Abstract
Thermal radiosensitization is believed to be mediated by an inhibition of double-strand break (DSB) repair, but the exact mechanism of radiosensitization remains to be elucidated. Previously, we demonstrated that proteins of the Mre11/Rad50/Nbs1 complex (MRN) translocate from the nucleus to the cytoplasm in cells have that been heated or heated and then irradiated; this finding led us to propose that heat radiosensitization was due at least in part to translocation of MRN. In the current study, we used leptomycin B to inhibit MRN translocation in heated, irradiated cells, but we found that heat radiosensitization was not altered. Thus enhanced radiosensitivity was not attributed to translocation of MRN proteins. To determine which of the MRN subunits contributed to heat radiosensitization, we compared the extent of heat radiosensitization in wild-type cells with that of cells hypomorphic for Mre11 or Nbs1 or cells in which the level of Rad50 was suppressed. We found that neither Nbs1 nor Rad50 is involved in heat radiosensitization, because a similar amount of heat radiosensitization was observed in cells deficient in those proteins compared to cells expressing normal levels. However, heat radiosensitization was not observed in A-TLD1 cells deficient in Mre11. Measurement of exonuclease activity of purified Mre11 heated at 42.5°C or 45.5°C indicated that the protein is very heat-labile. Immunoprecipitation of Mre11 from heated HeLa cells also revealed that hsp70 associates with Mre11 and that this association is maintained long after heating. Taken together, these findings implicate Mre11 as a target for heat radiosensitization and suggest that heat radiosensitization and inhibition of DSB repair may be mediated by heat-induced conformational changes in Mre11.
INTRODUCTION
The Mre11/Rad50/Nbs1 (MRN) complex plays a key role in the response of vertebrate cells to double-strand breaks (DSBs). MRN acts as a sensor of DSBs, links the DNA damage repair response with ATM-dependent cell cycle checkpoint regulation, and, in concert with other proteins (some of which the complex recruits, such as ATM and 53BP1), facilitates DSB repair with its DNA-binding, end-tethering, nuclease and unwinding activities (1–3). Deletion of NBS1, RAD50 or MRE11 is embryonic lethal in mice (4–6). In humans, hypomorphic mutations in NBS1 or MRE11 result in Nijmegen breakage syndrome (NBS) or ataxia telangiectasia-like disorder (ATLD), respectively; fibroblasts derived from individuals afflicted with these syndromes exhibit enhanced sensitivity to ionizing radiation and genetic instability due to defects in DSB repair [(7) and references therein].
In mammalian cells, two major pathways for the repair of radiation-induced DSBs have been identified: homologous recombination (HR) occurs preferentially during late S/G2, while non-homologous end joining (NHEJ) repairs DSBs throughout the cell cycle (8–10). Growing evidence supports the existence of two NHEJ subpathways: the conservative (C-NHEJ) “error-prone” pathway, which does not require microhomology, and the alternative deletional (D-NHEJ) pathway, which requires microhomology to join ends with small deletions (11). The MRN complex has long been known to be involved in HR, but its role in NHEJ has been ambiguous (12–14). Recent reports, however, indicate a role for Mre11 (and possibly the MRN complex) in the classical and alternative NHEJ pathways (15–17).
In human cells, the MRN complex is normally uniformly distributed in nuclei, but after exposure to ionizing radiation, MRN forms foci at the sites of DSBs (18). Mre11 functions as both an endonuclease that cleaves hairpin DNA structures and a 3′-5′ double-stranded DNA exonuclease that efficiently degrades double-stranded substrates with blunt or recessed 3′ ends (19, 20). These activities are increased when bound to Rad50. Mre11 nuclease activity appears to be required for processing of ends prior to ligation, when the MRN complex tethers the two strands at the site of the DSB. Mre11 also mediates resection and annealing of complementary single-stranded DNA molecules (21). Rad50 exhibits ATP-dependent double-stranded DNA binding and negatively regulates Mre11 endonucleolytic activity in yeast, prompting speculation that it regulates functional activity of the Mre11 complex (20, 22, 23). Nbs1 plays a role in the activation of ATM (24) and is also essential for the phosphorylation of Mre11, a prerequisite for focus formation (25). Nbs1 is involved in damage recognition, but the DNA damage-detection function of the MRN complex is not essential for the end-joining functions of the Rad50/Mre11 complex (26). The MRN complex possesses several activities that are not observed in the absence of Nbs1, including ATP-dependent partial unwinding of the DNA duplex, and cleavage of 3′ protruding strands at double-stranded/single-stranded transitions (20). Nbs1 phosphorylation in response to DNA damage is a prerequisite for activation of the S-phase checkpoint (27). Thus Nbs1 links the DSB repair response with cell cycle checkpoint control.
Since mammalian cells are sensitized to ionizing radiation if heated prior to or after irradiation, hyperthermia is being investigated for its use as an adjuvant to radiotherapy (28). Heat inhibits the repair of DSBs induced by ionizing radiation, and radiosensitization is believed to be mediated by inhibition of DSB repair. Several hypotheses have been advanced to explain the mechanism by which heat inhibits DSB repair and sensitizes cells to ionizing radiation, including masking of ionizing radiation-induced DNA damage, alterations of accessibility of damaged sites due to nonspecific protein aggregation, or inactivation or aggregation of DNA repair proteins by heat, which would be expected to affect one or more of the DSB repair pathways [(29–31) and references therein]. However, the exact mechanisms for heat radiosensitization have yet to be elucidated, and even the issue of which of the repair pathways may be affected has yet to be resolved conclusively (29).
In our previous studies, we found that heating human U-1 melanoma or HeLa cells at temperatures ranging from 42.5–45.5°C induced the translocation of Mre11, Rad50 and Nbs1 from the nucleus to the cytoplasm (32, 33). This translocation was increased significantly if cells were irradiated prior to heating. Similar findings were reported by Roti Roti and coworkers after heating at 41°C (34). These findings had led us and others to hypothesize that translocation of proteins of the Mre11 complex was responsible at least in part for heat radiosensitization and inhibition of DSB repair. In the current study, we show that translocation of MRN proteins cannot account for the thermal sensitization of irradiated cells. We also show that Mre11 represents a critical target for heat radiosensitization and that the protein is extremely heat-labile and associates with hsp70 long after the heat exposure. These data provide clues regarding potential mechanisms by which irradiated cells are sensitized by heat.
