Abstract
The enzymatic degradation of recalcitrant plant biomass is one of the key industrial challenges of the 21st century. Accordingly, there is a continuing drive to discover new routes to promote polysaccharide degradation. Perhaps the most promising approach involves the application of “cellulase-enhancing factors,” such as those from the glycoside hydrolase (CAZy) GH61 family. Here we show that GH61 enzymes are a unique family of copper-dependent oxidases. We demonstrate that copper is needed for GH61 maximal activity and that the formation of cellodextrin and oxidized cellodextrin products by GH61 is enhanced in the presence of small molecule redox-active cofactors such as ascorbate and gallate. By using electron paramagnetic resonance spectroscopy and single-crystal X-ray diffraction, the active site of GH61 is revealed to contain a type II copper and, uniquely, a methylated histidine in the copper's coordination sphere, thus providing an innovative paradigm in bioinorganic enzymatic catalysis.
Keywords: lignocellulose; bioethanol; posttranslational modification; cellulase, plant cell wall
Cellulose is Earth's most abundant biopolymer. Its exploitation as an energy source plays a critical role in the global ecology and carbon cycle. Industrial production of fuels and chemicals from this plentiful and renewable resource holds the potential to displace petroleum-based sources, thus reducing the associated economic and environmental costs of oil and gas production (1, 2) and promoting energy security as part of a balanced energy portfolio. However, despite the burgeoning potential of cellulose as a biofuel source, its remarkable recalcitrance to depolymerization has so far hindered the economical use of any form of lignocellulosic biomass as a feedstock for biofuel production (3, 4).
In addressing the issue of cellulose recalcitrance, much effort has been directed toward harnessing the known cellulose-degrading enzymatic pathways found in fungi. The consensus model of enzymatic degradation involves the concerted action of a consortium of different endoglucanases and “exo”-acting cellobiohydrolases (collectively termed “cellulases”); both enzyme classes perform classical glycoside hydrolysis through attack of water at the anomeric center of oligo/polysaccharide substrates (5–9). Necessarily as part of the overall enzymatic degradation of cellulose, the initial enzymatic step must overcome cellulose's inertness by disrupting the cellulosic structure, thus allowing attack by traditional cellulases. Originally, Reese et al. (10) suggested that undefined enzymes could play a major role in this step. This notion remained a hypothesis until very recently when, in a key paper, Harris et al. (11) demonstrated that inclusion of a novel enzyme class, currently termed GH61 glycoside hydrolases in the CAZy database of carbohydrate-active enzymes (12), greatly increases the performance of Hypocrea jecorina (Trichoderma reesei) cellulases in lignocellulose hydrolysis. From this work, it was suggested that GH61s act directly on cellulose rendering it more accessible to traditional cellulase action (11). Moreover, recent genomic sequencing of the brown rot fungi Postia placenta showed a number of GH61 genes in this organism (13–15), indicating the widespread nature of this family of enzymes in cellulose degradation. As such, GH61s likely hold major potential for industrial decomposition of cellulosic materials.
Notwithstanding this potential, the detailed biochemistry of GH61s has remained frustratingly recondite (11, 16). What is known is that GH61 activity is enhanced by the action of a noncellulosic component of biomass and that a metal cofactor is required, although the identity of neither the biomass component nor the metal was established (11). From a structural perspective, the 3D structures of GH61 enzymes show no similarity to classical sugar hydrolases, but instead display a predominantly β-sheet fold with an extended planar face, the center of which contains the N-terminal histidine where a metal ion is believed to bind (11, 16). Furthermore, a recent analysis of a structurally related, chitin-binding domain (CBP21) supported an oxidative function for CBP21, which, by analogy, might be extended to GH61 enzymes (17).
Here we demonstrate the nature of the enhancement of cellulase activity by GH61s. We describe in full its active site details and define a catalytic activity for the enzyme class. Using a recombinant GH61 originally from Thermoascus aurantiacus (termed herein TaGH61), we show direct degradation of cellulose, generating a distribution of both oxidized and nonoxidized cellodextrin products. We further identify copper as the metal directly involved in TaGH61 activity and demonstrate that copper binds to a classical type II copper site involving the N terminus of the enzyme. We also show a unique methyl modification of one of the metal-coordinating histidine residues for TaGH61.
