Abstract
In both immunocompromised and immunocompetent patients, Candida and Malassezia are causing or triggering clinical manifestations such as cutaneous infections and atopic eczema. The innate immune system provides rapid responses to microbial invaders, without requiring prior stimulation, through a sophisticated system of antimicrobial peptides (AMPs). High molecular weight kininogen (HMWK) and components of the contact system have previously been reported to bind to Candida and other pathogens, leading to activation of the contact system. A cutaneous Candida infection is characterized by an accumulation of neutrophils, leading to an inflammatory response and release of enzymatically active substances. In the present study we demonstrate that antifungal peptide fragments are generated through proteolytic degradation of HMWK. The recombinant domain 5 (rD5) of HMWK, D5-derived peptides, as well as hydrophobically modified D5-derived peptides efficiently killed Candida and Malassezia. Furthermore, the antifungal activity of modified peptides was studied at physiological conditions. Binding of a D5-derived peptide, HKH20 (His479-His498), to the fungal cell membrane was visualized by fluorescence microscopy. Our data disclose a novel antifungal activity of D5-derived peptides and also show that proteolytic cleavage of HMWK results in fragments exerting antifungal activity. Of therapeutic interest is that structurally modified peptides show an enhanced antifungal activity.
1. Introduction
The innate immune system provides rapid responses to microbial invaders without requiring prior stimulation. It is a sophisticated system comprising epithelial barriers, white blood cells, various peptides and proteins including antimicrobial peptides (AMPs), as well as lipids and other chemicals, such as reactive oxygen species [1–4]. At present, approximately 1600 AMPs of different origin have been identified (http://aps.unmc.edu/AP/main.php). A blend of endogenous AMPs, including the cathelicidin LL-37/hCAP18, β-defensins, RNase 7, various dermcidins, as well as psoriasin, are found in skin [5–9]. Peptides of the histatin family, like many other histidine-rich AMPs [10], are known to harbor antifungal activities. Histatin 5 is secreted by the salivary glands and exerts its activity by entering the fungal cytoplasm targeting the mitochondria [11, 12]. Bacterial, fungal, and mammalian cell surfaces have structural differences which may explain a certain degree of selectivity for AMP action. Specifically, bacterial surfaces contain many anionic components, including LPS and anionic lipids of Gram-negative bacteria, as well as teichoic and teichuronic acids of Gram-positive bacteria. Similarly, glucan, chitin, mannoprotein, and a blend of other cell wall proteins and polysaccharides contribute to a negative surface potential of fungal surfaces. Beyond the outer cell surface, AMPs interact with the plasma membrane. Furthermore, fungal membranes contain mostly zwitterionic lipids (e.g., phosphatidylcholine), but also ergosterol, whereas bacterial membranes comprise various acidic phospholipids (phosphatidylglycerol, phosphatidylserine, and cardiolipin), which confer a negative charge facilitating AMP binding and sometimes also defect formation [13–17]. AMPs may preferentially target fungi, bacteria, or mammalian cells, or combinations thereof, and the interaction and disruption of the target membrane is dependent on a number of parameters, including hydrophobic and ionic interactions, as well as influence of oligomerisation, the ionic environment, and the presence of plasma proteins [13–17]. Many endogenous AMPs are not only antimicrobial, however, but have many additional functions, including antiendotoxin or antitumor effects, anti-inflammatory properties, mediation of chemotaxis, or activation of the immune system [7]. Although AMPs display a range of conformations, such as α-helices and β-sheets, they all share common properties such as amphipathicity, cationicity, and the propensity to bind to negatively charged molecules such as negatively charged phospholipids, glycosaminoglycans, and heparin [18–21]. Concerning bacterial killing, the mechanisms of AMP action are complex and are often divided into membrane active and nonmembrane disruptive mechanisms [15, 22, 23].
Candida albicans is normally colonizing human skin as well as other epithelial surfaces. However, in both immunocompromised and immunocompetent patients, Candida may cause or trigger several clinical manifestations such as cutaneous infections, atopic eczema, oroesophageal candidiasis, candida vaginitis, urinary candidiasis, and disseminated candidiasis and candidemia [24–26]. C. albicans is the most prevalent yeast in clinical infections, but non-C. albicans species are increasingly causing severe infections such as candidemia, especially in patients suffering from endogenous or drug-induced immunosuppression and critically ill patients [27]. The yeast Malassezia is a member of the cutaneous microbial flora, and involved in several dermatologic diseases such as atopic dermatitis, pityriasis versicolor, and seborrheic dermatitis [28], and also reported to cause invasive infections in premature infants [29]. Importantly, some Candida isolates are known to have reduced susceptibility to antifungal drugs [24], which makes other strategies based on antifungal proteins and peptides interesting alternatives in the treatment of fungal infections.
High molecular weight kininogen (HMWK) and components of the contact system have previously been reported to have the capacity of binding to the surface of a blend of human pathogens, including Candida, leading to activation of the contact system [30–32]. The 120 kDa glycoprotein HMWK contains six domains with different functions. Domain 3 and 5 have been previously reported to harbor epitopes, that may function as antimicrobial peptides on release [33, 34]. Furthermore, activation and degradation of HMWK leads to kallikrein-mediated generation of bradykinin [35]. However, endogenous generation of active kinins can also be mediated by the cooperative action of mast-cell tryptase and neutrophil elastase. Kinins can also be proteolytically generated from plasma HMWK by the action of microbial and dust-mite proteases [36, 37].
In the present study, we have investigated whether rD5, D5-derived peptides, and structurally modified D5-derived peptides exert antifungal activity. In addition, the capability of these modified peptides to work as antifungal peptides at physiological conditions was elucidated. Our data disclose a novel antifungal activity of D5-derived peptides and also demonstrate that HMWK, when subjected to degradation by neutrophilic enzymes, may release fragments with antifungal activity.
2. Materials and Methods
2.1. Proteolytic Cleavage of HMWK and Detection of Fragments
To investigate the neutrophil elastase effects on HMWK, the protein (32 μg) was incubated at 37°C for 30 and 60 minutes with human leukocyte (neutrophil) elastase (HLE) (24 mU) or freeze/thaw-disrupted polymorphonuclear neutrophils (PMNs) (18 μL, 1×107 cells/mL) in a total volume of 40 μL. HMWK (32 μg) incubated at 37°C for 60 minutes was used as a control. Ten μL of the material was analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 16.5% Tris-Tricine gel (Bio-Rad Laboratories, Hercules, Calif, USA) under reducing conditions.
For detection of fragments generated by Cathepsin G, HMWK (16 μg) was incubated with Cathepsin G (9 μU) at 37°C for 30 minutes and analyzed by Western blot (Figure 1(c)). The proteins were further transferred onto nitrocellulose membranes (Hybond-C, Amersham Biosciences, UK). The membrane was blocked using 3% (w/v) skimmed milk (ICN Biomedicals Inc., Ohio, USA), washed, and then incubated for 1 h with rabbit polyclonal antibodies (Innovagen AB, Lund, Sweden) against the peptide HKH20 (at 1 : 1500), washed again and incubated with horseradish peroxidase-conjugated swine antirabbit secondary antibodies (DAKO A/S, Glostrup, Denmark) (1 : 1000). The proteins reacting with the antibodies were detected by using enhanced chemiluminescence ECL Western Blotting Detection Reagents Kit (Amersham Biosciences, Buckinghamshire, UK).
