Abstract
Molecular studies of adaptive evolution often focus on detecting selective sweeps driven by positive selection on a species-wide scale; however, much adaptation is local, particularly of ecologically important traits. Here, we look for evidence of range-wide and local adaptation at candidate genes for adaptive phenology in balsam poplar, Populus balsamifera, a widespread forest tree whose range extends across environmental gradients of photoperiod and growing season length. We examined nucleotide diversity of 27 poplar homologs of the flowering-time network—a group of genes that control plant developmental phenology through interactions with environmental cues such as photoperiod and temperature. Only one gene, ZTL2, showed evidence of reduced diversity and an excess of fixed replacement sites, consistent with a species-wide selective sweep. Two other genes, LFY and FRI, harbored high levels of nucleotide diversity and exhibited elevated differentiation between northern and southern accessions, suggesting local adaptation along a latitudinal gradient. Interestingly, FRI has also been identified as a target of local selection between northern and southern accessions of Arabidopsis thaliana, indicating that this gene may be commonly involved in ecological adaptation in distantly related species. Our findings suggest an important role for local selection shaping molecular diversity and reveal limitations of inferring molecular adaptation from analyses designed only to detect species-wide selective sweeps.
MOST species experience variable environments across their geographic range, and many show phenotypic variation that reflects local adaptation of ecologically important traits. However, molecular population genetic analyses of selection often focus on detecting selective sweeps driven by positive directional selection in a species-wide sample (Nielsen 2005). Only recently have studies started to scan patterns of gene nucleotide diversity for signatures of selection below the species level, particularly among human regional subpopulations (Tang et al. 2007; Nielsen et al. 2009; Pickrell et al. 2009; Chen et al. 2010).
The population genetic signatures of positive selection due to species-wide sweeps are quite different from those expected under local selection, used here to mean selection favoring different allelic variants in different parts of the range, even at broadly defined spatial scales. Sweeps due to positive selection can result in a species-wide sample that shows low levels of nucleotide variation (Maynard Smith and Haigh 1974), an excess of rare or derived polymorphisms (Tajima 1989), or reduced polymorphism relative to divergence (Hudson et al. 1987). In contrast, local selection may result in local sweeps within subpopulations and/or reduced effective migration of selected alleles between subpopulations (Charlesworth et al. 1997; Charlesworth 1998), leading to increased subpopulation differentiation, elevated polymorphism, or an excess of common variants in species-wide samples (i.e., positive Tajima’s D; Nordborg and Innan 2003; Innan and Kim 2008). Empirical evidence for local selection on candidate genes of ecological significance has been found by comparing nucleotide diversity and differentiation among subpopulations found in different selective environments to empirically derived or model-based neutral expectations (Le Corre 2005; Ingvarsson et al. 2006; Hancock et al. 2008; Moeller and Tiffin 2008; Storz and Kelly 2008; Wachowiak et al. 2009; Turner et al. 2010; Ma et al. 2010).
For species with geographic ranges that span seasonally variable environments, the timing of development for traits related to growth and reproduction is often subject to local selection favoring different phenotypic optima in different parts of the species’ range (Bradshaw et al. 2004). Among forest tree species, and plants in general, this is often manifest as variation among subpopulations in the seasonal timing of growth (i.e., phenology) in response to light and temperature, with strong local adaptation often observed at the phenotypic level (Savolainen et al. 2007).
In eudicots, phenological variation frequently maps to homologs of the Arabidopsis thaliana flowering-time gene network (Mouradov et al. 2002; Simpson and Dean 2002; Ehrenreich et al. 2009). This network functions via chromophore signaling of light to the circadian clock, which interprets critical daylengths and controls expression of downstream genes that integrate signals from interacting pathways to control meristem development (Jackson 2009). Comparative research across diverse taxa suggests that this gene network is responsible for variation in multiple phenological traits responsive to photoperiod, including flowering, tuberization, and seasonal dormancy (Lagercrantz 2009). Based upon transgenic and QTL mapping experiments in Populus, genes in this network are also associated with flowering and vegetative bud development in trees (Frewen et al. 2000; Chen et al. 2002; Böhlenius et al. 2006; Ingvarsson et al. 2008; Jackson 2009; Ma et al. 2010; Rohde et al. 2010).
A priori, we expect genes controlling phenology to be excellent candidates for the molecular basis of local adaptation. Surveys of natural allelic variation among A. thaliana accessions sampled from diverse latitudes have identified two flowering-time genes, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC), that show evidence of local adaptation with latitude (Caicedo et al. 2004; Stinchcombe et al. 2004; Le Corre 2005). In other species, local selection has targeted members of the phytochrome gene family (PHYE) in Cardamine japonica (Ikeda et al. 2009) and PHYB2 in Populus tremula (Ingvarsson et al. 2006; Ingvarsson et al. 2008) and components of the circadian clock (Böhlenius et al. 2006; Slotte et al. 2007; Brachi et al. 2010; Ma et al. 2010). These studies suggest that homologs of the A. thaliana flowering-time gene network underlie ecologically important adaptive variation in phenological responses in other species, making them promising candidates for studying the molecular basis of local adaptation, as well as testing the repeatability of evolution across taxa in genes that influence quantitative traits.
Balsam poplar (Populus balsamifera L.) is a widespread forest tree that has recolonized most of boreal North America in a postglacial range expansion during the past 10,000–18,000 years. On the basis of SNP data from 412 loci assayed from 474 trees sampled from across the species range, this expansion appears to have involved the spread of populations from a southwestern refugial population into the northwest and northeast (Keller et al. 2010). This spread is reflected in the current diversity forming three broadly distributed genetic clusters, currently situated in the northern, central, and eastern parts of the range. These clusters are weakly but significantly differentiated at SNP loci (FCT = 0.04), but show much stronger regional structure at multiple ecophysiological traits related to growth and phenology (Keller et al. 2011). The covariance between SNPs, ecophysiology, and environment strongly suggests that latitude or other climate-related aspects of the environment have driven adaptation in phenology traits during and since the northward expansion.
Here, we present analyses of the molecular evolution of 27 candidate genes homologous to the A. thaliana flowering-time network and associated pathways (hereafter referred to collectively as “phenology genes”) in balsam poplar. We test for evidence of nonneutral evolution in our species-wide sample by comparing (i) nucleotide diversity and the site frequency spectrum of phenology genes to a set of 219 background reference loci and to neutral expectations based on an approximate Bayesian computation (ABC) model of demographic history (Beaumont et al. 2002) and (ii) diversity and divergence of phenology genes against a reduced set of reference loci and to neutral coalescent expectations. Finally, we test for evidence of local adaptation by comparing nucleotide differentiation between high- and low-latitude accessions to neutral expectations simulated under a demographic model that reflects balsam poplar’s recent history of postglacial expansion.
