Abstract
Populations adapt physiologically using regulatory mechanisms and genetically by means of mutations that improve growth. During growth under selection, genetic adaptation can be rapid. In several genetic systems, the speed of adaptation has been attributed to cellular mechanisms that increase mutation rates in response to growth limitation. An alternative possibility is that growth limitation serves only as a selective agent but acts on small-effect mutations that are common under all growth conditions. The genetic systems that initially suggested stress-induced mutagenesis have been analyzed without regard for multistep adaptation and some include features that make such analysis difficult. To test the selection-only model, a simpler system is examined, whose behavior was originally attributed to stress-induced mutagenesis (Yang et al. 2001, 2006). A population with a silent chromosomal lac operon gives rise to Lac+ revertant colonies that accumulate over 6 days under selection. Each colony contains a mixture of singly and doubly mutant cells. Evidence is provided that the colonies are initiated by pre-existing single mutants with a weak Lac+ phenotype. Under selection, these cells initiate slow-growing clones, in which a second mutation arises and improves growth of the resulting double mutant. The system shows no evidence of general mutagenesis during selection. Selection alone may explain rapid adaptation in this and other systems that give the appearance of mutagenesis.
CLASSICAL experiments by Luria and Delbruck (1943) and Lederberg and Lederberg (1952) demonstrate that the bacterial mutants detected by stringent laboratory selections actually arise during the preceding nonselective growth period and are not induced in response to selection conditions. These experiments validate the laboratory use of stringent selection, which is essential to the practice of bacterial genetics. Later these experiments were interpreted more broadly as support for the idea that selective conditions never affect mutation rates. This broader interpretation has been called into question (Hall 1982; Shapiro and Brinkley 1984; Cairns et al. 1988; Cairns and Foster 1991).
Genetic adaptation can be very rapid in many situations. Examples are bacterial acquisition of resistance to antibiotics (Andersson and Hughes 2009; Sun et al. 2009) or host defenses (Matic et al. 1997), origins of cancer (Cairns 1998), and adaptive changes of beak size by Galapagos finches (Weiner 1994). Rapid adaptation has been seen in several laboratory genetic systems, in which bacteria were placed under selective conditions. The speed of adaptation in such systems suggested initially that selective stress might be mutagenic (Hall 1982; Shapiro and Brinkley 1984; Cairns et al. 1988; Steele and Jinks-Robertson 1992; Taddei et al. 1995, 1997; Torkelson et al. 1997).
Stress-induced mutagenesis, as discussed here, refers to genome-wide increases in mutation rate produced by cellular mechanisms suggested to have evolved because they speed genetic adaptation (Cairns et al. 1988; Foster 2007; Galhardo et al. 2007). This process has also been called adaptive mutation (Foster 1993, 1998). For this discussion, stress-induced mutagenesis is distinguished from mutagenesis by agents that damage DNA (e.g., UV, reactive oxygen species) and from mutagenesis caused by insufficiency of DNA repair systems. A theoretical difficulty with the idea of stress-induced general mutagenesis is that deleterious mutations are much more frequent than beneficial ones and many stresses cannot be relieved by any mutation (e.g., starvation in the absence of nutrients). The expected high cost of deleterious mutations makes it difficult to imagine how mechanisms of stress-induced general mutagenesis could evolve or be selectively maintained (Roth et al. 2006a). This cost could be avoided if stress could direct mutagenesis preferentially to sites that improve growth (Foster and Cairns 1992), a possibility that will be discussed here. The Cairns–Foster system is the only system for which genome-wide mutagenesis has been reported (Torkelson et al. 1997).
In the multiple genetic systems used to study the origin of new mutants under selective conditions, a population of bacterial cells is pregrown in nonselective liquid medium and then plated on solid selective medium that restricts growth of the majority cell type. During the ensuing several days, colonies appear and accumulate in number. The number of colonies is higher than the number of similar mutant cells present in the plated population, suggesting that growth limitation might be mutagenic. The idea of replication-independent mutagenesis is based in part on the assumption that the plated cell population does not grow but appears to give rise to new mutants over time (stationary phase mutagenesis). In part, the conclusion is also based on molecular differences between the mutations found before and after selection. Work on these systems has not ruled out the alternative possibility that revertant colonies are initiated by partially revertant cells present prior to selection and that secondary mutants arise within these developing colonies.
