Abstract
Sirtuin 1 (SIRT1), the mammalian ortholog of yeast Sir2, is a highly conserved NAD+-dependent protein deacetylase that has emerged as a key metabolic sensor that directly links environmental nutrient signals to animal metabolic homeostasis. SIRT1 is known to be involved in gluconeogenesis in the liver, fat mobilization in white adipose tissue, and insulin secretion in the pancreas. Recent studies have shown SIRT1 to regulate fatty acid oxidation in the liver, sense nutrient availability in the hypothalamus, influence obesity-induced inflammation in macrophages, and modulate the activity of circadian clock in metabolic tissues. The activity of SIRT1 also appears to be under control of AMPK and adiponectin. This review focuses on the involvement of SIRT1 in regulating metabolic diseases associated with obesity. It includes brief overviews of sirtuin signaling, with emphasis on SIRT1s role in the liver, macrophage, brain, and adipose tissue as it relates to obesity.
Introduction
Western societies are experiencing an alarming increase in the rate of metabolic syndrome, which consists of a collection of abnormalities including visceral obesity, type 2 diabetes, dyslipidemia, fatty liver, and a pro-inflammatory and prothrombotic state (1). Currently, approximately 27% of adults and 17% of children and adolescents in the United states have body mass index readings above 30 Kg/m2, which is considered the threshold of obesity (2). Moreover, one in four adults in the United States suffers from diseases associated with metabolic syndrome and worldwide estimates are over 2.1 billion (2). Ultimately, this epidemic threatens to overburden our healthcare system with weight-related diseases and reduce both human lifespan and quality of life.
The metabolic abnormalities caused by abdominal obesity can be further aggravated by additional factors including advancing age. Typically, aging is accompanied by gradual weight gain, and changes in the levels and distribution of fat (3). In the past, humans have shown increases in visceral fat of roughly 1 lb per year as they progress through adulthood (3). However, modern developments in agriculture and technologies have promoted the intake of high-calorie diets and sedentary lifestyles, causing accelerated weight gain in both children and adults. In obese individuals, excess adipose tissue releases increased levels of free fatty acids (NEFA). These excess fatty acids overload other metabolic tissues such as liver, muscle, and pancreatic β-cells with lipid, resulting in atherogenic dyslipidemia, insulin resistance, and hyperinsulinaemia (4). While the physiological implications of obesity remain an area of intense research, recent evidence has shown that the fat-storage depot white adipose tissue (WAT), has other functions besides fat storage (5). WAT is an endocrine organ, which produces hormones such as resistin, adiponectin, and leptin that are active throughout the entire body (6, 7). WAT can also become a source of inflammation through secretion of adipokines, and accumulation of cytokine producing adipose tissue macrophages (ATMs) (8, 9). In obese people, increased production of inflammatory cytokines and resistin seem to have a role in pro-inflammatory state and insulin resistance. Excess obesity also results in low adiponectin release from WAT which has been implicated in insulin resistance and fatty liver (4).
Recent studies in mice have demonstrated that SIRT1, a NAD+-dependant protein deacetylase, is involved in regulating fat cell accumulation and maturation, hepatic lipid metabolism, systemic inflammatory status, central nutrient sensing, as well as circadian regulation of metabolism. Understanding the role SIRT1 plays in visceral fat as well as tissues involved in fat metabolism will likely provide insights into developing treatments for obesity-induced metabolic disease.
1. SIRT1 links cell energy status to gene regulation
The sirtuin family of proteins is poised at the crossroads between nutritional status and longevity. Sirtuins are highly conserved NAD+-dependant protein deacetylases and/or ADP-ribosyl transferases that target histones, transcription factors, co-regulators, as well as metabolic enzymes to adapt gene expression and metabolic activity in response to the cellular energy state (10-12). Many members of this family, including the founder Sir2, have been shown to impact aging in species ranging from yeast to fly and it is believed these protective actions result from the beneficial regulation of stress management and energy homeostasis (13-15). SIRT1, like its yeast ortholog Sir2, is a NAD+-dependant protein deacetylase (Figure 1). SIRT1 is increasingly referred to as a master metabolic regulator due to its ability to modify and control numerous transcription factors involved in the whole body metabolic homeostasis (Figure 2).
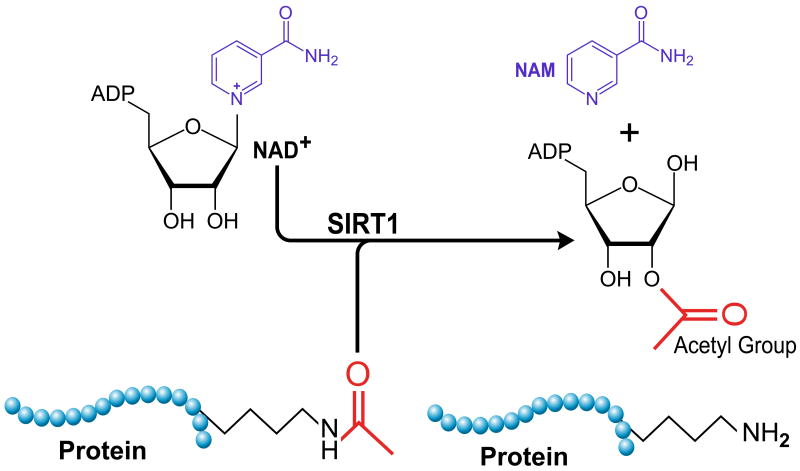
Figure 1.
SIRT1 uses NAD+ as a co-substrate to cleave acetyl groups from target proteins. Nicotinamide is a competitive inhibitor in the reverse reaction. Cellular energy as reflected in changes in NAD+ levels are thought to influence SIRT1 activation.
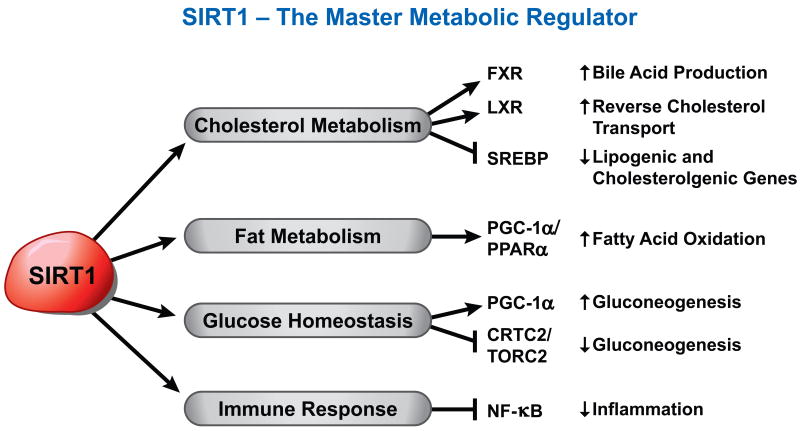
Figure 2.
SIRT1 targets both histone and non-histone proteins. SIRT1 is often referred to as a ‘Master Metabolic Regulator’ because of its ability to influence several transcriptions factors involved in energy homeostasis.
