Abstract
Granule neurons have a central role in cerebellar function via their synaptic interactions with other neuronal cell types both within and outside this structure. Establishment of these synaptic connections and its control is therefore essential to their function. Both intrinsic as well as environmental mechanisms are required for neuronal development and formation of neuronal circuits, and a key but poorly understood question is how these various events are coordinated and integrated in maturing neurons. In this review, we summarize recent work on the role of the Nuclear Factor I family in the transcriptional programming of cerebellar granule neuron maturation and synapse formation. In particular, we describe (1) the involvement of this family of factors in key developmental steps occurring throughout postmitotic granule neuron development, including dendrite and synapse formation and synaptic receptor expression, and (2) the mediation of these actions by critical downstream gene targets that control cell–cell interactions. These findings illustrate how Nuclear Factor I proteins and their regulons function as a “bridge” between cell-intrinsic and cell-extrinsic interactions to control multiple phases of granule neuron development.
Keywords: Neuronal differentiation, Transcription factor, Axonogenesis, Migration, Dendritogenesis, Synaptogenesis
Introduction
During central nervous system development, numerous neuronal subtypes are generated and become linked via synaptic interconnections into a complex network of neural circuits. In this process, maturing neurons proceed through multiple developmental stages, including migration, axon extension, dendritogenesis, and formation of functional synapses. In this process, distinct subsets of genes are expressed with varying spatiotemporal patterning. A central question is how these transcriptional events are coordinated to ensure the sequential expression of specific gene subsets within distinct neuronal populations. Little is currently known regarding the roles of specific trans-factors in determining the sequential completion of these different phases of neuronal development.
We have been investigating how different phases of postmitotic neuronal development are integrated in maturing cerebellar granule neurons (CGNs) and, specifically, the role of the Nuclear Factor I (NFI) family in coordinating these events. As outlined here, our findings demonstrate a central function of NFI proteins in regulating CGN maturation and developmental gene expression. Further, they illustrate how these proteins directly participate in the spatiotemporal organization of gene expression in differentiating CGNs. We also present a short overview of granule neuron function within the cerebellum in order to provide a physiological context, and we briefly discuss the value of these cells as an experimental model for neuronal development. Since our presentation of granule neuron development is of a more limited nature, we refer the reader to two excellent recent reviews of this topic [1, 2].
The Role of Granule Neurons in Cerebellar Circuitry
The cerebellum is crucial for sensorimotor control, including the coordination of movement (timing) and adaptive learning (plasticity) [3–5]. This structure is also being increasingly implicated in higher central nervous system (CNS) functions such as affect, cognition, working memory and attention via loop interactions with cerebral centers [4, 6–10]. CGNs play a pivotal role in the control of information flow between cerebellar inputs and outputs through synaptic interactions with mossy fibers and Purkinje neurons, respectively [11, 12]. In particular, CGNs have been implicated in cerebellar temporal processing and plasticity [11–13], the generation of oscillatory patterns [14–16], and in motor learning [11, 17, 18].
CGNs receive inhibitory inputs to their dendrites from type II Golgi interneurons that release gamma-aminobutyric acid (GABA) at synapses located within glomerular structures in the granule cell layer. These inputs regulate the responsiveness of CGNs to excitatory inputs from mossy fibers, and they are thought to be critical for the proper flow and storage of information within the cerebellar cortex [19]. In particular, feedforward inhibitory inputs from Golgi interneurons have been implicated in temporal, oscillatory, and information storage functions of CGNs [12, 15, 16]. GABA regulates CGN excitability by three main mechanisms: (1) fast-acting direct inhibitory postsynaptic currents due to local transmitter release within synapses; (2) indirect, delayed, and slowly inactivating currents caused by spillover of GABA from neighboring Golgi–CGN synapses [20]; and (3) a tonic inhibitory conductance induced by ambient GABA levels that is independent of action potentials [19, 21–23]. Of these, spillover- and tonically driven GABA inhibition play major roles in regulating CGN excitability, resulting in reduced gain for information processing through the mossy fiber–CGN–Purkinje cell pathway [19].
Granule Neuron Maturation and its Importance for the Development of Cerebellar Function
Within the maturing postnatal cerebellum, CGNs undergo a well-defined sequential program of differentiation characterized by progressive maturation stages (Fig. 1). In the mouse, CGN progenitors (CGNPs) proliferate within the outer portion of the external germinal layer (EGL) during the first 2 weeks. During this period, increasing numbers of CGNPs gradually exit the cell cycle and initiate differentiation by extending bipolar axons and migrating tangentially within the mid-region of the EGL [24]. Within the deeper pre-migratory zone (PMZ), immature CGNs extend long bipolar processes (≥100 μm) and project a third radial process [24]. With increasing age, CGNs then migrate radially from the PMZ through the forming molecular layer (ML) until they reach the internal granule cell layer (IGL). Upon onset of radial migration, axons emerging from immature CGNs within the PMZ/ML form fascicles of parallel fibers that are aligned along the longitudinal folial axis and perpendicular to Purkinje cell dendritic processes [25]. The final phase of CGN maturation then ensues with the formation of dendrites and synaptic connections with excitatory mossy fibers and inhibitory GABAergic terminals from Golgi type II neurons. The development of these latter inhibitory inputs coincides with the onset of eye opening and increased motor activity in rodent pups [26]. Thus, the timing of CGN maturation and GABA responsiveness is essential for the proper development of cerebellar functions.
Fig. 1.
