Abstract
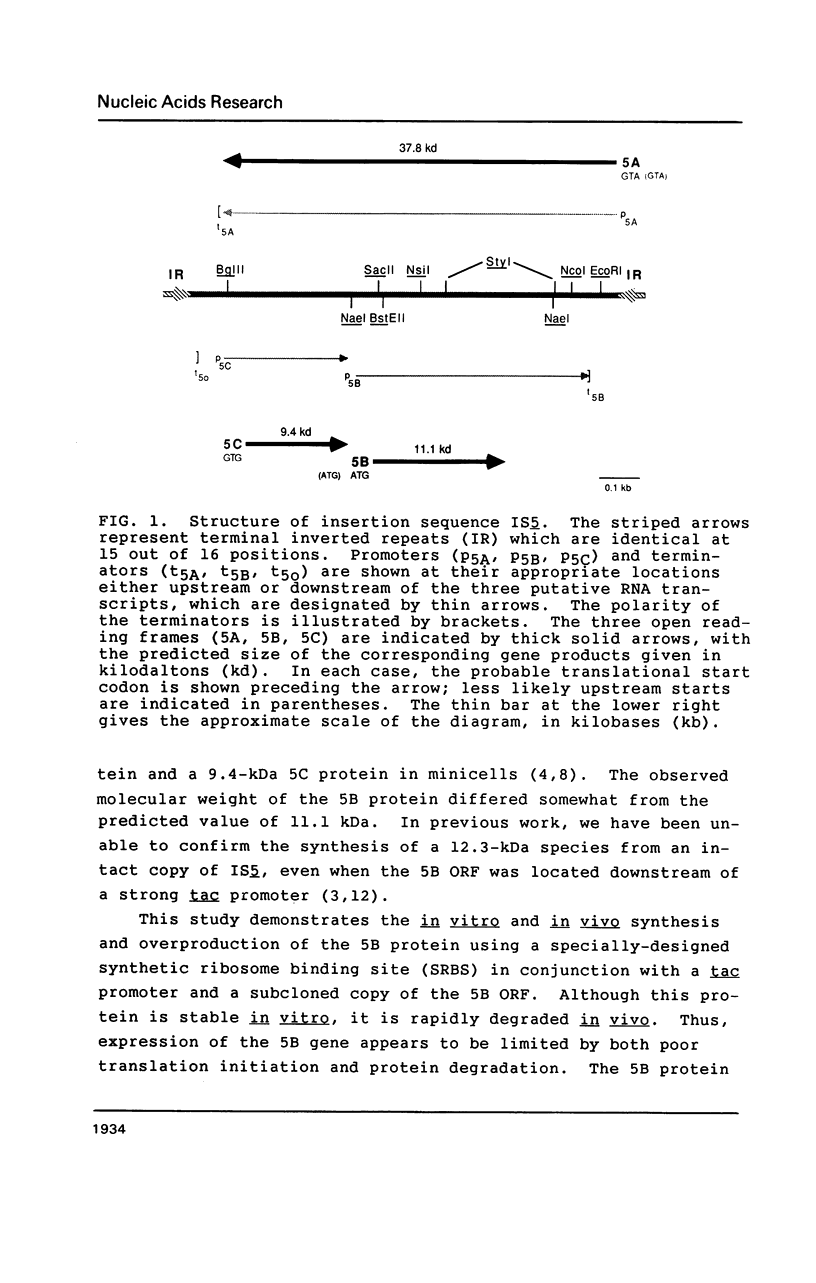
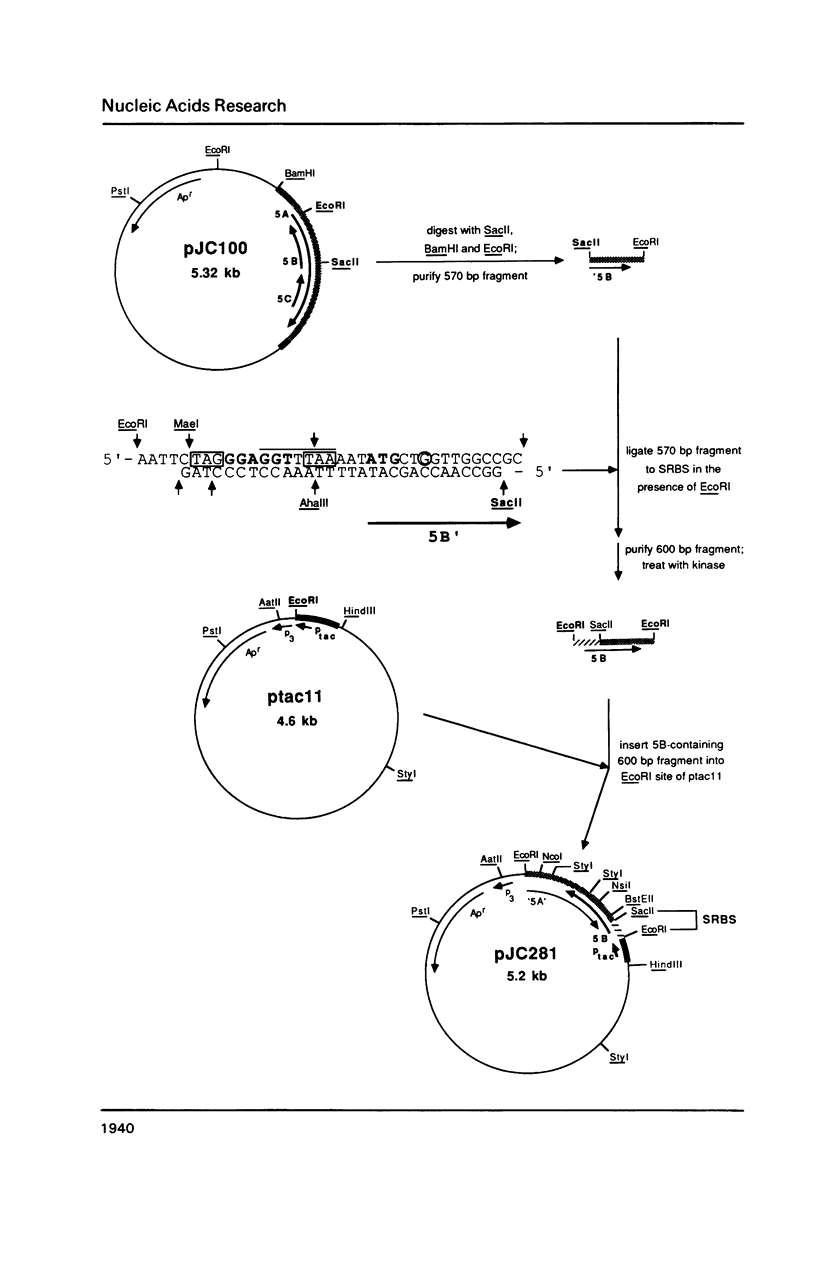
Insertion sequence IS5 is a bacterial transposable element which contains three open reading frames designated 5A, 5B and 5C. Although there was no detectable expression from the 5B open reading frame when it was preceded by the native promoter and ribosome binding site or by a tac promoter and the native ribosome binding site, we have overproduced a 5B protein both in vitro and in Escherichia coli cells by using a tac promoter and a specially-designed synthetic ribosome binding site. beta-galactosidase fusion studies suggested that the synthetic binding site is at least 150-fold more efficient than the native binding site. The 5B protein amounted to 80-85% of the total protein made in vitro and 20-25% of the total protein pulse-labelled in whole cells. It is stable in vitro but rapidly degraded in vivo. Thus expression of the 5B gene appears to be limited by both poor translation initiation and protein degradation.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland N. K., Lindquist B. H. Coliphage P2 late control gene ogr. DNA sequence and product identification. J Mol Biol. 1986 Apr 5;188(3):487–490. doi: 10.1016/0022-2836(86)90170-1. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Buell G., Schulz M. F., Selzer G., Chollet A., Movva N. R., Semon D., Escanez S., Kawashima E. Optimizing the expression in E. coli of a synthetic gene encoding somatomedin-C (IGF-I). Nucleic Acids Res. 1985 Mar 25;13(6):1923–1938. doi: 10.1093/nar/13.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernak J. M., Schlaffer E. J., Smith H. O. Synthesis and overproduction of the 5A protein of insertion sequence IS5. J Bacteriol. 