Abstract
Homing endonucleases have great potential as tools for targeted gene therapy and gene correction, but identifying variants of these enzymes capable of cleaving specific DNA targets of interest is necessary before the widespread use of such technologies is possible. We identified homologues of the LAGLIDADG homing endonuclease I-AniI and their putative target insertion sites by BLAST searches followed by examination of the sequences of the flanking genomic regions. Amino acid substitutions in these homologues that were located close to the target site DNA, and thus potentially conferring differences in target specificity, were grafted onto the I-AniI scaffold. Many of these grafts exhibited novel and unexpected specificities. These findings show that the information present in genomic data can be exploited for endonuclease specificity redesign.
Keywords: Computer Modeling, Enzyme Mutation, Gene Therapy, Nucleic Acid Enzymology, Protein-DNA Interaction, Computational Enzyme Design, Evolutionary Sequence Information, Genome Engineering, Homing Endonuclease
Introduction
Homing endonucleases (HEs)2 are DNA cleaving enzymes encoded by an open reading frame (ORF) located within an intron or intein. These enzymes are highly specific for target sites located on homologous alleles that lack the endonuclease ORF and intervening sequence. The homing endonuclease protein introduces a double-stranded break at this target site that then stimulates the homologous recombination DNA-repair pathway. Repair via recombination leads to duplication of the selfish mobile element containing the HE ORF (1, 2). Harnessing the natural potential of these enzymes through the reprogramming of their substrate specificity (3–5) will help drive forward the rapidly expanding areas of genome engineering and gene therapy (6).
Recent work on adapting homing endonucleases for use in such biotechnology applications has focused on the LAGLIDADG family, whose genes are primarily found within archaea, algal chloroplasts, and the mitochondria of fungi (7). These proteins are so named for the conserved sequence of amino acids in the α-helices that separate the N- and C-terminal halves of the pseudo-symmetric monomeric LAGLIDADG homing endonucleases (LHEs) and form the binding interface of homodimeric LHEs. These conserved helices include essential catalytic acidic residues that catalyze the cleavage of the target DNA between two scissile phosphates, separated by four base pairs referred to as the central four (2, 8). Crystal structures of both homodimeric and monomeric members of this protein family reveal a well conserved, canonical fold as well as a similar curvature of their ∼20–22-base pair long DNA substrate (9–11). Specific interactions with this target DNA are made by residues in two β-sheets that flank the central four bases and traverse the major groove of both the plus and minus DNA half-sites (1, 2). Homing endonuclease genes (HEGs) are selfish genetic elements, and their continued existence relies on their ability to sustain homing in the face of genetic drift and to invade new, albeit related, host organisms. The extended β-sheet topology for protein-DNA interactions provides this flexibility by allowing for low fidelity at some positions in the interface, most often at the wobble positions in the host gene sequence, whereas maintaining an overall high level of specificity due to the length of the DNA substrate (12, 13).
The pair of structurally characterized HE homologues, I-CreI and I-MsoI, provides a prime example of the extensive flexibility of this protein scaffold. These two enzymes cleave nearly identical target sites, yet they share only 38% sequence identity with only 5 of the 25 DNA contacting residues conserved (9). Such divergent evolution can facilitate the acquisition of alternative biochemical functionalities, often with a benefit to the host organism. For example, the endonuclease I-AniI can act as a maturase, assisting in the splicing of the intron containing its own coding HEG (14). As the selective pressure following HEG invasion is not high, genetic drift can also result in the loss of endonuclease function; the I-AniI homologue BI3 maturase acts exclusively as a maturase, having lost all endonuclease activity due to mutation of one of its catalytic glutamates to a lysine. Although the BI3 maturase no longer functions as an endonuclease, and only shares ∼50% sequence identity with I-AniI, it was observed to still bind the I-AniI target site DNA with high affinity (15).
In this era of genome-wide sequencing new LAGLIDADG ORFs are being identified rapidly (16, 17), including a large number of homologues of enzymes that are already being engineered for gene targeting applications (such as I-AniI). For most protein families, determining the native substrate and specificity of newly identified proteins is usually quite labor intensive. However, target site identification for homing endonucleases can be achieved with careful analysis of the sequence flanking the mobile element containing the HEG. As a result, we can compare amino acid substitutions and putative target sites between various homologues. Analysis of endonuclease sequence alignments revealed a number of amino acid differences between enzymes that were predicted to cleave similar target sites, leading us to question what effect these mutations have on endonuclease activity and specificity. The work described here addresses this question for one LAGLIDADG endonuclease, I-AniI.
I-AniI is one of the most thoroughly characterized homing endonucleases, and its particular biochemical properties make it a promising candidate for gene therapy applications. Importantly, degradation of the activity of the enzyme from genetic drift has been artificially reversed using directed evolution methods, resulting in an enzyme with high activity in human cell lines, an essential feature for a gene therapy reagent (18). Additionally, the specificity of the enzyme, and the effect of base pair substitutions on kinetic activity, has been acquired for every position of the target site, and variants with novel target site specificities have been generated by computational redesign of the protein-DNA interface (19, 20). In this study we dissect the differences in specificity between close homologues of I-AniI. Enzyme hybrids based on each homologue were made by transferring specific amino acids to the well characterized I-AniI scaffold, and the local specificity shifts resulting from these mutations were analyzed with in vitro DNA cleavage assays.
EXPERIMENTAL PROCEDURES
Identification of Homologues and Target Sites
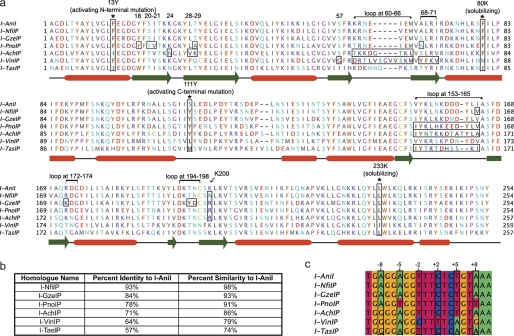
I-AniI homologues were identified by searches against the NCBI nonredundant data base with BLAST, blastp, and tblastn, using the I-AniI ORF as the query sequence (22). Target sites were identified by examination of the nucleotide sequence on either side of the intron containing the homologue ORF. The high similarity of the putative target sites to the I-AniI target supported these predictions. Multiple sequence alignments of both the homologues and their predicted target sites were constructed using Jalview (Fig. 1) (23). These putative endonucleases are denoted by the suffix P in accordance with nomenclature guidelines (24).
