Abstract
Developed in the early 1990's, PNA has emerged as a promising class of nucleic acid mimic because of its strong binding affinity and sequence selectivity towards DNA and RNA, and resistance to enzymatic degradation by proteases and nucleases; however, the main drawbacks, as compared to other classes of oligonucleotides, are water solubility and biocompatibility. Herein we show that installation of a relatively small, hydrophilic (R)-diethylene glycol (`miniPEG') unit at the γ-backbone transforms a randomly-folded PNA into a right-handed helix. Synthesis of optically pure R-MPγPNA monomers is described, which can be accomplished in a few simple steps from a commercially available and relatively cheap Boc-L-serine. Once synthesized, R-MPγPNA oligomers are preorganized into a right-handed helix and hybridize to DNA and RNA with greater affinity and sequence selectivity, and are more water soluble and less aggregating than the parental PNA oligomers. The results presented herein have important implications for the future design and application of PNA in biology, biotechnology and medicine, as well as in other disciplines including drug discovery and molecular engineering.
Introduction
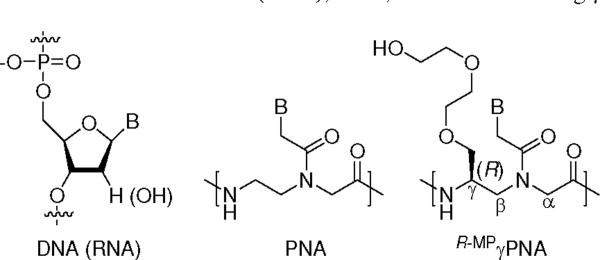
Peptide nucleic acid (PNA) is a promising class of nucleic acid mimic developed in the last two decades in which the naturally occurring sugar phosphodiester backbone is replaced with N-(2-aminoethyl)glycine units (Chart 1).1 PNA can hybridize to complementary DNA or RNA just as the natural counterpart, in accordance with the Watson-Crick base-pairing rules, but with higher affinity and sequence selectivity.2 Further, PNA can invade selected sequences of double-stranded DNA (dsDNA).3–5 The improvement in thermodynamic stability has been attributed, in part, to the lack of electrostatic repulsion in the backbone.1 The other contribution may come from counterion release upon hybridization, as opposed to condensation taking place with DNA and RNA, resulting in an increase in the overall entropy of the system.6 The enhancement in sequence selectivity, however, is less well understood.7 Structural studies suggested that hydration may play a key role in rigidifying the backbone of PNA upon hybridization to DNA or RNA (or PNA), making it less accommodating to structural mismatches. Structural or spinal water molecules have been observed in the X-ray structures of PNA-DNA8 and PNA-PNA9–11 duplexes bridging the amide proton in the backbone to the adjacent nucleobase. Besides hybridization properties, another appealing aspect of PNA is enzymatic stability. Because of its unnatural backbone, PNA is not easily degraded by proteases or nucleases.12 These properties, along with the ease13,14 and flexibility15 of synthesis, together make PNA an attractive reagent for many applications in biology, biotechnology and medicine.16,17
Chart 1.
Chemical structure of DNA (RNA), PNA, and MP-containing γPNA units.
PNA has so far been employed in a number of applications, from molecular tools for probing and manipulating nucleic acid structures and functions18–21 and regulation of gene expression,22,23 to recognition code for tagging molecules in drug discovery,24,25 amplifying genetic information in molecular evolution,26,27 and for organizing molecular self-assembly in materials science and nanotechnology.28–32 In spite of its many appealing features, however, PNA has one major setback as compared to other classes of oligonucleotides. Because of the charge-neutral backbone, PNA is only moderately soluble in aqueous solution. Further, it has a tendency to aggregate and adhere to surfaces and other macromolecules in a nonspecific manner.33,34 This inherent property posts a considerable technical challenge for the handling and processing of PNA—for instance, in the development of microarrays and related surface-bound applications, because of its propensity to aggregate and collapse onto the surface causing poor and nonspecific binding,35,36 and in biology and medicine, in part because of the concerns for off-target binding and cytotoxicity.34
In attempts to address this concern several approaches have been taken, including incorporation of charged amino acid residues, such as lysine at the termini37 or in the interior part of the oligomer;38,39 inclusion of polar groups in the backbone,40 carboxymethylene bridge41 and in the nucleobases;25,42 replacement of the original aminoethylglycyl backbone skeleton with a negatively-charged scaffold;43,44 conjugation of high molecular weight polyethylene glycol (PEG) to one of the termini;36,45 fusion of PNA to DNA to generate a chimeric oligomer,46–50 and redesign of the backbone architecture.51 These chemical modifications have led to improvement in solubility but it is often achieved at the expense of binding affinity and/or sequence specificity, not to mention the requirement of an elaborate synthetic scheme in some cases. Incorporation of cationic residues can improve water solubility, but it can also lead to nonspecific binding due to an increase in charge-charge interaction upon hybridization to DNA or RNA. Conjugation of PNA to DNA (or RNA) can improve water solubility as well, but it can compromise binding affinity due to an increase in charge-charge repulsion and conformational heterogeneity in the backbone.46,47,49 Herein we report the development of a new class of conformationally-preorganized, diethylene glycol-containing γPNAs that exhibit superior hybridization properties and water solubility as compared to the original PNA design or any other chiral γPNA that has been developed to date.
Results and Discussion
Design rationale
Diethylene glycol, commonly referred to as `miniPEG' or MP, was chosen as a chemical moiety for incorporation in the backbone of PNA because of its relatively small size and hydrophilic nature, and the fact it has been shown to be non-toxic and nonimmunogenic.52–54 MP and larger molecular-weight polyethylene glycol (PEG) have been incorporated into a number of macromolecular systems including peptides and proteins,55 nucleic acids,45,56–60 carbohydrates,61 synthetic polymers,62 dendrimers,63 liposomes,64,65 nanoparticles66,67 and self-assembled materials68—just to name a few, in attempts to improve their aqueous solubility, enzymatic stability, bioavailability, and pharmacokinetics as well as to minimize their immunogenicity. Incorporation of MP in the backbone of PNA is expected to confer similar beneficial effects, including improvements in water solubility and biocompatibility, along with reduction in aggregation and nonspecific binding. Inspection of the PNA backbone reveals three obvious sites for incorporation of this chemical moiety—α, β and γ (Chart 1). We selected the γ-position because prior studies from our group69–73 have revealed that installation of a chiral center at this position induces helical organization in the oligomer, providing an additional means for fine-tuning the thermodynamic stability of PNA. γPNAs have been made by other groups before but their emphasis has been mainly on synthesis74–77 and covalent attachment of PNA to other functional groups or molecular entities.78–80
The helical sense, i.e. whether it adopts a right-handed or left-handed helix, is determined by the stereochemistry. γPNAs prepared from L-amino acids adopt a right-handed helix, while those prepared from D-amino acids adopt a left-handed helix; however, only the right-handed helical γPNAs hybridize to DNA and RNA with high affinity and sequence selectivity. Further, we showed that the fully-modified, L-alanine-derived γPNAs (S-AlaγPNAs) can invade mixed-sequence double helical B-form DNA (B-DNA).81 Though they are promising as antisense and antigene reagents, S-AlaγPNAs are poorly soluble in water and have a tendency to aggregate, presumably due to the charge-neutral backbone and hydrophobic character of the methyl group at the γ-backbone position. We surmised that replacement of the methyl group with MP might ameliorate some of these issues, while maintaining the superior hybridization properties of the original γPNA design because of the helical preorganization in the backbone upon installation of MP at the γ-position (Chart 1).
Monomer synthesis
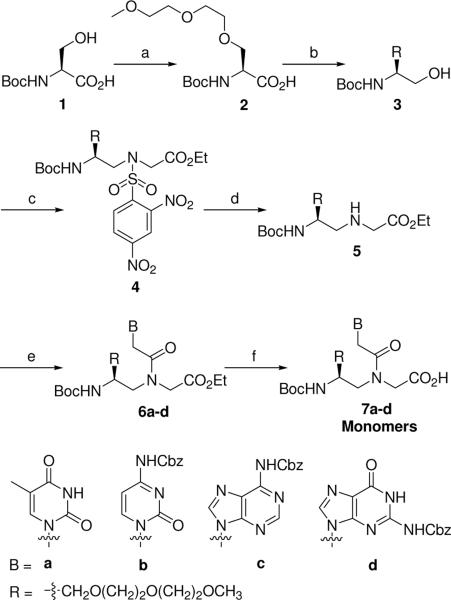
Boc-protected R-MPγPNA monomers containing all four natural nucleobases (A, C, G, T) were synthesized according to the procedures outlined in Scheme 1. Alkylation of Boc-protected L-serine (1) with 1-bromo-2-(2-methoxyethoxy)ethane under an optimized reaction condition: slow addition of 1 into a vigorously stirred, chilled solution of DMF containing 2 equivalents of sodium hydride, followed by addition of 1-bromo-2-(2-methoxyethoxy)ethane afforded compound 2 with high optical purity (see section below). The stoichiometry and order of addition were determined to be essential for obtaining optically pure product. Slow addition of sodium hydride was necessary to ensure complete deprotonation of the carboxyl group prior to removal of the hydroxyl proton. Formation of the carboxylate anion reduces the acidity of the α-proton making it less susceptible to deprotonation by base. Esterification of the alkylated product 2 followed by reduction with sodium borohydride yielded serinol 3. This chemical transformation, conversion of the carboxylic acid to alcohol, renders the α-proton inert to deprotonation and racemization in the subsequent reaction steps. Coupling 3 to N-(2,4-dinitrophenylsulfonamido)glycine ethyl acetate, which was prepared according to the published procedure,82 employing the Mitsunobu reaction followed by removal of the 2,4-dinitrobenzenesulfonyl group with a mild base yielded compound 5. DCC-mediated coupling of 5 with the appropriate carboxymethylnucleobases (A,83 C,14 G,83 T13) and hydrolysis of the resulting esters gave the desired monomers 7a–d.
Scheme 1.
Synthesis of R-MPγPNA Monomers Reagents and conditions: (a) NaH, BrCH2CH2OCH2CH2OCH3, DMF, 0 °C, 68%; (b) isobutyl chloroformate, NaBH4, NMM, DME, 0 °C→rt, 84%; (c) DIAD, 2,4-dinitrobenzenesulfonyl glycine ethyl ester, TPP, THF, 0 °C→rt, 56%; (d) n-propylamine, CH2Cl2, rt, 81%; (e) carboxymethylene nucleobase, DCC, DhObtOH, DMF, 50 °C, 67–85%; (f) 2M NaOH/THF (1:1), 0 °C, 85–98%.