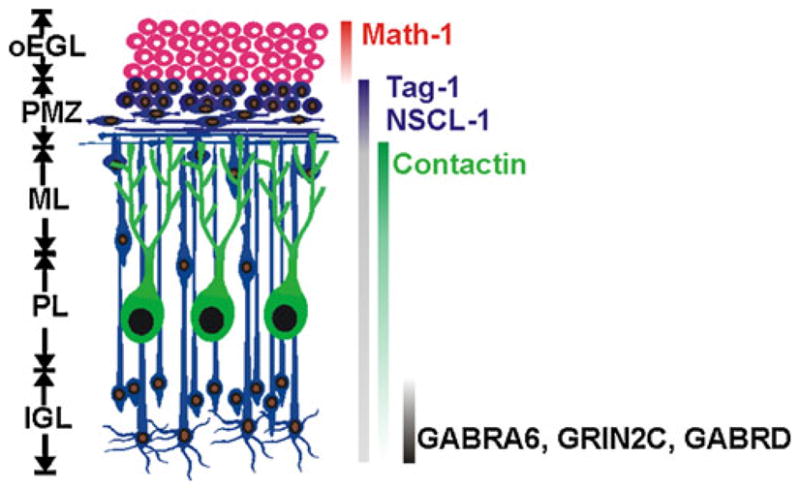
Granule neuron maturation within the developing cerebellum. Different cellular layers within the maturing postnatal cerebellum are shown on the left: oEGL outer external germinal layer, PMZ pre-migratory zone, ML molecular layer, PL Purkinje cell layer, and IGL internal granule cell layer. On the right are different genes that are expressed during specific stages of CGN maturation, with their developmental expression indicated by solid vertical bars (variable bar intensity indicates relative extents of expression for a given gene during development)
CGNs as a Model for Neuronal Development: Advantages and Experimental Approaches
Associated with the different morphological stages of CGN development is the sequential expression of numerous gene subsets in distinct temporal patterns [27]. Many of these genes are characteristic for a given stage of CGN maturation (Fig. 1). CGNs thus provide an excellent system to elucidate how various stages of neuronal development are regulated, including the functions of specific proteins as well as how the expression of various genes is controlled at different maturational steps. Further, the differential expression of various temporal gene subsets is reflected in their distinct patterns of spatial expression and organization. CGNs thus provide an opportunity to explore the importance of differential spatiotemporal patterning of gene expression in the integration of various neurodevelopmental phases.
CGN development is elaborated through the interaction of both cell-intrinsic and environmental mechanisms [28–33], for example, cell division and migration within the EGL are regulated by proteins produced by neighboring cells, including sonic hedgehog derived from Purkinje neurons and stromal cell-derived factor 1 released from the meninges [34–36]. At the same time, many aspects of CGN maturation appear to have a strong intrinsic component, including axon formation, migration, and dendritogenesis [37–39]. Accordingly, developmental patterning of gene expression in differentiating CGNs appears to be internally controlled to a substantial extent; for example, CGNPs purified from the postnatal rodent cerebellum recapitulate much of this developmental program as a “wave” upon plating and culture [32, 40, 41]. CGNs thus provide an excellent system for in vivo and in cellulo exploration of intrinsically driven regulation of gene transcription in maturing postmitotic neurons. A further advantage of this system is the ability to obtain purified CGNs and their progenitors in abundant amounts (~1–2×106 cells per P6 mouse cerebellum). This permits a highly detailed molecular analysis of a homogenous population of developing neurons. Further, because CGNs are by far the largest population of neurons within the cerebellum, it is possible to study molecular events (e.g., transcription factor–chromatin interactions (see below)) occurring in CGNs developing in vivo using intact cerebellar tissue.
A frequent limitation of neuronal cultures in defining regulatory mechanisms is the challenge of expressing exogenous proteins in a sufficient numbers of cells. Thus, one is often limited to the analysis of individual cells using morphological or histological endpoints. Since from the outset our goal has been to define in molecular detail how transcriptional mechanisms control CGN development, we initially explored approaches for obtaining quantitative transduction of CGN cultures (i.e., expression of a protein or RNA of interest in nearly all CGNs within a culture). It would then be possible to study how gene and protein expressions were regulated within a population of maturing neurons, including the analysis of transcription factor–gene interactions using chromatin immunoprecipitation (ChIP) assays. After exploring several approaches (e.g., biolistic and various viral vectors), we focused on the use of self-inactivating lentiviral vectors [42]. This virus offers several unique advantages, including the ability to transduce both proliferating and postmitotic neurons with high efficiency and low cytotoxicity, to harbor relatively large inserts, and to integrate its viral DNA into the host cell genome and provide continuous and stable expression. We found that these vectors provided quantitative transduction of CGN cultures during different stages of maturation (e.g., proliferating progenitors or fully postmitotic neurons), such that morphological and molecular perturbations could be characterized within a relatively large homogenous population of neurons [42–45].
The ability of lentivirus to integrate its DNA sequences into the host cell also provides another highly useful application in the study of transcriptional interactions: the ability to express and study the regulation of exogenous gene promoters within native chromatin [42, 45]. Thus, after ~24–48 h, the promoter of interest is expressed as a transgene within the neuronal culture since it is more or less randomly inserted into host cell genomic DNA. Promoter regulation can then be monitored as a function of neuronal development [42] (see also below).
NFI Proteins and Neuronal Development
The NFI family of transcriptional regulators is composed of four genes (NFIA, NFIB, NFIC, and NFIX) that are expressed in neurons throughout the CNS [46, 47]. Numerous splice variants exist for each NFI family member, and they function as either homo- or heterodimers to activate or repress transcription [48]. Thus, these factors may function in complex ways to regulate CNS development. Consistent with this, NFIA, NFIB, and NFIX have been specifically implicated in the development of neurons and/or glia within numerous structures within the mouse central nervous system, including basilar pontine and mossy fiber nuclei, the corpus callosum, forebrain, hippocampus, and spinal cord [49–57]. This body of work was recently comprehensively reviewed [58] and is not elaborated here. Further, haploinsufficiency of NFIA is associated with altered CNS formation in humans [59]. We have utilized the aforementioned experimental advantages of the CGN system to explore in detail the role of NFI proteins in neuronal development. In particular, our studies have identified multiple key roles for the NFI family in the coordination of the postmitotic differentiation of these cells.