Figure 1.
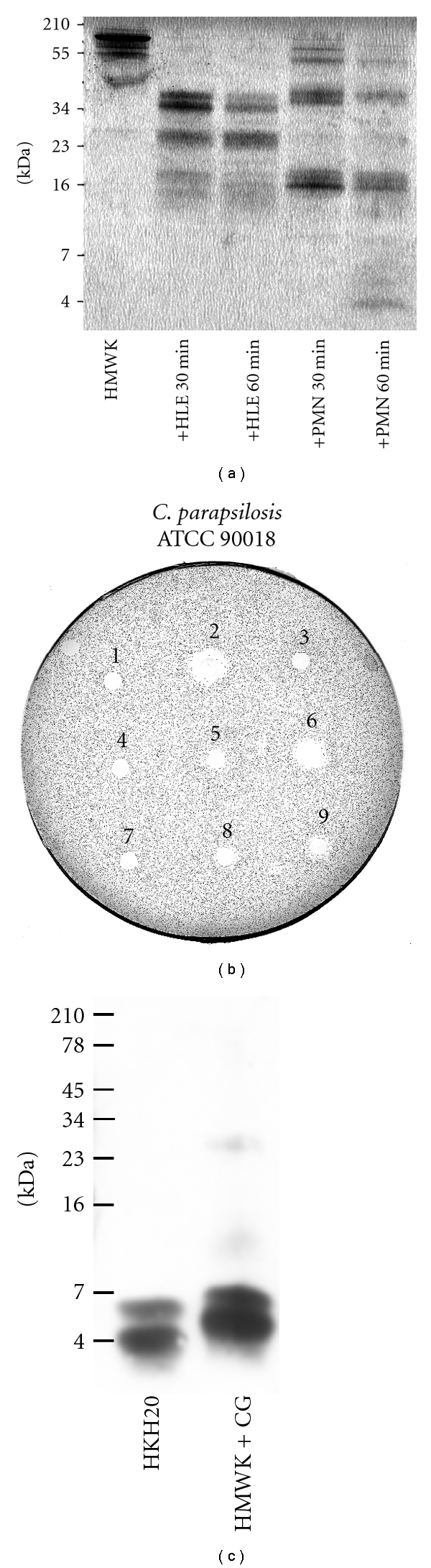
Degradation of HMWK generates antifungal peptides. (a) Intact HMWK and cleavage products from different incubations (indicated below, human leukocyte elastase, HLE, and polymorphonuclear neutrophils, PMN), were analyzed by SDS-PAGE (16.5% Tris-Tricine gel). (b) Antifungal activity of HMWK cleavage products is shown as clear zones corresponding to inhibition of fungal growth in RDA. Cleavage of HMWK was performed for 30 and 60 minutes at 37°C. (1) control, buffer (10 mM Tris, pH 7.4). (2) reference, LL-37 (6 μL at 100 μM). (3) control, HMWK. (4) control, human leukocyte elastase (HLE). (5) and (6) HMWK incubated with HLE for 30 and 60 minutes, respectively. (7) control, polymorphonuclear neutrophils (PMN). (8) and (9) HMWK incubated with PMN for 30 and 60 minutes, respectively. Data represents the results from a single representative experiment out of three. (c) HMWK was subjected to Cathepsin G, producing cleavage products recognized by polyclonal antibodies against HKH20 by Western blot analysis. In the panels, molecular mass markers are indicated on the left.
2.2. Peptides and Biological Materials
The peptides (Table 1), LL-37, histatin 5, and the synthetic peptides HKH20, GKH17, GGH20, GHG20, GHG21, KHN 20, GKH17WWW, GKH17WWWWW, and Texas Red-conjugated HKH20, were all synthesized by Innovagen AB (Lund, Sweden). The purity (>95%) and molecular weight of these peptides were confirmed by mass spectral analysis (MALDI-TOF Voyager, Applied Biosystems, Foster City, Calif, USA). Citrate plasma (fresh frozen) from healthy volunteers was obtained from the blood bank at Skåne University Hospital, Lund, Sweden. Neutrophils were prepared by routine procedures (Polymorphprep™, AXIS-SHIELD PoC AS, Oslo, Norway) from blood of healthy human donors. The cells were disrupted by freezethawing followed by the addition of 0.3% Tween 20. Human leukocyte elastase (4 mU μL−1) was obtained from Calbiochem (San Diego, Calif). The purification of recombinant domain 5 was done as previously described [33]. Briefly, the expression vector (pET25b) (Novagen, Inc., Madison, Wis) containing rD5 was expressed in Escherichia coli strain BL21 (DE3), generously provided by Herwald et al. [38]. E. coli was subjected to induction of protein production by 1 mM isopropyl thio-β-D-galactoside, and, after 3 hour incubation at 30°C, the bacteria were harvested by centrifugation. The pellet was resuspended in sonication buffer (50 mM phosphate, 300 mM NaCl, pH 8.0), and bacterial cells were lysed by repeated cycles of freeze thawing. The supernatant of the lysate was mixed with 2 mL of nickel-nitrilotriacetic acid-Sepharose (Qiagen GmbH, Hilden, Germany), equilibrated with sonication buffer, and incubated on rotation for 1 h at room temperature. The Sepharose gel was loaded into a column washed, and the rD5 protein was eluted as previously described [33].
Table 1.
D5-derived and control peptides investigated.
| Peptide | Sequence | Mw | pI1 | Net charge | Mean hydrophobicity2 |
|---|---|---|---|---|---|
| KHN20 | KHNLGHGHKHERDQGHGHQR | 2365.5 | 9.99 | +2 | −4.91 |
| GHG20 | GHGLGHGHEQQHGLGHGHKF | 2127.2 | 7.21 | 0 | −2.01 |
| GHG21 | GHGHKFKLDDDLEHQGGHVLD | 2354.5 | 5.63 | −3 | −2.32 |
| GGH20 | GGHVLDHGHKHKHGHGHGKH | 2169.3 | 9.70 | +2 | −3.44 |
| HKH20 | HKHGHGHGKHKNKGKKNGKH | 2151.5 | 10.78 | +7 | −5.91 |
| GKH17 | GKHKNKGKKNGKHNGWK | 1946 | 10.78 | +7 | −5.77 |
| GKH17WWW | GKHKNKGKKNGKHNGWKWWW | 2505 | 10.78 | +7 | −3.45 |
| GKH17WWWWW | GKHKNKGKKNGKHNGWKWWWWW | 2287 | 10.78 | +7 | −2.25 |
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | 4492 | 10.61 | +6 | −1.84 |
| Histatin-5 | DSHAKRHHGYKRKFHEKHHSHRGY | 3036 | 10.28 | +5 | −4.67 |
Mw: molecular weight. 1pI: theoretical isoelectric point calculated by using the Protparam tool available at http://us.expasy.org/tools/protparam.html. 2Mean hydrophobicity calculated using HydroMCalc (combined consensus hydrophobicity scale) available at http://www.bbcm.univ.trieste.it/~tossi/HydroCalc/HydroMCalc.html.
