Abstract
In this paper, we propose a novel method that combines PubMed knowledge and Electronic Health Records to develop a weighted Bayesian Network Inference (BNI) model for pancreatic cancer prediction. We selected 20 common risk factors associated with pancreatic cancer and used PubMed knowledge to weigh the risk factors. A keyword-based algorithm was developed to extract and classify PubMed abstracts into three categories that represented positive, negative, or neutral associations between each risk factor and pancreatic cancer. Then we designed a weighted BNI model by adding the normalized weights into a conventional BNI model. We used this model to extract the EHR values for patients with or without pancreatic cancer, which then enabled us to calculate the prior probabilities for the 20 risk factors in the BNI. The software iDiagnosis was designed to use this weighted BNI model for predicting pancreatic cancer. In an evaluation using a case-control dataset, the weighted BNI model significantly outperformed the conventional BNI and two other classifiers (k-Nearest Neighbor and Support Vector Machine). We conclude that the weighted BNI using PubMed knowledge and EHR data shows remarkable accuracy improvement over existing representative methods for pancreatic cancer prediction.
Keywords: Electronic Health Records, Pancreatic Neoplasms, Text Mining, Bayesian Method
1. Introduction
Every year, many people die of “silent killers”, those fatal diseases that are hard to diagnose and treat. Pancreatic cancer is one such disease. Early diagnosis is crucial to its successful treatment. In addition to searching for effective biomarkers [1-2] which can aid in early diagnosis, researchers have developed models to support disease risk prediction [3-4]. The Bayesian Network Inference (BNI) model, which uses Bayes’ theorem and represents probabilistic dependencies between disease-associated risk factors as a directed acyclic graph [5-6], has been a popular disease risk prediction model [7-8], especially for predicting breast cancer [9-17] and pancreatic cancer [18]. Several factors make the BNI model a better choice than other methods for disease risk prediction. First, whereas other classification methods, such as the k-Nearest Neighbor (KNN) and Support Vector Machine (SVM) methods, excel primarily in a high-dimensional feature space, the BNI model performs well in a low-dimensional feature space. Second, the BNI model can represent the joint probability distribution over interrelated hypotheses about disease risk factors using network topology, but other classification models cannot represent or use this valuable information. Third, heterogeneous or random variables can be combined to make predictions in the BNI model, while other classifiers often require variables of the same type. A recent advance in BNI modeling is weighted model counting, which uses a propositional knowledge base for improving prediction accuracy [19], although this method has rarely been used in clinical decision support.
In this paper, we propose a novel extension to the conventional BNI by combining text mining of PubMed knowledge with secondary use of clinical data from Electronic Health Records (EHR) to develop a weighted BNI model. We used PubMed, because as a rich public knowledge base, it contains official evidence of the associations between risk factors and diseases. We developed a text mining-based method which allows us to statistically weigh each of these associations. We make use of EHR clinical date because, with the expanding adoption of EHR worldwide, the rich clinical data they provide serves as additional practical evidence for disease modeling. We hypothesize that by combining PubMed and EHR, we can calculate the prior probabilities of a weighted BNI model for disease risk prediction. Next we present the design of such a weighted BNI model and its evaluation results. Note that while we used pancreatic cancer as a sample disease in our initial study, the method should generalize to other diseases.
2. Data Sources and Methods
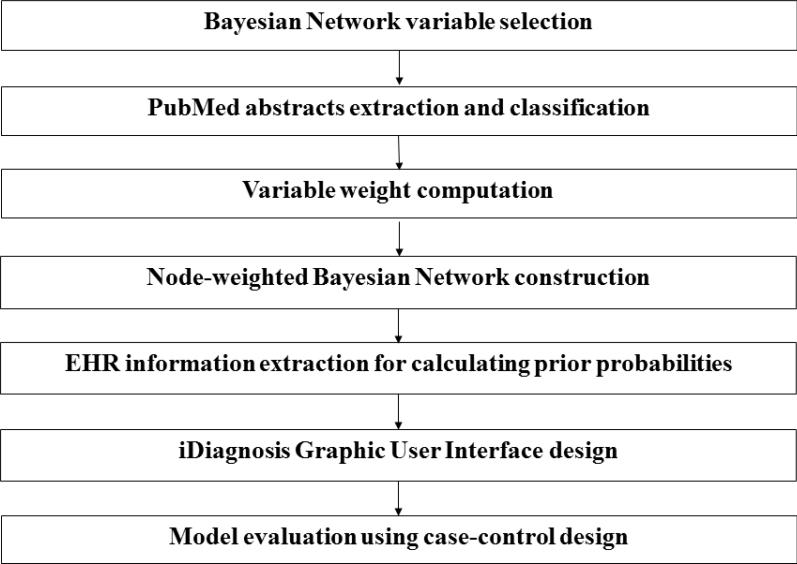
Figure 1 shows our process for developing a weighted BNI model for pancreatic cancer prediction. It consists of seven steps: (1) disease variable selection; (2) PubMed abstract mining and classification; (3) variable weight computation; (4) weighted Bayesian Network topology design; (5) EHR data extraction for prior probability calculation; (6) iDiagnosis Graphic User Interface (GUI) design; and (7) model evaluation.
Figure 1.
