Abstract
The three components of the mammalian nuclear SET domain containing protein (NSD) family have been implicated in multiple diseases and cancers, but very little is known about their mechanisms of action. NSD proteins are epigenetic regulators and methylate lysine side chains, particularly lysine 36 of histone H3 (H3K36), where they appear to deposit mono and/or dimethyl marks. This modification (H3K36Me) has been shown to be important in various processes including gene expression, alternative splicing and DNA repair. Here, we examine recent findings regarding the oncogenic role of NSD proteins and suggest that a de-regulated switch between H3K36Me and H3K27Me plays an important role in the oncogenic potential of NSD proteins.
Key words: NSD1, histone, methyltransferase, H3K36, H3K27
Introduction
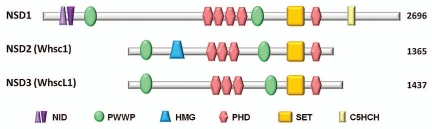
Using a two-hybrid screen with the retinoic acid receptor α as bait, Chambon and colleagues identified NSD1,1 a 2,676 residue protein in humans that possesses several important motifs known to function in chromatin biology (Fig. 1). NSD1 is an essential protein that binds nuclear receptors such as the retinoic acid receptor, estrogen receptor and the androgen receptor.1–3 Two other members of the NSD family have also been identified,4,5 but each of these lacks the nuclear receptor binding domains found in NSD1. Additionally, NSD2 (also known as Whsc1 or MMSET) has been shown to bind to the DNA binding domain of the androgen receptor through a high-motility group (HMG) domain.6 Each NSD family member can be spliced into various isoforms and the resulting transcripts encode proteins possessing various motifs known to function in chromatin regulation. This includes PHD fingers, PWWP motifs, and a catalytic lysine methyltransferase SET domain (Fig. 1). Each of the three NSD proteins appears non-redundant with each other as shown through mouse knockouts.2,7 For example, NSD1 defective mice exhibit embryonic lethality, but mice defective in NSD2 die shortly after birth from cardiac anomalies and phenotypes that resemble Wolf Hirschhorn syndrome.2,7 Surprisingly, to our knowledge, no knockout mouse phenotype has been reported for NSD3 (also known as WhscL1).
Figure 1.
Structural relationship between members of the NSD family. Major domains are highlighted: NID, nuclear receptor binding domains, ligand independent (light purple) or dependent (dark purple); PWWP, chromatin associated domains; HMG, high mobility group; PHD, plant homeo domain; SET, set domain with pre- and post-motifs; C5HCH, chromatin associated zinc fingers. Numbers represent the number of amino acids in each NSD protein. Not drawn to scale.
The mechanisms of action of NSD proteins are largely unknown, but likely proceed through regulating the levels of epigenetic methylation on histone tails. Although several discrepancies exist with respect to the catalytic specificity of NSD family members,7–9 the preponderance of data, particularly strong biochemical and structural studies from Reinberg and colleagues,9,10 indicate that these SET domains methylate nucleosomal histone H3 at lysine 36 (H3K36Me). A recent study has also shown that NSD1 methylates non-histone substrates, in this case the p65 subunit of NFκB.11 Histone lysines exist in up to four different forms: non-methylated and the mono-, di- and trimethylated forms (Me0, Me1, Me2, Me3, respectively), each of which elicits a biological signal.12,13 NSD family members deposit mono and dimethyl groups on nucleosomes at H3K36,9,10 and this is a substrate for Setd2/HYPB, the enzyme responsible for global trimethylation at H3K36 in mammals.14 The biological significance of H3K36Me in mammals and how it impacts chromatin function is still largely unknown, although it is most commonly associated with active or “open” chromatin. In various organisms, H3K36Me has been linked to development, transcription, alternative splicing and dosage compensation.12,15,16 Recent evidence has suggested that H3K36Me is mutually exclusive with H3K27Me at the level of the histone tail,17 generating a biological scenario where H3K36Me precludes methylation at H3K27, a repressive modification. This is consistent with previous reports in worms that showed antagonism between H3K27Me and H3K36Me.18
In contrast to higher eukaryotes, H3K36Me is non-essential in budding yeast, and a single enzyme (Set2) is responsible for all methylation at this site.12 In yeast, H3K36Me correlates with transcriptional activation and appears to suppress spurious, intragenic initiation in the wake of RNA Polymerase II (RNAP II) activity by enforcing a hypoacetylated chromatin structure through recruiting the Rpd3S complex.19,20 Therefore, although not proven, NSD family members will likely exert their oncogenic influence through H3K36Me1, 2 and this is relevant to initiation and/or elongation during gene expression. In support of this, an NSD1 transcriptional network was recently established in human HCT116 colorectal cancer cells. Here, NSD1 localizes to 5′ promoter elements where it regulates the local levels of H3K36Me.21 This is important for the recruitment of RNAP II to promoters and the coordination of H3K36Me with gene expression. This work suggested that NSD1 participates in initiation events by enforcing H3K36Me1, two levels that likely serve as a substrate for trimethylation.
