Abstract
Acquisition of the pluripotent state coincides with epigenetic reprogramming of the X-chromosome. Female embryonic stem cells are characterized by the presence of two active X-chromosomes, cell differentiation by inactivation of one of the two Xs, and induced pluripotent stem cells by reactivation of the inactivated X-chromosome in the originating somatic cell. The tight linkage between X- and stem cell reprogramming occurs through pluripotency factors acting on noncoding genes of the X-inactivation center. This review article will discuss the latest advances in our understanding at the molecular level. Mouse embryonic stem cells provide a standard for defining the pluripotent ground state, which is characterized by low levels of the noncoding Xist RNA and the absence of heterochromatin marks on the X-chromosome. Human pluripotent stem cells, however, exhibit X-chromosome epigenetic instability that may have implications for their use in regenerative medicine. XIST RNA and heterochromatin marks on the X-chromosome indicate whether human pluripotent stem cells are developmentally ‘naïve’, with characteristics of the pluripotent ground state. X-chromosome status and determination thereof via noncoding RNA expression thus provide valuable benchmarks of the epigenetic quality of pluripotent stem cells, an important consideration given their enormous potential for stem cell therapy.
Keywords: Epigenetics, Reprogramming, X-chromosome inactivation, Pluripotency, Stem cells, Noncoding RNAs
1. INTRODUCTION
This review article will discuss the tight linkage between X-chromosome and stem cell reprogramming. Recent studies have shown that this linkage is mediated by pluripotency factors acting specifically on noncoding genes of the X-inactivation center (Xic) to initiate or reverse X-chromosome inactivation (XCI), the mechanism of dosage compensation in mammals which leads to transcriptional inactivation of one X-chromosome in the female. XCI provides a classic model for noncoding RNA (ncRNA)-mediated epigenetic regulation [1–3]. These ncRNAs are located at the Xic, a regulatory hub that mediates the stepwise formation of Xi heterochromatin [4]. The onset of XCI corresponds with expression of the 17-kb noncoding Xist RNA, which coats the entire inactive X (Xi) chromosome in cis [5–11]. Xist mediates facultative heterochromatin on the Xi through recruitment and interaction with Polycomb group proteins [12], marking the Xi with histone H3 lysine 27 trimethylation (H3K27me3) [13–15]. Xist expression is regulated by three other ncRNAs, with two functioning in the activation of Xist (RepA, Jpx) [12, 16, 17] and one functioning to antagonize its activation (Tsix) [18–20].
Although this review will not focus on imprinted X-chromosome inactivation, it should be briefly mentioned that XCI can be subject to parental imprinting in marsupial mammals and also in the extraembryonic lineages of some eutherian mammals (e.g., mouse, cow) [21, 22]. Imprinted XCI occurs on the paternal X-chromosome and is believed to be the ancestral form of mammalian dosage compensation. In mice, the imprinted form of XCI is observed first during development in all cells, but persists only in the extraembryonic tissues after embryonic day 4.5, when imprint erasure and X-reactivation occur in the epiblast lineage [23–26]. Among ncRNAs involved in “random” XCI, Xist and Tsix are thus far the only ones known to also participate in imprinted XCI. Embryos lacking Tsix cannot protect the maternal X-chromosome from silencing [20, 27], and those lacking Xist cannot initiate genic silencing on the paternal X [10, 25].
Following reactivation of the paternal X-chromosome, cells of the epiblast lineage undergo random XCI and give rise to the embryo proper. From mouse and human embryos, it is possible to derive cells from this lineage and generate embryonic stem (ES) cells, a pluripotent cell type capable of differentiating into all three germ lineages (ectoderm, mesoderm, endoderm). ES cells have provided a valuable ex vivo system for the study of epigenetic reprogramming and the role of XCI and ncRNAs during cell differentiation [1–3, 28]. With the possibility of creating induced pluripotent stem (iPS) cells from adult somatic cells [29, 30] has come the opportunity to study how and whether reprogramming into pluripotent stem cells is accompanied by X-reactivation. These studies have shown that events on the X-chromosome and stem cell fate are indeed intimately connected. Below, we will focus on events surrounding cell differentiation and de-differentiation and the fate of the X-chromosome in ES and iPS cells, specifically those involving noncoding genes.
2. MOUSE X-CHROMOSOME REGULATION
2.1. Mouse ES cells
For random XCI studies, mouse ES [31] cells [31] have served as a powerful model system and enabled elucidation of function for many ncRNAs during this process. In undifferentiated female mES cells where parental epigenetic marks have been erased to be reprogrammed, both Xs remain active with very low levels of Xist expression. Cell differentiation then triggers XCI, initiated with Xist RNA upregulation on the future Xi. Although how Xist is regulated has yet to be fully understood, many studies have established the 40-kb Tsix ncRNA as a major regulator that antagonizes Xist induction in cis: deleting Tsix causes hypertranscription of Xist [19, 20, 27, 32], and overexpression of Tsix RNA prevents Xist upregulation [33, 34]. Various mechanisms are involved in Tsix-mediated repression of Xist: (1) Tsix modulates the chromatin state of Xist [35–38]; (2) it induces de novo CpG methylation and silencing of the Xist promoter [36, 37]; and (3) it recruits RNAi machinery to silence the Xist promoter [39–41]. Tsix transcription is positively regulated in cis by Xite, another non-coding gene that functions as an enhancer of Tsix transcription during mES cell differentiation [39, 42].