2.3. Determination of Antifungal Activity
C. albicans ATCC 90028 and C. parapsilosis ATCC 90018 were obtained from the Department of Bacteriology Lund at Skåne University Hospital. Cells were precultured overnight on Sabouraud-dextrose-agar (Difco, Detroit, Mich) at 28°C, inoculated in 10 mL yeast extract-peptone-dextrose (YPD) broth (Sigma-Aldrich, St. Louis, Mo USA) at 28°C, and grown to mid-logarithmic phase. The cells were washed three times and diluted in 10 mM Tris, pH 7.4. Fifty μL of a suspension containing 1 × 106 fungal colony-forming units (cfu)/mL was incubated at 28°C for two hours with rD5, HKH20 at concentrations ranging from 0.3 μM to 60 μM, or the peptides HKH20, GKH17, GKH17WWW, and GKH17WWWWW at a concentration of 30 μM. To quantify the fungicidal activity, serial dilutions of the incubation mixture were plated onto Sabouraud-dextrose-agar plates (Difco, Detroit, Mich) and incubated for 48 h at 28°C, and the number of cfu were thereafter determined by counting visible colonies. Activity of 30 μM HKH20, GKH17, GKH17WWW, and GKH17WWWWW peptides against Candida was also tested in physiological salt conditions (0.15 M NaCl in 10 mM Tris, pH 7.4) in presence of 20% as well as 50% human plasma, and 50% human serum (in 0.15 M NaCl, 10 mM Tris, pH 7.4) (Figure 3). The controls (Figure 3) defined the total (100%) survival of Candida cells incubated in the same buffer under identical conditions but without peptides. To test the time dependence of fungal killing, Candida was subjected to 30 μM of HKH20 peptide and incubated for 5, 10, 15, 30, 60, and 150 minutes (Figure 2(d)).
Figure 3.
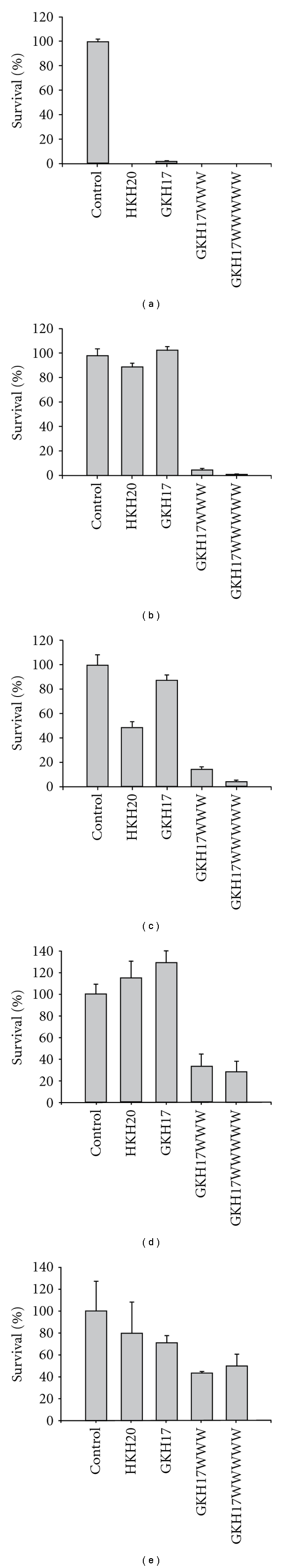
Antifungal effects of the D5-derived and the modified peptides. (a)–(e) The D5-derived peptides HKH20, GKH17, and modified variants of GKH17 were tested against C. parapsilosis ATCC 90018 in viable count assays at different physiological conditions. Candida cells were subjected to 30 μM of the peptides at (a) low salt conditions (10 mM Tris, pH 7.4), (b) physiological salt conditions (0.15 M NaCl in 10 mM Tris, pH 7.4), (c) in presence of 20% human plasma (0.15 M NaCl, 10 mM Tris, pH 7.4), (d) in presence of 50% human plasma (0.15 M NaCl, 10 mM Tris, pH 7.4), and (e) in presence of 50% human serum (0.15 M NaCl, 10 mM Tris, pH 7.4). Most of the antifungal activities of the D5-derived peptides HKH20 and GKH17 were lost, whereas the activities of the modified variants of GKH17 peptides were retained in physiological salt and presence of human plasma or serum. Data represents the mean ± (SD) of triplicate determinations from a single experiment representative of at least two experiments.
Figure 2.
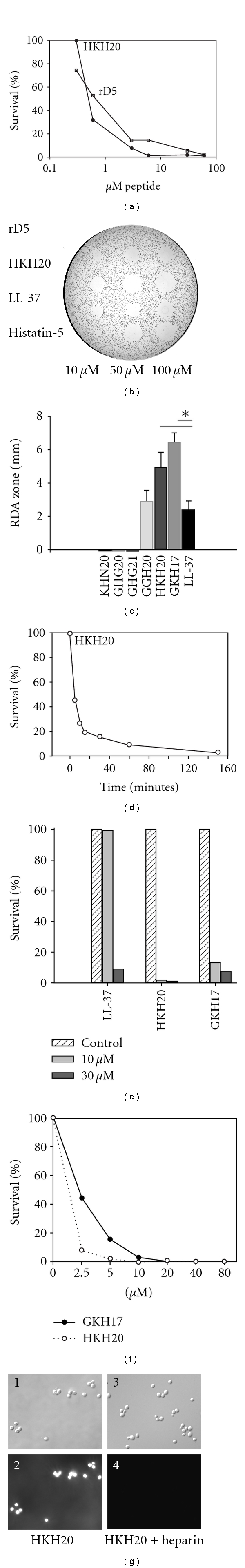
Antifungal activity of rD5 and the D5-derived peptide HKH20. (a) In viable count assay, antifungal activities of rD5 and HKH20 were detected. 1 ×106 cfu mL−1 of C. parapsilosis ATCC 90018 was incubated in 50 μL with rD5 or HKH20 at concentrations ranging from 0.3 μM to 60 μM. (b) C. parapsilosis ATCC 90018 was subjected to rD5, HKH20, and the reference peptides LL-37 and Histatin-5 in RDA, each well was loaded with 6 μl of peptide at the indicated concentrations. (c) The indicated D5-derived peptides (6 μL each at 100 μM) were tested in RDA in low salt conditions against C. albicans ATCC 90028. GGH20, HKH20, and GKH17 showed antifungal activity. For comparison, the reference peptide LL-37 is shown. * indicates a statistically significant difference (P < 0.05) in comparison to LL-37. (d) Time-dependent killing of C. parapsilosis ATCC 90018 by 30 μM HKH20 was analyzed by viable count assay. (e) M. furfur ATCC 44342 in a suspension of 1-2 × 105 cells/mL was added to buffer only (control, striped) or subjected to 10 μM (light gray) or 30 μM (dark gray) of the AMPs LL-37, HKH20, or GKH17 and incubated for 24 h. (f) M. furfur ATCC 44342 was subjected to increasing concentrations of GKH17 (filled circle) and HKH20 (open circle) and incubated for 24 h, and the percentage of survived yeast cells is shown. (g) Fluorescence microscopy shows binding of Texas Red-labeled HKH20 peptide (10 μM mL−1) to Candida cells. Panel 4 shows yeast cells incubated with heparin and Texas Red-labeled HKH20. Images in panel 2 and 4 were recorded using identical instrument settings. The corresponding Nomarski images are shown in panels 1 and 3. Data represents the mean of at least duplicate determinations from a single experiment representative of at least two experiments.