We have developed a general model by which growth limitation can enhance the number of fully adapted revertant colonies with no increase in mutation rate (Roth et al. 2006b; Roth 2010; Andersson et al. 2011). The basic features of this model are the following:
Mutation types that cause small increases in activity are often more common than one might expect based on experience with the large-effect mutations generally studied in the laboratory (Andersson et al. 2011). These frequent mutations include, but are not limited to duplications and amplifications.
Cells with small-effect mutations can initiate clones that grow slowly on selective medium.
The repeated acts of chromosome replication within these clones provide opportunities for secondary mutations that improve growth.
Cells with the secondary mutation(s) overgrow the colony.
When gene amplification improves growth of the initial cells, then opportunities for secondary changes are enhanced by increased number of mutation targets per cell as well as by an increased number of cells within each clone.
According to this model, the mutant colonies that arise above the lawn of plated parent cells are actually initiated by partially revertant cells within the plated population. The mutant colonies that accumulate above a nongrowing lawn seem to result from new mutant cells appearing during selection, but may be initiated by pre-existing small-effect mutants and improved by secondary mutations that are common under all conditions. We suggest that the selection-only model may explain (with no change in mutation rate) all of the systems initially interpreted as evidence for stress-induce mutagenesis.
Demonstrating the applicability of this model to any system requires identification of the proposed intermediates in the process and showing that their unenhanced formation rate and growth contribution are sufficient to explain the observed behavior. This task has been difficult, because the several systems used so far have features (e.g., Mu prophage, F′ plasmid) that complicate identification of the postulated intermediate cell types and measurement of their growth rates. The selection-only model can explain in principle the behavior of these pioneer systems, but has been difficult to test in these systems.
In the system of Shapiro, deletion mutations fuse silent lac genes to a nearby arabinose operon and modify or delete an intervening Mu prophage. (Shapiro 1984). The number of mutant colonies observed on selective plates is subject to fluctuation between parallel pregrowth liquid cultures, suggesting that revertants are initiated by cells that arise prior to plating (Foster and Cairns 1994). However, the molecular nature of the mutations recovered after selection is different from that of mutations found (by sib selection) in the unselected pregrowth cultures (Foster and Cairns 1994; Kim et al. 1997; Lamrani et al. 1999). These apparently contradictory observations can, in principle, be explained by the selection model above, if the cells arising prior to selection are partial or unstable revertants and the colonies arising under selection have acquired secondary changes that either improve growth or stability. In the RifR system (Taddei et al. 1995, 1997), the accumulation of RifR cells in an aging colony on LB medium was initially attributed to new mutations forming in stressed cells, but proved to reflect faster growth of pre-existing RifR mutants during the period of stress (Wrande et al. 2008). In the system of Hall (1982), cells unable to use lactose are plated on rich MacConkey lactose indicator plates, which favor growth of Lac+ revertants. On these plates, single Lac− parent cells form colonies that accumulate Lac+ papillae over several days. The Lac+ papillae carry multiple mutations and have been attributed to stress-induced mutagenesis directed to sites that improve growth. The behavior of these systems, could also be explained if one common small-effect mutant, formed during nonselective colony growth, initiated a subclone within which rarer secondary mutations lead to the observed Lac+ revertant (without mutagenesis).
In the system of Cairns and Foster (1991), reversion under selection depends heavily on having the lac region located on an F′ lac plasmid with an intact, constitutively expressed conjugation system, which may enhance selective amplification. It can now be shown that the number of revertant Lac+ colonies is dictated by rare cells with a lac amplification that are present in the unselected pregrowth culture (S. Maisnier-Patin, unpublished results; A. B. Reams, unpublished results; E. Sano, unpublished results). All of these systems behave in ways that are consistent with the selection model, but they make it hard to identify the essential intermediates, determine their growth rates, and test the selection model.
