Abstract
In eusocial insects the production of daughters is generally restricted to mated queens, and unmated workers are functionally sterile. The evolution of this worker sterility has been plausibly explained by kin selection theory [Hamilton W (1964) J Theor Biol 7:1–52], and many traits have evolved to prevent conflict over reproduction among the females in an insect colony. In honeybees (Apis mellifera), worker reproduction is regulated by the queen, brood pheromones, and worker policing. However, workers of the Cape honeybee, Apis mellifera capensis, can evade this control and establish themselves as social parasites by activating their ovaries, parthenogenetically producing diploid female offspring (thelytoky) and producing queen-like amounts of queen pheromones. All these traits have been shown to be strongly influenced by a single locus on chromosome 13 [Lattorff HMG, et al. (2007) Biol Lett 3:292–295]. We screened this region for candidate genes and found that alternative splicing of a gene homologous to the gemini transcription factor of Drosophila controls worker sterility. Knocking out the critical exon in a series of RNAi experiments resulted in rapid worker ovary activation—one of the traits characteristic of the social parasites. This genetic switch may be controlled by a short intronic splice enhancer motif of nine nucleotides attached to the alternative splice site. The lack of this motif in parasitic Cape honeybee clones suggests that the removal of nine nucleotides from the altruistic worker genome may be sufficient to turn a honeybee from an altruistic worker into a parasite.
Keywords: caste determination, cuticular protein 2-family, gene expression
The evolution of a sterile worker caste in eusocial hymenoptera has been plausibly explained by inclusive fitness theory (1). Generally, workers refrain from reproduction as a result of intracolonial reproductive hierarchies. Ovary activation in honeybee workers (Apis mellifera) is inhibited by the pheromones from the queen and the brood (2). In addition, the multiple mating of the queen and the coexistence of many half-sibling subfamilies in the colony facilitates worker policing, where workers remove eggs laid by other workers (3), leading to <1% of worker-laid offspring (4).
These mechanisms often fail, however, to control worker reproduction of the Cape honeybee (Apis mellifera capensis). In this subspecies, laying workers can function as social parasites invading foreign colonies, killing the resident queen and establishing themselves as pseudoqueens (5–7). The proximate mechanisms for this parasitic life history strategy are well understood (8). Parasitic pseudoqueen workers produce a queen-like pheromonal bouquet indicating the presence of a queen to the host workers (9–13). Moreover, the diploid offspring of the parasitizing workers produce brood pheromones, suggesting the presence of a laying queen to the host workers and preventing these workers from activating their ovaries. Finally, the diploid eggs laid by the parasitic workers are not policed as predicted by evolutionary theory (1, 14). A single locus termed thelytoky (th) is thought to control this parasitic life history strategy (15). Workers that are homozygous for the th allele produce parthenogenetic diploid offspring, have rapid ovary activation, and produce queen-like pheromones. The th locus, which has been mapped to a region spanning 11.4 cM comprising 15 genes on chromosome 13 (16), is thus in control of the three essential characters that facilitate social parasitism. Because transcription factors control several traits within one organism, the two transcription factors in this region [ATF2 (XM_393896) and CP2 (XM_001121158; XM_393898)] were considered to be prime candidate genes to control the parasitic worker phenotype in A. mellifera. Because the ATF2 homolog did not show differential expression between different castes (Fig. S1), and members of the CP2 transcription factor family were differentially expressed between queens and workers in both the honeybee (Fig. S1) and the stingless bee Melipona quadrifasciata (17), we focused on the CP2 transcription factor as the prime candidate for the th locus in honeybees. This transcription factor is homologous to the gemini (= genitalia missing) locus in Drosophila melanogaster (18), which was shown to interact with the Spindle-F protein CG12114 (19). This protein has a minus end-directed microtubule activity, and is involved in oocyte axis determination and oocyte microtubule cytoskeleton organization. This is of particular interest, as the restoration of diploidy in A. m. capensis worker eggs is caused by an abnormal spindle rotation during meiosis (20).
The functional diversity of several CP2 transcription factor family members is derived from alternative splicing leading to tissue- and stage-specific isoforms (21). We show that alternative splicing of the honeybee gemini homolog as a general mechanism for generating functional diversity of transcription factors (22) provides the genetic mechanism for the th locus to affect reproductive dominance of honeybee workers.
Results
Comparing the gemini splice patterns of laying and nonlaying thelytokous workers (A. m. capensis) with those of laying and nonlaying arrhenotokous workers (A. m. carnica) yielded different transcript isoforms at exon 5 and exon 7 (Fig. 1).
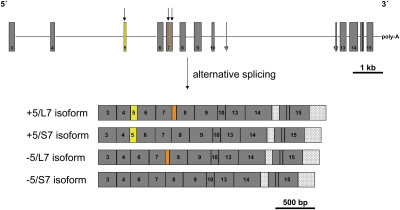
Fig. 1.