Radial diffusion assay (RDA) was performed as described by Lehrer et al. [39] with some minor modifications. Candida was grown for 16 h at 28°C in 10 mL YPD to obtain yeast form organisms. The culture was centrifuged at 2000 ×g for 10 min then washed three times with 10 mM Tris, pH 7.4. A volume containing 1 × 105 cfu was added to 5 mL of previously autoclaved, 42°C, underlay agarose gel that contained 1.5 mg of Tryptic Soy Broth (TSB) base, 50 mg of low-electroendosmosistype (Low-EEO) agarose type I (A-6013-25G) (Sigma-Aldrich Inc, St. Louis, Mo, USA), and 1 μL of Tween 20 (0.02% w/v) (Sigma Chemical Co.) dissolved in 10 mM Tris, pH 7.4. The fungi were directly dispensed into the agar and mixed with a vortex mixer for 10 seconds and then quickly poured out into a petri dish, 85 mm in diameter. Wells, 4 mm in diameter, were punched, and each well was then filled up with 6 μL of test sample. After incubation at 35–37°C for 3 h, 5 mL of overlay agarose gel, consisting of 300 mg TSB, 50 mg Low-EEO agarose dissolved in distilled sterilized water, was poured out on top of the underlay gel. The clear zone surrounding the wells, indicating antifungal activity, was measured after 18–24 h of incubation at 28°C. HMWK degradation products were tested for RDA inhibitory effects (Figure 1(b)). Human leukocyte (neutrophil) elastase (HLE) and freeze-thaw-disrupted polymorphonuclear neutrophils (PMNs) at equivalent concentrations as used in the HMWK degradation were incubated at 37°C for 60 minutes and added as controls (Figure 1(b)).
Malassezia furfur ATCC 44342 (CCUG 24230) was obtained from the Department of Bacteriology at Skåne University Hospital. The antifungal activity was tested in an assay described by Lopez-Gracia et al. [40], but with some minor modifications. Cells were cultured for 2 days at 37ºC in 10 mL medium containing 4% Bacto Malt Extract (Becton, Dickinson and Company, Md, USA), 1% glucose (VWR international), 1% Tween 80 (Sigma-Aldrich, St. Louis, Mo, USA) and 0.6% Bacto Peptone (Becton, Dickinson and Company, Md, USA). Cells were then washed in 1 mM sodium phosphate buffer, pH 7.4. M. furfur cells were grown in sterile 96-well microtiter plates (Costar, Corning Incorporated, Corning, USA) in a total volume of 100 μL, containing 60 μL of 1 mM sodium phosphate buffer, pH 7.4, 20 μL of culture medium, 10 μL of a 10x stock solution of the synthetic peptides, and a 10x stock solution of the fungal cell suspension, resulting in a final concentration of 1-2 × 105 cells/mL. Chloramphenicol was added to each well (16 μg/ml) to avoid contamination. The cells were incubated at 30°C for 24 h. To quantify the fungicidal activity, serial dilutions of the incubation mixture were plated onto agar plates and incubated 72 h at 37°C, and the number of cfu thereafter was determined by counting visible colonies. The minimal inhibitory concentration (MIC) was defined as the concentration which yielded at least 95% inhibition of fungal growth.
2.4. Fluorescence Microscopy
C. albicans ATCC 90028 was incubated for 18 hours in 10 mL YPD-broth at 28°C to obtain midlogarithmic phase organisms and washed three times, then diluted in 10 mM Tris, pH 7.4, to a concentration of 1 × 107 cfu/mL. Two hundred μL of the C. albicans suspension was incubated for 5 min on ice together with 1 μL Texas Red-conjugated HKH20 (2 mg/mL). Not only to block binding of HKH20 to the fungal membrane, but also to exclude the possibility that the Texas Red-label confers unspecific binding to fungal surfaces, 2 μL heparin (50 mg/mL) was added to the samples prior to incubation with Texas Red-conjugated HKH20. The pellets were washed three times in 10 mM Tris, pH 7.4. Cell FIX (Becton Dickinson Catalogue no. 340181) was added (to a final concentration of 4% formaldehyde), and the yeast cell pellets were incubated 15 min on ice and then 45 min at room temperature. Ethanol washed glass cover slips were coated with 0.25 mL of poly-lysine (0.2 mg/mL dissolved in water), dried, and then washed with distilled water. The samples were adhered to the polylysine glasses for 30 min and mounted on microscope slides for visual inspection. A Nikon Eclipse TE300 inverted fluorescence microscope was used.
2.5. Liposome Preparation and Leakage Assay
The liposomes investigated were zwitterionic DOPC/cholesterol (60/40 mol/mol) or DOPC/ergosterol (60/40 mol/mol). DOPC (1,2-Dioleoyl-sn-Glycero-3-Phosphatidylcholine) was from Avanti Polar Lipids (Alabaster, USA) and of >99% purity, while ergosterol and cholesterol (>99% purity) were from Sigma-Aldrich (St. Louis, USA). Due to the long, symmetric, and unsaturated acyl chains of DOPC, several methodological advantages are reached. In particular, membrane cohesion is good, which facilitates very stable unilamellar liposomes, allowing detailed values on leakage to be obtained. The lipid mixtures were dissolved in chloroform, after which solvent was removed by evaporation under vacuum overnight. Subsequently, 10 mM Tris buffer, pH 7.4, was added together with 0.1 M carboxyfluorescein (CF) (Sigma, St. Louis, USA). After hydration, the lipid mixture was subjected to eight freeze-thaw cycles consisting of freezing in liquid nitrogen and heated to 60°C. Unilamellar liposomes of about Ø140 nm were generated by multiple extrusions through polycarbonate filters (pore size 100 nm) mounted in a LipoFast miniextruder (Avestin, Ottawa, Canada) at 22°C. Untrapped CF was removed by two subsequent gel filtrations (Sephadex G-50, GE Healthcare, Uppsala, Sweden) at 22°C, with Tris buffer as eluent. CF release from the liposomes was determined by monitoring the emitted fluorescence at 520 nm from a liposome dispersion (10 μM lipid in 10 mM Tris, 5 mM glucose, pH 7.4). An absolute leakage scale was obtained by disrupting the liposomes at the end of each experiment through addition of 0.8 mM Triton X-100 (Sigma-Aldrich, St. Louis, USA). A SPEX-fluorolog 1650 0.22-m double spectrometer (SPEX Industries, Edison, USA) was used for the liposome leakage assay. Measurements were performed in at least duplicate at 37°C.
2.6. Statistical Analysis
Comparison of antifungal activity between the various peptides was performed using Kruskal-Wallis One-Way Analysis of Variance on Ranks and pair-ways multiple-comparison procedures (Tukey Test). P < 0.05 was considered as a significant difference. The statistical software used was SigmaStat (Systat Software Inc., Point Richmond, Calif, USA).