FIGURE 1.
I-AniI homologues and predicted cleavage sites. a, Multiple sequence alignment of ORFs encoding the putative (indicated by suffix P) LAGLIDADG homing endonuclease homologues of I-AniI that were experimentally explored in this study. Groups of amino acids that were transferred to the I-AniI scaffold are boxed with a black border and labeled above, as are the positions of activating and solubilizing mutations (both additionally denoted by an asterisk). Secondary structure elements are identified from the crystal structure definitions of Protein Data Bank code 2QOJ, with α-helices indicated by red boxes and β-sheets by green arrows. See supplemental Table S2 for a complete list of the mutations made for every hybrid protein. b, c, percent identity and similarity of each homologue to I-AniI are indicated and putative target sites identified for each homologue by examining the area surrounding the homologue ORF and comparing to the I-AniI target site. Positions in endonuclease target sites are identified in relationship to the predicted center of the site, with positions on the left, or (−) half-site, designated as −10 to −1 and positions on the right, or (+) half-site, designated as +1 to +10. Wobble positions in the cytochrome B gene are boxed in black and labeled.
Generation of Hybrid I-AniI Homologue Endonucleases
The amino acid sequence of each homologue of I-AniI was modeled onto the crystal structure of the wild-type I-AniI protein (Protein Data Bank code 2QOJ, Ref. 13). The I-AniI protein scaffold used for generating hybrid endonucleases contained point mutations that have been shown to increase either solubility or cleavage activity at physiological temperatures (13, 18). All C-terminal pocket transfers were made in the context of the activating mutation F13Y, whereas all N-terminal transfers, with the exception of the K24N/T29K enzyme pair and two variants derived from the I-VinIP homologue, were made in the context of S111Y (“Base Activity” column, Table 1). These two activating mutations were identified (18) during the course of this study and the choice of which activating mutations to include for the each hybrid protein was a function of experimental timing, the maintenance of consistency for C- and N-terminal transfers, and considerations regarding positions of the activating mutation in relationship to the transferred pocket. In all cases, the hybrids were compared with I-AniI with the corresponding activating mutation for consistency. See supplemental Table S2 for the complete list of mutations made for each hybrid.
TABLE 1.
Summary of altered specificities and activities
Variants are grouped into categories (C-terminal loops, K200, central 4 loops, core mutations) dependent on the location and theorized role of the mutations transferred to the I-AniI scaffold. The base activity column indicates whether the variant was made with either of the activating F13Y or S111Y mutations. Quantitative activities, cleavage plots, and additional information on each variant are available under supplemental Table S2 and Figs. S4–S8.
| Variant | Mutations from I-AniI | Base activity | Tested positions | Effect on specificity and activity |
|---|---|---|---|---|
| C-terminal loops | ||||
| I-PnoIP | V153I, N157H, D159E, D160E, I164V | F13Y | +8, +9, +10 | Specificity pattern similar to I-AniI; activity increased for most substitutions |
| I-AchIP | V153I, K155N, L156T, N157K, D160T, inserted A after 160, D161T, I164A | F13Y | +7, +8, +9, +10 | Novel +7A specificity; loss of specificity at +8, +9, and +10 |
| I-TasIP | K155T, L156R, N157T, K158 deletion, D160H, D161S, Y162S, L163K | F13Y | +7, +8, +9, +10 | Shifts favoring +7A, +8G, and +9C |
| K200 | ||||
| I-NfiIP | I164V, K200N | F13Y | +3, +4 | Novel +3C specificity; similar specificity to I-AniI at other substitutions |
| I-PnoIP | K200R | F13Y | +3, +4, +5 | Increased cleavage of +3G and +4T |
| Central 4 loops | ||||
| I-GzeIP | R172K, T196Y, N197D | F13Y | +2, +3 | Specificity pattern similar to I-AniI; activity increased for most substitutions |
| I-AchIP | K60T, R61K, N62K, E63D, inserted G after 63, I64T, E65K, M66L | S111Y | −2 | Purines preferred over pyrimidines |
| I-VinIP loop | S57G, K60D, N62T, E63L, I64V, SGVVS insert after 64, E65K, A68Y, L69F, R70K, I71V | S111Y | −2 | −2C favored over WT −2T; matches −2C target site prediction |
| Core mutations | ||||
| I-VinIP without core | A68Y, R70K | WT | −6, −5 | Minimal activity observed on −6 and −5 targets for variant with interface mutations only; activity recovered with addition of core mutations L69F and I71V |
| I-VinIP with core | A68Y, L69F, R70K, I71V | WT | ||
| I-VinIP-S111Y without core | A68Y, R70K | S111Y | −6, −5 | Novel −6T specificity; enhanced cleavage activity due to core mutations |
| I-VinIP-S111Y with core | A68Y, L69F, R70K, I71V | S111Y | ||
| I-VinIP Loop | S57G, K60D, N62T, E63L, I64V, SGVVS insert after 64, E65K, A68Y, L69F, R70K, I71V | S111Y | −6, −5 | Activity increased further relative to I-VinIP-S111Y w/core |
| K24N/L28V/T29K | K24N, L28V, T29K | WT | −8 | L28V core mutation enhances activity; computationally predicted K24N and T29K highly specific for −8G; matches −8G target site prediction |
| K24N/T29K | K24N, T29K, lacking L233K | WT | ||
| I-PnoIP N-terminal transfera | Y18F, S20T, I21V, T29A, A68S | S111Y | −6, −5 | −5T substitution preferred over WT −5A; matches −5T target site prediction |
a Quantitative data and cleavage plots for this variant, included as an additional example of transferred mutations conferring specificity towards the putative homologue target site, are given under supplemental data.