Oncogenic Role of NSD Family in Cancer
Accumulating evidence has revealed that various tumors from many tissue types have altered epigenetic profiles that contribute to the cancerous state through altering gene expression required for normal cellular homeostasis.22,23 Enzymes that mediate methylation at various histone sites such as H3K36 (NSD proteins)9 and H3K27 (EZH2)22 are among those epigenetic regulators that are often de-regulated in cancer. For example, one important modification that has been linked to prostate carcinogenesis is trimethylation of lysine 27 of histone H3 (H3K27Me3),22 an epigenetic mark characteristic of transcriptional repression. One enzyme responsible for this modification, EZH2, is overexpressed in prostate tumors relative to benign tissue.
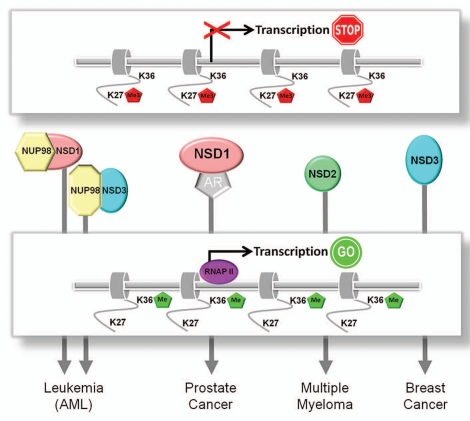
NSD proteins are disrupted and/or de-regulated in various diseases, including multiple types of cancer.4,8,24,25 NSD1 plays a causal role in acute myeloid leukemia (AML), a disease characterized by uncontrolled cellular proliferation of myeloid progenitor cells often accompanied by overexpression of Hox regulators.26 In this case, a chromosomal translocation generates an oncogenic fusion protein consisting of NUP98 and the C-terminal half of NSD1. NUP98-NSD3 fusions have also been shown to be involved in AML27 (Fig. 2). Kamps and colleagues have shown that a NUP98-NSD1 fusion protein causes marrow-derived progenitor cells to proliferate indefinitely as undifferentiated progenitor cells.25 In a mouse adoptive transfer experiment, NUP98-NSD1 was shown to induce AML in vivo and this was accomplished through the enforcement of an H3K36 methylation profile at targets such as MEIS1 and some HoxA genes.25 The NUP98-NSD1 fusion protein enforced an H3K36Me profile that appeared dominant to the H3K27Me-dependent repression that normally keeps Hox genes silent in these cells. How the oncogenic NSD1 fusion drives H3K36Me and the formation of AML (at the expense of H3K27Me) is unknown, but recent work has suggested that H3K36Me might be mutually exclusive with repressive H3K27Me at the level of the histone tail.5,17 Other histone modifications, including acetylation, may also be affected by the NSD1 fusion protein.25 Nevertheless, aberrant H3K36Me leads to progenitor cell immortalization through de-regulation of Hox gene expression and the development of AML in a mouse model through a reduction in H3K27-mediated repression. Thus, repressed (“shut”) chromatin is activated (“open”) via aberrant NSD1 function, leading to unscheduled expression of Hox genes.
Figure 2.
Role for NSD proteins as oncogenes in various cancer types. Overexpressed and/or deregulated NSD proteins (i.e., translocations) cause a switch from repressive H3K27 methylation to active H3K36 methylation. This leads to aberrant targeting of genes, causing unscheduled gene expression and subsequent tumorigenesis. AR, androgen receptor; RNAPII, RNA Polymerase II; AML, acute myeloid leukemia.
NSD1 is also involved in other cancers, i.e. prostate, where it has been found to be overexpressed in metastatic samples and correlates with prostate cancer progression.28 Such gene expression profiling identified NSD1 as a candidate gene in an epigenetic signature capable of discriminating between malignant and nonmalignant prostate tissue.28 How does the role of NSD1 as an oncogene in AML compare to its oncogenic role in other cancers such as prostate? We suggest that in prostate cancers that overexpress NSD1, the overexpressed protein acts as an oncogene by turning on gene sets normally silenced by H3K27Me (Fig. 2). How this is related to the previously described role of NSD1 as an androgen receptor co-regulator3 is unknown, but NSD1 could use H3K36Me to influence androgen receptor recruitment to its target genes. As a corollary to this, those prostate cancers that overexpress EZH2 might be expected to have genes that are normally active with H3K36Me be turned off through overexpression of H3K27Me.
The oncogenic nature of H3K36Me and its impact on H3K27Me has also been seen for NSD2.5 In an analogous manner to NSD1-driven AML, a significant number of multiple myelomas overexpress NSD2 via translocation between the immunoglobulin (Ig) locus on chromosome 14 and the NSD2 locus on chromosome 4. By placing NSD2 under the regulatory control of the Ig locus, the overexpressed protein behaves as an oncogene. As seen for NSD1, oncogenic NSD2 alters gene expression due to increased ratios of H3K36Me/H3K27Me5,24 (Fig. 2). How overexpressed NSD2 finds its targets in the oncogenic setting is unknown, but 60% of the cellular nucleosomes possess the H3K36Me3 mark after overexpression in multiple myeloma cells. These examples with NSD1 and NSD2 establish a strong and significant link between H3K36Me and oncogenesis and suggest a paradigm for the role for NSD family members as oncogenes.