The question of rapid adaptation is addressed here using a simpler system designed by Yang et al. (2001, 2006). As in previous systems, a lawn of Lac− parent cells gives rise to ∼100 Lac+ revertant colonies over 6 days and their appearance was initially interpreted as evidence for stress-induced general mutagenesis. While the plated population divides several times during the first day, it grows very little during the period of colony accumulation. In this system, all genetic events affect the chromosome (neither F′ plasmid nor Mu prophage are involved). Most importantly, all mutations are standard analyzable types. Here we show that no general mutagenesis is involved and suggest that the origin of revertants reflects selection alone.
Evidence is presented that two mutations are required to restore full growth on lactose. The double mutants are expected to be extremely rare, but nevertheless appear without any associated increase in general mutation rate. Instead, revertant colonies are initiated by pre-existing singly mutant cells that initiate slow-growing colonies on selective medium. As these colonies grow, the second mutation occurs and a new double mutant overgrows each colony. The secondary mutations are possible because a large cell population within the colony is subject to standard mutation rates. The delay in revertant colony appearance reflects the time required for slow growth of the initial clone and more rapid overgrowth of the colony by the secondary double mutant. Selection contributes to an increase in the number of revertant colonies with no increase in the general mutation rate.
Materials and Methods
Strains
All strains are derived from Salmonella enterica serovar Typhimurium, strain LT2 and are listed in Table 1. The tester strain used in the original description of this system was provided by Yang, Liu, and Wang (Yang et al. 2001, 2006) and has been given the strain number TT25154. The genotype of this strain listed in Table 1 was revealed in the course of this work.
Table 1 . Strain list.
| Strain | Genotype |
|---|---|
| TR10000 | Wild type Salmonella enterica serovar Typhimurium strain LT2 |
| TT12306 | purD2145::MudJ Parent of the tester, lacking the purD2380 (UGA) mutation. |
| TT12360 | zda-1891::Tn10dTc (90% linked to purR+) |
| TT25154 | Tester strain purO+purD2380(UGA), purD2145::MudJ purR+ |
| TT26169 | purD2380(UGA) purD2145::MudJ purR2379::Cm(sw) |
| TT26170 | purD2380(UGA) purD2145::MudJ purR+zda-1891::Tn10dTc |
| TT26172 | purD2380(UGA) purD2145::MudJ dinB1014::Rf(sw) |
| TT26174 | purD2380(UGA) purD2145::MudJ mDEL1 (DEL2070) |
| TT26175 | purD2380(UGA) purD2145::MudJ mDEL1 (DEL2070) purO2381(G3A) |
| TT26176 | purD2380(UGA) purD2145::MudJ purO2382(C14T) |
| TT26177 | purD2380(UGA) purD2145::MudJ mDEL1 (DEL2070) purR2379::Cm(sw) |
| TT26208 | purD2145::MudJ mDEL1 (DEL2070) |
| TT26209 | purD2380 (UGA) purD2145::MudJ purO2382(C14T) purR2379::Cm(sw) |
Media
Rich medium was either Luria–Bertani medium (LB; Difco Laboratories) or nutrient broth (NB; Difco Laboratories). Minimal medium was no-citrate-E medium (NCE) (Davis et al. 1980). These media were solidified with 1.5% agar (Baltimore Biological). Prior to selection, cells were grown overnight in liquid NCE minimum medium with 0.2% glycerol plus adenine and thiamine to satisfy the strains nutritional requirements. Revertants were selected on NCE minimal plates supplemented with 10 µg/ml adenine, 0.06 mM thiamine, and with 1% lactose (Yang et al. 2001). The chromogenic β-galactosidase substrate X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was added at 25 µg/ml in minimal medium or 40 µg/ml in rich medium. Antibiotic concentrations in rich medium were: 50 µg/ml kanamycin sulfate (Km), 20 µg/ml tetracycline HCl (Tc), and 10 µg/ml chloramphenicol (Cm).