Steps to construct a node-weighted BNI
2.1 Variable Selection
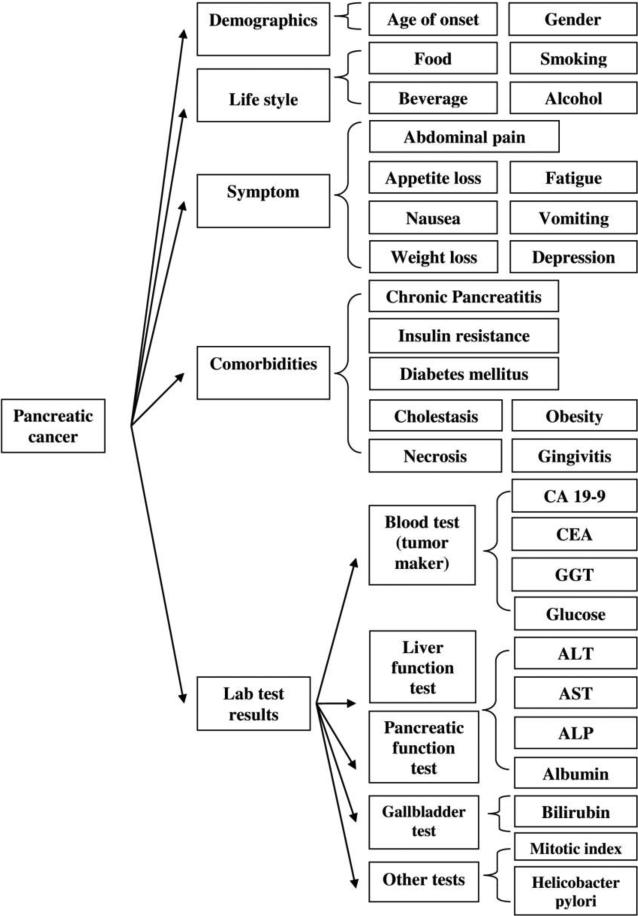
We identified 31 variables associated with pancreatic cancer by aggregating the results from a PubMed review, the recommendations by clinical experts on pancreatic cancer in our institution, and the risk factors associated with pancreatic cancer we had previously identified [20-21]. Figure 2 shows the class hierarchy of the risk factors, which fall into five categories: demographics, life style, symptoms, co-morbidities, and lab test results.
Figure 2.
Class hierarchy of pancreatic cancer variables
Since knowledge representation always involves making tradeoffs between tractability and expressiveness [22], the more variables used, the more complex the BNI model inference process. To curb complexity and improve efficiency for the BNI model, we considered two issues when selecting and aggregating a subset of these variables to construct the weighted BNI model: (1) the availability and quality of the information in EHR; for example, information about food intake is generally inaccessible or incomplete in EHR and hence is excluded; and (2) the importance of a variable according to the frequency of it being discussed in PubMed and the recommendation of clinical experts. In addition, to simplify the BNI model design, similar variables were manually grouped into one. For example, variable “alcohol abuse” and “cigarette abuse” were grouped into one variable “alcohol or cigarette abuse” since they were both “substance abuse”. Similarly, the variables “fatigue” and “asthenia” were grouped into one variable “fatigue or asthenia” since they are semantically similar or related symptoms.
Based on the above considerations, we identified the following 20 variables to design our BNI models: age, alcohol or cigarette abuse, abdominal pain, fatigue or asthenia, nausea, vomiting, weight loss, depression, appetite loss, diabetes mellitus, jaundice, carbohydrate antigen 19-9 (CA 19-9), carcinoembryonic antigen (CEA), gamma-glutamyl transferase (GGT), glucose, alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), albumin and bilirubin. All these variables are available in EHR, although half of them are in narrative format in free-text notes.
2.2 PubMed Knowledge Extraction and Processing
Many text-mining algorithms have already been developed to extract disease-related risk factors from PubMed. Learning from a popular rule-based method designed by Chen et al. to calculate associations among biological terms [23], we implemented a keyword-based method to automatically extract and classify PubMed abstracts that mentioned both any of the risk factors and pancreatic cancer together to calculate the weight of each risk factor. We implemented the program Entrez Programming Utilities (eUtils) [8], which provides direct access to the PubMed databases and supports terminology-based query term generation, information extraction and exportation from PubMed abstracts. Medical Subject Headings (MeSH) terms were used to generate query terms for each variable. When searching for information in PubMed, we used the eUtils ESearch tools. For each search, one MeSH term that represents a variable and one MeSH term representing pancreatic cancer were paired to issue a PubMed query searching for abstracts discussing this variable and pancreatic cancer together in the title. If the number of the retrieved abstracts was < 100, the query was expanded to search for the co-occurrence of the variable and pancreatic cancer in both the title and the abstract. If the number of retrieved citations was still < 100, the query was further expanded to search the full text. The classification accuracy of the included PubMed abstracts decreased as the search scope was expanded from the title to the full text since generally the co-occurrence of a variable and pancreatic cancer in the title indicates a stronger association than in the abstract. We used the eUtils Fetch tools to download all abstracts into a local MySQL database.
2.3 Weight Calculation
Each PubMed abstract was classified into one of the following three categories according to the association between the selected variable and pancreatic cancer: positive, negative, or neutral association. Each variable was assigned a set of keywords indicating the associations; details are shown in Appendix Table A.1. Keywords indicating a positive association typically include “risk”, “link”, “associated”, “association”, “influence” and hundreds of others. Keywords indicating a negative association typically includes “differentiation”, “comparison”, “discrimination”, “distinction”, “distinguish” and about a hundred more. Keywords such as “equal to”, “same to” and “sequential” were considered indicators of a neutral association. Abstracts that included co-occurring variables and pancreatic cancer without obvious associations were categorized as neutral associations. For example, a PubMed abstract may discuss biological molecules such as insulin receptor substrate-1 protein or insulin-like growth factor instead of the association between insulin and pancreatic cancer. To ensure high accuracy in the abstract classification phase, the machine classification results were further reviewed manually and corrected as appropriate, although the manual review was greatly enhanced by the text-mining algorithm since the sentences containing the keywords were automatically highlighted to ease the manual review.
For each risk factor, only the abstracts containing positive and negative associations between the risk factor and pancreatic cancer were used to calculate the original weights for each risk factor (woi). For variable Vi, Pi indicates the number of abstracts with positive associations, Ni denotes the number of abstracts with negative associations, and the original weight woi is the ratio between Pi and Ni, calculated as:
| (1.1) |
To avoid infinity, if Ni equals 0, woi is assigned the value 1. The rationale behind this design is that PubMed publications reflect collective evidence regarding the association generated by subject matter experts from all over the world and over time, who may disagree with one another; therefore, the ratio between Pi and Ni is a statistical summary of the collective evidence for the association between the risk factor and the disease. After the original weight woi is calculated, a procedure of normalization, wi, is defined as:
| (1.2) |
where woi is the original weight defined in Eq. (1.1), max(w1o, w2o ,.. w20o) is the maximum of the original weights woi, and wi is the normalized weight in the range [0, 1].