Determination of optical purities
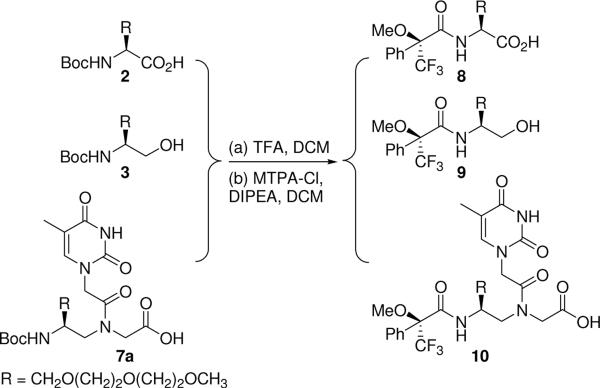
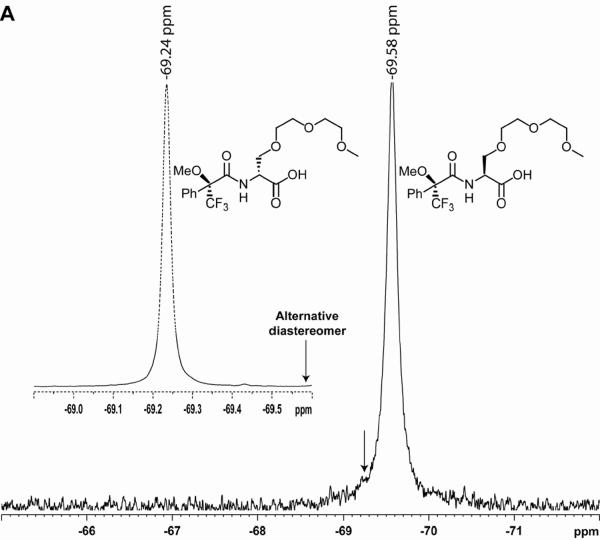
The optical purities of key intermediates and final monomers were determined by 19F-NMR following chemical derivatization (Scheme 2). Corradini and coworkers84 have previously shown that the enantiomeric excess (ee) of chiral α-PNA monomers and oligomers, with the latter obtained following acid hydrolysis, can be determined by GC/MS following chemical derivatization. We have discovered that 19F-NMR is a convenient and accurate alternative method for determining the ee values for γPNA monomers and prior intermediates following removal of the Boc-protecting group and subsequent coupling with Mosher's reagent, (+)-1-methoxy-1-(trifluoromethyl)phenylacetyl chloride (MTPA-Cl). Figure 1 shows representative spectra of MTPA-derivatized, MP-L-serine 2 (compound 8), MP-L-serinol 3 (compound 9), and thymine R-MPγPNA monomer 7a (compound 10). The corresponding diastereomers were prepared the same way starting from Boc-D-serine and are included for comparison. A close inspection of these spectra reveals no trace quantity of the other diastereomers, for the alkylated serine 8, serinol 9 or final monomer 7a, indicating that they were optically pure. If racemization were to occur, we would expect an additional peak in the spectrum, as indicated by the arrow, corresponding to the chemical shift of the other diastereomer. The ee values for serine 8, serinol 9 and final thymine monomer 7a were estimated to be at least 99%, within the detection limit of 19F-NMR.85 At this point it is not clear why the thymine R-MPγPNA monomer showed two rotamers (Figure 1C), at −68.80 and −68.95 ppm, while thymine S-MPγPNA showed only one (Figure 1C, Inset). (The existence of the two rotamers was determined by multinuclear and multidimensional NMR analyses as described in an earlier work;73 they differ from one another in the rotation around the C7-N4 tertiary amide bond.) We attribute the high optical purities of the intermediates and final monomers (the others not shown) to the optimized, initial alkylation step and the stereoselective conversion of carboxylic acid to alcohol in the second step, based on the procedure reported by Rodriguez and coworkers.86
Scheme 2.
Chemical derivatization for 19F-NMR analysis.
Figure 1.
19F-NMR spectra of MTPA-derived (A) MP-L-serine 2 (compound 8), (B) MP-L-serinol 3 (compound 9), and (C) thymine monomer 7a (compound 10). The corresponding diastereomers were prepared under identical conditions starting from Boc-D-serine and are included for comparison.
Oligomer design and synthesis
Three sets of PNA oligomers were prepared (Table 1). The first set, comprising PNA1 through 5, was designed to test the effects of MP on the conformation and hybridization properties of PNA. The second set, consisting PNA6 through 10, was designed to test the effect of MP on water solubility. The hexadecameric sequence was chosen for this study because it represents a statistical length that would be required to target a unique site within the mammalian genome or transcriptome. The third set, constituting PNA1 and 4, each separately linked to fluorescein (FITC) at the N-terminus (PNA1X and PNA4X) and tetramethylrhodamine (TAMRA) at the C-terminus (PNA1Y and PNA4), were designed to test the effect of MP on self-aggregation based on Förster Resonance Energy Transfer (FRET). Inclusion of a lysine residue at the C-terminus was necessary, since prior attempts to synthesize FITC- and TAMRA-containing PNA oligomers without the lysine residue resulted in sticky and poorly water-soluble materials that were difficult to purify and characterize.
Table 1.
Sequence of PNA oligomers
| Oligomer | Sequence | #MP units |
|---|---|---|
| PNA1 | H-GCATGTTTGA-NH2 | 0 |
| PNA2 | H-GCATGTTTGA-NH2 | 1 |
| PNA3 | H-GCATGTTTGA-NH2 | 3 |
| PNA4 | H-GCATGTTTGA-NH2 | 5 |
| PNA5 | H-GCATGTTTGA-NH2 | 10 |
|
| ||
| PNA6 | H-ACGGGTAGAATAACAT-NH2 | 0 |
| PNA7 | H-ACGGGTAGAATAACAT-NH2 | 1 |
| PNA8 | H-ACGGGTAGAATAACAT-NH2 | 3 |
| PNA9 | H-ACGGGTAGAATAACAT-NH2 | 5 |
| PNA10 | H-ACGGGTAGAATAACAT-NH2 | 8 |
|
| ||
| PNA1X | H-LOrn(X)-LLys-GCATGTTTGA-NH2 | 0 |
| PNA1Y | H-LLys-GCATGTTTGA-LOrn(Y)-NH2 | 0 |
| PNA4X | H-LOrn(X)-LLys-GCATGTTTGA-NH2 | 5 |
| PNA4Y | H-LLys-GCATGTTTGA-LOrn(Y)-NH2 | 5 |
Underlined letter indicates R-MP-γ-backbone modification. X = fluorescein (FITC), Y = tetramethylrhodamine (TAMRA).
All PNA oligomers, those with and without MP side-chains, were synthesized on solid-support according to the published procedures of Christensen and coworkers.87 Unlike modifications made at the α-backbone, which require further optimization of the reaction condition in order to minimize racemization during the on-resin coupling,88 no precaution was necessary for coupling of R-MPγPNA monomers on the resin. Upon completion of the last monomer coupling, the oligomers were cleaved from the resin and precipitated with ethyl ether. After being air-dried, the crude pellets were dissolved in water/acetonitrile mixture (80/20), and purified by reverse-phase HPLC and characterized by MALDI-TOF mass spectrometry.
Conformational analysis
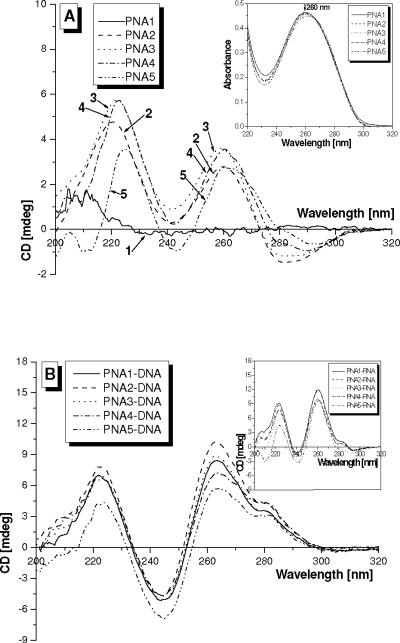
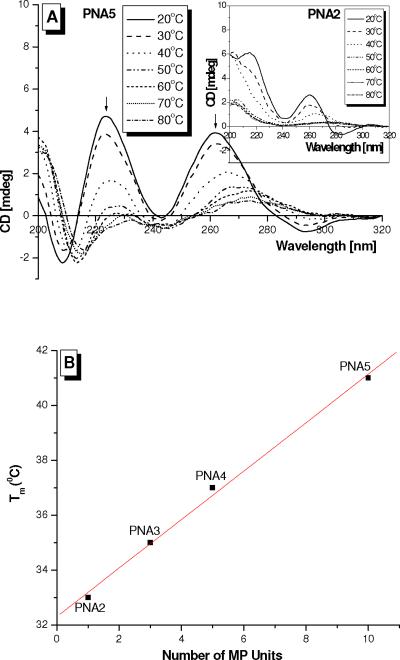
To determine the effect of MP on the conformation of PNA, we measured the CD spectra of PNA1 through 5. Consistent with the earlier finding,69 we did not observe noticeable CD signals in the nucleobase absorption regions for PNA1 (Figure 2A). This indicates one of two possibilities: either that the PNA does not adopt a helical conformation, or that it does but with an equal proportion of a right-handed and left-handed helix. The latter was suggested by MD simulations,89 but ruled out on the basis of NMR analyses.69 However, in the case of PNA2 through 5, we noticed distinct exciton coupling patterns, with minima at 242 and 280 nm and maxima at 220 and 260 nm, characteristic of a right-handed helix.90 The amplitude of the CD signals remained fairly constant as more MP units were added, but the profiles slightly shifted towards that of the PNA-DNA2 and PNA-RNA2 double helices (Figure 2B), especially at the 242 and 280 nm minima. A gradual dip at the 242 nm minimum generally indicates a tightening in the helical pitch of the oligomer,91 from one that resembles that of a PNA-PNA92 duplex with 18 base-pairs per turn to one that resembles that of a PNA-DNA93 duplex with 13 base-pairs per turn. Overall, the CD profiles of PNA2 through 5 are similar to those of the corresponding PNA-DNA and PNA-RNA hybrid duplexes (Figure 2B). The major difference however is in the amplitude, which is roughly doubled for the duplex as compared to the individual strand. This is because the concentration of bases in the hybrid duplex is twice that of the individual PNA strand. Taken together, these results show that a single, (R)-MP unit installed at the γ-backbone is sufficient to preorganize PNA into a right-handed helix. Incorporation of additional MP units did not further improve base-stacking, apparent from the similarities in the CD amplitudes, but it did help to tighten the helical pitch of the oligomers making them more rigid and compact. This is apparent from the temperature-dependent CD measurements which showed less dramatic reduction in the signal amplitude with temperature for PNA5 as compared to that of PNA2 (Figure 3A). Even at a temperature as high as 80 °C, a distinct CD profile still remained indicating that base-stacking still occurred at this temperature—whereas it was completely denatured in the case of PNA2. Overall the stability of the oligomers increases linearly with the number of MP units incorporated (Figure 3B). The fact that PNA5 adopts a helical conformation most closely resembling that of PNA-DNA and PNA-RNA duplexes suggests that it would hybridize to DNA and RNA more effectively than the other oligomers in this series.
Figure 2.
CD spectra of (A) the unhybridized (single-stranded) PNA oligomers, and (B) the corresponding PNA-DNA and PNA-RNA (Inset) hybrid duplexes at 5 μM strand concentration each. (A) Inset: UV-absorption spectra of the individual PNA oligomers at 90 °C, showing that they had the same concentration. This indicates that the differences in the CD spectra, both the amplitude and profile, are the intrinsic properties of the oligomer themselves and not the variations in concentrations. The samples were prepared in 10 mM sodium phosphate buffer at pH 7.4, and the CD spectra were recorded at room temperature. The complementary DNA strand used was: 5'-TCAAACATGC-3'.
Figure 3.
(A) CD spectra of PNA5 and PNA2 (Inset) as a function of temperature. Melting transition (Tm) of PNA2 through 5 as determined by CD, monitored at 260 nm. The oligomer concentration was 5 μM, prepared in 10 mM sodium phosphate buffer at pH 7.4.