Diagram of gemini (introns as lines, exons as boxes) and its four pre-mRNA splice products. Arrows indicate the alternatively spliced exons. +5, isoform with exon 5; –5, isoform lacking exon 5; L, long isoform of exon 7; S, short isoform of exon 7. Exon 5 (yellow) is present in the first two isoforms but absent in the other two. The length polymorphism in exon 7 is shown in orange. Spliced intronic regions are light gray. Exon numbering refers to the annotated transcript XM_001121158.1 (National Center for Biotechnology Information; Nucleotide Database).
Exon 5 is a cassette exon with two mRNA splice isoforms—the full-length transcript and a shorter transcript with a deletion of 78 bp. The deletion of these base pairs does not affect the ORF and leads to a putative protein shortened by 26 aa. Exon 7 contains two alternative 5′ splice sites. The first site causes a deletion of 59 bp with a shift in the reading frame. The resulting isoform contains a stop codon in exon 8 and has therefore an incomplete DNA-binding domain lacking the C-terminal protein domains. Quantification of this transcript was not possible in our samples due to its low abundance. The second splice site results in a deletion of 24 bp (S7 transcript form in Fig. 1) and the loss of 8 aa in the putative DNA-binding domain spanning from exon 7 to exon 9. Both splice forms of exon 5 and exon 7 were found in combination with each other (Fig. 1).
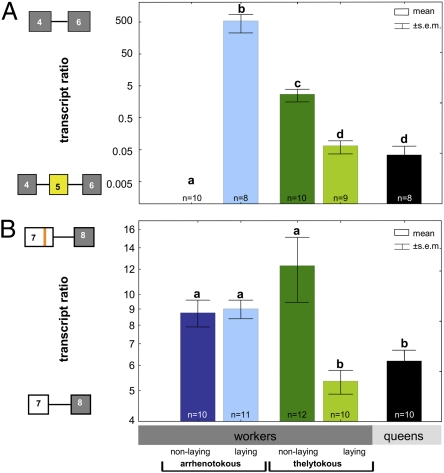
Semiquantitative RT-PCRs revealed that the splice patterns of exons 5 and 7 were characteristic of the different reproductive states of the tested queens and workers (Figs. 2 and 3, Fig. S2, and Table S3), and of the different modes of parthenogenesis. In laying arrhenotokous queens that served as positive controls for fully reproductive females, both the spliced and the full transcripts were produced for both exons (Figs. 2 and 3, Fig. S2, and Table S3). Arrhenotokous A. m. carnica workers showed a different pattern. Workers with undeveloped ovaries exclusively produced the full transcript containing exon 5, whereas laying workers almost exclusively produced the splice product lacking exon 5. Exon 7 predominantly showed the long splice variant in both reproductive states, although both isoforms were expressed at a higher level in laying workers. Thelytokous A. m. capensis workers (homozygous at the th locus), both laying and nonlaying, produced both transcript forms of exon 5 in a pattern similar to that of the queen. In this subspecies the different splice patterns of exon 7 rather than exon 5 matched the two reproductive states. When the splice pattern of thelytokous workers was similar to that of nonreproductive arrhenotokous workers, they did not have activated ovaries. However, when the exon 7 splicing pathway was similar to that of the queen, workers were egg layers.
Fig. 2.
Transcript ratios of alternate transcripts of gemini (mean values ± SE; log scale). (A) Exon 5-missing transcripts (−5) in relation to exon 5-containing transcripts (+5). (B) Unspliced exon 7 transcripts (L7) in relation to −24-bp deletion exon 7 transcripts (S7): arrhenotokous workers in blue (dark, nonlaying; light, egg laying), thelytokous workers in green, and arrhenotokous laying queens in black. Different letters indicate significant differences (P ≤ 0.05).
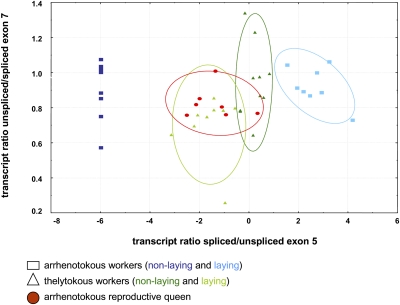
Fig. 3.
Transcript ratio 2D scatterplot with 0.6 confidence ellipses of all gemini isoforms. The ratio of exon 5-deficient transcripts in relation to exon 5-containing transcripts are plotted on the x axis, whereas the ratio of unspliced vs. spliced exon 7 transcript are plotted on the y axis. All of the values were log transformed before they were plotted. All of the tested groups apart from queen and reproductive A. m. capensis worker samples significantly differ from each other (one-way ANOVA F = 29.33; df = 4; P ≤ 0.001 and Newman–Keuls post hoc tests P ≤ 0.01). Every symbol stands for a single individual.