While Tsix mediates negative regulation of Xist, a recent study has revealed another ncRNA, Jpx, in the role of Xist activation [16]. Like Xist and Tsix, Jpx resides in the Xic [43–45] and is developmentally regulated, showing a 20- to 30-fold increase in its expression level prior to the initiation of XCI [16]. Deleting Jpx results in two major problems: defective XCI and female-specific lethality, specifically during differentiation of mES cells. Xist expression is severely attenuated in female mES cells, and embryoid body formation is disrupted during cell differentiation. However, the deletion has no effect in male mES cells, suggesting an essential and direct role for Jpx in the XCI process. Jpx RNA knockdown experiments using shRNAs recapitulates the deletion phenotype, thereby implicating Jpx RNA in the activation of Xist. Unlike other noncoding genes of the Xic, the Jpx deletion can be rescued by autosomal expression of a Jpx transgene, which implies that Jpx functions in trans as a diffusible ncRNA. Finally, truncating Tsix RNA in a Jpx deletion background also rescues Xist expression, indicating that the two regulatory ncRNAs work in parallel and antagonistic pathways to control Xist. Thus, Xist RNA levels during XCI are directly regulated by two ncRNA switches, Jpx and Tsix, which help designate the future Xi and active X (Xa) chromosomes.
Xist ncRNA accumulation on the Xi is almost immediately followed by the recruitment of Polycomb repressive complex 2 (PRC2) to catalyze H3K27me3 [13–15]. The search for mechanisms of PRC2 recruitment to the Xi led to identification of a novel ncRNA located within the 5′ end of Xist called RepA [12]. The 1.6-kb RepA is an independent transcription unit embedded within Xist that shares Repeat A, a conserved motif known to be important for X-chromosome silencing [17, 46]. RNA immunoprecipitation (RIP) and RNA electrophoretic mobility shift assay (EMSA) revealed that RepA directly interacts with Ezh2, a catalytic subunit of PRC2, via a secondary structure within Repeat A [12]. Autosomal RepA transgenes could increase recruitment of PRC2 upon induction, suggesting that RepA RNA is sufficient to recruit PRC2 to chromatin. Unlike Xist RNA, RepA is expressed prior to XCI, and its levels are not upregulated during cell differentiation. RepA RNA exhibits important functions in the pre-XCI state, where it plays a pivotal role in de novo recruitment of PRC2 to the Xic, perhaps aiding in the activation of Xist [12, 17] and enabling progression from pluripotency to differentiated cell states.
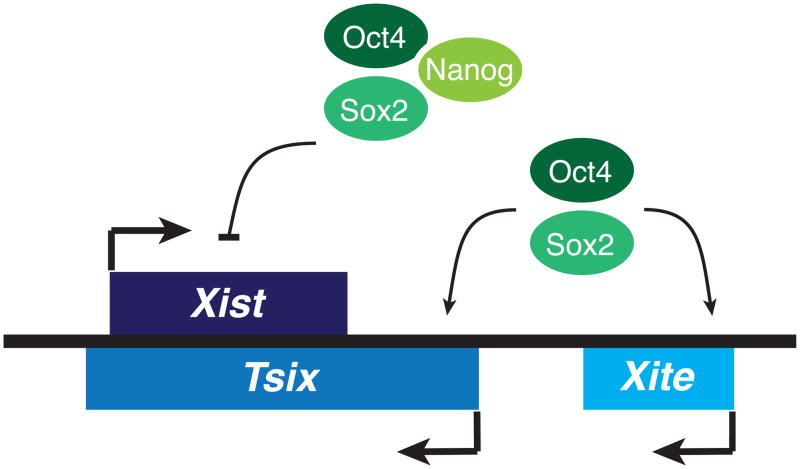
Studies using mES cells have yielded novel insights into the molecular circuitry that links XCI to pluripotency. Recent findings regarding the pluripotency factor Oct4 have uncovered its role as a master regulator of X-chromosome counting and pairing [47]. In mES cells, Oct4 directly binds the Tsix and Xite loci (Fig. 1), proximal to sites occupied by another regulator of X-chromosome pairing, Ctcf, which physically interacts with Oct4. A second pluripotency factor, Sox2, also directly binds Xite, while making indirect contact with Tsix through looping interactions between the Xite and Tsix domains [47]. Furthermore, Sox2 interacts with Yy1, a Tsix transactivator that regulates XCI choice. Because Yy1 is known to bind Ctcf [48], while Sox2 interacts with Oct4 as part of the core transcriptional circuitry that regulates pluripotency [49], it is likely that a multifactor complex comprised of Oct4, Sox2, Ctcf, and Yy1 directs the nascent stages of X-chromosome inactivation. In undifferentiated mES cells, biallelic occupancy of these factors is thought to promote expression of Tsix RNA, which in turn blocks the action of RepA and Xist RNAs in the initiation of X-chromosome silencing.
