Abstract
A long-debated question concerns the fate of archaic forms of the genus Homo: did they go extinct without interbreeding with anatomically modern humans, or are their genes present in contemporary populations? This question is typically focused on the genetic contribution of archaic forms outside of Africa. Here we use DNA sequence data gathered from 61 noncoding autosomal regions in a sample of three sub-Saharan African populations (Mandenka, Biaka, and San) to test models of African archaic admixture. We use two complementary approximate-likelihood approaches and a model of human evolution that involves recent population structure, with and without gene flow from an archaic population. Extensive simulation results reject the null model of no admixture and allow us to infer that contemporary African populations contain a small proportion of genetic material (≈2%) that introgressed ≈35 kya from an archaic population that split from the ancestors of anatomically modern humans ≈700 kya. Three candidate regions showing deep haplotype divergence, unusual patterns of linkage disequilibrium, and small basal clade size are identified and the distributions of introgressive haplotypes surveyed in a sample of populations from across sub-Saharan Africa. One candidate locus with an unusual segment of DNA that extends for >31 kb on chromosome 4 seems to have introgressed into modern Africans from a now-extinct taxon that may have lived in central Africa. Taken together our results suggest that polymorphisms present in extant populations introgressed via relatively recent interbreeding with hominin forms that diverged from the ancestors of modern humans in the Lower-Middle Pleistocene.
Keywords: H. sapiens, hybridization
It is now well accepted that anatomically modern humans (AMH) originated in Africa and eventually dispersed to all inhabited parts of the world. What is not known is the extent to which the ancestral population that gave rise to AMH was genetically isolated, and whether archaic hominins made a genetic contribution to the modern human gene pool. Answering these questions has important implications for understanding the way in which adaptations associated with modern traits were assembled in the human genome: do the genes of AMH descend exclusively from a single isolated population, or do our genes descend from divergent ancestors that occupied different ecological niches over a wider geographical range across and outside of the African Pleistocene landscape?
The introgression debate is typically framed in terms of interbreeding between AMH and Neandertals in Europe or other archaic forms in Asia. The opportunity for such hybridizations may have existed between 90 and 30 kya, after early modern humans dispersed from Africa and before archaic forms went extinct in Eurasia (1–5). Recent genome-level analyses of ancient DNA suggest that a small amount of gene flow did occur from Neandertals into the ancestors of non-Africans sometime after AMH left Africa (6) and that an archaic “Denisovan” population contributed genetic material to the genomes of present-day Melanesians (7). Given recent fossil evidence, however, the greatest opportunity for introgression was in Africa, where AMH and various archaic forms coexisted for much longer than they did outside of Africa (5, 8–11). Indeed, the fossil record indicates that a variety of transitional forms with a mosaic of archaic and modern features lived over an extensive geographic area from Morocco to South Africa between 200 and 35 kya (12–15).
Although sequencing the Neandertal and Denisovan genomes has provided evidence that gene flow between archaic and modern humans is plausible, it has not aided efforts to determine the extent of introgression in African populations. Here we use a different strategy to address the question of ancient population structure in Africa. Using multilocus DNA sequence polymorphism data from extant Africans, we analyze patterns of divergence and linkage disequilibrium (LD) to detect the signature of archaic admixture (16–18). Application of this approach to publicly available sequence data from the Environmental Genome Project found evidence of ancient population structure in both African and non-African populations (19). However, analyses of diversity in and around coding regions may be complicated by the effects of recent natural selection, which might contribute to unusual patterns of polymorphism. In this study we use a large resequencing dataset that includes 61 noncoding regions on the autosomes to test whether patterns of neutral polymorphism in three contemporary sub-Saharan African populations are better explained by archaic admixture. Although whole-genome polymorphism data are now available from hundreds of samples (20), they do not include individuals from African hunter-gatherer populations, which serve as important reservoirs of human genetic diversity. Our study includes two such populations (Biaka Pygmies and San), along with an agricultural population from West Africa (Mandenka). We use a model of historically isolated subpopulations (17, 21) to predict patterns of nucleotide variation expected as a consequence of no admixture (null hypothesis) vs. low levels of admixture (alternative hypothesis). We apply two complementary coalescent-based approaches, a two-population and a three-population model to test the null hypothesis, and then estimate three key parameters: the time of admixture (Ta), the ancestral split time (T0), and the admixture proportion (a).
Results
Two-Population Model.
