeast Pdi1p-Ero1p pathways might have different characteristics in the regulation and complementation because of differences in the organization of their disulfide bonds. A short form of yeast Ero1p (Fig. 3A), hereafter named sEro1p, which consisted of amino acids 56–424, was used to determine the crystal structure and was shown to have oxidase activity as previously reported (Fig. 3B) (12, 21, 22).
The oxygen consumption assay showed that, when yeast sEro1p was used as an oxidase, yeast Pdi1p was oxidized whereas human PDI was not (Fig. 3C). This effect was also demonstrated when the constitutively active sEro1p was used. Next, when human ERO1α was used as an oxidase, the yeast Pdi1p was oxidized less efficiently than human PDI (Fig. 3D). The same result was found for the constitutively active mutant ERO1α(C104A/C131A). These results indicated that ERO1 oxidizes PDI in a species-specific manner. In addition, we found that the binding affinity of yeast Pdi1p with human ERO1α was two orders of magnitude weaker than that of human PDI with ERO1α (Fig. 3E). Hence, the low catalytic activity of ERO1α with yeast Pdi1p could be partly explained by this weak binding affinity.
To confirm the correlation between the binding affinity and the oxygen consumption rate, a PDI mutant with decreased substrate-binding activity was constructed. Because both the F258W and I272A mutations in the b′ domain reduce the binding affinity for peptide substrates, we mutated these residues to obtain PDI(sub) (Fig. 4A) (23, 24). This mutant showed a marked decrease in the binding affinity for ERO1α that was three orders of magnitude weaker than that of wild-type PDI (Fig. 4B). The rate of oxidation of PDI(sub) by ERO1α was much decreased compared with that of wild-type PDI (Fig. 4C). Thus, the binding affinity directly affects the ERO1α oxidation reaction (11, 25).
Directional Intramolecular Electron Relay between PDI-active Domains
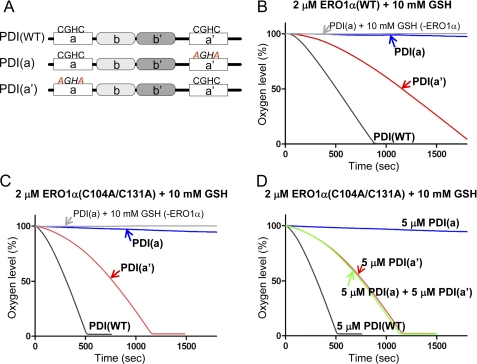
ERO1 preferentially oxidizes the a′ domain of human PDI (10, 20, 25). To examine whether the specificity of ERO1α toward the two active sites, a and a′ domains, of human PDI could be reproduced in our assay, we analyzed the activity of ERO1α in the presence of PDI mutants where the CGHC motif in the a or a′ domain was mutated to AGHA (Fig. 5A). The oxygen consumption assay showed that PDI(a′), with the intact CGHC motif in the a′ domain, was effectively oxidized by ERO1α. However, PDI(a) with the mutated AGHA motif in the a′ domain was not oxidized to any extent by ERO1α (Fig. 5B). Similar specific oxidation was also observed for the constitutively active ERO1α(C104A/C131A) mutant (Fig. 5C).
FIGURE 5.
Selective oxidation of ERO1α toward the a′ domain and the intramolecular electron relay within PDI. A, schematic representation of human PDI proteins with the -CGHC- active sites and the mutated -AGHA sites indicated. Kinetics of oxygen consumption by 2 μm wild-type ERO1α (B), or constitutively active ERO1α (C), during the reaction with 5 μm human PDI variants, as depicted in the figure, in the presence of GSH (10 mm). D, oxygen consumption by the 2 μm constitutively active ERO1α during the oxidation of 5 μm human PDI variants, including PDI(WT), PDI(a), and PDI(a′), or the mixture of PDI(a) and PDI(a′).