Thermal stability
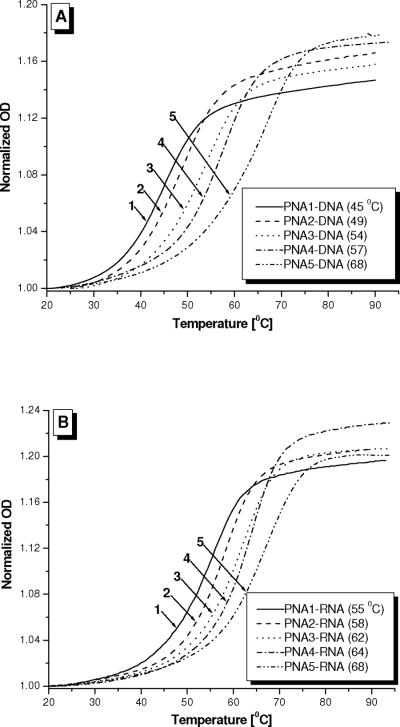
UV-melting experiments were performed to determine the effect of MP on the thermal stability of PNA upon hybridization to DNA and RNA. Our result shows that incorporation of a single MP side-chain stabilized a PNA-DNA duplex by 4 °C (Figure 4A). The extent of stabilization gradually increased with additional MP units added but tapered off to ~ 2.3 °C per unit for the fully-modified PNA5. A similar pattern was observed for RNA, but the extent of stabilization was less compared to that of DNA (Figure 4B). The enhancement was only 3 °C for the first MP unit incorporated and 1.2 °C per unit for PNA5, as compared to 2.3 °C/unit with DNA. It is interesting to note that the unmodified PNA1 binds more tightly to RNA than to DNA, with a differential Tm (ΔTm) of 10 °C. However, fully-modified PNA5 displayed identical thermal stability with RNA as well as with DNA. The lack of a preferential binding for PNA5 is not clearly understood at this point but it may reflect the rigidity of the backbone. Because PNA5 is more rigid and tightly wound as compared to PNA1, it may not have the flexibility or conformational freedom to accommodate the DNA and RNA template strands. Under such circumstance, the DNA and RNA strands would have to undergo conformational change of their own in order to accommodate the R-MPγPNA helix and in order for hybridization to take place. This may explain why S-AlaγPNA-DNA prefers a P-form helix,73 a structure that is intermediate between the A- and B-form DNA, an indication that DNA and RNA accommodate the γPNA exigencies rather than the other way around. The RNA strand is less accommodating to the RMPγPNA, when it is fully-modified, because the RNA backbone is more rigid as compared to that of DNA due to the presence of the chiral OH group at the 2'-position.94
Figure 4.
UV-melting profiles of (A) PNA-DNA and (B) PNA-RNA hybrid duplexes at 5 μM strand concentration each in 10 mM sodium phosphate buffer at pH 7.4. Both the heating and cooling runs were performed; they both had nearly identical profiles (only the heating runs are shown).
Thermodynamic analysis
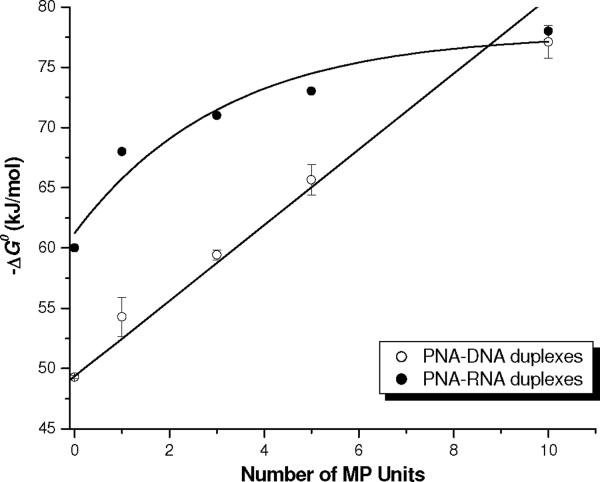
To gain greater insight into the contribution of MP to the stability of the PNA-DNA duplex, we used van't Hoff analysis to determine the thermodynamic parameters for PNA1 through 5 upon hybridization to complementary DNA and RNA strands.95 The data are tabulated in Table 2. Our results show that the Gibbs binding free energy (ΔG°) increases (to a rough approximation) linearly with the number of MP units for PNA-DNA, and follows a sigmoidal growth for PNA-RNA (Figure 5). Incorporation of a single MP unit resulted in a net gain in binding free energy of ~ 5 kJ/mol for PNA-DNA, whereas it is less for PNA-RNA—especially, at the higher end. A reduction in the equilibrium dissociation constant (Kd) by nearly five orders of magnitude for PNA5-DNA and three orders of magnitude for PNA5-RNA was observed, as compared to that of the respective PNA1-DNA and PNA1-RNA duplex. The binding free energy gain in this case appears to be predominantly enthalpically driven for both PNA-DNA and PNA-RNA, apparent from the gradual increase in the ΔH° term with the number of MP units concomitant with modest enthalpy-entropy compensation.
Table 2.
Thermodynamic parameters for PNA-DNA and PNA-RNA duplexes
| PNA-DNA† | PNA-RNA‡ | |||||||
|---|---|---|---|---|---|---|---|---|
| Oligo | −ΔH° (kJ/mol) | −TΔS° (kJ/mol) | −ΔG° (kJ/mol) | Kd | −ΔH° (kJ/mol) | −TΔS° (kJ/mol) | −ΔG° (kJ/mol) | Kd |
| PNA1 | 273±5 | 224±5 | 49±1* | 2.5×10−9 | 289 | 229 | 60 | 3.5×10−11 |
| PNA2 | 319±18 | 263±16 | 54±1 | 3.2×10−10 | 333 | 232 | 68 | 1.2×10−12 |
| PNA3 | 316±11 | 256±11 | 59±1* | 5.1×10−11 | 350 | 280 | 71 | 4.3×10−13 |
| PNA4 | 329±14 | 265±12 | 65±1 | 3.5×10−12 | 356 | 283 | 73 | 1.7×10−13 |
| PNA5 | 372±11 | 294±10 | 78±2 | 4.6×10−14 | 365 | 287 | 78 | 2.1×10−14 |
The averages of three trials (2 from concentration-dependence measurements + 1 from UV-melting curve fitting).
UV-melting curve fitting.
Standard deviation is less than 1 kJ/mol. Temperature = 298K.
Figure 5.
Correlation between Gibbs binding free energy (ΔG°) and number of MP units.
It might seem surprising that the enhanced affinity is not driven by reduced entropy, given the helical preorganization exhibited by the modified PNAs. However, it is important to recognize that unmodified PNA is not believed to be structurally flexible when free in solution. Rather, single-stranded PNA has been proposed to adopt a compact globular fold,34,96 presumably to minimize exposure of the hydrophobic core of nucleobases and the charge-neutral backbone to the aqueous solvent. Thus, an enthalpic penalty would be incurred for unfolding the collapsed PNA in order to adopt the helical structure needed to allow hybridization to a complementary DNA or RNA. Removal of this penalty by inducing a helical structure in the γ-modified PNA would then translate into a more favorable enthalpy change upon hybridization, as observed in Table 2. An additional enthalpic benefit of the modified backbone may be contributed by the formation of a network of structured water molecules that bridges the backbone amide protons to the adjacent nucleobases, stabilizing interactions that have been found to be more predominant in γPNA-DNA73 than in PNA-DNA8 or PNA-PNA.9–11 Additional studies will be required in order to further parse the thermodynamic contributions of the MP backbone modification to stabilization of PNA-DNA and PNA-RNA duplexes.
Kinetics of hybridization and dissociation
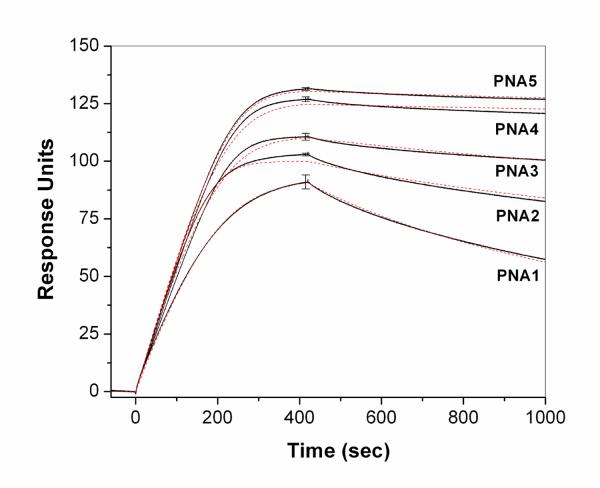
From a kinetic perspective, the higher apparent affinity of the R-MPγPNA oligomers indicated by the UV-melting curves, from which the equilibrium dissociation constants were extracted, could be due to faster on-rates and/or slower off-rates. We used surface plasmon resonance (SPR) analysis to study the hybridization kinetics. In most other examples of PNA-DNA hybridization studied by SPR, the PNA probe was immobilized to the chip while the DNA target was captured from solution.97–99 One exception was previous work from our group, in which a DNA guanine quadruplex target was immobilized and a homologous PNA probe hybridized to form a PNA-DNA heteroquadruplex.100,101 In the current studies, a biotinylated version of the DNA target was immobilized on a streptavidin-conjugated, carboxymethylated dextran chip. A relatively low surface density (ca. 100 response units) of DNA target was used in order to limit mass transport effects on the association kinetics. Solutions containing 10–50 nM PNA were allowed to flow over the chip for 420 seconds, at which point the flow was switched to a PNA-free buffer to allow net dissociation of the hybridized PNA.
Individual sensorgrams for the unmodified (PNA1) and R-MPγ-modified (PNA2 through 5) oligomers at 30 nM concentration are shown in Figure 6. Relatively minor variation is observed in the association kinetics, although singly modified PNA2 appears to bind approximately twice as fast as the unmodified PNA. Fitting the data to a 1:1 binding model yields association rate constants (ka) that range from 4.7 × 105 M−1s−1 to 9.7 × 105 M−1s−1 (Table 3). In contrast, significantly larger effects are seen in the dissociation phase of the experiment, with the dissociation rate constant (kd) varying by at least a factor of 50. Equilibrium dissociation constants (Kd) calculated from the ratio of the dissociation and association rate constants are also given in Table 3. Unmodified PNA1 and fully-modified PNA5 have Kd = 2.8 nM and 54 pM, respectively. Note that the Kd values for PNA1–3 determined by SPR (Table 3) are similar to those determined by UV melting experiments (Table 2). However, increasing divergences are observed for PNA4 and PNA5, with the SPR-derived values being 12- and 1200-fold greater, respectively. We attribute this discrepancy to the very small degrees of dissociation observed within the timescale of the SPR experiment. This introduces a large uncertainty into the fitting of the dissociation phases of the sensorgrams, translating into questionable Kd values. However, we also note that the free energy changes given in Table 2 are based on the assumption that the heat capacity change for PNA-DNA melting is zero, allowing the parameters determined at the high melting temperature to be extrapolated to lower temperatures. Future calorimetric measurements will allow us to determine whether there is a heat capacity change in these hybridization/melting transitions. Nevertheless, the SPR results clearly demonstrate that the enhanced affinity of the R-MPγPNAs is due almost entirely to the significantly slower dissociation kinetics. Thus, the helical preorganization of the modified PNA does not translate into significantly faster hybridization. This could be due to the fact that the complementary DNA strand is likely to require some structural reorganization of its own in order to hybridize to the PNA, negating the pre-organization of the latter. While the origin of the slower dissociation kinetics is not clear at this time, one possible contributor is the network of structured water molecules in the minor groove of the hybrid duplex, which would need to be disrupted in order to separate the two strands.
Figure 6.
SPR sensorgrams (solid black lines) and fits (red dotted lines) for hybridization of PNA probes to immobilized complementary DNA. Solutions contained 30 nM PNA. (Sensorgrams and kinetic fits for all PNA concentrations from 10–50 nM are provided in supporting information.) Error bars at t = 420 sec illustrate standard deviations for three separate trials.
Table 3.
The association rate constant (ka), dissociation rate constant (kd), and equilibrium dissociation constant (Kd) for hybridization of PNA probes with a complementary DNA target.
| Oligomer | ka (M−1s−1) | kd (s−1) | Kd (M) |
|---|---|---|---|
| PNA1 | 4.7×105 | 13.0×10−4 | 2.8×10−9 |
| PNA2 | 9.7×105 | 4.1×10−4 | 4.2×10−10 |
| PNA3 | 6.2×105 | 1.9×10−4 | 3.0×10−10 |
| PNA4 | 6.6×105 | 0.3×10−4† | 4.1×10−11† |
| PNA5 | 8.0×105 | 0.4×10−4† | 5.4×10−11† |
Indicates uncertainty due to the calculated value approaching the limits of detection of the instrument.