In this approach we follow a two-step strategy (18). First we estimate demographic parameters of the null model using summary statistics that quantify recent African population structure. Using these model parameters, we then test the hypothesis of no admixture using a different summary statistic that is designed to detect low levels of genetic exchange between modern and archaic humans. The null model of recent African population structure without archaic admixture incorporates divergence, migration, and recent population growth (Fig. 1A). We calculate a composite likelihood of the summarized data on a grid of parameter values (18) (SI Materials and Methods). Parameter estimates, along with simulation-based 95% confidence intervals (CIs), are given in Table S1. Two patterns emerge from these analyses: estimates of the start of growth are very recent, and estimates of the population split time are relatively old. Although the recent growth estimates are consistent with results of previous studies (23, 24), the estimate of a divergence time that predates the origin of modern humans based on fossil data (450 kya, Biaka–Mandenka comparison) was unexpected. There are several possible explanations for this observation. First, it is possible that the true divergence time is old and that AMH evolved within the context of a geographically structured population. Alternatively, it is possible that the true divergence time is younger and that the old estimate arose either by chance or by bias caused by model misspecification (i.e., the true demographic model is different from Fig. 1A). Our simulations suggest that the large divergence estimate might happen by chance roughly 2% of the time, if the true demographic model (i.e., without admixture) were as in Fig. 1A (SI Materials and Methods and Table S2).
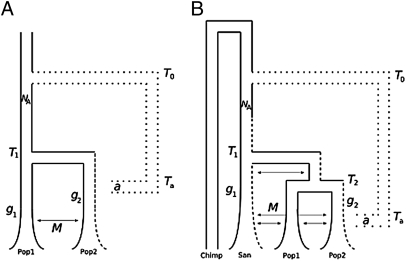
Fig. 1.
Schematic of the (A) two-population model and the (B) three-population model. Both demographic models test the fit of admixture with an archaic group (dotted lines) who split from the ancestors of modern humans at time T0 and a (%) of alleles introgressed into the modern gene pool at time Ta. The dashed lines represent all possible locations where admixture could occur. Both models begin with a single population of size Na, followed by a population split at time T1, with population growth beginning at times g1 and g2, and a constant symmetric migration rate M. For B, an additional population split at time T2 also occurs. This model also assumes that the ancestors of the San split first from those of the Mandenka and Biaka (22).
We then tested for archaic admixture using the estimated model parameters of the null model and a summary of LD (S*) that was specifically designed to be sensitive to archaic admixture (18, 19). The evidence for archaic admixture is extremely strong in the Biaka and the San (P < 10−4) but not in the Mandenka (P > 0.05). Quantile–quantile plots for the distribution of P values across loci are shown in Fig. S1.
Three-Population Model.
To complement the first approach, we also implemented an approximate-likelihood method to estimate admixture parameters under a three-population isolation and migration model (Fig. 1B). Because this is a new inferential strategy, we explain our approach in some detail. In an isolation and admixture model (17) we expect to find loci with both deep haplotype divergence (reflecting a long period of isolation for those haplotypes that trace to different subpopulations) and elevated levels of LD (reflecting a reduced time for diverged haplotypes to recombine). If levels of admixture are low, then one class of haplotypes is expected to be at low frequency (i.e., a small basal clade). In other words, low levels of recent admixture with an archaic human population are likely to produce data with a small subsample of sequences that are highly diverged over an extended region of the chromosome. With this in mind, we developed our three summary statistics as follows. For each locus, we identify the two most diverged sequences and then define two groups, G1 and G2, by genetic similarity to the two designated sequences. From this we set our three statistics for approximate likelihood: (i) D1, the fraction of polymorphisms that are shared between G1 and G2. D1 reflects the amount of recombination and thus is sensitive to the time of introgression, Ta. (ii) D2, the ratio of the number of differences between the two distinguished sequences described above and the number of fixed sequence differences between human and chimpanzee. D2 reflects the relative time-depth of the genealogy and thus is sensitive to the archaic split time, T0. (iii) D3, the size of the smaller of the groups, G1 and G2. D3 reflects the relative size of the two most basal clades and thus is sensitive to the amount of admixture, a.
Our approximate-likelihood protocol estimates the distribution of the summary statistics D1, D2, and D3 on the basis of the simulation of a large number of ancestral recombination graphs (ARGs). An important part of this protocol is the choice of tolerances or bin sizes δ1, δ2, and δ3 for their respective summary statistics. In general, we chose tolerances to maximize power for a = 1% (SI Materials and Methods).