MATERIALS AND METHODS
Cells and Culture Conditions
Asynchronous U-1 human melanoma cells, HeLa CCL-2 cells, or cells of various repair-defective cell lines were grown in monolayer in 25- or 175-cm2 tissue culture flasks. U-1 cells were obtained from Dr. James Mitchell (National Cancer Institute). HeLa cells were originally obtained from the American Type Culture Collection (Manassas, VA). U-1 and HeLa cells were grown in McCoy’s 5A medium [with 10% iron-supplemented calf serum (ISCS) (Hyclone, Inc., Logan, UT)]. For Rad50 knockdown experiments, HeLa cells were transfected with siRNA or scRNA using DharmaFECT 1 (Dharmacon, Lafayette, CO) for 4 h and used 72 h after transfection. At the time of irradiation, the expression of Rad50 in siRNA-transfected cells was 40% of the level in control cells.
Immortalized A-TLD1-vector and A-TLD1-Mre11 cells were obtained from Dr. M. Weitzman (Salk Institute for Biological Studies) and grown in DMEM with 20% fetal bovine serum and 1 mg/ml puromycin. A-TLD1 cells were immortalized by transduction with retroviruses expressing SV40T-antigen and hTERT. These cells were then transduced with either retrovirus expressing the wild-type Mre11 cDNA cloned into pLPC or the empty vector as described by Carson et al. (35).
Immortalized NBS cells transfected with either an empty vector (LXIN) or NBS cells transfected with the full-length NBS1 gene (NBS1 cells) that produce functional Nbs1 protein were obtained from Dr. P. Concannon (Virginia Mason Research Center) and grown in DMEM supplemented with 15% FBS and 500 μg/ml G418 (24). The recipient cell line for the empty vector and Nbs1 constructs was the ILB1 cell line, an SV40-transformed, immortal NBS fibroblast line established from primary cells from a patient carrying common founder mutation 657del5; this line was originally obtained and characterized by Dr. Margaret Zdzienicka (Leiden University) (36).
HeLa cells stably transfected with pTRE-hRAD50 (HeLa-Rad50 cells) or transfected with ptTA-hygro were established as described by Shin et al. (37). Briefly, after cells were transfected with the ptTA-hygro plasmid (Clontech Laboratories, Palo Alto, CA), surviving colonies in hygromycin-containing medium were isolated. pTRE-hRAD50 DNA and the pSVneo plasmid were cotransfected in the presence of tetracycline (Tc) (2 μg/ml) into one of the ptTA-expressing stable clones. Clones expressing hRAD50 were cultured in DMEM supplemented with 10% ISCS and containing 0.4 mg/ml G418, 0.1 mg/ml hygromycin, and 2 μg/ml Tc. Forty-eight hours prior to experiments, Tc was removed from the medium, resulting in a 1.5-fold increase in Rad50 content.
Irradiations, Hyperthermia, Drug Treatments and Measurement of Clonogenic Survival
Cells were either irradiated with various doses of 250 kVp X rays using a Siemans Stabilipan X-ray machine or irradiated and then heated in a water bath for 10 min to 2 h at 41.5–45.5°C (± 0.01°C). Medium was changed and flasks were placed on ice for 5 min prior to irradiation. All irradiations were done while cells were on ice (total time on ice was 15 min). After irradiation, medium was replaced with fresh 37°C medium. Some samples were then heated. After treatments, cells were either trypsinized immediately and plated into normal growth medium for analysis of clonogenic survival or returned to a 37°C incubator for various times prior to trypsinization. For leptomycin (LMB) experiments, cells were either incubated in the presence of LMB for 2 h prior to and during treatment or incubated in ethanol (vehicle). LMB was kindly provided by Dr. Minoru Yoshida (University of Tokyo) (38). Clonogenic cell survival was determined as described previously (39).
Immunoprecipitation and Western Blotting
Immunoprecipitation of whole cell lysates was performed with modifications to methods described by Xu et al. (34). After 72 h of asynchronous cell growth in 175-cm2 tissue culture flasks, 1 × 107 cells were heated at 42.5°C or 45.5°C and/or incubated at 37°C. Cells were trypsinized and centrifuged at 4°C for 7 min at 1,600 rpm, washed twice in cold PBS, incubated in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.2 mg PMSF, 1 μg/ml leupeptin) for 30 min on ice with intermediate mixing, and then centrifuged at 4°C for 15 min at 10,000g. Supernatants were collected and incubated with protein A/G sepharose beads (Calbiochem, Gibbstown, NJ) for 1 h on ice and sedimented for 10 min at 10,000g to separate and remove the sepharose. New supernatants were combined with 5 μg of anti-hMre11 antibody (rabbit polyclonal, Calbiochem, La Jolla, CA; catalogue number: PC388) and incubated overnight on a rocker at 4°C. Samples were then incubated for 4 h with 75 μl of protein A/G sepharose beads on a rocker at 4°C. The immunoprecipitates were collected by centrifugation at 10,000g for 30 s and then washed with 500 μl of lysis buffer four times. The immunocomplex-bound beads were boiled in SDS sample buffer, and supernatants were collected for quantification and electrophoresed in a 7.5% SDS-PAGE gel prior to immunoblotting. Nitrocellulose blots were probed for both Mre11 and Hsp70 (Stressgen, Ann Arbor, MI). Western blots were scanned and relative intensities of protein bands were obtained in each lane using NIH ImageJ. Intensities of Hsp70 bands were normalized to the unheated Hsp70 control band. Similarly, intensities of Mre11 were normalized against the unheated Mre11 control. This Mre11 normalization factor was then used to correct the Hsp70 values for loading error, since the amount of Mre11 does not change in heated samples of whole cell lysates (32).
Purification of Mre11 and Measurement of Mre11 Exonuclease Activity
Recombinant baculovirus driving expression of His6-tagged Mre11 was obtained from Dr. James Carney (University of Maryland, Baltimore County) and virus was amplified, titered and used to infect SF-9 insect cells. Forty-eight hours after infection, cells were collected by centrifugation and cell-free extracts were prepared as described previously by Hermanson and Turchi (40). Briefly, cells were allowed to swell in hypotonic buffer and lysed via Dounce homogenization followed by brief sonication. Insoluble material was removed via sedimentation at 12,0000g for 30 min and the supernatant was precleared on a phosphocellulose matrix at 1 M NaCl. The flow through was directly applied to a 2-ml Ni-NTA agarose column and washed extensively with buffer containing 10 mM imidazole. The bound protein was then eluted in buffer containing 350 mM imidazole and fractions containing MRE11 were identified by SDS-PAGE. Fractions were pooled and dialyzed in buffer containing 50 mM NaCl and further fractionated on a Q-Sepharose column. Bound proteins were eluted with a linear gradient from 50–750 mM NaCl. Active fractions were identified, pooled and dialyzed in buffer containing 50 mM NaCl and 10% glycerol.