Reversion experiment
Cells grown overnight on glycerol/adenine were pelleted and resuspended in 1.0 ml NCE medium. Aliquots (0.1 ml or ∼108 cells) were plated on selective medium (NCE 1% lactose, X-gal, adenine, thiamine) and plates were incubated at 30° (Yang et al. 2001). Revertant Lac+ colonies were counted daily for a period of 6 days. Each data point in the figures represents the mean of at least 10 independent measurements.
The population of parental tester cells in the lawn of the selection plated was determined by taking three agar plugs from the surface of the selection plates, avoiding any Lac+ colonies. Cells from the plugs were suspended on NCE salts, vortex mixed, and diluted. Dilutions were plated on NB plates containing X-gal and used to calculate the cell population on the selective plate.
Determining frequency of unselected mutants before and after selection
The basal frequency of mutants unrelated to lactose use in the Pur-Lac tester strain (TT25154) was first assessed under nonselective growth conditions. Cells were grown overnight on rich medium, dispensed into 96-well plates at a density of 104 cells per 200 μl, and grown. Each culture was then diluted and plated for single colonies on rich medium. A total of 2000 colonies were picked and patched onto NCE-glucose plates with an added mixture of amino acids, nucleic acid bases, and vitamins that cannot serve as a carbon source but can satisfy the nutritional requirements of various auxotrophs. This mixture includes histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, tyrosine, valine, adenine, guanine, thymine, uracil, and thiamine, added at previously described concentrations (Davis et al. 1980). The colony patches were replica plated onto NCE-glucose medium to assess the frequency of auxotrophs and onto MacConkey-fructose, MacConkey-maltose, MacConkey-propanediol, and MacConkey-xylose agar plates to identify fermentation mutations.
To test the frequency of unselected mutants among Lac+ revertants following selection, reversion experiments were done on NCE lactose plates containing the mixture of supplements described above. A total of 1000 independent Lac+ revertant colonies that first appeared on day 6 were single-colony isolated on the same medium and one clone from each revertant colony was patched to NCE-glucose plates containing the above supplements. The patches were then replica printed to NCE-glucose (adenine plus thiamine) and to the various MacConkey agar plates (described above) to indentify mutants.
Genetic linkage tests to identify mutations that cause the reduced lac expression seen in the tester
The tester strain TT25154 was transduced using phage grown on strain TT12360, which carries a Tn10dTc insertion 90% linked to purR+. Transductants (TetR) were screened for their level of LacZ expression on rich medium with X-gal. All transductants showed the same low level of LacZ expression, suggesting that the mutation that reduced expression was not in or near the purR gene. The purHD::MudJ region was tested for the responsible mutation by growing transducing phage on the tester strain TT25154 and using it as a donor in a transductional cross with a wild-type strain selecting for inheritance of the kanamycin resistance encoded in the MudJ element. Most recombinants (97%) showed low LacZ levels typical of the donor tester strain, but a few (3%) showed high expression characteristic of the original parent strain with the simple purD::lac insertion. This demonstrates that the mutation that reduces lac expression in the tester strain lies in the bacterial chromosome very near the site of the MudJ insertion. Sequencing of this region revealed the polar purD UGA mutation.
Crosses to identify mutations in new Lac+ revertants of the tester strain
Phage grown on a strain with a Tn10dTc insertion near purR+ (TT12360) were used to transduce the Lac+ revertant to TetR. If the Lac+ phenotype of the revertant is due to a purR mutation, ∼90% of transductants would lose their blue-colony phenotype on X-gal. To test for mutations affecting the purD::MudJ region, transducing phage grown on the Lac+ revertant were used to transduce kanamycin resistance (encoded by MudJ) into a wild-type recipient. Transductants (KanR) were screened for Lac expression on X-gal plates. If the revertant carries a purO mutation, 90% of KanR transductants will carry the nearby purO mutation and show high lac expression and 10% will retain the recipient purO+ region and show low expression. If the revertant has a deletion of the stem region of the MudJ element (mDEL), 100% of KanR transductants inherit this deletion with the MudJ element and show high lac expression.