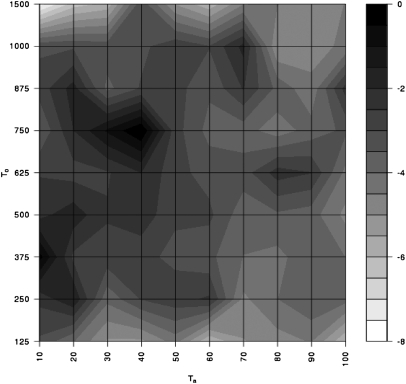
We find that the data are significantly unlikely under the null model of no admixture (i.e., the likelihood ratio test yields a bootstrapped P value of 0.0493). We note that this result is conservative because it is based on estimates of recombination rate that are biased downward and a tolerance that is less powerful in regions of high recombination (see below). Interestingly, we find evidence for two separate peaks in the maximum-likelihood surface: (i) an older peak with an archaic split time, T0 ≈ 700 kya, a time of admixture, Ta ≈ 35 kya, and an admixture proportion, a ≈ 2%; and (ii) a more recent peak with T0 ≈ 375 kya, Ta ≈ 15 kya, and a ≈ 0.5% (Fig. 2). Although our method has little power to infer the exact admixture proportion, we can place 95% CIs on the times of divergence (125 kya < T0 < 1.5 Mya) and admixture (Ta < 70 kya) (SI Materials and Methods). Note that T0 for the more recent peak is consistent with the Biaka–Mandenka split time estimates from the two-population model.
Fig. 2.
Approximate likelihood profile based on 60 loci for time of introgression and archaic split time. A log-likelihood difference of 3.92 defines the 95% confidence region (using the χ2 approximation). Likelihood estimates at each locus have at least 10 ARGs for both ψold and ψrecent.
Likelihood Ratios of Individual Loci.
The two inferential methods can also assess the evidence for archaic admixture at individual loci (SI Materials and Methods). Both methods identify the same locus on chromosome 4 (4qMB179) as a strong candidate for archaic admixture (P < 5 × 10−4 for each method). Table 1 describes the three loci exhibiting the lowest P value in the three-population model. Of the six individuals in the minimum clades, four are Biaka (4qMB179, 18qMB60) and two are San (13qMB107). Although both inferential methods identified the 13qMB107 locus as a likely candidate, the result is much more significant for the three-population (P < 0.001) vs. two-population (P = 0.049) model (Table 1). We note that the power of the two-population approach is reduced when evidence of introgression is limited to a short tract of DNA (as in the case of 13qMB107 where it is found only in the first subset; Discussion). For 18qMB60, the two-population method excludes singletons from the S* analysis. If they were included, the P value for 18qMB60 would be below 0.01 (Table 1).
Table 1.
Three loci that favor an alternative model
| Locus | Likelihood ratio | P value | T0 (Mya) | Ta (kya) | D1 | D2 | D3 | S* P value |
| 13qMB107 | 44.38 | <0.001 | 1 | 20 | 0.1 | 0.264 | 2 | 0.049 |
| 4qMB179 | 39.85 | <0.001 | 1.5 | 20 | 0 | 0.366 | 3 | <0.001 |
| 18qMB60 | 12.74 | 0.022 | 0.75 | 20 | 0 | 0.192 | 1 | >0.05† |
The likelihood ratio is defined to be maxψ{L(ψ|data)}/maxψ{L(ψ|data), ψ ∈ H0} and the P value determined with a parametric bootstrap. These values along with parameter values in columns 4–8 refer to results from the three-population model, whereas the S* P values in the last column refer to results from the two-population model.
†S* was originally calculated excluding all singletons from the analysis. When we recalculate S* including singletons, we obtain P < 0.01.
To address the question of whether some loci favor one maximum in the likelihood surface over the other (i.e., ψrecent vs. ψold), we compute the likelihood ratio (SI Materials and Methods) for each locus (Fig. S2). Notably, the four most extreme likelihood ratios include the three loci that individually favor ψold (Table 1).
Analysis of the 4qMB179 Region.