2.4 Node-weighted Bayesian Network Inference (BNI) Model Development
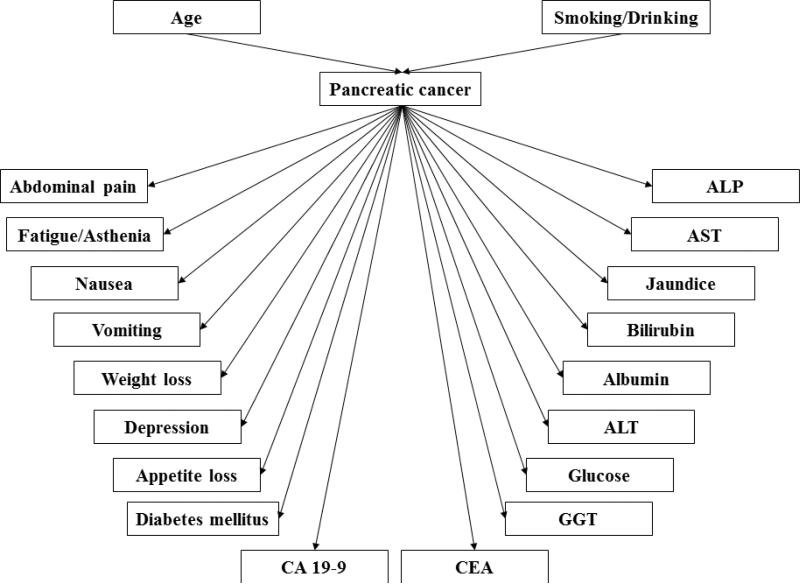
Figure 3 shows the topology of our weighted BNI model, which separates dependent variables (e.g., co-morbidities, symptoms, and lab test results that might be caused by pancreatic cancer) from independent variables (e.g., age, gender, smoking and alcohol abuse). Each risk factor is treated as a binary variable without considering the severity, degree, accumulative length, or other quantitative information of the risk factor. The value “true” represents the presence of a factor and the value “false” represents the absence of a risk factor. To integrate the normalized weights into the BNI model, we multiply the normalized weight, wi, of each variable to its corresponding prior probability P, as illustrated by the simple network in Figure 4 (a), where one node is pancreatic cancer and the other node is variable Vi. The function of the weighted prior probability is defined as:
| (2) |
where wi is the normalized weights in Eq. (1.2) with the range [0, 1]. Since the probability P(pancreatic cancer=true | Vi=true) is in the range [0. 1], the normalized weights are bounded in [0, 1], and the weighted prior probability Pw(Vi=true|pancreatic cancer=true) is in the range [0, 1]. The weighted posterior probability Pw(pancreatic cancer=true|Vi=true) can be calculated using the weighted prior probability Pw(Vi=true|pancreatic cancer=true) defined in Eq. (2).
Figure 3.
The topology of the Bayesian Network for predicting pancreatic cancer
Figure 4.
A simplified Bayesian Network for pancreatic cancer prediction: (a) two-node Bayesian Network (b) three-node Bayesian Network, where Vup is the set of all parent nodes of PC and Vdown is the set of all child nodes of pancreatic cancer.
Theorem One. The weighted posterior probability Pw(pancreatic cancer=true| Vi=true) is a probability function.
Proof. (PC is the abbreviation for Pancreatic Cancer hereafter.)
Given Bayes’ theorem and Eq. (2), the weighted posterior probability Pw(PC=true | Vi=true) can be calculated as:
| (3) |
Given Bayes’ theorem, the posterior probability P(PC=false | Vi=true) is:
| (4) |
Summarizing Eq. (3) and Eq. (4), we arrive at
Therefore, the posterior probability Pw(PC=true | Vi=true) is a probability function.
End proof.
Eq. (3) shows that for each variable Vi, the value of the posterior probability Pw(pancreatic cancer=true | Vi=true) depends on both the prior probability P(pancreatic cancer=true | Vi=true) and the normalized weight wi. If the variable and pancreatic cancer are positively associated, the normalized weight wi ≈ 1, and the weighted posterior probability Pw(pancreatic cancer=true | Vi=true) is approximately equal to the conventional posterior probability P(pancreatic cancer=true | Vi=true). This means that the posterior probability of pancreatic cancer increases if variable Vi is true. If the variable and pancreatic cancer are negatively associated, the normalized weight wi ≈ 0, and the posterior probability Pw(pancreatic cancer=true | Vi=true) is approximately equal to 0, which means that the posterior probability of pancreatic cancer barely increases.
Next we illustrate how wi can increase the BNI prediction accuracy for pancreatic cancer using a two-node Bayesian Network in Figure 4 (a). For example, if we use “vomiting” or “abdominal pain” to instantiate Vi to predict the risk of pancreatic cancer, according to the prior probabilities in Table 2, P(vomiting=true | pancreatic cancer=true) = 0.4592 and P(abdominal pain=true | pancreatic cancer=true) = 0.3980, respectively. To instantiate the model for illustration purposes only, we suppose the prior probability of pancreatic cancer is P(PC = true) = 10-3. According to the Bayes’ Theorem, using the two-node network in Figure 4 (a), we would arrive at similar posterior probabilities for the two risk factors: P(pancreatic cancer=true | vomiting = true) = 8.4924 × 10-4 and P(pancreatic cancer=true | abdominal pain = true) = 6.6135 × 10-4. However, looking at the calculated prior probabilities, one may infer that “vomiting” is associated with pancreatic cancer to the same degree as “abdominal pain”. However, the pure probability information is inconsistent with the existing medical knowledge that abdominal pain is more strongly associated with pancreatic cancer than vomiting. By using the normalized weights for pancreatic cancer risk factors in Table 1: wabdominal pain= 0.4828 or wvomiting =0.0172 and applying formula Eq. (2) and Eq. (3) to the two-node network in Figure 4(a), we obtain the weighted posterior probabilities Pw(pancreatic cancer=true | abdominal pain = true) = 6.6135 × 10-4 and Pw(pancreatic cancer=true | vomiting = true) = 1.4619 × 10-5. The former is significantly higher than the latter, which is consistent with our medical knowledge. Therefore, the weighted posterior probabilities are more realistic and consistent with prior knowledge about the risk factors and can correct possible errors introduced by pure statistics.
Table 2.