There was a large difference between the oxygen consumption rate of PDI(a′) and PDI(WT) even in the presence of constitutively active ERO1α(C104A/C131A), whereas both PDI(WT) and PDI(a′) contain the intact CGHC motif in the a′ domain (compare the red curve (PDI(a′)) with the gray curve (PDI(WT)) in Fig. 5C). This raised the possibility that the lack of CGHC motif in the a domain caused such a decrease in the oxygen consumption by ERO1α. Fig. 5C showed that ERO1α selectively oxidized the a′ domain of PDI and accepted electrons from that domain. Therefore, electrons must be supplied from some other donor, either from different molecules or from different domains in the same molecule, most probably from the a domain of PDI.
If intermolecular electron transfer can occur, electrons could be transferred from the a domain of one PDI to the a′ domain of the other PDI. To examine this possibility, PDI(a) and PDI(a′) were mixed in the presence of constitutively active ERO1α(C104A/C131A), and the oxygen consumption assay was performed. If intermolecular electron transfer can occur, that is, if the a′ domain accepted electrons from the a domain in different PDI molecules, the presence of both PDI(a) and PDI(a′) in the reaction mixture with ERO1α would result in the same oxygen consumption curve as that formed using wild-type PDI. However, the mixture of PDI(a) and PDI(a′) did not increase the oxygen consumption rate, and a rate almost similar to that found with PDI(a′) was observed. This suggested that intermolecular electron transfer did not occur during the oxidation of PDI by ERO1α.
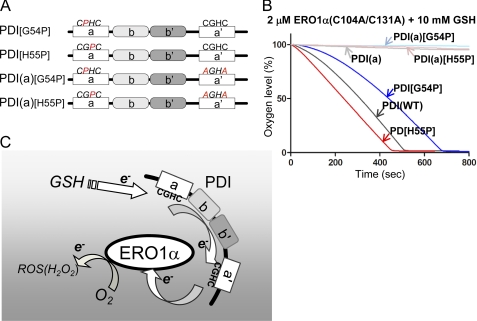
We further examined intermolecular electron transfer to determine whether changes in the reduction potential of the a domain affected reactivity with ERO1α. Altering the two residues between the cysteines in the Cys-X-X-Cys active motif in the thioredoxin domain has been reported to affect the reduction potential of this domain. For example, mutating the active motif from CGHC to CGPC in the a domain, here named PDI[H55P], was shown to lower its reduction potential, whereas mutation from CGHC to CPHC, here named PDI[G54P], was reported to increase its reduction potential (13, 20, 26). To evaluate the effect of each of these mutations, we mutated G54P or H55P in the presence of an AXXA mutation in the a′ domain, resulting in PDI(a)[G54P] and PDI(a)[H55P], respectively (Fig. 6A). The results of the oxygen consumption assay showed that PDI(a)[G54P] and PDI(a)[H55P] were minimally oxidized by constitutively active ERO1α, which was consistent with the data obtained with PDI(a) (Fig. 6B). However, PDI[H55P], which has an intact a′ domain, exhibited a significantly increased oxygen consumption rate, whereas PDI[G54P] had a decreased oxygen consumption rate (Fig. 6B). Although previous reports demonstrated no correlation between the reduction potential and the rate of oxidation, altering the reduction potential in the a domain, which is minimally oxidized by ERO1α, affects the net reactivity with ERO1α. This suggests the presence of an intramolecular electron relay between the a and a′ domains (13). Taken together, these results strongly suggest that electrons were transferred from the a domain to the a′ domain, within a single PDI molecule during oxidation by ERO1α.
FIGURE 6.
Modulation of the reduction potential of the a domain affects the net oxidation rate of PDI by ERO1α. A, schematic representation of human PDI proteins with the mutated -CPHC-, -CGPC-, and -AGHA- sites indicated. B, the kinetics of oxygen consumption by 2 μm constitutively active ERO1α in a reaction containing 5 μm of each human PDI variant, as indicated in the figure, in the presence of GSH (10 mm). C, schematic model illustrating the interaction of and electron transfer relays between ERO1α and PDI. GSH supplies the electron, probably to the a domain of PDI. Following internal electron transfer from the a to the a′ domain, ERO1α receives the electron from the a′ domain. Intermolecular electron transfer would be stochastically constrained.