As a member of class III family of histone deacetylase (HDACs), SIRT1 deacetylates histone proteins in addition to transcription factors and cofactors. As SIRT1 lacks a DNA-binding domain, it is recruited to target promoters by sequence-specific transcription factors to induce chromatin remodeling and subsequently regulation of gene expression (16). SIRT1 is also known to associate with heterochromatin regions and promote deacetylation of histone tails, recruitment and deacetylation of histone H1, and spreading of hypomethylated H3-K79 (17). These epigenetic modifications mediated by SIRT1 generally result in a silencing of gene transcription and are thought to play an integral role in organismal health and ultimately lifespan (18). Given its multiple functions, such as deacetylation, epigenetic modifications, and transcription factor modulation, SIRT1 have been proposed to provide a molecular link between cellular metabolic status, and adaptive transcriptional responses (3).
Not surprisingly, the activity of SIRT1 is tightly controlled in response to different environmental cues. One feeding regime that is thought to heighten sirtuin activation is Caloric Restriction (CR), which is a 20–40% reduction in calories consumed below ad libitum intake without malnutrition. CR has been shown to extend the median and maximum life span of numerous organisms including yeast, flies, worms, fish, and rodents and mammals (19). In mammals, CR can also ameliorate many of the pathologies associated with obesity and metabolic syndrome, such as reduction of body fat, lowering serum triglycerides and LDL cholesterol, increase of HDL cholesterol, and improvement of insulin sensitivity (20, 21). Sirtuins have been implicated as important mediators of CR-mediated lifespan extension. For example, in a number of lower model organisms, CR extends lifespan through activation of sirtuins (14, 22). In mice, SIRT1 regulates energy metabolism and physical responses to CR (23-25). SIRT1 has also been shown to participate in autophagy (26-29), which is a major mechanism by which a starving cell reallocates nutrients from unnecessary processes to more-essential processes (30, 31)
Whether sirtuins mediate the life-extending effects of CR in mammals is currently an area of intense research. A recent study reported that behavior associated with caloric restriction did not occur when SIRT1 knockout mice were put on a calorie restricted diet, implying that SIRT1 is necessary for mediating the effects of caloric restriction (23). Although biochemical parameters thought to mediate the lifespan extending effects of calorie restriction (reduced insulin, IGF1 and fasting glucose) were essentially the same in control mice as those lacking SIRT1, CR did not extend lifespan of full-body SIRT1 knockout mice (24). Moreover, transgenic mice overexpressing SIRT1 are leaner than controls, more metabolically active, and have reduced serum levels of cholesterol, adipokines, insulin, and glucose (32-34). Furthermore, activation of SIRT1 by the polyphenol resveratrol and several synthetic pharmacologic activators has been shown to protect against high-fat induced obesity and metabolic derangements (35-37).
Consistent with above observations, SIRT1 protein levels have been shown to be elevated during CR in the brain, WAT, muscles, liver, and kidney (38, 39). However, cellular NAD+ levels or the NAD+/NADH ratio, which have been claimed as the primary mechanisms regulating SIRT1 activity, fluctuate depending on tissue type during CR (40-42), suggesting that the activity of SIRT1 changes in different directions in different tissues during CR. Additional stimuli also modulate SIRT1 activity by altering cellular NAD+ levels or the NAD+/NADH ratio. For example, in skeletal muscle C2C12 cells, SIRT1 activity is enhanced by AMP-activated protein kinase (AMPK) and increasing cellular NAD+ levels (43, 44). SIRT1 activity in C2C12 cells also appears to be under control of adiponectin through Ca2+ signaling and changes of the NAD+/NADH ratio (45). What drives cellular NAD+/NADH levels and SIRT1 activity during different physiological conditions, and in different tissues remains unclear. For instance, it has been reported that cellular NAD+ levels increase in the liver upon starvation/fasting, but decrease upon refeeding (46). However, a more recent report by Escande et al. analyzed the levels of both SIRT1 protein and NAD+ concentration in the liver following a standard diet, starvation, and HFD feeding (47). Their data show that neither NAD+ levels nor SIRT1 protein levels fluctuate in the liver regardless of feeding patterns, which suggests that factors other than NAD+ changes, such as post-translational modifications or protein-protein interactions, regulate SIRT1 activity (47).
The notion that SIRT1 is a key mediator in longevity is not exempt of controversial data. For example, while overexpression of SIRT1 in mice improves healthy aging, these changes are not sufficient to affect their lifespan (48). In addition, the mechanisms of resveratrol's apparent protective effect on metabolic disorders and life span are not fully understood. Questions still remain whether or not resveratrol directly activates SIRT1 or functions through multiple signaling pathways (49, 50). Nevertheless, investigation of sirtuins as potential mediators of CR has revealed that these proteins participate in surprisingly diverse aspects of mammalian biology including cell survival, cell senescence, DNA repair, rDNA transcription, and numerous metabolic pathways (17). Given the many functions attributed to sirtuins, it is clear that their ability to regulate metabolism is essential to mammalian physiology.
2. SIRT1 signaling in the liver
The liver is a central metabolic organ that controls key aspects of lipid and glucose metabolism in response to nutritional and hormonal signals (51). In fasted conditions, the liver converts lipid and glycogen stores into available energy through the process of fatty acid β-oxidation and glycogenolysis/gluconeogenesis. In the fed condition, metabolic programs in the liver are switched on in order to store energy in the form of glycogen and lipid droplets. Recent reports have shown that SIRT1 is an important regulator of hepatic metabolism. For example, SIRT1 inhibits TORC2, a key mediator of early phase glucogenogenesis, during short-term fasting (52). Prolonged fasting leads to increased SIRT1 deacetylation and activation of PGC-1α as well as the lipid sensing nuclear receptor peroxisome proliferator-activated receptor (PPARα), resulting in increase in fatty acid oxidation and improved glucose homeostasis (46, 53, 54). SIRT1 also regulates hepatic cholesterol and bile acid homeostasis through direct modulation of the liver X receptor (LXR) and farnesoid X receptor (FXR), key nuclear receptors that function as cholesterol and bile sensors (Figure 3) (55-57). Additionally, SIRT1 acts downstream of AMPK signaling, promoting fatty acid catabolism (58).