Results
Identification of Components of Cellulosic Biomass That Potentiate GH61 Activity.
To identify those soluble factors that enhance the activity of GH61 enzymes, dilute acid pretreated corn stover (PCS), a major industrial substrate (18), was separated into its insoluble and soluble liquor phases. In the presence of 5% soluble PCS liquor, TaGH61 greatly increased the hydrolysis of microcrystalline cellulose by cellulases, from which it is possible to infer that a soluble component in the PCS liquor acts as a cofactor for TaGH61 (Fig. S1A). To probe which component in the highly heterogeneous PCS liquor potentiates GH61s, individual liquor components (19) were incubated together with TaGH61 and assessed for their ability to cleave cellulose. From these experiments, gallate (gallic acid) was shown to increase the activity of GH61 enzymes in the degradation of microcrystalline cellulose by H. jecorina cellulases (Fig. S1B) and the Aspergillus oryzae β-glucosidase (AoBG) (Fig.1). As a control, it was further shown that neither TaGH61 nor gallate alone have an impact on cellulose hydrolysis, but their combination significantly increased cellulose degradation by all cellulases tested (Fig. S1B). Indeed, the reaction where β-glucosidase (BG), but not GH61, was present gave insignificant release of glucose from the cellulose. During the time scale of the reaction (3 d), gallate was aerobically oxidized whether TaGH61 was present or not; because TaGH61 is required for cellulose degradation, these oxidized gallate species clearly did not act directly on the cellulose to liberate oligomers for the BG to hydrolyze (Fig. S1C).
Fig. 1.
PASC deconstruction by TaGH61A, AoBG, and various concentrations of gallate. 5 g/L PASC was incubated with the indicated gallate and TaGH61A concentrations at an AoBG concentration of 20 mg/g cellulose, 3-d reaction at 50 °C in 50 mM sodium acetate, pH 5, and 1 mM MnSO4. GH61 cleaved cellulose into cellodextrin, which was converted to glucose by AoBG and quantified by refractive index detection HPLC to calculate PASC conversion.
From these observations, we draw two conclusions. The first is that the requirement for the gallate indicates that TaGH61 needs a redox active cofactor. The second is that the synergy between TaGH61 and all of the major families of cellulases indicates an apparently “cellulase-independent” TaGH61 activity on cellulose. Together, these hypotheses lead to the general proposal that GH61s catalyze the direct cleavage of glycosidic bonds of cellulose to give exposed or soluble oligosaccharides, which are then more easily acted upon by cellulases, and that the mechanism of action of GH61 is principally one of an oxidoreductase rather than a classical glycoside hydrolase.
Identification of Products of TaGH61-Catalyzed Cellulose Cleavage.
To verify direct cleavage of cellulose by TaGH61, MALDI-TOF-MS was performed on products generated by incubation of phosphoric acid swollen cellulose (PASC) with TaGH61 in the presence of gallate (but in the absence of classical cellulases or BG). From these experiments, a series of molecular ions was observed in the MALDI-TOF-MS with m/z (M + Na+) corresponding to degree of polymerization (DP) 3 to DP8 cellodextrins (Fig. 2 and Fig. S2A), indicating a direct mode of action of TaGH61 on cellulose. Additionally, ions corresponding to various oxidized cellodextrin species were observed.
Fig. 2.
MALDI-TOF-MS analysis of permethylated products from 5 g/L PASC, 13 mg/g cellulose TaGH61A, and 3 mM gallate in 25 mM triethylammonium acetate and 2 mM CaCl2 incubated at pH 5.4 for 22 h. (Inset) Expanded view of spectra in the DP5 range.
Products formed from PASC by TaGH61 activity were further analyzed by permethylation and MALDI-TOF tandem MS (MS/MS), and molecular ions reflecting both diverse permethylated cellodextrins and their +30- and −16-Da species, derived from oxidized products, were observed. Comparison of the spectra of permethylated DP5 cellodextrin and the permethylated DP5 +30 Da species (Fig. S2 B and C) showed that the reducing end of this latter species is modified and is consistent with a reducing-end aldonic acid group. The fragmentation ions of the DP5 −16-Da permethylated species showed that this species is modified at the nonreducing end, most likely by oxidation of the 6C-alcohol to 6C-aldose (Fig. S2D). The diverse oxidation products suggest more than one mode of GH61 action, although not one of indiscriminate oxidation of the substrate.