DISCUSSION
The oxygen consumption assay is used in the analysis of the ERO1 oxidase activity. However, the experimental conditions were not well characterized, which could cause discrepancies in the data obtained (10, 11, 21, 25). We found a physical as well as functional pH dependence of ERO1α and PDI, with the apparent maximum catalytic rate and binding affinity observed at pH ∼7 (Fig. 1, C and D). This suggested that PDI is fine-tuned to catalyze oxidative folding near the physiological pH in the ER (27). This result is consistent with previous data, which suggests the isomerase and oxidase activities of PDI are most efficient at pH 7 when using RNase or a small peptide as model substrates (18, 19). The low pKa of the N-terminal catalytic cysteines of PDI (4.4–6.7) compared with the normal pKa of cysteines on the protein (∼8.3) may cause such a pH dependence (28–30). The pH dependence of the binding affinity might be due to the electrostatic properties of positive and negative charges of ERO1α and PDI, respectively, which are affected by the pH shift (5, 11).
Human ERO1α possesses regulatory disulfides, which serve as a redox-sensor to maintain the proper oxidative environment of the ER (14). It has not been clear whether the Cys85–Cys391 disulfide bond stabilizes the structure of ERO1α or regulates its activity (11, 14, 31). The results indicate that the Cys85–Cys391 disulfide was not involved in the regulation of ERO1α activity, but that Cys94–Cys104 and Cys99–Cys131 are the critical disulfide bonds that negatively regulate the enzymatic activity of ERO1α. Recently solved crystal structures of human ERO1α and yeast Ero1p illustrated the structural differences that underlie their distinct regulatory mechanisms (11, 12, 22). Yeast Ero1p contains a Cys150–Cys295 disulfide bond that innately controls its enzyme activation, but ERO1α does not have such a disulfide bond (7). Ero1p also has the regulatory disulfide bond Cys90–Cys349 that, when reduced, increases enzyme activity to a lesser extent. The corresponding disulfide bond in ERO1α is the Cys85–Cys391 bond, but this did not contribute to enzyme activation (Fig. 2, D and E, and supplemental Fig. S2). Regulatory disulfide bonds of Ero1p/ERO1α are probably determined by the redox states of Pdi1p or PDI and/or the redox buffer within the ER. In vitro complementation analysis showed that the substrate specificity with the highest enzyme activity was obtained using protein combinations from the same species (Fig. 3, C and D). However, even though the catalytic activity of ERO1α with yeast Pdi1p was less efficient in the in vitro analysis, the human ERO1α complemented the temperature-sensitive yeast Ero1p mutant in vivo. This apparent discrepancy would be an interesting issue to address in the future (31, 32).
The results also provide insights into the mechanism of how electrons are transferred within the PDI molecule during its oxidation by ERO1α. SPR analysis using point mutations in the b′ domain of PDI suggested that it is the b′ domain that contributes to the specific binding with ERO1α, a result that is consistent with previous reports (Fig. 4B) (11, 25, 33). If the single domains of each a and a′ domain were extracted, they showed the same oxygen consumption rate in the presence of constitutively active ERO1α (11). Domain-swapping mutants of PDI showed that the position of a′ domain was preferentially oxidized, suggesting the importance of the domain position (25). Recent structural analysis indicates that the b′ domain of PDI specifically interacts with a protruding β-hairpin of ERO1α (33). Hence, the specific binding site and domain architecture of PDI allow specific binding between PDI and ERO1α and define the preferred oxidation of the a′ domain by ERO1α.