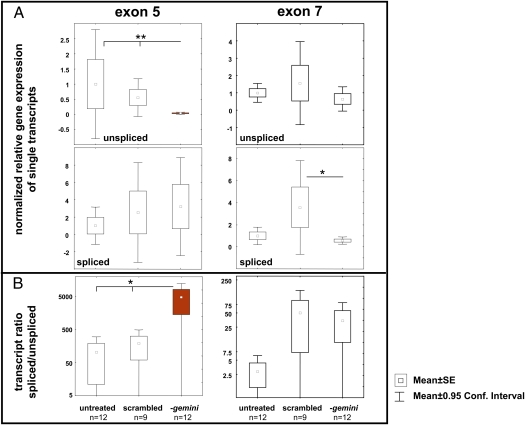
To rule out the possibility that the splice pattern of gemini was simply correlated with different reproductive states of workers, we knocked down the level of transcripts containing exon 5 in A. m. carnica worker bees by undertaking a series of siRNA feeding experiments to change the ratio of exon 5-containing/missing transcripts. Therefore, workers were fed daily with two siRNAs specific for the 3′ and 5′ end of exon 5 for 14 d. The abundance of transcripts containing exon 5 was reduced >90% (Fig. 4) by this treatment. Moreover, although the abundance of transcripts containing the shorter variant of exon 7 was lower in the treatment group compared with workers fed with nonsense siRNA, the ratio of exon 7-missing to exon 7-containing transcripts did not change significantly. Nevertheless, the exon 5 knockdown bees had a higher reproductive capacity, showing ovaries with swollen ovarioles and mature eggs that were significantly more frequent than in the various controls (Fig. 5).
Fig. 4.
Relative gene expression of all gemini transcripts after exon 5 knockdown by RNAi. (A and B Left) Gene expression of the transcripts containing or missing exon 5. (Right) Gene expression data on exon 7 . (A) Relative gene expression of different gemini transcripts. The transcript expressions were normalized to untreated bees whose transcript expression was set to 1. The asterisks indicate a significantly reduced expression of transcripts containing exon 5 after the feeding of exon 5-specific siRNAs (P < 0.01; Kruskal–Wallace ANOVA and multiple comparisons of mean ranks for all groups). (B) Expression ratios of spliced vs. unspliced transcripts shown on a logarithmic scale. The asterisk indicates a significantly shifted exon 5 transcript ratio toward the shorter isoforms of exon 5 (P < 0.05; Kruskal–Wallace ANOVA and multiple comparisons of mean ranks for all groups).
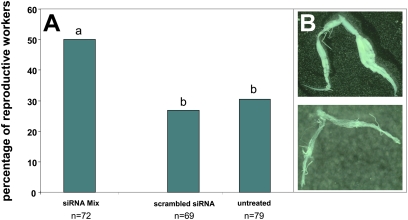
Fig. 5.
RNAi-mediated exon 5 knockdown and the resulting ovary activation in arrhenotokous A. m. scutellata workers from two different colonies treated with gemini-specific siRNA (n = 72), scrambled siRNA (n = 69), or sugar water (n = 79). (A) The frequency of reproductive workers in the three treatment groups. The knockdown by the combination of two transcript-specific siRNAs leads to a significant increase of reproductive workers shown by different letters (P ≤ 0.05, Mann–Whitney U test tests with P values corrected for multiple testing). (B) Dissected worker ovaries were classified as developed when the ovarioles were swollen and mature eggs visible (Upper) or as undeveloped (Lower).
Sequencing the coding regions of gemini in arrhenotokous and thelytokous workers and in arrhenotokous queens yielded no differences in the overall sequence. Hence these regions themselves cannot function as the allelic forms of the th locus. Therefore, we sequenced all noncoding regions of gemini gene, including the promoter region and the introns flanking the alternatively spliced exons. No consistent sequence differences were found in the promoter region and in the nonflanking introns. Furthermore, sequencing the flanking intronic regions of exon 7 did not reveal any consistent sequence differences between arrhenotokous and thelytokous subspecies. Only in the downstream intron flanking cassette exon 5 (the putative molecular thelytoky switch), a consistent deletion of 9 bp (5′-GAAACGATG-3′) was found in the parasitic Cape honeybee clone (Fig. S3). We named this deletion the thelytoky associated element 1 (tae1). Both arrhenotokous subspecies, the European A. m. mellifera and the African A. m. scutellata, show the identical intronic sequence in this region, confirming that it is not just a marker that is characteristic of African honeybees. This tae1 sequence shows a very high level of purines (77.8%) compared with the purine content of the whole intron (50.47%).