Calculated prior probabilities with the condition pancreatic cancer = true (data source: EHR, the 98-case dataset).
| Variable | Probability | Percent | Ranking |
|---|---|---|---|
| Glucose | P(Glucose=true|PC=true) | 0.8776 | 1 |
| Albumin | P(Albumin=true|PC=true) | 0.6536 | 2 |
| Nausea | P(Nausea = true|PC=true) | 0.5102 | 3 |
| Age >= 60 | P(Age>=60|PC=true) | 0.4592 | 4 |
| Vomiting | P(Vomiting=true|PC=true) | 0.4592 | 4 |
| Abdominal pain | P(Abdominal pain = true|PC=true) | 0.3980 | 6 |
| Weight loss | P(Weight loss=true|PC=true) | 0.3776 | 7 |
| Diabetes | P(Diabetes=true|PC=true) | 0.3367 | 8 |
| Smoking/Drinking | (UNION) P(Smoking or Alcohol = true|PC=true) | 0.3163 | 9 |
| Appetite loss | P(Appetite loss=true|PC=true) | 0.2347 | 10 |
| ALT | P(ALT=true|PC=true) | 0.1633 | 11 |
| Fatigue/Asthenia | P(Fatigue or Asthenia = true|PC=true) | 0.1531 | 12 |
| CEA | P(CEA=true|PC=true) | 0.1429 | 13 |
| Depression | P(Depression=true|PC=true) | 0.1327 | 14 |
| GGT | P(GGT=true|PC=true) | 0.1327 | 14 |
| CA 19-9 | P(CA 19-9=true|PC=true) | 0.1122 | 16 |
| AST | P(AST=true|PC=true) | 0.1122 | 16 |
| Bilirubin | P(Bilirubin=true|PC=true) | 0.0918 | 18 |
| ALP | P(ALP=true|PC=true) | 0.0816 | 19 |
| Jaundice | P(Jaundice=true|PC=true) | 0.0204 | 20 |
Table 1.
Calculated weights and rankings for the 20 risk factors (data source: PubMed).
| Variable | Positive | Negative | Original Weight | Normalized Weight | Ranking |
|---|---|---|---|---|---|
| Weight loss | 58 | 0 | 58 | 1 | 1 |
| Glucose | 51 | 0 | 51 | 0.8793 | 2 |
| CA 19-9 | 48 | 0 | 48 | 0.8276 | 3 |
| Diabetes | 74 | 2 | 37 | 0.6379 | 4 |
| Albumin | 33 | 0 | 33 | 0.5690 | 5 |
| Appetite loss | 32 | 0 | 32 | 0.5517 | 6 |
| CEA | 59 | 2 | 29.5 | 0.5086 | 7 |
| Bilirubin | 29 | 0 | 29 | 0.5000 | 8 |
| Abdominal pain | 28 | 0 | 28 | 0.4828 | 9 |
| Depression | 52 | 2 | 26 | 0.4483 | 10 |
| Jaundice | 12 | 3 | 4 | 0.0690 | 11 |
| Nausea | 2 | 0 | 2 | 0.0345 | 12 |
| Fatigue/Asthenia | 1 | 0 | 1 | 0.0172 | 13 |
| Vomiting | 1 | 0 | 1 | 0.0172 | 14 |
| GGT | 0 | 0 | 0 | 0 | 15 |
| ALP | 0 | 0 | 0 | 0 | 15 |
| AST | 0 | 0 | 0 | 0 | 15 |
| ALT | 0 | 0 | 0 | 0 | 15 |
Figure 4 (b) explains why the normalized weights wi can increase the BNI prediction accuracy for pancreatic cancer using a three-node Bayesian Network with the variable set V = (Vdown U Vup), where nodes Vup are the set of parent nodes of the decision node (“the risk of pancreatic cancer”) and nodes Vdown are the set of all children nodes of the decision node. After the prior probabilities are weighted, based on Theorem 3.1 in [24], we obtain:
| (5) |
where V = (Vdown U Vup). We calculate the posterior probability P(pancreatic cancer=true | Vup=true) using the method described in [24], and Pw(Vdown=true | pancreatic cancer=true) is calculated as follows:
| (6) |
where Vdown = (VL U VR). Substituting Eq. (6) into Eq. (5), we can see that the normalized weight wi contributes to the posterior probability P(pancreatic cancer=true | V=true).
2.5 EHR Information Extraction for Prior Probability Calculation
For a BNI model, the prior probability of each variable is often stored in a conditional probability table (CPT). We built such a CPT using de-identified EHR information for pancreatic cancer patients which had been extracted from our institutional research data warehouse. Two datasets were used to calculate the prior probabilities: one was a 98-sample dataset with patients who were manually confirmed pancreatic cancer cases, and the other was a 14971-sample dataset for patients who did not have ICD-9 diagnosis of pancreatic cancer.
As shown in Figure 3, in this BNI model, all 20 of the risk factor nodes had only one connection, which was to pancreatic cancer. The prior probability of each of the two independent variables (age and smoking/drinking) was obtained by querying the corresponding condition in the EHR. For example, to calculate the probability P (age ≥ 60), a search condition in EHR of “age at least 60 years old” was used. In contrast, for each of the 18 dependent variables, two search conditions were used. For example, to calculate the probability P (abdominal pain = true| pancreatic cancer = true), we queried the condition “abdominal pain = true” among the patients who had pancreatic cancer. The calculated prior probabilities for the twenty risk factors are described in Table 2 and 3. To calculate the prior probability P (pancreatic cancer = true| (age ≥ 60 AND smoking/drinking = true)), we first queried patients satisfying the two conditions: “is equal to or older than 60 years” and “is a smoker and a drinker”, among whom we searched for patients who had “pancreatic cancer”. Table 4 shows the prior probabilities for the pancreatic cancer node.
Table 3.