For ERO1α, the maximum rate of oxygen consumption required both the intact a and a′ domains, which suggests that electron exchange occurs between these domains. We also showed that the a′ domain of PDI was efficiently oxidized by ERO1α, whereas the a domain was not (Fig. 5, B and C). These results were reproduced using ERO1α containing a non-functional cysteine Cys166, which is thought to promote dimer formation of ERO1α, suggesting that Cys166 may not contribute to the preferential oxidation of the a′ domain (supplemental Fig. S4). Thus, the specific oxidation of the a′ domain is defined by the spatial binding of ERO1α to PDI, and the direct oxidation of the a domain of PDI by ERO1α is physically restricted. In addition, electron transfer occurred by way of intramolecular relay, but not through intermolecular transition (Fig. 5D). The occurrence of electron relay is further supported by the results of oxygen consumption assays using the reduction potential-altered mutants of the a domain. Although the a domain itself was minimally oxidized by ERO1α, the shift in reduction potential of the a domain dramatically affected the net oxidation rate, clearly indicating intra-electron relay between the a and a′ domains. Considering the fact that the a′ domain is preferentially oxidized by ERO1α, electrons are most probably transferred from the a domain to the a′ domain and, subsequently, to ERO1α (Fig. 6C). Recent data also indicated that the electron transfer occurred from the a to the a′ domain but did not distinguish between inter- or intramolecular electron transfer (20). A dynamic conformational change within PDI might allow direct electron transfer within the PDI molecule, or perhaps aromatic amino acid residues of PDI or ERO1α might allow intramolecular electron relays similar to those of photosynthesis or the mitochondrial respiratory complex (34–37). Additional studies are needed to understand the precise catalytic mechanism of PDI with its physiological substrates and ERO1α.
Supplementary Material
Acknowledgments
We thank Ryota Maeda and Yo-ichi Nabeshima for generously allowing us to use their SPR system. We also thank Kenji Inaba for valuable suggestions.
This work was supported by Grants-in-Aid for Creative Scientific Research (19G0314) and for Scientific Research on Priority Area (19058008) from Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to K. N.) and by a fellowship from the Japan Society for the Promotion of Science (to K. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
K. Araki and K. Nagata, unpublished observation.
- ER
- endoplasmic reticulum
- PDI
- protein disulfide isomerase
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Ellgaard L., Helenius A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 2. Hoseki J., Ushioda R., Nagata K. (2010) J. Biochem. 147, 19–25 [DOI] [PubMed] [Google Scholar]
- 3. Anelli T., Sitia R. (2008) EMBO J. 27, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Appenzeller-Herzog C., Ellgaard L. (2008) Biochim. Biophys. Acta 1783, 535–548 [DOI] [PubMed] [Google Scholar]
- 5. Hatahet F., Ruddock L. W., Ahn K., Benham A., Craik D., Ellgaard L., Ferrari D., Ventura S. (2009) Antioxid. Redox. Signal 11, 2807–2850 [DOI] [PubMed] [Google Scholar]
- 6. Freedman R. B., Klappa P., Ruddock L. W. (2002) EMBO Rep. 3, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sevier C. S., Kaiser C. A. (2008) Biochim. Biophys. Acta 1783, 549–556 [DOI] [PubMed] [Google Scholar]
- 8. Sevier C. S., Qu H., Heldman N., Gross E., Fass D., Kaiser C. A. (2007) Cell 129, 333–344 [DOI] [PubMed] [Google Scholar]