1988 Nov;170(11):5368–5370. doi: 10.1128/jb.170.11.5368-5370.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. Cell-free synthesis of proteins coding for mobilisation functions of ColE1 and transposition functions of Tn3. Gene. 1979 May;6(1):29–42. doi: 10.1016/0378-1119(79)90083-0. [DOI] [PubMed] [Google Scholar]
- DeVries J. K., Zubay G. DNA-directed peptide synthesis. II. The synthesis of the alpha-fragment of the enzyme beta-galactosidase. Proc Natl Acad Sci U S A. 1967 Apr;57(4):1010–1012. doi: 10.1073/pnas.57.4.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler J. A., van Bree M. P. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene. 1981 Aug;14(3):155–163. doi: 10.1016/0378-1119(81)90111-6. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Simons R. W., Way J. C., Walsh R. B., Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A., Hayflick J., Dinkelspiel K., de Boer H. A. Mutagenesis of the three bases preceding the start codon of the beta-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J. 1984 Mar;3(3):623–629. doi: 10.1002/j.1460-2075.1984.tb01858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Jakowec M., Prentki P., Chandler M., Galas D. J. Mutational analysis of the open reading frames in the transposable element IS1. Genetics. 1988 Sep;120(1):47–55. doi: 10.1093/genetics/120.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay G., Khoury G., Seth A. K., Jay E. Construction of a general vector for efficient expression of mammalian proteins in bacteria: use of a synthetic ribosome binding site. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5543–5548. doi: 10.1073/pnas.78.9.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- Klaer R., Kühn S., Tillmann E., Fritz H. J., Starlinger P. The sequence of IS4. Mol Gen Genet. 1981;181(2):169–175. doi: 10.1007/BF00268423. [DOI] [PubMed] [Google Scholar]