Calculated prior probabilities with the condition pancreatic cancer = false (data source: EHR, the 14971-control dataset).
| Variable | Probability | Percent | Ranking |
|---|---|---|---|
| Age | P(Age>=60|PC=false) | 0.4751 | 1 |
| Diabetes | P(Diabetes=true|PC=false) | 0.2458 | 2 |
| Depression | P(Depression=true|PC=false) | 0.2351 | 3 |
| AST | P(AST=true|PC=false) | 0.2100 | 4 |
| Albumin | P(Albumins=true|PC=false) | 0.2000 | 5 |
| ALP | P(ALP=true|PC=false) | 0.1900 | 6 |
| Vomiting | P(Vomiting=true|PC=false) | 0.1029 | 7 |
| Nausea | P(Nausea = true|PC=false) | 0.0855 | 8 |
| Fatigue/Asthenia | P(Fatigue or Asthenia = true|PC=false) | 0.0836 | 9 |
| Smoking/Drinking | (UNION) P(Smoking or Alcohol = true|PC=false) | 0.0626 | 10 |
| Weight loss | P(Weight loss=true|PC=false) | 0.0429 | 11 |
| CEA | P(CEA=true|PC=false) | 0.0300 | 12 |
| ALT | P(ALT=true|PC=false) | 0.0200 | 13 |
| Appetite loss | P(Appetite=true|PC=false) | 0.0129 | 14 |
| Jaundice | P(Jaundice=true|PC=false) | 0.0102 | 15 |
| CA 19-9 | P(CA 19-9=true|PC=false) | 0.0100 | 16 |
| Abdominal pain | P(Abdominal pain = true|PC=false) | 0.0013 | 17 |
| GGT | P(GGT=true|PC=false) | 0 | 18 |
| Glucose | P(Glucose=true|PC=false) | 0 | 18 |
| Bilirubin | P(Bilirubin=true|PC=false) | 0 | 18 |
Table 4.
Calculated prior probabilities of pancreatic cancer to be true under the conditions of age and substance (smoking or drinking) abuse (data source: EHR for the entire patient population).
| pancreatic cancer=true | Age >= 60 | Age < 60 |
|---|---|---|
| Smoking/Drinking =true | 0.00002 | 0.00001 |
| Smoking/Drinking =false | 0.0008 | 0.0003 |
2.6 Graphic User Interface Development
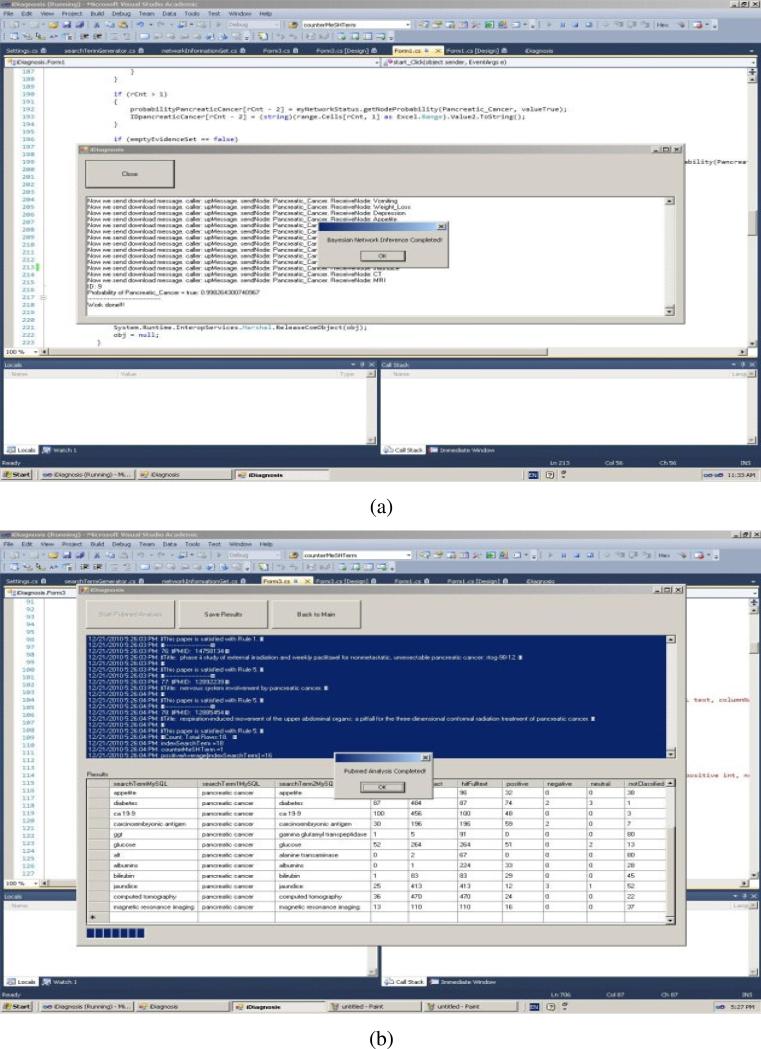
We developed a graphic user interface, iDiagnosis, for the weighted BNI model using the Professional Version of Microsoft Visual Studio 2010. iDiagnosis includes two main functions: the Bayesian function and the eUtils function. The Bayesian part realizes the inferences in a Bayesian Network using a Pearl's Message-Passing algorithm [6, 25-26]. The eUtils part is responsible for generating search terms, searching and fetching PubMed papers, and accessing MySQL data tables. Figure 5 (a) and Figure 5 (b) display the interface for the Bayesian part and the interface for the eUtils part, respectively.
Figure 5.
The interfaces of iDiagnosis for (a) the Bayesian function and (b) the eUtils function.
2.7 Model Evaluation
Our evaluation consisted of two parts: (1) comparing the weighted BNI model to the conventional BNI model using the data set that contained the 98 cases and 14,971 controls; and (2) comparing the weighted BNI model to two other popular classification models, KNN and SVM, in the open source Weka package [22]. Note that part one used only aggregated de-identified information for the 14,971 control set to calculate prior probabilities, as shown in Table 3, without requiring individual patient information for each of the 20 risk factors. However, KNN and SVM required more patient-level information than the BNI models for feature representation needed by machine-learning. Therefore, for part two, we reused the 98 cases but reduced the control group size from 14,971 to 196 since it was impractical to obtain the values of the 20 risk factors for each of the 14,971 controls. The 196-patient controls included 106 randomly selected patients without pancreatic cancer and 90 with symptoms similar to pancreatic cancer but without pancreatic cancer. We constructed a feature matrix with the size being 294 (patients) by 20 (risk factors) and divided the combined case and control, 294 in total, into two groups with the ratio between the training and the testing patients being 1 to 3 so that there were 73 training patients, consisting of 24 cases and 49 controls, and 221 testing patients, including 74 cases and 147 controls. When implementing SVM, the training data were centered on zero mean and scaled to a standard deviation of value 1. We selected the linear function (dot product) as the SVM kernel. To optimize the search for the separating hyper-plane by using quadratic programming, the interior point method was applied [27]. The soft margin was used by setting the value of the additional constraint C as 1.