3. Results
3.1. Proteolytic Cleavage of HMWK Generates Peptides with Antifungal Properties
In order to test whether antifungal peptides could be released after proteolytic digestion of HMWK in vivo, HMWK was incubated with neutrophil elastase or extracts of polymorphonuclear neutrophils. Analysis using SDS-PAGE revealed that HMWK was degraded into several fragments (Figure 1(a)). Furthermore, when the fragments were analysed in RDA, antifungal activity was observed (Figure 1(b)). In analogy to the above, HMWK incubated with Cathepsin G resulted in the generation of low molecular-weight fragments corresponding to the previously detected 20-mer peptide from D5, HKH20, as assessed by Western blot using polyclonal antibodies against HKH20 (Figure 1(c)).
3.2. Antifungal Effects of rD5 and the D5-Derived Peptide HKH20 from HMWK
To investigate whether domain 5 has antifungal activity, effects of purified rD5 on C. parapsilosis ATCC 90018 were tested in a fungicidal assay (concentrations ranging from 0.3 μM to 60 μM) and in RDA. The results showed a dose-dependent killing of Candida (Figure 2(a)), and, notably, rD5 exerted inhibitory activity on Candida in RDA similar to the reference peptides LL-37 and histatin-5 (Figure 2(b)). In order to determine possible antifungal peptides of the rD5, overlapping peptides spanning the D5-region were synthesized (Table 1). When HKH20 and GKH17 were tested in RDA, a significantly more potent inhibitory activity on Candida, in comparison to LL-37, was observed (Figure 2(c)). As shown, HKH20 possessed potent antifungal activity (Figure 2(a)), and >80% of the Candida cells were killed by 30 μM HKH20 within 15 minutes (Figure 2(d)). In RDA, HKH20 displayed significantly higher inhibitory activity on Candida, when compared with LL-37 (Figures 2(b) and 2(c)). Finally, M. furfur was tested in an antifungal assay. The yeast cells were subjected to 10 μM or 30 μM of LL-37, HKH20, or GKH17, and the results showed that particularly HKH20 and GKH17 were antifungal at 10 and 30 μM (Figure 2(e)). MIC values, defined as the concentration yielding at least 95% inhibition of fungal growth, for HKH20 and GKH17 were determined to be between 2.5–5 μM and 5–10 μM, respectively (Figure 2(f)).
3.3. The D5-Derived Peptide HKH20 Binds to the Surface of Candida
To elucidate whether the peptide HKH20 interacts with the fungal cell wall of Candida, HKH20, labeled with the fluorescent dye Texas Red, was incubated with the fungal cells. As demonstrated by fluorescence microscopy, the peptide bound to the surface of Candida and the binding were completely blocked by heparin, an anionic glycosaminoglycan previously shown to block the antibacterial activity of HKH20 [33] (Figure 2(g)).
3.4. Antifungal Effects of D5-Derived Peptides and Structural Modifications for an Optimized Effect at Physiological Conditions
Hydrophobic tagging with W (Trp) amino acid residues has previously been demonstrated to increase antibacterial activity against various pathogens, such as S. aureus and P. aeruginosa. Particularly, for short, highly positively charged, and hydrophilic peptides, this facilitates the design of potent AMPs that selectively bind microorganisms over human cells [41–43]. Thus, the GKH17 peptide was tagged with 3–5 W residues at the C-terminal and analysed for antifungal activity. As shown, the hydrophobic modification resulted in increased antifungal activity, which was observed when not only in low salt conditions, but also tested in physiological salt solution (0.15 M NaCl) as well as in the presence of human plasma and serum (20% and 50%) (Figure 3).
3.5. Membrane-Disruptive Potency of GKH17 and End-Tagged Variants
The membrane-disruptive potency for GKH17 and the end-tagged variant GKH17WWW were demonstrated to be higher for ergosterol-containing liposomes than for cholesterol-containing ones (Figure 4). This is in line with the membrane-condensing effect of cholesterol [44–48], which opposes membrane insertion of peptides, and the relatively smaller membrane condensation caused by ergosterol. Given this difference between cholesterol and ergosterol, identification of peptides which are selective for ergosterol-containing membranes may represent a means to achieve simultaneously antifungal and low-toxic therapeutics.
Figure 4.
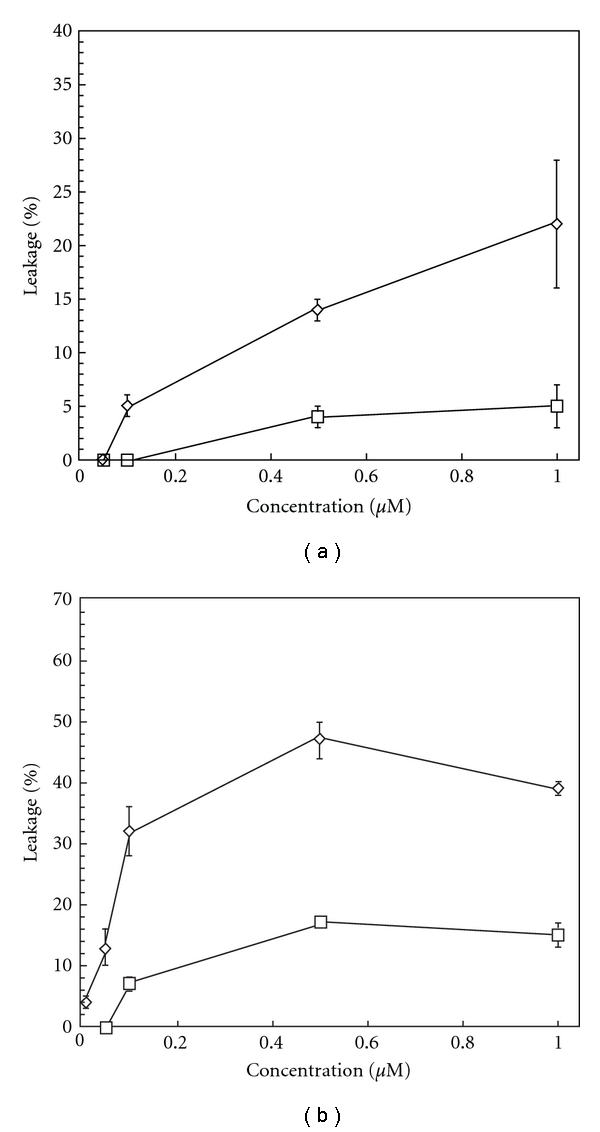
Effects of GKH17 (a) and GKH17WWW (b) on liposome permeability. For both peptides (at the doses indicated), permeabilization of ergosterol-containing liposomes (open circle) is demonstrated to be higher than that of cholesterol-containing (open square) ones.