NFI Proteins and the Control of GABA Receptor Maturation in CGNs
As noted earlier, GABA released from Golgi type II neurons plays a pivotal role in regulating CGN responsiveness and cerebellar circuitry involved in motor learning and other functions. GABAA receptors (GABAARs) are composed of a variety of different subunits, the nature of which strongly influences their GABA responses. During CGN development, GABAARs undergo a maturation-dependent change in their properties from mainly benzodiazepine-sensitive to a benzodiazepine-insensitive form [60]. Associated with this is a switch in GABAAR subunit expression in which the α2 and α3 subunits that confer benzodiazepine sensitivity are down-regulated while the α6 subunit is induced [60]. The α6 subunit is selectively expressed in granule neurons within the cerebellum [61, 62], where it promotes high-affinity, slow-inactivating, and benzodiazepine-insensitive GABA responsiveness. GABAARs containing this subunit predominate in mature CGNs [63] and are preferentially localized to extra-synaptic sites [64]. Accordingly, α6-containing receptors are primary targets of GABA-induced spillover and tonic inhibition within the granule layer and play an important role in cerebellar information processing and storage [19, 20]. Local confinement of GABA released from Golgi synapses to the glomerulus likely promotes its accumulation and activation of these extrasynaptic GABAARs [20].
In the developing cerebellum, expression of the GABAA α6 receptor subunit gene (Gabra6) occurs with a temporal delay in post-migratory CGNs within the IGL, increasing mainly during the second through fourth postnatal weeks in the mouse [65–67]. Thus, the Gabra6 gene provides an opportunity to investigate the control of dendritogenesis-related as well as CGN-specific gene expression. We therefore have explored the regulation of this gene in order to gain insight into transcriptional mechanisms controlling spatiotemporal regulation of neuronal development.
Using a combination of in vitro, in cellulo, and in vivo approaches, we found that the NFI family has a central role in Gabra6 gene expression [45]. NFI proteins were found to bind to a consensus site within the proximal Gabra6 promoter in vitro. Using lentiviral promoter constructs containing a 6-kb Gabra6 promoter fragment, this consensus NFI-binding site was shown to be critical for promoter activity in mature CGNs. Lentiviral vectors were also used to demonstrate that suppression of endogenous NFI activity in maturing CGN cultures by an NFI dominant repressor markedly reduced Gabra6 expression. Further, loss of NFIA in vivo caused a fourfold reduction in expression of this gene in the maturing mouse cerebellum. Thus, NFI family proteins, including NFIA, are involved in the switch from immature to mature GABAARs, and thereby GABA responsiveness, in CGNs within the IGL. These trans-factors are therefore predicted to be critical for tonic control of CGN excitability and the resultant reduced gain necessary for information processing through the mossy fiber–CGN–Purkinje cell pathway.
NFI Proteins Are Required for Multiple Stages of CGN Maturation
A further outgrowth of studies on Gabra6 gene regulation was the finding that NFI proteins are highly enriched in the mouse cerebellum [45]. These results suggested that the NFI family may play an important role in the maturation of CGNs more generally. To more fully explore this question, we first determined the expression of the different NFI family members in the postnatal cerebellum [44]. Based on indirect immunofluorescence, NFIA, NFIB, and NFIX were localized to the nucleus in immature CGNs within the PMZ as well as in fusiform, migrating cells in the ML and in post-migratory cells in the IGL. In contrast, immunostaining was generally weak and diffuse in the proliferative outer EGL containing CGNPs. Thus, NFI proteins are localized to the nucleus from the early onset of postmitotic CGN differentiation and throughout their subsequent development. This suggested a potential role for these factors at multiple stages of postmitotic CGN maturation.
Using lentiviral delivery of an NFI dominant repressor into CGN re-aggregate cultures, NFI trans-activation was shown to be dispensable for CGNP proliferation, but in contrast was critical for axon outgrowth [44]. Remarkably, this dependence of axon outgrowth on NFI was not observed in fully dissociated CGN cultures, suggesting a requirement for homotypic cell–cell interactions in these downstream actions. This was subsequently supported by identification of mediators of NFI effects on axonogenesis (see below). Besides axon extension, migration of post-mitotic CGNs from re-aggregates was severely inhibited by repression of NFI function. This migratory requirement was also supported by Transwell migration assays using the NFI dominant repressor lentivirus. Finally, the effects of NFI disruption on axon extension and migration were further confirmed in situ using retroviral delivery of the NFI dominant repressor into CGNPs within the EGL of early postnatal mouse cerebellar slices. The dominant repressor induced defasciculation of forming parallel fibers within the PMZ/ML, with many fibers extending aberrantly towards the EGL and IGL. This indicated a primary role for NFI trans-activation in both the extension as well as the orientation of parallel fibers. Further, disruption of NFI function also dramatically inhibited radial migration of CGNs from the EGL/PMZ to the IGL, consistent with cell culture findings.
In addition to the requirement for NFI function in these relatively early postmitotic differentiation events, dominant repressor studies also implicated this family in CGN dendrite formation [44]. Both the lengths as well as the number of dendritic processes were reduced in CGN cultures. Interestingly, these late-differentiation effects were not dependent on cell–cell contacts but occurred in fully dissociated cultures, in contrast to axon extension. Together, these findings strongly indicated a vital role for NFI proteins in numerous phases of CGN development, namely, axonogenesis, radial migration, and dendritogenesis.
Functional analyses were subsequently extended to the developing cerebellum using NFI knockout mice. These studies largely confirmed our culture findings and established the functional importance of specific NFI family members [44]. NFIA null mice survive to ~P20, permitting analysis of the major CGN developmental events. Grossly, foliation of the forming cerebellum was altered in these knockout mice, with lobules I–V being under-developed and lobules VI–VII poorly differentiated. Parallel fiber extension was also dramatically shortened in these mice and most axons were misoriented, indicating a central role for NFIA in parallel fiber extension as well as alignment. In addition, ectopic and radially migrating fusiform CGNs were still evident within the ML of the P17 NFIA null cerebellum, with a significant number of postmitotic CGNs remaining in a residual EGL/PMZ region that was not apparent in wild-type mice of the same age. This suggested a delay in the postmitotic maturation of CGNs, resulting in persistent radial migration.
Late maturation of CGNs within the IGL also was disrupted in NFIA knockout mice. In particular, dendrites within anterior cerebellar regions were few in number or were extremely short, as observed in dominant repressor culture studies [44]. More recent studies have extended NFIA function within the IGL to include synaptogenesis [43]. Specifically, maturation of mossy fiber–CGN synapses was defective in NFIA null mice, as evidenced by diminished staining intensity for the pre-synaptic marker synapsin I within the IGL and a reduction in the number of synapsin I-positive rosettes.