A 30-bp oligonucleotide substrate (SJC1.5-TCT CTG GCC TCT TCC CTT TCT TCC CCC TTT) was labeled with 32P at the 5′ end and was annealed to a 35-bp oligonucleotide (SJC1.5C-Xba-CTA GAA AGG GGG AAG AAA GGG AAG AGG CCA GAG A). This resulted in a 5′-labeled 3′-recessed double-stranded substrate.
Three micrograms of purified Mre11 (in a 50 mM NaCl/10% glycerol buffer) was heated at 42.5° or 45.5°C for various times in a water bath. After heating, protein was put on ice for 1 min and then centrifuged at 14,000 rpm for 1 min. Two micrograms was then taken and used for nuclease reactions. Reactions contained exonuclease buffer (50 mM Hepes, pH 7.5, 50 mM KCl, 1 mM DTT, 5 mM MnCl2, 0.01% NP-40), 200 pmol of DNA substrate, and Mre11 to a total volume of 20 μl. Reactions were incubated at 37°C for 60 min. Each reaction was mixed with 20 μl of 2× formamide loading buffer and then loaded onto a sequencing gel containing 15% acrylamide and 7 M urea. The gel was exposed on a phosphor-screen and scanned using a phosphorimaging system (GE Healthcare, Piscataway, NJ), and exonuclease activity was determined using Imagequant software (GE Healthcare).
RESULTS
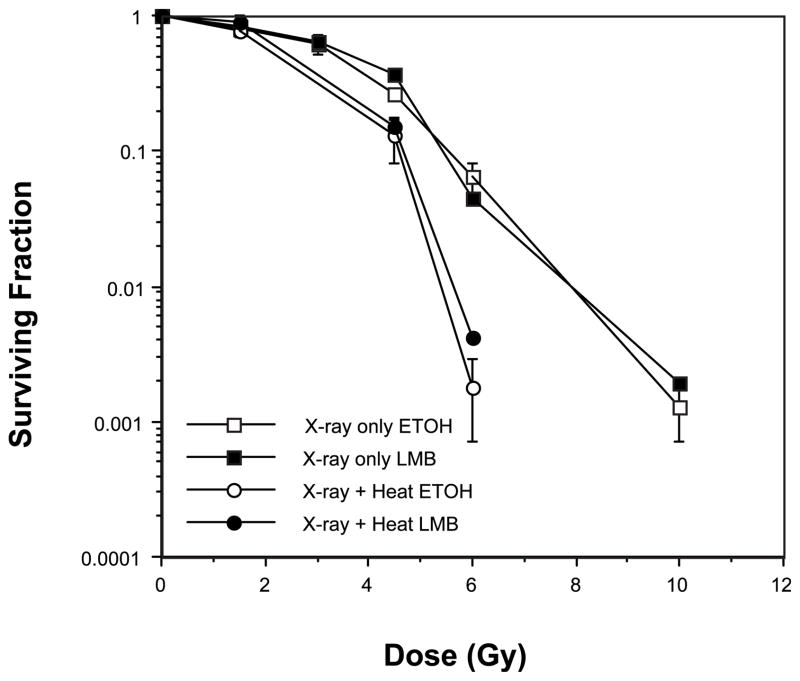
Leptomycin B (LMB) is a potent inhibitor of CRM1-mediated nuclear export of proteins containing a nuclear export sequence (38). We previously found that a nontoxic concentration (10 nM) of LMB inhibits translocation of Mre11, Rad50 and Nbs1 (from the nucleus to the cytoplasm) that normally occurs in U-1 cells that have been irradiated and then heated at 45.5°C for 15 min (32, 33). If translocation is involved, then LMB should protect cells from heat radiosensitization as determined using a clonogenic cell survival assay. To determine whether translocation of the proteins of the Mre11 complex correlates with heat radiosensitization, we compared clonogenic survival of U-1 cells that were irradiated with various doses of X rays and then heated at 45.5°C in the presence or absence of LMB (Fig. 1). The concentration of LMB used in this experiment was previously shown to be sufficient to inhibit the translocation of Mre11, Rad50 and Nbs1 of irradiated and heated cells. We reasoned that inhibition of translocation of proteins of the Mre11 complex with LMB should reduce heat radiosensitization if translocation was indeed involved in the phenomenon. However, LMB treatment did not reduce heat radiosensitization of irradiated, heated U-1 cells; a similar amount of heat radiosensitization was obtained in cells that had been irradiated and then heated, regardless of whether they were treated with LMB or vehicle (ethanol, ETOH). We therefore concluded that heat radiosensitization is not mediated through the translocation of one or more of the proteins of the Mre11 complex induced by hyperthermia treatment.
FIG. 1.
Inhibition of translocation of Mre11, Rad50 and Nbs1 does not protect against heat radiosensitization. U-1 melanoma cells either were irradiated with X rays or were heated for 10 min at 45.5°C after irradiation in the presence or absence of leptomycin B (LMB). LMB was dissolved in ethanol (ETOH), which was used as a control vehicle. Error bars represent SEM for the data from at least two or three experiments. The plating efficiencies of the unheated and heated cells were as follows: no heat (control), ETOH = 0.3 ± 0.028; no heat (control), LMB = 0.21 ± 0.026; heat only, ETOH = 0.013 ± 0.002; heat only, LMB = 0.006 ± 0.0006.
Our previous data indicating that much of the Mre11, Rad50 and Nbs1 content of the nuclei of heated, irradiated cells translocates to the cytoplasm, coupled with our recent study that showed that heat inhibited the formation of MRN repair foci and resulted in a decreased interaction between the three proteins (41), suggested that one or more of the individual proteins denatures or must undergo conformational changes after heating. Each of the proteins thus represented a potential target for heat inactivation. We therefore posited that inactivation of any or all of the MRN proteins might be expected to result in heat radiosensitization. A systematic evaluation of Mre11, Rad50 or Nbs1 as targets for heat radiosensitization was then undertaken by comparing the extent of heat radiosensitization, as determined from clonogenic cell survival assays, of normal cells to cells that contained hypomorphic mutations in Nbs1 or Mre11 or cells in which the level of Rad50 was modulated.