4. Discussion
The D5 domain of HMWK is multifunctional and may participate in cell binding [49], inhibit angiogenesis, proliferation, and migration of endothelial cells [50, 51], and act as an antibacterial agent [33]. The main result in the present study is that cleavage products of HMWK also act as antifungal peptides, paralleled by results showing that the D5 domain of HMWK and related amino-acid sequences exert an antifungal activity comparable with the well-known AMPs LL-37 and histatin 5. Innate immune responses, including small cationic peptides with antimicrobial activities, constitute the first line of defense and are crucial for rapid clearing of microbial invaders. Numerous antimicrobial and antifungal peptides have been identified to date, most of them acting by membrane disruptive mechanisms. As mentioned briefly in the Introduction, biological effects exerted by AMPs include growth stimulus and angiogenesis, protease inhibition, antiangiogenesis, and chemotaxis [52–54]. Conversely, cationic peptide motifs from proteins not previously considered as AMPs have been shown to exert antimicrobial activities. For example, complement C3 [55, 56], kininogen [34, 57], heparin-binding protein [58], heparin-binding epidermal growth factor and other growth factors [59], matrix proteins such as laminin, fibronectin and proline arginine-rich and leucine-rich repeat protein (PRELP) [60], prions [61], β2-glycoprotein [62], histidine-rich glycoprotein [10], thrombin [63], and tissue factor pathway inhibitor [64] may act either as holoproteins or smaller peptide derivatives or fragments thereof and also exert antimicrobial activities in vitro, and, in several cases, in vivo [10, 34, 57]. In general, these findings are compatible with the observation that consensus heparin-binding peptide sequences (Cardin and Weintraub motifs) XBBBXXBX or XBBXBX (where X represents hydrophobic or uncharged amino acids, and B represents basic amino acids), represented by multiples of the motifs ARKKAAKA or AKKARA [65], are antibacterial [20] and specifically interact with membranes [66].
The findings in this study therefore further underline the multifunctionality of AMPs and illustrate that the glycine- and histidine-rich part of D5, previously shown to interact with heparin, may bind to negatively charged fungal membranes, such as those of Candida and Malassezia [67–70]. It is also notable that HKH20, derived from the lysine- and histidine-rich region of D5, which comprises consensus sequences for heparin binding [20, 65], binds to the membrane of Candida, likely by interacting with negatively charged molecules at the fungal cell wall [71]. Interestingly, HKH20 resembles the H-and K-rich AMP histatin-5 (Table 1), an AMP known to mediate fungal killing. It is also worth noting that the light chain of HMWK is essential for the interaction of kininogen with the cell wall of Candida [32].
As previously demonstrated, end-tagging by hydrophobic amino acid stretches is a facile and flexible approach of general applicability, by which AMP boosting, particularly of linear cationic sequences, is obtained [72, 73]. The data demonstrating that the hydrophobically modified GKH17 peptides show enhanced activity at physiological conditions, and in presence of human plasma and serum are therefore well in line with these previous data, suggesting that increased hydrophobicity yields increased affinity for fungal membranes. Indeed, these data parallel the observations on liposomes, demonstrating specificity for ergosterol (fungi-like) liposomes compared with cholesterol (mammalian cell mimicking) liposomes.
In previous reports, it was demonstrated that fragments of HMWK are generated upon degradation with neutrophil elastase as well as bacterial enzymes [74]. The observation in the present study extend these previous findings and demonstrate that cathepsin G, a major neutrophilic enzyme, may also degrade HMWK, leading to release of AMPs (Figure 1(c)). Considering these data as well as the finding that PMNs may degrade HMWK and generate antifungal activity (Figure 1(b)), it is tempting to speculate that during a cutaneous Candida infection, neutrophils, recruited to the site of infection [75, 76], may upon activation release cathepsin G from azurophilic granules, leading to degradation of HMWK, present in blood or connective tissues. The cleavage products of HMWK can then act as antifungal AMPs and inhibit fungal cells at the site of infection. However, further investigations are clearly mandatory in order to further define the generation of D5 peptides in response to fungal infection.
In conclusion, we have shown that proteolysis of HMWK generates fragments with antifungal activities. Amino-acid sequences corresponding to D5-derived peptides exert antifungal effects, and, furthermore, the antifungal activity may be enhanced by tagging with hydrophobic amino-acid residues.
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Swedish Government Support for Clinical Research (ALF), Capio research foundation, Welander and Finsen Research Foundations, and the Royal Physiographic Society. Lise-Britt Wahlberg is gratefully acknowledged for technical support.
References
- 1.Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nature Reviews Microbiology. 2008;6(1):67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Goodarzi H, Trowbridge J, Gallo RL. Innate immunity: a cutaneous perspective. Clinical Reviews in Allergy and Immunology. 2007;33(1-2):15–26. doi: 10.1007/s12016-007-0037-4. [DOI] [PubMed] [Google Scholar]
- 4.Drake DR, Brogden KA, Dawson DV, Wertz PW. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. Journal of Lipid Research. 2008;49(1):4–11. doi: 10.1194/jlr.R700016-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nature Immunology. 2005;6(6):551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 6.Frohm M, Agerberth B, Ahangari G, et al. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. Journal of Biological Chemistry. 1997;272(24):15258–15263. doi: 10.1074/jbc.272.24.15258. [DOI] [PubMed] [Google Scholar]
- 7.Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. Journal of Leukocyte Biology. 2004;75(1):39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- 8.Harder J, Schröder JM. Psoriatic scales: a promising source for the isolation of human skin-derived antimicrobial proteins. Journal of Leukocyte Biology. 2005;77(4):476–486. doi: 10.1189/jlb.0704409. [DOI] [PubMed] [Google Scholar]
- 9.Dürr UHN, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochimica et Biophysica Acta. 2006;1758(9):1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Rydengård V, Shannon O, Lundqvist K, et al. Histidine-rich glycoprotein protects from systemic Candida infection. PLoS Pathogens. 2008;4(8) doi: 10.1371/journal.ppat.1000116. Article ID e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dale BA, Fredericks LP. Antimicrobial peptides in the oral environment: expression and function in health and disease. Current Issues in Molecular Biology. 2005;7(2):119–133. doi: 10.1093/jac/dki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clinical Microbiology Reviews. 2006;19(3):491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theis T, Stahl U. Antifungal proteins: targets, mechanisms and prospective applications. Cellular and Molecular Life Sciences. 2004;61(4):437–455. doi: 10.1007/s00018-003-3231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papo N, Shai Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides. 2003;24(11):1693–1703. doi: 10.1016/j.peptides.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Yount NY, Bayer AS, Xiong YQ, Yeaman MR. Advances in antimicrobial peptide immunobiology. Biopolymers. 2006;84(5):435–458. doi: 10.1002/bip.20543. [DOI] [PubMed] [Google Scholar]