Defects in early CGN development also were observed in NFIB knockout mice. At E18, NFIB null mice were previously shown to have altered foliation in the cerebellum [55]. Subsequent studies showed axon formation was also markedly reduced in the forming EGL/PMZ region of these mice [44]. Evaluation of postnatal maturation events in NFIB null mice was precluded by their perinatal death. Studies are currently ongoing to examine postnatal CGN development in conditional NFIB knockout mice. Based on morphological analyses, NFIC-null mice do not exhibit significant morphological phenotypes in the cerebellum (W. Wang, D. Kilpatrick, and R. Gronostajski, unpublished findings), although molecular changes have not been fully investigated.
Mediators of NFI in CGN Development: An Interplay of Cell-Intrinsic and Cell–Cell Interactions
The functioning of NFI proteins throughout postmitotic CGN development was consistent with their expression patterns (see above). These actions likely involved the regulation of multiple downstream genes that together help to elaborate the various stages of CGN differentiation. This was addressed by examining potential regulators of various phases of CGN development. Cell adhesion molecules (CAMs) are versatile proteins that can function in axon extension and fasciculation, neuronal migration, and dendrite and synapse formation. We initially identified two CAMs that were markedly down-regulated in NFI dominant repressor-treated cultures and in NFIA null mice: N cadherin and ephrin B1 [44]. Further, ChIP assays confirmed that NFI proteins occupied NFI consensus sites within the promoters of these two genes in purified CGN cultures and in the mouse cerebellum, implicating these CAMs as direct targets and downstream mediators of NFI trans-activation. This was subsequently confirmed using antagonists of ephrin B1 and N cadherin function [44]. In both cases, axon extension, migration, and dendritogenesis by CGNs were severely reduced or altered by relevant inhibitors using re-aggregate as well as cerebellar slice cultures, recapitulating findings with the NFI dominant repressor.
More recently, another CAM was identified as a downstream target of NFI, transient axonal glycoprotein 1 (Tag-1/contactin-2) [43]. Tag-1 is transiently and highly up-regulated in pre-migratory mouse CGNs during the first postnatal week, where it is expressed on both cell bodies and elongating parallel fibers just prior to onset of radial migration [68–70]. Tag-1 expression was markedly reduced in the forming cerebellum of E18 NFIB null mice. Interestingly, Tag-1 expression was unaltered in NFIA null mice at P8, when Tag-1 expression within the PMZ is robust. Thus, Tag-1 expression during CGN axon formation appears to be selectively dependent on NFIB. Further, NFI proteins bind to two regions spanning NFI sites within the mouse Tag-1 promoter region in immature mouse CGNs and in the P6 mouse cerebellum [44]. NFI proteins also directly stimulated the promoter for the human homologue of the Tag-1 gene in co-transfection studies. Thus, Tag-1 appears to be an important direct target of NFI in early maturing CGNs.
The pattern of Tag-1 expression in forming parallel fibers suggested a role in axon extension and alignment, with soma expression possibly reflecting regulation of the timing of radial migration onset [68, 69] and of tangential migration within the EGL/PMZ [71]. Further, Tag-1 was recently implicated in regulation of parallel fiber alignment in the developing chick cerebellum [72]. Evidence for a direct role for Tag-1 in parallel fiber formation by mouse CGNs has not been previously demonstrated. Tag-1-null mice showed no obvious cerebellar phenotype at P2, likely reflecting compensatory mechanisms [73]. Using Tag-1 blocking antibodies, we recently found evidence that Tag-1 is required for axon formation as well as migration in both CGN re-aggregate cultures as well as in situ in P5 mouse cerebellar slices, but not in dissociated cultures (W. Wang, D. Karagogeos, and D. Kilpatrick, unpublished studies). Thus, Tag-1 may be another important mediator of NFI-dependent cell–cell interactions during parallel fiber extension and onset of radial migration from the PMZ.
As noted earlier, NFIA regulates the expression of synapsin I in pre-synaptic endings within synaptic rosettes in the IGL. How are these effects mediated? Previous studies demonstrated that synapsin I clustering and accumulation at mossy fiber synapses within glomerular rosettes was regulated by Wnt7a released from CGNs [74]. Based on these observations, we examined whether Wnt7a was a target of NFI proteins in maturing CGNs. Both Wnt7a transcripts and protein were markedly reduced in the cerebellum of P17 NFIA null mice [43], and CGNs are the major site of Wnt7a expression in the developing postnatal cerebellum [75]. Further, the mouse Wnt7a gene contains an NFI consensus site that is strongly bound by NFI proteins in nuclei from both mature CGN cultures and in P21 mouse cerebellum [43]. Thus, Wnt7a is an apparent direct target of the NFI family that also mediates important downstream cell–cell interactions controlled by these factors. In this case, these interactions are heterotypic, affecting mossy fiber pre-synaptic remodeling via synapsin I expression within rosettes.
A Model for NFI Regulation of CGN Development
Together, these findings indicate that NFI proteins are essential regulators of CGN maturation, controlling numerous events: axonogenesis, radial migration, dendritogenesis, and synaptogenesis (Fig. 2). These effects are mediated by multiple downstream targets that act in part via intercellular interactions. CAMs appear to be major downstream mediators of NFI in maturing postmitotic CGNs. This is fully consistent with the requirement for homotypic cell contact in NFI actions on parallel fiber extension and fasciculation. Our results also suggest that onset of radial migration is controlled by NFI via multiple CAMs, including N cadherin, ephrin B1, and possibly Tag-1. Ephrin B proteins were previously implicated in onset of CGN radial migration [76]. In this case, they are thought to promote departure from the EGL by overcoming the chemoattractant effects of SDF-1 released from the overlying meninges [76]. Further, NFI proteins regulate heterotypic interactions controlling mossy fiber remodeling via the secreted protein Wnt7a. Thus, the NFI family and its downstream targets function as a bridge between cell-intrinsic (gene transcription) and homotypic and heterotypic cell contact-dependent mechanisms. In addition, NFI is implicated in the control of Golgi–CGN extra-synaptic interactions, and thereby CGN excitability, via regulation of Gabra6 expression and responsiveness to the inhibitory transmitter GABA within the granule cell layer.
