Abstract
Genome-wide association studies (GWAS) can identify common alleles that contribute to complex disease susceptibility. Despite the large number of SNPs assessed in each study, the effects of most common SNPs must be evaluated indirectly using either genotyped markers or haplotypes thereof as proxies. We have previously implemented a computationally efficient Markov Chain framework for genotype imputation and haplotyping in the freely available MaCH software package. The approach describes sampled chromosomes as mosaics of each other and uses available genotype and shotgun sequence data to estimate unobserved genotypes and haplotypes, together with useful measures of the quality of these estimates. Our approach is already widely used to facilitate comparison of results across studies as well as meta-analyses of GWAS. Here, we use simulations and experimental genotypes to evaluate its accuracy and utility, considering choices of genotyping panels, reference panel configurations, and designs where genotyping is replaced with shotgun sequencing. Importantly, we show that genotype imputation not only facilitates cross study analyses but also increases power of genetic association studies. We show that genotype imputation of common variants using HapMap haplotypes as a reference is very accurate using either genome-wide SNP data or smaller amounts of data typical in fine-mapping studies. Furthermore, we show the approach is applicable in a variety of populations. Finally, we illustrate how association analyses of unobserved variants will benefit from ongoing advances such as larger HapMap reference panels and whole genome shotgun sequencing technologies.
Keywords: imputation, haplotyping, sequencing
INTRODUCTION
Most ongoing genome-wide association studies (GWAS) rely on a commercial SNP genotyping panel that directly assays only a small fraction of SNPs in the human genome [Carlson et al., 2003; The International HapMap Consortium 2005]. In these scans, the majority of SNPs in the genome must be evaluated indirectly using one or more of the genotyped SNPs as proxies [Barrett and Cardon, 2006; Pe’er et al., 2006]. Despite the ability of individual genome-wide association scans to identify common alleles that make large contributions to disease risk and a subset of the loci with smaller effect [Hirschhorn and Daly, 2005], many alleles that contribute to complex disease can only be identified through the meta-analysis of multiple genome-wide scans [for specific examples, see Lettre et al., 2008; Sanna et al., 2008; Willer et al., 2008, 2009]. Although it is possible to assign SNPs genotyped in each study as proxies for SNPs genotyped in the other studies [Carlson et al., 2004; de Bakker et al., 2005; Lin et al., 2004; Nicolae, 2006; Zaitlen et al., 2007], meta-analyses of GWAS conducted in this manner would be cumbersome because of the limited overlap between the different commercial panels and because different choices of proxies for a particular SNP might lead to somewhat different conclusions.
GENOTYPE IMPUTATION
A much more attractive approach for cross study analyses is to combine genotypes generated by the International HapMap Consortium, [The International HapMap Consortium, 2005] with genotypes from individual studies, and then use a haplotyping algorithm that can handle genome scale data to impute genotypes at untyped markers in each study [Scheet and Stephens, 2006]. This strategy results in a situation where all studies are “genotyped” at all the markers examined by the HapMap consortium (albeit some markers are genotyped using conventional means and others are genotyped in silico [Burdick et al., 2006]). The approach relies on the intuition that even two apparently “unrelated” individuals can share short stretches of haplotype inherited from distant common ancestors. Once one of these stretches is identified using genotypes for a few SNPs, alleles for intervening SNPs that are measured in one of the individuals, but not the other, can be imputed. Provided shared haplotype stretches are identified correctly, imputed genotypes will be accurate unless they have been disrupted by gene conversion or mutation events.
INITIAL EVALUATION OF IMPUTED GENOTYPES AND HAPLOTYPES
Here, we systematically evaluate the genotype imputation approach outlined in the paragraph above using our Markov Chain Haplotyping algorithm (MaCH 1.0; see Appendix for implementation details). To estimate haplotypes, our approach starts by randomly generating a pair of haplotypes that is compatible with observed genotypes for each sampled individual. These initial haplotype estimates are then refined through a series of iterations. In each iteration, a new pair of haplotypes is sampled for each individual in turn using a Hidden Markov Model (HMM) that describes the haplotype pair as an imperfect mosaic of the other haplotypes. Model parameters that characterize the probability of change in the mosaic pattern between every pair of consecutive markers and the probability of observing an imperfection in the mosaic at each specific point are also updated. After many iterations (typically 20–100), a consensus haplotype can be constructed by merging the haplotypes sampled in each round.
HAPLOTYPING
Our approach was inspired by the Markov models commonly used for pedigree analysis [for examples, see Abecasis et al., 2002; Kruglyak et al., 1996; Lander and Green, 1987] and shares several features with other HMMs used to describe sampled haplotypes as a mosaic of a set of reference haplotypes [Daly et al., 2001; Li and Stephens, 2003; Mott et al., 2000; Stephens and Scheet, 2005a]. In order to evaluate its performance, we simulated two sets of 100 1 Mb regions that mimic the degree of linkage disequilibrium (LD) in the HapMap CEU and YRI samples [Schaffner et al., 2005]. In each region, we simulated genotypes for ~200 markers, ascertained to mimic HapMap I allele frequency patterns [Marchini et al., 2006], in 90 individuals with 2% of the genotypes missing at random. We then used our method to reconstruct individual haplotypes and tallied three measures of haplotyping quality [Marchini et al., 2006]: (1) the number of incorrectly imputed missing genotypes, (2) among heterozygous sites, the number of consecutive sites that are phased incorrectly with respect to each other (this is the number of “flips” required to transform estimated haplotypes into the true haplotypes, after masking incorrectly imputed sites), and (3) the number of perfectly inferred haplotypes. The three measures were averaged over all 100 regions and the results are summarized in Table I. For comparison, the table also includes results for PHASE [Stephens and Scheet, 2005b; Stephens et al., 2001] and fastPHASE [Scheet and Stephens, 2006], two state of the art haplotyping algorithms [Marchini et al., 2006], and for BEAGLE [Browning, 2006] and PL-EM [Qin et al., 2002], two alternative haplotyping algorithms that are very computationally efficient. Table I clearly shows that our method is competitive in all three measures: our method results in slightly fewer incorrectly imputed genotypes, requires slightly fewer flips to transform imputed haplotypes into the true haplotypes, and produces slightly more correctly inferred haplotypes over the entire 1 Mb stretch than PHASE, which was the second best method. Furthermore, note that estimates of haplotypes and missing genotypes obtained in 5–20 min using our method are comparable in quality to those produced by PHASE runs averaging ~1 day.
TABLE I.
Quality of haplotypes and missing genotypes estimates
| Dataset mimicking HapMap CEU | Dataset mimicking HapMap YRI | |||||||
|---|---|---|---|---|---|---|---|---|
| Method | # Iterations | Computation time | # Errors | # Flips | # Perfect | # Errors | # Flips | # Perfect |
| MaCH | 20 | ~2 min | 11.6 | 216 | 26.5 | 17.9 | 256 | 22.6 |
| 60 | ~5 min | 10.8 | 200 | 28.4 | 16.6 | 232 | 24.1 | |
| 200 | ~15 min | 10.6 | 192 | 29.1 | 16.3 | 222 | 25.1 | |
| 1,000 | ~1.4 hr | 10.6 | 182 | 29.3 | 16.3 | 218 | 25.5 | |
| 3,000 | ~3.9 hr | 10.5 | 178 | 29.7 | 16.1 | 214 | 25.7 | |
| PHASE | – | ~25 hr | 12.6 | 201 | 25.3 | 19.8 | 270 | 19.9 |
| fastPHASE | – | ~17 min | 12.9 | 220 | 20.1 | 22.9 | 331 | 11.7 |
| PL-EM | – | ~3 sec | 23.2 | 356 | 13.8 | 36.8 | 521 | 7.2 |
| BEAGLE | – | ~2 sec | 13.9 | 230 | 21.1 | 23.1 | 332 | 13.1 |
The table summarizes results from the analysis of two sets of 100 simulated 1 Mb regions. The two sets reflect the degree of LD in the HapMap CEU and YRI samples, respectively. In each region, ~200 markers were ascertained to mimic HapMap allele frequency spectra and 2% missing data was introduced at random. The data were then analyzed with one of five haplotypers (MaCH, PHASE, fastPHASE, PL-EM and BEAGLE) and the quality of haplotype solutions and imputed genotypes evaluated. The number of missing genotypes imputed incorrectly (# Errors), the number of switches in haplotype phase required to convert the estimated haplotypes into the simulated haplotypes (# Flips) and the number of perfectly estimated haplotypes (# Perfect) was recorded. Averages of these three quantities are tabulated. Mach 1.0 was run with default settings and different numbers of iterations. PHASE version 2.1.1 [Stephens and Scheet, 2005b; Stephens et al., 2001] was run with default settings, as recommend by Matthew Stephens. fastPHASE version 1.3 [Scheet and Stephens, 2006] was run with default settings, as recommended by Paul Scheet. Beagle 2.1.3 was run with default settings, as recommend by Brian Browning. PL-EM was run with settings recommended by Steve Qin. All timings refer to a 2.33GHz Pentium Xeon.
GENOTYPE IMPUTATION FOR UNTYPED MARKERS
Encouraged by these initial results, we proceeded to apply our method to impute genotypes for untyped markers in the Finland United States Investigation of NIDDM genetics (FUSION) GWAS [Scott et al., 2007]. Since a previous analysis suggested LD patterns in the HapMap CEU and in FUSION are similar [Willer et al., 2006], we used genotypes for 290,690 autosomal markers with allele frequency >5% in the Illumina 317K SNP chip and haplotypes for 2.5M polymorphic markers in the phased HapMap CEU chromosomes as input. After running the haplotyping procedure described above, we estimated the most likely genotype at each position (taking a majority vote across all iterations) and the expected number of copies of the minor allele at each position (a fractional value between 0 and 2) for each individual. We obtained similar results running the haplotyping procedure for 50–100 iterations or using only a smaller number of iterations (10–20) to estimate model parameters and then calculating maximum likelihood estimates for the missing genotypes and allele counts. Different chromosomes were analyzed in parallel and, overall, imputing genotypes for 2,335 unrelated individuals took <2 days for each of the largest chromosomes on a 2006 vintage 2.40GHz Pentium Xeon processor. In total, we imputed genotypes for 2,266,562 SNPs per individual. On average, our method used stretches of ~150 kb from the HapMap CEU panel to reconstruct haplotypes for individuals in the FUSION sample.
IMPUTATION IN THE FUSION GENOMEWIDE ASSOCIATION STUDY
To evaluate the quality of imputed genotypes, we contrasted our estimates of the most likely genotypes and the expected number of copies of the minor allele with actual genotype data for three sets of markers: 521 SNP markers in a region of chromosome 14 previously examined to fine-map a candidate linkage region [Willer et al., 2006], 1,234 SNP markers selected to augment coverage of the Illumina 317K panel in regions surrounding 222 candidate genes [Gaulton et al., 2008] and 12,702 markers with MAF <5% not included in the set of 290,690 markers used for imputation. We expected the last two panels of markers to be harder to impute, because they represent SNPs that are not well tagged by the Illumina 317K SNP chip or that have lower MAF. We observed that 98.60% of imputed alleles matched actual genotyped alleles in the fine-mapping panel, 96.24% in the candidate gene panel, and 98.73% in the low MAF SNP panel. Furthermore, the average r2 between imputed genotypes and actual genotypes was 90.4, 79.1, and 74.0% in the three SNP panels, respectively. This represents an improvement of 14–39% compared to the best available single marker tags, which provided an average r2 of 76.5, 52.8, and 35.5% in the three SNP panels, respectively.
MEASURES OF IMPUTATION QUALITY
Our Markov Chain produces three estimates of imputation quality and these can be used to focus analyses on subsets of high-quality genotypes. First, it produces a quality score that estimates the accuracy of each imputed genotype and is simply the proportion of iterations where the final imputed genotype (by taking a majority vote across all iterations) was selected. Second, it produces an overall measure of the accuracy of imputation for each marker, which is the genotype quality score averaged across all individuals. Finally, by comparing the distribution of sampled genotypes in each iteration with the estimated allele counts that result from averaging over all iterations, it produces an estimate of the r2 between imputed and true genotypes (see Methods for more details). Quality measures for individual genotypes were good predictors of imputation accuracy (Supplementary Figure 1, Right Panel) and show that most imputed genotypes are called with a high degree of confidence (Supplementary Figure 1, Left Panel). For example, as measured by their quality scores, the top 95% of genotypes had average quality scores of 98.9% and actually matched experimental genotypes 98.6% of the time. Most of the errors affect a single allele so that, when measured on a per allele basis, concordance increases to 99.3%.