Methods
Sampling and DNA sequencing
We isolated DNA from a range-wide sample of 24 accessions (Figure 1) and sequenced each sample for a set of 27 phenology-associated candidate genes: 24 genes from the flowering-time network and 3 abscisic acid (ABA) genes that cosegregate with QTL for dormancy phenotypes in a P. trichocarpa × P. deltoides mapping population (Frewen et al. 2000). The 24 sampled accessions are part of the AgCanBaP collection growing in Indian Head, Saskatchewan, Canada (for collection details, see Soolanayakanahally et al. 2009). We identified P. balsamifera homologs of A. thaliana phenology genes using BLASTN searches against the P. trichocarpa genome (Tuskan et al. 2006). Primers (supporting information, Table S2) were designed to amplify and sequence the exons of each gene, as these are most likely to contain causative polymorphisms for adaptive protein evolution, but for most genes some intron sequence was also captured. The majority of sequence data were obtained by direct sequencing of PCR products. For two genes (PiF3.1 and TOC1), some samples were sequenced from PCR products that had been cloned into pGemT vectors. For the cloned sequences, all variants present in only one sample were assumed to be due to misincorporation during amplification if they could not be verified by direct sequencing of PCR products from an independent reaction. For TOC1, the first four exons were exclusively from cloned sequence, while sequences from exons 5–6 were direct sequenced from PCR products. This necessitated phasing one part of the alignment and not the other (see below); therefore, we report analyses for these two regions separately.
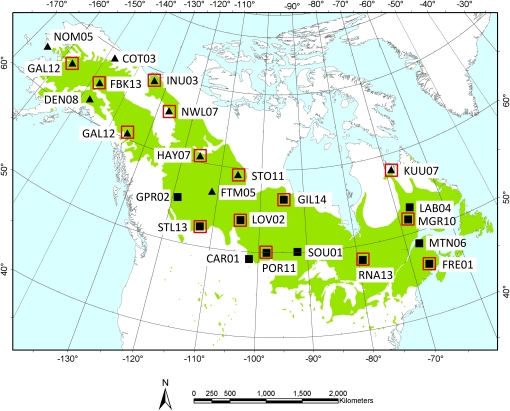
Figure 1 .
Sampling locations and IDs for accessions sequenced in this study. Accessions in the northern group are symbolized by triangles, and those in the southern groups as squares. Red boxes denote those accessions included in the reference gene sequences (from Olson et al. 2010) used for generating the empirical distributions of diversity. The geographic range of P. balsamifera is shown in green.
To conduct analyses that require an outgroup, we obtained sequence data for 24 of the candidate genes as well as 16 reference loci from either previous studies (Ingvarsson et al. 2006; Hall et al. 2007; Neiman et al. 2009; Ma et al. 2010) or PCR sequences from one to four P. tremula individuals. For sites that were polymorphic among P. tremula alleles, we assumed the ancestral state was identical to the base found in the P. balsamifera sample.
Sequences were trimmed of poor flanking sequence, aligned, and edited manually in Aligner v. 3.0.0 (Codon Code Corporation, Dedham, MA). Sites that were heterozygous, polymorphic, or of low sequence quality were visually examined for base call quality. Insertion–deletion polymorphisms were reconstructed from forward and reverse sequences and were removed from the alignments prior to further analysis. One gene, HY2.2, showed an extremely divergent haplogroup suggestive of introgression from another Populus species (Figure S1); thus we confined analysis of HY2.2 to a reduced set of samples that omitted the divergent haplogroup. No other genes showed evidence of introgression. Gene annotations were based on the P. trichocarpa genome build v. 2.0 available at Phytozome (http://www.phytozome.net/poplar.php). All new sequences have been deposited in GenBank (accession numbers in Table S1)
Data analysis
Diploid sequences were disambiguated into haplotypes using Phase v. 2.1 (Stephens et al. 2001; Stephens and Scheet 2005). We ran 10,000 iterations of the Bayesian MCMC chain, sampling every 10 iterations after a burn-in of 100 iterations. We then used the phased haplotypes to estimate the number of segregating sites, nucleotide diversity as Watterson’s θW, and pairwise divergence (π) at replacement, synonymous, and silent (synonymous and noncoding) sites, and the site frequency spectra based on Tajima’s D (Tajima 1989) and Fay and Wu’s H (Fay and Wu 2000), using P. tremula as the outgroup for calculating H. Summary statistics were calculated using DNASP v. 5 (Librado and Rozas 2009).
To characterize range-wide diversity and search for evidence of selection in the species-wide sample, we compared synonymous nucleotide diversity (θW and π) and site frequency spectra (using Tajima’s D) for phenology genes to both genome-wide empirical distributions of synonymous diversity and simulated neutral distributions. The empirical distributions were derived from a set of nuclear reference loci (Olson et al. 2010) sequenced in 15 accessions that represent a subset of the 24 accessions sequenced for phenology genes (Figure 1). We computed distributions of summary statistics based on synonymous sites, restricting the analysis to just the 219 reference loci with ≥100 synonymous sites (mean length = 130.3 synonymous sites). Defining expected patterns of diversity from empirical distributions of reference genes has the advantage of being free from specifying a particular model of demographic history or defining a priori how indirect selection may affect the variance among sites across the genome (Tenaillon and Tiffin 2008; Garrigan et al. 2010). However, because our reference loci were considerably shorter than our phenology genes (mean ± SD = 617 ± 63 bp compared to 2535 ± 1137 bp) direct comparison of the phenology to reference loci may be confounded by sampling effects, causing the reference loci to have greater variance of summary statistics and a large number of sequences with zero segregating sites. For this reason, we also used ABC ( Beaumont et al. 2002) to develop a demographic model that captured the pattern of diversity found at reference loci and could subsequently be used to generate distributions of expected neutral diversity for loci that were the same average length as our phenology genes.