Nature of Metal Ion at Active Site of TaGH61.
In addition to soluble redox-active cofactors, GH61s further require a metal ion for maximal activity (11). Determining the identity and role of this metal ion has been confused, with reports (11, 16, 17) suggesting that many diverse metals could enhance catalysis in simple EDTA/metal ion addition experiments and further complicated by different assignments of structural metal ions in the various crystal structures of GH61 enzymes (16). Therefore, to identify the metal ion and the nature of the active site of GH61s, we performed isothermal titration calorimetry (ITC), electron paramagnetic resonance (EPR), and single crystal X-ray diffraction studies on TaGH61.
ITC experiments at room temperature showed that general metal ion binding to previously demetallated TaGH61 is weak or immeasurable. In dilute acetate buffer at pH 5, no significant binding was detected for Mg2+, Ca2+, Mn2+, Co2+, Ni2+, or Zn2+. In marked contrast, however, Cu2+ bound very strongly with a clear 1:1 metal:TaGH61 stoichiometry (Fig. S3A). Indeed, the binding affinity of Cu2+ to TaGH61 was so high that we were unable to determine an accurate dissociation constant value from the ITC experiments, suggesting a KD tighter than 1 nM. The strength of copper binding was further demonstrated by observing very slow formation of copper–EDTA from copper-loaded TaGH61 (Fig. S3B) upon the addition of Na2EDTA at pH 5. From the EPR experiments, the first-order rate constant for metal dissociation from copper–TaGH61 could be roughly estimated to be <0.001 s−1; by making the assumption that the on-rate for copper binding to TaGH61 is near diffusion limited, we would further estimate the dissociation constant of Cu-TaGH61 to be <1 pM. It should be noted that such selective and high affinity for copper introduces significant complications for the study of GH61s, insofar as, even in ostensibly demetallated systems, any trace copper from the substrate or solution would be strongly bound by the enzyme; therefore, a background activity would nearly always be seen, regardless of the specific addition of different metal ions.
EPR analysis of apo-TaGH61 treated with stoichiometric amounts of copper(II) at pH 5 gave an unambiguous signal for a copper ion in a single, well-defined site with tetragonal coordination geometry (Fig. 3B). The EPR parameters for Cu-TaGH61 were very similar to those found for classical type II copper oxygenases (Table S1). Along with the ITC data, this finding strongly indicates that Cu-TaGH61 is a natural copper-dependent enzyme with a type II copper ion at the active site.
Fig. 3.
The structure of the active site and the EPR spectrum of Cu-TaGH61. (A) 3D structure of the primary metal site as observed in a T. aurantiacus GH61A crystal soaked in 10 mM CuNO3 for 30 min. Maps shown are the maximum likelihood/σA weighted 2Fobs − Fcalc density (contoured at 0.4 electrons per Å3) in blue, and the ACORN unbiased direct methods Emap contoured at 1.1 electrons per Å3 (3.3σ) in red. Also shown is a small molecule modeled as a PEG fragment that bonds in the equatorial position. Unmodeled density (at ∼1/5 the Cu site) lies ∼1.6 Å from the Cu; it is unclear whether this residual electron density is a counterion or a low occupancy of a copper ion in a different position due to oxidation state or protonation state differences. Images were drawn with CCP4Mg (34). (B) X-band EPR spectrum (140 K) of copper-loaded TaGH61A in 10 mM acetate buffer and ∼15% (vol/vol) glycerol. (C) Schematic diagram of the copper site in GH61, depicting histidine brace.