Fig. 2.
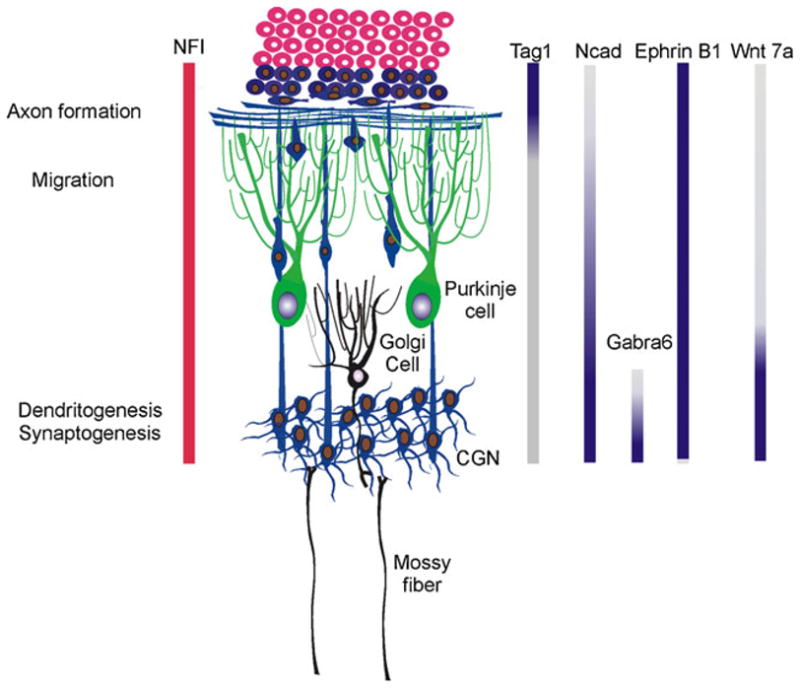
Summary of the actions of NFI proteins and their effector target genes during CGN development. Different events subject to NFI regulation in maturing CGNs are shown on the left, along with a portrayal of the developmental pattern of NFI protein expression. The temporal patterns of expression for currently identified NFI target genes are shown by vertical bars on the right. The NFI targets N cadherin (Ncad) and ephrin B1 regulate axon formation and fasciculation via homotypic cell interactions, as well as CGN migration and dendritogenesis. Tag-1, Wnt-7a, and Gabra6 exhibit more restricted expression patterns and have more selective functions either in pre-migratory or post-migratory CGNs
The developmental expression patterns of this diverse set of downstream mediators also help to explain the ability of NFI proteins to regulate multiple stages of CGN maturation (Fig. 2). The CAMs N cadherin and ephrin B1 are expressed throughout CGN postmitotic development [44], and correspondingly, they regulate both early and later differentiation events (Fig. 2). In contrast, the CAM Tag-1 and the secreted proteins Wnt7a and Gabra6 are more highly expressed and function during specific stages (Fig. 2).
As discussed earlier, CGNs provide an excellent system for exploring the link between spatiotemporal patterning of gene expression and various stages of neuronal development. Our findings highlight the importance of coordinated spatiotemporal gene expression by a single family of transcriptional regulators for the integration of different maturational steps into a coherent developmental program.
Future Directions: A Question of Timing
As noted earlier, Tag-1 is transiently up-regulated in pre-migratory CGNs undergoing axon extension, while Gabra6 is expressed in post-migratory CGNs within the IGL. Further, both N cadherin and Wnt7a are gradually up-regulated as CGNs mature, while ephrin B1 is more constitutively expressed (W. Wang and D. Kilpatrick, unpublished observations) (Fig. 2). This raises an important question: how are NFI proteins able to regulate gene targets having divergent patterns of developmental expression in maturing CGNs? The question of timing and sequential gene expression is clearly an important one for neuronal development. In the case of CGNs, intrinsic timing mechanisms have been directly implicated in the regulation of their migration [38] as well as for onset of expression of the Gabra6 gene [30, 32, 67]. Such temporal regulation is in turn critical for CGN function; for example, up-regulation of Gabra6 expression in CGNs within the IGL coincides with GABAergic synapse formation, motor control and learning [77], and tonic GABAA receptor-mediated conductance within the IGL [22, 26, 78]. Thus, temporal regulation of Gabra6 expression is important for proper developmental responsiveness of maturing CGNs to inhibitory GABA inputs and, in turn, information processing within the developing and mature cerebellum. Similar temporal considerations undoubtedly apply to other NFI targets, including transient up-regulation of Tag-1 expression within the PMZ and elevated expression of Wnt7a within the IGL.
One mechanism by which the NFI family may regulate distinct genes at different stages of development is via selective actions of different family members; for example, NFIB appears to affect CGN maturation at an early stage based on analyses of E18 null mice. In contrast, developmental defects were evident in P17 NFIA knockout mice but not at P8, suggesting distinct functional time frames for these two family members. What might account for such developmentally distinct actions? We have observed no apparent differences in the stage-dependent expression of NFI family gene isoforms in maturing CGNs, although differential expression of novel splice variants or post-translational modifications cannot be entirely ruled out at this time. Alternatively, different NFI family proteins may have unique interactions with promoter-specific co-regulators that have temporally distinct activities in developing CGNs.
A primary question is the nature and expression patterns of the gene subsets lying downstream of the different NFI family proteins. Ongoing analyses of NFIA null mice and NFIB conditional knockout mice will serve to shed light on this question. Assuming NFI family specific gene targets are identified, more detailed analyses of trans-factor/promoter interactions for representative targets should ultimately provide valuable insight into how NFI family members collaborate at the transcriptional level to direct CGN development. At the same time, comparison of the cerebellar phenotypes for NFIB conditional and NFIA mice will reveal the degree to which these two family members have overlapping or complementary roles in CGN morphological and functional maturation.
Acknowledgments
This work was supported by the Public Health Service grants R01 NS063047 (to DLK) and by core resources provided by the Diabetes Endocrinology Research Center grant DK32520.
Footnotes
Conflict of Interest Statement The authors have no financial or personal relationships, dual commitments, or competing interests or loyalties that bias the work presented here.