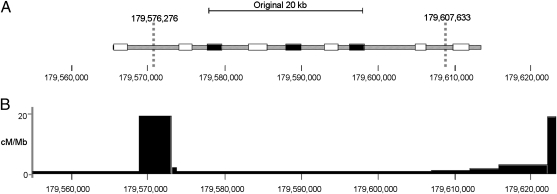
We now turn to a focused analysis of the 4qMB179 region, a region characterized by no evidence of recombination between the major clades and deep haplotype divergence. In the ≈20-kb region that was initially surveyed (Fig. 3A), we identified 20 SNPs (and one insertion) that separate three Biaka haplotypes (B1–B3; Table S3) from all of the remaining African sequences. To determine the full length of the unusual pattern of SNPs, we gathered additional DNA sequence data from all individuals in our panel (Fig. 3A) and identified a 31.4-kb region with 37 completely linked sites where the Biaka haplotypes are 0.3% diverged from the other sequences in our sample. Using a simple model of isolation followed by recent mixing, we next developed likelihood-based methods for estimating a split time and admixture time for the locus (SI Materials and Methods). We estimated an initial split time of 1.25 Mya (95% CI, 0.7–2.1 Mya) and an admixture time of 37 kya (95% CI, 1–137 kya) (Fig. S3).
Fig. 3.
(A) Schematic of the original (filled bars) and extended sequence data (open bars) for the 4qMB179 locus. The unusual Biaka haplotype extends for ≈31.4 kb between the vertical dotted lines. (B) Recombinational landscape as inferred from HapMap Phase I data.
Geographic Surveys.
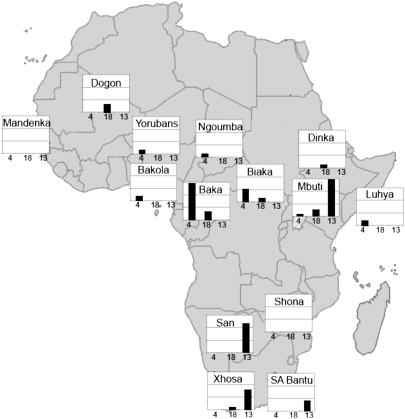
A survey of the insertion that is diagnostic for the divergent haplotype at 4qMB179 (i.e., at position 179,598,847 in Table S3) in 502 individuals from West, East, central, and southern Africa reveals that it reaches its highest average frequency (3.6%) in Pygmy groups from west-central Africa (Fig. 4). The variant is also found at low average frequencies (0.8%) in some non-Pygmy groups from West and East Africa. An A→G mutation that marks the divergent haplotype at 18qMB60 shows a similar distribution—also reaching its highest average frequency in the Pygmy groups—although it is found at slightly lower frequencies than the variant at 4qMB179 (i.e., 1.6% vs. 3.6%, respectively). This variant is also found in some non-Pygmy groups, exhibiting similar average frequencies as the 4qMB179 variant in West Africans (0.8%), East Africans (0.8%), and southern Africans (0.5% vs. 0.0%, respectively) (Fig. 4). Interestingly, the distribution of the G→A variant marking the divergent haplotype at 13qMB107 exhibits a somewhat different geographic distribution, reaching its highest average frequency in our sample of southern Africans (6.3%, and especially in the San at a frequency of 11.9%) rather than in central African Pygmies (average of 5.2%). However, it is important to note that its presence in our sample of central Africans is entirely limited to the Mbuti, where it has a frequency of 14.8%.
Fig. 4.
Frequency of introgressive variants within three sequenced regions in an expanded sample of ≈500 sub-Saharan Africans (SI Materials and Methods). The filled bar represents the frequency of a variant marking the divergent haplotype at 4qMB179 (Left), 18qMB60 (Center), and 13qMB179 (Right) in each of 14 population samples. Each horizontal line on the bar charts represents a frequency of 5%.
Discussion
Our inference methods reject the hypothesis that the ancestral population that gave rise to AMH in Africa was genetically isolated and point to several candidate regions that may have introgressed from an archaic source(s). For example, we identified a ≈31.4-kb region within the 4qMB179 locus with highly diverged haplotypes, one of which is found at low frequency in several Pygmy groups in central Africa. We hypothesize that the unusual haplotype descends from an archaic DNA segment that entered the AMH population via admixture. The observed haplotype structure is highly unusual (P < 5 × 10−5), even when we account for recent population structure or uncertainty in the underlying recombination rate (Table S4). It is noteworthy that the two ends of the archaic haplotype correspond to recombinational hotspots in the 4qMB179 region (Fig. 3B), suggesting that an initially much longer block of archaic DNA was whittled down by frequent recombination in the hotspots.