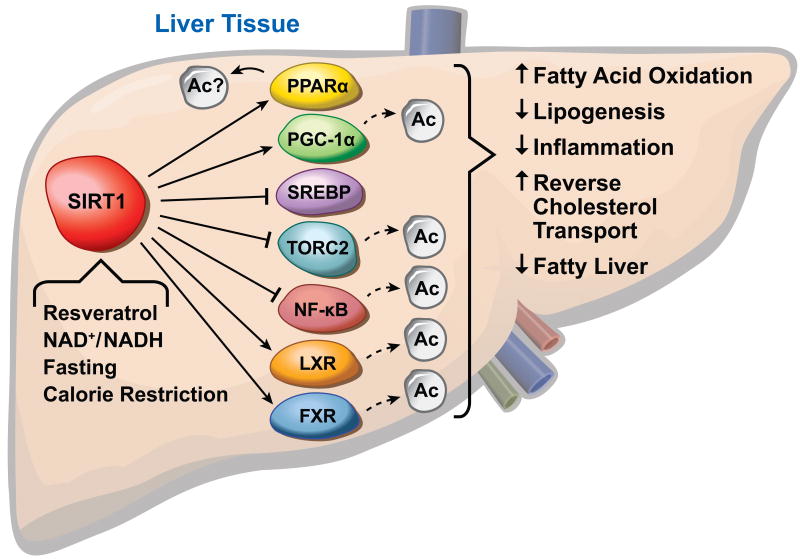
Figure 3.
SIRT1 is involved in many functions in the liver. It controls key aspects of lipid and glucose metabolism through interaction with transcription factors. SIRT1 is activated in response to fasting, calorie restriction, changes in NAD+/NADH levels and by the polyphenol resveratrol.
The biological consequence of SIRT1's ability to control hepatic lipid metabolism is evidenced in several liver-specific transgenic mouse studies. Using an adenovirus expressing short hairpin RNA to knock down SIRT1, Rogers et al. showed that adenoviral knockdown of SIRT1 reduces expression of fatty acid β-oxidation genes in the liver of fasted mice (59). Using liver-specific knockout mice (SIRT1 LKO), our group showed that deletion of SIRT1 in the liver impairs PPARα signaling and decreases fatty acid β-oxidation, whereas over-expression of SIRT1 induces expression of PPARα target genes (53). As a result, SIRT1 LKO mice are susceptible to high-fat diet induced dyslipidemia and hepatic steatosis (53). Our data further suggest that SIRT1 likely modulates PPARα activity through PGC-1α, a key coactivator for PPARα signaling and a direct target of SIRT1 (46, 60). SIRT1 activates PGC-1α primarily though its ability to deacetylate this coactivator.
In addition to fatty acid metabolism, SIRT1 has also been shown to regulate cholesterol metabolism through deacetylation of LXRs (55). Activation of LXR is beneficial in that it not only inhibits intestinal cholesterol uptake and promotes reverse cholesterol transport but also exerts potent anti-inflammatory effects that involve transrepression (61). SIRT1 can directly deacetylate LXRs, resulting in increased LXR turnover and target gene expression (55). Disruptions in LXR signaling are observed in whole-body SIRT1 deficient mice, which have lower levels of HDL cholesterol, while low-density lipoprotein (LDL) cholesterol is unaffected (55). Since cholesterol efflux is reduced in macrophages and hepatocytes from these mice, it is likely that SIRT1 positively controls LXR activity to stimulate reverse cholesterol transport in vivo. An independent study from Rodgers et al. has shown that another LXR target gene CYP7A1 is reduced in the absence of hepatic SIRT1. This evidence suggests that bile acid synthesis, in addition to cholesterol efflux, is enhanced by SIRT1 (59). Interestingly, the regulation of CYP7A1 was also found to be dependent on the coregulator PGC-1α.
FXR is also a target of hepatic SIRT1 in metabolic regulation (56). FXR is an important regulator of cholesterol and bile acid homeostasis that regulates expression of numerous bile acid-responsive genes in the liver and intestine (57). Acetylation of FXR inhibits its activity and is dynamically regulated by p300 and SIRT1 under normal conditions (56). Downregulation of hepatic SIRT1 increases FXR acetylation, causing deleterious metabolic outcomes. Additionally, FXR acetylation levels were shown to be elevated in two mouse models of metabolic disorders, ob/ob mice and mice chronically fed a western-style diet. Treatment of mice with resveratrol or overexpression of SIRT1 substantially reduced FXR acetylation levels in these disease models (56). These findings provide an intriguing correlation between elevated FXR acetylation and decreased SIRT1 activity, and provide further evidence of benefits for therapeutic targeting of SIRT1.
Another means by which SIRT1 may regulate hepatic lipid metabolism is through deacetylation of the sterol regulatory element binding protein (SREBP) family of transcription factors. SREBPs are critical regulators of lipid metabolism, which promote expression of lipogenic and cholesterolgenic genes involved in lipid storage (62). Two recent reports revealed that SIRT1 can directly deacetylate SREBP, and that SIRT1 activity is important in the fasting-dependent attenuation of SREBP (63, 64). In addition, chemical activators of SIRT1 inhibit SREBP target gene expression in vitro and in vivo, correlating with decreased hepatic lipid and cholesterol levels and attenuated liver steatosis in diet-induced and genetically obese mice. In summary, these findings imply that hepatic SIRT1 plays a critical role in metabolic regulation and activation of SIRT1 in the liver may prove beneficial in treating obesity-associates diseases.
Again, studies on the function of SIRT1 in the liver have yielded controversial and even contradictory observations. For example, we have observed that SIRT1 LKO mice gain weight and develop hepatic steatosis when fed a high-fat high-cholesterol diet (53). However, using a similar hepatic-specific knockout mouse model, Chen et al. (42) reported a reduction in weight gain and liver fat accumulation in LKO mice when fed a western-style diet. One possible factor contributing to the discrepancy between our observations and those of Chen et al. may be the difference in age of animals at which the feeding was initiated and data were collected. The varied responses of SIRT1 LKO mice to a western-style diet at different ages raises the possibility that hepatic SIRT1 may selectively regulate alternative metabolic pathways at multiple stages of development. An inducible SIRT1 knockout model will be helpful to dissect age-dependent effects of SIRT1.
In addition to hepatic lipid metabolism, SIRT1 has been implicated as an important regulator of hepatic gluconeogenesis (52, 59). In fact, one of concerns regarding applying SIRT1 activators in lipid metabolism diseases is that they may worsen obesity-associated diabetes due to increased hepatic gluconeogenesis. However, we have failed to observe significant changes of gluconeogenesis in our SIRT1 LKO mice in response to a 16-h fasting (53). Gluconeogenesis is regulated by a complex interplay between transcription factor and hormonal and co-regulator signaling. Given the fact that SIRT1 deacetylates and represses two key transcriptional factors that are involved in the early and late fasting phases, FOXO1 (65) and TORC2 (52), while deacetylates and activates another key gluconeogenesis co-activator, PGC-1α (46), it is not surprising that the net effect of loss of SIRT1 on the gluconeogenesis is minimized by the complex compensation patterns of these factors.
3. SIRT1 and inflammatory disease
Macrophage activation and infiltration into resident tissues is known to mediate local inflammation and is a hallmark feature of metabolic syndrome (66-68). This infiltration activates local inflammation which has been increasingly recognized as a causal factor leading to the development of the cluster of diseases surrounding metabolic syndrome (69). Macrophages are phagocytc cells that engulf cellular debris and pathogens as part of the innate immune response. They quiescently monitor the bloodstream and tissue for signs of infection or damage. Upon stimulation, macrophages infiltrate resident tissue, perpetuating local inflammation and contribute to the development of insulin resistance and metabolic derangements (70).