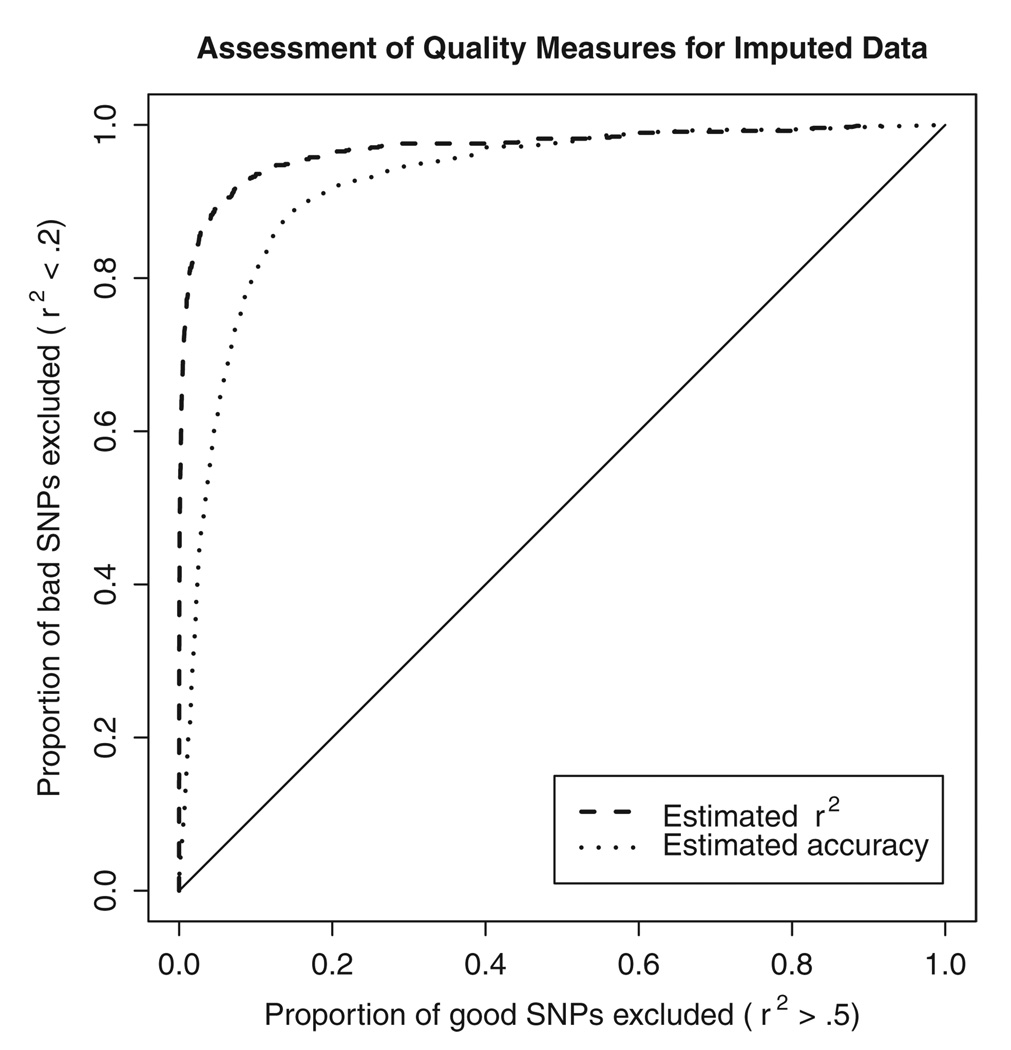
To avoid preferential removal of rare genotypes or alleles at each marker, we recommend using the per marker quality scores to select a subset of imputed SNPs for analysis, instead of the per genotype quality scores. Overall, we saw a correlation of 0.77 between the estimated and actual accuracy of imputed genotypes for each marker. We also saw a correlation of 0.84 between the r2 estimated by our method and the actual r2 that resulted from comparing experimentally derived allele counts with their imputed estimates. Figure 1 shows the ROC curve [Pepe, 2003] for the two quality measures, showing that the estimated r2 measure is a more effective way to identify poorly imputed markers. In the FUSION GWAS scan [Scott et al., 2007], we used an r2 threshold of 0.30 to decide which markers were well imputed and should be included in further analyses, and which were not. At this threshold, we expect to remove 70% of poorly imputed markers (those where r2 with experimental genotypes is <20%) but only 0.50% of better imputed markers (those where r2 with experimental genotypes is >50%).
Fig. 1.
ROC curve comparing two measures of data quality. For imputed SNPs on chromosome 14, where both imputed and actual genotypes were available, we evaluated the ability of two different measures of data quality (the estimated concordance between imputed and true genotypes and the estimated r2 between imputed and true genotypes) to discriminate between poor and well imputed SNPs. Both estimates of imputation quality are calculated without using the actual observed genotypes.
IMPUTATION OF STRONGLY ASSOCIATED SNPS
The results summarized so far compare a variety of imputed genotypes with experimentally derived counterparts. However, a more interesting comparison focuses on imputed genotypes that appear to show strong evidence for association, as those might motivate further downstream experiments. To evaluate the accuracy of imputed genotypes for these “strongly associated SNPs,” we compared imputed and experimental genotypes in regions that were only selected for follow-up genotyping after imputation (for example, because imputed genotypes resulted in strong evidence for association but nearby genotyped markers did not). Table II summarizes the comparison of allele frequencies, association test statistics, and individual genotype calls between imputed genotypes and actual genotypes later determined by genotyping. Overall, it is clear that even among these strongly associated SNPs imputation provided accurate estimates of the true P-values. The largest observed discrepancies were for rs17384005, rs11646114, and rs4812831, which were also the three markers for which our imputation approach estimated lower r2 with actual genotypes. Imputation is particularly useful because it allows evidence for association at SNPs with no reliable proxies to be evaluated more accurately. For instance, after imputation, average r2 increased from 0.22 to 0.66 in the set of SNPs whose best genotyped proxy had r2<0.30 and from 0.33 to 0.75 in the set of SNPs whose best genotyped proxy had r2<0.5 [for specific examples of disease susceptibility loci that would be missed without imputation, see Li et al., 2009b].
TABLE II.
Comparison of imputed and experimental genotypes for a subset of SNPs showing strong association in FUSION [Scott et al., 2007]
| SNP | FUSION allele frequency |
Analysis of imputed data |
Analysis of actual genotypes |
Max. r2 w/GWAS SNPs |
Imputed vs. Actual genotypes, r2 |
Observed allelic concordance |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Imputed | Genotyped | P-value | OR | P-value | OR | Actual | Estimated | |||
| rs12910827 | 0.035 | 0.033 | 2.5 × 10−6 | 2.57 | 6.3 × 10−6 | 2.20 | 0.39 | 0.843 | 0.720 | 0.994 |
| rs1449725 | 0.579 | 0.573 | 5.3 × 10−6 | 1.33 | 1.1 × 10−5 | 1.31 | 0.90 | 0.965 | 0.977 | 0.990 |
| rs17081352 | 0.075 | 0.078 | 7.3 × 10−6 | 1.70 | 5.5 × 10−6 | 1.68 | 0.87 | 0.989 | 0.954 | 1.000 |
| rs11616188 | 0.502 | 0.545 | 1.5 × 10−5 | 1.40 | 4.8 × 10−5 | 1.27 | 0.27 | 0.755 | 0.585 | 0.919 |
| rs10837766 | 0.138 | 0.152 | 1.5 × 10−5 | 1.49 | 8.6 × 10−5 | 1.40 | 0.46 | 0.822 | 0.930 | 0.975 |
| rs11036627 | 0.080 | 0.071 | 1.7 × 10−5 | 1.67 | 1.9 × 10−5 | 1.66 | 0.75 | 0.876 | 0.901 | 0.987 |
| rs17384005 | 0.175 | 0.149 | 1.9 × 10−5 | 1.84 | .011 | 1.15 | 0.11 | 0.241 | 0.309 | 0.874 |
| rs7750445 | 0.138 | 0.158 | 2.0 × 10−5 | 1.47 | 4.1 × 10−5 | 1.41 | 0.50 | 0.836 | 0.965 | 0.977 |
| rs2267339 | 0.640 | 0.643 | 2.8 × 10−5 | 1.33 | 4.5 × 10−6 | 1.34 | 0.72 | 0.951 | 0.873 | 0.990 |
| rs17356414 | 0.580 | 0.715 | 3.0 × 10−5 | 1.30 | 8.0 × 10−4 | 1.25 | 0.34 | 0.562 | 0.920 | 0.878 |
| rs1800774 | 0.664 | 0.696 | 3.9 × 10−5 | 1.39 | 7.3 × 10−6 | 1.35 | 0.29 | 0.861 | 0.617 | 0.972 |
| rs175200 | 0.476 | 0.479 | 6.6 × 10−5 | 1.28 | 5.5 × 10−5 | 1.28 | 0.85 | 0.989 | 0.976 | 0.997 |
| rs6103716 | 0.371 | 0.371 | 7.3 × 10−5 | 1.28 | 4.8 × 10−5 | 1.29 | 0.33 | 0.996 | 0.978 | 0.999 |
| rs13297268 | 0.059 | 0.062 | 7.5 × 10−5 | 1.72 | 9.0 × 10−5 | 1.65 | 0.28 | 0.973 | 0.916 | 0.998 |
| rs11646114 | 0.119 | 0.092 | 9.1 × 10−5 | 1.66 | .0020 | 1.38 | 0.13 | 0.687 | 0.512 | 0.956 |
| rs2021966 | 0.609 | 0.603 | 9.1 × 10−5 | 1.32 | 2.6 × 10−4 | 1.25 | 0.46 | 0.811 | 0.769 | 0.937 |
| rs1270874 | 0.231 | 0.224 | 1.4 × 10−4 | 1.33 | 3.9 × 10−4 | 1.30 | 0.24 | 0.933 | 0.954 | 0.988 |
| rs4812831 | 0.165 | 0.129 | 1.6 × 10−4 | 1.53 | .0055 | 1.28 | 0.45 | 0.587 | 0.516 | 0.944 |
| rs4402960 | 0.683 | 0.681 | 1.7 × 10−4 | 1.27 | 1.2 × 10−4 | 1.28 | 1.00 | 0.994 | 1.026 | 0.998 |
| rs9402346 | 0.669 | 0.646 | 4.5 × 10−4 | 1.26 | .0014 | 1.22 | 0.62 | 0.881 | 0.915 | 0.965 |
| rs10019985 | 0.629 | 0.619 | 4.8 × 10−4 | 1.25 | 4.2 × 10−4 | 1.25 | 0.66 | 0.990 | 0.953 | 0.998 |
| rs2466291 | 0.579 | 0.618 | 6.3 × 10−4 | 1.26 | .0016 | 1.22 | 0.47 | 0.829 | 0.830 | 0.935 |
| rs1409184 | 0.671 | 0.646 | 8.2 × 10−4 | 1.26 | .0011 | 1.22 | 0.58 | 0.865 | 0.873 | 0.963 |
| rs8079544 | 0.091 | 0.106 | 8.9 × 10−4 | 1.50 | .013 | 1.27 | 0.22 | 0.707 | 0.731 | 0.961 |
| rs1801282 | 0.165 | 0.165 | 9.5 × 10−4 | 1.31 | .0011 | 1.30 | 1.00 | 0.999 | 1.002 | 1.000 |
| rs3802177 | 0.372 | 0.371 | 9.9 × 10−4 | 1.23 | .0012 | 1.22 | 1.00 | 0.996 | 1.015 | 0.999 |
The table shows a comparison of the results from analysis of imputed data with results for actual genotyping for a subset of the SNPs that reached a P-value of <10−3 in our analysis of the FUSION data. Successive columns include SNP name, estimated allele frequency in FUSION cases and controls, using either imputed data or actual genotype data, P-value and odds ratio for association test comparing allele frequencies in cases and controls using imputed genotypes, P-value and odds ratio for association test comparing allele frequencies in cases and controls using experimentally derived genotypes, r2 between the best single marker tag in the GWAS panel and this SNP, r2 between imputed and observed genotypes (actual r2 and estimated from our method as a measure of imputation quality) and, finally, proportion of alleles matching between imputed and actual genotypes. Note that because these are all imputed SNPs that show strong association in the FUSION data, they are subject to a “winner’s curse” effect. Thus, SNPs where imputation resulted in inflated P-values were more likely to be selected for follow-up in this analysis. Not all imputed SNPs showing association at this significance level were genotyped experimentally. Rather, a subset of SNPs was selected for genotyping either because (a) they showed substantially stronger evidence for association than other nearby genotyped SNPs and stronger evidence for association than nearby imputed SNPs or (b) they were selected to improve coverage of the genome in and around 222 candidate genes [Gaulton et al., 2008]. All SNPs with a P-value <10−3 in the imputed data and which were subsequently genotyped are tabulated.