Luria–Delbruck fluctuation assay—scored after 2 days under selection
A single culture of the Pur-Lac tester strain was grown to saturation in 0.2% glycerol minimal medium supplemented with 10 μg/ml adenine and 0.06 mM thiamine overnight at 30° C. Cells were diluted to ∼100 cells/100 mL in 0.2% glycerol minimal medium (supplemented with adenine and thiamine) and distributed into 50 tubes. Each culture was grown overnight at 30° C. Cells from all 50 cultures were pelleted and resuspended in 0.2 ml 1× NCE salts solution. Aliquots of 0.1 ml from each culture (109 cells) were plated on 1% lactose minimal medium, supplemented with 10 μg/ml adenine, and 0.06 mM thiamine, and X-gal. Plates were incubated at 30°. Lac+ colonies were counted 44 hr after plating.
Luria–Delbruck fluctuation assay—scored after 3 and 6 days under selection
Twenty-five independent cultures were grown to saturation in 0.2% glycerol minimal medium supplemented with 10 μg/ml adenine and 0.06 mM thiamine. The cells were pelleted and resuspended in 1.0 ml 1× NCE salts solution. Aliquots of 0.1 ml (108 cells) were plated on 1% lactose minimal medium, supplemented with 10 μg/ml adenine, and 0.06 mM thiamine, and X-gal. Lac+ colonies were counted at the 3rd and 6th day after incubation. The total cell number was calculated by plugging the agar medium between the revertant colonies just like in the reversion experiment methodology described above.
Determining the reversion rate in the Pur-Lac system
Mutation rates can be calculated by using the distribution of mutant numbers in parallel cultures from a Luria–Delbruck fluctuation assay (Luria and Delbruck 1943; Rosche and Foster 2000, 2006). The mean or most likely number of mutational events per culture (m) was estimated by the curve fitting method (Rosche and Foster 2000). In this method, the mutant distribution is plotted as the log of the fraction of cultures with more than “x” mutants vs. the log of the mutants/tube (x). When the data describe a straight line of slope approaching a value of −1, m is estimated graphically as the intercept of this curve. When log(x) = 0 the log of the probability of tubes with >x mutations equals log(2m). The value of m is then used to calculate the mutation rate (μ) by dividing the observed number of mutational events (m) per culture by the average of total number of cells (N) of the parallel cultures as μ = m/N (Luria and Delbruck 1943).
Enzyme assays (β-galactosidase)
Cells were grown overnight on NCE 0.2% glycerol adenine medium at 37°. Overnight cultures were diluted 1/100 in fresh medium and grown to mid-log phase. Cells were washed with chilled Z-buffer and β-galactosidase activity was determined for permeabilized whole cells as described previously (Miller 1972).
DNA sequencing
The purR coding region or the purO region of the purHD operon were PCR amplified. A different set of primers was used to amplify the region of purD and MudJ genes that includes the stem structure. Strains were routinely screened by examining the size of the PCR product obtained from this region, but several fragments were sequenced to reveal the exact deletion end points. Products of PCR amplification were purified using the QIAquick PCR purification kit (cat. no. 28106) and sequenced at the College of Biological Sciences UCDNA Sequencing Facility (http://dnaseq.ucdavis.edu/).
Results
The genotype of the Pur-Lac tester strain
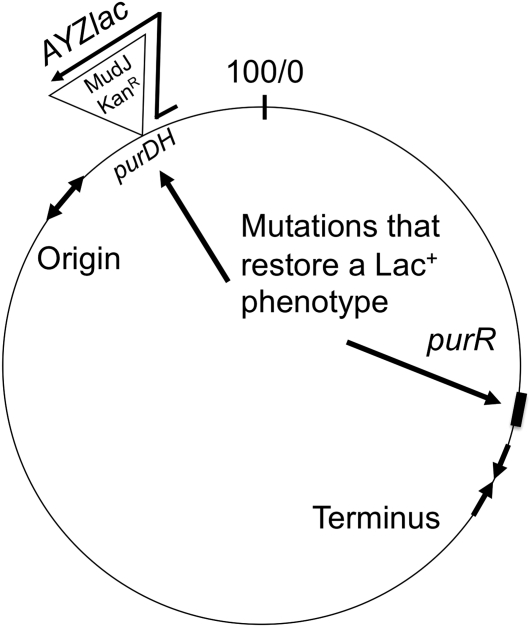
The genotype of the tester strain is diagrammed in Figure 1. The lac operon is inserted into the chromosomal purHD operon of Salmonella as part of a MudJ element (Casadaban and Cohen 1979), whose transposition functions are deleted. The lac genes are transcribed from the purHD promoter and expression is repressed when the purine repressor protein (PurR) binds the operator site (purO) (He et al. 1990). The hybrid purHD::lac operon is located in the bacterial chromosome far from the purR gene.