To define the structure of the active site more completely, a structure of TaGH61 that had been Cu2+-soaked over a period of 30 min was determined at 1.25-Å resolution. The structure exhibited significant electron density at the putative N-terminal active center. Unbiased electron density, in terms of the ACORN (20) direct phase map or the anomalous difference (Δƒ″), revealed clear electron density (40σ in the Δƒ″ synthesis), which is reliably modeled by a single copper ion. The immediate coordination geometry of which has near-D4h symmetry, with the square basal plane formed by the oxygen atom of a PEG molecule (2.1 Å) and three nitrogen atoms of the N-terminal histidine (1.9 Å), the amino terminus (2.2 Å), and a second histidine group (2.1 Å), herein referred to as the “histidine brace.” Oxygen atoms of a tyrosine/ate (2.9 Å) and a further water molecule (2.9 Å) occupy the apical positions and completed the immediate coordination sphere (Fig. 3 A and C). The overall coordination geometry exhibits the classical Jahn–Teller axial distortion expected for a copper(II) ion and is consistent with the EPR data. (At much lower contour levels, ∼1/5 of the main peak, there was evidence for a second anomalously scattering site ∼1.6 Å from the main copper position. This observation is indicative either of slight contamination of the active site with copper in a different oxidation state or, potentially, different counteranions coordinating to the copper.)
Intriguingly, the closest structural match was found with the putative active site of copper methane mono-oxygenase (Cu-MMO), where the histidine brace was also observed (21, 22), albeit at a coordination site that, informed by extended X-ray absorption fine structure and EPR studies, was modeled with two copper ions separated by ∼2.6 Å (22). Notwithstanding this difference, the structural homology between the actives sites of Cu-TaGH61A and Cu-MMO is striking, even extending beyond the immediate histidine brace coordination sphere of the copper ion to potential distal residues that may be important in O2 activation (Fig. S4A). Indeed, in this context, both enzymes carried out oxidative chemistry, indicating that the copper histidine brace represents a special subclass of copper oxidases. The possibility that Cu-GH61 and Cu-MMO use a monomeric copper(III)-oxo [or tyrosyl-copper(II)-oxo in the case of GH61] stabilized by a deprotonated amino terminus now warrants detailed investigation.
GH61 Active Site Contains Unique N-methylated Histidine Motif.
Further close examination of the Cu-TaGH61 structure reveals a previously undescribed feature that went unmodeled in the previous structure determinations of both H. jecorina (HjGH61B; Fig. S4B) and Thelavia terrestris (TtGH61E) (11). In both of the previous structures, the Nε2 atom of the N-terminal histidine shows clear electron density consistent with a covalent modification at this atom. In the structure of Cu-TaGH61 presented here, the residual difference electron density is also present at the Nε2 atom. The difference density is best modeled as a methyl group and gives the 2Fo − Fc electron density map shown in Fig. 3A and Fig. S4B for HjGH61B. The methylation of the terminal histidine in TaGH61 was confirmed by intact protein MS, high-accuracy MS analysis of N-terminal peptides, and sequential N-terminal Edman degradation (Fig. S5).
Although natural modification of copper-binding amino acids including histidine has been observed before (e.g., in galactose oxidase and hemocyanin), we believe that natural methyl modification of a histidine group that binds to copper has not been described previously. His methylation has been reported in the Protein Data Bank (PDB), where the modified histidine has the code HIC; however, out of the 51 PDB HIC-containing entries, 47 are for actin. The remaining modified Nε2 histidines are part of an unusual cyclic peptide in RNA methyltransferases. It is worth noting that nonnatural histidine methylation of the copper coordinating residues of the Alzheimer disease amyloid-β peptide (incorporated by in vitro peptide synthesis) leads to four times higher rate of H2O2 than the unmodified peptide (23). Precedence thus exists for histidine methylation modulating the reactivity of metals in biological systems. Nevertheless, given that the modification seen in GH61s would define a previously undescribed paradigm in copper oxidases [although one consistent with small molecule studies (24)], the importance of the methylation for catalytic competency and for the stability of the enzyme must be investigated further.
Reactivity Studies.