Both inferential methods also identified the 13qMB107 locus as a likely introgression candidate; however, only ≈7 kb of the surveyed region contains SNPs that are in high LD, all of which are found at the 5′ end of the sequenced region in two San individuals. To determine whether the length of the unusual pattern of SNPs extends beyond our sequenced region at 13qMB107, we examined public full genome sequence data (25). We identified a San individual (!Gubi) who carried one copy of the unusual 13qMB107 haplotype and noted a run of heterozygous sites that extended an additional ≈7 kb to the 5′ side of our sequenced region. Like the case of 4qMB179, the two ends of the unusual haplotype correspond to recombinational hotspots, and analysis of 13qMB107 yields an estimated divergence time of ≈1 Mya and a recent introgression time (≈20 kya) (Table 1). The geographic distribution of the introgressive variant at 18qMB60, a third candidate identified in the three-population model (Table 1), is very similar to that of 4qMB179, albeit consistently found at lower frequencies (Fig. 4). On the other hand, the distribution of the introgressive variant at 13qMB107 is distinguished from that of the other two candidate loci by its presence in the San and the southern African Xhosa, as well as in Mbuti from the Democratic Republic of Congo. Interestingly, the Mbuti represent the only population in our survey that carries the introgressive variant at all three candidate loci, despite the fact that no Mbuti were represented in our initial sequencing survey. Given that the Mbuti population is known to be relatively isolated from other Pygmy and neighboring non-Pygmy populations (26), this suggests that central Africa may have been the homeland of a now-extinct archaic form that hybridized with modern humans.
We have relied on an indirect approach to detect ancient admixture in African populations because there are no African ancient DNA sequences to make direct comparisons with our candidate loci. As proof of principle that an indirect approach can be useful, we reexamined the RRM2P4 pseudogene on the X chromosome. Using a similar approximate-likelihood methodology, it was previously posited that a divergent allele at the pseudogene introgressed from an archaic taxon in Asia (27, 28). We compared human and Neandertal RRM2P4 sequences and found that the three derived sites that define the non-African basal lineage are shared with Neandertal (Fig. S4). Thus, we verified that this unusual human sequence, which is characterized by a deep haplotype divergence and a small basal clade, is indeed shared with an archaic form. Further genome-level (i.e., multilocus) analysis will also shed light on the process of archaic admixture, which is likely to be more complicated than we have modeled. For instance, the multimodal likelihood surface in Fig. 2 suggests that gene flow among strongly subdivided populations in Africa may characterize multiple stages of human evolution in Africa.
Our results are consistent with earlier inferences supporting the role of archaic admixture in sub-Saharan Africa based on analyses of coding regions (19) and the Xp21.1 noncoding region (16). Although our estimates of isolation and admixture dates are tentative, the results point to relatively recent genetic exchange with an unknown archaic hominin that diverged from the ancestors of modern humans in the Lower-Middle Pleistocene and remained isolated for several hundred thousand years. Despite a fragmentary African fossil record, there are plenty of candidates for the source(s) of this introgression. Beginning ≈700 kya, fossil evidence from many parts of Africa indicate that Homo erectus was giving way to populations with larger brains, a change that was accompanied by several structural adjustments to the skull and postcranial skeleton (14). By ≈200 kya, individuals with more modern skeletal morphology begin to appear in the African record (8, 14). Despite these signs of anatomical and behavioral innovation, hominins with a combination of archaic and modern features persist in the fossil record across sub-Saharan Africa and the Middle East until after ≈35 kya (12, 14). Although there is currently a major debate about the meaning of this piecemeal or mosaic-like appearance of modern traits for taxonomic classification (12, 29), the evidence presented here and elsewhere suggests that long-separated hominin groups exchanged genes with forms that either were in the process of evolving fully modern features, or were already fully modern in appearance. The emerging geographic pattern of unusual variants discovered here suggests that one such introgression event may have taken place in central Africa (where there is a very poor fossil record). Interestingly, recent studies attest to the existence of Late Stone Age human remains with archaic features in Nigeria (Iwo Eleru) and the Democratic Republic of Congo (Ishango) (30–32). The observation that populations from many parts of the world, including Africa, show evidence of introgression of archaic variants (6, 16, 19) suggests that genetic exchange between morphologically divergent forms may be a common feature of human evolution. If so, hybridization may have played a key role in the de novo origin of some our uniquely human traits (33).
Materials and Methods
Samples and Regions Sequenced.