Sequence selectivity
Based on the CD, NMR,69 and X-ray73 data which showed that γPNAs derived from L-amino acids adopt a right-handed helix, and that the helix becomes more rigid as more γ-chiral units are added in the backbone, one would expect a fully-modified PNA5 to hybridize to DNA and RNA targets with greater sequence selectivity than PNA1. To verify this prediction, we determined the thermal stabilities of PNA5-DNA and PNA5-RNA duplexes containing perfectly-matched (PM) and single-base mismatched (MM) targets and compared to those from an earlier study with PNA1-DNA and PNA1-RNA.69 Our results showed that despite the strong binding affinity, PNA5 is able to discriminate between closely related sequences. The ΔTm ranges from −17 to −21 °C for PNA5-DNA and −16 to −20 °C for PNA5-RNA containing a single-base mismatch (X=C, G, T), as compared to −10 to −14 °C for PNA1-DNA and −11 to −18 for PNA1-RNA (Table 4). The level of sequence discrimination is greater for PNA5-DNA than for PNA1-DNA, and similar, if not slightly better, for PNA5-RNA as compared to PNA1-RNA. This result is consistent with PNA5 adopting a more rigid helical motif, which is less accommodating to structural mismatches as compared to PNA1.
Table 4.
Sequence mismatch discrimination
| PNA1: H-GCATGTTTGA-LLys-NH2 | ||||
|---|---|---|---|---|
| PNA5: H-GCATGTTTGA-LLys-NH2 | ||||
| DNA: 3'-CGTACAXACT-5, X = A, C, G, T | ||||
| RNA: 3'-CGUACAXACU-5, X = A, C, G, U | ||||
| Tm (°C) | Tm (°C) | |||
| X-T | PNA1-DNA* | PNA5-DNA | PNA1-RNA* | PNA5-RNA |
| A-T | 45 | 68 | 55 | 68 |
| C<>T | 31 (−14)† | 47 (−21) | 37 (−18) | 48 (−20) |
| G<>T | 31 (−14) | 48 (−20) | 44 (−11) | 52 (−16) |
| T(U)<>T | 35 (−10) | 51 (−17) | 40 (−15) | 48 (−20) |
The data for PNA1-DNA and PNA1-RNA mismatched binding was taken from reference.69
The value in the parenthesis indicates ΔTm between the perfect match and mismatch.
Water solubility
To determine whether inclusion of MP in the backbone has an effect on water solubility, we determined the saturating concentrations of PNA6 through 10 (Table 1) in aqueous solution using UV-spectroscopy. It is interesting to note that incorporation of a single MP unit enhanced the solubility of PNA6 by nearly 2-fold (Table 5). The solubility of the oligomers is further improved, albeit to a smaller extent, with additional MP units. For PNA8, which contained 8 MP units, we could not determine the saturating concentration of this oligomer because addition of a minimum amount of water (1 μL) into a vial containing 2.7 mg of fluffy material resulted in complete dissolution. On the basis of this result, we estimated the saturating concentration to be at least 500 mM—more than an order of magnitude higher than that of the unmodified PNA6. Solvation of the MP side-chain likely contributes to the enhanced solubility. The other contributing factor may come from helical organization. Stacking the nucleobases on top of one another in a helical arrangement would shield the hydrophobic core from exposure to solvent. As such, only the heteroatoms at the periphery of nucleobases are exposed to and interact with the water molecules. This may explain the nonlinear relationship between the number of MP units and the saturating concentrations of PNA oligomers.
Table 5.
Saturated concentrations of PNA oligomers
| Oligomer | # MP units | Sat. conc. (mM) |
|---|---|---|
| PNA6 | 0 | 39 |
| PNA7 | 1 | 76 |
| PNA8 | 3 | 108 |
| PNA9 | 5 | 350 |
| PNA10 | 8 | >500 |
This suggestion was corroborated by additional experiments (Figure S1, Supplemental Information) which showed that incorporation of a single methyl group instead of MP at the γ-backbone of PNA resulted in nearly 1.2-fold improvement in water solubility, while the reversed trend was observed with additional methyl groups. This is because addition of the alkyl group beyond the first unit does not help to preorganize PNA any more than it already has but instead introduces additional hydrophobic character to the backbone. This would exacerbate self-aggregation, leading to formation of large molecular weight complexes and precipitation of the aggregates out of the solution.
Self-aggregation
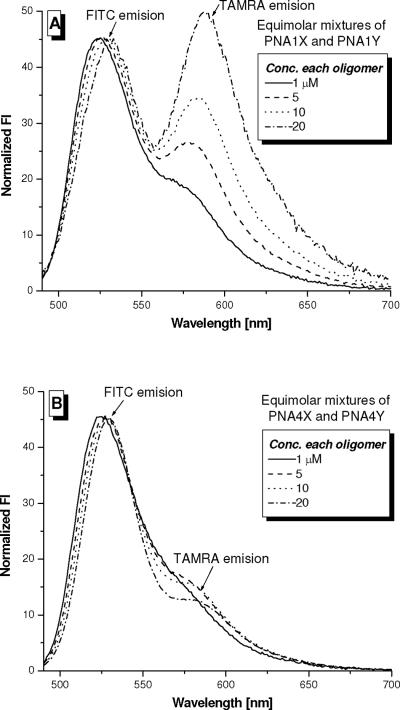
Next, we performed a FRET study to determine whether incorporation of MP in the backbone of PNA would also help reduce aggregation. Different concentrations of unmodified PNA1X/PNA1Y and homologous γ-modified PNA4X/PNA4Y pairs (Table 1) were prepared by mixing equimolar ratios of the individual oligomers in sodium phosphate buffer. The samples were excited at 475 nm, the λmax of FITC, and the emission was recorded from 480 to 700 nm. Upon aggregation, in which the oligomers bearing FITC and TAMRA come into contact with one another, excitation at 475 nm would lead to energy transfer from FITC to TAMRA because of the proximity of the two chromophores. Comparison of the FRET efficiencies of the two systems at different concentrations should provide an assessment of the effect of MP on the intermolecular interaction of PNA.
Inspection of Figure 7A reveals that at a concentration as low as 1 μM of each PNA oligomer, a small but noticeable emission appeared at 580 nm, an indication of FRET and correspondingly aggregation between PNA1X and PNA1Y. The extent of aggregation was further intensified with increasing concentrations of oligomers, apparent from the fluorescent intensity of TAMRA at ~ 580 nm upon excitation of the FITC donor at 475 nm. In contrast, at 20 μM, the point at which nearly 70% FRET efficiency was observed for PNA1X/PNA1Y, only ca. 5% FRET efficiency was observed for PNA4X/PNA4Y (Figure 7B). This indicates that the γ-modified PNA pair does not interact with each other as much as the modified PNAs. The distinction is apparent from photographs of the samples illuminated with a short-wavelength (254 nm), hand-held UV-lamp (Figure 8). The PNA1X/PNA1Y solution displayed a light orange emission at room temperature and yellow-green hue at 90 °C, an indication of the aggregate dissociating upon heating. In contrast, the PNA4X/PNA4Y solution displayed the same color, yellow-green, at room temperature as well at 90 °C, indicating that the oligomers were well dispersed even at room temperature. Thus, the R-MP-γ-modification imparts not only enhanced solubility to PNA, but also suppresses aggregation.
Figure 7.
Fluorescent spectra of (A) PNA1X/PNA1Y and (B) PNA4X/PNA4Y pairs at different concentrations. The samples were prepared by mixing equimolar ratios of the oligomers in 10 mM sodium phosphate buffer (pH 7.4). The samples were excited at 475 nm (FITC λmax) and the emissions were recorded from 480 to 700 nm. The spectra were normalized with respect to the FITC emission.
Figure 8.
Photographs of (A) PNA1X/PNA1Y and (B) PNA4X/PNA4Y samples at different temperatures upon excitation with a short-wavelength (254 nm), hand-held UV-lamp. The sample indicated “90 °C” was incubated in a heating block at 90 °C for 5 min prior to placing on the UV-lamp, after which the photographs were immediately taken.
Off-target binding
It has been documented that at moderate concentrations, PNA tends to aggregate and stick to surfaces and other macromolecules in a nonspecific manner.33 Such interactions could lead to off-target binding and cytotoxic effects, when employed in the cellular context. Among the macromolecules that PNA is known to interact nonspecifically with are nucleic acids and proteins.34 To assess the extent of off-target binding of PNA and R-MPγPNA, we performed a gel-shift assay. In this case a DNA fragment (PCR product), 171bp in length, was incubated with different concentrations of PNA6 and PNA10 (Table 1) in 10 mM sodium phosphate buffer at 37 °C for 16 hr. The two oligomers contained identical nucleobase sequence but differed from another at the γ-backbone; PNA6 was unmodified, whereas PNA10 was modified at every other position with R-MP-γ side-chain. Following incubation the samples were separated on non-denaturing polyacrylamide gel and stained with SYBR-Gold.
Since the target did not contain a complementary sequence to the oligomers, we did not expect binding to take place, in which case the intensity of the DNA band should remain fairly constant, independent of the PNA6 and PNA10 concentrations. Instead, we observed a drastic reduction in the intensity of the DNA band with increasing concentrations of PNA6 (Figure 9). At 10 μM (corresponding to a PNA/DNA ratio of 25:1) or higher, the DNA band completely disappeared from the gel. We neither observed a shifted band, indicating formation of a stable complex, nor staining in the wells, which would indicate formation of globular complexes too large to traverse through the polyacrylamide gel. One possible explanation for the disappearance of the DNA band could be that once formed, the PNA/DNA complex precipitated from solution and/or adhered to the walls of the Eppendorf tubes in which the samples were incubated. On the other hand, for γ-modified PNA10, the intensity of the DNA bands remained fairly constant even at a concentration as high as 20 μM (PNA/DNA ratio of 50:1). This result is consistent with the solubility and FRET data, indicating that incorporation of MP at the γ-backbone not only improves the hybridization properties and water solubility of PNA but also helps reduce nonspecific binding with other macromolecules as well—in this case, DNA.
Figure 9.
Result of a gel-shift assay following incubation of a 171bp, linear double-stranded DNA with different concentrations of PNA6 and PNA10 in 10 mM sodium phosphate buffer (pH 7.4) at 37 °C for 16 hr, followed by electrophoretic separation on nondenaturing-gel and SYBR-Gold staining. Note that this particular DNA fragment, which was PCR-amplified from a Psuper plasmid vector, does not have sequence complementary to PNA6 or PNA10 (both contained the same nucleobase sequence). The concentration of the 171bp DNA fragment was 0.4 μM.
Conclusion
In summary, we have shown that a randomly-folded PNA can be preorganized into a right-handed helix by installing an (R)-MP side-chain at the γ-backbone. The MP-containing γPNA monomers can be prepared in a few simple steps starting from a commercially available and relatively cheap Boc-L-serine. We attribute the high optical purities of the backbone intermediates and final monomers to the optimized alkylation condition in the first step and stereoselectivity of the reduction of carboxylic acid to alcohol in the second step. In contrast to the aminoethylglycine backbone prepared via the reductive amination route which degrades over time (unpublished data), compound 4 can be stored over a prolonged period without signs of deterioration. Another appealing feature of γ-backbone modification is that it is configurationally stable. After the reduction step, i.e. conversion of the carboxylic acid to alcohol, the γ-proton is inert to deprotonation by base. Thus, there is no need for concern for racemization in the subsequent reaction steps. Compared to the previous attempts to improve water solubility, which is often achieved at the expense of binding affinity and/or sequence selectivity, incorporation of MP in the γ-backbone enhances the hybridization properties as well as the water solubility of PNA. Improvements in thermodynamics are attributed to slower dissociation, while enhanced sequence selectivity is the result of backbone preorganization and helical base-stacking arrangement. Improvement in water solubility is the result of solvation of the MP side-chains as well as helical nucleobase arrangement. Helical induction presumably permits greater shielding of the hydrophobic cores of nucleobases and better hydrogen-bonding interaction between heteroatoms at the periphery of nucleobases and solvent molecules.