- 9. Appenzeller-Herzog C., Riemer J., Christensen B., Sørensen E. S., Ellgaard L. (2008) EMBO J. 27, 2977–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker K. M., Chakravarthi S., Langton K. P., Sheppard A. M., Lu H., Bulleid N. J. (2008) EMBO J. 27, 2988–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Inaba K., Masui S., Iida H., Vavassori S., Sitia R., Suzuki M. (2010) EMBO J. 29, 3330–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heldman N., Vonshak O., Sevier C. S., Vitu E., Mehlman T., Fass D. (2010) Protein Sci. 19, 1863–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vitu E., Kim S., Sevier C. S., Lutzky O., Heldman N., Bentzur M., Unger T., Yona M., Kaiser C. A., Fass D. (2010) J. Biol. Chem. 285, 18155–18165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tavender T. J., Bulleid N. J. (2010) Antioxid. Redox. Signal 13, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 15. Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 16. Clark L. C., Jr., Wolf R., Granger D., Taylor Z. (1953) J. Appl. Physiol. 6, 189–193 [DOI] [PubMed] [Google Scholar]
- 17. Berntsson M., Tengberg A., Hall P. O., Josefson M. (1997) Anal. Chim. Acta 355, 43–53 [Google Scholar]
- 18. Wang Y. H., Narayan M. (2008) Protein J. 27, 181–185 [DOI] [PubMed] [Google Scholar]
- 19. Alanen H. I., Salo K. E., Pirneskoski A., Ruddock L. W. (2006) Antioxid. Redox. Signal 8, 283–291 [DOI] [PubMed] [Google Scholar]
- 20. Chambers J. E., Tavender T. J., Oka O. B., Warwood S., Knight D., Bulleid N. J. (2010) J. Biol. Chem. 285, 29200–29207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gross E., Sevier C. S., Heldman N., Vitu E., Bentzur M., Kaiser C. A., Thorpe C., Fass D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gross E., Kastner D. B., Kaiser C. A., Fass D. (2004) Cell 117, 601–610 [DOI] [PubMed] [Google Scholar]
- 23. Koivunen P., Salo K. E., Myllyharju J., Ruddock L. W. (2005) J. Biol. Chem. 280, 5227–5235 [DOI] [PubMed] [Google Scholar]
- 24. Pirneskoski A., Klappa P., Lobell M., Williamson R. A., Byrne L., Alanen H. I., Salo K. E., Kivirikko K. I., Freedman R. B., Ruddock L. W. (2004) J. Biol. Chem. 279, 10374–10381 [DOI] [PubMed] [Google Scholar]
- 25. Wang L., Li S. J., Sidhu A., Zhu L., Liang Y., Freedman R. B., Wang C. C. (2009) J. Biol. Chem. 284, 199–206 [DOI] [PubMed] [Google Scholar]
- 26. Mössner E., Huber-Wunderlich M., Glockshuber R. (1998) Protein Sci. 7, 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim J. H., Johannes L., Goud B., Antony C., Lingwood C. A., Daneman R., Grinstein S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2997–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hawkins H. C., Freedman R. B. (1991) Biochem. J. 275, 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kortemme T., Darby N. J., Creighton T. E. (1996) Biochemistry 35, 14503–14511 [DOI] [PubMed] [Google Scholar]
- 30. Ruddock L. W., Hirst T. R., Freedman R. B. (1996) Biochem. J. 315, 1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bertoli G., Simmen T., Anelli T., Molteni S. N., Fesce R., Sitia R. (2004) J. Biol. Chem. 279, 30047–30052 [DOI] [PubMed] [Google Scholar]
- 32. Cabibbo A., Pagani M., Fabbri M., Rocchi M., Farmery M. R., Bulleid N. J., Sitia R. (2000) J. Biol. Chem. 275, 4827–4833 [DOI] [PubMed] [Google Scholar]
- 33. Masui S., Vavassori S., Fagioli C., Sitia R., Inaba K. (2011) J. Biol. Chem. 286, 16261–16271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang C., Chen S., Wang X., Wang L., Wallis A. K., Freedman R. B., Wang C. C. (2010) J. Biol. Chem. 285, 26788–26797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dempsey J. L., Winkler J. R., Gray H. B. (2010) Chem. Rev. 110, 7024–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson N., Yocum C. F. (2006) Annu. Rev. Plant Biol. 57, 521–565 [DOI] [PubMed] [Google Scholar]
- 37. Tian G., Xiang S., Noiva R., Lennarz W. J., Schindelin H. (2006) Cell 124, 61–73 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.