- 16.Pasupuleti M, Walse B, Svensson B, Malmsten M, Schmidtchen A. Rational design of antimicrobial C3a analogues with enhanced effects against staphylococci using an integrated structure and function-based approach. Biochemistry. 2008;47(35):9057–9070. doi: 10.1021/bi800991e. [DOI] [PubMed] [Google Scholar]
- 17.Bechinger B. A dynamic view of peptides and proteins in membranes. Cellular and Molecular Life Sciences. 2008;65(19):3028–3039. doi: 10.1007/s00018-008-8125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gennaro R, Zanetti M. Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers. 2000;55(1):31–49. doi: 10.1002/1097-0282(2000)55:1<31::AID-BIP40>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 19.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 20.Andersson E, Rydengård V, Sonesson A, Mörgelin M, Björck L, Schmidtchen A. Antimicrobial activities of heparin-binding peptides. European Journal of Biochemistry. 2004;271(6):1219–1226. doi: 10.1111/j.1432-1033.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 21.Baranska W, Sonesson A, Nowicki R, Schmidtchen A. Glycosaminoglycans inhibit the antibacterial activity of LL-37 in biological fluids. Journal of Antimicrobial Chemotherapy. 2006;57(2):260–265. doi: 10.1093/jac/dki460. [DOI] [PubMed] [Google Scholar]
- 22.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nature Reviews Microbiology. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 23.Strömstedt AA, Ringstad L, Schmidtchen A, Malmsten M. Interaction between amphiphilic peptides and phospholipid membranes. Current Opinion in Colloid and Interface Science. 2010;15(6):467–478. [Google Scholar]
- 24.Gomez-Lopez A, Zaragoza O, Rodriguez-Tudela JL, Cuenca-Estrella M. Pharmacotherapy of yeast infections. Expert Opinion on Pharmacotherapy. 2008;9(16):2801–2816. doi: 10.1517/14656566.9.16.2801. [DOI] [PubMed] [Google Scholar]
- 25.Odds FC. Candida and Candidosis. London, UK: Bailliére Tindall; 1988. [Google Scholar]
- 26.Savolainen J, Lammintausta K, Kalimo K, Viander M. Candida albicans and atopic dermatitis. Clinical and Experimental Allergy. 1993;23(4):332–339. doi: 10.1111/j.1365-2222.1993.tb00331.x. [DOI] [PubMed] [Google Scholar]
- 27.Trick WE, Fridkin SK, Edwards JR, Hajjeh RA, Gaynes RP. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clinical Infectious Diseases. 2002;35(5):627–630. doi: 10.1086/342300. [DOI] [PubMed] [Google Scholar]
- 28.Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL., Jr. Skin diseases associated with Malassezia species. Journal of the American Academy of Dermatology. 2004;51(5):785–798. doi: 10.1016/j.jaad.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 29.Devlin RK. Invasive fungal infections caused by Candida and Malassezia species in the neonatal intensive care unit. Advances in Neonatal Care. 2006;6(2):68–77. doi: 10.1016/j.adnc.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Ben Nasr A, Herwald H, Muller-Esterl W, Bjorck L. Human kininogens interact with M protein, a bacterial surface protein and virulence determinant. Biochemical Journal. 1995;305:173–180. doi: 10.1042/bj3050173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herwald H, Mörgelin M, Olsen A, et al. Activation of the contact-phase system on bacterial surfaces—a clue to serious complications in infectious diseases. Nature Medicine. 1998;4:298–302. doi: 10.1038/nm0398-298. [DOI] [PubMed] [Google Scholar]
- 32.Rapala-Kozik M, Karkowska J, Jacher A, et al. Kininogen adsorption to the cell surface of Candida spp. International Immunopharmacology. 2008;8(2):237–241. doi: 10.1016/j.intimp.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Nordahl EA, Rydengård V, Mörgelin M, Schmidtchen A. Domain 5 of high molecular weight kininogen is antibacterial. Journal of Biological Chemistry. 2005;280(41):34832–34839. doi: 10.1074/jbc.M507249200. [DOI] [PubMed] [Google Scholar]
- 34.Frick IM, Åkesson P, Herwald H, et al. The contact system—a novel branch of innate immunity generating antibacterial peptides. EMBO Journal. 2006;25(23):5569–5578. doi: 10.1038/sj.emboj.7601422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan AP, Ghebrehiwet B. The plasma bradykinin-forming pathways and its interrelationships with complement. Molecular Immunology. 2010;47(13):2161–2169. doi: 10.1016/j.molimm.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Kozik A, Moore RB, Potempa J, Imamura T, Rapala-Kozik M, Travis J. A novel mechanism for bradykinin production at inflammatory sites. Diverse effects of a mixture of neutrophil elastase and mast cell tryptase versus tissue and plasma kallikreins on native and oxidized kininogens. Journal of Biological Chemistry. 1998;273(50):33224–33229. doi: 10.1074/jbc.273.50.33224. [DOI] [PubMed] [Google Scholar]
- 37.Maruo K, Akaike T, Inada Y, Ohkubo I, Ono T, Maeda H. Effect of microbial and mite proteases on low and high molecular weight kininogens. Generation of kinin and inactivation of thiol protease inhibitory activity. Journal of Biological Chemistry. 1993;268(24):17711–17715. [PubMed] [Google Scholar]
- 38.Herwald H, Mörgelin M, Svensson HG, Sjöbring U. Zinc-dependent conformational changes in domain D5 of high molecular mass kininogen modulate contact activation. European Journal of Biochemistry. 2001;268(2):396–404. doi: 10.1046/j.1432-1033.2001.01888.x. [DOI] [PubMed] [Google Scholar]
- 39.Lehrer RI, Rosenman M, Harwig SS, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. Journal of Immunological Methods. 1991;137(2):167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Garcia B, Lee PH, Gallo RL. Expression and potential function of cathelicidin antimicrobial peptides in dermatophytosis and tinea versicolor. Journal of Antimicrobial Chemotherapy. 2006;57(5):877–882. doi: 10.1093/jac/dkl078. [DOI] [PubMed] [Google Scholar]
- 41.Avrahami D, Shai Y. Conjugation of a magainin analogue with lipophilic acids controls hydrophobicity, solution assembly, and cell selectivity. Biochemistry. 2002;41(7):2254–2263. doi: 10.1021/bi011549t. [DOI] [PubMed] [Google Scholar]
- 42.Murillo LA, Lan CY, Agabian NM, Larios S, Lomonte B. Fungicidal activity of a phospholipase A2-derived synthetic peptide variant against Candida albicans. Revista Espanola de Quimioterapia. 2007;20(3):330–333. [PubMed] [Google Scholar]
- 43.Schmidtchen A, Ringstad L, Kasetty G, Mizuno H, Rutland MW, Malmsten M. Membrane selectivity by W-tagging of antimicrobial peptides. Biochimica et Biophysica Acta. 2011;1808(4):1081–1091. doi: 10.1016/j.bbamem.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 44.Mouritsen OG, Zuckermann MJ. What’s so special about cholesterol? Lipids. 2004;39(11):1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 45.Henriksen J, Rowat AC, Brief E, et al. Universal behavior of membranes with sterols. Biophysical Journal. 2006;90(5):1639–1649. doi: 10.1529/biophysj.105.067652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azouzi S, El Kirat K, Morandat S. The potent antimalarial drug cyclosporin a preferentially destabilizes sphingomyelin-rich membranes. Langmuir. 2010;26(3):1960–1965. doi: 10.1021/la902580w. [DOI] [PubMed] [Google Scholar]