Copper as the metal ion necessary for GH61 activity is confirmed from reactivity studies, thereby completing a structure–activity relationship for Cu-TaGH61 and for the GH61 family in general. PASC solutions were treated with apo-TaGH61 or copper-loaded TaGH61 in the presence of ascorbate (as another example of a redox cofactor), and the supernatants were subjected to polysaccharide analysis by using carbohydrate gel electrophoresis (PACE) and MALDI-TOF analyses (Fig. 4). To conduct this experiment, it was necessary to remove fully from solution any transition metal ions that would otherwise catalyze the rapid and futile aerobic oxidation of the reducing cofactor, thus rendering the catalyst inactive. This complication is a particular consequence for experiments that use insoluble substrates (in this case PASC), from which it is very difficult to remove metal ions quantitatively in advance. This complication, coupled with the extremely high affinity of GH61s for copper, bedevils this area of research and can lead to ostensibly contradictory findings, as evidenced by the numerous previous suggestions for the metal ion cofactor or the observation that added metal ions inhibit GH61 activity in GH61s and in CBP21. Mindful of this issue, we decided to take advantage of the very high kinetic and thermodynamic stability of Cu-TaGH61 by adding EDTA directly to the reaction mixture. The EDTA chelated all free metal ions, including in the PASC, thus reducing the futile oxidation of the cofactor (25), but the EDTA did not remove the copper from the enzyme over the timescale of the experiment (as described above and in Fig. S3B).
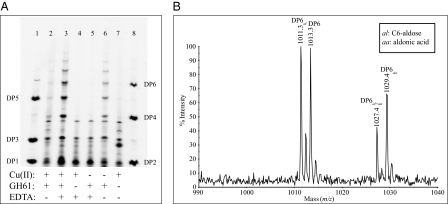
Fig. 4.
Copper-loaded TaGH61A is catalytically active as seen by PACE (A) and MALDI-TOF (B) analyses of saccharide products formed during the reaction of 4 mg/g cellulose of TaGH61A with 5 g/L PASC in the presence of 10 mM ascorbate for 1 h at 50 °C at pH 5, with 25 mM triethylammonium acetate. TaGH61A had previously been demetallated and, where indicated, loaded with Cu(II). A shows the spectrum of saccharide products with an unmodified reducing end, and B shows the full spectrum of DP6 products. The product profile is similar to that from the reaction of TaGH61 with undefined metal load with PASC using gallate as the reducing agent (as seen in Fig. 2).
Under these conditions, after 1 h of reaction, high levels of oligosaccharides were detected only in the supernatant of the reaction of copper-loaded TaGH61 of PASC with EDTA added (Fig. 4A, lane 3). No activity at all was seen with apo-TaGH61 and EDTA. Notably, in the absence of added copper and EDTA (Fig. 4A, lane 6), low levels of activity were seen. This activity was presumably due to the presence of adventitious copper, especially in the PASC [cellulose is known to have a high capacity for binding copper (26)], which could bind to TaGH61. The pattern of cellodextrins and oxidized oligomers generated by Cu-TaGH61 is similar to the pattern seen in previous experiments where no additional copper had been added to the enzyme or the reaction mixture (Fig. 4 A and B).
Discussion
The recalcitrance of crystalline cellulose to enzymatic degradation is well established. The first challenge is that the glycosidic bond is intrinsically resistant to hydrolytic attack; indeed, it has been calculated that the uncatalyzed half-life of cellulose is some 5 million years (27). A second factor that renders plant cell wall polysaccharides challenging is that they are themselves insoluble, reflecting extensive intrachain hydrogen bonding. In planta, these substrates are often embedded in highly complex composites with other polysaccharides and with lignin (3, 8, 15, 28). These problems amplify each other, making cellulose degradation a major biotechnological challenge of the 21st century. Therefore, effective enzymatic cleavage of cellulose would benefit greatly from a strong thermodynamic driving force—one provided here with oxygen as the ultimate oxidizing agent. Moreover, the stability can only be overcome with a catalyst that employs highly reactive oxygen species similar to those found in other metalloenzyme oxidases. In this context, TaGH61's, and by analogy other GH61s’, use of copper–oxygen species—as opposed to classical acid/base facilitated hydrolysis—to initiate and promote polysaccharide breakdown overcomes the major hurdle of extracting and distorting a single polysaccharide chain in the active center of a classical glycoside hydrolase. The GH61 product profile can be contrasted with that resulting from the action of the structurally related CBP21 enzyme on chitin, where only even-numbered DP oligosaccharides with aldonic acid termini are produced (17). Additionally, although it is likely that both GH61s and CBP21 act as oxidative enzymes, they may differ in their detailed modes of action.