Discussion
In eusocial insects, caste-specific phenotypic plasticity and worker reproduction are controlled by differential gene expression (23, 24) maintained by epigenetic modifications (25, 26) and caused by differential feeding of queens with royal jelly (25, 27). This study reveals another genetic mechanism that controls reproductive dominance in honeybees: different transcript isoforms of a single transcription factor generated by alternative splicing, to control the reproductive physiology of this eusocial insect.
Characterization and quantification of honeybee gemini isoforms revealed that the abundance of four processed transcripts corresponded with different ovarian states of workers and also with different castes. Furthermore, knockdown experiments in A. m. carnica workers revealed exon 5 to be causative in regulating the ovarian development in arrhenotokous workers. This knockdown leads to an alteration in the ratio of exon 5-containing and exon 5-missing transcripts, which subsequently controls the onset of egg laying. We did not observe such a shift in the exon 7-containing/missing transcripts, suggesting that exon 7 is not involved in the control of ovary activation in arrhenotokous bees.
In addition to the control of ovary activation, the mapped th locus controls the production of queen-like amounts of 9-ODA, the queen substance (16). This pheromone suppresses worker reproduction in arrhenotokous subspecies and ensures the reproductive dominance of the queen. Furthermore, the very same locus is responsible for the thelytokous production of diploid worker-laid eggs (16). Because the alternative splice pattern of gemini is characteristic for both the queen pheromonal phenotype and the thelytokous mode of parthenogenesis in workers, we suggest that gemini and its altered transcripts are also important for these traits (although we did not prove this by knockout experiments as for ovary activation). As we showed that exon 5 directly affects ovarian development, we expect the shorter exon 7 in combination with a specific expression ratio of exon 5 to be responsible for both the caste-specific reproductive dominance in arrhenotokous honeybee species and the thelytokous production of diploid offspring. One possible interpretation of the data (among many others) is a simplistic two-exon regulatory model (Fig. 6) to explain this complex network. We suggest that splicing of exon 5 and a certain threshold ratio of unspliced vs. spliced transcripts directly controls the shift of ovary activation in arrhenotokous subspecies. To be consistent with our model, splicing of exon 5 alone should be insufficient to exhibit the full pseudoqueen phenotype in thelytokous Cape honeybees. The abundance of transcripts missing exon 5 differs slightly between laying and nonlaying thelytokous workers, but is well below the threshold that governs ovary activation of arrhenotokous workers; it may not sufficiently vary to serve as a switch for ovary activation in thelytokous workers. As thelytokous worker groups produce both splice variants of exon 5 qualitatively similar to that of arrhenotokous queens, we propose this queen-like level of exon 5 to determine the mode of parthenogenesis (thelytoky). Additionally, the differences in the splice products of exon 7 between laying and nonlaying thelytokous workers suggest that alternative splicing of exon 7 is involved in ovary activation in thelytokous workers. Reproductive thelytokous workers may reduce the relative abundance of the long exon 7 and 5 when exploiting a host, thereby completely resembling the queen splice pattern for both exon 5 and exon 7 (red queen line in Fig. 6).
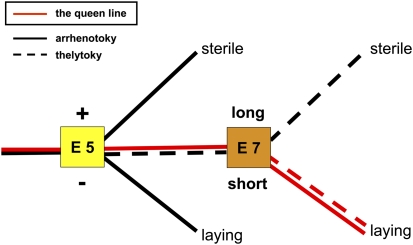
Fig. 6.
Double-exon regulatory model for controlling worker reproduction by alternative splicing of exon 5 (E5) and exon 7 (E7) of gemini. The expression of different transcript isoforms is dependent on the worker's reproductive status and the mode of worker parthenogenesis (solid line, arrhentoky; dotted line, thelytoky). The red line represents the splice pattern of the queen (queen line) that expresses both transcript isoforms for both exons. Minus (−) or plus (+) signs indicate the absence or presence of exon 5. Long or short indicates the unspliced or spliced isoform of exon 7.
The missing protein domains of the spliced transcripts may essentially modify the access of gemini to different DNA-binding domains of other genes, which eventually result in the observed phenotypic differences. Exon 7 is part of the CP2 DNA-binding domain, and the deletion of 8 aa is expected to have an impact on the DNA-binding capacity of the resulting protein and therefore on the worker phenotype. Similar effects have been shown in human LBP-1d (UBP-1; another CP2 member) transcript isoforms that have a deletion in the putative DNA-binding domain, generated by alternative splicing. Those transcripts are no longer able to bind DNA and repress DNA binding of other isoforms by forming heteromers (28). Because exon 5 is not part of the CP2 DNA-binding domain, a mechanism other than DNA binding must control the molecular switch controlling the onset of egg laying. A common mechanism for controlling the action of transcription factors is the alteration of dimerization domains that cause isoform variability (22). For example, in grainyhead, another member of the CP2 family, a mutant lacking the N-terminal activation domain binds to the full-length grainyhead isoform and hence inhibits its ability to homodimerize (29).