Over the past few years, sirtuins have been identified as important regulators of the immune system, with several studies showing that SIRT1 can repress inflammation in multiple tissues including the macrophage (71-74). Some reports have demonstrated that artificial overexpression of SIRT1 leads to suppression of the inflammatory response, whereas deletion of SIRT1 in hepatocytes results in increased local inflammation (34, 53). It appears that in immune signaling SIRT1's inhibitory actions work through at least two mechanistically distinct pathways. On one hand, sirtuins may diminish histone acetylation by inactivating histone acetyltransferase (HAT) enzymatic activity. In support of this notion, SIRT1 directly interacts with p300, the CREB-binding protein (CBP), and other HATs to inhibit the acetylation status of these enzymes (75). Alternatively, SIRT1 has recently been reported to deacetylate the RelA/p65 subunit of NF-κB at lysine 310 in vitro using overexpression systems (75). Deacetylation of K310 of RelA/p65 leads to decreases in NF-κB transcription activity, reducing production of proinflammatory cytokines and anti-apoptotic genes (75). In addition, it was recently reported that moderate overexpression of SIRT1 in mice leads to down-regulated NF-κB activity (34).
As secretory cells, macrophages are vital to the regulation of immune responses and the development of inflammation (76). Activation of NF-κB in macrophages leads to production and release of a wide array of proinflammatory cytokines and chemokines (77). Several studies indicate that the beneficial effect of SIRT1 on metabolic disorders is due in part by its ability to suppress NF-κB activity and thus cytokine production of macrophages. For example, knockdown of SIRT1 in the mouse macrophage RAW264.7 cell line and in intraperitoneal macrophages increases LPS-stimulated TNFα secretion (73). Moreover, gene expression profiles reveal that SIRT1 knockdown leads to an increase in inflammatory gene expression. Interaction between SIRT1 and NF-κB is also diminished by cigarette smoke, which causes increased acetylation of and activation of NF-κB proinflammatory response in human macrophages (71). Using a macrophage specific knockout mouse (Mac-SIRT1KO), our group has recently provided in vivo evidence that SIRT1 deacetylates the nuclear RelA/p65 subunit of NF-κB and attenuates NF-κB-mediated gene transcription (74). Genetic deletion of SIRT1 in myeloid cells not only leads to hyperactive NF-κB signaling but also predisposes mice to the development of insulin resistance and metabolic disorders (Figure 4). Indeed, other studies have shown that SIRT1-mediated deacetylation of of NF-κB inhibits iNOS and cytokine-mediated beta-cell cell damage in the isolated rat islets. Together, these findings demonstrate that SIRT1 activity in the macrophage directly regulates immune response and suggests that activators of SIRT1 may play an important therapeutic role in the treatment of chronic inflammatory diseases.
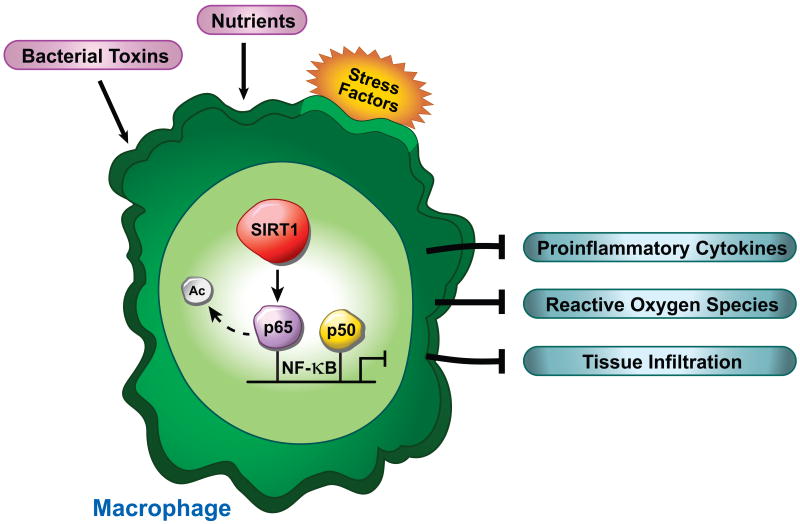
Figure 4.
SIRT1 inhibits inflammatory signaling by deacetylating the p65 subunit of the NF-κB transcription factor. Loss of SIRT1 in the macrophage leads to increased production and release of proinflammatory cytokines, reactive oxygen species, and macrophage infiltration into resident tissue. These factors contribute to a state of chronic inflammation, which is thought to exacerbate visceral obesity (115).
4. SIRT1 and adipose tissues
Adipose tissue functions both to store fat and as a conduit for hormone signaling. WAT secretes hormones such as leptin and adiponectin, which control energy balance, glucose regulation, and fatty acid catabolism. In addition, adipose tissue macrophages (ATMs) are prone to secrete high levels of TNFα, IL-6 and iNOS, resulting in a heightened inflammatory status particularly under obesity condition (78). Therefore, an increase in adiposity elevates the risk of developing metabolic disease, while decreased levels of WAT are presumed to reduce metabolic risk factors. Among numerous factors involved in adipose tissue differentiation, the nuclear receptor PPARγ plays an essential role in modulating fatty acid storage and glucose metabolism (79). PPARγ is integral in regulating the maturation of preadipocytes into mature fat cells. In mature fat cells, PPARγ regulates the induction of genes involved in free fatty acid uptake and triglyceride synthesis, thereby increasing the capacity of WAT for lipid storage (80). SIRT1 has been shown to repress PPARγ, by docking to the negative cofactors of the nuclear receptor, and thereby downregulating genes such as the mouse aP2 gene (38). Thus, when mice are starved, SIRT1 is induced to bind the aP2 promoter in WAT, repress gene expression, and promote mobilization of fat into the blood. Moreover, in differentiated adipose cells, upregulation of SIRT1 leads to decreased fat storage and increased lipolysis. Consistent with these findings, treatment of mice on a high fat diet with resveratrol was shown to reduce weight gain (35, 36). These findings demonstrate that SIRT1 acts in concert with lipid regulating transcription factors to adapt gene transcription to changes in nutrient levels.
Obese individuals have an increase in quantity and size of adipocytes, whereas individuals under dietary restriction display decrease in body weight and WAT mass. The repression of PPARγ activity by SIRT1 in WAT therefore implies that SIRT1 may regulate adipocyte formation and function in response to dietary restriction (38). As a reduction in adipose tissue is sufficient to extend murine lifespan (81), these findings provide a possible molecular pathway connecting calorie restriction to life extension in mammals.