- Kröger M., Hobom G. Structural analysis of insertion sequence IS5. Nature. 1982 May 13;297(5862):159–162. doi: 10.1038/297159a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65(1):363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- Morisato D., Way J. C., Kim H. J., Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983 Mar;32(3):799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- Pato M. L., Reich C. Instability of transposase activity: evidence from bacteriophage mu DNA replication. Cell. 1982 May;29(1):219–225. doi: 10.1016/0092-8674(82)90106-4. [DOI] [PubMed] [Google Scholar]
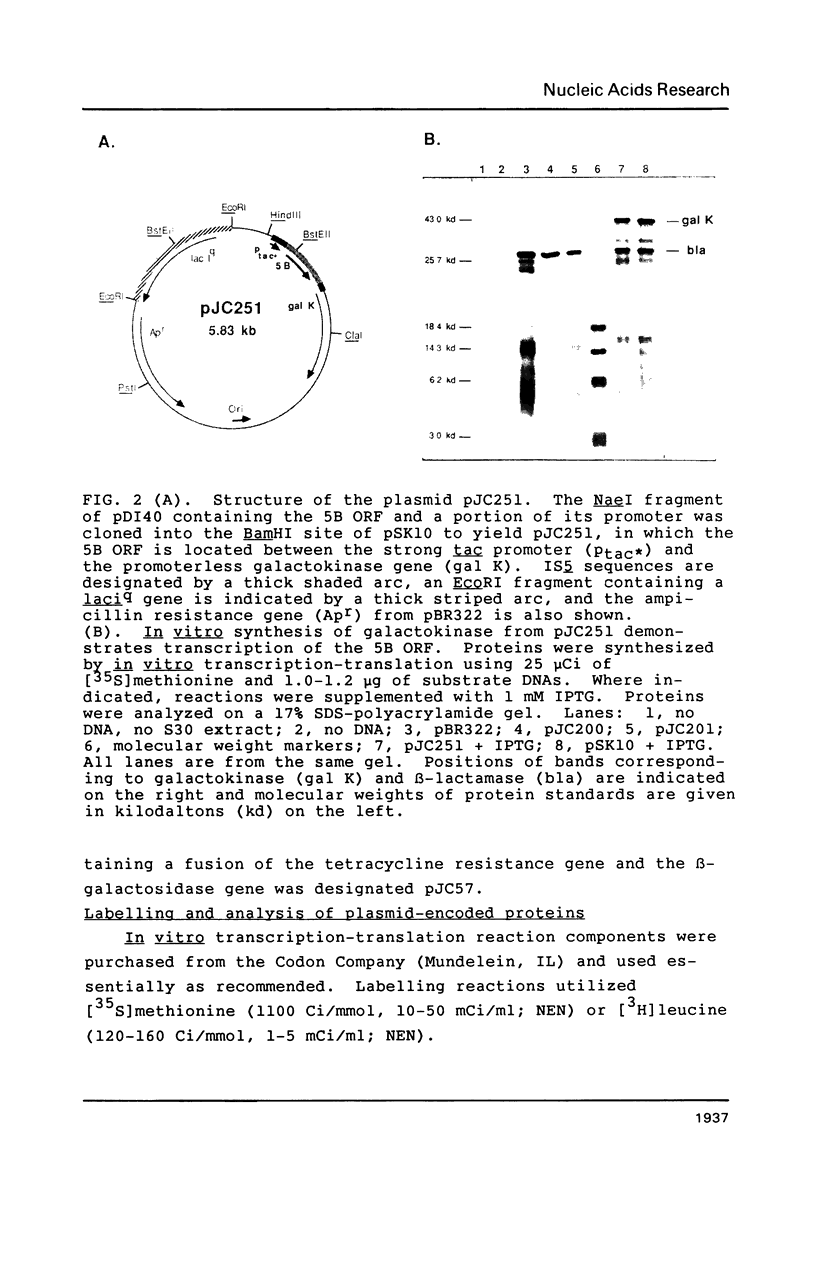
- Rak B., Lusky M., Hable M. Expression of two proteins from overlapping and oppositely oriented genes on transposable DNA insertion element IS5. Nature. 1982 May 13;297(5862):124–128. doi: 10.1038/297124a0. [DOI] [PubMed] [Google Scholar]
- Rak B., von Reutern M. Insertion element IS5 contains a third gene. EMBO J. 1984 Apr;3(4):807–811. doi: 10.1002/j.1460-2075.1984.tb01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Kleckner N. Quantitation of insertion sequence IS10 transposase gene expression by a method generally applicable to any rarely expressed gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1787–1791. doi: 10.1073/pnas.83.6.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. R., Bennett G. N. Construction and analysis of in vivo activity of E. coli promoter hybrids and promoter mutants that alter the -35 to -10 spacing. Gene. 1982 Dec;20(2):231–243. doi: 10.1016/0378-1119(82)90042-7. [DOI] [PubMed] [Google Scholar]