NSD3 overexpression appears to be an important driver in some breast cancers.4,29 Amplification of 8p11-12 is associated with about 15% of breast cancers.4,29 As NSD3 is located in this region, it might be expected that overexpression of NSD3 would be a major driver in 8p11-12 breast cancers. Indeed, NSD3 has been recently shown to act as a potent transforming oncogene and knockdown of NSD3 via lentiviral-mediated shRNAs results in cell death.29 In this scenario, the role of NSD3 as an oncogene in breast cancer would be comparable to the NSD2-driven AML and NSD2-driven multiple myeloma. We advocate a model where overexpression of NSD3 through gene amplification of 8p11-12 activates gene sets through changing the balance between H3K36Me and H3K27Me (Fig. 2). Thus, genes that are normally turned off would be activated in response to overexpressed, oncogenic NSD3, ultimately leading to cellular proliferation and the transformed phenotype. Such genes are expected to be different than those regulated by NSD1 despite the fact that both enzymes catalyze similar reactions. This is because of the non-redundancy between NSD family members, cell type-dependent targets, and the possible distinct roles for NSD proteins in initiation and/or elongation.7,21,30
Very little is known about the global spectrum of direct NSD3 targets or even its mechanistic action within them. One recent paper identified NSD3 as a component of an LSD2 demethylase complex that was found associated with elongation factors within the body of target genes in HeLa cells.30 This is interesting because both NSD1 and NSD2 appear to reside at the 5′ end of genes.7,21 A second paper used lentiviral-mediated shRNAs targeted against NSD3 followed by microarrays to identify its targets.29 Although informative, this study failed to discern direct vs. indirect targets and, importantly, did not investigate the role of histone modifications (i.e., H3K36Me vs. H3K27Me) in NSD3-mediated activities.
Although clearly acting as an oncogene in various contexts, one report has also shown that NSD1 acts as tumor suppressor in some neuroblastomas and gliomas.8 In this case, NSD1 expression is reduced through transcriptional silencing caused by CpG promoter hypermethylation. The authors noted that this leads to diminished methylation at H3K36 and H4K20, although global levels of these modifications, as well as the status of H3K27Me, was not described. Interestingly, NSD1-depleted cells show elevated levels of the MEIS1 oncogene, a Hox regulator that has previously been described as an NSD1 target in a separate study.25 Thus, NSD1 appears to be capable of activating and repressing various target genes, an observation consistent with early studies with NSD1 that described the protein as both a co-activator and a co-repressor in reporter assay-based experiments.1 In the context of NSD1 playing a role in a switch between H3K36Me and H3K27Me, perhaps the lack of NSD1 function in tumor suppression favors a balance in repressive H3K27Me and shuts off critical gene targets.
Conclusions
Although NSD proteins have been well described to function in oncogenesis, little is known about their targets in transcriptional activation or repression although they influence H3K36Me and probably other modifications as well. Clearly, it will be important to define the global spectrum of NSD targets during development and in various tumor types. Moreover, the generation of mouse models with tissue specific overexpression of NSD1 (i.e., prostate), or in combination with its fusion partners should reveal important mechanistic clues regarding the relationship between NSD function and oncogenesis. Studies such as this should identify therapeutic targets that when combined with the future development of much-anticipated NSD family inhibitors will pose a potent threat to NSD-mediated diseases.
Acknowledgments
We apologize to our colleagues whose work could not be cited due to space limitations. Research in the Carpenter lab has been supported by the NIH and the Welch Foundation (AU-1569).
References
- 1.Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, et al. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, et al. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003;22:3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Yeh S, Wu G, Hsu CL, Wang L, Chiang T, et al. Identification and characterization of a novel androgen receptor coregulator ARA267-alpha in prostate cancer cells. J Biol Chem. 2001;276:40417–40423. doi: 10.1074/jbc.M104765200. [DOI] [PubMed] [Google Scholar]
- 4.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2010;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang HB, Choi Y, Lee JM, Choi KC, Kim HC, Yoo JY, et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009;583:1880–1886. doi: 10.1016/j.febslet.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 7.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 8.Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA. 2009;106:21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiao Q, Li Y, Chen Z, Wang M, Reinberg D, Xu RM. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem. 2010;286:8361–8368. doi: 10.1074/jbc.M110.204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, et al. Regulation of NFkappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, et al. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, et al. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 21.Lucio-Eterovic AK, Singh MM, Gardner JE, Veerappan CS, Rice JC, Carpenter PB. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc Natl Acad Sci USA. 2010;107:16952–16957. doi: 10.1073/pnas.1002653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4:4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 24.Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–4069. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 27.Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99:3857–3860. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- 28.Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, Thomas M, et al. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 2010;19:2611–2622. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- 29.Yang ZQ, Liu G, Bollig-Fischer A, Giroux CN, Ethier SP. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res. 2010;70:8487–8497. doi: 10.1158/0008-5472.CAN-10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]