USING IMPUTATION TO ESTIMATE PAIRWISE DISEQUILIBRIUM
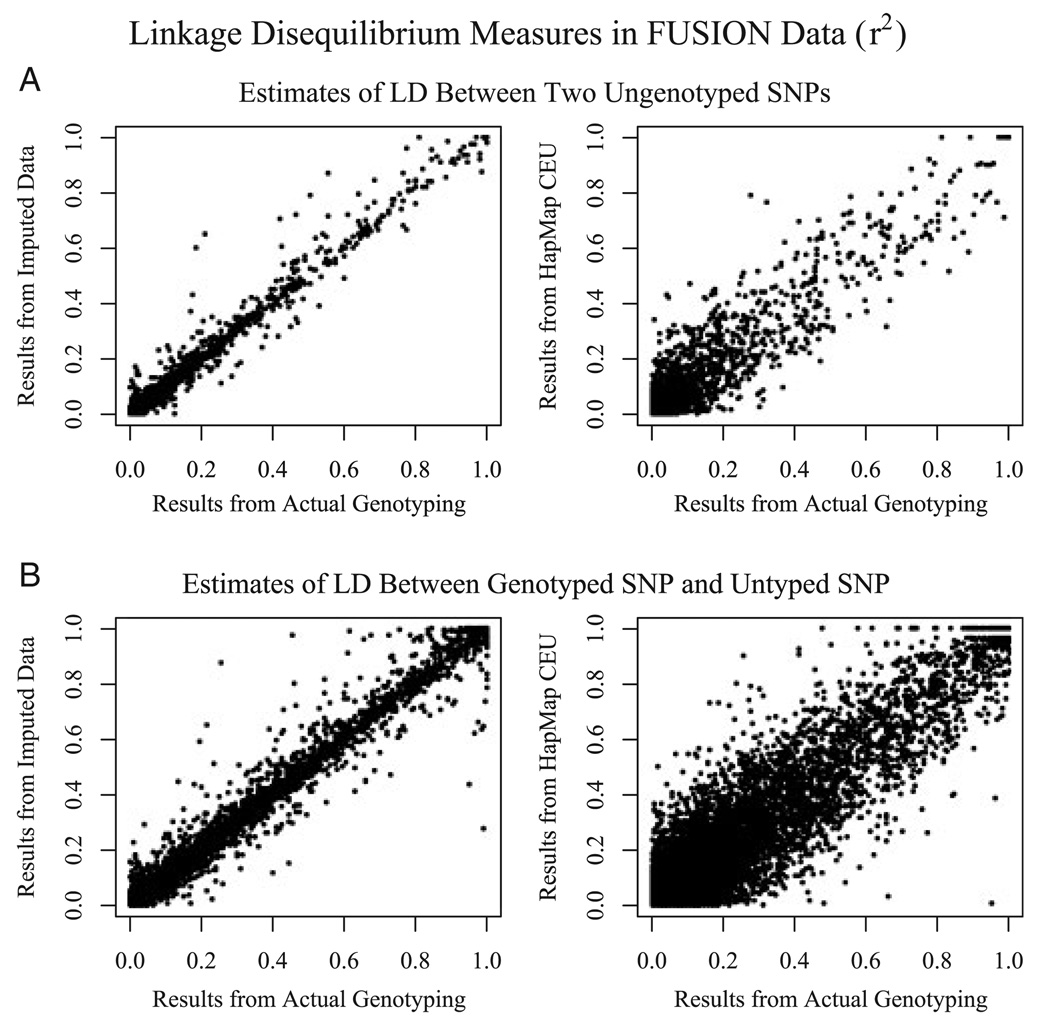
Remarkably, we observed that imputed genotypes could also be used to obtain very accurate estimates of LD between pairs of untyped markers, or of LD between a genotyped marker and an untyped marker. As shown in Figure 2, estimates of LD between two SNPs obtained using imputed data are much closer to the results obtained by actually genotyping the two SNPs than estimates obtained by looking up the two markers in the HapMap CEU database (Supplementary Figure 2 shows a similar comparison for D’ estimates). Even with some imprecision in estimates of individual genotypes, the increased sample size compensates to reduce variation in the estimated LD measures.
Fig. 2.
Imputation improves quality of LD estimates. For imputed SNPs on chromosome 14, the figure compares estimates of LD obtained by genotyping both SNPs (“Results from Actual Genotyping,” X axis) with estimates of LD obtained by imputing genotypes for both SNPs using markers on the 317K marker chip (“Results from Imputed Data,” Y axis, Top left), obtained by imputing genotypes for one of the SNPs (“Results from Imputed Data,” Y axis, Bottom Left) or obtained from the HapMap CEU panel (“Results from HapMap CEU,” Y axis, Top and Bottom Right).
COMPARISON OF DIFFERENT GENOTYPING PLATFORMS
Our experience with the FUSION GWAS, summarized above, shows that imputation can be an effective way to estimate unobserved genotypes and/or allele counts. These genotypes can then be used in a variety of downstream analyses, including logistic regression analyses for discrete traits and linear regression analyses for quantitative traits, and to facilitate meta-analysis of studies based on different platforms. A key issue when considering imputation-based approaches is whether similarly accurate estimates of unobserved data points can be obtained with different genotyping panels or in different populations [Clark and Li, 2007], and to evaluate this we conducted two additional experiments.
In the first experiment, we used genotype data generated by the International HapMap Consortium. We considered each of the HapMap samples in turn and masked available genotypes so as to mimic an experiment using one of several commercially available chips. For example, to evaluate the Affymetrix 500K SNP chip, we marked genotypes for all markers that are not on the chip as missing for the individual being considered. We then used haplotypes for the remaining individuals on the same HapMap analysis panel (either YRI, CEU, or JPT+CHB) to impute the missing genotypes. The results are summarized in Table III and clearly show that a large number of SNPs can be imputed very accurately using any of the commercially available panels (e.g. with r2>0.80 to experimental genotypes) and that, compared to relying on single marker tagging, imputation results in improved coverage of the genome.
TABLE III.
Coverage of the Phase II HapMap with commercial genotyping panels, before and after imputation
| Coverage by single-marker tags | Coverage by imputed SNPs | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # Panel SNPs | # Imputed SNPs | MAF<5% | MAF≥5% | MAF<5% | MAF≥5% | |||||||||
| Used | Lost | MAF<5% | MAF≥5% | r2 | r2>0.8 | r2 | r2>0.8 | r2 | r2>0.8 | Error (%) | r2 | r2>0.8 | Error (%) | |
| CEU | ||||||||||||||
| A100 | 100,844 | 1,609 | 259,261 | 2,086,690 | 0.36 | 0.22 | 0.50 | 0.31 | 0.47 | 0.32 | 1.80 | 0.63 | 0.46 | 7.85 |
| A250S | 195,864 | 4,393 | 251,807 | 2,002,214 | 0.48 | 0.31 | 0.65 | 0.44 | 0.61 | 0.46 | 1.33 | 0.79 | 0.65 | 4.12 |
| A250N | 216,747 | 4,836 | 250,364 | 1,983,146 | 0.50 | 0.34 | 0.67 | 0.48 | 0.63 | 0.49 | 1.26 | 0.80 | 0.68 | 3.94 |
| A500 | 412,611 | 9,229 | 234,049 | 1,809,352 | 0.61 | 0.44 | 0.77 | 0.61 | 0.73 | 0.60 | 0.93 | 0.89 | 0.82 | 2.12 |
| A1000 | 676,182 | 87,766 | 209,636 | 1,580,321 | 0.71 | 0.57 | 0.86 | 0.76 | 0.79 | 0.68 | 0.73 | 0.93 | 0.91 | 1.23 |
| I300 | 305,050 | 3,115 | 267,573 | 1,871,586 | 0.30 | 0.08 | 0.84 | 0.74 | 0.70 | 0.54 | 1.08 | 0.93 | 0.90 | 1.39 |
| I550 | 513,779 | 238 | 254,183 | 1,681,501 | 0.59 | 0.40 | 0.90 | 0.85 | 0.79 | 0.67 | 0.76 | 0.95 | 0.94 | 0.90 |
| I650 | 578,864 | 14,627 | 244,431 | 1,630,298 | 0.66 | 0.48 | 0.91 | 0.86 | 0.80 | 0.68 | 0.72 | 0.95 | 0.94 | 0.88 |
| I1000 | 779,800 | 130,014 | 225,439 | 1,456,134 | 0.70 | 0.54 | 0.93 | 0.89 | 0.81 | 0.71 | 0.67 | 0.96 | 0.95 | 0.73 |
| YRI | ||||||||||||||
| A100 | 100,627 | 3,223 | 326,772 | 2,320,439 | 0.21 | 0.08 | 0.33 | 0.14 | 0.35 | 0.18 | 2.22 | 0.50 | 0.26 | 10.32 |
| A250S | 210,242 | 4,698 | 318,680 | 2,220,904 | 0.30 | 0.13 | 0.47 | 0.22 | 0.50 | 0.29 | 1.73 | 0.69 | 0.44 | 6.05 |
| A250N | 231,026 | 4,971 | 317,321 | 2,201,821 | 0.32 | 0.15 | 0.49 | 0.26 | 0.53 | 0.33 | 1.64 | 0.71 | 0.49 | 5.68 |
| A500 | 441,268 | 9,669 | 300,455 | 2,013,203 | 0.41 | 0.21 | 0.60 | 0.36 | 0.65 | 0.46 | 1.24 | 0.83 | 0.69 | 3.30 |
| A1000 | 737,369 | 91,811 | 275,794 | 1,749,271 | 0.54 | 0.31 | 0.73 | 0.54 | 0.74 | 0.58 | 0.92 | 0.90 | 0.83 | 1.91 |
| I300 | 271,991 | 15,346 | 315,631 | 2,163,803 | 0.33 | 0.15 | 0.52 | 0.26 | 0.60 | 0.39 | 1.42 | 0.79 | 0.60 | 3.97 |
| I550 | 474,049 | 19,355 | 301,391 | 1,981,088 | 0.42 | 0.20 | 0.68 | 0.46 | 0.70 | 0.51 | 1.09 | 0.88 | 0.80 | 2.13 |
| I650 | 573,953 | 28,487 | 300,785 | 1,881,962 | 0.46 | 0.23 | 0.75 | 0.56 | 0.73 | 0.55 | 0.98 | 0.90 | 0.85 | 1.72 |
| I1000 | 788,503 | 149,152 | 274,766 | 1,702,039 | 0.53 | 0.30 | 0.78 | 0.60 | 0.76 | 0.59 | 0.88 | 0.92 | 0.88 | 1.47 |
| JPT+CHB | ||||||||||||||
| A100 | 95,521 | 1,994 | 299,643 | 1,919,001 | 0.35 | 0.22 | 0.47 | 0.28 | 0.44 | 0.32 | 1.68 | 0.60 | 0.42 | 8.86 |
| A250S | 186,411 | 4,368 | 290,265 | 1,840,510 | 0.49 | 0.33 | 0.63 | 0.42 | 0.58 | 0.45 | 1.23 | 0.76 | 0.61 | 5.00 |
| A250N | 205,274 | 4,713 | 288,661 | 1,823,236 | 0.51 | 0.36 | 0.65 | 0.46 | 0.59 | 0.48 | 1.17 | 0.77 | 0.64 | 4.70 |
| A500 | 391,685 | 9,081 | 268,427 | 1,663,552 | 0.62 | 0.47 | 0.76 | 0.60 | 0.69 | 0.59 | 0.84 | 0.87 | 0.80 | 2.60 |
| A1000 | 638,817 | 86,838 | 239,528 | 1,455,644 | 0.72 | 0.59 | 0.85 | 0.75 | 0.75 | 0.67 | 0.65 | 0.92 | 0.89 | 1.50 |
| I300 | 274,751 | 12,851 | 287,456 | 1,755,289 | 0.54 | 0.38 | 0.75 | 0.58 | 0.69 | 0.57 | 0.88 | 0.88 | 0.82 | 2.25 |
| I550 | 467,073 | 13,322 | 269,299 | 1,587,153 | 0.67 | 0.52 | 0.87 | 0.78 | 0.75 | 0.67 | 0.64 | 0.93 | 0.91 | 1.26 |
| I650 | 531,807 | 23,155 | 259,962 | 1,534,915 | 0.71 | 0.57 | 0.88 | 0.80 | 0.76 | 0.69 | 0.61 | 0.94 | 0.92 | 1.19 |
| I1000 | 728,837 | 136,560 | 239,252 | 1,365,519 | 0.73 | 0.61 | 0.90 | 0.85 | 0.78 | 0.70 | 0.57 | 0.95 | 0.94 | 0.91 |
For each platform, the table lists the number of SNPs in the platform that overlap with the phased HapMap chromosomes (release 21a). The number of SNPs that were not in the phased HapMap are also listed, most of these were monomorphic. This number is followed by the number of SNPs that we attempted to impute, either with minor allele frequency <5% or >5%. We did not attempt to impute singletons. Coverage statistics using conventional single-marker tagging are provided and refer to the maximum r2 between a HapMap SNP not on the panel and its best tag on the panel. Coverage statistics using imputation are also tabulated and refer to the relationship between imputed genotypes for each SNP and true genotypes for the same SNP. To evaluate the coverage of each genotyping platform using imputation, we focused on the markers that overlapped between the platform and the Phase II HapMap. We then considered each HapMap founder in turn and masked all genotypes for all markers not present in the commercial platform being evaluated. Finally, we used the remaining (unmasked) genotypes together with haplotypes for the other HapMap founders to impute the masked genotypes. The proportion of alleles that were imputed incorrectly, together with the correlation between imputed genotypes and actual genotypes are tabulated for each platform.