The results reported herein have important implications for the future design and utility of PNA in biology, biotechnology and medicine. The improvements in hybridization properties may enableRMPγPNAs to invade double helical DNA and structured RNA that may not be permissible with other classes of oligonucleotide mimics. The former has already been demonstrated in a recent study.81 The enhancements in water solubility will facilitate the handling and processing of PNA while lessening the concerns for nonspecific binding and cytotoxic effects. Improvements in these areas, along with the flexibility of synthesis whereby other chemical functionalities can be installed at the γ-backbone with ease, will further expand the utility of PNA into other burgeoning disciplines, including drug discovery and nanotechnology.
Experimental Section
Monomer synthesis
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine (2)
To a stirred, ice-cold (0 °C) solution of NaH (60 % suspended in mineral oil, 1.7 g, 42.6 mmol) in dry DMF (80 ml) under an inert atmosphere was added Boc-L-ser-OH (4.05 g, 19.6 mmol) in DMF (30 ml) dropwise over a period of 3 hr. To this mixture, while still in the ice-bath, was added 1-bromo-2-(2-methoxyethoxy)ethane (6.4 ml, 42.6 mmol) at once. The ice-bath was then removed and the reaction was allowed to gradually warm to room temperature and stirred for another 3 hr. The reaction mixture was quenched with H2O (100 ml) at 0 °C. The solvents (DMF and water) were evaporated under reduced pressure at room temperature. Water (20 ml) was added to the crude residue and acidified with 5 % HCl to pH ~ 3 at 0 °C. The aqueous layers were extracted with ethyl acetate (5 × 100 ml) and dried over Na2SO4. The solvent was evaporated under reduced pressure and the crude mixture was purified by column chromatography to afford a colorless liquid (4.1 g, 13.3 mmol). Yield 68 %; 1H-NMR (CDCl3, 300 MHz) δ 6.84 (br s, 1H), 5.57 (d, J = 7.4 Hz, 1H), 4.46-4.38 (m, 1H), 3.93 (dd, J1 = 3.9 Hz, J2 = 3.5 Hz, 1H), 3.90-3.49 (m, 9H), 3.37 (s, 3H), 1.41 (s, 9H); 13C-NMR (CDCl3, 75 MHz) δ 28.2, 53.8, 58.7, 70.2, 70.7, 70.9, 71.7, 80.0, 155.8, 173.4; HRMS (ESI/MSm/z) Mcalc for C13H25NO7Na 330.1529, found 330.1546.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine-ol (3)
To a stirred solution of Boc-(2-(2-methoxyethoxy)ethyl)-L-serine (2) (4 g, 13.0 mmol) in 20 ml DME in an ice-bath was added NMM (1.43 ml, 13.0 mmol) dropwise under an inert atmosphere, followed by isobutyl chloroformate (1.76 ml, 13.0 mmol). The reaction mixture was stirred at the same temperature for another 0.5 hr. The cold solution was filtered, and the precipitate was washed with DME (2×10ml). To the combined filtrate stirred in an ice-bath was added NaBH4 (0.741 g, 19.5 mmol in 10 ml water) slowly. A strong effervescence occurred. The aqueous layer was extracted 3× with ethyl acetate and the combined organic layers were washed with brine and dried over Na2SO4. The solvent was evaporated under reduced pressure and the oily residue was purified by column chromatography to give a colorless liquid (3.2 g, 10.9 mmol). Yield 84 %; 1H-NMR (CDCl3, 300 MHz) δ 5.26 (br s, 1H), 3.84-3.50 (m, 12 H), 3.36 (s, 3H), 2.93 (br s, 1H), 1.42 (s, 9H); 13C-NMR (CDCl3, 75 MHz) δ 28.3, 51.5, 58.9, 63.2, 70.3, 70.4, 70.5, 71.3, 71.8, 79.5, 156.0; HRMS (ESI/MSm/z) Mcalc for C13H27NO6Na 316.1736, found 316. 1719.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine-ψ[CH2N(o,p-diNBS)]Gly-OEt (4)
To a stirred, cold solution of o,p-diNBS-Gly-OEt (3.53 g, 10.5 mmol), triphenyl phosphine (2.73 g, 10.5 mmol) and compound 3 (3.1 g, 10.5 mmol) in dried THF (20 ml) under an inert atmosphere was added DIAD (1.48 ml, 10.5 mmol) dropwise over a period of 0.5 hr. The reaction mixture was allowed to warm to room temperature and then stirred overnight. The solvent was evaporated and the oily residue was purified by column chromatography to afford a yellow solid (3.6 g, 5.91 mmol). Yield 56 %; 1H-NMR (CDCl3, 300 MHz) δ 8.51 (dd, J = 2.1 Hz, 1H), 8.41 (d, J = 2.1 Hz, 1H), 8.32 (d, J = 8.7 Hz, 1H), 5.16 (d, J = 8.6 Hz, 1H), 4.34 (dd, J = 18.6 Hz, 2H), 4.10 (2×q, J = 3.6 Hz, 2H), 4.01-3.87 (m, 1H), 3.70-3.49 (m, 12H), 3.39 (s, 3H), 1.43 (s, 9H), 1.23 (t, J = 7.1 Hz, 3H); 13C-NMR (CDCl3, 75 MHz) δ 14.0, 28.2, 48.3, 48.7, 49.6, 58.9, 61.6, 70.0, 70.3, 70.4, 70.8, 71.9, 79.7, 119.4, 126.0, 132.0, 138.4, 147.9, 149.5, 155.6, 168.6; HRMS (ESI/MSm/z) Mcalc for C23H36N4O13SNa 631.1898, found 631.1840.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine-ψ[CH2N]Gly-OEt (5)
To a stirred solution of 4 (2.6 g, 4.2 mmol) in dichloromethane (20 ml) was added n-propyl amine (7.0 ml, 85.4 mmol) under an inert atmosphere. The reaction mixture was stirred at room temperature for another 20 min. The solvent was evaporated and the crude mixture was purified by column chromatography to afford 5 as a light yellow liquid (1.3 g, 3.4 mmol). Yield 81 %; 1H-NMR (CDCl3, 300 MHz) δ 5.13 (br s, 1H), 4.12 (q, J = 7.0 Hz, 2H), 3.78-3.63 (m, 1H), 3.62-3.33 (m, 12H), 3.32 (s, 3H), 2.71 (2×dd, J1 = J2 = 5.9 Hz, 2H), 1.80 (br s, 1H), 1.39 (s, 9H), 1.22 (t, J = 7.3 Hz, 3H); 13C-NMR (CDCl3, 75 MHz) δ 14.2, 28.3, 49.9, 50.3, 51.0, 58.9, 60.5, 70.4, 70.6, 71.4, 71.9, 79.1, 155.6, 172.4; HRMS (ESI/MSm/z) Mcalc for C17H34N2O7Na 401.2264, found 401.2278.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine Thymine ethyl ester (6a)
To a stirred solution of thymine acetic acid (0.287 g, 1.56 mmol) in dried DMF (15 ml) under an inert atmosphere was added DCC (0.324 g, 1.56 mmol) and DhbtOH (0.254 g, 1.56 mmol). The resulting mixture was stirred at room temperature for 1 hr. Compound 5 (0.5 g, 1.32 mmol) was dissolved in dried DMF (10 ml) and added to the above reaction mixture, and stirred at 50 °C for 24 hr. The solvent was removed under reduced pressure and the remaining residue was dissolved in ethyl acetate (100 ml), washed with saturated solution of NaHCO3 (100 ml) followed by 10% KHSO4 (100 ml). The organic layer was washed with brine (50 ml) and dried over Na2SO4. The solvent was removed under reduced pressure. The crude product was purified by column chromatography to afford a white solid (0.480 g, 0.88 mmol). Yield 67 %; 1H-NMR (DMSO d6, 300 MHz) δ 7.27-7.19 [2×d (Rot1,2), J1 = J2 = 1.0 Hz, 1H], 6.86-6.57 [2×d (Rot1,2)*, J1 = J2 = 8.6 Hz, 1H], 4.80-4.36 [2×ABq (Rot1,2), J1 = J2 = 16.8 Hz, 2H], 4.33-3.65 (m, 5H), 3.58-3.25 (m, 12H), 3.22-3.15 [2×s(Rot1,2), 3H], 1.73 (s, 3H), 1.35 (br s, 9H), 1.27-1.11 [2×t(Rot1,2), J1 = J2 = 7.1 Hz, 3H]; 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 12.3, 14.4, 28.6, 47.9, 48.1, 48.6, 49.5, 58.5, 60.9, 70.0, 70.3, 71.7, 78.6, 108.5, 142.4, 151.4, 155.8, 164.8, 168.0, 169.3; HRMS (ESI/MSm/z) Mcalc for C24H40N4O10Na 567.2642 found, 567.2651. *Two rotamers were found, as determined by multinuclear and multidimensional NMR experiments.73
Adenine ester 6b, guanine ester 6c, and cytosine ester 6d were prepared, purified, and characterized the same way as described for 6a.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine Adenine(Cbz) ethyl ester (6b)
1H-NMR (DMSO d6, 300 MHz) δ 10.56 (br s, 1H), 8.59-8.55 [2×s (Rot1,2), 1H], 8.30-8.24 [2×s (Rot1,2), 1H], 7.55-7.20 (m, 5H), 7.04-6.53 [2×d (Rot1,2), J1 = J2= 8.5 Hz, 1H], 5.35 [ABq (Rot1), J = 17.2 Hz, Rot2 appears as a br s at 5.15 ppm, 2H], 5.20 (s, 2H), 4.54-3.87 (m, 5H), 3.81-3.31 (m, 12H), 3.23-3.10 [2×s (Rot1,2), 3H], 1.36 (s, 9H), 1.30-1.10 [2×t (Rot1,2), J1 = J2 = 7.1 Hz, 3H]; 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 14.4, 28.6, 44.3, 48.7, 49.3, 49.9, 58.4, 60.9, 66.7, 70.0, 70.2, 70.4, 71.0, 71.6, 78.7, 123.4, 128.3, 128.4, 128.8, 136.8, 145.5, 149.9, 151.9, 152.7, 152.9, 155.8, 167.4, 169.2; HRMS (ESI/MSm/z) Mcalc for C32H45N7O10Na 710.3126, found 710.3110.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine PNA Guanine(Cbz) ethyl ester (6c)
1H-NMR (DMSO d6, 300 MHz) δ 7.78-7.74 [2×s (Rot1,2), 1H], 7.45-7.28 (m, 5H), 6.98-6.59 [2×d (Rot1,2), J1 = J2= 8.5 Hz, 1H], 5.23 (s, 2H), 5.20-4.84 [ABq (Rot1), J = 17.1 Hz, m (Rot2), 2H], 4.45-3.77 (m, 5H), 3.76-3.33 (m, 12H), 3.22-3.11[2×s (Rot1,2), 3H], 1.38-1.28 [2×s (Rot1,2), 9H], 1.28-1.10 [2×t (Rot1,2), J1 = J2 = 7.2 Hz, 3H]; 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 14.4, 28.5, 44.1, 48.8, 49.4, 49.8, 58.4, 61.0, 67.7, 70.0, 70.1, 70.2, 70.4, 71.6, 78.4, 119.6, 128.5, 128.6, 128.8, 128.9, 135.9, 140.8, 147.8, 149.9, 155.0, 155.6, 155.8, 167.4, 169.2; HRMS (ESI/MSm/z) Mcalc for C32H45N7O11Na 726.3075, found 726.3070.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine PNA Cytosine(Cbz) ethyl ester (6d)