Fig. 1.
Xist regulation by the core pluripotency factors. Oct4 and Sox2 bind the noncoding Tsix and Xite loci, upregulating the expression of Tsix. Xist levels are also controlled by direct binding of Oct4, Sox2, and Nanog to Xist intron 1.
Through its intrinsic developmental specificity, Oct4 triggers changes in Xic behavior during the process of mES cell differentiation. Loss of Oct4 during cell differentiation is thought to induce homologous pairing between the two X-chromosomes [47], an act mediated by Tsix and Xite and associated with the regulatory steps of X-chromosome counting and choice that occur prior to the initiation of XCI [50, 51]. Knockdown of either Oct4 or Ctcf prevents pairing interactions from occurring [47, 52]. Deleting either Tsix or Xite also interferes with X-chromosome pairing, and insertion of Tsix and Xite sequences into an autosomal locus results in ectopic pairing between the autosome and an X-chromosome [51]. These results support the idea that a complex of Oct4, Ctcf, and Tsix/Xite sequences underlies the pairing interaction between the X-chromosomes. It is presently unknown whether ncRNAs transcribed from Tsix and Xite are required for pairing. However, inhibition of RNA polymerase II by Actinomycin D or α-amanitin disrupts the pairing interaction. As differentiation proceeds, the progressive loss of Oct4 may cause dissolution of the complex and dissociation of the X-chromosomes, which may result in redistribution of the Tsix transcription factors Ctcf, Oct4, and Yy1 from both alleles to one allele, due to the highly cooperative binding of factors [47, 51, 53]. By this model, the persistent binding of transcription factors on the Xa allele sustains Tsix expression exclusively on that chromosome. Interestingly, Oct4 knockdown has also been shown to result in biallelic Xist expression, indicating misregulation of X-chromosome counting [47]. Oct4 is thus the first known trans-factor that regulates X-chromosome counting.
Pluripotency factors also intersect the within the gene body of Xist/Tsix. Nanog binding sites are found within Xist intron 1 (Fig. 1), and co-occupancy by Oct4 and Nanog can repress Xist expression, either directly repressing Xist or indirectly repressing it via Tsix, which overlaps Xist in this region [54]. Nanog-null male mES cells display elevated levels of Xist RNA with no change in steady state levels of Tsix. It is thus proposed that Nanog may function independently of Tsix as a repressor of Xist. In Tsix-truncated male mES cells, Nanog remains bound to Xist intron 1 [54]. Of note, Oct4 and Sox2 also remain associated with Xist intron 1 in Nanog-null male mES cells. In Oct4-null male mES cells, however, Sox2 and Nanog binding to Xist intron 1 is compromised. Additionally, a small fraction of Oct4-null male mES cells display Xist upregulation, suggesting that Oct4 exhibits a more prominent role than Nanog in Xist regulation and X-chromosome reprogramming.
Together, Tsix, Oct4, and Nanog serve as important regulators of Xist expression during mES cell differentiation. The idea that Tsix and Oct4 might regulate Xist independently is supported by the fact that different mES cell differentiation methods affect Xist expression differentially when Tsix is deficient [55]. When TsixΔCpG male mES cells are differentiated in the absence of all-trans retinoic acid (RA), only a minute percentage of differentiated cells exhibit Xist clouds. However, in the presence of RA, partial Xist clouds appear in almost one-third of TsixΔCpG male mES cells (the Xist clouds are generally dispersed and do not necessarily result in genic silencing). The use of RA to differentiate ES cells was shown to accelerate downregulation of the general pool of Oct4 mRNA, as well as to accelerate loss of Oct4 binding to Xist intron 1. In the presence of a functional Tsix allele, however, the use of RA during male mES cell differentiation does not lead to ectopic Xist cloud formation. These results indicate that Tsix is sufficient for proper Xist regulation, irrespective of Oct4 binding to Xist intron 1. While Tsix serves as the primary regulator of Xist, Oct4 may compensate for the absence of Tsix when male mES cells are differentiated without RA, given the low incidence of ectopic Xist cloud formation. These results indicate that Oct4 and Tsix act in both coordinated and independent pathways to regulate Xist levels in mES cells.