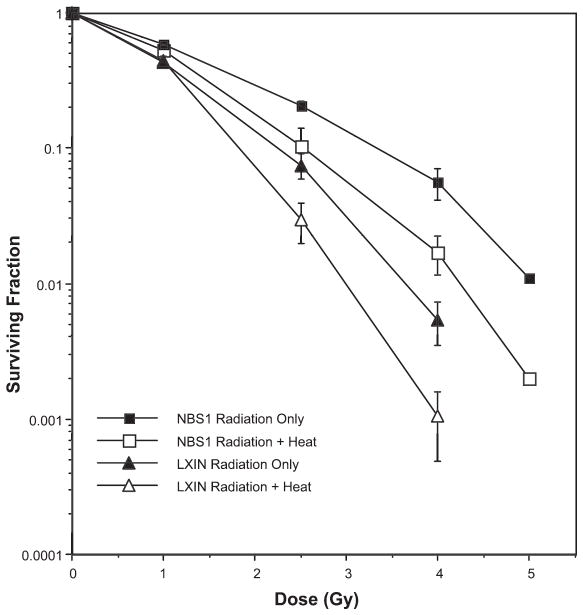
Radiosensitive NBS fibroblasts (designated as LXIN) that fail to express Nbs1 protein (but in which Mre11 and Rad50 still form a complex) (24) and NBS cells in which a normal copy of the NBS1 gene was introduced (and that express full-length Nbs1 protein at levels comparable to normal fibroblasts, designated as NBS1) were either irradiated with various doses of X rays or irradiated prior to being heated for 10 min at 45.5°C (Fig. 2). LXIN cells lacking Nbs1 showed enhanced radiosensitivity compared to Nbs1-complemented NBS1 cells, as expected. Both cell lines were also sensitized to ionizing radiation by the postirradiation hyperthermia treatment. Indeed, a similar amount of heat radiosensitization was observed when the respective lines of irradiated cells were compared to cells that where heated after irradiation. That is, when survival curves were obtained for irradiated or irradiated and heated fibroblasts lacking Nbs1 and compared with survival curves obtained for cells expressing normal levels of Nbs1, we found that heat radiosensitization was not reduced in the cells deficient in Nbs1. These results indicate that Nbs1 is not essential for heat radiosensitization. If Nbs1 were a target for or played a role in heat radiosensitization, the survival curves of irradiated LXIN cells compared to irradiated and heated LXIN cells would be superimposable, or nearly so; that is, a significant reduction of radiosensitization would have been observed in cells deficient in Nbs1 compared to cells expressing near-normal amounts of Nbs1 (at a surviving fraction of 0.01, a thermal enhancement ratio, or TER, of 1.2 was calculated for both cell lines).
FIG. 2.
Lack of involvement of Nbs1 in heat radiosensitization. NBS cells that were transfected with an empty vector (Nbs1-deficient LXIN cells) or NBS cells transfected with the full-length NBS1 gene (NBS1 cells) either were irradiated or were irradiated and then heated for 10 min at 45.5°C prior to plating for clonogenic survival. Error bars represent the SEM for the data from at least two or three experiments. The plating efficiencies of the unheated and heated cells were as follows: LXIN, no heat = 0.491 ± 0.13; LXIN, heat only = 0.135 ± 0.016; NBS1, no heat = 0.583 ± 0.085; NBS1, heat only = 0.269 ± 0.08.
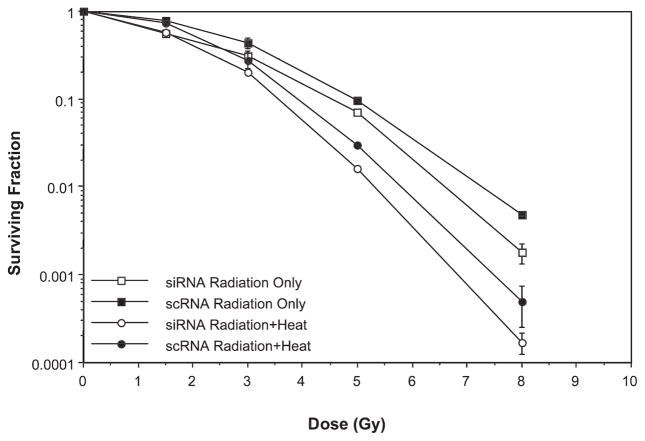
Experiments in which the level of Rad50 protein was modulated in HeLa cells yielded similar results in two distinct series of experiments (Fig. 3 and Supplementary Fig. 1; http://dx.doi.org/10.1667/RR2594.1.S1). In one set of experiments, heat radiosensitization of HeLa cells that had been rendered deficient in Rad50 using a small interfering RNA (siRNA) approach was compared to cells expressing normal levels of Rad50 (Fig. 3). A knockdown approach was employed due to the lack of availability of Rad50-null cell lines. A reduction of Rad50 in siRNA-treated cells to 40% of the level found in nuclei of control cells was achieved; this level was nearly equivalent to the amount of Rad50 remaining in nuclei of cells after the heat-induced translocation of MRN proteins to the cytoplasm noted in our previous studies (33). Cells were then either irradiated or irradiated and then heated for 2 h at 41.5°C. The reduction in Rad50 alone resulted in slight radiosensitization of irradiated siRNA-transfected cells compared to cells transfected with scrambled RNA (scRNA) at higher radiation doses. The lack of significant radiosensitization with the level of knockdown achieved in this experiment would appear to corroborate the conclusion reached in Fig. 1 that it is not the reduction of Rad50, mediated by heat-induced translocation, that results in heat radiosensitization. In Fig. 3, we also show that the extent of heat radiosensitization of cells in which Rad50 protein expression was reduced after transfection of cells with Rad50 siRNA was similar to that in cells that had been transfected with scRNA over the total range of radiation doses. Thus, as with Nbs1, these data suggest that Rad50 is not a target for heat radiosensitization (at a surviving fraction of 0.01, an identical TER of 1.2 was calculated for cells expressing normal levels of Rad50 and cells in which Rad50 was suppressed). In a second set of experiments, HeLa cells induced to express a 1.5-fold higher level of Rad50 (as determined by quantitative Western blotting; data not shown) after removal of Tc, designated as Tc (−) cells, showed an identical amount of heat radiosensitization after irradiation and heating for 10 min at 45.5°C as cells that expressed normal levels of Rad50 [Tc (+) cells] (Supplementary Fig. 1; http://dx.doi.org/10.1667/RR2594.1.S1), thus supporting our conclusion from Fig. 3 for cells irradiated or irradiated and then heated at 41.5°C. It is worth noting that overexpression of Rad50 did not result in an increase in radioresistance in cells that were only irradiated.