To further understand the mechanisms controlling the observed splice patterns, we sequenced all noncoding regions of gemini, including the promoter region, which may also comprise sequence stretches necessary for accurate splicing (30–32). We did not observe any consistent sequence differences in the nonflanking intronic regions or in introns flanking exon 7. However, all parasitic Cape honeybee workers share a common deletion of 9 bp in the flanking intron of exon 5—the exon that controls ovary activation in arrhentotokous honeybees and that is differentially spliced in thelytokous honeybees. Both European A. m. mellifera and African A. m. scutellata show the identical sequence without the 9-bp deletion in this flanking intron, confirming that it is not only a marker of African honeybees but perhaps the functional site controlling the mode of worker parthenogenesis. Interestingly, and similar to other intronic splice enhancer (ISE) motifs, this thelytoky associated element 1 (tae1) sequence is purine rich and short in length. Differential splicing has repeatedly been shown to be controlled by such ISE motifs located downstream of alternate exons (33, 34). The 9-bp deletion in A. m. capensis may therefore have far-reaching effects on the splice-site recognition of the upstream laying cassette exon 5. As this was the only consistent sequence difference we found in the four flanking introns of the spliced exons, it might be possible that tae1 not only affects the exon 5 switch but also the splicing of exon 7, as it has been shown that more distant splice regulatory motifs can control splicing of non-neighbor exons (32). Predictions of the secondary structure of intron 5_6 of gemini suggest that this sequence is part of a stem/loop conformation (Mfold) (35). With the deletion of this motif, the stem integrating the relevant bases is predicted to disappear. This conformational change might influence the pre-mRNA splicing process via different mechanisms (36). Either structural elements might hinder the accessibility of cis regulatory sequences by splicing factors, or the altered secondary structure might interfere with the regulatory mechanisms required for successful and accurate splicing. Furthermore, it remains possible that the deletion itself is directly involved in the assembly of putative splice factors. Serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) have been shown to interact with such purine-rich motifs in mammals (33) and hence directly control the altered splice pattern of gemini in honeybees.
Irrespective of the actual molecular switch mechanisms, we have found a gene that is alternatively spliced in a caste and type of parthenogenesis-specific manner and central to the regulation of worker fertility. If the 9-bp deletion with the ISE motif (tae 1) flanking the ovary activation exon in nonparasitic honeybee workers proves to be the allelic form of the thelytoky gene in future studies, it would take as little as nine nucleotides to turn an altruistic worker into one that can become a social parasite.
Material and Methods
Honeybee Samples for gemini Expression.
A. m. capensis workers were reared from brood produced by social parasitic workers in an A. m. scutellata host colony at the University of Pretoria and are therefore homozygous for the thelytoky allele. The brood frames were incubated at 35 °C and ∼60% relative humidity. Emerging bees were kept in hoarding cages containing 120 individuals with food and water ad libitum. After 16 d of incubation, the bees were killed and the ovaries dissected and stored in RNAlater until RNA preparation. A. m. carnica workers and queens were held at the apiary of the Martin-Luther-Universität Halle-Wittenberg. Sealed brood frames with young nurse bees and emerging brood were removed from the colony and kept queenless. After 14 d, ∼200 workers (aged ∼7–21 d) were randomly sampled from the frames and immediately flash-frozen in liquid nitrogen. A. m. carnica queens were reared using standard apicultural techniques and kept in small colonies of ∼1,000 worker bees until day 7 after the onset of egg laying. All sampled bees were stored at −80 °C. Worker ovaries were classified using the categories of Hess (37) by pooling classes 1 and 2 as undeveloped and 3–5 as developed.
RNA Preparation.
Ovaries from each individual were manually homogenized, and RNA extraction followed the standard TRIzol (Invitrogen) protocol with subsequent DNase (Promega) digestion. RNA quality and quantity was photometrically assessed. Equal amounts of RNA were immediately reverse-transcribed with M-MLV H− Point Mutant Reverse Transcriptase (Promega) using oligo(dT) primer (0.5 μg/μL; Promega) according to the manufacturer's instructions.
Splice Variant Detection.
Because exon 1 was never transcribed in any of the samples, cDNA was amplified using primers 4I and 18II (Table S1) spanning all exons except the first one (resulting in a PCR product of 2,299 bp; Amel_4.0) (38) with long-range PCR Enzymes (Finnzymes and Fermentas) following the manufacturers’ instructions. The PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and thereafter cloned into the pGEM-T Vector using the TA cloning kit (Invitrogen). Transformation was done in JM109 competent cells according to the manufacturer's instructions (Promega). Plasmids were purified and inserts sequenced and aligned using CLC Free Workbench 3 (CLC Bio). The annotated exons 11 and 12 are not transcribed, and because the annotated exons 14 and 15 are not separated by an intron, they are fused to exon 14. Moreover, the transcripts contain 179 bp at the 3′ end, which had been originally annotated to be untranscribed genomic DNA in the Amel_4.0 genome sequence.