2.2. Mouse iPS cells
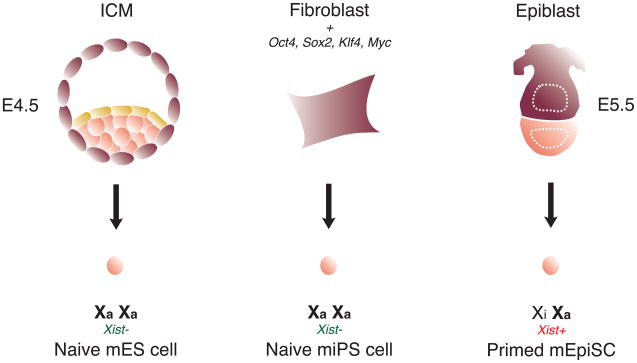
Mouse induced pluripotent stem (miPS) cells are generated from somatic cells through ectopic expression of the transcription factors Oct4, Sox2, Klf4, and c-Myc [29]. Interestingly, converting somatic female cells into miPS cells results in extensive X-chromosome reprogramming [56]. The Xi in female miPS cells is reactivated, and Xist expression becomes undetectable upon direct reprogramming (Fig. 2). Tsix and X-linked gene expression become biallelic, while Xite is also expressed. The reactivated X-chromosome loses H3K27me3 and Polycomb group protein enrichment, creating a transcriptionally permissive chromatin environment. Furthermore, the Xist, Tsix, and Xite ncRNAs are reprogrammed to their pluripotent, mES-like expression state. When induced to differentiate, miPS cells behave equivalently to mES cells with respect to XCI: Tsix RNA is downregulated, Xist RNA is upregulated and cytologically coats one X-chromosome, and the Xi is decorated by Polycomb proteins and the hallmark H3K27me3 modification [13, 14]. These findings further underscore the tight linkage between X-chromosome state and stem cell pluripotency.
Fig. 2.
X-chromosome state in mouse pluripotent stem cells. Naïve mES and miPS cells represent the ground state of pluripotency, as evidenced by the presence of two active X-chromosomes and the absence of Xist RNA. Primed mEpiSCs have already undergone X-chromosome inactivation and represent a developmentally more advanced state.
During the induction of pluripotency by defined factors, X-chromosome reactivation is a late event in the reprogramming process [57]. Epigenetic reprogramming of the X-chromosome is a hallmark of bona fide female miPS cells, along with the reactivation of endogenous pluripotency genes and telomerase. Sox2 and Oct4 are reactivated with faster kinetics than the silent X-chromosome, although these processes are also considered relatively late events during the reprogramming process. Assessment of endogenous Sox2 reactivation after ~18 days of fibroblast reprogramming indicates that a majority of cells express Sox2, while only a small fraction of cells at analogous time points show X-chromosome reactivation. Expression of endogenous Sox2 and Oct4 may subsequently facilitate silencing of Xist expression through Tsix and Xite activation, as well as direct binding to Xist intron 1. The molecular mechanism underlying X-chromosome reactivation during direct reprogramming, however, remains an area for further investigation, and miPS cells provide an excellent model in which to investigate the linkage between the epigenetic status of the X-chromosome and pluripotency.
2.3. Mouse EpiSCs
Mouse pluripotent stem cells are also derived from the epiblast layer of post-implantation embryos (d5.5) and are referred to as epiblast stem cells (mEpiSCs) [58, 59]. The epigenetic state of mEpiSCs differs from mES cells, with their pluripotency signaling pathways, cellular morphology, and gene expression patterns being more analogous to human ES cells [60]. mEpiSCs express the pluripotency factors Oct4, Nanog, and Sox2 and differentiate into the three germ layers, but grow as monolayer colonies in a similar manner to hES cells. Unlike mES cells, mEpiSCs are Rex1-negative, express FGF-5 and Nodal, and fail to incorporate into pre-implantation embryos. mEpiSCs have already undergone XCI (Fig. 2), as evidenced by H3K27me3 on the Xi and similar to many hES cell lines (discussed in section 3.1). Of note, Klf2, Klf4, and Klf5 levels are downregulated in mEpiSCs when compared to mES cells [61], suggesting that Klf family members might be involved in X-chromosome reprogramming. Adding exogenous Klf4 to mEpiSCs, however, fails to induce X-chromosome reactivation and convert these cells into mES cells [62]. Interestingly, culturing mEpiSCs with exogenous Klf4 in the presence of LIF and small molecule inhibitors of Mek/Erk MAP kinase signaling and glycogen synthase kinase (2i) induces their transition to a mES-like state, along with epigenetic reprogramming of the X-chromosome and efficient chimeric contribution [62]. The efficiency of converting mEpiSCs to mES cells using this procedure is ~0.1%, comparable to the efficiencies observed when reprogramming fibroblasts to miPS cells using defined factors (Oct4/Sox2/c-Myc/Klf4) [63]. In part, low efficiencies may stem from the potential requirement for stochastic events to facilitate the epigenetic reprogramming process [64].