1H-NMR (DMSO d6, 300 MHz) δ 7.84 (d, J = 7.3 Hz, 1H), 7.46-7.26 (m, 5H), 6.99 (d, J = 7.3 Hz, 1H), 6.87-6.57 [2×d (Rot1,2), J1 = J2= 8.4 Hz, 1H], 5.17 (s, 2H), 4.93-4.52 [2×ABq (Rot1,2), J1 = J2 = 15.9 Hz, 2H], 4.37-3.68 (m, 5H), 3.63-3.29 (m, 12H), 3.22-3.15 [2×s (Rot1,2), 3H], 1.35 (s, 9H), 1.27-1.09 [2×t (Rot1,2), J1= J2= 7.1 Hz, 3H]; 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 14.4, 28.6, 48.7, 49.9, 58.5, 60.9, 66.9, 70.0, 70.3, 71.7, 78.6, 94.3, 128.3, 128.6, 128.9, 136.4, 151.1, 153.6, 155.4, 155.7, 163.6, 167.9, 169.2; HRMS (ESI/MSm/z) Mcalc for C31H45N5O11Na 686.3013, found 686.3006.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine Thymine monomer (7a)
To a stirred, cold solution of 6a (0.480 g, 0.88 mmol) in THF (10 ml) was added 2N NaOH (10 ml) dropwise over a period of 15 min. The resulting mixture was stirred at the same temperature for another 0.5 hr. Upon completion of the reaction, as confirmed by TLC, H2O (20 ml) was added and the resulting mixture was extracted with ethyl acetate (2×25 ml). The aqueous layers were combined and acidified with 5 % HCl to pH ~ 4 at 0 °C, and then extracted with ethylacetate (4×25ml) and dried over Na2SO4. The solvent was evaporated in vacuo and the crude product was purified by column chromatography to afford a colorless solid (0.400 g, 4.5 mmol). Yield 87%; 1H-NMR (DMSO d6, 300 MHz) δ 7.27-7.18 [2×s (Rot1,2), 1H], 6.88-6.53 [2×d (Rot1,2), J1 = J2 = 8.2 Hz, 1H], 4.75-4.33 [ABq (Rot1), J = 17.5 Hz, m (Rot2), 2H], 4.01-3.59 (m, 3H), 3.55-3.25 (m, 12H), 3.22-3.19 [2×s (Rot1,2), 3H], 1.73 (s, 3H), 1.35 (s, 9H); 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 12.3, 28.7, 48.1, 49.3, 51.9, 58.5, 70.0, 70.2, 70.3, 71.0, 71.7, 78.2, 78.5, 108.5, 142.6, 151.5, 155.7, 164.8, 167.5, 168.5; HRMS (ESI/MSm/z) Mcalc for C22H35N4O10Na2 561.2148, found 561.2115.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine Adenine(Cbz) monomer (7b)
1H- NMR (DMSO d6, 300 MHz) δ 8.57-8.53 [2×s (Rot1,2), 1H], 8.30-8.16 [2×s (Rot1,2), 1H], 7.47-7.24 (m, 5H), 7.04-6.50 [2×d (Rot1,2), J1 =J2= 8.7 Hz, 1H], 5.22 [ABq (Rot1), J = 16.4 Hz, Rot2 appears as br s at 5.10 ppm, 2H], 5.20 (s, 2H), 4.50-3.65 (m, 5H), 3.63-3.43 (m, 5H), 3.41-3.21 (m, 5H), 3.20-3.14 [2×s (Rot1,2),3H], 1.41-1.27 [2×s (Rot1,2),9H]; 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 28.6, 44.5, 49.3, 58.5, 63.4, 66.7, 70.0, 70.2, 70.4, 71.0, 71.7, 78.2, 78.6, 123.3, 126.8, 128.3, 128.4, 128.8, 136.8, 145.7, 149.7, 151.8, 152.9, 155.7, 167.8; HRMS (ESI/MSm/z) Mcalc for C30H40N7O10Na2 704.2632, found 704.2620.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine Guanine(Cbz) monomer (7c)
1H-NMR (DMSO d6, 300 MHz) δ 7.78-7.72 [2×s (Rot1,2), 1H], 7.48-7.23 (m, 5H), 7.00-6.61 [2×d (Rot1,2), J1 =J2= 8.6 Hz, 1H], 5.22-5.19 [2×s (Rot1,2), 2H], 5.03-4.84 [ABq (Rot1), J = 17.3 Hz, br s (Rot2), 2H], 3.98-3.62 (m, 4H), 3.59-3.40 (m, 7H), 3.41-3.23 (m, 4H), 3.22-3.12 [2×s (Rot1,2), 3H], 1.35-1.31 [2×s (Rot1,2), 9H]; 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 28.6, 44.4, 49.3, 52.7, 58.5, 67.4, 70.0, 70.1, 70.2, 70.4, 71.0, 71.7, 78.2, 119.4, 128.4, 128.5, 128.6, 128.9, 136.1, 140.8, 147.9, 150.2, 155.7, 166.7, 167.9, 171.9; HRMS (ESI/MSm/z) Mcalc for C30H40N7O11Na2 720.2581, found 720.2581.
Boc-(2-(2-methoxyethoxy)ethyl)-L-serine Cytosine(Cbz) monomer (7d)
1H- NMR (DMSO d6, 300 MHz) δ 7.84 (d, J = 7.2 Hz, 1H), 7.43-7.24 (m, 5H), 7.01-6.93 [2×d (Rot1,2), J1 =J2= 7.3 Hz, 1H], 6.87-6.58 [2×d (Rot1,2), J1 =J2= 8.73 Hz, 1H], 5.17 (s, 2H), 4.9-4.5 [ABq (Rot1), J = 15.8 Hz, br s (Rot2), 2H], 4.01-3.61 (m, 3H), 3.58-3.43 (m, 7H), 3.43-3.23 (m, 5H), 3.22-3.16 [2×s (Rot1,2), 3H], 1.35 (s, 9H); 13C-NMRMajor rotamer (DMSO d6, 75 MHz) δ 28.1, 48.6, 48.9, 49.7, 52.2, 58.0, 66.4, 69.5, 69.7, 69.8, 70.4, 71.2, 77.8, 94.0, 127.9, 128.1, 128.4, 135.9, 150.9, 153.1, 155.1, 155.2, 163.0, 167.8, 172.2; HRMS (ESI/MSm/z) Mcalc for C29H40N5O11Na2 680.2520, found 680.2546.
Determination of Optical Purities
A general procedure for preparing MTPA-derivatives
Compound 8. To a stirred, cold solution of 2 (100 mg) in DCM (5 mL) was added 5 mL of TFA/m-cresol solution (95:5). The ice-bath was removed and the reaction mixture was stirred at room temperature overnight. The solvent was evaporated under reduced pressure and triturated with diethyl ether (3 × 5 ml). The remaining residue was dried under high vacuum overnight and used in the next coupling step without further purification. The crude residue was dissolved in DCM (5 mL) and chilled at 0 °C. DIPEA (2.0 equiv.) and (S)-(+)-α-Methoxy-α-trifluoromethylphenylacetyl chloride (MTPA-Cl, 1.1 equiv.) were added and the reaction mixture was stirred at room temperature overnight. After completion, the solution was diluted with DCM and washed with water (2× 10 mL), and then brine solution. The organic layer was dried on Na2SO4, concentrated under reduced pressure and purified by column chromatography. Compounds 9 and 10 were prepared the same way.
3-[2-(2-Methoxy-ethoxy)-ethoxy]-2-(3,3,3-trifluoro-2-methoxy-2-phenylpropionylamino)-propionic acid (8)
1H-NMR (300 MHz, CDCl3): 7.78 (brs, 1H), 7.56 (m, 2H), 7.37 (m, 3H), 4.47 (m, 1H), 4.03-3.7 (m, 2H), 3.68-3.45 (m, 8H), 3.40 (s, 3H), 3.30 (s, 3H); 13CNMR (DMSO d6, 75 MHz): δ 42.7, 52.1, 54.8, 55.6, 57.9, 60.1, 68.9, 69.5, 69.6, 71.2, 72.3, 83.8, 127.2, 127.5, 128.1, 129.4, 165.3, 174.7; HRMS (ESI/MSm/z) Mcalc for C18H24F3NO7Na 446.1403, found 446.1359; 19F-NMR (300 MHz, CDCl3): −69.58 (s, L-stereomer), −69.24 (s, D-stereomer).
3,3,3-Trifluoro-N-{2-hydroxy-1-[2-(2-methoxy-ethoxy)-ethoymethyl]-ethyl}-2-methoxy-2-phenyl-propionamide (9)
1H-NMR (300 MHz, CDCl3): 7.55 (m, 2H), 7.44 (brs, 1H), 7.40 (m, 3H), 4.13 (m, 1H), 3.83 (m, 1H), 3.70-3.60 (m, 9H), 3.54 (m, 2H), 3.41(m, 3H), 3.37 (s, 3H), 3.07 (brs, 1H); 13C-NMR (CDCl3, 75 MHz) δ 50.5, 54.3, 58.4, 62.2, 69.8, 69.89, 69.9, 70.2, 71.3, 72.1, 83.4, 121.4, 125.2, 127.2, 128.0, 128.9, 132.0, 165.9; HRMS (ESI/MSm/z) Mcalc for C18H26F3NO6Na 432.1610 found 432. 1595; 19F-NMR (300 MHz, CDCl3): −69.43 (s, L-stereomer), −69.33 (s, D-stereomer).
{[3-[2-(2-Methoxy-ethoxy)-ethoxy]-2-(3,3,3-trifluoro-2-methoxy-2-phenylpropionylamino)-propyl]-[2-thymine-acetyl]-amino}-acetic acid (10)
1H-NMR (300 MHz, DMSO-d6): 11.22 (brs, 1H), 8.33 & 8.25 (2×d, J= 8.5 Hz, 1H), 7.54-7.35 (m, 5H), 7.09 & 6.96 (2×s,1H), 4.52-4.26 (ABq, J= 16.8 Hz, 2H), 4.26-4.16 (m, 1H) 3.6-3.34 (m, 16H), 3.21 & 3.20 (2×s, 3H), 3.10 (ABq, J = 14.0 Hz, 2H), 1.70 & 1.68 (2×s, 3H); 13C-NMR Major rotamer (DMSO d6, 75 MHz) δ 11.8, 47.5, 48.0, 48.4, 52.2, 54.8, 58.0, 69.2, 69.5, 69.6, 69.9, 71.2, 83.7, 107.9, 127.0, 127.2, 128.3, 129.3, 133.3, 142.0, 151.0, 164.3, 165.3, 168.2, 171.6; HRMS (ESI/MSm/z) Mcalc for C27H35F3N4O10Na 655.2305 found 655.2299; 19F-NMR (300 MHz, CDCl3): −68.80 & −68.95 (s, L-stereomer), −68.70 (s, D-stereomer).
Oligomer synthesis
Unmodified, Boc-protected PNA monomers were purchased from Applied Biosystems (they are no longer available). The oligomers were synthesized on MBHA-resin according to the published protocol.87 Upon completion of the last monomer coupling, the oligomers were cleaved from the resin (and the protecting groups were simultaneously removed) by immersing the resin in a cocktail containing m-cresol/thioanisole/TFA/TFMSA (150/150/900/300μL—for 100mg resin) for 2 hr. The crude mixture was eluted and precipitated in ethyl ether, dissolved in water/acetonitrile mixture (80/20), purified by reversed-phase HPLC, and characterized by MALDI-TOF mass spectrometry. A solution of α-cyano-4-hydroxycinnamic acid (10 mg α-cyano-4-hydroxycinnamic acid in 500 mL water with 0.1 %TFA and 500 mL acetonitrile with 0.1 %TFA) was used as the matrix for MALDI-TOF analysis. Concentrations of the oligomers were determined by UV-absorption at 260 nm at 90 °C in water using the following extinction coefficients: 13,700 M−1cm−1 (A), 11,700 M−1cm−1 (G), 6,600 M−1cm−1 (C), and 8,600 M−1cm−1 (T).