Expression and Purification of Proteins
Genes for each homologue-based variant of I-AniI were assembled, cloned, sequence verified, and transformed into BL21 Star cells (Invitrogen). A half-liter or 1 liter culture of autoinduction media (25) was then inoculated and grown at 37 °C for 8–12 h until approximate saturation, after which expression at 18 °C was allowed for 20–24 h. Cells were then harvested and resuspended in 20 mm Tris, pH 7.5, 30 mm imidazole, and 1.0 m NaCl prior to lysis via a freeze-thaw cycle, sonication, and the addition of lysozyme.
Proteins were isolated from the soluble fraction with nickel affinity chromatography. The purified proteins were concentrated, buffer exchanged in 20 mm Tris, pH 7.5, and 500 mm NaCl, and stored in 50% (v/v) glycerol. Purity of about >95% for all samples was confirmed by SDS-PAGE and the mass and purity were additionally verified by mass spectrometry. Protein concentration for each sample was determined by measuring absorbance at 280 nm.
DNA Cleavage Assays
Plasmid DNA substrates, containing single base pair substitutions from the I-AniI wild-type target site, were constructed by site-directed mutagenesis according to methods described elsewhere (19, 26) and linearized with the restriction endonuclease ScaI. For optimized enzyme activity and stability (19), the reaction buffer contained final concentrations of 170 mm KCl, 10 mm MgCl2, and 20 mm Tris, pH 9.0. For every I-AniI variant assayed, 8 serial 2-fold dilutions of enzyme were performed in 1.25 × reaction buffer, ranging from 5 to 1500 nm depending on the experiment, and each dilution was incubated with 100 ng (about 5 nm) of linearized substrate for 30 min at 37 °C. Wild-type I-AniI, containing the same activating mutations as each hybrid, was tested in parallel under the same reaction conditions. Reactions were quenched with about 17 nm EDTA, followed by 60 °C incubation for 5–10 min. The resulting cleavage products were separated by gel electrophoresis on 1.2% agarose TBE gel and were visualized by staining with ethidium bromide.
The data were analyzed as previously described (19) by quantifying the spectral density of substrate and product bands using ImageJ. The percent cleavage was calculated by dividing the sum of the two product band densities by the sum of the densities of all three bands and was plotted versus enzyme concentration in GraphPad Prism. At least two independent determinations of each enzyme cleavage profile were performed and inspected for substrate degradation and experimental error prior to being reported here.
To estimate the concentrations (nm) of enzyme corresponding to half-maximal cleavage of the target site (EC0.5max), data were fit to a sigmoid function as follows,
f([endonuclease]) is the fraction of DNA site cleavage corresponding to endonuclease concentration in nanomolar as denoted by [endonuclease]. fmax is the maximal fraction of site cleavage, with 1 being its greatest allowable value indicating complete cleavage of the substrate with no remaining uncut fraction. Although the value of the Hill coefficient (H) is 1 for a simple hyperbolic formula, setting H to 1.5 allowed a sigmoid function that consistently achieved a better fit to the data. EC0.5max could then be determined by solving the equation.
RESULTS
Identification of I-AniI Homologues and Their Putative Target Sites
BLAST searches against the NCBI nonredundant data base using the I-AniI sequence identified a group of homologues. The ORFs encoding homologues with greater than 47% identity to I-AniI (supplemental Fig. S1) were identified from a variety of fungal species, at insertion sites within an intron of the same host gene as I-AniI (cytochrome B). Because residence in that particular intron suggests that at one point these enzymes were able to cleave the same or a closely related intronless allele, it is likely that they have activity on a target site similar to that of I-AniI. We found that in alignments with less than 40% identity it was significantly more challenging to identify the flanking target sites, as these enzymes were inserted in a different location in the mitochondrial genome and the exact target site boundaries were unclear. We therefore focused our analysis on homologues that were unambiguously determined to reside in the same intron as the I-AniI ORF. A multiple sequence alignment of all such I-AniI homologues available at the time of publication is shown under supplemental Fig. S1 and the subset of these enzymes that were analyzed in this study are shown in Fig. 1a.
Examination of the DNA flanking this intron in homologues reveals nucleotide sequences that closely match that of the I-AniI substrate (Fig. 1c and supplemental Fig. S2, showing the relationship between the intronic sequence, target site, and endonuclease ORF). However, aligning these putative sites with that of the wild-type reveals some variation, mainly at the wobble positions in the codons of the cytochrome B protein. There are a number of positions in these homologues at which the amino acid identity differs from that of I-AniI. Some of these residues have the potential to directly interact with the substituted bases in these putative target sites, whereas others are non-conservative and do not neighbor any predicted base changes. To understand the effect of these mutations on the specificity and activity of I-AniI, we constructed hybrid endonucleases by transferring amino acids observed in the homologue alignments to I-AniI.
Generation of Hybrid I-AniI Homologue Endonucleases
The homologues selected for testing (Fig. 1b) have sequence identities ranging from 57 to 93% relative to I-AniI (Fig. 1a and supplemental Table S1) and are predicted to cut very similar sites (Fig. 1c). Thus, a large proportion of their protein mutations are likely to be neutral and the result of genetic drift rather than arising from selective pressure to act on different substrates. We hypothesized that any specificity differences between members of the I-AniI subfamily might correspond to changes in individual DNA binding residues that have arisen during their evolutionary divergence, and that these changes might illustrate new strategies to alter the recognition specificity of LHEs. To test this concept, we chose to transfer subsets of amino acids from the homologue sequences to I-AniI and to experimentally test the activity and specificity of the resulting hybrids.
The expression of four full-length homologues was attempted to compare their specificity and activity to that of the hybrid endonucleases. Of these four homologues, only I-NfiIP was soluble and its specificity profile is available under supplemental Fig. S3. Specificity profiles for the other three endonucleases (I-AchIP, I-VinIP, and I-TasIP) could not be obtained due to poor expression, with all expressed proteins observed to be insoluble by SDS-PAGE. This challenge of reliably characterizing many diverse homologues can potentially be alleviated by hybrid generation methods, where mutations predicted to affect properties of interest are transferred onto a stable, well studied protein scaffold.