FIG. 3.
Lack of involvement of Rad50 in heat radiosensitization. HeLa CCL2 cells were transfected with scrambled RNA (scRNA) or SMART pool siRNA directed against Rad50 for 72 h at 37°C. Cells were then irradiated or irradiated and then heated for 2 h at 41.5°C prior to plating for clonogenic survival. Error bars represent the SEM for the data from at least two or three experiments. The plating efficiencies of the unheated and heated cells were as follows: siRNA, no heat = 0.345 ± 0.015; siRNA, heat only = 0.25 ± 0.005; scRNA, no heat = 0.33 ± 0.02; scRNA, heat only = 0.22 ± 0.008.
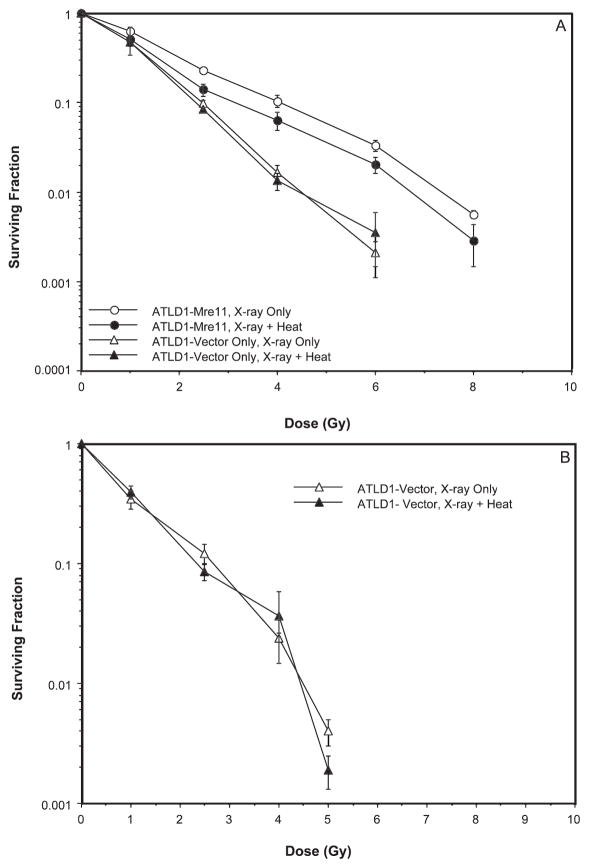
Since neither Rad50 nor Nbs1 appears to be essential for heat radiosensitization, we next determined whether Mre11 played a role in heat radiosensitization. Cells obtained from patients with ataxia telangiectasia-like disorder produce a truncated hypomorphic form of the Mre11 protein; subcellular distribution of Mre11 is aberrant, and formation of MRN foci in response to ionizing radiation is abrogated (7). A-TLD1 cells transduced with wild-type Mre11 cDNA and that produce levels of functional Mre11 protein comparable to normal cells (designated in Fig. 4 as ATLD-Mre11) were much more radioresistant than A-TLD1 cells transduced with vector only (designated ATLD-Vector in Fig. 4). Heat radiosensitization was observed in ATLD-Mre11 cells, most notably at radiation doses of 4 Gy or greater (Fig. 4A). However, we found that the survival curves for ATLD-Vector cells lacking Mre11 that were irradiated or that were irradiated and heated for either 2 h at 41.5 or 10 min at 45.5°C were almost superimposable (Fig. 4A and B). From interpolation of the data in Fig. 4A for cells heated at 41.5°C, ATLD-Vector cells showed no radiosensitization by heat; for a surviving fraction of 0.01, a TER of 1.00 was obtained. The same result (TER = 1.00) was also obtained for ATLD-Vector cells heated for 10 min at 45.5°C. Thus the Mre11 protein appears to be the key component of the MRN complex involved in heat radiosensitization. With Mre11 implicated in heat radiosensitization, we sought to determine the effects of hyperthermia on the Mre11 protein to gain insight into the mechanisms by which heat radiosensitization might be mediated. Since the bulk of our earlier studies to determine the role of MRN proteins in heat radiosensitization were performed using 42.5°C (moderate or chronic) or 45.5°C (acute) heat treatments, these temperatures were used for the remainder of the experiments.
FIG. 4.
Involvement of Mre11 in heat radiosensitization. ATLD1 cells that express wild-type hMRE11 (Mre11) and ATLD1 cells transfected with an empty vector (Vector) were either irradiated or irradiated and then heated for 2 h at 41.5°C (panel A) or irradiated and then heated for 10 min at 45.5°C (panel B) prior to plating for clonogenic survival. Error bars represent the SEM for data from at least four experiments. The plating efficiencies of the unheated and heated cells were as follows: ATLD1-Mre11, no heat = 0.077 ± 0.002; ATLD1-Mre11, heat only = 0.03 ± 0.01; ATLD1-Vector, no heat = 0.042 ± 0.01; ATLD1-Vector, heat only = 0.039 ± 0.005.
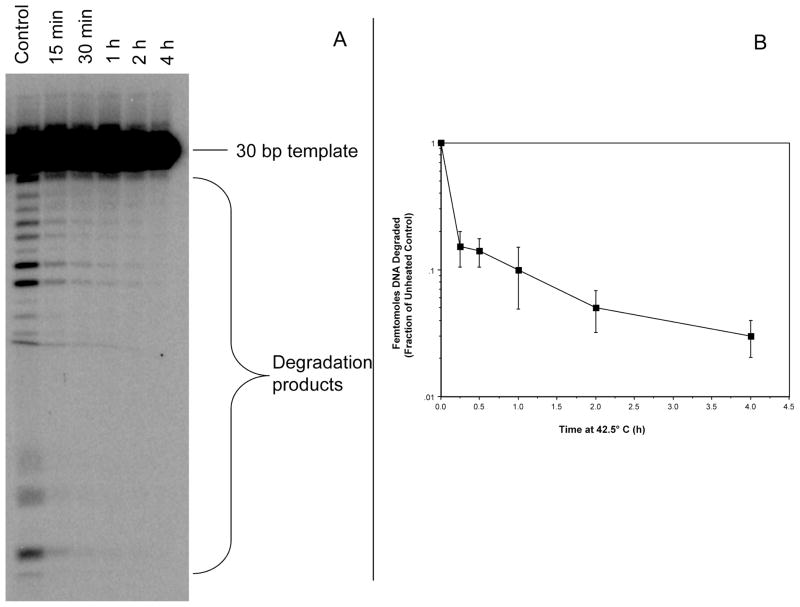
Since heat may cause structural and functional changes in proteins, we hypothesized that the exonuclease activity of Mre11 may be altered by heat shock. We therefore subjected purified Mre11 to hyperthermia treatment and assessed its ability to degrade a 5′-labeled 3′-recessed double-stranded substrate. Heating at 42.5°C induced a significant and rapid decrease in Mre11activity as indicated by a reduction in the degradation of the DNA substrate by the purified protein (Fig. 5). A quantitative analysis of DNA degradation revealed that 1 h of heating of purified Mre11 at 42.5°C reduced exonuclease activity to approximately 10% of the unheated control protein. Rapid loss of exonuclease activity was also observed when Mre11 was heated at 45.5°C (Supplementary Fig. 2; http:dx.doi.org/10.1667/RR2594.1.S1). Exonuclease activity of purified Mre11 was greatly reduced (compared to unheated control Mre11 protein samples) after just 5 min of heating (representative gel shown in Supplementary Fig. 2).