Depending on the commercial panel and population being investigated, coverage of HapMap SNPs (proportion of SNPs with r2>0.80) increased by 10–30% for low MAF alleles (MAF<5%) and by 10–20% for more common alleles (MAF>5%). In agreement with this result, the average r2 between each untyped SNP and imputed genotypes was up to 40% higher on average when using imputed genotypes than when using the best available single marker proxy. Imputation remained valuable even for panels with ~1 million directly genotyped SNPs. In practice, the results shown in Table III are likely to represent an upper bound on the performance of our method in real settings, because additional errors will result from discrepancies in genotyping protocols between individual laboratories and the HapMap and from differences in LD patterns between the HapMap and the samples being studied. Nevertheless, they suggest our method is likely to be helpful for a variety of currently available commercial SNP panels.
IMPUTATION IN DIVERSE POPULATIONS
In a second experiment, we evaluated the performance of our method in 927 samples from 52 populations in the Human Genome Diversity Project (HGDP). In a previous evaluation of tag SNP portability, these 927 samples were genotyped for 1,864 SNPs in 32 autosomal regions (average minor allele frequency 0.15–0.24, depending on population) [Conrad et al., 2006]. The regions were selected to represent regions of high and low LD across the genome. Each region spanned ~330 kb, including a central “core” region of ~90 kb, where ~60 SNPs were attempted, and two ~120 kb flanking regions on either side, where ~12 SNPs were attempted. To evaluate the performance of genotype imputation across these diverse populations, we selected a thinned marker set including 872 SNPs spaced ~10 kb apart across all 32 regions. We then used these SNPs to impute genotypes for the remaining 992 SNPs and evaluated our approach.
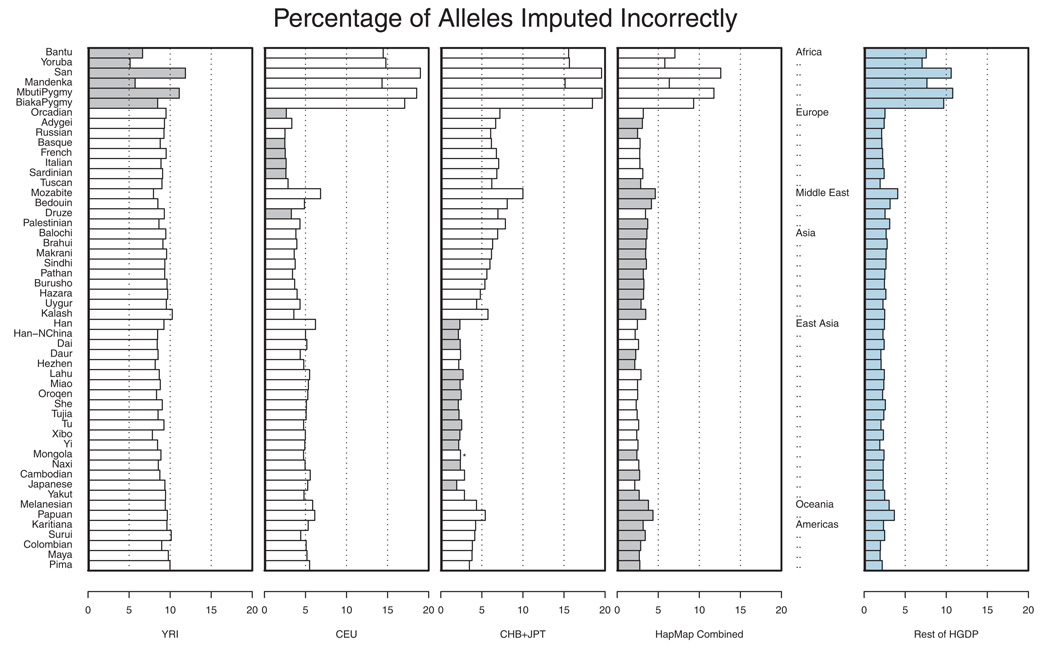
Figure 3 shows the proportion of incorrectly imputed alleles in each of the populations. Results are presented using a single HapMap analysis panel as a reference (either the CEU, YRI, or CHB+JPT) or using all HapMap samples as a larger reference panel. For each of the populations, the reference panel that resulted in the smallest overall error rate is highlighted. Overall, African samples were the most difficult to impute, with error rates ranging between 5.13% for the Yoruba and 11.86% for the San when the HapMap YRI panel was used as a reference. In other parts of the world, we generally observed that the HapMap CEU provided a good reference panel for European populations and that the HapMap CHB+JPT provided a good reference panel for East Asian populations, resulting in error rates of <3.34 and <2.89%, respectively. Outside Europe and East Asia, when imputation was applied to populations from the Middle East, Central and South Asia, the Americas or Oceania, it was generally better to use the combined HapMap sample as a reference than to use any single HapMap analysis panel as a reference. It is interesting to note that, in all cases, combining the three HapMap panels into a single reference set was either the best option or the second best option. Furthermore, in situations where this combined reference panel reduced imputation accuracy, it resulted in an average increase of only 0.15% in error rates. Our results are consistent with those of Huang et al. [Huang et al., 2009] who showed, in a smaller subset of HGDP populations and a different set of genotyped SNPs, that combined reference panels could outperform panels that included only one population. The figure also illustrates that, when a large number of individuals are genotyped in study samples, it may be possible to bypass the HapMap reference panel altogether. In the last panel, rather than using the HapMap genotypes to impute missing data, we used a combined dataset including all other HGDP populations.
Fig. 3.
Evaluation of imputation accuracy across HGDP panels. For each of 52 populations in the Human Genome Diversity Panel (HGDP) a set of 872 SNPs distributed evenly across 32 regions, each ~330 kb in length, was used to impute 992 other SNPs. The 992 imputed SNPs were located near the middle of each imputed region. Imputation was done using either the HapMap YRI, CEU, CHB+JPT, or a combination of three HapMap panels (first four panels, best panel is shaded in gray) or using the remaining HGDP samples as a reference. In each case, the proportion of correctly imputed alleles is tabulated. The figure is based on a re-analysis of data of Conrad et al. [2006].
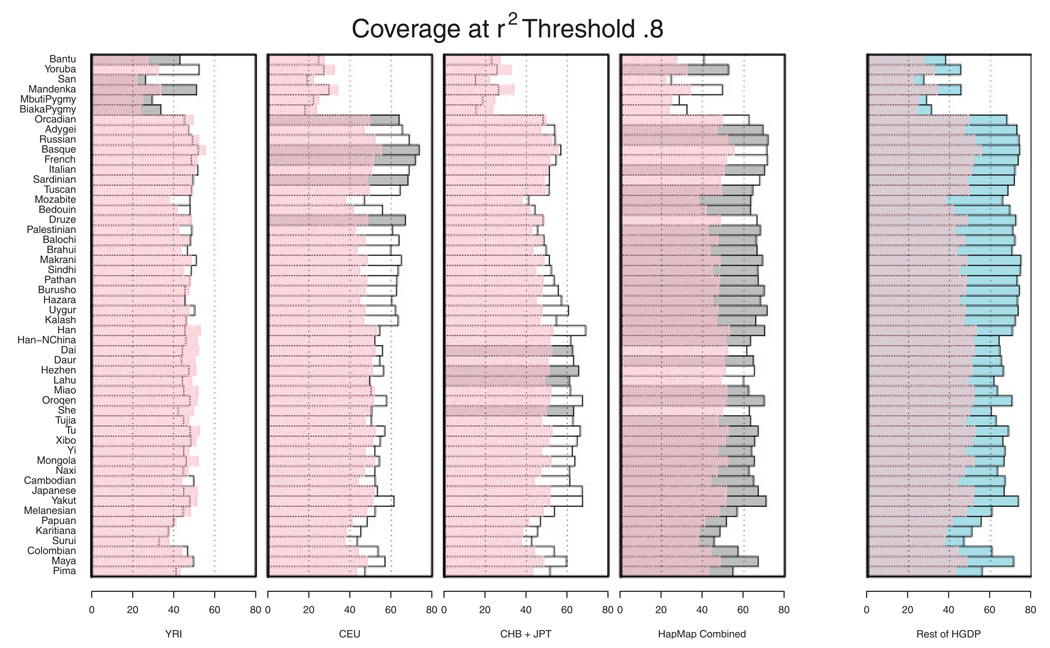
Figure 4 focuses on the estimated r2 between imputed and observed allele counts. In each stripe, accuracy of imputation is assessed using a different reference panel. Superimposed in pink is the coverage that would be provided by single marker tagging approaches. Broadly, it is clear that imputation using an appropriate reference panel will improve coverage. Using an inappropriate reference panel (for example using the HapMap CEU to impute genotypes for one of the African populations), can result in imputed genotypes and allele counts that are not as strongly correlated with the true genotypes as the best available single marker tag but, even then, the loss appears to be small. Importantly—in all cases—combining the three HapMap panels resulted in substantial improvements in coverage over single marker tagging—suggesting that this might be a cautious approach when the choice of reference panel is unclear. Combining the three HapMap panels is also a good choice for genotype imputation in admixed populations [Mathias et al., 2010] where, depending on the ancestry of each stretch of the genome, the best matching haplotype will likely originate from a different HapMap reference panel. Our conclusion that the combined panel is a sensible reference for all populations facilitates practical decision making on the choice of reference panel. The conclusion is also supported by Huang et al. [Huang et al., 2009]. Although their aim was to find an optimal population-specific reference panel for each HGDP sample, their Figure 6 shows that a combined panel, including all HapMap haplotypes is the best compromise choice, in the sense that it performs almost optimally in each of the 39 HGDP populations examined. In the future, we expect that imputation methods that weigh the different reference panels could further improve imputation quality.
Fig. 4.
Evaluation of imputation accuracy across HGDP panels. Genotypes for a set of 992 SNPs were imputed in the HGDP and then compared with actual genotypes. For each pair of true and imputed genotypes an r2 coefficient was calculated and averaged for each population. The best set of HapMap reference individuals for each population is shaded. The coverage obtained by using the best available tag SNP (rather than imputed genotypes) is overlaid in pink. See Figure 3 legend for further details.
IMPACT OF IMPUTATION ON POWER OF ASSOCIATION STUDIES
Our evaluation of imputed genotypes in the FUSION, HapMap, and HGDP samples clearly shows that imputation can be very accurate in a variety of populations. In this way, we believe it will be an important tool for combining results across studies that rely on different marker panels. To investigate whether using imputed genotypes might also improve power in individual studies, we carried out a simulation experiment. As previously described [Schaffner et al., 2005], we simulated 10,000 chromosomes for a series of 1 Mb regions. Within each region, simulated LD patterns mimicked the HapMap CEU or YRI [Schaffner et al., 2005]. We then used a subset of 120 simulated chromosomes to generate a region specific “HapMap.” As described in the methods, we then picked the minor allele for a randomly selected polymorphic site in each region as the “disease susceptibility allele” and simulated a set of 500 case and 500 control individuals using the remaining chromosomes. The susceptibility allele varied in frequency between 2.5 and 50%, with larger simulated effect sizes assigned to rarer alleles to ensure comparable power in a hypothetical fully genotyped sample. We also simulated 2,000 datasets where the disease allele had no effect to calibrate region-wide type I error rates for each approach.