To identify possible mutations for transfer from homologues to I-AniI, the amino acid sequence of each homologue was threaded onto the I-AniI crystal structure, and mutations were grouped into four categories, the protein surface distant from the bound DNA, direct interface contacts between DNA and protein, the protein core, and the peptide linker between protein domains, depending on their location. The most interesting of these groups, from the perspective of specificity and activity-altering potential, are those that are located in the protein-DNA interface and those in the enzyme core that are immediate neighbors to interface changes. We selected a subset of these substitutions (Fig. 1 and Table 1) for transfer to the I-AniI scaffold. The groups of amino acids chosen were either adjacent to a change in the putative target site of the homologue or were nonconservative substitutions directly in the protein-DNA interface that we hypothesized could alter the interaction of the enzyme with the DNA. The following sections describe the activities and specificities of the hybrid endonucleases that were characterized, with the results summarized in Table 1.
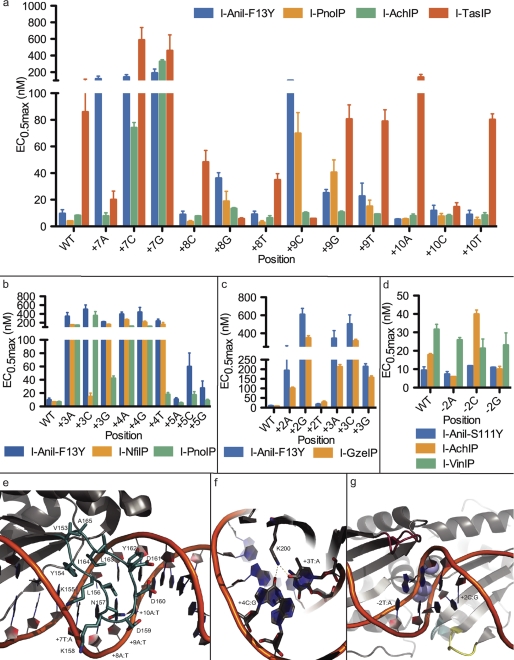
Loop Transfers at the Edge of the DNA Target Site
A surface-exposed, C-terminal loop near one end of the I-AniI protein scaffold contacts the final 4 base pairs (corresponding to positions +7 to +10, in the (+) half of the target site (Fig. 2e)), and extends, approximately, from residue 153 to 165. I-AniI displays relatively low specificity in this region compared with the rest of the target site, showing little preference for one nucleotide over another, especially at positions +8 to +10 (Fig. 2a). This reduced specificity is presumably due both to the heightened flexibility of the loop (which displays relatively few packing interactions against the underlying core of the protein) and also to the lower number of direct protein-DNA interactions. However, subtle effects on activity are still observed for different substitutions at these positions (19). Furthermore, it has been previously shown that specificity can be significantly increased at the +8 position with structure-based computational redesign of the enzyme, indicating that it is possible to generate enzymes with alternative and higher specificities in this area.
FIGURE 2.
Transfer of loops and interface residues from homologues results in new specificities and activities. Error bars in all panels are mean ± S.E. See supplemental Table S2 for a complete list of the mutations made for every variant protein. a, cleavage efficiencies were quantified by EC0.5max (nm) at the +7 to +10 positions contacted by the C-terminal distal loop. Loops transferred from the I-AchIP and I-TasIP homologues are shown to significantly alter the specificity profile of I-AniI-F13Y, most notably introducing new activity for +7A. b, cleavage efficiencies at +3, +4, and +5 for the Lys-200 variants were tested. In comparison to I-AniI-F13Y, transferring the K200R found in I-PnoIP markedly improves +3G and +4T cleavage, whereas K200N from I-NfiIP improves +3C cleavage. c, results from homologue I-GzeIP-derived transfers of the two C-terminal loops near the central four nucleotides show increased activity at positions +2 and +3. d, N-terminal loop transfers from I-AchIP and I-VinIP demonstrate altered specificities at the −2 position. The I-VinIP loop was tested in the context of additional mutations from the I-VinIP homologue that were incorporated for a related experiment to analyze the effects of core mutations on position −6 specificity and activity (see Fig. 3a, panel 5). The reduced activity on all target site substitutions at the −2 position is presumably due to a lower activity on the wild-type site arising from these other amino acid changes. e, the C-terminal distal loop between Val-153 and Ala-165 as seen in the solved I-AniI crystal structure (Protein Data Bank code 2QOJ, Ref. 13) is colored in blue. f, residue Lys-200 forms direct hydrogen bonds with wild-type target site bases +3T and +4C. g, three loops in I-AniI that contact the central four target site positions are shown. The loop in the N terminus roughly spans residues 60 to 66 (red) and contacts the −2 position. Two C-terminal loops potentially affect positions +2 and +3: one from 172 to 174 (teal), a second from 194 to 198 (yellow). Significant variation in both the sequence and length of these loops is seen among I-AniI homologues.
This loop is one of the more variable regions of the protein-DNA interface among the selected homologues. Three different loops were transferred to the I-AniI scaffold and tested for their effect on the ability of the enzyme to discriminate between target sites with single base pair substitutions in the +7 to +10 range (Fig. 2a and supplemental Fig. S4). These loops, derived from the I-PnoIP, I-AchIP, and I-TasIP endonucleases, have varying levels of similarity to the I-AniI loop and all three are different lengths, as is highlighted in the sequence alignment of this region in Fig. 1a. The hybrid proteins were made in the context of the F13Y mutation in the I-AniI scaffold (a sequence change that increased catalytic activity, discovered during directed evolution experiments on that protein (18)). The relative activities of the enzyme hybrids using various substrates were therefore compared with I-AniI-F13Y activity on the same substrates.
I-PnoIP has the highest sequence identity to I-AniI of the three enzymes, both overall and in the transferred C-terminal loop. This loop is the same length as the analogous I-AniI loop, but has changes in 5 of the 12 amino acids. V153I is a conservative substitution that points into the core of the protein, D159E and D160E do not appear to interact with the substrate, and N157H and I164V are located within contact distance to the bound DNA and thus are most likely to alter the specificity of the interface (Fig. 2e). We tested activity of the I-PnoIP hybrid (in which the sequence of the I-AniI loop was changed to that of I-PnoIP) on the singly substituted target sites from +8 to +10 and found to have a very similar pattern of specificity to I-AniI-F13Y, but shows an overall slight increase in activity on the majority of tested substrates (Fig. 2a). I-AniI-F13Y cleaves the +8T substitution identified in the putative I-PnoIP target site (Fig. 1c) and the few changes made to the loop of this hybrid enzyme did not improve specificity for the thymine nucleotide.