The role of SIRT1 in brown adipose tissue (BAT) is still not clear. A study by Timmons et al. has suggested that SIRT1 may promote BAT differentiation through repression of MyoD-mediated myogenic gene expression signature and stimulation of PGC-1α-mediated mitochondrial gene expression (82). Interestingly, it has been recently shown that SIRT1 in the propiomelanocortin (POMC) expressing neurons selectively controls perigonadal WAT-to-BAT-like remodeling to increase energy expenditure in female mice (83). POMC neuron-specific deletion of SIRT1 in mice causes hypersensitivity to diet-induced obesity due to reduced energy expenditure (83). Consistent with these observations, Activation of SIRT1 by a specific SIRT1 activator, SRT1720, enhances oxidative metabolism in skeletal muscle, liver, and BAT (84). Together, these data indicate that SIRT1 regulates BAT differentiation and functions through both cell autonomous and non-cell autonomous mechanisms.
5. SIRT1 in the brain
The brain is essential in controlling whole-body metabolism through both neurologic and endocrine functions. The hypothalamus is the primary brain center that interprets adiposity or nutrient related inputs to regulate energy homeostasis, and secretion of growth hormone (GH) and several satiety/hunger hormones such as α-melanocyte-stimulating hormone (α-MSH), cocaine-and-amphetamine-regulated transcript (CART) peptides, neoropeptide Y (NPY), and agouti-related protein (ARrP). A recent study by Cohen et al. demonstrated that mice lacking SIRT1 in the brain show specific defects in cells in the anterior pituitary (25). Moreover, SIRT1 activity in the brain is required to mediate changes in pituitary signaling and physical activity that occur in response to CR (25). This finding suggests that SIRT1 in the brain may function as a potential link between the pituitary hormones and CR longevity pathways in mammals.
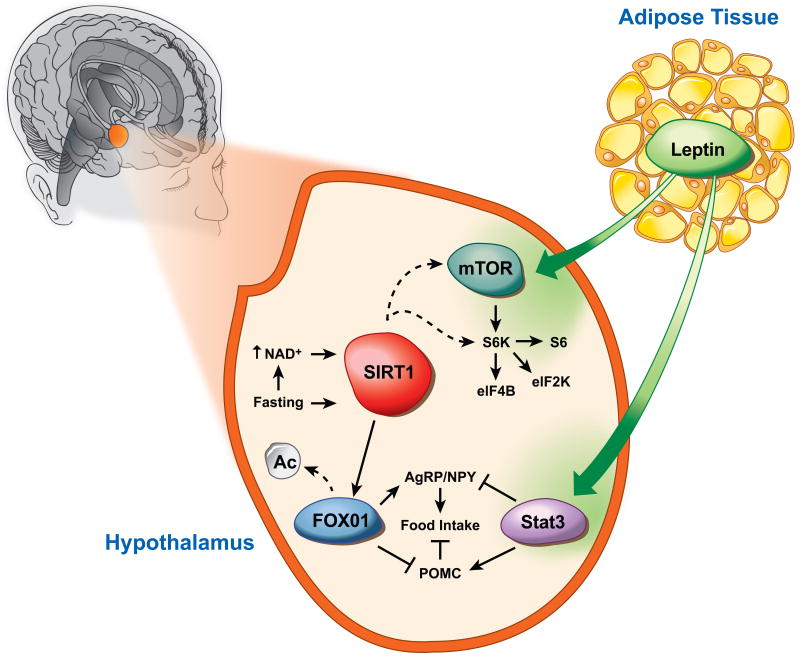
The anorexigenic POMC expressing neurons and the orexigenic agouti-related protein (AgRP) expressing neurons of the hypothalamus are the major regulators of feeding and energy expenditure (85). The POMC neurons produce satiety peptides thereby inhibiting food intake after feeding, while the AgRP neurons promote feeding in response to fasting and CR. Studies have shown that CR and fasting enhance SIRT1 expression and activity in the hypothalamus (86, 87). However, in contrast to its anti-obesity functions in the periphery tissues, increased hypothalamic SIRT1 appears to associate with increased food intake and subsequent body-weight gain. A study by Çakir et al. revealed that ablation or knockdown of SIRT1 in the hypothalamic region results in decreased food intake and body weight gain (Figure 6, (86)). Moreover, fasting increases hypothalamic NAD+ levels and SIRT1 protein content, while inhibition of hypothalamic SIRT1 activity reverses the fasting induced decrease of FOXO1 acetylation, resulting in increased POMC and decreased AgRP expressions (86). Furthermore, recent studies have demonstrated that brain-specific SIRT1 transgenic mice displays enhanced neural activity in hypothalamus (87), and AgRP neurons-specific deletion of SIRT1 decreases AgRP neuronal activity, thereby alleviating the inhibitory tone on the POMC neurons, resulting in decreased food intake and body weight (88). However, the apparent dichotomy of role of SIRT1 action in the periphery vs the hypothalamus is not all together surprising, given the fact that both fasting and CR induce fatty acid oxidation in periphery tissues but feeding behavior through central regulation. These observations confirm that SIRT1 is an essential element in the periphery-central feedback circuits that mediate normal responses to nutrient deprivation.
Figure 6.
Hypothalamic SIRT1 controls central adaptive response to nutrient signals. Fasting increases NAD+ levels and SIRT1 activity. SIRT1 deacetylates and activates FOXO1, and thus regulates POMC and AgRP expression. Hypothalamic SIRT1 also influences S6k signaling, however, it is not known whether this activation occurs upstream or down-stream of mTOR. Figure is an adaptation from Çakir et al. (86).
In addition, the role of SIRT1 in the central control of whole body energy homeostasis appears to be complicated. A recent study has revealed that POMC neurons-specific deletion of SIRT1 in mice causes hypersensitivity to diet-induced obesity due to a blunted response to leptin signaling and reduced energy expenditure (83). Importantly, deletion of SIRT1 in POMC neurons does not lead to overt dysfunctions of the CNS melanocortin system, and the reduced energy expenditure is not associated with changes in food intake (83). Therefore, it is possible that chronic losses of SIRT1 in different neurons lead to different compensation patterns in the hypothalamus. An inducible cell type-specific SIRT1 knockout model will be helpful to further dissect the roles of SIRT1 in regulating central adaptive response to nutrient signals.
Given the important role of SIRT1 in regulation of nutrient sensing, targeting its activity in the brain may show promise for the treatment of obesity and associated metabolic disorders. Indeed, long-term intracerebroventricular infusion of resveratrol normalizes hyperglycemia and greatly improves hyperinsulinemia in mice with diet-induced obesity and diabetes (89). These effects were observed without significant systemic changes in body weight, food intake, and circulating leptin levels. These findings are in line with other studies showing that resveratrol fed mice were resistant to diet-induced obesity and insulin resistance (35, 36). In addition, central neural system resveratrol delivery reduces hypothalamic NF-κB inflammatory signaling by lowering acetylated-RelA/p65 and total RelA/p65 protein contents, as well as mRNA levels of NF-κB inhibitor of IκB kinase β (89). In summary, SIRT1 activity appears to be important in the regulation of the brain involved in metabolic homeostasis. As little is known at this stage, this area of sirtuin biology may yield important discoveries in coming years.