FIG. 5.
Loss of Mre11 exonuclease activity after 42.5°C heat treatment. Panel A: 3 μg of Mre11 was heated at 42.5°C for indicated times. After heat treatment 2 μg of Mre11 was incubated with 200 pmol of DNA substrate and then run on a sequencing gel (representative gel shown). Panel B: Quantitative analysis of Mre11 exonuclease activity. Degradation products were quantified and then graphed as percentage of control. Error bars represent SEM from at least three experiments.
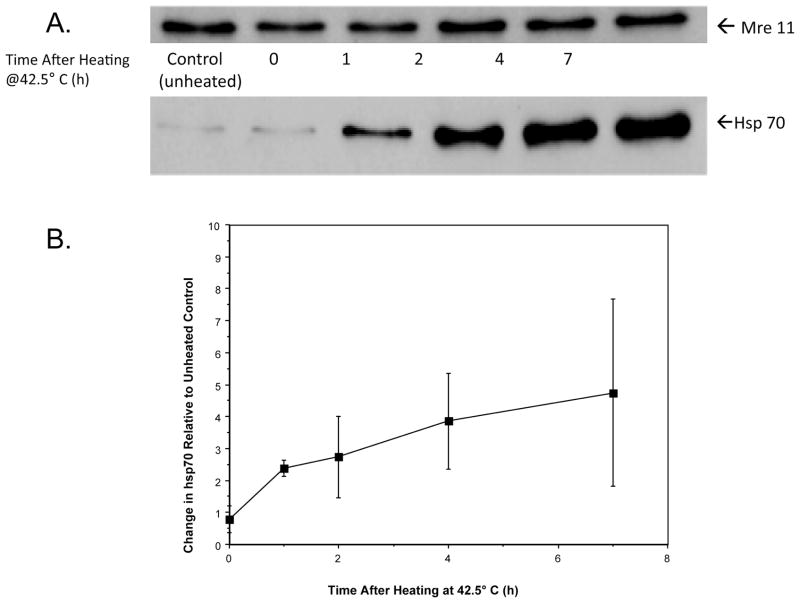
After a moderate (42.5°C, 2 h) hyperthermia treatment, hsp70 associates and then remains associated with Mre11 in HeLa cells long after the actual heat exposure (Fig. 6). Mre11 levels (as measured from whole cell lysates) remained relatively constant, while the amount of hsp70 associated with Mre11 increased throughout the 7-h period of post-heat incubation at 37°C. Similar results were obtained after heating cells for 15 min at 45.5°C (Supplementary Fig. 3; http://dx.doi.org/10.1667/RR2594.1.S1) [a nearly equitoxic exposure (data not shown)].
FIG. 6.
Heated-induced association between Hsp70 and Mre11. HeLa cells were heated for 2 h at 42.5°C and incubated at 37°C for various times, and then Mre11 was immunoprecipitated from whole cell lysates prior to SDS-PAGE. Representative immunoblots probed for Mre11 and Hsp70 are shown (panel A). To quantitatively assess the interaction between Mre11 and Hsp70 as a function of time after heating, immunoblots were analyzed and Hsp70 content was normalized against the Mre11 protein content of the unheated control (panel B). The change in the amount of Hsp70 protein immunoprecipitated with Mre11 increased with time after heating.
DISCUSSION
Data from several laboratories support the notion that heat radiosensitization may be attributed to decreased accessibility of repair proteins to sites of DNA damage induced by ionizing radiation (due at least in part to masking of damaged sites by nonspecific protein aggregation, chromatin changes or substrate modification), translocation of repair proteins (specifically the MRN proteins) out of the nucleus, or inactivation of critical DNA repair proteins by heat. Our data indicate that heat radiosensitization is seemingly unrelated to the heat-induced translocation of a large amount of nuclear MRN proteins to the cytoplasm of irradiated cells (Fig. 1) and support the latter scenario. By performing clonogenic cell survival assays on various cell lines that are hypomorphic or deficient in each of the MRN proteins to determine extent of heat radiosensitization, we systematically eliminated Rad50 and Nbs1 as targets, but we implicated a role for Mre11 in heat radiosensitization (Figs. 2–4). These results are consistent with those of Roti Roti and coworkers (42), who found that the heat-induced reduction in the availability of nuclear Mre11 for potential participation in the repair of radiation-induced DNA damage contributes at least in part to enhanced sensitivity to ionizing radiation in heated human adenocarcinoma cells. They demonstrated that under conditions where Mre11 is knocked down using siRNA, the unheated cells are substantially radiosensitized, but the Mre11-deficient cells fail to show heat radiosensitization compared to cells expressing normal levels of Mre11. Our results also reconcile well with the data of Xu et al. (43), which demonstrated that for several human tumor cell lines, the greater the endogenous Mre11 content, the greater the heating time required to elicit radiosensitization in that particular cell line. Curiously, Ohnishi et al. (44) showed that cells deficient in Nbs1 displayed enhanced sensitivity to heat, and given that the protein appears to be required for damage sensing, it is somewhat perplexing that Nbs1 is not a target for heat radiosensitization.