All four models were applied to classify each patient. We compared performances by measuring sensitivity, specificity, and accuracy. The definition of accuracy is provided below:
We drew the ROC curves for the weighted BNI, the conventional BNI, KNN and SVM, and compared area under curve (AUC), standard error (SE) and 95% confidence interval (CI) for each to evaluate each one's performance.
3. Results
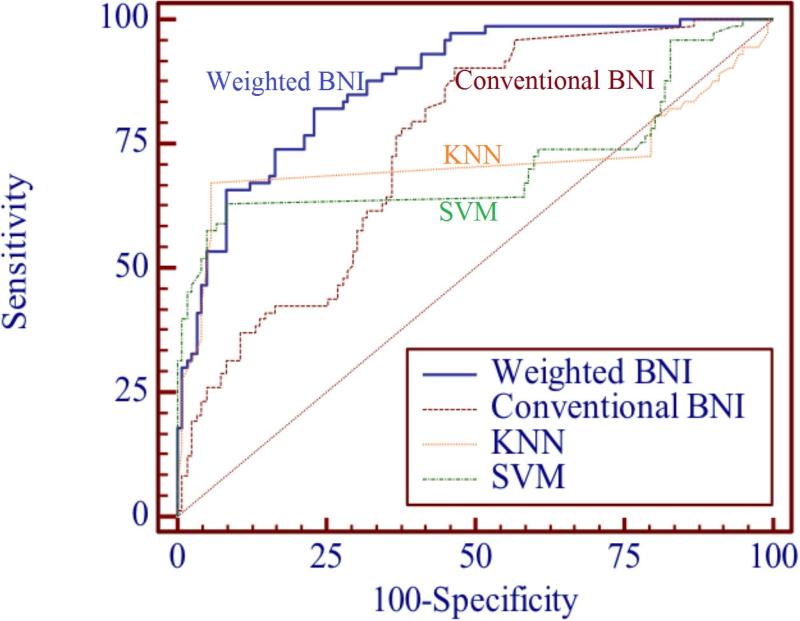
Table 1 shows the resulting normalized weights and variable rankings. The top three variables associated with pancreatic cancer, ranked by importance, were: weight loss, abnormal glucose, and abnormal CA 19-9. According to the PubMed weights, these variables weigh about 50 times more than the most weakly associated variables (GGT and ALT). Table 2 shows the prior probabilities of P(Vi=true | pancreatic cancer=true) for each of the 20 risk factors and their frequencies in pancreatic cancer patients. Note the strength of an association is measured by the weights, or the frequency of the PubMed citation of the association. In contrast to the PubMed weighting results, the top three most frequent variables appearing in pancreatic cancer patients EHR were: glucose, albumin, and nausea. The most frequent variable, glucose, is about 50 times more frequent than the least frequent variable, jaundice. Table 3 shows the prior probabilities of P(Vi=true | pancreatic cancer=false) for patients without pancreatic cancer. The three least frequent variables in patients without pancreatic cancer were: GGT, glucose, and bilirubin. Table 5 shows the sensitivity, specificity, accuracy, and ROC curve of the weighted BNI, the conventional BNI, KNN and SVM. The accuracy indicated by the AUC value of the weighted BNI (0.910) is significantly higher than that of the conventional BNI (0.806), KNN (0.718) and SVM (0.727) with P<0.0001. Figure 6 shows the ROC curves for the weighted BNI, the conventional BNI, KNN and SVM. All ROC curves are on the upper-left side of ROC space, but the ROC curve of the weighted BNI is higher than that of the conventional BNI, KNN and SVM, indicating a better performance is achieved by the weighted BNI for pancreatic cancer prediction. Results in Figure 6 and Table 5 suggest that the weighted BNI is significantly more accurate than the conventional BNI, KNN and SVM for pancreatic cancer prediction (P<0.0001).
Table 5.
Comparative performance of the weighted BNI, the conventional BNI, KNN and SVM: the weighted BNI model is 10% more accurate than the conventional BNI model for pancreatic cancer prediction.
| Sensitivity | Specificity | Accuracy | ROC |
|||
|---|---|---|---|---|---|---|
| AUC | SE | 95% CI | ||||
| Weighted BNI | 0.847 | 0.852 | 0.850 | 0.910 | 0.021 | (0.869, 0.951) |
| Conventional BNI | 0.796 | 0.704 | 0.735 | 0.806 | 0.029 | (0.750, 0.863) |
| KNN | 0.650 | 0.332 | 0.389 | 0.718 | 0.046 | (0.654, 0.776) |
| SVM | 0.768 | 0.396 | 0.534 | 0.727 | 0.043 | (0.663, 0.785) |
Figure 6.
Comparison of ROC curves of the weighted BNI, the conventional BNI, KNN and SVM: the weighted BNI model is more accurate than the conventional BNI, KNN and SVM for pancreatic cancer prediction.
4. Discussion
In this paper, we developed a weighted BNI model for pancreatic cancer prediction by combining PubMed knowledge and EHR data to calculate the ratio between the positive and negative evidence for the associations between each risk factor and the target disease. The evaluation results indicate that the weighted BNI significantly outperformed the conventional BNI and two other classification models for pancreatic cancer prediction. This result can be explained by the following characteristics of the weighted BNI model. First, the posterior probabilities of the weighted BNI are determined by two data sources, the ratio between the positive-negative evidence for the association between the risk factor and the disease and the prior probability of each risk factor in EHR, both being important empirical evidence for disease risk prediction. The more frequently a risk factor can be found in EHR, the higher the posterior probability of the risk factor. The weighted BNI can tell clinically relevant variables from clinically irrelevant variables and weigh the relevant variables according to PubMed evidence. The conventional BNI can recommend risk factors only by using high posterior probability of statistical significance. Moreover, some approaches simply eliminate irrelevant variables; however, we keep seemingly irrelevant variables in the model but use PubMed knowledge to avoid abusing their prior probability. Our design seems to be more realistic and sensitive than a simplified model that disregards such variables. To our knowledge, the weighted BNI is a novel approach to handling clinically irrelevant variables for disease risk prediction. Our results in Table 5 and Figure 6 confirm our hypothesis that the weighted BNI model can overcome the limitations in the conventional BNI based on pure probabilities.