To analyze each region, we thinned SNPs in the simulated HapMap to match the density and allele frequency spectrum of the Phase II HapMap [The International HapMap Consortium, 2007]. Using the thinned data, we selected a panel of 100 tag SNPs for each region that included the 90 tag SNPs with the largest number of proxies and 10 additional SNPs selected at random among the remaining tags. This approach resulted in panels that captured ~78% of the common variants (MAF>5%) in the simulated CEU HapMap, similar to the real life performance of the Illumina 317K SNP genotyping chip. Finally, we analyzed each of the simulated datasets using the selected marker panel and one of three analysis strategies: (a) single marker chi-squared association tests, (b) single and multi-marker association tests [Pe’er et al., 2006] as suggested by the PLINK [Purcell et al., 2007] program based on the simulated HapMap, or (c) tests using imputed allele counts for all the markers in the simulated HapMap. Results are summarized in Table IV. The first row in the table shows the significance thresholds used for each analysis (since approaches (b) and (c) both increase the total number of tests, note that the P-value threshold increases slightly when multi-marker tests are used and increases further when imputation is used). Subsequent rows summarize power for markers of different allele frequencies. In populations with strong LD, it is clear that for common susceptibility alleles the single marker tests provide high power and that imputation or multi-marker analyses provide only small gains in power. However, for rarer alleles (such as those with frequencies <5%), imputation can provide dramatic increases in power. For instance, power increased from 24.4 to 56.2% when the disease allele frequency was 2.5% and imputation was used in the panel with CEU-like LD. As large genome scans and meta-analyses that are well-powered to evaluate rarer variants with modest effects are completed, we believe that imputation will become an increasingly important primary analysis and there are now examples of confirmed disease susceptibility loci that would have been missed without genotype imputation [Li et al., 2009b].
TABLE IV.
Imputed genotypes result in increased power
| Power (LD mimics CEU) | Power (LD mimics YRI) | |||||
|---|---|---|---|---|---|---|
| Single marker tags |
Multi marker tags |
Imputed genotypes |
Single marker tags |
Multi marker tags |
Imputed genotypes |
|
| Empirical P-value threshold | 0.00081 | 0.00071 | 0.00030 | 0.00067 | 0.00067 | 0.00017 |
| MAF = 2.5% | 24.4% | 25.0% | 56.2% | 21.2% | 22.6% | 43.6% |
| MAF = 5% | 55.8% | 56.4% | 74.0% | 35.6% | 36.0% | 55.0% |
| MAF = 10% | 77.4% | 78.4% | 87.8% | 62.4% | 63.8% | 73.0% |
| MAF = 20% | 85.6% | 86.2% | 91.4% | 68.8% | 70.6% | 78.2% |
| MAF = 50% | 93.0% | 93.6% | 96.4% | 75.4% | 77.4% | 86.6% |
The table summarizes results from the analysis of two sets of 100 simulated 1Mb regions. For each region, we generated a simulated HapMap including ~200 SNPs and used this panel to pick 100 tag SNPs that provided good coverage of the region (average r2~0.8 in CEU, ~0.6 in YRI). We then simulated and analyzed a series of case control studies, each with 500 cases and 500 controls. Association tests were carried out at each tag SNP marker (“Single Marker Tags”), initially. These results were then augmented with the analysis of multi-marker tags as suggested by PLINK [Pe’er et al., 2006; Purcell et al., 2007] (“Multi Marker Tags”) or with the analysis of imputed genotypes (“Imputed Genotypes”). In each case, we first simulated and analyzed 2,000 null (20 per region) datasets by assigning a pair of random chromosomes to each case and control. These analysis were used to establish the empirical P-value threshold that, when applied to the most significant result in each region, resulted in a type I error rate of 5%. Then, for each tabulated minor allele frequency (MAF), we simulated 500 case-control datasets (5 per region, 500 cases and 500 controls each) where a variant with the specified MAF was associated with susceptibility. Power refers to the proportion of replicates where the top P-value exceeds the empirical P-value threshold. Note that the susceptibility variant was picked at random among all simulated SNPs with the requisite MAF and was not necessarily included in the tag SNP set or in the markers ascertained for each region specific HapMap. To ensure comparable power across varying MAF, we increased genotype relative risk for rarer SNPs. Specifically, we set GRR = 2.500, 2.020, 1.715, 1.530, and 1.440 for SNPs with MAF = 2.5, 5, 10, 20 and 50%, respectively. These settings correspond to 80% power for single marker tests when the susceptibility variant is typed and a P-value threshold of 0.0005 is used (0.05/100, corresponding to a Bonferroni threshold that assumes 100 independent SNPs are tested).
PRACTICAL CONSIDERATIONS
A key ingredient for any imputation-based approach is to ensure that alleles are consistently labeled across studies. In our evaluation of FUSION and HGDP samples, using the HapMap as a reference, we were fortunate that a subset of the HapMap individuals were genotyped in each study for quality control. Contrasting the genotypes for these quality control samples with those generated by the HapMap Consortium made the usually laborious process of ensuring consistent allele labeling across labs much easier. We strongly recommend that all labs conducting GWAS genotype a small number of HapMap individuals for this purpose.
Another practical consideration arises when integrating data from studies that use diverse genotyping platforms. Superficially, it is tempting to first impute missing genotypes in each sample and to then conduct a pooled analysis of all available data. However, this is almost never a good idea, as illustrated by a particularly extreme case where a set of cases and controls have been genotyped on two different platforms and a marker of interest has been genotyped in cases but must be imputed in controls. If the marker of interest cannot be well predicted by flanking markers, imputation will default to suggesting that the genotype distribution at that marker matches the reference panel—but this could be a very poor assumption if the reference panel and study sample have drifted apart, potentially resulting in spurious association. Even if the marker can be well predicted by flanking markers, it is possible that the reference panel and the case sample used different genotyping assays that, for technical reasons such as the presence of a polymorphism that overlaps assay primers, give consistently distinct results—again resulting in spurious association. To avoid these sources of spurious association, we recommend that, when analyzing genotype data generated using different platforms, different versions of the same platform, or using the same platform but with experiments carried out at different labs, an initial round of association analysis should be carried out using data from each platform/version/site combination. The results from this initial round of analysis can then be meta-analyzed, minimizing the risk of artifacts. This recommendation does preclude analyses where all cases are genotyped at one site, and all controls are genotyped at a different site.
In the experiments described so far, we illustrated the accuracy of genotype imputation that relies on existing resources (such as the Phase II HapMap) and genotyping technologies (including a variety of commercial genotyping chips). It is likely that both these resources and technologies will continue to evolve rapidly and it is interesting to consider how these developments might impact imputation-based approaches. For example, it is clear that genotyping chips of the future will be able to examine an ever larger number of tag SNPs in a cost-effective manner. Extrapolating from Table III, it is clear these should provide improved genomic coverage, eventually allowing investigators to impute nearly all HapMap SNPs with near perfect accuracy. Nevertheless, it is also clear from Table III that when coupled with imputation-based analyses even relatively low-density SNP chips can provide excellent coverage of the genome in populations with LD patterns similar to the CEU, JPT, and CHB. Thus, we expect the main advantages of new higher-density chips will be in the study of populations with less extensive LD, such as the YRI, and in the analysis of rarer variants.
THE FUTURE: LARGER REFERENCE PANELS
Another interesting possibility to consider is the impact of larger HapMap reference panel on imputation or, similarly, the utility of using extra genotype data on a subset of individuals in a study to aid imputation in the remaining individuals in the study. To evaluate these possibilities, we generated a reference panel with varying numbers of Finnish individuals (between 30 and 500, see Table V) and used these reference panels to impute genotypes for 521 SNPs in an independent set of 500 individuals from the FUSION study of type II diabetes. Imputation accuracy and genomic coverage increase noticeably with the larger reference panels, with overall discrepancy rates between typed and untyped alleles as low as 0.40% when a reference panel of 500 unrelated individuals is available. One of the reasons for this increase in accuracy is that the length of haplotypes shared between individuals in the reference panel and those in the study sample increases gradually as the size of the reference panel increases. For example, mosaic fragments used to reconstitute the FUSION samples using the individuals in the 500-sample reference panel were slightly >1 Mb long on average. These long stretches are easier for our Markov model to identify and are also likely to descend from a more recent common ancestor. This means they will have undergone fewer rounds of gene conversion and mutation, which gradually erode haplotype similarities and reduce the quality of our imputed genotypes. Overall, our results suggest that either genotyping a number of the study samples for markers of interest or increasing the size of the public reference panels will greatly improve the quality of genotype imputation.
TABLE V.
Effect of increasing reference panel size on imputation accuracy
| Genotypic matching error rates | |||||||
|---|---|---|---|---|---|---|---|
| # Reference panel size |
Major allele homozygote (%) |
Heterozygote (%) |
Minor allele homozygote (%) |
Overall (%) | Allelic matching error (%) |
Mean r2 (%) |
Median r2 (%) |
| All SNPs | |||||||
| 30 | 2.18 | 16.43 | 16.27 | 5.56 | 2.95 | 84.7 | 94.6 |
| 60 | 1.23 | 7.99 | 8.44 | 2.54 | 1.31 | 91.5 | 97.5 |
| 100 | 0.95 | 4.31 | 5.48 | 1.73 | 0.88 | 93.6 | 98.2 |
| 200 | 0.56 | 3.17 | 3.25 | 1.03 | 0.52 | 96.1 | 98.8 |
| 500 | 0.46 | 1.96 | 2.41 | 0.79 | 0.40 | 97.1 | 99.1 |
| Best single SNP tag r2<0.5 | |||||||
| 30 | 3.54 | 46.55 | 43.96 | 11.00 | 5.94 | 63.5 | 66.2 |
| 60 | 2.09 | 28.29 | 25.64 | 5.73 | 2.99 | 76.4 | 82.5 |
| 100 | 1.93 | 14.17 | 19.32 | 3.89 | 2.00 | 82.0 | 87.9 |
| 200 | 1.35 | 11.96 | 13.19 | 2.68 | 1.37 | 87.5 | 91.3 |
| 500 | 1.04 | 6.76 | 9.06 | 2.00 | 1.02 | 91.0 | 94.6 |
| MAF≥20% | |||||||
| 30 | 3.18 | 10.14 | 11.92 | 6.96 | 3.74 | 85.4 | 94.7 |
| 60 | 1.83 | 4.03 | 5.50 | 3.13 | 1.62 | 92.7 | 97.7 |
| 100 | 1.32 | 2.75 | 3.63 | 2.15 | 1.10 | 95.2 | 98.7 |
| 200 | 0.82 | 1.68 | 2.36 | 1.30 | 0.66 | 97.0 | 98.9 |
| 500 | 0.69 | 1.31 | 1.60 | 1.01 | 0.51 | 97.7 | 99.2 |
| MAF>20% | |||||||
| 30 | 1.07 | 23.47 | 21.80 | 3.99 | 2.06 | 83.9 | 94.3 |
| 60 | 0.56 | 12.43 | 12.16 | 1.88 | 0.96 | 90.0 | 96.7 |
| 100 | 0.54 | 6.05 | 7.84 | 1.26 | 0.64 | 91.5 | 97.4 |
| 200 | 0.26 | 4.83 | 4.38 | 0.72 | 0.36 | 94.9 | 98.7 |
| 500 | 0.20 | 2.68 | 3.44 | 0.54 | 0.27 | 96.4 | 98.9 |
To evaluate the impact of a larger reference panel on the accuracy of genotype imputation, we used different numbers of individuals from the FUSION study genotype for markers on the Illumina 317K SNP chip and also 521 SNPs on a candidate region of chromosome 14 [Willer et al., 2006] to impute genotypes for an independent set of 500 FUSION individuals on whom only the Illumina 317K SNP chip genotypes were available. The imputation procedure converged after ~300 rounds with panel size = 30 and 60, ~200 rounds with panel size = 100, and <100 rounds for panel sizes = 200 or 500 individuals. Imputed genotypes were compared with experimental genotypes to determine accuracy at the genotype and allele level and to evaluate the r2 between true and imputed genotypes. Imputation accuracy measures are broken down by relatively common and rare SNPs using a MAF (minor allele frequency cutoff of 20%) and imputation error rates at the genotypic level are evaluated both overall and by genotype categories.
THE FUTURE: COMBINING IMPUTATION WITH NEW SEQUENCING TECHNOLOGIES
With the rapid development of very high-throughput re-sequencing technologies [Bentley, 2006], it is oft proposed that genotyping-based approaches will soon become outdated. Re-sequencing-based approaches capture variants that are absent from public databases including, potentially, population specific variants. Our haplotyping approach can use whole genome re-sequencing data as input. In this setting, it uses information from individuals with similar haplotypes to reconstruct patterns of variation in regions where deep coverage is not available. In principle, the approach could be useful to help describe regions that, due to chance, are poorly covered in a particular sequencing experiment or to allow for economical evaluation of many individuals. To evaluate the possibilities, we simulated data for ten 1 Mb regions and simulated shotgun sequence data for each region. We simulated reads that were only 32 base pairs long and with a per base-pair error rate of 0.2%. Very roughly, these correspond to the performance of early versions of next generation re-sequencing technologies; newer versions of these technologies can generate longer and more accurate reads and should thus outperform the simulations presented here. We then re-sequenced between 100 and 400 individuals at different depths and used our approach to reconstruct haplotypes and genotypes for each individual. Note that the simulated reads are typically too short to include useful information on phase (because they will generally include only zero or one sites that truly differ from the reference sequence). In addition, given the large number of bases examined, they will also suggest a large number of false polymorphic sites. To control false-positive variant calls, it is imperative to confirm true polymorphic sites either by examining overlapping similar reads from the same individual or, potentially, from other individuals who share a similar haplotype.