Contributor Information
Daniel L. Kilpatrick, Email: Daniel.kilpatrick@umassmed.edu, Department of Microbiology and Physiological Systems, and Program in Neuroscience, University of Massachusetts Medical School, 55 Lake Ave N., Worcester, MA 01655, USA
Wei Wang, Stem Cell & Regenerative Medicine International, 33 Locke Drive, Marlborough, MA 01752, USA.
Richard Gronostajski, Department of Biochemistry and Program in Neuroscience, State University of New York, Buffalo, NY 14214-3000, USA.
E. David Litwack, Department of Anatomy and Neurobiology and Program in Neuroscience, Baltimore, School of Medicine, University of Maryland, Baltimore, MD, USA.
References
- 1.Carletti B, Rossi F. Neurogenesis in the cerebellum. Neuroscientist. 2008;14:91–100. [Google Scholar]
- 2.Chedotal A. Should I stay or should I go? Becoming a granule cell. Trends Neurosci. 2010;33:163–72. doi: 10.1016/j.tins.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Eccles JM, Ito M, Szentagothai J. The cerebellum as a neuronal machine. Berlin: Springer-Verlag; 1967. [Google Scholar]
- 4.Schmahmann JD, Caplan D. Cognition, emotion and the cerebellum. Brain. 2006;129:290–2. doi: 10.1093/brain/awh729. [DOI] [PubMed] [Google Scholar]
- 5.Ito M. Cerebellar circuitry as a neuronal machine. Prog Neurobiol. 2006;78:272–303. doi: 10.1016/j.pneurobio.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–34. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- 7.Stoodley CJ, Schmahmann JD. Functional topography in the human cerebellum: a meta-analysis of neuroimaging studies. Neuroimage. 2009;44:489–501. doi: 10.1016/j.neuroimage.2008.08.039. [DOI] [PubMed] [Google Scholar]
- 8.Ito M. Movement and thought: identical control mechanisms by the cerebellum. Trends Neurosci. 1993;16:448–50. doi: 10.1016/0166-2236(93)90073-u. discussion 453-4. [DOI] [PubMed] [Google Scholar]
- 9.Sacchetti B, Scelfo B, Tempia F, Strata P. Long-term synaptic changes induced in the cerebellar cortex by fear conditioning. Neuron. 2004;42:973–82. doi: 10.1016/j.neuron.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Habas C. Functional imaging of the deep cerebellar nuclei: a review. Cerebellum. 2010;9:22–8. doi: 10.1007/s12311-009-0119-3. [DOI] [PubMed] [Google Scholar]
- 11.Medina JF, Garcia KS, Nores WL, Taylor NM, Mauk MD. Timing mechanisms in the cerebellum: testing predictions of a large-scale computer simulation. J Neurosci. 2000;20:5516–25. doi: 10.1523/JNEUROSCI.20-14-05516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Angelo E, De Zeeuw CI. Timing and plasticity in the cerebellum: focus on the granular layer. Trends Neurosci. 2009;32:30–40. doi: 10.1016/j.tins.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Fujita M. Adaptive filter model of the cerebellum. Biol Cybern. 1982;45:195–206. doi: 10.1007/BF00336192. [DOI] [PubMed] [Google Scholar]
- 14.D’Angelo E, Koekkoek SK, Lombardo P, Solinas S, Ros E, Garrido J, et al. Timing in the cerebellum: oscillations and resonance in the granular layer. Neuroscience. 2009;162:805–15. doi: 10.1016/j.neuroscience.2009.01.048. [DOI] [PubMed] [Google Scholar]
- 15.Maex R, De Schutter E. Synchronization of golgi and granule cell firing in a detailed network model of the cerebellar granule cell layer. J Neurophysiol. 1998;80:2521–37. doi: 10.1152/jn.1998.80.5.2521. [DOI] [PubMed] [Google Scholar]
- 16.Medina JF, Mauk MD. Computer simulation of cerebellar information processing. Nat Neurosci. 2000;3(Suppl):1205–11. doi: 10.1038/81486. [DOI] [PubMed] [Google Scholar]
- 17.Marr D. A theory of cerebellar cortex. J Physiol. 1969;202:437–70. doi: 10.1113/jphysiol.1969.sp008820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyrrell T, Willshaw D. Cerebellar cortex: its simulation and the relevance of Marr’s theory. Philos Trans R Soc Lond B Biol Sci. 1992;336:239–57. doi: 10.1098/rstb.1992.0059. [DOI] [PubMed] [Google Scholar]
- 19.Hamann M, Rossi DJ, Attwell D. Tonic and spillover inhibition of granule cells control information flow through cerebellar cortex. Neuron. 2002;33:625–33. doi: 10.1016/s0896-6273(02)00593-7. [DOI] [PubMed] [Google Scholar]
- 20.Rossi DJ, Hamann M. Spillover-mediated transmission at inhibitory synapses promoted by high affinity alpha6 subunit GABA (A) receptors and glomerular geometry. Neuron. 1998;20:783–95. doi: 10.1016/s0896-6273(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 21.Brickley SG, Revilla V, Cull-Candy SG, Wisden W, Farrant M. Adaptive regulation of neuronal excitability by a voltage-independent potassium conductance. Nature. 2001;409:88–92. doi: 10.1038/35051086. [DOI] [PubMed] [Google Scholar]
- 22.Wall MJ, Usowicz MM. Development of action potential-dependent and independent spontaneous GABAA receptor-mediated currents in granule cells of postnatal rat cerebellum. Eur J Neurosci. 1997;9:533–48. doi: 10.1111/j.1460-9568.1997.tb01630.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaneda M, Farrant M, Cull-Candy SG. Whole-cell and single-channel currents activated by GABA and glycine in granule cells of the rat cerebellum. J Physiol. 1995;485(Pt 2):419–35. doi: 10.1113/jphysiol.1995.sp020739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komuro H, Yacubova E, Rakic P. Mode and tempo of tangential cell migration in the cerebellar external granular layer. J Neurosci. 2001;21:527–40. doi: 10.1523/JNEUROSCI.21-02-00527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altman J. Postnatal development of the cerebellar cortex in the rat. 3. Maturation of the components of the granular layer. J Comp Neurol. 1972;145:465–513. doi: 10.1002/cne.901450403. [DOI] [PubMed] [Google Scholar]