The loop transferred from I-AchIP is 1 amino acid longer than the I-AniI loop and only 4 of the 13 amino acids are conserved between the two regions. This hybrid showed activity equivalent to that of I-AniI-F13Y (Fig. 2a), however, its specificity profile is significantly altered. Positions +8 to +10 no longer display even the low levels of specificity observed for I-AniI and position +7 now allows an adenine nucleotide in addition to the wild-type thymine. This new specificity observed at position +7 was not previously accomplished using the I-AniI protein during either computational redesign or directed evolution experiments. Therefore, the information derived from the identification and exploitation of homologous endonucleases can be used to create enzyme variants with additional functional capabilities.
The loop derived from the I-TasIP homologue has only 1 residue in common (Tyr-154) with that of I-AniI and is 1 residue shorter. The hybrid protein shows remarkably high specificity that differs significantly from that of the wild-type I-AniI. Like the I-AchIP loop hybrid, this I-TasIP transfer also displays activity against the substrate with an adenine at position +7 that is comparable with the wild-type enzyme with the wild-type +7T substrate (Fig. 2a). However, unlike the I-AchIP loop hybrid, it displays significantly reduced activity on the wild-type thymine at +7, as well as on the other two +7 singly substituted sites. In addition, with the inclusion of the I-TasIP amino acid changes to the loop, the +8G and +9C substitutions shift from being the least favored nucleotides (with the wild-type loop) to the most favored. Both the I-TasIP and the untested I-VinIP homologues contain an L156R mutation in their C-terminal loop. This mutation is likely one source for the preference of a guanine at the +8 position, found in the putative target site of both homologues. Due to the difficulty of modeling protein-DNA interactions in a flexible loop, identification of such loop changes that confer novel specificities is key to designing endonucleases.
Sequence Changes in the Middle of the Protein-DNA Half-site
Surface-exposed loops are generally considered to be more flexible than secondary structure elements, such as β-sheets. The high degree of variation between homologues in the C-terminal loop described in the previous section, as well as the low level of substrate specificity observed under I-AniI loops (19, 20), attest to this flexibility and tolerance to mutation in both partners of the protein-DNA interface. In contrast, the identities of protein residues located in the relatively inflexible β-sheet region of a LAGLIDADG interface are predicted to be more influential on target site activity and specificity.
Lysine 200 of I-AniI is located in the center of the β-sheet in the C-terminal domain of I-AniI. This lysine forms direct hydrogen bonds with positions +3 and +4 of the target site (Fig. 2f) and likely contributes to the high specificity of I-AniI at both positions. As indicated in Fig. 1a, four of the eight homologues studied have a mutation to arginine at position 200, and one, I-NfiIP, has a mutation to asparagine. I-NfiIP has only one other mutation in the C-terminal interface, and the effect of a K200N mutation in I-AniI was tested in the context of this I164V mutation. This I164V mutation was also present in the I-PnoIP C-terminal loop, where it was found to have little effect on activity in the context of the other loop mutations (Fig. 2a).
I-AniI hybrids with either mutation to lysine 200 maintain some activity on the wild-type substrate, albeit to different degrees (Fig. 2b). The arginine substitution, seen in I-PnoIP and three other homologues, yields slightly more activity on all tested target sites than the corresponding I-AniI-F13Y enzyme, however, it also results in an overall reduced specificity; in particular the cleavage of the +3G and +4T targets is significantly enhanced (Fig. 2b and supplemental Fig. S5). This observed relaxation of specificity might result from the increased length of the arginine side chain. The K200N substitution derived from I-NfiIP increased the activity on the +3C site by more than 30-fold, to levels close to that of the wild-type enzyme on the wild-type +3T site (Fig. 2b). For the remaining sites tested, the I-NfiIP K200N variant had a similar specificity pattern to I-AniI-F13Y. The comparatively low levels of cleavage activity on the purine bases at position +3, independent of the identity of the residue tested at position 200, may reflect the requirements of sequence-dependent DNA bending on catalysis.
Loop Transfers at the Center of the DNA Target Site
The central four bases of many LAGLIDADG endonuclease target sites are distorted away from B-form DNA, due to a greater degree of bending at these nucleotide positions. This region of the DNA is not in direct contact with the enzyme, yet it displays significant sequence preferences that are presumably the result of indirect readout of DNA conformational requirements. Although certain DNA sequences within this region of the target may be completely disallowed because of their inability to deform to the necessary conformation, the protein sequence surrounding this region should have some influence on the specificity of the base substitutions that are conformationally accessible. Identifying areas in the protein that can influence the specificity of the central 4 base pairs is valuable not only to increase the pool of potential starting targets for further engineering, but also because increasing our understanding of how to control specificity in this region is important for more general redesign of homing endonuclease cleavage specificities.
Loops on both domains of the endonuclease contact the central four positions: one in the N terminus that extends from approximately position 60 to 66 in I-AniI, one in the C terminus from positions 172 to 174 that interacts with the phosphate backbone of the central four, and a second C-terminal loop that spans approximately positions 194 to 198 (Fig. 2g). There is significant variation in the sequence, as well as the length, of these three loops between homologous enzymes (Fig. 1a). Three loop transfer variants were produced: one incorporating sequence changes from both of the C-terminal loops together, made in the F13Y background, and one from each of the two homologues with N-terminal loop changes, both made in the S111Y background. The effects of the C-terminal transfer were tested on positions +2 and +3, whereas the N-terminal transfers were analyzed on −2.
I-GzeIP was chosen for transfer of C-terminal loop mutations because, whereas its putative target site is identical to I-AniI, the amino acid changes in this homologue are nonconservative and structurally proximal to the central 4 base pairs. Amino acid mutations made to the two C-terminal loops, R172K, T196Y, and N197D, resulted in an enzyme with slightly increased activity on the wild-type substrate, with the wild-type cytosine at position +2 still preferred over other substitutions (Fig. 2c and supplemental Fig. S6). This hybrid enzyme showed a modest reduction in activity for a thymine substitution at this position, the next most-favored nucleotide for the wild-type enzyme. Both purine substitutions remain unfavorable for this mutant, however, the I-GzeIP hybrid shows higher activity against these sites compared to the wild-type enzyme. The I-GzeIP enzyme was also tested on the +3 singly substituted targets, and although it maintains the same order of substrate reactivity, it has an overall increased activity on all three alternative substrates.