Figure 1 .
Genetic map of tester strain (TT25154). Growth on lactose is limited by low expression of the chromosomal lacZYA operon. The promoter of the purHD operon, which expresses the lac genes, is repressed by the PurR protein. Mutations that restore a Lac+ phenotype affect either the purR gene or several sites in the purHD::lac region (see arrows). These mutations are described in the text.
In the original description of this system (Yang et al. 2001, 2006), the tester strain carried the MudJ-lac insertion plus a second mutation added to impair growth on lactose. This second mutation was reported to be a purRS super-repressor mutation. Genetic tests and sequencing revealed no evidence of a purRS mutation, but showed a UGA nonsense mutation within the purD coding sequence (UGG 253 UGA) with a strong polar effect on lac expression. It should be noted that the original purD::MudJ-lac fusion strain (lacking the UGA mutation) produces a surprisingly high level of β-galactosidase (180 units) and the purD UGA mutation reduces this to ∼10 units. The purD UGA mutation was present in the tester strain used in all of the experiments described below.
Behavior of the Pur-Lac tester strain under selection
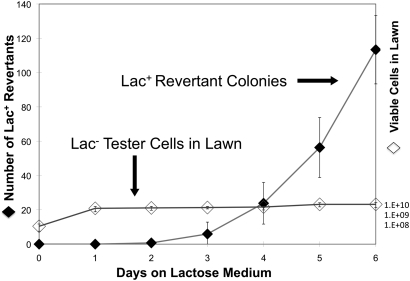
A typical reversion experiment is described in Figure 2. Cells of the tester strain are pregrown on minimal glycerol (adenine) medium and plated (108 cells) on minimal lactose (adenine) plates. The first Lac+ revertant colonies appear on day 3 and accumulate to ∼100 by day 6. The lawn of Lac− tester cells grows a few divisions during the first day, presumably using nutrients other than lactose that contaminate the agar, but no further tester growth occurs during the period in which revertant colonies accumulate. This behavior is strikingly similar to that of the Cairns system, in which a mutant lac operon carried by an F′ plasmid regains function under selection (Cairns and Foster 1991). An important difference is that the Pur-Lac system involves only chromosomal genes. Another difference is that the plated population goes through ∼6 divisions during day 1 on the selection plate, using nutrients that contaminate the agar. (This will be analyzed later.)
Figure 2 .
Time course of a reversion experiment. Tester cells (108) were grown on minimal glycerol medium, washed, and plated on minimal lactose medium. Colony (Lac+) number increases over 6 days. The lawn of tester cells increases for 1 day and then remains constant.
Looking for evidence of general mutagenesis
All revertant colonies appear after day 3, yet cells in these colonies (when retested under selection) are able to form a colony above a tester lawn within 2 days. (This evidence is described below.) This result suggested initially that the revertants formed on the plate and might be stress induced (Yang et al. 2001, 2006) as proposed for the Cairns system (Torkelson et al. 1997; Rosche and Foster 1999).
To determine whether general mutagenesis occurs in the Pur-Lac system, a reversion experiment was performed on medium that contained a mixture of amino acids, vitamins, and bases that would allow growth of a variety of auxotrophic mutants but cannot serve as a source of carbon and energy. One thousand Lac+ revertants appearing on day 6 were single-colony isolated and tested for auxotrophic requirements. These revertants were also tested for defects in use of fructose, maltose, propanediol, and xylose. This procedure reveals loss-of-function mutations in any of 100 chromosomal genes and was used previously to demonstrate that mutagenesis does occur during selection in the Cairns system (Slechta et al. 2002; Kofoid et al. 2003).