3. HUMAN X-CHROMOSOME REGULATION
3.1. Human ES cells
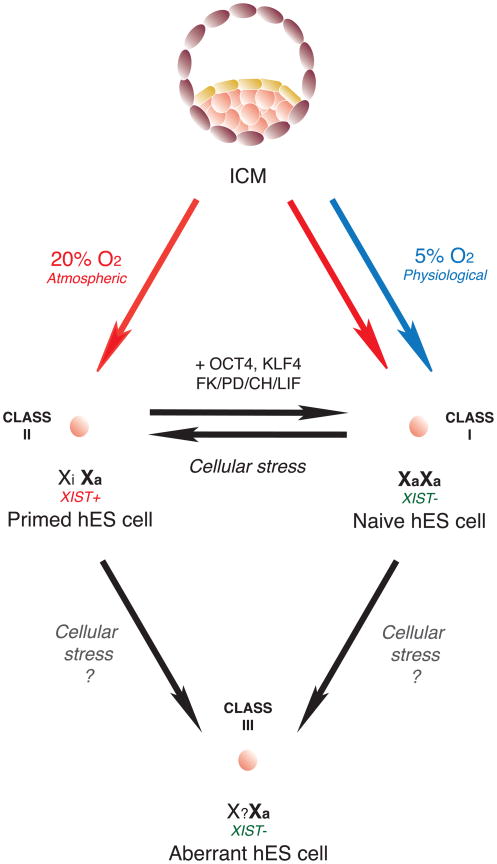
Assessing XIST ncRNA and XCI status in hES cells provides a measure of their epigenetic stability, which is an important consideration for their potential applications in regenerative medicine. Existing hES cell lines exhibit diverse patterns of XIST expression, indicative of both pre- and post-XCI states [65–70]. Because of similarities between mEpiSCs and hES cells (morphology, Activin/Nodal signaling for pluripotency, bFGF growth requirements), it was hypothesized that hES cells represent post-XCI cells, similar to mEpiSCs. Numerous female hES cell lines have been extensively characterized with respect to XCI, revealing the existence of three distinct classes of XCI status (Fig. 3) [70, 71]. Lines of hES cells designated as ‘Class I’ show relatively low levels of XIST expression in the undifferentiated state, and upon differentiation, XIST is upregulated and the number of cells with large XIST RNA nuclear foci increases. In ‘Class II’ lines, XIST expression levels are comparable in both the undifferentiated and differentiated states, indicating that XCI has already occurred in these hES cell lines. ‘Class III’ lines no longer express XIST ncRNA, whether in the undifferentiated or differentiated state, yet maintain monoallelic X-linked gene expression, suggestive of an aberrant epigenetic state. Interestingly, Class III hES cells appear to have at least partially undergone XCI, suggesting that XIST expression was lost after XCI was established. While expression of X-linked repetitive elements remain mostly suppressed, there may be partial reactivation of a small number of X-linked genes [66, 69, 70]. Of note, Class I and II cells readily transition into Class III cells with prolonged culture, demonstrating the highly dynamic and unstable nature of the epigenetic state in hES cells (Fig. 3) [70].
Fig. 3.
X-chromosome state in human ES cells. Deriving hES cells under physiological O2 (5%) generates hES cells that are more likely to be Class I, lacking XIST expression and other markers of XCI (‘naïve’), whereas derivation under hyperoxic conditions more likely yields hES cells of the Class II type, having already undergone XCI (‘primed’). However, ambient oxygen levels can also yield Class I cells. Cellular stress converts Class I and Class II hES cells into Class III.
3.3. Naïve human ES cells
Current research efforts are aimed at establishing and maintaining hES cells in conditions that would enhance their epigenetic stability. A recent study hypothesized that replicating physiological oxygen concentrations of the early embryo (hypoxic; 5% O2) would be beneficial for the derivation of epigenetically stable hES cells [68]. Of note, XCI status was the most sensitive measure for distinguishing between lines generated and maintained in either ambient or hypoxic oxygen concentrations. Importantly, hES cells in a pre-XCI state were only generated when early embryos and dissected ICM cells were cultured at physiological oxygen levels. Using physiological O2 concentration resulted in the derivation developmentally ‘naïve’ [72] hES cells that display two active X chromosomes [68] (Fig. 3), as indicated by the use of XIST and XCI markers as diagnostic tools. These lines upregulated XIST upon differentiation and formed cytologically visible XIST clouds, suggesting that naïve hES cells in fact resemble mES cells instead of mEpiSCs. hES cells derived in ambient oxygen had already undergone XCI, similar to the majority of hES cells derived previously. Another recent study isolated pre-XCI Class I hES cells in ambient oxygen, but these lines quickly became Class II and III lines within 15–20 passages [71]. Both groups observed increased DNA methylation at the XIST promoter in pre-XCI lines, suggestive of XIST silencing on both alleles. hES cells grown in ambient O2 conditions (20%), however, displayed ~50% reduction in XIST promoter methylation, along with activation of one XIST allele. Biallelic expression of X-linked genes was also observed in Class I pre-XCI hES cells, further indicating the presence of two active X chromosomes. Class III hES cell lines grown in ambient oxygen continued to display monoallelic X-linked gene expression despite the lost of XIST RNA. Loss of XIST RNA in primed hES cells resulted in varying degrees of X-linked gene derepression, and genes that remain silenced might be repressed by H3K27me3, which was present on the Xi despite the absence of XIST [68] [note: H3K27me3 foci are not observed in Class III hiPS and hES cells [70, 73]].
Naïve hES cells (Class I) convert to the primed state (Class II) upon exposure to hyperoxic conditions, undergoing irreversible XCI. Additionally, harsh freeze-thaw cycles, as well as treatment of naïve hES cells with compounds that induce cellular stress, result in XIST gene activation, indicating that naïve hES cells are acutely sensitive to various types of cellular stress. Treatment of naïve hES cells with antioxidants confers protection against XCI and XIST activation when cells are exposed to hyperoxic conditions, indicating that oxidative stress is a key determinant of precocious XCI in the undifferentiated, naive state.