Rapid Amplification of cDNA Ends (RACE)-PCR.
To complete the transcript sequences, RACE-PCRs were conducted. A 3′ end cDNA amplification was done according to Scotto-Lavino et al. (39), and the nested PCRs were adapted to the BiothermTM DNA Polymerase (Genecraft) using 0.2 mM dNTPs, 0.3 μM of each primer (gemini-specific forward primers: sp7.1I; 7.2I and 7.3I in PCR1, 15I in PCR2; Table S1) and 0.5 units polymerase in a total of 10 μL. The PCR programs consisted of 5 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 54 °C, and 60 s at 72 °C, and a final extension of 20 min at 72 °C. The PCR products were either directly sequenced after gel elution or cloned (see above) if the quality of the PCR products was insufficient for direct sequencing. Plasmids having the right insert were purified and sequenced as mentioned above.
For the amplification of 5′cDNA ends the protocol of Scotto-Lavino et al. (40) was modified. After reverse transcription and poly(A)-tailing poly(T) gemini-specific primers (QgemI; Qgem4I; Table S1) were used for amplification. Tailing the primers with 15 thymine residues ensured that these primers were only bound to the 5′ end of the tailed cDNAs. The transcript-specific primers sp5II (which spans the boundaries of exon 5 and 6) and 5II (which is situated in exon 5) were used as reverse primers. Purified PCR products were sequenced.
RT-PCR.
ATF2, gemini, glycerol-phosphate-dehydrogenase (GPDH), ribosomal protein 49 (rp49), and elongation factor 1α (EF) quantification was conducted with real-time PCR using SYBR Green (Bio-Rad) assays following the manufacturer's instructions. The four different splice products were analyzed separately, because the PCRs of the combined altered transcript isoforms (+5.L7; +5.S7; −5.L7; −5S7) did not yield adequate products. Each isoform was quantified with primers specific for the two exons and their spliced counterpart (Table S1). Each sample was run in duplicate. The real-time PCR cycling profile consisted of a 3-min incubation at 95 °C, followed by 39 cycles of 15 s at 95 °C and 30 s at 54 °C for annealing, and 30 s at 72 °C for extension and data collection. Melting curve analyses were performed between 50 °C and 90 °C, reading the fluorescence at 1 °C increments. Additionally, the purity of the PCR products was visually verified on 1.8% agarose gels. After baseline subtraction using the global minimum trend option, C(t) values were calculated by the Opticon Monitor 3 software (Bio-Rad) using a single SD over cycle range.
Data Analysis.
The relative gene expression of ATF2 was calculated as the efficiency-corrected gene expression of ATF2 compared with the efficiency-corrected gene expression of the housekeeping genes GPDH (NM001014994), rp49 (AF441189.1), and EF (NM001014993).
The expression ratios of the different transcript forms of gemini were calculated from the efficiency-corrected gene expressions for each gemini amplicon. The efficiency-corrected relative expression of the individual transcript isoforms was calculated by normalizing the gemini gene expression to the two housekeeping genes GPDH and EF. PCR efficiencies of every PCR product were determined by pooling cDNA from every sample used in the calculations. PCR efficiencies were determined using serial dilutions covering a 104-fold template dilution range (41). Nonparametric Mann–Whitney U tests were used for statistical comparisons adjusting the P values for replicate tests.
gemini (Exon 5) Knockdown by RNAi.
Our aim was to activate ovaries in arrhenotokous workers by reducing the abundance of specific splice variants. Because transcripts missing exon 5 were not detected by qPCR in nonreproductive arrhenotokous workers, we did not conduct a silencing experiment to even further reduce this already rare transcript. Instead we targeted transcripts containing exon 5, which were highly abundant in all arrhenotokous workers, to substantially alter the ratio of unspliced vs. spliced transcripts and eventually activate the ovaries of A. m. carnica workers. Hence, two gemini-specific siRNA template sequences within the cassette exon 5 (78 bp) were selected using the siRNA target designer version 1.6 (Promega), which also scrambled the target-specific siRNA sequences resulting in siRNAs to be used as negative controls. The selected siRNA sequences were blasted against the honeybee genome to avoid potential off-target effects in genes other than gemini. None of the siRNAs shared sequence similarities longer than 14 nt (16- to 18-nt–long stretches of homology are suggested as the maximum acceptable length in RNAi studies per Ambion siRNA design guidelines). Furthermore, none of the genes with the highest sequence similarities was found twice when blasting both siRNAs individually. The two siRNAs obtained (for sequences, see Table S2) cover the 3′ and the 5′ regions of the template. Using this siRNA mixture decreased the chance of affecting unintended targets. siRNA was synthesized using the T7 Ribomax Express RNAi System (Promega) according to the manufacturer's instructions but with an extended incubation time of up to 2 h. The siRNA quality and quantity was assessed by capillary gel electrophoresis and photometry.