Conclusions
We have demonstrated that TaGH61 is activated by two cofactors: a soluble redox active agent exemplified here by ascorbate or gallate and a copper ion. The activated enzyme, in concert with traditional cellulases, significantly enhances cellulose degradation. The active site is best described as a type II copper site and is very likely to be the place of oxygen activation and subsequent oxidation of cellulose. The site has an unprecedented methylated histidine, thereby offering a unique paradigm in copper bioinorganic chemistry. As such, Cu-TaGH61, and by analogy the GH61 family of proteins and structurally related polypeptides, falls into the class of copper oxidoreductases, all of which are capable of performing powerful oxidation chemistry. As part of the consortium of cellulose-degrading enzymes, GH61 action renders the substrate far more prone to attack by the classical endoglucanases and cellobiohydrolases and thus provides a major breakthrough in enzymatic biomass conversion—one that opens up avenues in the continuing drive toward environmentally friendly and secure energy.
Materials and Methods
Reagents and buffers were reagent-grade or better. PCS from the U.S. National Renewable Energy Laboratory was ground, sieved, and adjusted to pH 5. PASC was prepared from Avicel (FMC, PH101) (29). The enzyme mixture used for cellulose hydrolysis contained H. jecorina cellulases (GH7A, GH6A, GH7B, and GH5A) and A. oryzae GH3A (AoBG). AoBG and T. aurantiacus GH61A (TaGH61A) were expressed and purified as reported (11). Enzymatic reactions with PCS, Avicel, and PASC were carried out as reported (11). Permethylation of cellodextrin before MALDI-TOF-MS or MS/MS was accomplished as described (30). MALDI-TOF-MS and MS/MS were performed with a 4700 Proteomics Analyzer (Applied Biosystems) using a 2,5-DHB matrix.
For EPR, the sample of GH61 was demetallated as described (31). Approximately 10 μL of 10-mM aqueous solutions of copper(II) nitrate was added to create a 1:1 metal:protein stoichiometry at a concentration of ∼0.5 mM. Continuous-wave EPR spectra were obtained as frozen glasses in 10–20% glycerol solutions at 140 K on a Bruker EMX spectrometer at 9.28 GHz.
ITC was performed at 25 °C with a Microcal AUTO-ITC by using either 194 or 58 μM TaGH61A following extensive dialysis into 10 mM sodium acetate, pH 5.0. The metal ions were diluted to 2 mM in the dialyzate buffer and pH-adjusted. 25 × 4-μL injections of each metal were made, and data were fit with a single-site binding model.
TaGH61A enzyme activity dependence of Cu(II) was evaluated by incubating 0.5% PASC with 0.28 μM TaGH61A in the presence of 10 mM ascorbic acid in 25 mM triethylammonium acetate at pH 5. The reactions were incubated at 50 °C in an Eppendorf Thermomixer at 1,400 rpm. Stock solutions of enzyme, buffer, and ascorbate were demetallated as described (24), and the PASC was washed with ultrapure water (Sigma-Aldrich). Cu(II) solution was prepared from Cu(NO3)2 (Sigma-Aldrich) and added either to the enzyme in 3/4 the molar amount of GH61 or to a final concentration of 0.21 mM in the final reaction mixtures.. EDTA was prepared from the Na–EDTA salt (Merck) and added to a concentration of 10 mM where indicated. The enzymatic reactions were terminated by heating to 99 °C for 10 min. The samples were briefly centrifuged, and 50 μL of the supernatants were dried down. PACE, involving derivatization of reducing carbohydrates by 8-aminonaphthalene-1,3,6-trisulfonic acid, was performed as described (32).
Crystallization, Structure Determination, and Refinement.
T. aurantiacus GH61 protein produced recombinantly in A. oryzae and deglycosylated was concentrated to 15 mg/mL in 20 mM Na-acetate, pH 5.5. X-ray quality rod-shaped crystals were obtained in a 0.4-μL drop with 0.2 M NaCl, 0.1 M Hepes, pH 8.0, and 25% (wt/vol) PEG 3350 as precipitant.
Data Collection and Processing.
The crystal was cryoprotected with a mixture of glycerol, ethylene glycol, D(+)-sucrose, and D(+)-glucose before flash freezing in liquid nitrogen. Data were collected at Beamline I911-2 of MAX-lab (Lund, Sweden) to a maximum resolution of 1.5 Å and processed in MOSFLM and SCALA (33).