Purified Mre11 rapidly loses its exonuclease activity after exposure to hyperthermia (Fig. 5, Supplementary Fig. 2; http://dx.doi.org/10.1667/RR2594.1.S1). Such a finding (loss of activity of a critical repair enzyme after heat shock) would not be without precedent. Although Ku-dependent NHEJ is not involved in heat radiosensitization (39), the kinase and DNA-binding activity of DNA-PK and both Ku subunits, respectively, are known to decrease rapidly during hyperthermia treatment (45, 46).
It is attractive to speculate that heat radiosensitization results from the loss of Mre11 exonuclease activity mediated by conformational changes to Mre11. Two of our previous findings suggest that heat induces conformational changes in Mre11. We observed a decreased interaction between Mre11 and other components of Mre11 complexes found in the nuclei of heated, irradiated cells as evidenced by reduced colocalization of Mre11 and Rad50, and inhibition of Mre11 focus formation in cells that were irradiated and heated compared to cells that have only been irradiated (41). In addition, we found different kinetics of translocation of individual MRN proteins from the nucleus to the cytoplasm of irradiated and heated cells (33). Both suggest that the MRN complex disassociates and that the NES becomes readily accessible for facilitation of CRM1-mediated nuclear export. Laszlo and Fleischer (47) have recently suggested that 53BP1 [a protein required for ATM-dependent NHEJ as well as alternate NHEJ pathways (e.g., Ku70-independent) (48)] plays a role in heat radiosensitization; since the formation of the γ-H2AX/MDC1/53BP1 complex is delayed in heated-irradiated cells, it is likely that 53BP1 or the other repair proteins of that complex undergo heat-induced conformational changes that alter their ability to interact with each other.
While a reduction in exonuclease activity might account at least in part for inhibition of repair of radiation-induced DSBs by hyperthermia, further studies are necessary to determine the effect of heat on other DNA processing functions of Mre11, ideally when the protein has been isolated from cell lysates. It is possible that the reduction in degradation of DNA that we observed is indicative of a true loss in Mre11 exonuclease activity, possibly due to conformational changes or post-translational modification after heating; however, the reduction in the ability to degrade a DNA substrate after heating may also reflect an inability of Mre11 to bind to DNA. Within the cell, this could possibly be due to binding of other proteins, such as HSPs (see below). An investigation of changes in endonucleolytic processing by Mre11 after heating is also warranted.
The notion that Mre11 may become denatured or aggregated with other proteins was strengthened by the finding that Hsp70 associates with Mre11 during heating (Fig. 6, Supplementary Fig. 3; http://dx.doi.org/10.1667/RR2594.1.S1), and that changes in the association of hsp70 with Mre11 correlate well with the radiosensitization of HeLa cells by 41°C (49). Dote et al. (50) found that Hsp90 associates with the MRN complex and facilitates interaction between Nbs1 and ATM under normal conditions (e.g., cells maintained at 37°C). Treatment of cells with the Hsp90 inhibitor 17DMAG reduced the ability of MRN to form foci after irradiation. It is possible that the association of Mre11 with hsp70, while potentially assisting in the refolding of Mre11 or in preventing its denaturation, alters the interaction of the MRN complex with, contributes to inhibition of MRN focus formation after heat shock, and also interferes with its ability to effect DSB repair in cells that have been irradiated and heated. However, it is also possible that the heat-induced binding of hsp70 to Mre11 may compromise its ability to function as an exonuclease that acts on DSBs or impede other critical functions of Mre11 in irradiated cells.
Since we have previously shown that Ku-dependent NHEJ is not involved in heat radiosensitization (39), by deduction one might assume that either D-NHEJ (alternative NHEJ) or HR, or both pathways, must also be implicated in heat radiosensitization, since Mre11 is known to function in both pathways. Using an siRNA approach, cells in which Mre11 was knocked down to 30–40% of control levels were significantly radiosensitized (42). This observation, coupled with the development of drugs such as mirin that inhibit Mre11 exonuclease activity (51), suggests that Mre11 may be an attractive target to be exploited in radiotherapy.
Supplementary Material
Acknowledgments
The authors thank Jody Bethel, Eric Bittner and Joy Garrett for technical support, and Drs. Matthew Weitzman, Pat Concannon, Margaret Zdzienicka, James Carney and Minoru Yoshida for providing cell lines and reagents for this work. This work was supported by grants from the National Institutes of Health [CA108582 (JRD) and CA082741 (JJT)].
Footnotes
Note. The online version of this article (DOI: 10.1667/RR2594.1) contains supplementary information that is available to all authorized users.
References
- 1.Yuan J, Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J Biol Chem. 2010;285:1097–104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rupnik A, Lowndes NF, Grenon M. MRN and the race to the break. Chromosoma. 2010;119:115–35. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- 3.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 4.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–91. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–9. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 6.Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci U S A. 1999;96:7376–81. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–87. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 8.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Critchlow SE, Jackson SP. DNA end-joining from yeast to man. Trends Biochem Sci. 1998;23:394–8. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 10.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–53. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 11.Iliakis G. Backup pathways of NHEJ in cells of higher eukaryotes: cell cycle dependence. Radiother Oncol. 2009;9:310–5. doi: 10.1016/j.radonc.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–74. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong Q, Boyer TG, Chen PL, Lee WL. Deficient non-homologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 2002;62:3966–70. [PubMed] [Google Scholar]