Honeybees and siRNA Feeding.
Newly emerged A. m. carnica workers (arrhenotokous, 1–2 d old) were sampled from a single brood frame and divided into three groups: untreated bees, bees treated with scrambled siRNA (which has no sequence similarity to any bee-specific gene), and a gemini-specific siRNA treatment group. Bees in the latter group were fed with a mixture of equal amounts of the two gemini-specific siRNAs. In each case, 35–40 newly emerged bees were put in wooden cages (10.5 × 13 cm) provided with a small piece of comb and pollen ad libitum at 34 °C. The bees were fed daily with 1.5 mL 50% sugar water containing 1 μg siRNA per individual. After 14 d, a time span after which ovary activation can easily be detected, bees were killed and screened for ovary activation in a double-blind test. The samples were stored in coded tubes by a person with no knowledge of the treatments, to allow for an unbiased examination of the ovaries by a second person without any prior knowledge of the experimental groups. Ovaries were classified as developed when ovarioles contained mature eggs. This procedure was repeated in two consecutive trials, using two brood frames from two different colonies. The quantification of the transcript changes were conducted as discussed previously.
Sequencing of Noncoding Regions of gemini and Characterization of tae1.
The putative promoter region and genomic sequences between the exons were analyzed using phenol/chloroform-extracted genomic (gDNA) from parasitic A. m. capensis clone workers and A. m. scutellata workers from the University of Pretoria. PCRs were run with gDNA from one individual, each with intron-spanning primers (Table S1). Sequencing of fragments <1 kb were done by standard PCRs using the BiothermTM DNA Polymerase (Genecraft), following the manufacturer's instructions. Products were directly sequenced. Fragments >1 kb were processed by long-range PCRs using the Kapa2G Robust PCR Kit (Peqlab). The PCR programs consisted of an initial heating at 95 °C for 3 min, followed by 40 cycles of 30 s at 95 °C, 30 s at 54 °C, and 2 min at 72 °C and ended with a final elongation at 72 °C for 20 min. PCR products were purified (QIAquick PCR Purification Kit; Qiagen) and cloned (see above). Clones were sequenced by primer walking, and sequences were compared with the annotated honeybee genome (Amel_4.0). Sequence differences in intron 5_6 (tae1) in A. m. capensis workers were thereafter confirmed by PCRs spanning the region of interest using primers I2500I and I21000II and subsequent sequencing in four workers of each subspecies. Length differences caused by the deletion of the tae1 were thereafter confirmed in 130 A. m. capensis workers and 10 A. m. scutellata workers by PCRs using primers gem_taeI and gem_taeII.
Supplementary Material
Acknowledgments
We thank Berit Langer, Christoph Eller, Denise Kleber, and Petra Leibe for their technical assistance. We are also grateful for the constructive comments of two unknown reviewers, which substantially improved the manuscript. Financial support was granted by the Deutsche Forschungsgemeinschaft (R.F.A.M.) and the National Research Foundation of South Africa.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109343108/-/DCSupplemental.
References
- 1.Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 2.Winston ML, Slessor KN. Honeybee primer pheromones and colony organization: Gaps in our knowledge. Apidologie (Celle) 1998;29:81–95. [Google Scholar]
- 3.Ratnieks FLW. Reproductive harmony via mutual policing by workers in eusocial hymenoptera. Am Nat. 1988;132:217–234. [Google Scholar]
- 4.Visscher PK. A quantitative study of worker reproduction in honey bee colonies. Behav Ecol Sociobiol. 1989;25:247–254. [Google Scholar]
- 5.Sakagami SF. The false-queen: Fourth adjustive response in dequeened honeybee colonies. Behaviour. 1958;13:280–296. [Google Scholar]
- 6.Onions GW. South African “fertile-worker bees”. S Afr Agric J. 1912;1:720–728. [Google Scholar]
- 7.Anderson RH. The laying worker in the cape honey-bee Apis mellifera capensis. J Apic Res. 1963;2:85–92. [Google Scholar]
- 8.Neumann P, Hepburn HR, Radloff SE. Modes of worker reproduction, reproductive dominance and brood cell construction in queenless honeybee (Apis mellifera l.) colonies. Apidologie (Celle) 2000;31:479–486. [Google Scholar]
- 9.Crewe RM, Velthuis HHW. False queens: A consequence of mandibular gland signals in worker honeybees. Naturwissenschaften. 1980;67:467–469. [Google Scholar]
- 10.Wossler TC. Pheromone mimicry by Apis mellifera capensis social parasites leads to reproductive anarchy in host Apis mellifera scutellata colonies. Apidologie (Celle) 2002;33:139–163. [Google Scholar]