For each site, we counted the number of times that the reference base or an alternative base was sequenced for each individual. For computational convenience, we only considered sites where both bases were observed several times (see Appendix for detailed methods and implementation details) in downstream analyses and assigned the most frequently sampled base to all other sites. On this scale, the shotgun re-sequencing approach typically characterized ~4,000 polymorphic sites across the sampled individuals - ~4 × the SNP density of the Phase II HapMap. Even relatively light shotgun re-sequencing provided very accurate haplotypes for each individual. For example, when 400 individuals were sequenced at 4 × depth, there were only 18.97 errors per individual on average (over 1,000,000 base-pairs). Across ~980,000 sites that were monomorphic in the population only 82 false polymorphisms were called on average. Accuracy was also excellent at sites that were polymorphic in the population. For example, 3,558 of the 3,641 simulated polymorphic sites with MAF>0.5% were identified and, at these sites, alleles were called with an accuracy of 99.93% (see Tables VI and VII). For any given depth, imputed accuracy increased with the number of sequenced individuals (for example, accuracy at sites with MAF >0.5% was ~98.8% when 100 individuals were sequenced at 2 × coverage but increased to ~99.7% when 400 individuals were sequenced at the same depth; the number of errors per individual decreased similarly from 106.3 per individual to 40.3 per individual). In addition, the depth required to achieve a given accuracy decreased as the number of sequenced individuals increased: achieving 99.9% accuracy for sites with population MAF >0.5% requires ~8 × depth for 100 individuals, ~6 × depth in 200 individuals and only 4 × depth in 400 individuals. In each case, note that error rates are higher at heterozygous sites than at homozygous sites. Again, performance of the approach with larger numbers of individuals improves because the mosaic fragments described by our model increase in length and, thus, become easier to find. This is also reflected in the accuracy of estimated haplotypes, which—when compared with simulated haplotypes—have ~1 switch per 50 kb when 100 individuals are examined, but ~1 switch per 500 kb when 400 individuals are examined. We expect that combining shotgun re-sequencing of whole genomes with imputation-based approaches such as ours will allow economical association studies that evaluate SNP variation in large numbers of individuals even more exhaustively than is currently possible. Furthermore, we expect that whatever the characteristics of the re-sequencing technology used, it will be possible to improve the quality of estimated genotypes and haplotypes at each site by combining information across individuals, rather than simply increasing the depth at which each individual is sequenced.
TABLE VI.
Accuracy of imputed genotypes using shotgun sequence data as input
| 20,358 polymorphic sites, segregated according to population frequency | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1,000,000 bases | 979,642 monomorphic sites |
16716 sites with MAF<.5% |
510 sites with MAF 0.5–1% |
425 sites with MAF 1–2% |
590 sites with MAF 2–5% |
2116 sites with MAF>5% |
|||||||||
| Sequencing depth |
Total investment |
Average length (in Kb) per flip |
Average errors per individual |
False positives SNPs |
Allelic accuracy (%) |
Detected SNPs |
Allelic accuracy (%) |
Detected SNPs |
Accuracy (%) |
Detected SNPs |
Allelic accuracy (%) |
Detected SNPs |
Allelic accuracy (%) |
Detected SNPs |
Allelic accuracy (%) |
| n = 100 individuals resequenced using a shotgun approach | |||||||||||||||
| 2 × | 200 × | 75.15 | 106.32 | 99 | 97.11 | 176 | 98.79 | 90 | 98.78 | 176 | 98.81 | 465 | 98.78 | 2,109 | 98.84 |
| 4 × | 400 × | 56.62 | 45.01 | 43 | 98.42 | 440 | 99.51 | 188 | 99.62 | 286 | 99.60 | 550 | 99.62 | 2,115 | 99.66 |
| 8 × | 800 × | 53.89 | 12.90 | 59 | 99.30 | 995 | 99.92 | 309 | 99.94 | 369 | 99.94 | 582 | 99.94 | 2,115 | 99.94 |
| 12 × | 1200 × | 55.46 | 4.60 | 42 | 99.41 | 1,310 | 99.98 | 357 | 99.98 | 395 | 99.99 | 585 | 99.99 | 2,116 | 99.98 |
| 16 × | 1600 × | 51.39 | 2.19 | 33 | 99.49 | 1,432 | 99.99 | 368 | 100.00 | 397 | 100.00 | 585 | 100.00 | 2,116 | 99.99 |
| ∞ | ∞ | – | – | – | 1,526 | – | 384 | – | 405 | – | 589 | – | 2,116 | – | |
| n = 200 individuals resequenced using a shotgun approach | |||||||||||||||
| 2 × | 400 × | 250.74 | 57.52 | 219 | 98.87 | 365 | 99.56 | 186 | 99.52 | 295 | 99.46 | 565 | 99.41 | 2,116 | 99.59 |
| 4 × | 800 × | 248.05 | 25.18 | 52 | 99.47 | 734 | 99.84 | 310 | 99.85 | 378 | 99.85 | 587 | 99.86 | 2,116 | 99.90 |
| 6 × | 1200 × | 209.44 | 14.36 | 45 | 99.67 | 1,270 | 99.95 | 386 | 99.95 | 405 | 99.94 | 590 | 99.94 | 2,116 | 99.96 |
| 8 × | 1600 × | 199.30 | 9.29 | 34 | 99.68 | 1,654 | 99.98 | 425 | 99.97 | 415 | 99.97 | 590 | 99.97 | 2,116 | 99.97 |
| ∞ | ∞ | – | – | – | 2,829 | – | 485 | – | 423 | – | 590 | – | 2,116 | – | |
| n = 400 individuals resequenced using a shotgun approach | |||||||||||||||
| 1 × | 400 × | 470.53 | 84.95 | 212 | 99.02 | 183 | 99.50 | 149 | 99.32 | 296 | 99.15 | 570 | 98.94 | 2,116 | 99.14 |
| 2 × | 800 × | 548.85 | 40.34 | 243 | 99.44 | 532 | 99.72 | 307 | 99.68 | 393 | 99.64 | 589 | 99.65 | 2,116 | 99.77 |
| 3 × | 1200 × | 553.40 | 25.98 | 143 | 99.65 | 906 | 99.86 | 389 | 99.84 | 413 | 99.83 | 590 | 99.84 | 2,116 | 99.89 |
| 4 × | 1600 × | 519.38 | 18.97 | 82 | 99.77 | 1,258 | 99.92 | 431 | 99.90 | 421 | 99.91 | 590 | 99.91 | 2,116 | 99.94 |
| ∞ | ∞ | – | – | – | 4,729 | – | 509 | – | 425 | – | 590 | – | 2,116 | – | |
We simulated 1Mb regions in individuals with HapMap-like degrees of LD. Then, we generated shotgun sequence data for a subset of individuals (n = 100, 200 or 400) at varying depths (1 × –16 ×). The depths were selected to represent a total investment of between 400 × and 1,600 × coverage of the region (200 × coverage was also examined for n = 100). Simulated reads were 32-bp long and had a per base error rate of 0.2%. Read counts at sites where multiple copies of each alternative base were observed (see Methods) were then provided as input to our haplotyper, MaCH 1.0. The column with average errors per individual summarizes the overall haplotyping accuracy, across polymorphic and monomorphic sites. For example, when 400 individuals were sequenced at 4 × depth, an average of 18.97 imputed genotypes differed from the actual simulated genotypes in each individual. The next several columns summarize results for positions where the haplotyper called a polymorphism. The number of false-positive sites is listed together with the accuracy of bases at those sites. Typically, only a few false positive polymorphisms were called (in 400 individuals at 4 × depth, 82 false-positive polymorphisms were observed). The next columns summarize results for sites that were truly polymorphic in the population and these are grouped by frequency (calculated from a sample of N = 10,000 chromosomes). For each frequency class, information is provided on the number of polymorphic sites identified and the overall base calling accuracy at those sites. Note that, especially for rarer SNPs, many sites are not scored as polymorphic simply because they are invariant in the set of individuals selected for sequencing.
TABLE VII.
Accuracy of imputed genotypes using shotgun sequence data as input, additional quality metrics
| 20,358 polymorphic sites, segregated according to population frequency | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16,716 sites with MAF<0.5% |
510 sites with MAF 0.5–1% |
425 sites with MAF 1–2% |
590 sites with MAF 2–5% |
2116 sites with MAF>5% |
||||||||||||
| Sequencing depth |
Total investment |
All allelic accuracy (%) |
Het allelic accuracy (%) |
Dosage r2(%) |
All allelic accuracy (%) |
Het allelic accuracy (%) |
Dosage r2 (%) |
All allelic accuracy (%) |
Het allelic accuracy (%) |
Dosage r2 (%) |
All allelic accuracy (%) |
Het allelic accuracy (%) |
Dosage r2 (%) |
All allelic accuracy (%) |
Het allelic accuracy (%) |
Dosage r2 (%) |
| n = 100 individuals resequenced using a shotgun approach | ||||||||||||||||
| 2 × | 200 × | 98.79 | 98.42 | 50.58 | 98.78 | 95.48 | 61.41 | 98.81 | 93.94 | 68.43 | 98.78 | 93.99 | 76.61 | 98.84 | 97.87 | 92.78 |
| 4 × | 400 × | 99.51 | 99.19 | 68.99 | 99.62 | 98.15 | 80.80 | 99.60 | 97.75 | 85.35 | 99.62 | 98.14 | 91.17 | 99.66 | 99.51 | 97.79 |
| 8 × | 800 × | 99.92 | 99.86 | 91.94 | 99.94 | 99.52 | 95.58 | 99.94 | 99.54 | 97.12 | 99.94 | 99.60 | 98.42 | 99.94 | 99.90 | 99.63 |
| 12 × | 1200 × | 99.98 | 99.96 | 98.10 | 99.98 | 99.87 | 98.78 | 99.99 | 99.87 | 99.30 | 99.99 | 99.90 | 99.62 | 99.98 | 99.96 | 99.89 |
| 16 × | 1600 × | 99.99 | 99.99 | 99.39 | 100.00 | 99.95 | 99.70 | 100.00 | 99.96 | 99.80 | 100.00 | 99.98 | 99.90 | 99.99 | 99.98 | 99.97 |
| n = 200 individuals resequenced using a shotgun approach | ||||||||||||||||
| 2 × | 400 × | 99.56 | 96.47 | 61.17 | 99.52 | 94.17 | 71.39 | 99.46 | 94.16 | 77.95 | 99.41 | 95.97 | 85.94 | 99.59 | 99.32 | 96.98 |
| 4 × | 800 × | 99.84 | 98.32 | 78.18 | 99.85 | 97.69 | 87.34 | 99.85 | 98.28 | 91.83 | 99.86 | 99.22 | 95.84 | 99.90 | 99.86 | 99.19 |
| 6 × | 1200 × | 99.95 | 99.18 | 89.31 | 99.95 | 98.79 | 94.31 | 99.94 | 99.32 | 96.27 | 99.94 | 99.69 | 98.37 | 99.96 | 99.94 | 99.66 |
| 8 × | 1600 × | 99.98 | 99.58 | 94.58 | 99.97 | 99.34 | 97.07 | 99.97 | 99.56 | 98.20 | 99.97 | 99.82 | 99.19 | 99.97 | 99.96 | 99.81 |
| n = 400 individuals resequenced using a shotgun approach | ||||||||||||||||
| 1 × | 400 × | 99.50 | 89.99 | 46.49 | 99.32 | 86.16 | 56.68 | 99.15 | 86.45 | 64.32 | 98.94 | 90.07 | 75.41 | 99.14 | 98.22 | 93.87 |
| 2 × | 800 × | 99.72 | 93.20 | 60.22 | 99.68 | 93.32 | 74.76 | 99.64 | 95.06 | 82.82 | 99.65 | 97.64 | 91.00 | 99.77 | 99.63 | 98.18 |
| 3 × | 1200 × | 99.86 | 95.13 | 71.97 | 99.84 | 95.85 | 84.67 | 99.83 | 97.58 | 90.40 | 99.84 | 99.05 | 95.47 | 99.89 | 99.84 | 99.14 |
| 4 × | 1600 × | 99.92 | 96.69 | 80.67 | 99.90 | 97.30 | 89.59 | 99.91 | 98.81 | 94.46 | 99.91 | 99.51 | 97.46 | 99.94 | 99.91 | 99.50 |
This table is analogous to Table VI, but presents additional quality metrics that separate accuracy at heterozygous and homozygous sites. In addition, the estimated r2 between imputed genotype calls and true genotypes is also tabulated.