The ABC modeling process involved simulating data under three demographic models: a standard neutral model, an instantaneous size change model, and an exponential growth model. We refrained from testing more complicated models that include population subdivision and migration among regional groups (e.g., Keller et al. 2010) because our initial simple models captured the major features of diversity found in the range-wide sample of 15 individuals (see Results). In other words, these ABC models should not be viewed as accurate descriptors of the demographic history as much as simplified models that capture the patterns of nucleotide diversity found in the reference loci from a widespread sample of a limited number of individuals. For each model, we generated 106 simulated data sets of the neutral coalescent with recombination using Hudson’s ms (Hudson 2002). Each simulated data set consisting of 219 loci containing 130.3 neutrally evolving (synonymous) sites, a fragment length for recombination of 617 bp, and a sample size of 28 chromosomes (values reflect averages for the reference loci). Demographic parameters were drawn at random from prior distributions. For all three models, the population mutation rate per locus (θ = 4Neμ) had a uniform prior of θ ∼ [0.1–4] and values for the population recombination rate (ρ = 4Ner) were drawn from a uniform prior of ρ ∼ [0–0.25]. For the instantaneous size change model, looking backward in time we estimated the first size change (TB) going from the current population size (N0) to the previous size (NB), and then the second size change at time (TA) to the ancestral population size (NA). Prior distributions on timing and size parameters were uniform, and on the intervals of TB ∼ [0.01–4], NB ∼ [0.01–2], TA ∼ [TB + 0.01–1], and NA∼[NB.+ 0.5–4]. Because the instantaneous size change model allows for either increases or decreases in population size at each time this model can recreate, but is not limited to, a traditional bottleneck model. The exponential growth model followed the form Nt = N0-αt, where N0 is the current effective population size, t is time in 4N0 generations, and α is the growth parameter (positive values indicate increasing population size looking forward in time). Prior values on growth were drawn from a uniform distribution of α ∼ [1–20], and the timing of the start of growth (TG) was drawn from a uniform prior of TG ∼ [0.01–10].
We compared the results from the simulated data to the empirical data from our 219 reference loci using five summary statistics based on only synonymous sites: mean and standard deviation of π, mean and standard deviation of Tajima’s D, and the proportion of loci with zero segregating sites. From the pooled set of 3 × 106 simulations, we retained the 3000 simulations (i.e., 0.1% of the pooled set) that were closest to the summary statistics from our reference loci using the Euclidian rejection algorithm, msreject, distributed with the MSBayes package (Hickerson et al. 2007), and calculated the percentage of simulations contributed by each model as our estimate of that model’s posterior probability. We then used ABCREG (Thornton 2009) with the tangent transformation to perform local regression estimation of the posterior distributions of model parameters. From these posteriors, we obtained the Bayesian most probable estimate of each parameter from the median value of the distribution. These estimates were then used in a final round of coalescent simulations where the simulated length was adjusted to match the phenology genes by increasing the per-gene θ and fragment length for recombination to reflect the longer candidate gene sequences (average of 302 synonymous sites and a total length of 2535 sites). Under these conditions, we simulated 500,000 replicates of the coalescent and obtained the shape of the distribution of neutral diversity under the best fitting demographic model. Simulated distributions under neutrality were then compared with the observed values for the phenology genes. Note that although the simulated distributions were generated using the best-fit parameters obtained from the ABC model of the reference loci, because we simulated regions that are approximately four times longer than the length of the loci used to fit the model, we no longer expect the new simulated distributions to match the reference loci. In particular, because of the longer length of the simulated sequences, we no longer expect the inflation of loci with zero polymorphism that is observed in the reference loci data. The full ms command line input for ABC model development and the final simulated distributions are provided in File S1.
In addition to examining patterns of intraspecific diversity, we looked for signatures of selection in our range-wide samples using two approaches for comparing patterns of nucleotide diversity within P. balsamifera to divergence from P. tremula. First, we used J. Hey’s multilocus HKA program to evaluate whether any of the candidate genes deviated from neutral expectations of diversity and divergence, using 10,000 coalescent simulations for significance testing. Second, we used the hierarchical Bayesian implementation of the MacDonald–Kreitman Poisson random fields model (MKPRF) to estimate the strength of selection (γ = 2Nes) on amino acid changes (Bustamante et al. 2002, 2005). We defined a hierarchical model that allowed selection to act differently on the phenology genes than the reference loci. For the MKPRF, we omitted eight phenology genes that had fewer than four replacement site mutations (either polymorphic or fixed differences) because they would provide little statistical power for detecting selection (e.g., Bustamante et al. 2005), leaving 16 phenology genes and 16 reference loci. We ran 10 independent mcmc chains, drawing 6000 samples from each following a burn-in of 1000 steps, and sampling every 10 steps. Input parameter values were set to default values except for ρ, the ratio of Ne in species 2 relative to species 1, which we set to 5.0 on the basis of previous work that showed θ (= 4Neμ) is roughly five times larger in P. tremula than P. balsamifera (Ingvarsson 2008; Olson et al. 2010). For both HKA and MKPRF analyses, we used sequences from P. tremula, instead of P. trichocarpa, as an outgroup, because P. trichocarpa and P. balsamifera are recently diverged and share abundant ancestral polymorphism (Levsen et al., unpublished data).
To test if nucleotide diversity was structured by local adaptation to environmental conditions that covary with latitude (e.g., photoperiod, growing season length, summer and winter temperatures; Keller et al. 2011), we assigned each of the 24 individuals into either northern or southern subgroups, depending on whether individuals had been sampled from high (>56° N) or low (<56° N) latitude sites. The cutoff between north and south was chosen from the median latitude among our sampled individuals, rather than an a priori expectation of a threshold latitude effect. We then tested if the empirical differentiation between subgroups was greater than expected in the absence of selection, using an index of absolute nucleotide differentiation, πT-S = πT − πS, in which πT is the total nucleotide diversity across the pooled sample and πS is the average nucleotide diversity within each of the two latitudinal groups (Charlesworth 1998). Because we lacked the sampling to explicitly model a demographic scenario of latitudinal divergence among three regional clusters (Keller et al. 2010), we simulated the expected distribution of πT-S under a neutral model that captured the major biological features of balsam poplar’s northward expansion from a southern refugial population. Our coalescent model consisted of a single ancestral population of N0 = 20,000 that split into two equal sized subpopulations (N1 = N2 = 10,000) 650 generations before present. Assuming a 15-year generation time (Ingvarsson 2008), this corresponds to ∼10,000 years ago, when a large proportion of the glaciated area of P. balsamifera’s current range started to become ice free and available for colonization. For three reasons our demographic model should produce conservative estimates of differentiation for testing local adaptation: there was no migration between subpopulations (allowing that migration narrowed the confidence intervals on πT-S; results not shown), unaccounted-for population substructure will elevate πS, thereby lowering the empirical estimates of πT-S, and finally the northern subgroup we used for our empirical data contained individuals from all three regional genetic clusters (Keller et al. 2010), which should increase empirical estimates of πS and lower πT-S over expectations from a simple north–south demographic split. We simulated 50,000 replicates of the coalescent with Hudson’s MS, conditioned on the total number of segregating sites for each phenology gene, and compared the observed πT-S to the 95% confidence interval under neutrality. In addition, we scanned for regions of elevated differentiation within genes by conducting sliding window analyses of πT-S using 300-bp windows with 100-bp step increment.