- 26.Tia S, Wang JF, Kotchabhakdi N, Vicini S. Developmental changes of inhibitory synaptic currents in cerebellar granule neurons: role of GABA(A) receptor alpha 6 subunit. J Neurosci. 1996;16:3630–40. doi: 10.1523/JNEUROSCI.16-11-03630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–82. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 28.Gao WO, Heintz N, Hatten ME. Cerebellar granule cell neurogenesis is regulated by cell-cell interactions in vitro. Neuron. 1991;6:705–15. doi: 10.1016/0896-6273(91)90168-y. [DOI] [PubMed] [Google Scholar]
- 29.Gao WQ, Hatten ME. Neuronal differentiation rescued by implantation of weaver granule cell precursors into wild-type cerebellar cortex. Science. 1993;260:367–9. doi: 10.1126/science.8469990. [DOI] [PubMed] [Google Scholar]
- 30.Gao B, Fritschy JM. Cerebellar granule cells in vitro recapitulate the in vivo pattern of GABAA-receptor subunit expression. Brain Res Dev Brain Res. 1995;88:1–16. doi: 10.1016/0165-3806(95)00062-i. [DOI] [PubMed] [Google Scholar]
- 31.Kawaji K, Umeshima H, Eiraku M, Hirano T, Kengaku M. Dual phases of migration of cerebellar granule cells guided by axonal and dendritic leading processes. Mol Cell Neurosci. 2004;25:228–40. doi: 10.1016/j.mcn.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Lin X, Bulleit RF. Cell intrinsic mechanisms regulate mouse cerebellar granule neuron differentiation. Neurosci Lett. 1996;220:81–4. doi: 10.1016/s0304-3940(96)13214-6. [DOI] [PubMed] [Google Scholar]
- 33.Lu Q, Sun EE, Flanagan JG. Analysis of PDZ-RGS3 function in ephrin-B reverse signaling. Methods Enzymol. 2004;390:120–8. doi: 10.1016/S0076-6879(04)90008-0. [DOI] [PubMed] [Google Scholar]
- 34.Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–8. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 35.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–14. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 36.Klein RS, Rubin JB, Gibson HD, DeHaan EN, Alvarez-Hernandez X, Segal RA, et al. SDF-1 alpha induces chemotaxis and enhances Sonic hedgehog-induced proliferation of cerebellar granule cells. Development. 2001;128:1971–81. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- 37.Powell SK, Rivas RJ, Rodriguez-Boulan E, Hatten ME. Development of polarity in cerebellar granule neurons. J Neurobiol. 1997;32:223–36. doi: 10.1002/(sici)1097-4695(199702)32:2<223::aid-neu7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 38.Yacubova E, Komuro H. Intrinsic program for migration of cerebellar granule cells in vitro. J Neurosci. 2002;22:5966–81. doi: 10.1523/JNEUROSCI.22-14-05966.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trenkner E, Smith D, Segil N. Is cerebellar granule cell migration regulated by an internal clock? J Neurosci. 1984;4:2850–5. doi: 10.1523/JNEUROSCI.04-11-02850.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bahn S, Wisden W, Dunnett SB, Svendsen C. The intrinsic specification of gamma-aminobutyric acid type a receptor alpha6 subunit gene expression in cerebellar granule cells. Eur J Neurosci. 1999;11:2194–8. doi: 10.1046/j.1460-9568.1999.00662.x. [DOI] [PubMed] [Google Scholar]
- 41.Raetzman LT, Siegel RE. Immature granule neurons from cerebella of different ages exhibit distinct developmental potentials. J Neurobiol. 1999;38:559–70. [PubMed] [Google Scholar]
- 42.Wang W, Qu Q, Smith FI, Kilpatrick DL. Self-inactivating lentiviruses: versatile vectors for quantitative transduction of cerebellar granule neurons and their progenitors. J Neurosci Methods. 2005;149:144–53. doi: 10.1016/j.jneumeth.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Crandall JE, Litwack ED, Gronostajski RM, Kilpatrick DL. Targets of the nuclear factor I regulon involved in early and late development of postmitotic cerebellar granule neurons. J Neurosci Res. 2010;88:258–65. doi: 10.1002/jnr.22199. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Mullikin-Kilpatrick D, Crandall JE, Gronostajski RM, Litwack ED, Kilpatrick DL. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 2007;27:6115–27. doi: 10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W, Stock RE, Gronostajski RM, Wong YW, Schachner M, Kilpatrick DL. A role for nuclear factor I in the intrinsic control of cerebellar granule neuron gene expression. J Biol Chem. 2004;279:53491–7. doi: 10.1074/jbc.M410370200. [DOI] [PubMed] [Google Scholar]
- 46.Chaudhry AZ, Lyons GE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–25. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 47.Plachez C, Lindwall C, Sunn N, Piper M, Moldrich RX, Campbell CE, et al. Nuclear factor I gene expression in the developing forebrain. J Comp Neurol. 2008;508:385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- 48.Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 49.Kumbasar A, Plachez C, Gronostajski RM, Richards LJ, Litwack ED. Absence of the transcription factor Nfib delays the formation of the basilar pontine and other mossy fiber nuclei. J Comp Neurol. 2009;513:98–112. doi: 10.1002/cne.21943. [DOI] [PubMed] [Google Scholar]