DISCUSSION
In summary, we have described and evaluated a very effective model for haplotyping and genotype imputation in whole genome studies. The idea of genotype imputation is not new and was outlined as early as 2006 [Scheet and Stephens, 2006]. Here, we evaluate the practical performance of imputation based on a variety of genotyping platforms and populations, using both simulations and real data. We show that our model leads to imputed genotypes whose quality improves as more data becomes available, either because a larger reference panel is used or because study samples are genotyped in finer detail. Similarly, haplotype estimates improve in quality as more individuals are genotyped. Furthermore, we have introduced novel approaches for the analysis of short read shotgun sequencing data, which is likely to become extremely important as human geneticists move beyond chip-based genotyping to resequencing (as in the 1,000 Genomes Project, whose initial design was partly based on the simulations summarized in our Table VI, see http://www.1000genomes.org for more details).
Other approaches for genotype imputation have been developed independently [Marchini et al., 2007; Servin and Stephens, 2007]. We expect that our results demonstrating the utility of larger reference panels, showing that the three HapMap analysis panels can be combined to better impute genotypes in populations that are genetically distant from the HapMap analysis panels, illustrating the ability of imputation-based approaches to estimate LD between untyped markers, and comparing the relative performance of imputation-based approaches for different commercial marker panels will apply when these alternative approaches for genotype imputation are used. The approaches differ in the precise details of how they search for shared haplotype stretches and also in the efficiency of their computational implementations. For example, whereas [Marchini et al., 2007] rely on recombination rates generated by the HapMap Consortium and assume a uniform mutation/error rate for all markers, we estimate “recombination rates” within each dataset and allow “mutation rates” to vary. These parameters capture not only intrinsic characteristics of the markers and regions being examined, but also—for example—the genetic distance between the samples being imputed and the reference panel (which can impact apparent “recombination rates”) and differences in genotyping protocols between the two samples (which can impact apparent “mutation rates”).
We expect that, in small samples, the use of external recombination rate estimates (as in IMPUTE) might be beneficial, but that with large sample sizes or in the presence of genotyping error our approach, which uses available data to model “recombination” and “mutation” rates should become advantageous. We performed two sets of preliminary comparisons of MaCH and IMPUTE. In the first experiment, we applied IMPUTE [Marchini et al., 2007] to the FUSION GWAS data for chromosome 14 and estimated genotypes for 521 previously genotyped markers [Willer et al., 2006]. Genotypes estimated by IMPUTE and MaCH were identical in 99.2% of cases. In the cases where the two estimates differed, IMPUTE matched experimental genotypes 44.6% of the time, MaCH matched experimental genotypes 52.3% of the time, and both estimates were wrong 3.06% of the time. For the second experiment, we applied IMPUTE to the HGDP data of Conrad et al. [2006]. Table VIII tabulates the proportion of markers imputed with r2>0.80 in each population using either MaCH or IMPUTE (in each case, we selected the HapMap reference panel that provided the best imputed genotypes). Overall, the two methods perform similarly. MaCH slightly outperforms IMPUTE in 37 out of 52 populations, slightly underperforms in 13 populations and the two methods are tied in the remaining two populations. Our results are consistent with other published comparisons [Biernacka et al., 2009; Pei et al., 2008], which include detailed comparisons of the performance of MaCH and IMPUTE with each other and with alternative imputation approaches such as BEAGLE and fastPHASE.
TABLE VIII.
MaCH vs. IMPUTE in the HGDP populations
| Population | Continental group | Best MaCH (%) |
Best IMPUTE (%) |
|---|---|---|---|
| Bantu | Africa | 42.64 | 42.64 |
| BiakaPygmy | Africa | 33.06 | 33.17 |
| Mandenka | Africa | 51.31 | 50.71 |
| MbutiPygmy | Africa | 29.74 | 30.44 |
| San | Africa | 25.81 | 25.91 |
| Yoruba | Africa | 52.42 | 53.93 |
| Colombian | Americas | 58.06 | 57.46 |
| Karitiana | Americas | 49.19 | 49.09 |
| Maya | Americas | 67.14 | 66.43 |
| Pima | Americas | 54.54 | 53.73 |
| Surui | Americas | 45.56 | 45.36 |
| Balochi | Central South Asia | 65.42 | 64.31 |
| Brahui | Central South Asia | 66.63 | 65.02 |
| Burusho | Central South Asia | 69.56 | 69.46 |
| Hazara | Central South Asia | 69.05 | 68.15 |
| Kalash | Central South Asia | 67.14 | 64.92 |
| Makrani | Central South Asia | 69.56 | 67.44 |
| Pathan | Central South Asia | 67.04 | 67.14 |
| Sindhi | Central South Asia | 67.54 | 66.23 |
| Uygur | Central South Asia | 70.77 | 69.35 |
| Cambodian | East Asia | 65.83 | 64.52 |
| Dai | East Asia | 62.90 | 62.60 |
| Daur | East Asia | 64.42 | 64.92 |
| Han | East Asia | 69.25 | 69.46 |
| Han-NChina | East Asia | 63.21 | 62.00 |
| Hezhen | East Asia | 65.42 | 65.02 |
| Japanese | East Asia | 67.54 | 66.73 |
| Lahu | East Asia | 60.69 | 61.09 |
| Miao | East Asia | 62.80 | 61.69 |
| Mongola | East Asia | 64.92 | 65.83 |
| Naxi | East Asia | 62.30 | 62.10 |
| Oroqen | East Asia | 69.76 | 69.35 |
| She | East Asia | 62.80 | 63.10 |
| Tu | East Asia | 67.24 | 66.94 |
| Tujia | East Asia | 63.41 | 63.21 |
| Xibo | East Asia | 65.12 | 64.72 |
| Yakut | East Asia | 70.67 | 69.86 |
| Yi | East Asia | 63.71 | 62.20 |
| Adygei | Europe | 69.25 | 67.04 |
| Basque | Europe | 73.39 | 72.68 |
| French | Europe | 71.47 | 71.27 |
| Italian | Europe | 70.67 | 70.26 |
| Orcadian | Europe | 64.01 | 64.52 |
| Russian | Europe | 72.58 | 71.88 |
| Sardinian | Europe | 67.94 | 67.94 |
| Tuscan | Europe | 64.01 | 64.92 |
| Bedouin | Middle East | 63.71 | 62.40 |
| Druze | Middle East | 66.73 | 66.83 |
| Mozabite | Middle East | 63.71 | 63.51 |
| Palestinian | Middle East | 67.94 | 66.03 |
| Melanesian | Oceania | 56.75 | 56.15 |
| Papuan | Oceania | 52.12 | 49.50 |
For each of the 52 HGDP populations, we picked the reference panel (CEU, JPT+CHB, YRI, or three combined) that resulted in the highest coverage (measured by r2 between experimental genotypes and imputed fractional counts) for MaCH and IMPUTE v0.5.0.
Our method uses an HMM to describe genetic variation along each haplotype. It is clear that when HMM models are applied to genetic data, many opportunities for identifying computational efficiencies exist [Abecasis et al., 2002; Gudbjartsson et al., 2000; Idury and Elston, 1997; Kruglyak and Lander, 1998; Lander and Green, 1987]. In the methods section we describe several optimizations that we have already implemented, including a general strategy for reducing memory requirements for the Baum algorithm [Baum, 1972; Wheeler and Hughey, 2000]. We expect that further efficiencies will be forthcoming. Our model is implemented in the MaCH package (freely available with C++ source code from our website, see http://www.sph.umich.edu/csg/abecasis/mach/). Our implementation can be used to carry out all the analyses described in this paper. Specifically, it can estimate haplotypes, impute missing genotypes in a variety of populations, using the HapMap sample or another set of densely genotyped individuals as a reference, analyze shotgun re-sequencing data from high-throughput technologies now being developed, and carry out simple tests of association.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mike Boehnke, Karen Mohlke, and the other FUSION investigators for helpful discussions. This research was supported by research grants from the NIMH, NHLBI, and the NHGRI to GRA.
APPENDIX
DETAILED METHODS DESCRIPTION
HIDDEN MARKOV MODEL
Our model resolves a set of unphased genotypes G into an imperfect mosaic of several template haplotypes. We assume that H template haplotypes are each genotyped at L loci and let Tj(i) denote the allele observed at locus j in reference haplotype i. Furthermore we define a series of indicator variables S1, S2, …, SL that denote an hypothetical (and unobserved) mosaic state underlying the unphased genotypes. At a specific position j there are H2 possible states. A specific state, such as Sj = (xj, yj), indicates that the first chromosome uses haplotype xj as a template, whereas the second chromosome uses haplotype yj as a template.
We are interested in making inferences about the sequence of mosaic states S that best describe the observed genotypes. Knowledge of S will implicitly order alleles at heterozygous sites and suggest an allele for each untyped location. We define the joint probability of the observed genotypes and an underlying haplotype state as:
In the model above, P(S1) denotes the prior probability of the initial mosaic state and is usually assumed to be equal for all possible configurations, P(Sj | Sj−1) denotes the transition probability between two mosaic states and reflects the likelihood of historical recombination events in the interval between j and j−1, P(Gj|Sj) denotes the probability of observed genotypes at each position conditional on the underlying mosaic state and reflects the combined effects of gene conversion, mutation, and genotyping error. Interestingly, note that, whereas, our model and IMPUTE both use a large number of haplotypes as templates, fastPHASE [Scheet and Stephens, 2006] uses a smaller set of estimated haplotype “groupings” as templates in an otherwise similar HMM, resulting in improved computational efficiency at the cost of some fuzziness in haplotype templates.
MONTE-CARLO HAPLOTYPING PROCEDURE
To estimate haplotypes in a sample of genotyped individuals we first assign a random pair of haplotypes to each individual, consistent with the observed genotypes. This involves randomly ordering alleles at each heterozygous site and sampling alleles at untyped sites according to population frequencies. Then, we update the haplotypes for each individual in turn by using the current set of haplotype estimates for all individuals as templates and sampling S proportional to the likelihood L(S|G) ∝ P(G,S). Note that since the Sj define a Markov Chain this sampling can be done conveniently using Baum’s forward and backward algorithm [Baum, 1972]. A new set of haplotypes for an individual is then defined according to sampled mosaic and edited to ensure it matches the observed genotypes. We repeat the update procedure several times, looping over all individuals (more updates result in gradual refinement of the estimated haplotypes, but very accurate haplotype estimates can often be obtained in ~20 rounds, see Table I). After a pre-specified number of rounds are completed, we generate a pair of consensus haplotypes for each individual. This consensus haplotype pair is defined as the pair that minimizes total switch error when compared to the haplotypes sampled at each round.
PARAMETER ESTIMATION
Key ingredients in the above procedure are the transition probabilities P(Sj | Sj−1) and emission probabilities P(Gj | Sj). We define the transition probabilities as a function of the crossover parameter θj:
The possible values of P(Sj | Sj−1) reflect both the overall rate of changes in the mosaic for the interval, given by θj, and the fact that when a change occurs a new mosaic state is selected at random among all possible states.
We let T(Sj) = T(xj)+T(yj) denote the genotype implied by state Sj and define the emission probabilities P(Gj | Sj) as a function of the error parameter εj:
Initially, we let set θj = θ = 0.01 and εj = ε = 0.01 or some other suitable constant. As we sample a new mosaic state for each individual we keep track of the number and location of change points in the mosaic and of the number of times that the genotype implied by the sampled mosaic state matches the observed genotype (or not). These quantities are then used to update the θj and εj parameters for the next iteration. It is important to avoid setting either θj = 0 or εj = 0, as that could make it difficult for our Markov sampler to investigate different mosaic configurations. To avoid this, a combined crossover parameter is estimated for intervals with a small number of sampled changes in mosaic state and an analogous procedure is employed for markers with a small number of observed mismatches between the mosaic and observed genotypes.
Overall, we expect the θj will reflect a combination of population recombination rates and the relatedness between the haplotypes being resolved and the true underlying haplotypes (for example, if CEU chromosomes are used as templates to resolve CHB genotypes we expect, on average, higher θ estimates than when other CHB individuals are used as templates). We considered using distance between flanking markers to inform estimates of θj (since θ’s are generally larger in larger intervals), but did not find noticeable improvements. Overall, we expect that εj will reflect a combination of genotyping error, gene conversion events, recurrent mutation and, when genotype data from multiple platforms or laboratories is used, assay inconsistencies between different platforms. We observed slightly lower data quality measures (completeness, duplicate concordance, Hardy–Weinberg test statistics) for markers with large estimates of εj in the FUSION GWAS.