The weighted BNI also outperformed KNN and SVM for pancreatic cancer prediction. This may be because of two reasons. Firstly, the BNI model better serves risk prediction than other classification models by using a small number of variables. Our model contains only 20 variables. KNN and SVM, on the other hand, usually excel in a high-dimensional feature space, such as highly dimensional microarray datasets and do not show advantages in low-dimensional feature space. Secondly, the weighted BNI obtains information of the association between the variables and pancreatic cancer from the topology of Bayesian Network and the weights from PubMed and EHR, while KNN and SVM do not have such knowledge to support accurate prediction.
The combination of PubMed and EHR knowledge and information for weighing risk factors can be used to generate hypotheses about the clinical significance of an association between a variable and pancreatic cancer by a combined analysis of its frequency in patients with pancreatic cancer (Table 2) and in patients without pancreatic cancer (Table 3). For example, glucose is top ranked in Table 2, but appears at the bottom in Table 3. This result indicates a positive association between glucose and pancreatic cancer, which is consistent with scientific knowledge in that glucose is the second most frequently studied variable in pancreatic cancer research due to the association between diabetes and pancreatic cancer.
According to Eq (1.1) and (1.2), the weight woi represents the ratio between the number of PubMed abstracts of positive association (Pi) and the number of PubMed abstracts of negative association (Ni) for each risk factor i. If Pi is bigger than Ni, then the original weight woi is bigger than 1, which means there is more positive than negative evidence showing variable Vi is associated with pancreatic cancer; if Pi is smaller than Ni, then the original weight woi is smaller than 1, which means that there is more negative than positive evidence indicating that variable Vi is associated with pancreatic cancer. In Table 1, almost all the original weights woi are > 1, which may imply that most PubMed publications about risk factors are positive results and negative results are rare. Because we cannot tell if there is a publication bias toward only positive association, this warrants further study to guide the use of PubMed evidence.
We identified several tasks as future work to continuously improve the weighted BNI model for disease risk prediction. First, a highly accurate dataset is crucial to realizing the full potential of the weighted BNI and the software iDiagnosis for pancreatic cancer risk prediction. In this paper, we reused a dataset of manually reviewed 98 cases [21]; however, we faced significant challenges when it came to verifying the completeness and accuracy of the information for the larger sample population, the 14,971-patient control group. Each variable entails laborious information extraction and summarization from PubMed and EHR. The same variable may be reflected in multiple formats in different data sources (e.g., ICD-9 codes, various types of notes, and other structured data sources such as lab results) in EHR. Our unstructured EHR data in the research data warehouse were pre-processed by one of the best medical natural language processing software, MedLEE [10, 28-31], but the data accuracy was not close to 100%. Time was an issue in this study as for the smaller case sample we used manual review to compensate for the NLP limitations, which was time consuming. We also lacked a method to reconcile the inconsistencies between structured and unstructured data sources. Development, validation, and reuse of sophisticated phenotyping algorithms in the EHR are much needed to improve the efficiency and accuracy of EHR phenotyping.
Second, although the weighted BNI model improves the accuracy for pancreatic cancer prediction over conventional BNI and the other two popular classification methods, it can be improved in multiple aspects, including the efficiency for variable generation and selection, prior probabilities calculation, and variable weights calculation. In this study, we selected 20 variables to predict pancreatic cancer risks. It is possible that unknown variables related to pancreatic cancer have not been included in our model. It is beyond our current capacity to define a model with hundreds or thousands of fine-grained phenotypic features related to pancreatic cancer. Therefore, efficient discovery of unknown disease features is a challenging research topic that needs more future work.
Moreover, in this study, we used the batch processing mode to obtain data to calculate prior probabilities from EHR. It would be more efficient to support prior probability calculation using a real-time data warehouse to automatically update the parameters of the model online as the warehouse receives updates. An advanced analytical framework based on efficient EHR-phenotyping algorithms can be developed to increase the efficiency of dynamic prior probability calculation for each risk factor in vivo.
Finally, in this weighted BNI network, we weighted nodes only. As an alternative, the causal edges between nodes can be weighted. In [32], Zhou et al. developed a causal edge weighted BNI for visual tracking, and the authors achieved better recognition results than when using a conventional BNI. One of our future works is to investigate the efficacy of weighing causal edges for improving the predictive accuracy of BNI.
5. Conclusion
We developed a weighted BNI using both PubMed knowledge and EHR data to weigh network nodes for pancreatic cancer prediction. We demonstrated that the weighted BNI model showed remarkable improvement in prediction accuracy over the conventional BNI for pancreatic cancer prediction (P<0.0001). We conclude that an integration of a statistical summary of PubMed knowledge and real-world evidence collected from EHR data can improve weighting of the variables in a BNI model and improve its disease predictive accuracy. More studies are warranted to generalize the findings here to allow modeling of other diseases based on the integration of PubMed and EHR knowledge.