- 50.Campbell CE, Piper M, Plachez C, Yeh YT, Baizer JS, Osinski JM, et al. The transcription factor Nfix is essential for normal brain development. BMC Dev Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Driller K, Pagenstecher A, Uhl M, Omran H, Berlis A, Grunder A, et al. Nuclear factor I X deficiency causes brain malformation and severe skeletal defects. Mol Cell Biol. 2007;27:3855–67. doi: 10.1128/MCB.02293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, Macklin WB, et al. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci USA. 1999;96:11946–51. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piper M, Moldrich RX, Lindwall C, Little E, Barry G, Mason S, et al. Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. 2009;4:43. doi: 10.1186/1749-8104-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–12. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steele Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–98. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–68. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 57.Wong YW, Schulze C, Streichert T, Gronostajski RM, Schachner M, Tilling T. Gene expression analysis of nuclear factor I-A deficient mice indicates delayed brain maturation. Genome Biol. 2007;8:R72. doi: 10.1186/gb-2007-8-5-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mason S, Piper M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- 59.Lu W, Quintero-Rivera F, Fan Y, Alkuraya FS, Donovan DJ, Xi Q, et al. NFIA haploinsufficiency is associated with a CNS malformation syndrome and urinary tract defects. PLoS Genet. 2007;3:e80. doi: 10.1371/journal.pgen.0030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wisden W, Korpi ER, Bahn S. The cerebellum: a model system for studying GABAA receptor diversity. Neuropharmacology. 1996;35:1139–60. doi: 10.1016/s0028-3908(96)00076-7. [DOI] [PubMed] [Google Scholar]
- 61.Kato K. Novel GABAA receptor alpha subunit is expressed only in cerebellar granule cells. J Mol Biol. 1990;214:619–24. doi: 10.1016/0022-2836(90)90276-r. [DOI] [PubMed] [Google Scholar]
- 62.Laurie DJ, Seeburg PH, Wisden W. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. II. Olfactory bulb and cerebellum. J Neurosci. 1992;12:1063–76. doi: 10.1523/JNEUROSCI.12-03-01063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quirk K, Gillard NP, Ragan CI, Whiting PJ, McKernan RM. Model of subunit composition of gamma-aminobutyric acid A receptor subtypes expressed in rat cerebellum with respect to their alpha and gamma/delta subunits. J Biol Chem. 1994;269:16020–8. [PubMed] [Google Scholar]
- 64.Nusser Z, Sieghart W, Somogyi P. Segregation of different GABAA receptors to synaptic and extrasynaptic membranes of cerebellar granule cells. J Neurosci. 1998;18:1693–703. doi: 10.1523/JNEUROSCI.18-05-01693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng T, Santi MR, Bovolin P, Marlier LN, Grayson DR. Developmental expression of the alpha 6 GABAA receptor subunit mRNA occurs only after cerebellar granule cell migration. Brain Res Dev Brain Res. 1993;75:91–103. doi: 10.1016/0165-3806(93)90068-l. [DOI] [PubMed] [Google Scholar]
- 66.Varecka L, Wu CH, Rotter A, Frostholm A. GABAA/benzodiazepine receptor alpha 6 subunit mRNA in granule cells of the cerebellar cortex and cochlear nuclei: expression in developing and mutant mice. J Comp Neurol. 1994;339:341–52. doi: 10.1002/cne.903390304. [DOI] [PubMed] [Google Scholar]
- 67.Mellor JR, Merlo D, Jones A, Wisden W, Randall AD. Mouse cerebellar granule cell differentiation: electrical activity regulates the GABAA receptor alpha 6 subunit gene. J Neurosci. 1998;18:2822–33. doi: 10.1523/JNEUROSCI.18-08-02822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pickford LB, Mayer DN, Bolin LM, Rouse RV. Transiently expressed, neural-specific molecule associated with premigratory granule cells in postnatal mouse cerebellum. J Neurocytol. 1989;18:465–78. [Google Scholar]
- 69.Yamamoto M, Hassinger L, Crandall JE. Ultrastructural localization of stage-specific neurite-associated proteins in the developing rat cerebral and cerebellar cortices. J Neurocytol. 1990;19:619–27. doi: 10.1007/BF01188031. [DOI] [PubMed] [Google Scholar]
- 70.Wolfer DP, Henehan-Beatty A, Stoeckli ET, Sonderegger P, Lipp HP. Distribution of TAG-1/axonin-1 in fibre tracts and migratory streams of the developing mouse nervous system. J Comp Neurol. 1994;345:1–32. doi: 10.1002/cne.903450102. [DOI] [PubMed] [Google Scholar]
- 71.Bailly Y, Kyriakopoulou K, Delhaye-Bouchaud N, Mariani J, Karagogeos D. Cerebellar granule cell differentiation in mutant and X-irradiated rodents revealed by the neural adhesion molecule TAG-1. J Comp Neurol. 1996;369:150–61. doi: 10.1002/(SICI)1096-9861(19960520)369:1<150::AID-CNE11>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 72.Baeriswyl T, Stoeckli ET. Axonin-1/TAG-1 is required for pathfinding of granule cell axons in the developing cerebellum. Neural Dev. 2008;3:7. doi: 10.1186/1749-8104-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fukamauchi F, Aihara O, Wang YJ, Akasaka K, Takeda Y, Horie M, et al. TAG-1-deficient mice have marked elevation of adenosine A1 receptors in the hippocampus. Biochem Biophys Res Commun. 2001;281:220–6. doi: 10.1006/bbrc.2001.4334. [DOI] [PubMed] [Google Scholar]
- 74.Hall AC, Lucas FR, Salinas PC. Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell. 2000;100:525–35. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 75.Lucas FR, Salinas PC. WNT-7a induces axonal remodeling and increases synapsin I levels in cerebellar neurons. Dev Biol. 1997;192:31–44. doi: 10.1006/dbio.1997.8734. [DOI] [PubMed] [Google Scholar]
- 76.Lu Q, Sun EE, Klein RS, Flanagan JG. Ephrin-B reverse signaling is mediated by a novel PDZ-RGS protein and selectively inhibits G protein-coupled chemoattraction. Cell. 2001;105:69–79. doi: 10.1016/s0092-8674(01)00297-5. [DOI] [PubMed] [Google Scholar]
- 77.Takayama C. Formation of GABAergic synapses in the cerebellum. Cerebellum. 2005;4:171–7. doi: 10.1080/14734220510008012. [DOI] [PubMed] [Google Scholar]
- 78.Brickley SG, Cull-Candy SG, Farrant M. Development of a tonic form of synaptic inhibition in rat cerebellar granule cells resulting from persistent activation of GABAA receptors. J Physiol. 1996;497(Pt 3):753–9. doi: 10.1113/jphysiol.1996.sp021806. [DOI] [PMC free article] [PubMed] [Google Scholar]