DNA samples used in this study for resequencing were taken from publicly available cell lines administered by the Centre d’Étude du Polymorphisme Humain Human Genome Diversity Panel (34), while samples used for geographic surveys were described in refs. 22 and 27. Individual identifiers for each of these samples are described in Wall et al. (35). Our updated resequence dataset consists of 61 loci in autosomal intergenic regions (36), each ≈20 kb of primarily single-copy noncoding (i.e., putatively nonfunctional) DNA in regions of medium or high recombination (ρ ≥ 0.9 cM/Mb) at least 100 kb away from the nearest gene (37). We used a locus trio design, sequencing three fragments of ≈2 kb spaced evenly across the region (35). The sample was composed of ≈16 individuals from each population (with the exception of the San, which included nine samples), nearly all of which were males. Although the Hammer et al. (36) dataset includes 30 X-linked loci, we chose not to include them in the current analysis because of the much smaller number of X chromosomes and the need to make assumptions about sex-biased processes. Exact locations are available at http://hammerlab.biosci.arizona.edu/ArchadData/PNAS.archad.locusLocationInfo.xls, and genotyping assays and primers are available at http://hammerlab.biosci.arizona.edu/ArchadData/PNAS.primers.doc.
Inferential Approach.
Here we implement two types of models, denoted “two-population” and “three-population” model (Fig. 1). Because the two-population model is simpler, it has the advantage of using a broader array of summary statistics and allows evaluation over a finer grid of parameter space. The more complex three-population model is much more computationally expensive, yet has the advantage of considering all three sampled populations simultaneously. Our approximate-likelihood approach allows us to investigate the entire grid of parameter space. In contrast, full-likelihood methods require computationally intensive techniques (e.g., Markov chain Monte Carlo) that limit analysis of parameter space to regions near local maxima, whereas Bayesian methods suffer from the need to use priors that may be poorly justified in this study.
Coalescent-Based Model Testing.
Two-population model.
For each pair of sub-Saharan African populations we consider the demographic model described in Fig. 1A and use a previously published composite-likelihood methodology (18, 19) to estimate parameters ψ = (g1, g2, T1, M) for growth, split time, and migration rate (see SI Materials and Methods for details of model and methodology). This method uses information from levels of diversity and the joint frequency spectrum, but not LD for estimating (composite) likelihoods. For each pair of African populations, we then use the parameters estimated above as a null model and test for the presence of additional ancient population structure (19). If archaic admixture occurs at a locus, then “archaic” SNPs on introgressed sequences would be in strong pairwise LD. Simulations suggest that both the number of such SNPs and the total distance spanned by such SNPs are elevated when archaic admixture occurs (21). To exploit these two observations and to account for the effects of intragenic recombination (18), we calculate, for each locus, a statistic, S*, shown to be sensitive to archaic admixture (18). S* looks for population-specific SNPs that are in strong LD (e.g., the square correlation r2≈1). We determine the significance of S* values from the actual data by running simulations using the previously estimated demographic parameters to obtain a distribution of S* values under the null hypothesis of no (archaic) admixture. Significantly high S* values are interpreted as departures from the null model in the direction of some unknown ancient population structure. The P values across loci are combined (assuming independence) using the method of Fisher (38).
Three-population model.
The three-population model has nine parameters, three of which (Ta, T0, and a) are the key parameters for inference (SI Materials and Methods). Ancestral recombination graphs under a grid of parameter space are created using the software tool ms (39) to approximate distributions under several tolerances or bin sizes. All inference is based on approximate-likelihood computations for the three key summary statistics, D1, D2, D3, as described in SI Materials and Methods. Because the parameter space of our null model of no admixture is a subspace of the entire parameter space, we can make inference using a likelihood ratio test.
Approximate-likelihood surfaces are generated in two stages. First, we simulate 5,000 ARGs over a coarse grid of parameter space. This allows us to reduce the parameter space to the null space and to those values within the 99% CI of the coarse grid estimates. We then run simulations using 100,000 ARGs for each parameter value, storing approximations of the summary statistic distribution using reduced tolerances. This allows us to perform bootstrap and goodness of fit (GOF) tests for larger tolerances by adding empirical probabilities from the simulations.
To test the null model of no admixture, we used tolerances of δ1 = 0.06, δ2 = 0.05, and δ3 = 2 to estimate the approximate likelihood for each locus using local-scale estimates of recombination rate, which yielded an estimate of Ta = 40 kya, T0 = 750 kya, and a = 1% and a log-likelihood ratio of −2.01. To estimate the significance of this value we drew 10,000 points from the maximum-likelihood location under H0 using our 3D histogram and tabulated the probability of observing a log-likelihood ratio as small (or smaller) than −2.01 with an archaic split time no more recent than 750 kya.