Another study recently showed that HDAC inhibitors help promote epigenetic stability and decrease cellular differentiation during routine culture of hES cells [76]. Treatment of the H9 female hES cell line, containing a mixed population of XIST+ and XIST− cells, resulted in complete loss of XIST RNA in the undifferentiated state, with upregulation observed during cell differentiation. HDAC inhibitor treatment, which reversibly altered expression of several hundred genes and increased cell cycle growth, also reversed the XCI state and rendered this cell line more Class I-like [76]. However, since the authors only examined one female hES cell line known to have epigenetic variability between different passages and laboratories, this effect may not be universal for all hES cell lines.
It has also been proposed that priming hES cells with a specific cocktail of small molecules and transgenes carrying pluripotency factors can convert the epigenetically abnormal hES cells into naïve hES cells [74]. Specifically, adding ERK1/2 inhibitor PD0325901 (PD), GSK3 inhibitor CHIR99021 (CH), and Klf4/Klf2 regulator Forskolin [75], as well as providing Oct4 and Klf4 expression, converts primed hES and hiPS cells into a naïve state after ~10 days in culture (Fig. 3). These converted Class I hES cells lack XIST clouds in the undifferentiated state, and unlike their parental Class II state, can be passaged as single cells with trypsin, similar to mES cells and without the acquistion of chromosomal abnormalities. Moreover, converted naïve hES cells exhibit biallelic XIST promoter methylation, indicating XIST silencing, and display global gene expression patterns that are characteristic of naïve mES cells. Upon differentiation, naïve hiPS cells upregulate XIST RNA, which coats the Xi (experiments were carried out in ambient oxygen conditions; differentiated naïve hES cells were not examined). Given the inherent epigenetic instability of hES cells, defined small molecule cocktails might help stabilize the naïve hES-state. Interestingly, global gene expression patterns for naïve hES and hiPS cells resemble those of mES cells, suggesting that these naïve human cell types might serve as true representations of the inner cell mass in human embryos.
These studies highlight the permissiveness of X-chromosome epigenetic reprogramming in established hES cell lines using defined culture conditions, while also highlighting the usefulness of ncRNA markers in the study of regenerative medicine. While XIST ncRNA now appears to be an excellent marker for determining epigenetic stability in hES cells, additional studies are needed to learn about other ncRNAs that regulate XCI, particularly those that have been shown to play crucial roles in the mouse system. Given that early mouse and human development share more similarities with respect to XCI than previously thought, it is likely that human TSIX [77] and JPX [78], both with unknown functions in pluripotent cells, might also be harnessed as diagnostic tools for assessing hES cell epigenetic stability.
3.3. Human iPS cells
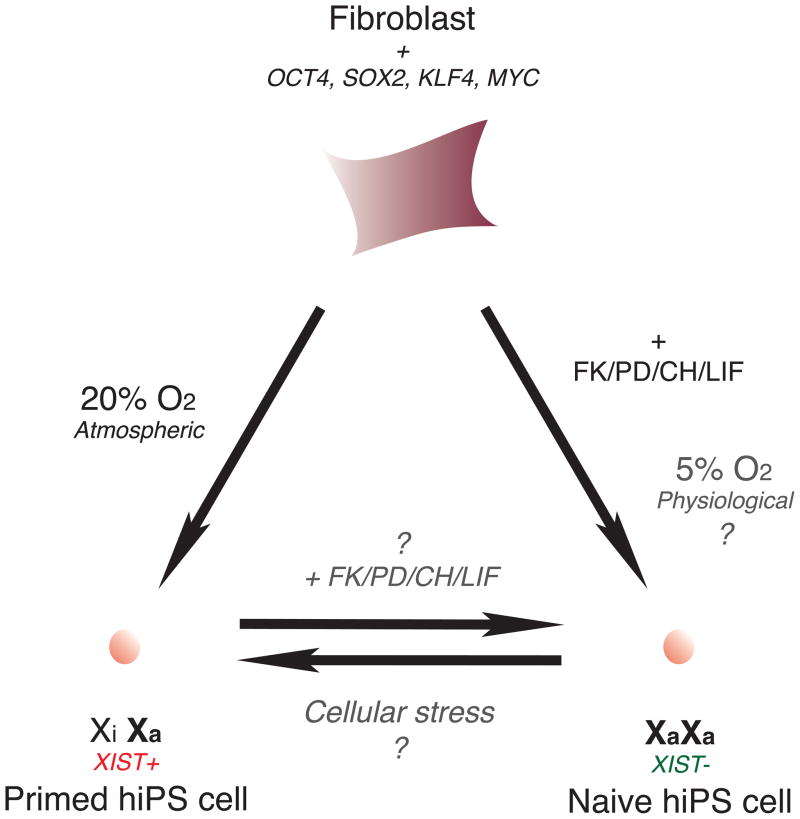
Studies of human iPS (hiPS) cells are currently being pursued with great interest, given their enormous potential in personalized regenerative medicine [79]. The ectopic expression of Oct4, Sox2, Klf4, and c-Myc in somatic cells yields hiPS cells with high degrees of molecular and functional similarities to hES cells [80]. Assessing the X-reactivation state provides valuable insight into the epigenetic status of hiPS cells and indicates whether acquisition of the pluripotent ground state has been achieved. Two recent studies on hiPS cells examined whether induction of pluripotency using defined factors resulted in epigenetic reprogramming of the X-chromosome, leading to different conclusions. One study found that hiPS cells, generated using either lentiviral or retroviral reprogramming vectors, exhibited similar genome-wide expression profiles to hES cells, while forming