Results
Nucleotide diversity at phenology genes vs. expectations from demographic history
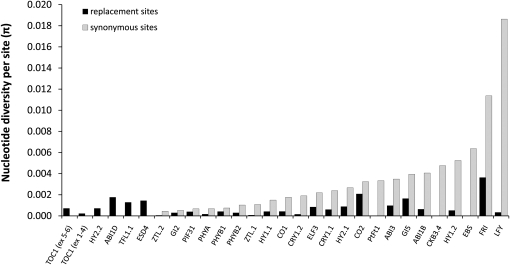
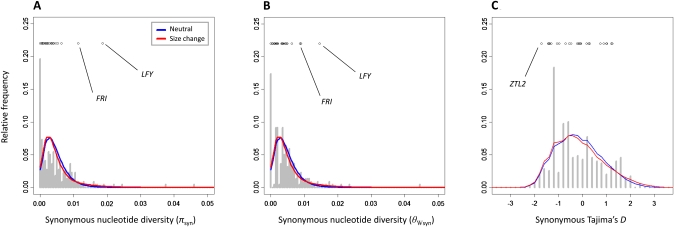
We sequenced 876–5434 bp from each of the 27 phenology candidate genes, resulting in ∼71 kb of sequence from each of 24 individuals, sampled across the range of P. balsamifera (Figure 1). Nucleotide diversities based on segregating sites and pairwise divergence were highly variable among phenology genes (θW = 0.000 – 0.0146; π = 0.000 – 0.0186; Figure 2 and Table S3) and most phenology genes had synonymous site θW, π, and D that were well within the range of values from the genome-wide empirical distributions of reference loci (Figure 3 and File S2). However, two phenology genes, LFY and FRI, had high pairwise diversity (πsyn = 0.0186 and πsyn = 0.0114, respectively, Figure 3A) within the upper 2% and 7% tail of the empirical distribution. These two genes also harbored more segregating sites (θsyn) than other candidates and were in the upper tail of the empirical distribution (LFY θsyn = 0.0146, upper 4%; FRI θsyn = 0.0088, upper 17.5% tail; Figure 3B). In contrast, four phenology genes had zero synonymous polymorphism (ABI1D, HY2.2, TFL1.1, and TOC1). Because a large number of reference loci lacked polymorphism, these were not outliers in the empirical distributions (Figure 3). Only one phenology gene, ZTL2, exhibited an excess of rare variants (Dsyn = −1.72) relative to reference loci (lower 3% tail).
Figure 2 .
Phenology gene nucleotide diversity from pairwise divergence (π) estimated for replacement and synonymous sites. Genes are ranked from lowest to highest on the basis of πsyn.
Figure 3 .
Comparison of nucleotide diversity (A and B) and site frequency spectrum (C) at synonymous sites for phenology genes (points), background reference loci (shaded bars), and coalescent simulations from approximate Bayesian computation (ABC) demographic models (colored lines). The two ABC models with high posterior probability are shown: a standard neutral model (blue) and a size change model (red).
Because reference loci sequences were considerably shorter than our phenology genes, the empirical distribution was skewed toward values of zero diversity (Figure 3). For this reason, we generated distributions of expected neutral diversity using an ABC-derived demographic model. After rejecting all but 0.1% of the coalescent simulations that most closely matched our empirical data, 57.2% of our ABC simulations were from a sequential size change model, 42.2% from a standard neutral model, and 0.6% from a growth model. While the growth model was clearly rejected, the standard neutral model with a current effective population size of Ne = 32,000 was almost as good a fit to the reference loci as the size change model, which described a large ancestral population of Ne = 148,000, bottlenecked ∼1.7 MYA to an Ne = 36,600, and then reduced in size again ∼639 KYA to an Ne = 32,200 (Table 1). Because the timing of the oldest bottleneck far exceeds the expected coalescent time within P. balsamifera, as well as the estimated divergence time between P. balsamifera and P. trichocarpa (Levsen et al., unpublished data), this model likely is affected by the recent speciation of P. trichocarpa and P. balsamifera and possibly introgression of alleles from more distantly related Populus species.
Table 1. Approximate Bayesian computation (ABC) analysis of the two best fitting models of species-wide demographic history in P. balsamifera.
| Standard neutral model | Bottleneck model | |
|---|---|---|
| % of retained simulations | 42.2 | 57.2 |
| Theta (πs) × 10−3 | 4.79 (4.34–5.30) | 4.83 (4.10–6.12) |
| Recombination (ρs) × 103− | 0.66 (0.028–1.86) | 1.11 (0.078–1.89) |
| Current population size | 32,000 (28,900–35,300) | 32,200 (27,300–40,800) |
| Time to size changea | – | 639,000 (503,000–3,320,000) |
| Size change population size | – | 36,600 (7,100–50,500) |
| Time to ancestral population1 | – | 1,740,000 (637,000–4,490,000) |
| Ancestral population size | – | 148,000 (76,700–194,000) |
A third model of exponential growth provided a poor fit to the observed data (0.6% of retained simulations), and so was not included for parameter estimation.
Time estimates were converted from coalescent units (= 4N0 generations) to absolute time (in years), assuming a neutral mutation rate per site per year of 2.5 × 10−9 (Tuskan et al. 2006).
Distributions of nucleotide diversity from ABC demographic models were still skewed toward low values, similar to the empirical distributions, but there was no longer a peak at zero diversity, reflecting that the simulated sequences were considerably longer than the reference loci. Moreover, LFY and FRI remained outliers in the simulated distributions; values of synonymous nucleotide diversity at LFY and FRI were in the upper 0.5% and 5% tails of πsyn, respectively, and in the upper 1.5% and 11.5% tails of θsyn (Figure 3, A and B). The four phenology genes with zero synonymous polymorphism (ABI1D, HY2.2, TFL1.1, and TOC1) fell within the lower 1% of simulated values of πsyn and θsyn. Similar to the empirical distribution, only ZTL2 showed significantly negative Tajima’s D, falling in the lower 2.5% tail of simulated values (Figure 3C).
Tests of selection based on comparisons of diversity and divergence
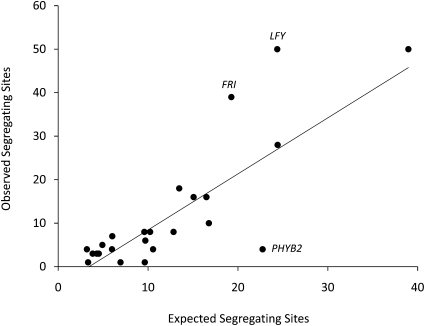
The average silent site divergence of phenology genes (KS) was 0.033, and the ratio of replacement to silent site divergence (KA/KS) was 0.29 (Table S4). Four phenology genes (CRY1.1, EBS, ELF3, and ZTL2) showed strongly negative values of H (all values P < 0.05) indicating an excess of high frequency derived sites consistent with positive selection (Table S4). HKA tests comparing diversity within P. balsamifera to divergence from P. tremula revealed significant heterogeneity among phenology genes (χ2 = 67.49, d.f. = 23, P < 0.001), with the largest deviances from neutrality driven by an excess of polymorphism in LFY and FRI, and a deficit of polymorphism in PHYB2, relative to divergence from P. tremula (Figure 4 and Table S4).