- 11.Simon UE, Moritz RFA, Crewe RM. The ontogenetic pattern of mandibular gland components in queenless worker bees (Apis mellifera capensis Esch.) J Insect Physiol. 2001;47:735–738. doi: 10.1016/s0022-1910(00)00167-0. [DOI] [PubMed] [Google Scholar]
- 12.Dietemann V, Neumann P, Härtel S, Pirk CWW, Crewe RM. Pheromonal dominance and the selection of a socially parasitic honeybee worker lineage (Apis mellifera capensis Esch.) J Evol Biol. 2007;20:997–1007. doi: 10.1111/j.1420-9101.2007.01303.x. [DOI] [PubMed] [Google Scholar]
- 13.Malka O, Shnieor S, Hefetz A, Katzav-Gozansky T. Reversible royalty in worker honeybees (Apis mellifera) under the queen influence. Behav Ecol Sociobiol. 2007;61:465–473. [Google Scholar]
- 14.Greeff JM. Effects of thelytokous worker reproduction on kin-selection and conflict in the cape honeybee, Apis mellifera capensis. Philos T Roy Soc B. 1996;351:617–625. [Google Scholar]
- 15.Lattorff HMG, Moritz RFA, Fuchs S. A single locus determines thelytokous parthenogenesis of laying honeybee workers (Apis mellifera capensis) Heredity. 2005;94:533–537. doi: 10.1038/sj.hdy.6800654. [DOI] [PubMed] [Google Scholar]
- 16.Lattorff HMG, Moritz RFA, Crewe RM, Solignac M. Control of reproductive dominance by the thelytoky gene in honeybees. Biol Lett. 2007;3:292–295. doi: 10.1098/rsbl.2007.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Judice CC, et al. Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol Biol. 2006;15:33–44. doi: 10.1111/j.1365-2583.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoskins RA, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 20.Verma S, Ruttner F. Cytological analysis of the thelytokous parthenogenesis in the cape honeybee (Apis mellifera capensis Escholtz) Apidology. 1983;14:41–57. [Google Scholar]
- 21.Kang HC, et al. Erythroid cell-specific alpha-globin gene regulation by the CP2 transcription factor family. Mol Cell Biol. 2005;25:6005–6020. doi: 10.1128/MCB.25.14.6005-6020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López AJ. Developmental role of transcription factor isoforms generated by alternative splicing. Dev Biol. 1995;172:396–411. doi: 10.1006/dbio.1995.8050. [DOI] [PubMed] [Google Scholar]
- 23.Evans JD, Wheeler DE. Expression profiles during honeybee caste determination. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0001. 10.1186/gb-2000-2-1-research0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grozinger CM, Fan Y, Hoover SER, Winston ML. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera) Mol Ecol. 2007;16:4837–4848. doi: 10.1111/j.1365-294X.2007.03545.x. [DOI] [PubMed] [Google Scholar]
- 25.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 26.Elango N, Hunt BG, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci USA. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473:478–483. doi: 10.1038/nature10093. [DOI] [PubMed] [Google Scholar]
- 28.Yoon JB, Li G, Roeder RG. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol. 1994;14:1776–1785. doi: 10.1128/mcb.14.3.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attardi LD, Tjian R. Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev. 1993;7(7B):1341–1353. doi: 10.1101/gad.7.7b.1341. [DOI] [PubMed] [Google Scholar]
- 30.Pagani F, Stuani C, Zuccato E, Kornblihtt AR, Baralle FE. Promoter architecture modulates CFTR exon 9 skipping. J Biol Chem. 2003;278:1511–1517. doi: 10.1074/jbc.M209676200. [DOI] [PubMed] [Google Scholar]
- 31.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenasi T, Peterlin BM, Dovc P. Distal regulation of alternative splicing by splicing enhancer in equine β-casein intron 1. RNA. 2006;12:498–507. doi: 10.1261/rna.7261206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hastings ML, Wilson CM, Munroe SH. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA. 2001;7:859–874. doi: 10.1017/s1355838201002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy EM, Phillips JA., III Characterization of an intron splice enhancer that regulates alternative splicing of human GH pre-mRNA. Hum Mol Genet. 1998;7:1491–1496. doi: 10.1093/hmg/7.9.1491. [DOI] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hess G. On the influence of the fertility of queenless and vitamin E on the ovaries of worker bees (Translated from German) Beih Schweiz Bienen Ztg. 1942;1:33–110. [Google Scholar]
- 38.Consortium HGS. Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scotto-Lavino E, Du G, Frohman MA. 3′ end cDNA amplification using classic RACE. Nat Protoc. 2006;1:2742–2745. doi: 10.1038/nprot.2006.481. [DOI] [PubMed] [Google Scholar]
- 40.Scotto-Lavino E, Du G, Frohman MA. 5′ end cDNA amplification using classic RACE. Nat Protoc. 2006;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.