6. SIRT1 and circadian rhythm
Circadian rhythm, the cyclical 24-hour period of biological processes of living entities on Earth, depends on internal circadian clocks that generate rhythms in physiology and behavior in part through chromatin remodeling and epigenetic control of gene expression (90). Recent studies have revealed an intriguing association between the circadian clock and cellular metabolism. For example, circadian disruption in mice has been linked to metabolic dysfunctions (91, 92), while high-fat diet feeding alters both behavioral and molecular circadian rhythms (93, 94).
Recent studies have linked SIRT1 function to the regulation of the circadian rhythm. The mammalian clock is controlled by negative-feedback loops mediated by the heterodimeric transcription factors CLOCK-BMAL1 and their transcriptional targets, including the PER and CRY proteins that directly repress CLOCK-BMAL1 activity, as well as REV-ERB and ROR nuclear receptors that control BMAL1 expression (95). Interestingly, the CLOCK protein itself is a transcriptional activator that functions as a histone acetyltransferase (HAT) (96), indicating that chromatin remodeling plays an important role in the regulation of circadian gene expression. Two earlier studies have shown that SIRT1 interacts with CLOCK-BMAL1 to directly regulate the amplitude of circadian clock-controlled gene expression through deacetylation of PER2 and/or BMAL1 (97, 98). However, whether it is the amount of SIRT1 or its activity that is cyclically regulated by circadian clock remains under debate. Two recent studies appear to favor the circadian regulation of SIRT1 activity (99, 100). The expression of NAMPT, an enzyme that controls a rate-limiting step in NAD+ biosynthesis, is directly controlled by CLOCK-BMAL1 (Figure 7). It has been proposed that this regulation leads to circadian oscillation of cellular NAD+ levels, resulting in a cyclical regulation of SIRT1 activity (99, 100). Together, these studies add a new feedback loop in the circadian clock that involves CLOCK-BMAL1, NAMPT, NAD+, and SIRT1 (Figure 7), and provide an important link between the circadian clock and cellular metabolism.
Figure 7.
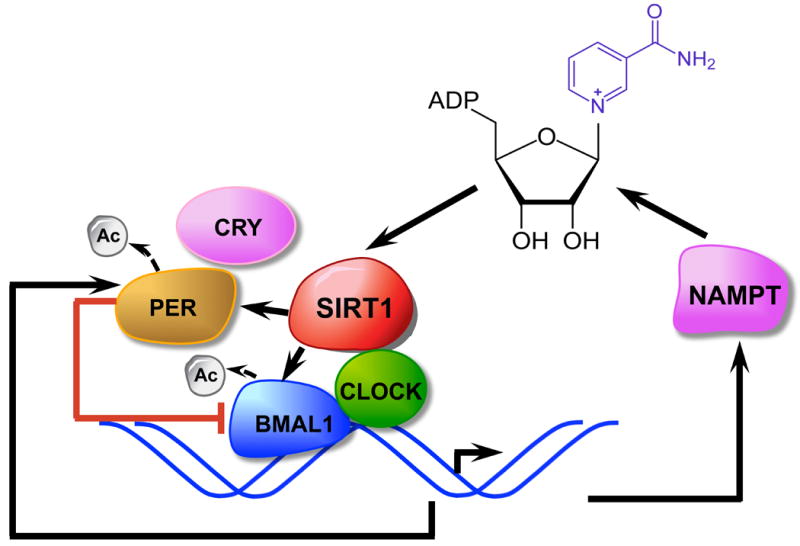
SIRT1 as an essential element of a feedback loop that links cellular metabolism to circadian clock. The CLOCK-BMAL1 transcript complex directly controls the expression of NAMPT, which encodes the rate-limiting enzyme in NAD+ biosynthesis. NAD+ then regulates SIRT1 activity and modulates CLOCK-BMAL1-mediated transcription. SIRT1 also directly regulates the activity of PER, a negative regulator of CLOCK-BMAL1 transcription, through deacetylation.
However, it remains to be determined whether the observed oscillation of cellular NAD+ levels (99, 100) is indeed responsible for the oscillation of SIRT1 activity (98). It is interesting to note that the activity of immuno-purified endogenous SIRT1 proteins display a circadian oscillating pattern when measured in vitro with fixed amount of exogenous NAD+ (Figure 1D in (98)). This finding suggests that post-translational modifications, or protein-protein interactions also play a role in the circadian regulation of SIRT1 activity. Recent studies have shown that SIRT1 activity can be directly inhibited (101, 102) or activated (103, 104) by several protein factors. Its activity can also be modulated by phosphorylation modifications (105-107). Particularly, we have recently shown that SIRT1 can be phosphorylated and activated by DYRK1A (108), an essential clock component that governs the rhythmic phosphorylation and degradation of CRY2 protein (109). Therefore, exploring the possible role of these factors in the circadian regulation of SIRT1 activity may provide novel insights into its function in the circadian rhythm.
Conclusions and Therapeutic Implications
The disruption of hepatic lipid metabolism, immune signaling, fat maturation, and circadian gene expression in SIRT1-deficient mouse models suggests that pharmacological modulation of SIRT1 could be of interest to curb diseases associated with obesity. Interestingly, small molecule activators of sirtuins, such as the polyphenol resveratrol and closely related derivatives, have shown promise as a therapeutic agent for the treatment of metabolic diseases (37, 110). Whether these small-molecule drugs are direct activators of SIRT1 that function to prevent obesity and diabeties (49, 50), is a topic of intense debate (50). However, some studies have demonstrated that mice fed a high-fat diet along with resveratrol remain lean and healthy compared to over-weight control animals (reviewed in (111)). Additionally, resveratrol significantly increased aerobic capacity, as evidenced by increased running time and elevated oxygen consumption in muscle fibers. It is proposed that these drugs activate SIRT1 and thus act as calorie restriction mimetics by increasing fat mobilization, catabolism, and altering cholesterol homeostasis (111).
Consistent with the importance of SIRT1 in the regulation of metabolic homeostasis in animal models, three single nucleotide polymorphisms (SNPs) in the SIRT1 gene were found to be correlated with weight gain in a study of elderly human subjects (112). The study reported that common variants in SIRT1 are associated with lower BMI in two independent Dutch populations (112). Carriers of these variants displayed 13–18% decreased risk of obesity and gain less weight over time. A separate case/control study of Belgian obese patients showed that a minor SIRT1 SNP was associated with decreased risk of obesity (113). Interestingly, this variant allele was also associated with increased visceral fat in obese men (113). Since increase visceral adiposity has been paradoxically implicated in increased insulin sensitivity (114), SIRT1 may help to improve insulin sensitivity in obese males by coupling a protective effect against obesity with increased visceral adiposity. Furthermore, if activation of SIRT1 can result in loss of body fat without decreasing caloric intake, this could open the door for novel treatment and prevention strategies for obesity and related diseases.