GENOTYPE IMPUTATION AND ESTIMATES OF IMPUTATION QUALITY
Genotype imputation analyses proceed similarly to the haplotyping analyses described above, but do not require each sampled haplotype configuration to be stored. Instead, after each iteration, a series of counters is updated to indicate the number of times each genotype was sampled at a particular position. Once all iterations are completed, these counters give an indication of the relative probability of observing each possible genotype and can be used to impute the most likely genotype and to calculate various measures of the quality of imputed genotypes.
Without loss of generality, consider a SNP with alleles A and B. Let nA/A, nA/B, and nB/B be the number of times each possible genotype was sampled after I = nA/A + nA/B+nB/B iterations. For downstream analysis of imputed alleles, we typically consider either the most likely genotype or the expected number of copies of allele A. The most likely genotype is simply the genotype that was sampled most frequently. The expected number of counts of allele A is the genotype score g = (2nA/A + nA/B)/I. Both of these quantities can be conveniently incorporated into a variety of analysis, including regression-based association analysis of discrete or quantitative traits.
To measure the accuracy of imputation for a single imputed genotype IG, we define the genotype quality score Q = nIG/I. This quantity can be averaged over all genotypes for a particular marker to quantify the average accuracy of imputation for that marker. We have found that a better measure of imputation quality for a marker is the estimated r2 between true allele counts and estimated allele counts (Fig. 1). This quantity can be estimated by comparing the variance of the estimated genotype scores with what would be expected if genotype scores were observed without error. For a given SNP, let Var(g) be the variance of estimated genotype and let p = Mean(g)/2 be the estimated frequency of allele A. The estimated r2 with true genotypes can then be defined as
An alternative definition is
Empirically, we have found that while both definitions lead to similar conclusions, the first definition appears to be marginally better.
ASSOCIATION ANALYSIS USING IMPUTED GENOTYPES
When analyzing the FUSION data [Scott et al., 2007], we included imputed genotype scores as predictors in a logistic regression that also included age, sex, and geographic origin as covariates. For analyzing simulated case-control data, we simply used a t-test to compare the average genotype scores in cases and controls. Other approaches to the analysis of imputed data are possible but, in our experience, the imputed genotype scores provide a good balance between computationally demanding multiple imputation procedures [Servin and Stephens, 2007] and analyses that simply use the most likely genotype.
HMM FOR SHOTGUN SEQUENCE DATA (SIMULATIONS)
When shotgun re-sequencing, or another single molecule re-sequencing technology, is used on diploid individuals, genotypes are not directly observed. In this case, we assume the data consists of counts Aj and Bj indicating how many times base A (or B) was observed at site j. We then define our HMM as
Here, we sum over possible genotypes at each site and calculate the probability of the observed traits for each possible genotype set. In addition, we define the probability of observing a specific set of traces given the underlying genotype as
The parameter δ denotes the per base sequencing error rate and can be separated from the effects of mutation and gene conversion captured in ε, unless the re-sequencing depth is very low.
In principle, the method could be applied to all sites where an alternative base call is observed at least once. However, since we simulated many short reads and an error rate of 0.2%, the minor allele was observed at least once at nearly every simulated position. For reasons of computational efficiency, we applied the MaCH 1.0 haplotyper only to positions were the minor allele was observed multiple times. Specifically, we defined mkj as the number of traces where the minor allele was observed at position j in individual k. Then, we defined the score wj = Σk mkj(mkj + 1)/2 and applied our haplotyping algorithm to all sites where wj exceeded a predefined threshold (other sites were assumed to contain the major allele). The score gives higher weight to sites where the minor allele is observed multiple times in the same individual. We used thresholds for wj of 5, 7, 9, 11, 13 depending on whether the total coverage (defined as depth × individuals) was 200, 400, 800, 1,200, or 1,600 ×. When the number of individuals sequenced was 400, these thresholds were reduced to 4, 6, 8, 10, and 12, respectively. This means that, for example, when 400 individuals were re-sequenced at 4 × depth (total depth = 1,600 ×) we considered only sites where the minor allele was observed in at least 12 traces from different individuals or slightly fewer traces concentrated in one or more individuals.
HMM FOR SHOTGUN SEQUENCE DATA (SIMULATIONS)
The model described above is convenient for the analysis of simulated data where the per base error rate is constant. For analyses of real data, where base quality scores are associated with individual bases, we adapted our implementation to use P(base calls, quality scores | G) as stored in Genotype Likelihood Files generated by samtools [Li et al., 2008, 2009a].
COMPUTATIONAL EFFICIENCY
A number of optimizations are possible to increase the computational efficiency of our model. For example, since haplotype states are unordered only H(H+1)/2 distinct states must be considered at each location, rather than H2 distinct states. Below, we summarize some of the other efficiencies that we identified and how these are implemented in MaCH.
TRANSITION MATRICES
When sampling a mosaic state S conditional on the observed genotypes G, we rely on the Baum algorithm. The algorithm requires a series left and right conditioned probability vectors which provide an indication of the relative probability of a specific state at a given location conditional observed genotypes at markers to its left (or right). For example, the probability of observing state (x,y) at location j conditional on all preceding genotypes is simply:
The calculation of these probabilities can be sped up by taking advantage of the regular patterns in the transition matrices. Specifically, we define the quantities:
Then, the previous definition becomes:
When this updated definition is used to calculate left conditional probabilities for each possible state, computational requirements become O(H2) rather than O(H4) using the original definition, provided that C(a) and C are pre-computed. An analogous speed up is available for right-conditioned probabilities.
MEMORY EFFICIENCY
One large computational constraint when applying our algorithm on a genomic scale is the storage required to track left-conditioned probabilities. Typically, this requires storage of L vectors each with H2 elements (or, as noted above H(H+1)/2 elements). It is clear that this requirement becomes cumbersome as the number of polymorphic sites increases. We devised a solution that requires storage of only 2*sqrt(L) vectors. For notational convenience let K = sqrt(L). Our algorithm pre-allocates 2K vectors and organizes these into two groups: a framework set of K vectors, and a working set of another K vectors. When left-conditional probabilities are first calculated, proceeding left to right, we store every Kth vector in the framework set and discard other intermediate results. Then, as these vectors are used in the second pass of the chain (which combines left and right conditional probabilities, proceeding right to left), we recalculate K of these vectors at a time (starting from the nearest vector in the framework set) and store them in the working set of vectors. Completing the full chain requires calculation of all L vectors of left conditional probabilities, recalculation of K of these vectors L/K times, and calculation of L vectors of right conditional probabilities. Overall, our solution no more than doubles computing time (since each vector of left conditional probabilities must be calculated twice), but reduces memory requirements from O(L) to O(L1/2). The solution is general and can be applied to many other HMMs (see also [Wheeler and Hughey, 2000]).
REDUCING THE NUMBER OF TEMPLATES
If all available chromosomes are used as templates, the complexity of our algorithm will increase cubically with sample size (because the cost of each update increases quadratically and the number of updates increases linearly with sample size). One way to avoid this is to restrict the size of the template pool. When there are more than a pre-specified number of potential templates (say H > 300), we typically select a random subset of these for each update. With this restriction, the complexity of our algorithm increases only linearly with sample size (because the cost of each update now remains fixed and only the number of updates to be performed grows). Furthermore, even though each update is based on only a random sample of the available haplotypes, the overall quality of solutions still increases with sample size. When the focus is on genotype imputation, rather than haplotyping, an alternative is to use as templates individuals who have been genotyped for the markers being imputed (e.g. the HapMap reference samples). Both of the above solutions are heuristics that trade-off some accuracy for computational efficiency. An alternative strategy for reducing the size of the template pool is to consider local similarities and redundancies among the haplotypes in the pool. These redundancies are already exploited to increase computational efficiency in the handling of other Markov models [Abecasis et al., 2002; Markianos et al., 2001], and our preliminary implementations suggest that speed-ups of 5–10 × are possible for our haplotyping model.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- Baum LE. An inequality and associated maximization technique in statistical estimation for probabilistic functions of Markov processes. Inequalities. 1972;3:1–8. [Google Scholar]
- Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Biernacka JM, Tang R, Li J, McDonnell SK, Rabe KG, Sinnwell JP, Rider DN, de Andrade M, Goode EL, Fridley BL. Assessment of genotype imputation methods. BMC Proc. 2009;3:S5. doi: 10.1186/1753-6561-3-s7-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR. Multilocus association mapping using variable-length Markov chains. Am J Hum Genet. 2006;78:903–913. doi: 10.1086/503876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Smith JD, Kruglyak L, Nickerson DA. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet. 2003;33:518–521. doi: 10.1038/ng1128. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Li J. Conjuring SNPs to detect associations. Nat Genet. 2007;39:815–816. doi: 10.1038/ng0707-815. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SE, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Gaulton KJ, Willer CJ, Li Y, Scott LJ, Conneely KN, Jackson AU, Duren WL, Chines PS, Narisu N, Bonnycastle LL, Luo J, Tong M, Sprau AG, Pugh EW, Doheny KF, Valle TT, Abecasis GR, Tuomilehto J, Bergman RN, Collins FS, Boehnke M, Mohlke KL. Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes. 2008;57:3136–3144. doi: 10.2337/db07-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idury RM, Elston RC. A faster and more general hidden Markov model algorithm for multipoint likelihood calculations. Hum Hered. 1997;47:197–202. doi: 10.1159/000154413. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES. Faster multipoint linkage analysis using Fourier transforms. J Comput Biol. 1998;5:1–7. doi: 10.1089/cmb.1998.5.1. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Boehnke M, Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009a;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Sanna S, Abecasis GR. Genotype imputation. Annu Rev Genomics Hum Genet. 2009b;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat Genet. 2004;36:1181–1188. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- Marchini J, Cutler D, Patterson N, Stephens M, Eskin E, Halperin E, Lin S, Qin ZS, Munro HM, Abecasis GR, Donnelly P International HapMap Consortium. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 2006;78:437–450. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Markianos K, Daly MJ, Kruglyak L. Efficient multipoint linkage analysis through reduction of inheritance space. Am J Hum Genet. 2001;68:963–977. doi: 10.1086/319507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C, Gao P, Togias A, Hansel NN, Diette G, Adkinson NF, Liu MC, Faruque M, Dunston GM, Watson HR, Bracken MB, Hoh J, Maul P, Maul T, Jedlicka AE, Murray T, Hetmanski JB, Ashworth R, Ongaco CM, Hetrick KN, Doheny KF, Pugh EW, Rotimi CN, Ford J, Eng C, Burchard EG, Sleiman PM, Hakonarson H, Forno E, Raby BA, Weiss ST, Scott AF, Kabesch M, Liang L, Abecasis G, Moffatt MF, Cookson WO, Ruczinski I, Beaty TH, Barnes KC. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–346. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL. Testing untyped alleles (TUNA)-applications to genome-wide association studies. Genet Epidemiol. 2006;30:718–727. doi: 10.1002/gepi.20182. [DOI] [PubMed] [Google Scholar]
- Pe’er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- Pei YF, Li J, Zhang L, Papasian CJ, Deng HW. Analyses and comparison of accuracy of different genotype imputation methods. PLoS One. 2008;3:e3551. doi: 10.1371/journal.pone.0003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford, UK: Oxford University Press; 2003. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, Shen H, Timpson NJ, Lettre G, Usala G, Chines PS, Stringham HM, Scott LJ, Dei M, Lai S, Albai G, Crisponi L, Naitza S, Doheny KF, Pugh EW, Ben-Shlomo Y, Ebrahim S, Lawlor DA, Bergman RN, Watanabe RM, Uda M, Tuomilehto J, Coresh J, Hirschhorn JN, Shuldiner AR, Schlessinger D, Collins FS, Davey Smith G, Boerwinkle E, Cao A, Boehnke M, Abecasis GR, Mohlke KL. Common variants in the GDF5 region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005a;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005b;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2005;437:1299–1320. [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R, Hughey R. Optimizing reduced-space sequence analysis. Bioinformatics. 2000;16:1082–1090. doi: 10.1093/bioinformatics/16.12.1082. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Scott LJ, Bonnycastle LL, Jackson AU, Chines P, Pruim R, Bark CW, Tsai YY, Pugh EW, Doheny KF, Kinnunen L, Valle TT, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. Tag SNP selection for Finnish individuals based on the CEPH Utah HapMap database. Genet Epidemiol. 2006;30:180–190. doi: 10.1002/gepi.20131. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJF, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN The Genetic Investigation of ANthropometric Traits (GIANT) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Kang HM, Eskin E, Halperin E. Leveraging the HapMap correlation structure in association studies. Am J Hum Genet. 2007;80:683–691. doi: 10.1086/513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.