The structure was solved by molecular replacement (MR) and partially built by using the TrGH61B structure (ref. 16; PDB ID code 2VTC; sequence identity of 45%) as search model to find the two molecules (related by pseudotranslatonal symmetry) in the asymmetric unit. Refinement (see Table S2 for statistics) was carried with manual rebuilding in COOT (35) for all structures. In final refinement rounds, protein atoms were refined anisotropically. Figures were made in PYMOL (www.pymol.org). The final model has good geometry with 98% of residues in the favored regions of the Ramachandran plot. For more details see SI Materials and Methods.
Another crystal (SI Materials and Methods) was soaked in 10 mM Cu(NO3)2, and another dataset was collected. The structure was solved by MR using the initial TaGH61A structure as a search model. Density was found at the metal binding site, both in the structure solved in the absence of divalent metal cations in the crystallization conditions and when the crystal had been soaked in a copper-containing solution (see Table S2 for details and statistics).
Protein Mass Spectrometry Analyses.
Intact protein MS was performed by using a Bruker microTOF focus electrospray mass spectrometer (Bruker Daltonik). High-accuracy MS analysis of N-terminal peptides was performed with an LTQ Orbitrap Velos nano-LC-MS/MS system (Thermo Scientific). N-terminal Edman degradation was performed with a Procise system (Applied Biosystems). For further details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Keith McCall, Kim Borch, Janne E. Tønder, Leonardo De Maria, Rune N. Monrad, Kim B. Andersen, Anne Sofie U. Larsen, Charlotte G. B. Veng, Jimmi O. Kristiansen, Kim Brown, K. C. McFarland, Elena Vlasenko, Hanshu Ding, Armindo R. Gaspar, Hui Xu, Don Higgins, Lars Anderson, Ani Tijerian, Jim Langston, and Paul Harris (Novozymes) for technical assistance and discussion; Robert L. Starnes and Claus C. Fuglsang (Novozymes) for critical reading of the manuscript; Dr. Victor Chechik (University of York) for collecting the EPR spectra; Dorthe Boelskifte and Veronika Karlsson (University of Copenhagen) for help with crystallization; and the MAX-lab synchrotron for beam time. Synchrotron usage was supported by the DANSCATT program (Danish Natural Science Research Council). Research on plant cell wall degrading enzymes in the G.J.D. laboratory is funded by the Biotechnology and Biological Sciences Research Council through Grant BB/I014802. G.J.D. is a Royal Society/Wolfson Research Merit award recipient. This material is based upon work supported by U.S. Department of Energy Award DE-FC36-08GO18080.
Footnotes
Conflict of interest statement: R.J.Q., M.D.S., K.S.J., K.B.R.M.K., C.I.J., M.T., A.A., C.P.W., and F.X. are employees of Novozymes, which is a major enzyme producing company.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2YET and 3zud).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105776108/-/DCSupplemental.