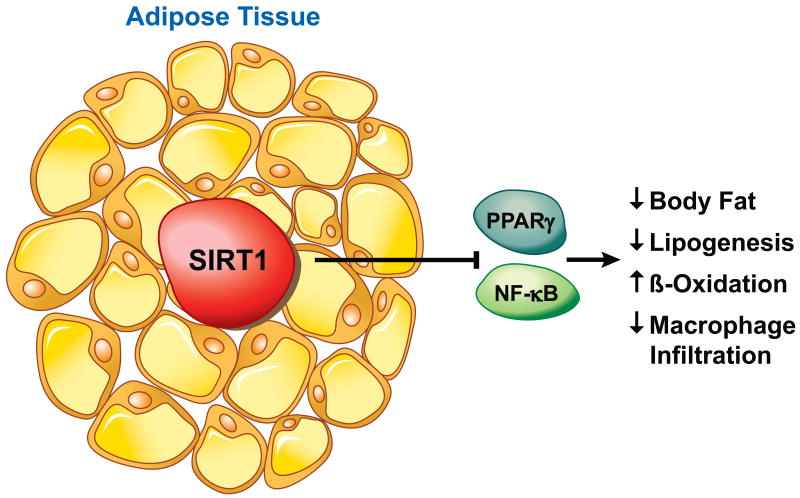
Figure 5.
SIRT1 controls many aspects of lipid metabolism and fat cell maturation. SIRT1 represses the PPARγ nuclear receptor, thus down-regulating adipocyte differentiation and maturation. Loss of SIRT1 leads to increased adipose tissue macrophages and elevated inflammation.
Key Messages.
SIRT1 is a metabolic sensor that directly couples cellular metabolic status to chromatin structure and gene expression.
SIRT1 regulates a number of targets involved in lipid metabolism at different tissues.
The activity of SIRT1 is regulated in response to environmental stimuli.
Acknowledgments
We thank Drs. Anton Jetten, Huiming Gao, and John Cidlowski for critical reading of the manuscript; NIEHS Multimedia Services Department for the cartoon graph of Figures. The work related to this article was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to X.L. (Z01 ES102205).
Footnotes
Conflict of Interests Statement: The authors in this manuscript have no conflict of interests to declare.
Contributor Information
Thaddeus T. Schug, Email: Schugt@niehs.nih.gov.
Xiaoling Li, Email: Lix3@niehs.nih.gov.
References
- 1.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005 Apr 16-22;365(9468):1415–28. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 2.Hedley AA, Ogden CL, Johnson CL, Carroll MD, Curtin LR, Flegal KM. Prevalence of overweight and obesity among US children, adolescents, and adults, 1999-2002. Jama. 2004 Jun 16;291(23):2847–50. doi: 10.1001/jama.291.23.2847. [DOI] [PubMed] [Google Scholar]
- 3.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005 Apr;6(4):298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 4.Grundy SM. Drug therapy of the metabolic syndrome: minimizing the emerging crisis in polypharmacy. Nat Rev Drug Discov. 2006 Apr;5(4):295–309. doi: 10.1038/nrd2005. [DOI] [PubMed] [Google Scholar]
- 5.Vazquez-Vela ME, Torres N, Tovar AR. White adipose tissue as endocrine organ and its role in obesity. Arch Med Res. 2008 Nov;39(8):715–28. doi: 10.1016/j.arcmed.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Steppan CM, Lazar MA. The current biology of resistin. J Intern Med. 2004 Apr;255(4):439–47. doi: 10.1111/j.1365-2796.2004.01306.x. [DOI] [PubMed] [Google Scholar]
- 7.Meier U, Gressner AM. Endocrine regulation of energy metabolism: review of pathobiochemical and clinical chemical aspects of leptin, ghrelin, adiponectin, and resistin. Clin Chem. 2004 Sep;50(9):1511–25. doi: 10.1373/clinchem.2004.032482. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Mariman E, Renes J, Keijer J. The secretory function of adipocytes in the physiology of white adipose tissue. J Cell Physiol. 2008 Jul;216(1):3–13. doi: 10.1002/jcp.21386. [DOI] [PubMed] [Google Scholar]
- 9.Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, Vats D, Morel CR, Goforth MH, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008 Jun;7(6):496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000 Feb 17;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 11.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000 May 23;97(11):5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6658–63. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–35. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 14.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004 Nov 9;101(45):15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SJ, Guarente L. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2006 Mar;2(3):e33. doi: 10.1371/journal.pgen.0020033. author reply e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nature cell biology. 2008 Apr;10(4):385–94. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 17.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular cell. 2004 Oct 8;16(1):93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 18.Sinclair DA, Oberdoerffer P. The ageing epigenome: damaged beyond repair? Ageing Res Rev. 2009 Jul;8(3):189–98. doi: 10.1016/j.arr.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomb DJ, Laurent G, Haigis MC. Sirtuins regulate key aspects of lipid metabolism. Biochim Biophys Acta. Aug;1804(8):1652–7. doi: 10.1016/j.bbapap.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 20.Kim C, Park J, Kang E, Ahn C, Cha B, Lim S, et al. Comparison of body fat composition and serum adiponectin levels in diabetic obesity and non-diabetic obesity. Obesity (Silver Spring) 2006 Jul;14(7):1164–71. doi: 10.1038/oby.2006.133. [DOI] [PubMed] [Google Scholar]
- 21.Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci U S A. 2004 Apr 27;101(17):6659–63. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000 Sep 22;289(5487):2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005 Dec 9;310(5754):1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 24.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PloS one. 2008;3(3):e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009 Dec 15;23(24):2812–7. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008 Mar 4;105(9):3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009 May;15(5):217–24. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Goligorsky MS. SIRTing out the link between autophagy and ageing. Nephrol Dial Transplant. 2010 Aug;25(8):2434–6. doi: 10.1093/ndt/gfq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang JW, Chung S, Sundar IK, Yao H, Arunachalam G, McBurney MW, et al. Cigarette smoke-induced autophagy is regulated by SIRT1-PARP-1-dependent mechanism: implication in pathogenesis of COPD. Arch Biochem Biophys. 2010 Aug 15;500(2):203–9. doi: 10.1016/j.abb.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushik S, Singh R, Cuervo AM. Autophagic pathways and metabolic stress. Diabetes Obes Metab. 2010 Oct;12 2:4–14. doi: 10.1111/j.1463-1326.2010.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Navarro JA, Cuervo AM. Autophagy and lipids: tightening the knot. Semin Immunopathol. 2010 Aug 22; doi: 10.1007/s00281-010-0219-7. [DOI] [PubMed] [Google Scholar]
- 32.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007 Dec;6(6):759–67. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 33.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, et al. SirT1 Gain of Function Increases Energy Efficiency and Prevents Diabetes in Mice. Cell metabolism. 2008;8:333–41. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008 Jul 15;105(28):9793–8. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006 Nov 16;444(7117):337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006 Dec 15;127(6):1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 37.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007 Nov 29;450(7170):712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004 Jun 17;429(6993):771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005 Oct 14;310(5746):314–7. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 40.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006 Dec 14;444(7121):868–74. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 41.Hagopian K, Ramsey JJ, Weindruch R. Influence of age and caloric restriction on liver glycolytic enzyme activities and metabolite concentrations in mice. Exp Gerontol. 2003 Mar;38(3):253–66. doi: 10.1016/s0531-5565(02)00203-6. [DOI] [PubMed] [Google Scholar]