To better characterize the alternative model we used a two-tiered approach. First we examined a more refined grid of parameter space, and then we ran two levels of simulations. In the initial pass we generated 5,000 ARGs per parameter value, and in the second pass we took all of the values within the 99% CI and computed 3D histograms of summary statistics for each parameter value in the manner described above. We then used a parametric bootstrap to address GOF as described in SI Materials and Methods.
Estimating recombination rates.
The three-population methodology is highly sensitive to recombination rates. For this reason, we chose to estimate local-scale recombination and favored these estimates in our inference over the much larger-scale estimates of Kong et al. (37). To this end, we used Phase 2.1 (40, 41) using two qualitatively different strategies. The first uses HAPMAP Yoruba data (42), and the second estimates ρ using the major clade of each locus in our own resequencing data (SI Materials and Methods). We chose the major clade because recently introgressed archaic lineages, if they exist, serve as a barrier to recombination and thus will bias estimates of ρ downward.
Locus-specific analyses.
To infer the split and admixture times for individual introgressive candidates, we calculate the probability of observing the number of completely correlated sites for the relevant population data (e.g., the Biaka for 4qMB179), assuming panmixia, as a function of the underlying scaled recombination parameter ρ (Table S4). To estimate admixture time (Ta), we first estimate the minimum length of the diverged haplotype. Using the genetic map of Kong et al. (37), we then estimated the total recombination rate for the diverged haplotype. Given an admixture event g generations ago, the distribution of lengths of inherited chromosomal segments roughly follows an exponential distribution with mean genetic distance 1/g. It follows that the maximum-likelihood estimate for the time of admixture is Ta = 1/g generations ago, with 95% CI (0.0253 Ta–3.69 Ta) generations. We assume a mean generation time of 25 y. Note that we have not accounted for the additional uncertainty in estimating ρ.
We use polymorphism data along with an outgroup (orang-utan) sequence to estimate the split time T0. We assume that exactly one archaic sequence was introduced into the modern gene pool, leaving the observed number of descendant sequence(s) in the divergent haplotypes. Our general approach was to tabulate polymorphic sites and fixed differences, noting whether the SNPs were polymorphic (or fixed) in the archaic or the modern sequences. We then run coalescent simulations (39) to estimate the likelihood of observing the actual numbers of fixed differences and polymorphisms of different categories, as a function of T0, Ne,, μ (the mutation rate), ρ, and Ta. By simulating over a grid of values with increments 0.25 Myr for T0, 1,000 for Ne, 1 × 10−9/bp for μ, 2.5 × 10−9 for ρ, and 0.04 Ne, generations for Tα, we estimate a profile likelihood curve for T0. A total of 104 replicates for each parameter combination were sufficient to accurately estimate the likelihood.
Supplementary Material
Acknowledgments
We thank J. Cahill and collaborators, including G. Destro-Bisol, T. Jenkins, H. Soodyall, and L. Louie who donated DNA samples. This research was funded by National Science Foundation HOMINID Grant BCS-0423670 (to M.F.H. and J.D.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109300108/-/DCSupplemental.
References
- 1.Coppa A, Grun R, Stringer C, Eggins S, Vargiu R. Newly recognized Pleistocene human teeth from Tabun Cave, Israel. J Hum Evol. 2005;49:301–315. doi: 10.1016/j.jhevol.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Grün R, et al. ESR and U-series analyses of enamel and dentine fragments of the Banyoles mandible. J Hum Evol. 2006;50:347–358. doi: 10.1016/j.jhevol.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Klein RG. Darwin and the recent African origin of modern humans. Proc Natl Acad Sci USA. 2009;106:16007–16009. doi: 10.1073/pnas.0908719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morwood MJ, et al. Archaeology and age of a new hominin from Flores in eastern Indonesia. Nature. 2004;431:1087–1091. doi: 10.1038/nature02956. [DOI] [PubMed] [Google Scholar]
- 5.Stringer C. The origin and dispersal of Homo sapiens: Our current state of knowledge. In: Mellars P, Boyle K, Bar-Yosef O, Stringer C, editors. Rethinking the Human Revolution. Cambridge, UK: McDonald Institure for Archaeological Research; 2007. [Google Scholar]
- 6.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 9.Stringer C. The chronological and evolutionary position of the Broken Hill cranium. Am J Phys Anthropol. 2011;144(Suppl 52):287. [Google Scholar]
- 10.Tattersall I. Once we were not alone. Sci Am. 2003;(Summer):20–27. [Google Scholar]