Circular Dichroism (CD)
All CD data were recorded at room temperature otherwise stated. All spectra represent an average of at least 15 scans, recorded from 320 to 200 nm at the rate of 100 nm/min. A 1-cm path-length cuvette was used, and the temperature was maintained at 22 °C. All spectra were processed using Origin software, baseline-subtracted and, unless otherwise noted, smoothed using a five-point-adjacent averaging algorithm.
Melting temperature (Tm)
All hybridization experiments were performed by measuring the change in the absorbance at 260 nm as a function of temperature. All samples were prepared by mixing stoichiometric amount of each strand (5 μM) in a 10 mM sodium phosphate at pH 7.4 and annealed prior to each measurement. The recorded spectra were smoothed using a five-adjacent-point averaging algorithm unless otherwise specified. The melting transitions were determined by taking the first derivative of the absorbance-temperature profile.
Thermodynamic analysis
Thermodynamic parameters were determined from concentration dependent Tm measurements following van't Hoff analysis.95 ΔH° and ΔS° were obtained from a plot of 1/Tm (K−1) vs. lnCT, where CT is the total strand concentration. ΔH° was obtained from the slope of the plot, which should be a linear function, using the relationship: slope = (n−1)R/ΔH, where n is the molecularity of the association interaction (in this case n = 2) and R is the gas constant (R = 8.314 J/mol·K). ΔS° was determined from the y-intercept (lnCT = 0), where for a nonself-complementary oligonucleotide the following relationship stands: slope = [ΔS° − (n−1)R·ln2n]/ΔH°. ΔG° was calculated at 298.15 K using the equation, ΔG° = ΔH° − TΔS°, and K using the equation, ΔG° = −RT ln K.
SPR analysis
DNA and PNA concentrations were calculated by measuring sample absorbance at 260 nm and 90 °C. At this temperature the bases are unstacked, resulting in the extinction coefficient of the oligomer being equal to the sum of the extinction coefficient of its bases. The DNA extinction coefficients were obtained from the literature, while the PNA extinction coefficients were provided by Applied Biosystems.
SPR experiments were conducted on a Biacore T100 with a four-channel carboxymethylated dextran-coated gold sensor chip (CM5) at 25 °C. Approximately 6300 response units (RU) of streptavidin (Promega) were immobilized on the chip's surface at a flow rate of 5 μL/min using standard N-hydroxysuccinimide/1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (NHS/EDC) coupling followed by capping with ethanolamine. Then, 100 RU of 5'-biotinylated DNA were attached to the chip via noncovalent capture at a flow rate of 2 μL/min and a sample concentration of 25 nM. HBS-EP (Biacore, 0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, and 0.005% Surfactant P-20, pH=7.4) was used as the running buffer for both the streptavidin and biotinylated DNA immobilizations.
Kinetic information was obtained from the SPR studies by flowing a dilution series (10–50 nM in 10 nM increments) of PNA across the chip. All samples were run in triplicate at a flow rate of 50 μL/min with a running buffer consisting of 10 mM sodium phosphate, pH= 7.4, 100 mM NaCl, 0.1 mM EDTA, and 0.005% Surfactant P-20. The chip was regenerated with two 30 s pulses of 10 mM NaOH/1 M NaCl. Buffer injections were conducted before each sample injection to flush any lingering regeneration solution. All samples were reference-corrected to account for nonspecific binding by subtracting off the signal resulting from flowing sample over a streptavidin-coated (i.e. no DNA) flow cell. Kinetic constants were calculated using a 1:1 binding global fit model in Biacore T100 Evaluation Software.
Solubility
A saturated solution was prepared for each oligomer by dissolving the dried pellet in a minimum amount of water so that after heating at 95 °C for 5 min and thorough mixing, undissolved material still remained in the solution. The samples were centrifuged at 12,000g for 5 min. The concentrations of the saturated solutions were determined by measuring the UV-absorption at 260 nm following a serial dilution.
Self-aggregation
Samples containing different concentrations of PNA1X/PNA1Y or PNA4X/PNA4Y pair at equimolar ratios were prepared in 10 mM sodium phosphate buffer at pH 7.4. The samples were allowed to equilibrate for 1 hr. The fluorescence spectrum of each sample was recorded at room temperature on a Varian Cary Eclipse fluorescence spectrometer from 490 to 700 nm upon excitation at 475 nm (FITC). All spectra were subtracted from the buffer baseline and smoothed using a five-point adjacent averaging algorithm on the Origin software. FRET efficiencies were determined using the relationship: E = 1 − F'D/FD, where F'D and FD are the fluorescence intensities with and without the acceptor, respectively. The donor in this case is FITC (X). For the PNA1X/PNA1Y pair, F'D = F'PNA1X/PNA1Y and FD = FPNA1X; and for the PNA4X/PNA4Y pair, F'D = F'PNA4X/PNA4Y and FD = FPNA4X, at the respective concentration.
Off-target binding
A PCR product, 171bp in length (0.4 μM) was separately incubated with 4, 10 and 20 μM of PNA6 and PNA10 in sodium phosphate buffer at 37 °C for 16 hr. Following incubation, the samples were separated on nondenaturing polyacrylamide gel and stained with SYBR-Gold. The gel was washed 3 times with 1× TBE buffer and imaged using the BioDoc-It gel documentation system.
Supplementary Material
Acknowledgements
Financial support for this work was provided by the National Institutes of Health (GM076251) and the National Science Foundation (CHE-1012467) to D.H.L. SPR instrumentation was purchased with support from NSF MRI award 0821296. K.J.Z gratefully acknowledges support from a DOD, Air Force Office of Scientific Research NDSEG Fellowship 32 CFR168a.
Footnotes
Supporting Information Available: UV-absorption spectra of PNA11–13, 1H and 13C-NMR spectra of compounds 1–10, HPLC and MALDI-TOF MS spectra of PNA1–10, UV-melting curves of PNA5-DNA and PNA5-RNA duplexes containing perfectly-matched and single-base mismatched targets, CD melting curves of PNA2–5, and SPR sensorgrams & kinetic fits for PNA1–5. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Nielsen PE, Egholm M, Berg RH, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- (2).Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- (3).Nielsen PE. Acc. Chem. Res. 1999;32:624–630. [Google Scholar]
- (4).Bentin T, Larsen HJ, Nielsen PE. Biochemistry. 2003;42:13987–13995. doi: 10.1021/bi0351918. [DOI] [PubMed] [Google Scholar]
- (5).Kaihatsu K, Shah RH, Zhao X, Corey DR. Biochemistry. 2003;42:13996–14003. doi: 10.1021/bi035194k. [DOI] [PubMed] [Google Scholar]
- (6).Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen PE, Norden B, Graeslund A. J. Am. Chem. Soc. 1996;118:5544–5552. [Google Scholar]
- (7).Ratilainen T, Holmen A, Tuite E, Nielsen PE, Norden B. Biochemistry. 2000;39:7781–7791. doi: 10.1021/bi000039g. [DOI] [PubMed] [Google Scholar]
- (8).Menchise V, De Simone G, Tedeschi T, Corradini R, Sforza S, Marchelli R, Capasso D, Saviano M, Pedone C. Proc. Natl. Acad. Sci. USA. 2003;100:12021–12026. doi: 10.1073/pnas.2034746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Rasmussen H, Kastrup JS, Nielsen JN, Nielsen JM, Nielsen PE. Nat. Struct. Biol. 1997;4:98–101. doi: 10.1038/nsb0297-98. [DOI] [PubMed] [Google Scholar]
- (10).Haaima G, Rasmussen H, Schmidt G, Jensen DK, Kastrup JS, Stafshede PW, Norden B, Buchardt O, Nielsen PE. New J. Chem. 1999;23:833–840. [Google Scholar]
- (11).Eldrup AB, Nielsen BB, Haaima G, Rasmussen H, Kastrup JS, Christensen C, Nielsen PE. Eur. J. Org. Chem. 2001:1781–1790. [Google Scholar]
- (12).Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sonnichsen SH, Nielsen PE. Biochem. Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- (13).Dueholm KL, Egholm M, Behrens C, Christensen L, Hansen HF, Vulpius T, Petersen KH, Berg RH, Nielsen PE, Buchardt O. J. Org. Chem. 1994;59:5767–5773. [Google Scholar]
- (14).Thomson SA, Josey JA, Cadilla R, Gaul MD, Hassman CF, Luzzio MJ, Pipe AJ, Reed KL, Ricca DJ, Wiethe RW, Noble SA. Tetrahedron. 1995;51:6179–6194. [Google Scholar]
- (15).Beck F, Nielsen PE. Artificial DNA: methods and applications. Vol. 2003. CRC Press; Boca Raton: 2003. pp. 91–114. [Google Scholar]
- (16).Ray A, Norden B. FASEB Journal. 2000;14:1041–1060. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- (17).Nielsen PE. Mol. Biotech. 2004;26:233–248. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- (18).Veselkov AG, Demidov VV, Frank-Kamenetskii MD, Nielsen PE. Nature. 1996;379:214. doi: 10.1038/379214a0. [DOI] [PubMed] [Google Scholar]
- (19).Komiyama M, Aiba Y, Yamamoto Y, Sumaoka J. Nat. Protoc. 2008;3:655–622. doi: 10.1038/nprot.2008.7. [DOI] [PubMed] [Google Scholar]
- (20).Chin JY, Kuan JY, Lonkar PS, Krause DS, Seidman MM, Peterson KR, Nielsen PE, Kole R, Glazer PM. Proc. Nat. Acad. Sci. USA. 2008;105:13514–13519. doi: 10.1073/pnas.0711793105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Singer A, Wanunu M, Morrison W, Kuhn H, Frank-Kamenetskii M, Meller A. Nano Lett. 2010;10:738–742. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mollegaard NE, Buchardt O, Egholm M, Nielsen PE. Proc. Natl. Acad. Sci. USA. 1994;91:3892–3895. doi: 10.1073/pnas.91.9.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR. Nat. Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- (24).Winssinger N, Ficarro S, Schultz PG, Harris JL. Proc. Natl. Acad. Sci. USA. 2002;99:11139–11144. doi: 10.1073/pnas.172286899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Debaene F, Da Silva JA, Pianowski Z, Duran FJ, Winssinger N. Tetrahedron. 2007;63:6577–6586. [Google Scholar]
- (26).Bruno Y, Birnbaum ME, Kleiner RE, Liu DR. Nat. Chem. Biol. 2010;6:148–155. doi: 10.1038/nchembio.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ura Y, Beierle JM, Leman LJ, Orgel LE, Ghadiri MR. Science. 2009;325:73–77. doi: 10.1126/science.1174577. [DOI] [PubMed] [Google Scholar]