Figure 4 .
HKA test of genetic hitchhiking. Values are observed numbers of segregating sites at each gene vs. expected values based on 10,000 neutral coalescent simulations in J. Hey’s HKA software. The line is the best fit excluding genes showing evidence of selection (labeled).
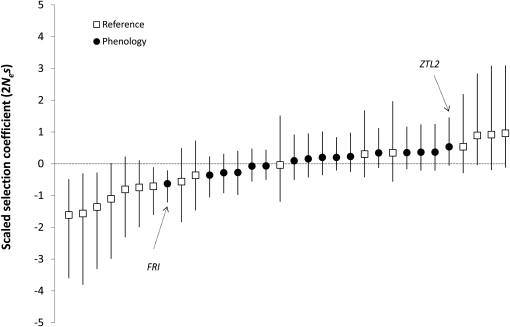
Estimates of the scaled selection coefficient on replacement sites γ (2Nes) revealed similar patterns of replacement site polymorphism relative to divergence at phenology and reference genes (Figure 5). Two phenology genes showed evidence of nonneutral evolution at replacement sites. FRI had a significantly negative selection coefficient (γ = −0.626, P < 0.001), indicating an abundance of replacement site polymorphism relative to neutral expectations, likely caused by either balancing/local selection maintaining polymorphism or segregating weakly deleterious mutations. The other gene, ZTL2, showed an excess of fixed replacement sites between P. balsamifera and P. tremula (γ = 0.533, P = 0.045) consistent with positive selection.
Figure 5 .
Strength of selection (γ = 2Nes) on replacement site variation. Error bars are 95% credible intervals from a hierarchical Bayesian MKPRF analysis. Genes showing significant deviation from zero are labeled with arrows.
Local selection and latitudinal adaptation
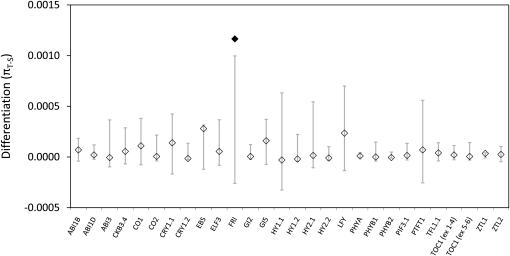
The elevated range-wide diversity at LFY and FRI suggests the possibility of geographically variable local selection. To test this, we compared differentiation between high and low latitude accessions (πT-S) for the phenology genes relative to expectations from a neutral model of range expansion following Pleistocene glacial retreat. This demographic model produced null distributions that frequently overlapped empirical estimates of πT-S from phenology genes. At the whole-gene level, only FRI showed πT-S in excess of the demographic model, falling in the upper 1% of neutral values (Figure 6).
Figure 6 .
Coalescent analysis of nucleotide differentiation between high and low latitude samples. Shaded bars denote 95% confidence intervals around the simulated distribution from the neutral demographic model. Diamonds are observed values of differentiation, with values falling outside of the simulated distributions depicted by solid diamonds.
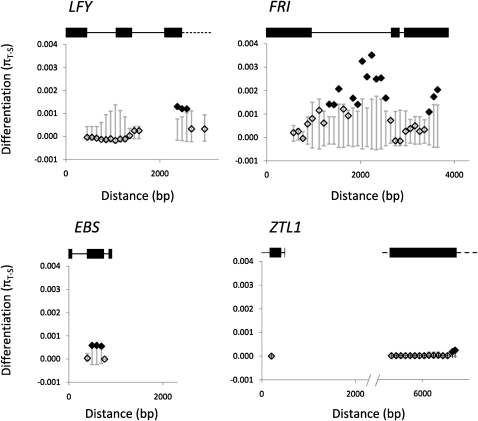
Sliding window analyses of πT-S identified windows of elevated πT-S within four phenology genes. These included 13 windows in FRI (upper 0.2–2% of simulated values) which also showed excess north–south differentiation in the whole-gene analysis, as well as LFY (three windows, upper 0.2–2%), EBS (two windows, upper 2%), and ZTL1 (two windows, upper 1–2%) (Figure 7 and Figure S2). Due to the large number of comparisons, some of these windows may be significant by chance alone. We reiterate, however, that our tests of differentiation should be conservative since our demographic null model simulated drift between subgroups with no migration and no population structure within subgroups. Many windows of elevated differentiation were composed predominately, or at times exclusively, of polymorphisms at synonymous or intron sites that are typically assumed to be silent with regard to selection. For example, in FRI, latitudinal differentiation was most pronounced across the first intron (Figure 7). Similarly, in EBS and ZTL1, latitudinal differentiation was exclusively at synonymous SNPs in the regions sequenced.
Figure 7 .
Sliding window analysis of latitudinal differentiation within phenology candidate genes. At the top of each plot is shown the gene boundaries, including exons (rectangles), introns (thin solid lines), and UTR. Note that the current gene model from v. 2.0 of the P. trichocarpa genome divides ZTL1 into two separate coding regions separated by intergenic space. See Figure 6 legend for additional details.
Discussion
Molecular adaptation of phenology network genes
Among the 27 candidate genes for adaptive phenology, FRI was repeatedly identified as a potential target of selection. In particular, FRI harbored high intraspecific nucleotide diversity, was among the upper tail of diversity values from the empirical genome-wide and simulated ABC distributions, and showed elevated nucleotide differentiation between northern and southern accessions. The functional role of FRI in temperature-induced phenological responses makes this gene a particularly strong candidate for local adaptation to regional differences in growing season length. Among natural accessions of A. thaliana, FRI is a major regulator of flowering-time variation by controlling expression of FLC to produce a vernalization requirement in high latitude accessions, permitting flowering only after a prolonged exposure to cold temperatures (Johanson et al. 2000; Mouradov et al. 2002; Putterill et al. 2004). In balsam poplar, our analyses implicate FRI in the response to local selection at different latitudes, probably reflecting a role of FRI in the temperature-sensitive timing of seasonal development. Because flowering-time homologs in Populus can affect a range of developmental traits, including flowering and bud dormancy (Böhlenius et al. 2006; Ingvarsson et al. 2008; Lagercrantz 2009; Ma et al. 2010), the link between allelic variation in FRI and variation in specific phenotypes in poplar awaits further study.
Interestingly, perhaps the strongest evidence for local selection in FRI, excess north–south differentiation, is largely due to polymorphism at synonymous and intron sites within our sequenced region, and these are separated from the nearest nonsynonymous polymorphism by windows of nonsignificant differentiation. It is possible that some of the silent site variation in FRI results in alternate splice variants, changes to gene regulatory regions, or RNA processing. In this context, it is relevant to note that much of the natural phenotypic variation in Arabidopsis flowering time is associated with loss of function mutants that result in premature truncation of the FRI protein, rather than nonsynonymous polymorphisms (Johanson et al. 2000; LeCorre 2002).