teratomas in vivo that contained cell types from all three germ layers [73]. Both lentiviral- and retroviral-derived Nanog-positive hiPS cells exhibited an X-chromosome coated with XIST RNA in ~88% of NANOG-positive cells (Fig. 4) – an unanticipated result given prior findings that X-reactivation accompanies reprogramming into iPS cells in the mouse system. The XIST-coated X-chromosomes were also transcriptionally silent, evidenced by the lack of X-linked gene expression. All cells within a selected hiPS cell clone exhibited XCI on the same X-chromosome, indicating that a single fibroblast had clonally-expanded to generate the hiPS cell line without undergoing X-reactivation. In control fibroblasts, the Xi was coated with H3K27me3, but not the Polycomb group proteins that mediate this histone mark. However, in fibroblast-derived hiPS cells, the Xi displayed enrichment for Polycomb group proteins, but only after endogenous NANOG has been activated. Contrary to these findings, a second group recently observed that some hiPS cell lines exhibited complete X-reactivation in the undifferentiated state, with XIST cloud formation subsequently occurring in neurosphere-derived neurons [81].
Fig. 4.
X-chromosome state in human iPS cells. Direct reprogramming of fibroblasts under hyperoxic conditions results in hiPS cells that have not undergone X-reactivation. Deriving hiPS cells with added small molecules results in epigenetic reprogramming of the X-chromosome.
During prolonged passaging of hiPS cells, the Xi loses XIST coating, as seen in Class III hES cells. Additionally, Class III hiPS cells are able to maintain monoallelic expression of X-linked genes despite the absence of XIST, while RNA polymerase II is excluded from the Xi, similar to Class III hES cells [70]. The epigenetic silencing of XIST expression is achieved via DNA methylation of its promoter region, and Polycomb group proteins are also lost from the Xi for Class III hiPS cells. These findings reveal that prolonged periods in culture result in distinct epigenetic changes on the Xi, which may also indicate global epigenetic perturbations in the hiPS cell population as a whole.
Given the conflicting results on human X-chromosome reactivation following direct reprogramming, we can speculate that optimal reprogramming conditions that result in consistent and uniform X-reactivation, as observed for the mouse system, may be achievable in the near future. However, unlike in the mouse system, the human iPS system may exhibit an unexpected uncoupling of X-chromosome reprogramming and acquisition of the pluripotent state. Whether this presents a natural uncoupling or is an epigenetic aberration of hiPS cell derivation conditions is an important question that will have clinical implications relevant to the epigenetic stability and safety of hiPS cells overall.
4. CONCLUSIONS
The review presented here recapitulates how XCI is achieved by noncoding genes (Xist, Tsix, Xite, RepA, and Jpx) in mouse pluripotent stem cells and provides evidence for a tight linkage between these noncoding elements and core pluripotency factors in the control of XCI. Because hES cells can be isolate in a pre-XCI state, the mechanisms of XCI between human and mouse might be more similar than previously thought. Deciphering the molecular mechanisms underlying X-chromosome reprogramming may yield new insights into the acquisition of the pluripotent ground state. While analysis of hES and hiPS cells have somewhat lagged behind that of their mouse counterparts, it is already clear – based on analysis of XIST expression and other XCI markers – that hES and hiPS cells demonstrate a degree of epigenetic fluidity that far exceeds that observed in the mouse system. Defining how and why the changes occur in human cells will be crucial prior to their use as vehicles in human stem cell therapy and regenerative medicine. One might predict that discoveries involving ncRNAs will provide answers to critical questions in stem cell biology.