- (28).Myers CP, Williams ME. Coordin. Chem. Rev. 2010;254:2416–2428. [Google Scholar]
- (29).Chakrabarti R, Klibanov AM. J. Am. Chem. Soc. 2003;125:12531–12540. doi: 10.1021/ja035399g. [DOI] [PubMed] [Google Scholar]
- (30).Janssen PGA, Meeuwenoord N, van der Marel G, Jabbari-Farouji S, van der Schoot P, Surin M, Tomovic Z, Meijer EW, Schenning APHJ. Chem. Comm. 2010;46:109–111. doi: 10.1039/b913307k. [DOI] [PubMed] [Google Scholar]
- (31).Riaz U, Ahmad S, Ashraf SM. J. Nanopart. Res. 2008;10:1209–1214. [Google Scholar]
- (32).Popescu DL, Parolin TJ, Achim C. J. Am. Chem. Soc. 2003;125:6354–6355. doi: 10.1021/ja029714v. [DOI] [PubMed] [Google Scholar]
- (33).Braasch DA, Corey DR. Methods. 2001;23:97–107. doi: 10.1006/meth.2000.1111. [DOI] [PubMed] [Google Scholar]
- (34).Tackett AJ, Corey DR, Raney KD. Nucleic Acids Res. 2002;30:950–957. doi: 10.1093/nar/30.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Masuko M. Nucleic Acids Res. Suppl. 2003:145–146. doi: 10.1093/nass/3.1.145. [DOI] [PubMed] [Google Scholar]
- (36).Cattani-Scholz A, Pedone D, Blobner F, Abstreiter G, Schwartz J, Tornow M, Andruzzi L. Biomacromolecules. 2009;10:489–496. doi: 10.1021/bm801406w. [DOI] [PubMed] [Google Scholar]
- (37).Egholm M, Buchardt O, Nielsen PE, Berg RH. J. Am. Chem. Soc. 1992;114:1895–1897. [Google Scholar]
- (38).Haaima G, Lohse A, Buchardt O, Nielsen PE. Angew. Chem. Int. Ed. 1996;35:1939–1941. [Google Scholar]
- (39).Sforza S, Tedeschi T, Corradini R, Marchelli R. Eur. J. Org. Chem. 2007:5879–5885. [Google Scholar]
- (40).Boyarskaya NP, Kirillova YG, Stotland EA, Prokhorov DI, Zvonkova EN, Shvets VI. Doklady Chemistry. 2006;49:57–60. [Google Scholar]
- (41).Gildea BD, Casey S, MacNeill J, Perry-O'Keefe H, Sorensen D, Coull JM. Tetrahedron Lett. 1998;39:7255–7258. [Google Scholar]
- (42).Hudson RHE, Liu Y, Wojciechowski F. Can. J. Chem. 2007;85:302–312. [Google Scholar]
- (43).Peyman A, Uhlmann E, Wagner K, Augustin S, Breipohl G, Will DW, Schafer A, Wallmeier H. Angew. Chem. Int. Ed. 1996;35:2636–2638. [Google Scholar]
- (44).Efimov VA, Choob MV, Buryakova AA, Kalinkina AL, Chakhmakhcheva OG. Nucleic Acids Res. 1998;26:566–575. doi: 10.1093/nar/26.2.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Bonora GM, Drioli S, Ballico M, Faccini A, Corradini R, Cogoi S, Xodo L. Nucleos. Nucleot. Nucl. 2007;26:661–664. doi: 10.1080/15257770701490548. [DOI] [PubMed] [Google Scholar]
- (46).Petersen KH, Jensen DK, Egholm M, Nielsen PE, Buchardt O. Bioorg. Med. Chem. Lett. 1995;5:1119–1124. [Google Scholar]
- (47).Bergmann F, Bannwarth W, Tam S. Tetrahedron Lett. 1995;36:6823–6826. [Google Scholar]
- (48).Uhlmann E, Will DW, Breipohl G, Langner D, Ryte A. Angew. Chem. Int. Ed. 1996;35:2632–2635. [Google Scholar]
- (49).Finn PJ, Gibson NJ, Fallon R, Hamilton A, Brown T. Nucleic Acids Res. 1996;24:3357–3363. doi: 10.1093/nar/24.17.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).vander Laan AC, Brill R, Kuimelis RG, KuylYeheskiely E, vanBoom JH, Andrus A, Vinayak R. Tetrahedron Lett. 1997;38:2249–2252. [Google Scholar]
- (51).Kuwahara M, Arimitsu M, Sisido M. J. Am. Chem. Soc. 1999;121:256–257. [Google Scholar]
- (52).Harris JM, Chess RB. Nat. Rev. Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- (53).Veronese FM, Pasut G. Drug. Discov. Today. 2005;10:1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- (54).Knop K, Hoogenboom R, Fischer D, Schubert US. Angew. Chem. Int. Ed. 2010;49:6288–6308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- (55).Roberts MJ, Benley MD, Harris JM. Adv. Drug. Deliv. Rev. 2002;54:459–476. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- (56).Wang Y, Chang F, Zhang Y, Liu N, Liu G, Gupta S, Rusckowski M, Hnatowich DJ. Bioconj. Chem. 2001;12:807–816. doi: 10.1021/bc0100307. [DOI] [PubMed] [Google Scholar]
- (57).Rapozzi V, Cogoi S, Spessotto P, Risso A, Bonora GM, Quadrifoglio F, Xodo LE. Biochemistry. 2002;41:502–510. doi: 10.1021/bi011314h. [DOI] [PubMed] [Google Scholar]
- (58).Bonora GM. J. Bioact. Compatib. Polym. 2002;17:375389. [Google Scholar]
- (59).Zhao H, Greenwald RB, Reddy P, Xia J, Peng P. Bioconj. Chem. 2005;16:758–766. doi: 10.1021/bc049804k. [DOI] [PubMed] [Google Scholar]
- (60).Liu B, Burdine L, Kodadek T. J. Am. Chem. Soc. 2006;128:15228–15235. doi: 10.1021/ja065794h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kikkeri R, Lepenies B, Adibekian A, Laurino P, Seeberger PH. J. Am. Chem. Soc. 2009;131:2110–2112. doi: 10.1021/ja807711w. [DOI] [PubMed] [Google Scholar]
- (62).Konig HM, Gorelik T, Kolb U, Kilbinger AFM. J. Am. Chem. Soc. 2006;129:704–708. doi: 10.1021/ja0672831. [DOI] [PubMed] [Google Scholar]
- (63).Gopin A, Ebner S, Attali B, Shabat D. Bioconj. Chem. 2006;17:1432–1440. doi: 10.1021/bc060180n. [DOI] [PubMed] [Google Scholar]
- (64).Xiong S, Li H, Yu B, Wu J, Lee RJ. J. Pharmaceut. Sci. 2010;99:5011–5018. doi: 10.1002/jps.22210. [DOI] [PubMed] [Google Scholar]
- (65).Zhao H, Duong HHP, Yung LYL. Macromol. Rapid. Comm. 2010;31:1163–1169. doi: 10.1002/marc.200900876. [DOI] [PubMed] [Google Scholar]
- (66).Nagahori N, Abe M, Nishimura S-I. Biochemistry. 2009;48:583–594. doi: 10.1021/bi801640n. [DOI] [PubMed] [Google Scholar]
- (67).Li J, Chen Y-C, Tseng Y-C, Mozumdar S, Huang L. J. Control Release. 2010;142:416–421. doi: 10.1016/j.jconrel.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Arnaud A, Belleney J, Boue F, Bouteiller L, Carrot G, Wintgens V. Angew. Chem. Int. Ed. 2004;43:1718–1721. doi: 10.1002/anie.200353434. [DOI] [PubMed] [Google Scholar]
- (69).Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. J. Am. Chem. Soc. 2006;128:10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- (70).Rapireddy S, He G, Roy S, Armitage BA, Ly DH. J. Am. Chem. Soc. 2007;129:15596–15600. doi: 10.1021/ja074886j. [DOI] [PubMed] [Google Scholar]
- (71).Chenna V, Rapireddy S, Sahu B, Ausin C, Pedroso E, Ly DH. ChemBioChem. 2008;9:2388–2391. doi: 10.1002/cbic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Sahu B, Chenna V, Lathrop KL, Thomas SM, Zon G, Livak KJ, Ly DH. J. Org. Chem. 2009;74:1509–1516. doi: 10.1021/jo802211n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Yeh JI, Shivachev B, Rapireddy S, Crawford MJ, Gil RR, Du S, Madrid M, Ly DH. J. Am. Chem. Soc. 2010;132:10717–10727. doi: 10.1021/ja907225d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Kosynkina L, Wang W, Liang TC. Tetrahedron Lett. 1994;35:5173–5176. [Google Scholar]
- (75).Falkiewicz B, Kowalska K, Kolodziejczyk AS, Wisniewski K, Lankiewicz L. Nucleos. Nucleot. 1999;18:353–361. [Google Scholar]
- (76).Wu Y, Xu JC. Tetrahedron. 2001;57:8107–8113. [Google Scholar]
- (77).Tedeschi T, Sforza S, Corradini R, Marchelli R. Tetrahedron Lett. 2005;46:8395–8399. [Google Scholar]
- (78).Dose C, Seitz O. Org. Lett. 2005;7:4365–4368. doi: 10.1021/ol051489+. [DOI] [PubMed] [Google Scholar]
- (79).Englund EA, Appella DH. Org. Lett. 2005;7:3465–3467. doi: 10.1021/ol051143z. [DOI] [PubMed] [Google Scholar]
- (80).Englund EA, Appella DH. Angew. Chem. Int. Ed. 2007;46:1414–1418. doi: 10.1002/anie.200603483. [DOI] [PubMed] [Google Scholar]
- (81).He G, Rapireddy S, Bahal R, Sahu B, Ly DH. J. Am. Chem. Soc. 2009;131:12088–12090. doi: 10.1021/ja900228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Fukuyama T, Cheung M, Jow C-K, Hidai Y, Kan T. Tetrahedron Lett. 1997;38:5831–5834. [Google Scholar]
- (83).Debaene F, Winssinger N. Org. Lett. 2003;5:4445–4447. doi: 10.1021/ol0358408. [DOI] [PubMed] [Google Scholar]
- (84).Corradini R, Di Silvestro G, Sforza S, Palla G, Dossena A, Nielsen PE, Marchelli R. Tetrahedron-Asymmetry. 1999;10:2063–2066. [Google Scholar]
- (85).Seco JM, Quinoa E, Riguera R. Chem. Rev. 2004;104:17–117. doi: 10.1021/cr2003344. [DOI] [PubMed] [Google Scholar]
- (86).Rodriguez M, Llinares M, Doulut S, Martinez J. Tetrahedron Lett. 1991;32:923–926. [Google Scholar]
- (87).Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, Egholm M, Buchardt O, Nielsen PE, Coull J, Berg RH. J. Pept. Sci. 1995;1:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- (88).Tedeschi T, Corradini R, Marchelli R, Pushl A, Nielsen PE. Tetrahedron-Asymmetry. 2002;13:1629–1636. [Google Scholar]
- (89).Sen S, Nilsson L. J. Am. Chem. Soc. 2001;123:7414–7422. doi: 10.1021/ja0032632. [DOI] [PubMed] [Google Scholar]
- (90).Johnson WC. In Circular Dichroism: Principles and Applications. Wiley-VCH; New York: 2000. [Google Scholar]
- (91).Nielsen EB, Schellman JA. J. Phys. Chem. 1967;71:2297–2304. doi: 10.1021/j100866a051. [DOI] [PubMed] [Google Scholar]
- (92).Wittung P, Nielsen PE, Buchardt O, Egholm M, Norden B. Nature. 1994;368:561–563. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]
- (93).Eriksson M, Nielsen PE. Nat. Struct. Biol. 1996;3:410–413. doi: 10.1038/nsb0596-410. [DOI] [PubMed] [Google Scholar]
- (94).Cheatham TEI, Kollman PA. J. Am. Chem. Soc. 1997;119:4805. [Google Scholar]
- (95).Marky L, Breslauer KJ. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- (96).Gildea BD, Coull JM, Hyldig-Nielsen JJ, Fiandaca MJ. 1999. [DOI] [PMC free article] [PubMed]
- (97).Ananthanawat C, Vilainvan T, Hoven V, Su X. Biosens. Bioelectron. 2010;25:1064–1069. doi: 10.1016/j.bios.2009.09.028. [DOI] [PubMed] [Google Scholar]
- (98).Lao AI, Su X, Aung KMM. Biosens. Bioelectron. 2009;24:1717–1722. doi: 10.1016/j.bios.2008.08.054. [DOI] [PubMed] [Google Scholar]
- (99).Sawata S, Kai E, Ikebukuro K, Iida T, Honda T, Karube I. Biosens. Bioelectron. 1999;14:397–404. doi: 10.1016/s0956-5663(99)00018-4. [DOI] [PubMed] [Google Scholar]
- (100).Roy S, Tanious FA, Wilson WD, Ly DH, Armitage BA. Biochemistry. 2007;46:10433–10443. doi: 10.1021/bi700854r. [DOI] [PubMed] [Google Scholar]
- (101).Lusvarghi S, Murphy CT, Roy S, Tanious FA, Sacui I, Wilson W, Ly DH, Armitage BA. J. Am. Chem. Soc. 2009;131:18415–18424. doi: 10.1021/ja907250j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.