Three other genes, LFY, ZTL1, and EBS, also showed evidence of local selection. LFY, a transcription factor that is part of the integrative pathway downstream from FLC and FRI that is responsible for floral meristem and organ development (Moyroud and Tichtinsky 2009), harbored an excess of intraspecific diversity. Regions of exon 3 in LFY also showed greater than expected latitudinal differentiation. Multiple functional studies have shown that LFY affects flowering and other developmental phenotypes, including in Populus (Rottman et al. 2000). In contrast to the very high levels of segregating variation in Populus LFY, Arabidopsis LFY harbors low diversity consistent with a recent selective sweep (Olsen et al. 2002). The protease-encoding EARLY BOLTING IN SHORT DAYS (EBS) that regulates the transition from juvenile to reproductive maturity in Populus through transcriptional control of FT (Böhlenius et al. 2006) also showed an excess of north–south differentiation.
Finally, both ZEITLUPE paralogs (ZTL1 and ZTL2) showed evidence of selection across multiple analyses. ZTL1 showed a combination of low diversity (θsyn = 0.00057; <5% of values from ABC simulations), and evidence of a balanced polymorphism (Dsyn = 1.24; >85% of values from ABC simulations). This was due to a single synonymous SNP at the end of exon 2 segregating at high frequency, which also showed differentiation between northern and southern accessions (Figure 7). This could reflect genetic hitchhiking during local selective sweeps, or drift during expansion (Excoffier and Ray 2008). Diversity at ZTL2 was consistent with a recent selective sweep – including an excess of rare polymorphisms and derived variants (negative Tajima’s D and Fay and Wu’s H) and evidence of positive selection on replacement sites (positive γ from MKPRF). ZEITLUPE genes are hypothesized to be blue light photoreceptors, known to interact functionally with the circadian clock genes GIGANTEA (GI) and TIMING OF CAB 1 (TOC1) (Somers et al. 2000; Kim et al. 2007).
Several recent studies have investigated evidence for adaptation in plant genomes (Gossman et al. 2010), including specifically on the flowering-time gene network (Flowers et al. 2009, Hall et al. 2011), and have found scant evidence for adaptive evolution (but see Ingvarsson 2010 and Hall et al. 2011 for Populus, and Slotte et al. 2010 for Capsella). However, the generally employed sampling schemes and analytical methods in these and similar studies are designed to detect positive selection on a species-wide scale, and less attention has been paid to scanning sequence data for signatures of local selection within a species (Tenaillon and Tiffin 2008; Siol et al. 2010). In our species-wide tests, we found only weak evidence of selective sweeps in P. balsamifera. This is in contrast to P. tremula, which shows evidence of positive selection at a large fraction of substitutions (Ingvarsson 2010). Thus, had we confined our analyses in P. balsamifera to just the species-wide scale, there would have been little to suggest adaptation in phenology genes, despite their prominent role in controlling important life-history traits. Theoretical studies of geographically variable selection on quantitative traits indicate that local adaptation may be difficult to detect from QTL or DNA sequence data, unless adaptation involves genes of major effect (Latta 2003; Kelly 2006). Our findings of local selection in Populus, along with several recent studies in other plant species, contribute empirical evidence that geographically variable selection can be detected at the level of DNA sequence (LeCorre 2002; Ingvarsson et al. 2006; Moeller and Tiffin 2008; Wachowiak et al. 2009; Ma et al. 2010; Turner et al. 2010), and may constitute a major cause of adaptive evolution in plant genes that underlie ecologically important traits. Local selection on poplar phenology genes is also consistent with previous results showing elevated phenotypic differentiation along a latitudinal gradient for traits such as bud set and height growth (Keller et al. 2011).
Common targets of selection on plant phenology genes
In addition to our study, the molecular population genetics of the phenology network have been investigated in P. tremula (25 genes; Hall et al. 2011) and A. thaliana (52 genes; Flowers et al. 2009), and many of the individual genes involved in developmental phenology have been investigated in related species in the Brassicaceae. These data provide an opportunity to investigate the repeatability of ecological adaptation at the molecular level (Stern and Orgogozo 2009), and in a gene network that is known to control important life-history phenotypes (Mouradov et al. 2002; Putterill et al. 2004; Lagercrantz 2009). In both P. balsamifera and A. thaliana, the strongest candidate for local adaptation to conditions that covary with latitiude is FRI, which harbors high polymorphism as well as elevated subpopulation differentiation between northern and southern accessions in both species (LeCorre 2002, 2005; Caicedo et al. 2004; Stinchcombe et al. 2004; this study). FRI alleles also display an excess of long-range haplotypes in A. thaliana, consistent with a recent history of selection (Flowers et al. 2009). Population genetic analysis of FRI has not yet been done in P. tremula. Two aspects of FRI may explain why it has been the target of independent local selection in these phylogenetically distant species. First, FRI shows relatively low “connectedness” with other genes in the network, especially compared to core circardian clock genes that participate in many epistatic interactions, which may lessen the amount of constraint of FRI’s rate of evolution (Rausher et al. 1999; Yukelivich et al. 2008; Stern and Orgogozo 2009). Studies have shown that FRI interacts directly with FLC to affect flowering in Arabidopsis, but FRI does not appear to interact with any other core components of the network. Second, at least in A. thaliana, loss-of-function mutations hasten development and produce earlier flowering and potentially provide adaptive phenotypic variation (Johanson et al. 2000; Scarcelli and Kover 2009). Given that loss-of function mutations are much more common than gain-of function, this may result in an evolutionary bias toward ecologically relevant allelic variation in FRI underlying adaptation to local variation in growing season. At this stage, it is not possible to identify which trait(s) mediate local selection in FRI, as multiple different developmental phenotypes are probably affected by the outputs from this network, including, but not limited to, flowering time and bud traits.
Members of the phytochrome family of light-signaling genes also appear to be repeat targets of local selection in some taxa, although they do not appear to be under local selection in P. balsamifera. In P. tremula, PHYB2 shows excess polymorphism as well as strong latitudinal clines (Ingvarsson et al. 2006; Hall et al. 2007; Ma et al. 2010; Hall et al. 2011) and in Cardamine nipponica, a close relative of Arabidopsis, PHYE shows elevated replacement site divergence between northern and southern populations, and a significant excess of intermediate frequency polymorphism in species-wide samples (i.e., positive Tajima’s D) (Ikeda et al. 2009). In P. tremula, a role in local adaptation is bolstered by an association between PHYB2 alleles and adaptive variation in bud set—a key phenotypic trait that controls local adaptation to photoperiod (Ingvarsson et al. 2008; Ma et al. 2010). In contrast, the phytochromes in P. balsamifera (this study) show low nucleotide diversity and little evidence of positive selection (Table S4 and Figure 2), nor do phytochrome genes cosegregate with bud set QTL in Populus trichocarpa × P. deltoides mapping populations (Chen et al. 2002; Rohde et al. 2010). Although congenerics, P. balsamifera and P. tremula, are geographically isolated and estimated to have last shared a common ancestor ∼40 million years ago (Hamzeh and Dayanandan 2004); thus they have presumably migrated and adapted to northern latitude environments independently. The difference in selection on phenology genes suggests that despite showing convergent phenotypic clines in bud set with latitude, these two poplar species differ in the genetic basis of local adaptation.