- Sancar A., Wharton R. P., Seltzer S., Kacinski B. M., Clarke N. D., Rupp W. D. Identification of the uvrA gene product. J Mol Biol. 1981 May 5;148(1):45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer G. F., Walkinshaw M. D., Arnott S., Morré D. J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980 Sep 11;8(17):3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner B., Kahn M. The nucleotide sequence of IS5 from Escherichia coli. Gene. 1981 Aug;14(3):165–174. doi: 10.1016/0378-1119(81)90112-8. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K., Gilberg W. Construction of plasmid vectors with unique PstI cloning sites in a signal sequence coding region. Gene. 1980 Dec;12(3-4):235–241. doi: 10.1016/0378-1119(80)90105-5. [DOI] [PubMed] [Google Scholar]
- Trinks K., Habermann P., Beyreuther K., Starlinger P., Ehring R. An IS4-encoded protein is synthesized in minicells. Mol Gen Genet. 1981;182(2):183–188. doi: 10.1007/BF00269656. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Jakowec M., Prentki P., Galas D. J., Chandler M. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 1987 Oct;6(10):3163–3169. doi: 10.1002/j.1460-2075.1987.tb02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer H. A., Hui A., Comstock L. J., Wong E., Vasser M. Portable Shine-Dalgarno regions: a system for a systematic study of defined alterations of nucleotide sequences within E. coli ribosome binding sites. DNA. 1983;2(3):231–235. doi: 10.1089/dna.1983.2.231. [DOI] [PubMed] [Google Scholar]