Acknowledgments
This research was funded under NLM grant R01LM009886, R01LM010815, and CTSA award UL1 RR024156. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. We thank Dr. Carol Friedman for making free-text symptoms searchable using MedLEE, which was sponsored by the NLM grant R01LM008635. We thank the two anonymous reviewers for their thoughtful suggestions to improve the quality of this paper. We also thank Dr. Zhihui Luo for providing programming advice to implement the iDiagnosis software.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chakraborty S, Baine MJ, Sasson AR, Batra SK. Current status of molecular markers for early detection of sporadic pancreatic cancer. BBA-Rev Cancer. 2011;1815(1):44–64. doi: 10.1016/j.bbcan.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma M. Pancreatic cancer biomarkers and their implication in cancer diagnosis and epidemiology. Cancers. 2010;2(4):1830–1837. doi: 10.3390/cancers2041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerstung M, Baudis M, Moch H, Beerenwinkel N. Quantifying cancer progression with conjunctive Bayesian Networks. Bioinform. 2009;25(21):2809–2815. doi: 10.1093/bioinformatics/btp505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim DJ, Rockhill B, Colditz GA. Validation of the Harvard Cancer Risk Index: a prediction tool for individual cancer risk. J Clin Epidemiol. 2004;57(4):332–340. doi: 10.1016/j.jclinepi.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Heckerman D. A tutorial on learning with Bayesian Networks. Microsoft Research Tech Report. 1995:57. [Google Scholar]
- 6.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. Inference in Bayesian Networks. Nat Biotech. 2006;24(1):51–53. doi: 10.1038/nbt0106-51. [DOI] [PubMed] [Google Scholar]
- 7.Jiang X, Wallstrom G, Cooper GF, Wagner MM. Bayesian prediction of an epidemic curve. J Biomed Inform. 2009;42(1):90–99. doi: 10.1016/j.jbi.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Shen Y, Cooper GF. A Bayesian biosurveillance method that models unknown outbreak diseases. Proceedings of the 2nd NSF conference on intelligence and security informatics: Biosurveillance.; New Brunswick, NJ, USA. 2007; Springer-Verlag; pp. 209–215. [Google Scholar]
- 9.Charles EK, Linda MR, Katherine AS, Peter H. Construction of a Bayesian network for mammographic diagnosis of breast cancer. Comput Biol Med. 1997;27(1):19–29. doi: 10.1016/s0010-4825(96)00039-x. [DOI] [PubMed] [Google Scholar]
- 10.Jain NL, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. AMIA Annu Fall Symp. 1997:829–833. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X-H, Zheng B, Good WF, King JL, Chang Y-H. Computer-assisted diagnosis of breast cancer using a data-driven Bayesian Belief Network. Int J Med Inform. 1999;54(2):115–126. doi: 10.1016/s1386-5056(98)00174-9. [DOI] [PubMed] [Google Scholar]
- 12.Burnside ES, Rubin DL, Fine JP, Shachter RD, Sisney GA, Leung WK. Bayesian Network to predict breast cancer risk of mammographic microcalcifications and reduce number of benign biopsy results: initial experience. Radiol. 2006;240(3):8. doi: 10.1148/radiol.2403051096. [DOI] [PubMed] [Google Scholar]
- 13.Nicandro C-R, Héctor Gabriel A-M, Humberto C-C, Luis Alonso N-F, Rocío Erandi B-M. Diagnosis of breast cancer using Bayesian Networks: a case study. Comput Biol Med. 2007;37(11):1553–1564. doi: 10.1016/j.compbiomed.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Velikova M, de Carvalho Ferreira N, Lucas P. Bayesian network decomposition for modeling breast cancer detection. In: Bellazzi R, Abu-Hanna A, Hunter J, editors. Artificial Intelligence in Medicine. Heidelberg; Springer Berlin: 2007. pp. 346–350. [Google Scholar]
- 15.Velikova M, Samulski M, Karssemeijer N, Lucas P. Toward expert knowledge representation for automatic breast cancer detection. In: Dochev D, Pistore M, Traverso P, editors. Artificial Intelligence: Methodology, Systems, and Applications. Heidelberg; Springer Berlin: 2008. pp. 333–344. [Google Scholar]
- 16.Gadewadikar J, Kuljaca O, Agyepong K, Sarigul E, Zheng Y, Zhang P. Exploring Bayesian Networks for medical decision support in breast cancer detection. African Journal of Mathematics and Computer Science Research. 2010;3(10):7. [Google Scholar]
- 17.Stojadinovic A, Eberhardt C, Henry L, Eberhardt J, Elster EA, Peoples GE, et al. Development of a Bayesian classifier for breast cancer risk stratification: a feasibility study. J Plast Surg. 2010;10(e25) [PMC free article] [PubMed] [Google Scholar]
- 18.De Icaza E, López-Cervantes M, Arredondo A, Robles-Díaz G. Likelihood ratios of clinical, laboratory and image data of pancreatic cancer: Bayesian approach. J Eval Clin Pract. 2009;15(1):62–68. doi: 10.1111/j.1365-2753.2008.00955.x. [DOI] [PubMed] [Google Scholar]
- 19.Chavira M, Darwiche A. On probabilistic inference by weighted model counting. Artificial Intelligence. 2008;172(6-7):772–799. [Google Scholar]
- 20.Botsis T, Anagnostou VK, Hartvigsen G, Hripcsak G, Weng C. Modeling prognostic factors in resectable Pancreatic Adenocarcinomas. Cancer Inform. 2010;2009(7):281. doi: 10.4137/cin.s3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botsis T, Anagnostou VK, Hartvigsen G, Hripcsak G, Weng C. Developing a multivariable prognostic model for pancreatic endocrine tumors using the clinical data warehouse resources of a single institution. Applied Clinical Informatics. 2010;1(1):12. doi: 10.4338/ACI-2009-12-RA-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levesque HJ, Brachman RJ. A fundamental tradeoff in knowledge representation and reasoning. In: Brachman R, Levesque H, editors. Readings in Knowledge Representation. Morgan Kaufmann; 1985. pp. 41–70. [Google Scholar]
- 23.Chen H, Sharp B. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinform. 2004;5(1):147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neapolitan RE. Learning Bayesian Networks. illustrated edition Prentice Hall; 2003. [Google Scholar]
- 25.Pearl J. Probabilistic reasoning in intelligent systems: networks of plausible inference. Morgan Kaufmann Publishers Inc.; San Francisco, CA: 1988. [Google Scholar]
- 26.Wei S, Chang KC. Message passing for hybrid Bayesian Networks: representation, propagation, and integration. IEEE T Aero Elec Sys. 2009;45(4):1525–1537. [Google Scholar]
- 27.Zhao D. Non-negative matrix factorization to speed up interior point method of SVM training, in Stanford 50: State of the Art and Future Directions of Computational Mathematics and Numerical Computing. Stanford University; 2007. [Google Scholar]
- 28.Friedman C. Towards a comprehensive medical language processing system: methods and issues. AMIA Annu Fall Symp. 1997:595–599. [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman C. A broad-coverage natural language processing system. AMIA Symp. 2000:270–274. [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman C, Hripcsak G, Shablinsky I. An evaluation of natural language processing methodologies. AMIA Symp. 1998:855–859. [PMC free article] [PubMed] [Google Scholar]
- 31.Jain NL, Knirsch CA, Friedman C, Hripcsak G. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. AMIA Annu Fall Symp. 1996:542–546. [PMC free article] [PubMed] [Google Scholar]
- 32.Yue Z, Huang TS. Weighted Bayesian Network for visual tracking.. Pattern Recognition, 2006. ICPR 2006. 18th International Conference on..2006. pp. 523–526. [Google Scholar]