- 14.Di Virgilio M, Gautier M. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J Cell Biol. 2005;171:765–71. doi: 10.1083/jcb.200506029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–8. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang J, Jiang G, Willers H, Xia F. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem. 2009;284:30565–73. doi: 10.1074/jbc.M109.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–24. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 18.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–2. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 19.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–79. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 20.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–88. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jager M, Dronkert MLG, Modesti M, Beerens CE, MT, Kanaar R, van Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–25. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosal G, Muniyappa K. The characterization of Saccharomyces cerevisiae Mre11/Rad50/Xrs2 complex reveals that Rad50 negatively regulates Mre11 endonucleolytic but not the exonucleolytic activity. J Mol Biol. 2007;372:864–82. doi: 10.1016/j.jmb.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Raymond WE, Kleckner N. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–6. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerosaletti KM, Desai-Mehta A, Yeo TC, Kraakman-Van Der Zwet M, Zdzienicka MZ, Concannon P. Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis. 2000;15:281–6. doi: 10.1093/mutage/15.3.281. [DOI] [PubMed] [Google Scholar]
- 25.Dong Z, Zhong Q, Chen P. The Nijmegen breakage syndrome protein is essential for Mre11 phosphorylation upon DNA damage. J Biol Chem. 1999;274:19513–6. doi: 10.1074/jbc.274.28.19513. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Weng YC, Yuan SS, Lin YT, Jsu HC, Lin SC, et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–7. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 27.Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–6. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 28.Dewey WC. Arrhenius relationships from the molecule and cell to the clinic. Int J Hyperthermia. 2009;25:3–20. doi: 10.1080/02656730902747919. [DOI] [PubMed] [Google Scholar]
- 29.Kampinga HH, Dynlacht JR, Dikomey E. Mechanisms of radiosensitization by hyperthermia (>43°C) as derived from studies with DNA repair defective mutant cell lines. Int J Hyperthermia. 2004;20:131–9. doi: 10.1080/02656730310001627713. [DOI] [PubMed] [Google Scholar]
- 30.Pandita TK, Pandita S, Bhaumik SR. Molecular parameters of hyperthermia for radiosensitization. Crit Rev Eukaryot Gene Expr. 2009;19:235–51. doi: 10.1615/critreveukargeneexpr.v19.i3.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roti Roti JR, Wright WD, VanderWaal R. The nuclear matrix: a target for heat shock effects and a determinant for stress response. Crit Rev Eukaryot Gene Expr. 1997;7:343–60. doi: 10.1615/critreveukargeneexpr.v7.i4.30. [DOI] [PubMed] [Google Scholar]
- 32.Zhu WG, Seno JD, Beck BD, Dynlacht JR. Translocation of MRE11 from the nucleus to the cytoplasm as a mechanism of radiosensitization by heat. Radiat Res. 2001;156:95–102. doi: 10.1667/0033-7587(2001)156[0095:tomftn]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 33.Seno JD, Dynlacht JR. Intracellular redistribution and phosphorylation of proteins of the Mre11/Rad50/Nbs1 repair complex following irradiation and heat-shock. J Cell Physiol. 2004;199:157–70. doi: 10.1002/jcp.10475. [DOI] [PubMed] [Google Scholar]
- 34.Xu M, Myerson RJ, Straube WL, Moros EG, Lagroye I, Wang LL, et al. Radiosensitization of heat resistant human tumour cells by 1 hour at 41. 1 degrees C and its effect on DNA repair. Int J Hyperthermia. 2002;18:385–403. doi: 10.1080/02656730210146908. [DOI] [PubMed] [Google Scholar]
- 35.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–20. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraakman-van der Zwet M, Overkamp WJ, Friedl AA, Klein B, Verhaegh GW, Jaspers NG, et al. Immortalization and characterization of Nijmegen breakage syndrome fibroblasts. Mutat Res. 1999;434:17–27. doi: 10.1016/s0921-8777(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 37.Shin BA, Ahn KY, Kook H, Koh JT, Kang IC, Lee HC, et al. Overexpressed human RAD50 exhibits cell death in a p21(WAF1/CIP1)-dependent manner: its potential utility in local gene therapy of tumor. Cell Growth Differ. 2001;12:243–54. [PubMed] [Google Scholar]
- 38.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–7. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 39.Dynlacht JR, Bittner ME, Bethel JA, Beck BD. The Non-homologous end-joining pathway is not involved in the radiosensitization of mammalian cells by heat shock. J Cell Physiol. 2003;196:557–64. doi: 10.1002/jcp.10334. [DOI] [PubMed] [Google Scholar]
- 40.Hermanson IL, Turchi JJ. Overexpression and purification of human XPA using a baculovirus expression system. Protein Expr Purif. 2000;19:1–11. doi: 10.1006/prep.2000.1224. [DOI] [PubMed] [Google Scholar]
- 41.Gerashchenko BI, Gooding G, Dynlacht JR. Hyperthermia alters the interaction of proteins of the Mre11 complex in irradiated cells. Cytometry A. 2010;77:940–52. doi: 10.1002/cyto.a.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu M, Myerson RJ, Hunt C, Kumar S, Moros EG, Straube WL, et al. Transfection of human tumor cells with Mre11 siRNA increases radiation sensitivity and reduces heat induced radiosensitization. Int J Hyperthermia. 2004;20:157–62. doi: 10.1080/02656730310001625986. [DOI] [PubMed] [Google Scholar]
- 43.Xu M, Myerson RJ, Xia Y, Whitehead T, Moros EG, Straube WL, et al. The effects of 41°C hyperthermia on the DNA repair protein, MRE11, correlate with radiosensitizatioin in four human tumor cell lines. Int J Hyperthermia. 2007;23:343–51. doi: 10.1080/02656730701383007. [DOI] [PubMed] [Google Scholar]
- 44.Ohnishi K, Scuric Z, Yau D, Schiestl RH, Okamoto N, Takahashi A. Heat-induced phosphorylation of NBS1 in human skin fibroblast cells. J Cell Biochem. 2006;99:1642–50. doi: 10.1002/jcb.20995. [DOI] [PubMed] [Google Scholar]
- 45.Matsumoto Y, Suzuki N, Morimatsu A, Murofushi H. A possible mechanism for hyperthermic radiosensitization mediated through hyperthermic lability of Ku subunits in DNA-dependent protein kinase. Biochem Biophys Res Commun. 1997;234:568–72. doi: 10.1006/bbrc.1997.6689. [DOI] [PubMed] [Google Scholar]
- 46.Burgman P, Ouyang H, Peterson S, Chen DJ, Li GC. Heat inactivation of Ku autoantigen: Possible role in hyperthermic radiosensitization. Cancer Res. 1997;57:2847–50. [PubMed] [Google Scholar]
- 47.Laszlo A, Fleischer I. Heat-induced perturbations of DNA damage signaling pathways are modulated by molecular chaperones. Cancer Res. 2009;69:2042–9. doi: 10.1158/0008-5472.CAN-08-1639. [DOI] [PubMed] [Google Scholar]
- 48.Iwabuchi K, Hashimoto M, Matsui T, Kurihara T, Shimizu H, Adachi N, et al. 53BP1 contributes to survival of cells irradiated with X-ray during G1 without Ku70 or Artemis. Genes Cells. 2006;11:935–48. doi: 10.1111/j.1365-2443.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 49.Dynlacht JR, Xu M, Pandita RK, Wetzel EA, Roti Roti JL. Effects of heat shock on the Mre11/Rad50/Nbs1 complex in irradiated or unirradiated cells. Int J Hyperthermia. 2004;20:144–56. doi: 10.1080/02656730410001660634. [DOI] [PubMed] [Google Scholar]
- 50.Dote H, Burgan WE, Camphausen K, Tofilon P. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66:9211–20. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- 51.Dupré A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–25. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.