In summary, we found evidence for local selection driving patterns of polymorphism and latitudinal differentiation in at least one and up to four members of the phenology gene network of Populus balsamifera, while evidence for species-wide selection was confined to a single gene. The finding of local selection on the phenology gene network of both balsam poplar and European aspen, along with the apparently different genic targets of selection in each species, suggests that Populus may be a promising system for understanding the genetic basis of local adaptation, and how different forms of selection, along with the historical contingencies of population demographic history, combine to affect the evolution of genes underlying ecologically important phenotypes.
Acknowledgments
We thank Bill Schroeder for kindly sharing tissue samples for DNA extraction, Jennifer Reese and Riya Jayachandran for assistance with sequencing, Amanda Robertson and Naoki Takebayashi for discussions and programming advice, and computational support from the Life Science Informatics cluster at the University of Alaska, Fairbanks, and the Minnesota Supercomputing Institute at the University of Minnesota. We also appreciate the comments of Outi Savolainen and two anonymous reviewers. This work was funded by National Science Foundation Plant Genome award DBI-0701911 to M.S.O. and P.T., and the Swedish Research Council to P.K.I.
Literature Cited
- Beaumont M. A., Zhang W. Y., Balding D. J., 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H., Huang T., Charbonnel-Campaa L., Brunner A. M., Jansson S., et al. , 2006. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Brachi B., Faure N., Horton M., Flahauw E., Vazquez A., et al. , 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw W. E., Zani P. A., Holzapfel C. M., 2004. Adaptation to temperate climates. Evolution 58: 1748–1762 [DOI] [PubMed] [Google Scholar]
- Bustamante C. D., Nielsen R., Sawyer S. A., Olsen K. M., Purugganan M. D., et al. , 2002. The cost of inbreeding in Arabidopsis. Nature 416: 531534. [DOI] [PubMed] [Google Scholar]
- Bustamante C. D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M. T., et al. , 2005. Natural selection on protein-coding genes in the human genome. Nature 437: 1153–1157 [DOI] [PubMed] [Google Scholar]
- Caicedo A. L., Stinchcombe J. R., Olsen K. M., Schmitt J., Purugganan M. D., 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101: 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15: 538–543 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Nordborg M., Charlesworth D., 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70: 155–174 [DOI] [PubMed] [Google Scholar]
- Chen T. H. H., Howe G. T., Bradshaw H. D., 2002. Molecular genetic analysis of dormancy-related traits in poplars. Weed Sci. 50: 232–240 [Google Scholar]
- Chen H., Patterson N., Reich D., 2010. Population differentiation as a test for selective sweeps. Genome Res. 20: 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Hanzawa Y., Chou L., Roe J. L., Kover P. X., et al. , 2009. Candidate gene association mapping of Arabidopsis flowering time. Genetics 183: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Ray N., 2008. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 23: 347–351 [DOI] [PubMed] [Google Scholar]
- Fay J. C., Wu C.-I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers J. M., Hanzawa Y., Hall M. C., Moore R. C., Purugganan M. D., 2009. Population genomics of the Arabidopsis thaliana flowering time gene network. Mol. Biol. Evol. 26: 2475–2486 [DOI] [PubMed] [Google Scholar]
- Frewen B. E., Chen T. H. H., Howe G. T., Davis J., Rohde A., et al. , 2000. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154: 837–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Lewontin R., Wakeley J., 2010. Measuring the sensitivity of single-locus “neutrality tests” using a direct perturbation approach. Mol. Biol. Evol. 27: 73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossman T. I., Song B.-H., Windsor A. J., Mitchell-Olds T., Dixon C. I., et al. , 2010. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27: 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D., Luquez V., Garcia M. V., St Onge K. R., Jansson S., et al. , 2007. Adaptive population differentiation in phenology across a latitudinal gradient in European Aspen Populus tremula, L.: A comparison of neutral markers, candidate genes and phenotypic traits. Evolution 61: 2849–2860 [DOI] [PubMed] [Google Scholar]
- Hall D., Ma X. F., Ingvarsson P. K., 2011. Adaptive evolution of the Populus tremula photoperiod pathway. Mol. Ecol. 20: 1463–1474 [DOI] [PubMed] [Google Scholar]
- Hamzeh M., Dayanandan S., 2004. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. Am. J. Bot. 91: 1398–1408 [DOI] [PubMed] [Google Scholar]
- Hancock A. M., Witonsky D. B., Gordon A. S., Eshel G., Pritchard J. K., et al. , 2008. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 4: e32 10.1371/journal.pgen.0040032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson M. J., Stahl E., Takebayashi N., 2007. msBayes: a flexible pipeline for comparative phylogeographic inference using approximate Bayesian computation (ABC). BMC Bioinformatics 8: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright–Fisher neutral model. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguade M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Fujii N., Setoguchi H., 2009. Molecular evolution of phytochromes in Cardamine nipponica Brassicaceae suggests the involvement of PHYE in local adaptation. Genetics 182: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson P. K., 2008. Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson P. K., 2010. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol. Biol. Evol. 27: 650–660 [DOI] [PubMed] [Google Scholar]
- Ingvarsson P., Garcia M. V., Hall D., Luquez V., Jansson S., 2006. Clinal variation in phyB2, a candidate gene for daylength-induced growth cessation and bud set, across a latitudinal gradient in European Aspen Populus tremula. Genetics 172: 1845–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson P. K., Garcia M. V., Luquez V., Hall D., Jansson S., 2008. Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen Populus tremula, Salicaceae. Genetics 178: 2217–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kim Y., 2008. Detecting local adaptation using the joint sampling of polymorphism data in the parental and derived populations. Genetics 179: 1713–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. D., 2009. Plant responses to photoperiod. New Phytol. 181: 517–531 [DOI] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., et al. , 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Keller S. R., Olson M. S., Silim S., Schroeder W., Tiffin P., 2010. Genomic diversity, population structure, and migration following rapid range expansion in the Balsam Poplar, Populus balsamifera. Mol. Ecol. 19: 1212–1226 [DOI] [PubMed] [Google Scholar]
- Keller S. R., Soolanayakanahally R. Y., Guy R. D., Silim S. N., Olson M. S., et al. , 2011. Climate-driven local adaptation of ecophysiology and phenology in balsam poplar, Populus balsamifera L. (Salicaceae). Am. J. Bot. (in press). [DOI] [PubMed] [Google Scholar]