The I-AniI-S111Y wild-type enzyme has low specificity at position −2, and homologue N-terminal loops neighboring this position were transferred to test for sequence effects on specificity in this region. Both N-terminal transferred loops are longer than the wild-type I-AniI loop; the I-AchIP derived loop is extended by 1 residue and the I-VinIP loop by 5. The I-AchIP loop displays a preference for purines over pyrimidines at the −2 position, cleaving the wild-type thymine with slightly reduced activity than the wild-type enzyme and cleaving the cytosine substitution significantly more poorly than any other nucleotide (Fig. 2d and supplemental Fig. S6). Activity on the guanine and adenine counterparts was maintained across the hybrid and wild-type enzymes. Of all analyzed homologues, the I-VinIP ORF is the only one with substitutions in the central four positions of the predicted target site (Fig. 1c), with a switch of thymine to cytosine at position −2. The hybrid protein incorporating the I-VinIP loop was found to favor a cytosine nucleotide at position −2, thus corroborating the target site prediction.
Mutations to Residues in the Protein Core
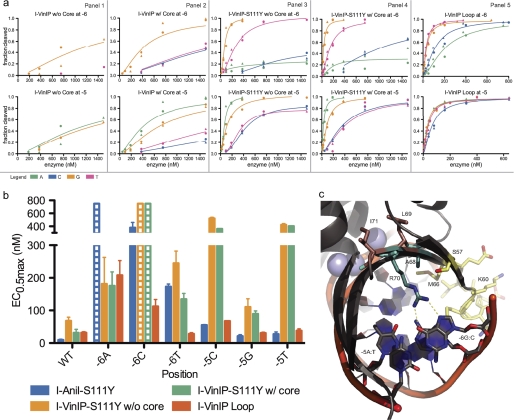
Although interface mutations are most likely responsible for specificity and activity shifts, core mutations proximal to these could also contribute to these properties. In our final experiments, we tested whether core mutations could have a substantial effect on enzyme activity and lead to shifts in specificity that are not achieved with interface mutations alone (Fig. 3).
FIGURE 3.
Transfer of homologue-derived core substitutions results in increased activity. See supplemental Table S2 for a complete list of the mutations made for every variant protein. a, cleavage profiles for each of the five variants derived from the homologue I-VinIP are given in panels 1–5. The upper plot of each panel displays the activity of the variant on the singly substituted −6 target site position, whereas the lower plots show analogous data for position −5. Curves are colored by the substituted base: adenine (green), cytosine (blue), guanine (yellow), or thymine (pink). b, the associated EC0.5max values of the five I-VinIP variants represent further quantitative assessment of the data in a. Bars with dashed lines indicate substitutions where some cleavage was observed, but EC0.5max was too high (>750 nm) to allow accurate quantitative determination. The incorporation of core mutations directly adjacent to the interface mutations surrounding the −6 and −5 positions demonstrated a striking increase in activity. Extending the loop in this region further increased activity and established a preference for −6T over the wild-type −6C. c, the relevant substitutions and target site positions are colored by mutation type in this view of the I-AniI crystal structure. The interface substitutions (A68Y and R70K) are shown in cyan, core substitutions (L69F and I71V) are shown in brown, and the positions where additional mutations were incorporated in the I-VinIP Loop variant (a, panel 5) are shown in yellow.
Two I-AniI protein-DNA interface mutations, A68Y and R70K, were transferred from I-VinIP. Arg-70 interacts with a guanine nucleotide at position −6 (Fig. 3c). The activity of this enzyme on variants at positions −6 and −5 is increased by the addition of two core mutations, L69F and I71V. Although the inclusion of these core mutations in hybrids both with and without the S111Y activating substitution led to activity enhancement, the effect was more dramatic with the latter variant (Fig. 3a). The hybrid containing both interface and core changes has relaxed specificity at position −6 compared with I-AniI-S111Y, now cleaving the previously inaccessible −6T in addition to the wild-type −6G. An additional hybrid was made by transferring a loop (spanning residues 60–66 in I-AniI) and nearby mutation S57G from the I-VinIP homologue to the protein containing both the interface and core changes, as well as the activating S111Y. This hybrid showed an increase in activity for all substitutions at the −5 and −6 positions and a further increase in activity against the −6T (Fig. 3, a, panel 5, and b).
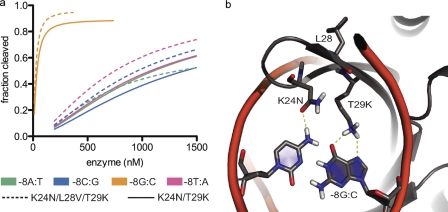
A second example of core influences on enzyme activity involves transfers from I-AchIP. This homologue contains two interface mutations, K24N and T29K, which were previously predicted computationally by Rosetta (19) and confirmed experimentally to be highly specific for a guanine at position −8 (Fig. 4a). Consistent with these results, the predicted homologue target site contains the −8G substitution (Fig. 1c). However, in addition to these two mutations, I-AchIP also contains the L28V core substitution. Incorporation of this very conservative mutation resulted in increased activity on 3 of the 4 possible nucleotides at position −8 (computationally modeled contacts are visualized in Fig. 4b).
FIGURE 4.
Previously designed variant identified in homologue and enhanced by core substitution. See supplemental Table S2 for a complete list of the mutations made for every variant protein. a, cleavage profiles for the I-AchIP-based variants K24N/T29K and K24N/L28V/T29K on position −8 confirm previously predicted computational mutations for −8G (19). Introducing the L28V core substitution increased activity slightly at three of the four possible bases at this position. b, the computationally predicted model for these amino acid mutations (19). Asn-24 forms a hydrogen bond with −8C, whereas Lys-29 is able to form two hydrogen bonds with −8G.