- 11.Wood B. Hominid revelations from Chad. Nature. 2002;418:133–135. doi: 10.1038/418133a. [DOI] [PubMed] [Google Scholar]
- 12.Brauer G. The origin of modern anatomy: By speciation or intraspecific evolution? Evol Anthropol. 2008;17:22–37. [Google Scholar]
- 13.Klein RG. The Earlier Stone Age of southern Africa. S Afr Archaeol Bull. 2000;55:107–122. [Google Scholar]
- 14.Rightmire GP. Out of Africa: Modern human origins special feature: Middle and later Pleistocene hominins in Africa and Southwest Asia. Proc Natl Acad Sci USA. 2009;106:16046–16050. doi: 10.1073/pnas.0903930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trinkaus E. Early modern humans. Annu Rev Anthropol. 2005;34:207–230. [Google Scholar]
- 16.Garrigan D, Mobasher Z, Kingan SB, Wilder JA, Hammer MF. Deep haplotype divergence and long-range linkage disequilibrium at xp21.1 provide evidence that humans descend from a structured ancestral population. Genetics. 2005;170:1849–1856. doi: 10.1534/genetics.105.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordborg M. In: On Detecting Ancient Admixture. Genes, Fossils and Behaviour: An Integrated Approach to Human Evolution, NATO Science Series: Life Sciences. Donnelly P, Foley RA, editors. Vol 310. Amsterdam: IOS Press; 2001. [Google Scholar]
- 18.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wall JD, Lohmueller KE, Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol Biol Evol. 2009;26:1823–1827. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durbin RM, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wall JD. Detecting ancient admixture in humans using sequence polymorphism data. Genetics. 2000;154:1271–1279. doi: 10.1093/genetics/154.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veeramah KR, et al. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal re-sequencing data. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr212. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox MP, et al. Autosomal resequence data reveal Late Stone Age signals of population expansion in sub-Saharan African foraging and farming populations. PLoS ONE. 2009;4:e6366. doi: 10.1371/journal.pone.0006366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voight BF, et al. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci USA. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster SC, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patin E, et al. Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox MP, et al. Testing for archaic hominin admixture on the X chromosome: Model likelihoods for the modern human RRM2P4 region from summaries of genealogical topology under the structured coalescent. Genetics. 2008;178:427–437. doi: 10.1534/genetics.107.080432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrigan D, Mobasher Z, Severson T, Wilder JA, Hammer MF. Evidence for archaic Asian ancestry on the human X chromosome. Mol Biol Evol. 2005;22:189–192. doi: 10.1093/molbev/msi013. [DOI] [PubMed] [Google Scholar]
- 29.Tattersall I, Schwartz JH. The morphological distinctiveness of Homo sapiens and its recognition in the fossil record: Clarifying the problem. Evol Anthropol. 2008;17:49–54. [Google Scholar]
- 30.Allsworth-Jones P, Harvati K, Stringer C. The archaeological context of the Iwo Eleru cranium from Nigeria, and preliminary results of new morphometric studies. In: Allsworth-Jones P, editor. West African Archaeology, New Developments, New Perspectives. Oxford: British Archaeological Reports International Series S2164; 2010. pp. 29–42. [Google Scholar]
- 31.Crevecoeur I, Semal P, Cornelissen E, Brooks AS. The Late Stone Age human remains from Ishango (Democratic Republic of Congo): Contribution to the study of the African Late Pleistocene modern human diversity. Am J Phys Anthropol. 2010;141(Suppl 50):87. [Google Scholar]
- 32.Stringer C, Harvati K, Allsworth-Jones P, Grün R, Adebayo Folorunso C. New research on the Iwo Eleru cranium from Nigeria. Am J Phys Anthropol. 2010;141(Suppl 50):225–226. [Google Scholar]
- 33.Arnold ML, Sapir Y, Martin NH. Review. Genetic exchange and the origin of adaptations: Prokaryotes to primates. Philos Trans R Soc Lond B Biol Sci. 2008;363:2813–2820. doi: 10.1098/rstb.2008.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cann HM, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 35.Wall JD, et al. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammer MF, et al. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat Genet. 2010;42:830–831. doi: 10.1038/ng.651. [DOI] [PubMed] [Google Scholar]
- 37.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 38.Mosteller F, Fisher RA. Questions and answers #14. The American Statistician. 1948;2:30–31. [Google Scholar]
- 39.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 40.Crawford DC, et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 41.Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frazer KA, et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.