- 42.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008 Jul 1;22(13):1753–7. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009 Apr 23;458(7241):1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010 Mar 3;11(3):213–9. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010 Apr 29;464(7293):1313–9. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 46.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005 Mar 3;434(7029):113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 47.Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, Levine J, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 2010 Feb 1;120(2):545–58. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010 Apr 12;1(1):1–8. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, et al. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009 Dec;74(6):619–24. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 50.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010 Mar 12;285(11):8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van den Berghe G. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. J Inherit Metab Dis. 1991;14(4):407–20. doi: 10.1007/BF01797914. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008 Oct 5; doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009 Apr;9(4):327–38. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominy JE, Jr, Lee Y, Gerhart-Hines Z, Puigserver P. Nutrient-dependent regulation of PGC-1alpha's acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim Biophys Acta. 2010 Aug;1804(8):1676–83. doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Molecular cell. 2007 Oct 12;28(1):91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 56.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009 Nov;10(5):392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995 Dec 15;83(6):841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 58.Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008 Jul 18;283(29):20015–26. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007 Jul 31;104(31):12861–6. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, Liu C, Li N, Hao T, Han T, Hill DE, et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008 Aug;8(2):105–17. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feige JN, Auwerx J. DisSIRTing on LXR and cholesterol metabolism. Cell Metab. 2007 Nov;6(5):343–5. doi: 10.1016/j.cmet.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009 Nov 15;23(22):2578–91. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010 Jul 1;24(13):1403–17. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285(44):33959–70. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004 Feb 20;116(4):551–63. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 66.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006 Jul;116(7):1793–801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006 Dec 14;444(7121):860–7. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 68.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007 Jun 28;447(7148):1116–20. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suganami T, Ogawa Y. Adipose tissue macrophages: their role in adipose tissue remodeling. J Leukoc Biol. 2010 Jul;88(1):33–9. doi: 10.1189/jlb.0210072. [DOI] [PubMed] [Google Scholar]
- 70.Iyer A, Fairlie DP, Prins JB, Hammock BD, Brown L. Inflammatory lipid mediators in adipocyte function and obesity. Nat Rev Endocrinol. Feb;6(2):71–82. doi: 10.1038/nrendo.2009.264. [DOI] [PubMed] [Google Scholar]
- 71.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008 Apr 15;177(8):861–70. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, et al. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. 2009 Mar;29(5):1363–74. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010 Mar;298(3):E419–28. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schug TT, Xu Q, Gao H, Peres-da-Silva A, Draper DW, Fessler MB, et al. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol Cell Biol. 2010 Jul 19; doi: 10.1128/MCB.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004 Jun 16;23(12):2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, et al. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol. 2005 Oct;19(10):2466–77. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- 77.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003 Jan;3(1):23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 78.Odegaard JI, Chawla A. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat Clin Pract Endocrinol Metab. 2008 Nov;4(11):619–26. doi: 10.1038/ncpendmet0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 80.Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004 Apr;10(4):355–61. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 81.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003 Jan 24;299(5606):572–4. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 82.Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4401–6. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramadori G, Fujikawa T, Fukuda M, Anderson J, Morgan DA, Mostoslavsky R, et al. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010 Jul 4;12(1):78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008 Nov;8(5):347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 85.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006 Sep 21;443(7109):289–95. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 86.Cakir I, Perello M, Lansari O, Messier NJ, Vaslet CA, Nillni EA. Hypothalamic Sirt1 regulates food intake in a rodent model system. PloS one. 2009;4(12):e8322. doi: 10.1371/journal.pone.0008322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010 Jul 28;30(30):10220–32. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dietrich MO, Antunes C, Geliang G, Liu ZW, Borok E, Nie Y, et al. Agrp neurons mediate Sirt1's action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J Neurosci. 2010 Sep 1;30(35):11815–25. doi: 10.1523/JNEUROSCI.2234-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramadori G, Gautron L, Fujikawa T, Vianna CR, Elmquist JK, Coppari R. Central administration of resveratrol improves diet-induced diabetes. Endocrinology. 2009 Dec;150(12):5326–33. doi: 10.1210/en.2009-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–48. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 91.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005 May 13;308(5724):1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004 Nov;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007 Nov;6(5):414–21. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 94.Barnea M, Madar Z, Froy O. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology. 2009 Jan;150(1):161–8. doi: 10.1210/en.2008-0944. [DOI] [PubMed] [Google Scholar]
- 95.Wijnen H. Circadian rhythms. A circadian loop asSIRTs itself. Science. 2009 May 1;324(5927):598–9. doi: 10.1126/science.1174132. [DOI] [PubMed] [Google Scholar]
- 96.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006 May 5;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 97.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008 Jul 25;134(2):317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 98.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008 Jul 25;134(2):329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009 May 1;324(5927):654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009 May 1;324(5927):651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008 Jan 31;451(7178):587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008 Jan 31;451(7178):583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 103.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Molecular cell. 2007 Oct 26;28(2):277–90. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 104.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nature cell biology. 2007 Nov;9(11):1253–62. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, et al. Phosphorylation regulates SIRT1 function. PloS one. 2008;3(12):e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PloS one. 2009;4(8):e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS one. 2009;4(12):e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010 Apr 23;285(17):13223–32. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kurabayashi N, Hirota T, Sakai M, Sanada K, Fukada Y. DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol Cell Biol. 2010 Apr;30(7):1757–68. doi: 10.1128/MCB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003 Sep 11;425(6954):191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 111.Camins A, Sureda FX, Junyent F, Verdaguer E, Folch J, Pelegri C, et al. Sirtuin activators: Designing molecules to extend life span. Biochim Biophys Acta. 2010 Jun 23; doi: 10.1016/j.bbagrm.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 112.Zillikens MC, van Meurs JB, Rivadeneira F, Amin N, Hofman A, Oostra BA, et al. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009 Dec;58(12):2828–34. doi: 10.2337/db09-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peeters AV, Beckers S, Verrijken A, Mertens I, Roevens P, Peeters PJ, et al. Association of SIRT1 gene variation with visceral obesity. Hum Genet. 2008 Nov;124(4):431–6. doi: 10.1007/s00439-008-0567-8. [DOI] [PubMed] [Google Scholar]
- 114.Johansson LE, Lindblad U, Larsson CA, Rastam L, Ridderstrale M. Polymorphisms in the adiponutrin gene are associated with increased insulin secretion and obesity. Eur J Endocrinol. 2008 Nov;159(5):577–83. doi: 10.1530/EJE-08-0426. [DOI] [PubMed] [Google Scholar]
- 115.Gupta AK, Johnson WD. Prediabetes and prehypertension in disease free obese adults correlate with an exacerbated systemic proinflammatory milieu. J Inflamm (Lond) 2010;7:36. doi: 10.1186/1476-9255-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]