DISCUSSION
Generation of Novel Specificities for Endonuclease Engineering
The use of homing endonucleases for applications such as gene therapy and genome engineering is limited by our ability to target DNA sequences of interest. The endonucleases identified at present recognize only a limited set of DNA targets and thus new approaches are required to access novel DNA target sequences (17). Although computational design has proven successful in generating variants for targets with as many as 3 base pair changes (27, 28), these methods are still limited to small numbers of substitutions and typically require subsequent, time consuming, experimental selection (29) to optimize enzyme activity. Information garnered from endonuclease homologues can be used for further design with a 2-fold benefit: by gathering a set of previously uncharacterized nucleases with altered binding and cleaving properties to be used as starting scaffolds, and by identifying pockets of mutations that can be grafted to activate the enzyme toward local DNA substitutions.
Grafting of mutations from homologues onto I-AniI resulted in a number of expected and novel specificities (Table 1). The majority of the substitutions identified in the predicted homologue target sites are cleaved, to some extent, by the wild-type enzyme. However, it was hypothesized that amino acid mutations from homologues that neighbor the substituted bases would improve enzymatic activity against these sites. Indeed, several hybrid enzymes were found to have improved activity toward the nucleotide substitutions in their putative target site over the wild-type nucleotide (Fig. 1c and Table 1): I-PnoIP N-terminal transfer prefers −5T over −5A, I-VinIP central 4 loop prefers −2C over −2T, and the I-AchIP derived K24N and T29K prefers −8G over −8A.
Conversely, a number of hybrid proteins yielded unexpected specificities given the sequence of their predicted target site. As described under “Results,” a single lysine to asparagine mutation at position 200 opens up specificity at position +3 to now allow a cytosine, previously the least favored +3 nucleotide (Fig. 2b). Transfer of both the I-AchIP and I-TasIP C-terminal distal loops resulted in activity on the +7A target site that has never before been achieved with either the wild-type I-AniI or any of its designed variants (Fig. 2a), and transfer of the I-VinIP interface and core residues neighboring the central four positions allowed for cleavage of the −6T substitution (Fig. 3). These specificity shifts were the most pronounced of all shifts observed for the hybrid enzymes characterized in this study, with all three enzymes tolerating nucleotide substitutions that are not cleaved by wild-type I-AniI. However, all three of these new enzymes have activity against the wild-type I-AniI target indicating that the target site assessments are likely accurate despite the unforeseen specificity changes. For I-AniI, and likely all other endonucleases, neutral drift (30, 31), the accumulation of non-deleterious mutations with adaptive potential, has resulted in the acquisition of new substrate specificities.
Identifying mutations that result in novel specificities, such as the cases discussed above, can directly aid in sequence-specific targeting and modification of disease-causing genes. Supplemental Table S3 displays five cleavage site sequences in close proximity to chromosomal loci of interest that contain +3C, +7A, and −6T relative to the native I-AniI target site, which were substitutions that could not be cleaved prior to this study. These genes have also recently demonstrated promise as potential therapeutic targets in animal models and perhaps even clinical trials; additional information is given under supplemental data.
The Role of Core Mutations in Endonuclease Activity
The contribution of surface residues to the activity and specificity of homing endonucleases is generally straightforward, whereas the role of core mutations on the same has been less well understood. Mutations to the hydrophobic core of I-AniI can yield dramatic effects on activity (15). We find that transfer of a patch of both core and interface mutations from the I-AniI homologue I-VinIP results in greater activity against the −6 position compared with the interface mutations alone (Fig. 3). The addition of these core mutations also cause shifts to the specificity at the neighboring DNA bases. In contrast, addition of a single core mutation to the I-AchIP interface mutations K24N and T29K has only a modest effect on activity and almost none on specificity (Fig. 4). Further understanding of how core residue changes alter homing endonuclease properties will be important for engineering designs against currently inaccessible target sites.
A New Source of Data for Computational Modeling and Design
Although gathering information from homing endonuclease homologues can aid in identifying mutations that allow us to access novel targets, more extreme methods may be required to produce enzymes that cleave sequences that differ further from that of the parent endonuclease. The homologues characterized in this paper have evolved to cleave very similar DNA substrates to that of I-AniI, as is apparent when we compare their putative insertion sites (Fig. 1c). Transferring pockets of mutations from more divergent homologues is complicated by the challenges of target site identification and identification of residues critical to the switch in specificity. Accessing the vast information in these homologues will require either laborious experimentation or modeling of the homologue in complex with its putative target site.
Computational modeling has yielded accurate predictions for specificity-causing changes, such as the K24N and T29K substitutions that were originally determined by computational prediction to cleave the −8G position (19) and are further validated by their presence in homologues targeting sites with this substitution. By incorporating information from homologue alignments into standard design protocols it should be possible to engineer enzymes toward more divergent sites. In particular, the modeling of loops (32), how core changes result in protein backbone shifts (33), and sequence preferences of DNA bending (21) are computationally challenging. Improving our understanding of how the amino acid variation alters the specificity and activity of enzymes in relationship to these challenges can provide insight into ways to improve computational design methodologies.
Conclusions
Cross-species sequencing provides a useful repository of information on how protein sequence relates to function. For closely related homing endonucleases, it is possible to determine the target DNA of these homologues and to identify amino acid mutations that likely influence target site specificity. We show here that transferring these variable residues to a scaffold protein can be used to determine their effect on both specificity and activity, and that such methods can facilitate the engineering of variant endonucleases with novel target site specificities. This study focuses on aspects of endonuclease structure that are particularly difficult to model, such as flexible loops, the indirect readout of the central four DNA bases, and how core residues influence a protein backbone. Our novel sequence mining approach will likely aid future engineering efforts and provide data that can improve tools for computational prediction, with the benefits not only limited to endonucleases and protein-DNA interactions, but broadly applicable to many enzyme-substrate redesign challenges.
Supplementary Material
Acknowledgments
We thank Barry Stoddard for critical reading of the manuscript, Kyle Jacoby, Andrew Scharenberg, and Jordan Jarjour for providing helpful discussion, and S. Arshiya Quadri for assistance with plasmid substrate preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants GM084433 and RL1CA133832, the Foundation for the National Institutes of Health through the Gates Foundation Grand Challenges in Global Health Initiative, and the Howard Hughes Medical Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S8.