Acknowledgments
We thank all laboratory members for valuable discussions. D.H.K. is supported by a Damon Runyon Cancer Research Foundation Fellowship (DRG-#2027-09) and the Beckman Fellows Program at Caltech, Y.J. by a Korean Research Foundation grant (C00069) and a Discovery grant from MGH ECOR, M.C.A. by NIH-T32CA009216, and J.T.L. by NIH-GM58839. J.T.L is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–72. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 2.Wutz A, Gribnau J. X inactivation Xplained. Curr Opin Genet Dev. 2007;17(5):387–93. doi: 10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Starmer J, Magnuson T. A new model for random X chromosome inactivation. Development. 2009;136(1):1–10. doi: 10.1242/dev.025908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–42. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borsani G, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351(6324):325–9. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 6.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–26. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 7.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 8.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–42. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 9.Penny GD, et al. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 10.Marahrens Y, et al. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11(2):156–66. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 11.Clemson CM, et al. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132(3):259–75. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 14.Silva J, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4(4):481–95. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 15.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2(7):E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoki Y, et al. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development. 2009;136(1):139–46. doi: 10.1242/dev.026427. [DOI] [PubMed] [Google Scholar]
- 18.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21(4):400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 19.Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99(1):47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 20.Sado T, et al. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128(8):1275–86. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 21.Sharman GB. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature. 1971;230(5291):231–2. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- 22.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256(5519):640–2. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 23.Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426(6968):857–62. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 24.Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303(5658):666–9. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 25.Namekawa SH, et al. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol. 2010;30(13):3187–205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto I, et al. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303(5658):644–9. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 27.Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103(1):17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 28.Navarro P, Avner P. An embryonic story: analysis of the gene regulative network controlling Xist expression in mouse embryonic stem cells. Bioessays. 2010;32(7):581–8. doi: 10.1002/bies.201000019. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465(7299):704–12. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seandel M, et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449(7160):346–50. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vigneau S, et al. An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proc Natl Acad Sci U S A. 2006;103(19):7390–5. doi: 10.1073/pnas.0602381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luikenhuis S, Wutz A, Jaenisch R. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol. 2001;21(24):8512–20. doi: 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci U S A. 2001;98(18):10232–7. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro P, et al. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19(12):1474–84. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9(1):159–65. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21(5):617–28. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 38.Ohhata T, et al. Tsix-deficient X chromosome does not undergo inactivation in the embryonic lineage in males: implications for Tsix-independent silencing of Xist. Cytogenet Genome Res. 2006;113(1–4):345–9. doi: 10.1159/000090851. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11(3):731–43. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 40.Nesterova TB, et al. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin. 2008;1(1):2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanellopoulou C, et al. X chromosome inactivation in the absence of Dicer. Proc Natl Acad Sci U S A. 2009;106(4):1122–7. doi: 10.1073/pnas.0812210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stavropoulos N, Rowntree RK, Lee JT. Identification of developmentally specific enhancers for Tsix in the regulation of X chromosome inactivation. Mol Cell Biol. 2005;25(7):2757–69. doi: 10.1128/MCB.25.7.2757-2769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chow JC, Brown CJ. Forming facultative heterochromatin: silencing of an X chromosome in mammalian females. Cell Mol Life Sci. 2003;60(12):2586–603. doi: 10.1007/s00018-003-3121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chureau C, et al. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12(6):894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston CM, et al. Enox, a novel gene that maps 10 kb upstream of Xist and partially escapes X inactivation. Genomics. 2002;80(2):236–44. doi: 10.1006/geno.2002.6819. [DOI] [PubMed] [Google Scholar]
- 46.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30(2):167–74. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 47.Donohoe ME, et al. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460(7251):128–32. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donohoe ME, et al. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25(1):43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 49.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 50.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8(3):293–9. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 51.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311(5764):1149–52. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 52.Xu N, et al. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39(11):1390–6. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 53.Nicodemi M, Prisco A. Symmetry-breaking model for X-chromosome inactivation. Phys Rev Lett. 2007;98(10):108104. doi: 10.1103/PhysRevLett.98.108104. [DOI] [PubMed] [Google Scholar]
- 54.Navarro P, et al. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321(5896):1693–5. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 55.Ahn JY, Lee JT. Retinoic acid accelerates downregulation of the Xist repressor, Oct4, and increases the likelihood of Xist activation when Tsix is deficient. BMC Dev Biol. 10:90. doi: 10.1186/1471-213X-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1(1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 57.Stadtfeld M, et al. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2(3):230–40. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 59.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 60.Migeon BR, et al. Low-copy-number human transgene is recognized as an X inactivation center in mouse ES cells, but fails to induce cis-inactivation in chimeric mice. Genomics. 2001;71(2):156–62. doi: 10.1006/geno.2000.6421. [DOI] [PubMed] [Google Scholar]
- 61.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10(3):353–60. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 62.Guo G, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136(7):1063–9. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24(20):2239–63. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460(7251):49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 65.Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS One. 5(6):e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall LL, et al. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol. 2008;216(2):445–52. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoffman LM, et al. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23(10):1468–78. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 68.Lengner CJ, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 141(5):872–83. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 69.Shen Y, et al. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A. 2008;105(12):4709–14. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva SS, et al. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105(12):4820–5. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS One. 2010;5(6):e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–92. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 73.Tchieu J, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 7(3):329–42. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanna J, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 107(20):9222–7. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Luijn JC, Gribnau FW, Leufkens HG. Availability of comparative trials for the assessment of new medicines in the European Union at the moment of market authorization. Br J Clin Pharmacol. 2007;63(2):159–62. doi: 10.1111/j.1365-2125.2006.02812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ware CB, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4(4):359–69. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Migeon BR, et al. Identification of TSIX, encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: implications for X inactivation. Am J Hum Genet. 2001;69(5):951–60. doi: 10.1086/324022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chow JC, et al. Characterization of expression at the human XIST locus in somatic, embryonal carcinoma, and transgenic cell lines. Genomics. 2003;82(3):309–22. doi: 10.1016/s0888-7543(03)00170-8. [DOI] [PubMed] [Google Scholar]
- 79.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137(1):13–7. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 80.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–23. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 143(4):527–39. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]