- Kelly J., 2006. Geographical variation in selection, from phenotypes to molecules. Am. Nat. 167: 481–495 [DOI] [PubMed] [Google Scholar]
- Kim W.-Y., Fujiwara S., Suh S.-S., Kim J., Kim Y., et al. , 2007. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Lagercrantz U., 2009. At the end of the day: A common molecular mechanism for photoperiod responses in plants? J. Exp. Bot. 60: 2501–2515 [DOI] [PubMed] [Google Scholar]
- Latta R., 2003. Gene flow, adaptive population divergence and comparative population structure across loci. New Phytol. 161: 51–58 [Google Scholar]
- Le Corre V., 2002. DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol. Biol. Evol. 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Le Corre V., 2005. Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Mol. Ecol. 14: 4181–4192 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Ma X. M., Hall D., St. Onge K. R., Jansson S., Ingvarsson P., 2010. Genetic differentiation, clinal variation and phenotypic associations with growth cessation across the Populus tremula photoperiodic pathway. Genetics 10.1534/genetics.110.120873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. Camb. 23: 23–35 [PubMed] [Google Scholar]
- Moeller D. A., Tiffin P., 2008. Geographic variation in adaptation at the molecular level: a case study of plant immunity genes. Evolution 62: 3069–3081 [DOI] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G., 2002. Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyroud E., Tichtinsky G., 2009. The LEAFY floral regulators in Angiosperms: conserved proteins with diverse roles. J. Plant Biol. 52: 177–185 [Google Scholar]
- Neiman M., Olson M. S., Tiffin P., 2009. Selective histories of poplar protease inhibitors: elevated polymorphism, purifying selection, and positive selection driving divergence of recent duplicates. New Phytol. 183: 740–750 [DOI] [PubMed] [Google Scholar]
- Nielsen R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218 [DOI] [PubMed] [Google Scholar]
- Nielsen R., Hubisz M. J., Hellmann I., Torgerson D., Am. Andrés A., et al. , 2009. Darwinian forces affecting human protein coding genes. Genome Res. 19: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Innan H., 2003. The genealogy of sequences containing multiple sites subject to strong selection in a subdivided population. Genetics 163: 1201–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen K. M., Womack A., Garrett A. R., Suddith J. I., Purugganan M. D., 2002. Contrasting evolutionary forces in the Arabidopsis thaliana floral developmental pathway. Genetics 160: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. S., Robsertson A. L., Takebayashi N., Silim S., Schroeder W. R., et al. , 2010. Nucleotide diversity and linkage disequilibrium in balsam poplar Populus balsamifera. New Phytol. 186: 526–536 [DOI] [PubMed] [Google Scholar]
- Pickrell J. K., Coop G., Novembre J., Kudaravalli S., Li J. Z., et al. , 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19: 826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Laurie R., Macknight R., 2004. It’s time to flower: the genetic control of flowering time. BioEssays 26: 363–373 [DOI] [PubMed] [Google Scholar]
- Rausher M. D., Miller R. E., Tiffin P., 1999. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Mol. Biol. Evol. 16: 266–274 [DOI] [PubMed] [Google Scholar]
- Rohde A., Storme V., Jorge V., Gaudet M., Vitacolonna N., et al. , 2011. Bud set in poplar: genetic dissection of a complex trait in natural and hybrid populations. New Phytol. 189: 106–121 [DOI] [PubMed] [Google Scholar]
- Rottman W. H., Meilan R., Sheppard L. A., Brunner A. M., Skinner J. S., et al. , 2000. Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 22: 235–245 [DOI] [PubMed] [Google Scholar]
- Savolainen O., Pyhäjärvi T., Knurr T., 2007. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 38: 595–619 [Google Scholar]
- Scarcelli N., Kover P. X., 2009. Standing genetic variation in FRIGIDA mediates experimental evolution in flowering time in Arabidopsis. Mol. Ecol. 18: 2039–2049 [DOI] [PubMed] [Google Scholar]
- Simpson G. G., Dean C., 2002. Arabidopsis, the Rosetta Stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Siol M., Wright S. I., Barrett S. C. H., 2010. The population genomics of plant adaptation. New Phytol. 188: 313–332 [DOI] [PubMed] [Google Scholar]
- Slotte T., Holm K., McIntyre L. M., Lagercrantz U., Lascoux M., 2007. Differential expression of genes important for adaptation in Capsella bursa-pastoris (Brassicaceae). Plant Physiol. 145: 160–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Foxe J. P., Hazzouri K. M., Wright S. I., 2010. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 27: 1813–1821 [DOI] [PubMed] [Google Scholar]
- Somers D. E., Schultz T. F., Milnamow M., Kay S. A., 2000. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Soolanayakanahally R. Y., Guy R. D., Silim S. N., Drewes E. C., Schroeder W. R., 2009. Enhanced assimilation rate and water use efficiency with latitude through increased photosynthetic capacity and internal conductance in balsam poplar (Populus balsamifera L.). Plant Cell Environ. 32: 1821–1832 [DOI] [PubMed] [Google Scholar]
- Stephens M., Scheet P., 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 76: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Smith N. J., Donnelly P., 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. L., Orgogozo V., 2009. Is genetic evolution predictable? Science 323: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe J. R., Weinig C., Ungerer M., Olsen K. M., Mays C., et al. , 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. USA 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F., Kelly J. K., 2008. Effects of spatially varying selection on nucleotide diversity and linkage disequilibrium: insights from deer mouse globin genes. Genetics 180: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K., Thornton K. R., Stoneking M., 2007. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5: 1587–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon M. I., Tiffin P., 2008. The quest for adaptive evolution: a theoretical challenge in a maze of data. Curr. Opin. Plant Biol. 11: 110–115 [DOI] [PubMed] [Google Scholar]
- Thornton K., 2009. Automating approximate Bayesian computation by local linear regression. BMC Genet. 10: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Bourne E. C., Von Wettberg E. J., Hu T. T., Nuzhdin S. V., 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42: 260–263 [DOI] [PubMed] [Google Scholar]
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., et al. , 2006. The genome of the black cottonwood, Populus trichocarpa Torr. and Gray. Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Wachowiak W., Balk P., Savolainen O., 2009. Search for nucleotide diversity patterns of local adaptation in dehydrins and other cold-related candidate genes in Scots pine Pinus sylvestris L. Tree Genet. Genomes 51: 117–132 [Google Scholar]
- Yukelivich R., Lachance J., Aoki F., True J. R., 2008. Long-term adaptation of epistatic genetic networks. Evolution 62: 2215–2235 [DOI] [PubMed] [Google Scholar]