- HE
- homing endonuclease
- LHE
- LAGLIDADG homing endonuclease
- HEG
- homing endonuclease gene.
REFERENCES
- 1. Chevalier B. S., Stoddard B. L. (2001) Nucleic Acids Res. 29, 3757–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stoddard B. L. (2005) Q. Rev. Biophys. 38, 49–95 [DOI] [PubMed] [Google Scholar]
- 3. Ulge U. Y., Baker D. A., Monnat R. J., Jr. (2011) Nucleic Acids Res. 39, 4330–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Redondo P., Prieto J., Muñoz I. G., Alibés A., Stricher F., Serrano L., Cabaniols J. P., Daboussi F., Arnould S., Perez C., Duchateau P., Pâques F., Blanco F. J., Montoya G. (2008) Nature 456, 107–111 [DOI] [PubMed] [Google Scholar]
- 5. Chevalier B. S., Kortemme T., Chadsey M. S., Baker D., Monnat R. J., Stoddard B. L. (2002) Mol. Cell 10, 895–905 [DOI] [PubMed] [Google Scholar]
- 6. Pâques F., Duchateau P. (2007) Curr. Gene Ther. 7, 49–66 [DOI] [PubMed] [Google Scholar]
- 7. Stoddard B. L. (2011) Structure 19, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chevalier B., Monnat R. J., Jr., Stoddard B. L. (2005) in Homing Endonucleases and Inteins (Belfort M., Derbyshire V., Wood D., Stoddard B. L. eds) pp. 33–47, Springer Verlag, Berlin/Heidelberg [Google Scholar]
- 9. Chevalier B., Turmel M., Lemieux C., Monnat R. J., Jr., Stoddard B. L. (2003) J. Mol. Biol. 329, 253–269 [DOI] [PubMed] [Google Scholar]
- 10. Moure C. M., Gimble F. S., Quiocho F. A. (2003) J. Mol. Biol. 334, 685–695 [DOI] [PubMed] [Google Scholar]
- 11. Bolduc J. M., Spiegel P. C., Chatterjee P., Brady K. L., Downing M. E., Caprara M. G., Waring R. B., Stoddard B. L. (2003) Genes Dev. 17, 2875–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gimble F. S. (2001) Nucleic Acids Res. 29, 4215–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scalley-Kim M., McConnell-Smith A., Stoddard B. L. (2007) J. Mol. Biol. 372, 1305–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ho Y., Kim S. J., Waring R. B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8994–8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Longo A., Leonard C. W., Bassi G. S., Berndt D., Krahn J. M., Hall T. M., Weeks K. M. (2005) Nat. Struct. Mol. Biol. 12, 779–787 [DOI] [PubMed] [Google Scholar]
- 16. Grishin A., Fonfara I., Alexeevski A., Spirin S., Zanegina O., Karyagina A., Alexeyevsky D., Wende W. (2010) J. Bioinform. Comput. Biol. 8, 453–469 [DOI] [PubMed] [Google Scholar]
- 17. Barzel A., Privman E., Peeri M., Naor A., Shachar E., Burstein D., Lazary R., Gopha U., Pupko T., Kupiec M. (2011) Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takeuchi R., Certo M., Caprara M. G., Scharenberg A. M., Stoddard B. L. (2009) Nucleic Acids Res. 37, 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thyme S. B., Jarjour J., Takeuchi R., Havranek J. J., Ashworth J., Scharenberg A. M., Stoddard B. L., Baker D. (2009) Nature 461, 1300–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jarjour J., West-Foyle H., Certo M. T., Hubert C. G., Doyle L., Getz M. M., Stoddard B. L., Scharenberg A. M. (2009) Nucleic Acids Res. 37, 6871–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becker N. B., Wolff L., Everaers R. (2006) Nucleic Acids Res. 34, 5638–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 23. Clamp M., Cuff J., Searle S. M., Barton G. J. (2004) Bioinformatics 20, 426–427 [DOI] [PubMed] [Google Scholar]
- 24. Roberts R. J., Belfort M., Bestor T., Bhagwat A. S., Bickle T. A., Bitinaite J., Blumenthal R. M., Degtyarev S. Kh., Dryden D. T., Dybvig K., Firman K., Gromova E. S., Gumport R. I., Halford S. E., Hattman S., Heitman J., Hornby D. P., Janulaitis A., Jeltsch A., Josephsen J., Kiss A., Klaenhammer T. R., Kobayashi I., Kong H., Krüger D. H., Lacks S., Marinus M. G., Miyahara M., Morgan R. D., Murray N. E., Nagaraja V., Piekarowicz A., Pingoud A., Raleigh E., Rao D. N., Reich N., Repin V. E., Selker E. U., Shaw P. C., Stein D. C., Stoddard B. L., Szybalski W., Trautner T. A., Van Etten J. L., Vitor J. M., Wilson G. G., Xu S. Y. (2003) Nucleic Acids Res. 31, 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Studier F. W. (2005) Protein Exp. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 26. Kunkel T. A., Roberts J. D., Zakour R. A. (1987) Methods Enzymol. 154, 367–382 [DOI] [PubMed] [Google Scholar]
- 27. Ashworth J., Havranek J. J., Duarte C. M., Sussman D., Monnat R. J., Jr., Stoddard B. L., Baker D. (2006) Nature 441, 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ashworth J., Taylor G. K., Havranek J. J., Quadri S. A., Stoddard B. L., Baker D. (2010) Nucleic Acids Res. 38, 5601–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doyon J. B., Pattanayak V., Meyer C. B., Liu D. R. (2006) J. Am. Chem. Soc. 128, 2477–2484 [DOI] [PubMed] [Google Scholar]
- 30. Amitai G., Gupta R. D., Tawfik D. S. (2007) HFSP J. 1, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bloom J. D., Romero P. A., Lu Z., Arnold F. H. (2007) Biol. Direct 2, 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy P. M., Bolduc J. M., Gallaher J. L., Stoddard B. L., Baker D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9215–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kellogg E. H., Leaver-Fay A., Baker D. (2011) Proteins 79, 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.