- 47.Sood R, Domanov Y, Pietiainen M, Kontinen VP, Kinnunen PK. Binding of LL-37 to model biomembranes: insight into target vs host cell recognition. Biochimica et Biophysica Acta. 2008;1778(4):983–996. doi: 10.1016/j.bbamem.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 48.Ohvo-Rekila H, Ramstedt B, Leppimaki P, Slotte JP. Cholesterol interactions with phospholipids in membranes. Progress in Lipid Research. 2002;41(1):66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
- 49.Hasan AA, Cines DB, Herwald H, Schmaier AH, Muller-Esterl W. Mapping the cell binding site on high molecular weight kininogen domain 5. Journal of Biological Chemistry. 1995;270(33):19256–19261. doi: 10.1074/jbc.270.33.19256. [DOI] [PubMed] [Google Scholar]
- 50.Zhang JC, Claffey K, Sakthivel R, et al. Two-chain high molecular weight kininogen induces endothelial cell apoptosis and inhibits angiogenesis: partial activity within domain 5. FASEB Journal. 2000;14(15):2589–2600. doi: 10.1096/fj.99-1025com. [DOI] [PubMed] [Google Scholar]
- 51.Colman RW. Role of the light chain of high molecular weight kininogen in adhesion, cell-associated proteolysis and angiogenesis. Biological Chemistry. 2001;382(1):65–70. doi: 10.1515/BC.2001.011. [DOI] [PubMed] [Google Scholar]
- 52.Beisswenger C, Bals R. Functions of antimicrobial peptides in host defense and immunity. Current Protein and Peptide Science. 2005;6(3):255–264. doi: 10.2174/1389203054065428. [DOI] [PubMed] [Google Scholar]
- 53.Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annual Review of Immunology. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- 54.Elsbach P. What is the real role of antimicrobial polypeptides that can mediate several other inflammatory responses? Journal of Clinical Investigation. 2003;111(11):1643–1645. doi: 10.1172/JCI18761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nordahl EA, Rydengård V, Nyberg P, et al. Activation of the complement system generates antibacterial peptides. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(48):16879–16884. doi: 10.1073/pnas.0406678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonesson A, Ringstad L, Nordahl EA, Malmsten M, Mörgelin M, Schmidtchen A. Antifungal activity of C3a and C3a-derived peptides against Candida. Biochimica et Biophysica Acta. 2007;1768(2):346–353. doi: 10.1016/j.bbamem.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 57.Nordahl EA, Rydengård V, Mörgelin M, Schmidtchen A. Domain 5 of high molecular weight kininogen is antibacterial. Journal of Biological Chemistry. 2005;280(41):34832–34839. doi: 10.1074/jbc.M507249200. [DOI] [PubMed] [Google Scholar]
- 58.Pereira HA. CAP37, a neutrophil-derived multifunctional inflammatory mediator. Journal of Leukocyte Biology. 1995;57(6):805–812. doi: 10.1002/jlb.57.6.805. [DOI] [PubMed] [Google Scholar]
- 59.Malmsten M, Davoudi M, Walse B, et al. Antimicrobial peptides derived from growth factors. Growth Factors. 2007;25(1):60–70. doi: 10.1080/08977190701344120. [DOI] [PubMed] [Google Scholar]
- 60.Malmsten M, Davoudi M, Schmidtchen A. Bacterial killing by heparin-binding peptides from PRELP and thrombospondin. Matrix Biology. 2006;25(5):294–300. doi: 10.1016/j.matbio.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Pasupuleti M, Roupe M, Rydengaård V, et al. Antimicrobial activity of human prion protein is mediated by its N-terminal region. PLoS ONE. 2009;4(10) doi: 10.1371/journal.pone.0007358. Article ID e7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsson M, Wasylik S, Mörgelin M, et al. The antibacterial activity of peptides derived from human β-2 glycoprotein I is inhibited by protein H and M1 protein from Streptococcus pyogenes. Molecular Microbiology. 2008;67(3):482–492. doi: 10.1111/j.1365-2958.2007.05974.x. [DOI] [PubMed] [Google Scholar]
- 63.Papareddy P, Rydengård V, Pasupuleti M, et al. Proteolysis of human thrombin generates novel host defense peptides. PLoS Pathogens. 2010;6(4) doi: 10.1371/journal.ppat.1000857. Article ID e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papareddy P, Kalle M, Kasetty G, et al. C-terminal peptides of tissue factor pathway inhibitor are novel host defense molecules. Journal of Biological Chemistry. 2010;285(36):28387–28398. doi: 10.1074/jbc.M110.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9(1):21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 66.Ringstad L, Schmidtchen A, Malmsten M. Effect of peptide length on the interaction between consensus peptides and DOPC/DOPA bilayers. Langmuir. 2006;22(11):5042–5050. doi: 10.1021/la060317y. [DOI] [PubMed] [Google Scholar]
- 67.Cadena RAD, Colman RW. The sequence HGLGHGHEQQHGLGHGH in the light chain of high molecular weight kininogen serves as a primary structural feature for zinc-dependent binding to an anionic surface. Protein Science. 1992;1(1):151–160. doi: 10.1002/pro.5560010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Björk I, Olson ST, Sheffer RG, Shore JD. Binding of heparin to human high molecular weight kininogen. Biochemistry. 1989;28(3):1213–1221. doi: 10.1021/bi00429a039. [DOI] [PubMed] [Google Scholar]
- 69.Pixley RA, Lin Y, Isordia-Salas I, Colman RW. Fine mapping of the sequences in domain 5 of high molecular weight kininogen (HK) interacting with heparin and zinc. Journal of Thrombosis and Haemostasis. 2003;1(8):1791–1798. doi: 10.1046/j.1538-7836.2003.00291.x. [DOI] [PubMed] [Google Scholar]
- 70.Kunapuli SP, Cadena RAD, Colman RW. Deletion mutagenesis of high molecular weight kininogen light chain. Identification of two anionic surface binding subdomains. Journal of Biological Chemistry. 1993;268(4):2486–2492. [PubMed] [Google Scholar]
- 71.Ringstad L, Kacprzyk L, Schmidtchen A, Malmsten M. Effects of topology, length, and charge on the activity of a kininogen-derived peptide on lipid membranes and bacteria. Biochimica et Biophysica Acta. 2007;1768(3):715–727. doi: 10.1016/j.bbamem.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 72.Schmidtchen A, Pasupuleti M, Mörgelin M, et al. Boosting antimicrobial peptides by hydrophobic oligopeptide end tags. Journal of Biological Chemistry. 2009;284(26):17584–17594. doi: 10.1074/jbc.M109.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pasupuleti M, Schmidtchen A, Chalupka A, Ringstad L, Malmsten M. End-tagging of ultra-short antimicrobial peptides by W/F stretches to facilitate bacterial killing. PLoS ONE. 2009;4(4) doi: 10.1371/journal.pone.0005285. Article ID e5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stuardo M, Gonzalez CB, Nualart F, et al. Stimulated human neutrophils form biologically active kinin peptides from high and low molecular weight kininogens. Journal of Leukocyte Biology. 2004;75(4):631–640. doi: 10.1189/jlb.1103546. [DOI] [PubMed] [Google Scholar]
- 75.Odds FC. Pathogenesis of Candida infections. Journal of the American Academy of Dermatology. 1994;31(3):S2–S5. doi: 10.1016/s0190-9622(08)81257-1. [DOI] [PubMed] [Google Scholar]
- 76.Ray TL, Wuepper KD. Activation of the alternative (properdin) pathway of complement by Candida albicans and related species. Journal of Investigative Dermatology. 1976;67(6):700–703. doi: 10.1111/1523-1747.ep12598581. [DOI] [PubMed] [Google Scholar]