References
- 1.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen H, Kristensen JB, Felby C. Enzymatic conversion of lignocellulose into fermentable sugars: Challenges and opportunities. Biofuels Bioproducts and Biorefining. 2007;1:119–134. [Google Scholar]
- 3.Himmel ME, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 4.Service RF. Is there a road ahead for cellulosic ethanol? Science. 2010;329:784–785. doi: 10.1126/science.329.5993.784. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DB. Cellulases and biofuels. Curr Opin Biotechnol. 2009;20:295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Viikari L, Alapuranen M, Puranen T, Vehmaanpera J, Siika-aho M. In: Biofuels. Olsson L, editor. Berlin/Heidelberg: Springer; 2007. pp. 121–145. [Google Scholar]
- 7.Brumer H. Carbohydrases. New York: John Wiley & Sons; 2002. [Google Scholar]
- 8.Henrissat B, Driguez H, Viet C, Schulein M. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Nat Biotechnol. 1985;3:722–726. [Google Scholar]
- 9.Vocadlo DJ, Davies GJ. Mechanistic insights into glycosidase chemistry. Curr Opin Chem Biol. 2008;12:539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Reese ET, Siu RGH, Levinson HS. The biological degradation of soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J Bacteriol. 1950;59:485–497. doi: 10.1128/jb.59.4.485-497.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PV, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry. 2010;49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 12.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 13.Arantes V, Milagres AM, Filley TR, Goodell B. Lignocellulosic polysaccharides and lignin degradation by wood decay fungi: The relevance of nonenzymatic Fenton-based reactions. J Ind Microbiol Biotechnol. 2011;38:541–555. doi: 10.1007/s10295-010-0798-2. [DOI] [PubMed] [Google Scholar]
- 14.Varela E, Tien M. Effect of pH and oxalate on hydroquinone-derived hydroxyl radical formation during brown rot wood degradation. Appl Environ Microbiol. 2003;69:6025–6031. doi: 10.1128/AEM.69.10.6025-6031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez D, et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karkehabadi S, et al. The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J Mol Biol. 2008;383:144–154. doi: 10.1016/j.jmb.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 18.Tucker MP, Kim KH, Newman MM, Nguyen QA. Effects of temperature and moisture on dilute-acid steam explosion pretreatment of corn stover and cellulase enzyme digestibility. Appl Biochem Biotechnol. 2003;105:165–177. doi: 10.1385/abab:105:1-3:165. [DOI] [PubMed] [Google Scholar]
- 19.Du B, et al. Effect of varying feedstock-pretreatment chemistry combinations on the formation and accumulation of potentially inhibitory degradation products in biomass hydrolysates. Biotechnol Bioeng. 2010;107:430–440. doi: 10.1002/bit.22829. [DOI] [PubMed] [Google Scholar]
- 20.Dodson EJ, Woolfson MM. ACORN2: New developments of the ACORN concept. Acta Crystallogr D Biol Crystallogr. 2009;65:881–891. doi: 10.1107/S0907444909016515. [DOI] [PubMed] [Google Scholar]
- 21.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramanian R, et al. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tickler AK, et al. Methylation of the imidazole side chains of the Alzheimer disease amyloid-beta peptide results in abolition of superoxide dismutase-like structures and inhibition of neurotoxicity. J Biol Chem. 2005;280:13355–13363. doi: 10.1074/jbc.M414178200. [DOI] [PubMed] [Google Scholar]
- 24.Santagostini L, Gullotti M, Pagliarin R, Monzani E, Casella L. Enantio-differentiating catalytic oxidation by a biomimetic trinuclear copper complex containing L-histidine residues. Chem Commun (Camb) 2003;(17):2186–2187. doi: 10.1039/b307349a. [DOI] [PubMed] [Google Scholar]
- 25.Kuzuya M, et al. Role of lipoprotein-copper complex in copper catalyzed-peroxidation of low-density lipoprotein. Biochim Biophys Acta Lipids Lipid Metab. 1992;1123:334–341. doi: 10.1016/0005-2760(92)90015-n. [DOI] [PubMed] [Google Scholar]
- 26.Druz N, Andersone I, Andersons B. Interaction of copper-containing preservatives with wood. Part 1. Mechanism of the interaction of copper with cellulose. Holzforschung. 2001;55:13–15. [Google Scholar]
- 27.Wolfenden R, Lu X, Young G. Spontaneous hydrolysis of glycosides. J Am Chem Soc. 1998;120:6814–6815. [Google Scholar]
- 28.Selig MJ, Knoshaug EP, Adney WS, Himmel ME, Decker SR. Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresour Technol. 2008;99:4997–5005. doi: 10.1016/j.biortech.2007.09.064. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y-HP, Cui J, Lynd LR, Kuang LR. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: Evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules. 2006;7:644–648. doi: 10.1021/bm050799c. [DOI] [PubMed] [Google Scholar]
- 30.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 31.Carrer C, et al. Removing coordinated metal ions from proteins: A fast and mild method in aqueous solution. Anal Bioanal Chem. 2006;385:1409–1413. doi: 10.1007/s00216-006-0603-2. [DOI] [PubMed] [Google Scholar]
- 32.Goubet F, Jackson P, Deery MJ, Dupree P. Polysaccharide analysis using carbohydrate gel electrophoresis: A method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal Biochem. 2002;300:53–68. doi: 10.1006/abio.2001.5444. [DOI] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.