In the Pur-Lac system, only 1 of 1000 Lac+ revertants showed an associated auxotrophy. This frequency is roughly that predicted for a nonmutagenized culture. A control set of 2000 unselected nonrevertant clones were isolated following growth of the tester on rich medium and were tested for the same list of mutant phenotypes. Only 2 of the 2000 clones showed an associated mutation in any of the 100 genes tested. We conclude that no general mutagenesis occurs during selection in the Pur-Lac system. Later we will discuss this conclusion in more detail.
The DinB error-prone polymerase does not contribute to the yield of revertants in the Pur-Lac system
The DinB protein is central to several current models for stress-induced mutagenesis (Foster 2004; Galhardo et al. 2009; Cohen and Walker 2010). These models suggest that stress signals SOS induction of DinB, an error-prone repair polymerase, which then causes the mutagenesis.
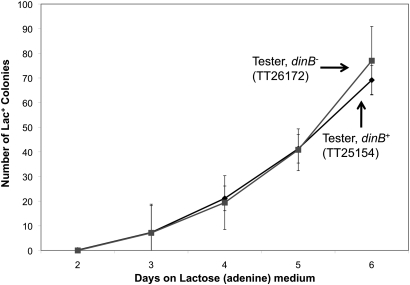
To determine whether DinB plays a role in the Pur-Lac system, the course of a reversion experiment was compared for strains with and without a dinB mutation. The results (Figure 3) show that accumulation of Lac+ revertants does not depend on DinB. In the Pur-Lac system, dinB is not located near the lac operon. Appearance of Lac+ revertants is not associated with mutagenesis and shows no dependence on DinB. It should be noted that reversion experiments in the Pur-Lac system are done at 30° so the lack of any DinB effect is in striking contrast to the strong effect of dinB on revertant number seen in the Cairns system at 30° (Cohen and Walker 2010).
Figure 3 .
Effect of a dinB mutation on reversion in the Pur-Lac system. Pur-Lac tester strain (TT25154) and an isogenic variant that carries a dinB mutation (TT26172) were plated on lactose (adenine) medium and the appearance of Lac+ colonies was scored for a period of 6 days.
Some Lac+ revertants carry an unexpected mutation
Selective reversion in the Pur-Lac system was originally described (Yang et al. 2001) as a simple, one-step process in which starved nongrowing tester cells acquire a mutation in either the purR gene (1000 bp) or the purO operator sequence (16 bp). Among the 44-day 6-revertant colonies analyzed here, an additional mutation type was encountered. Each revertant colony was single-colony isolated and a subclone was characterized by genetic crosses and DNA sequencing. The entire colony from which each subclone was derived was saved for future reference, since the colony population reflects the history of the revertant clone.
Of the single colony subclones isolated initially from 44 Lac+ revertant colonies, 18 carried a purR mutation and 4 a purO mutation. The sequence changes in these mutations are listed in Appendix 1. These two types were expected on the basis of the initial description of the system (Yang et al. 2001). The higher frequency of purR mutants was consistent with the larger mutational target of purR (∼1000 bp) compared to purO (16 bp). Unexpectedly, half of the 44 Lac+ revertant subclones had neither a purR nor a purO mutation. Genetic linkage tests showed that these revertants owed their Lac+ phenotype to a mutation (other than purO) linked to the purD::lac region.
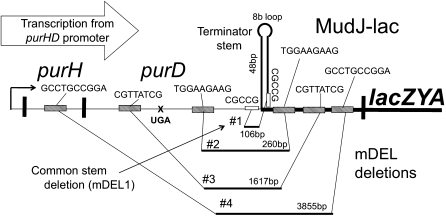
All of these subclones carried a deletion that extends across the junction between the purD coding sequence and the inserted MudJ element and removes an interrupted palindrome located just within MudJ. Of 22 sequenced stem deletion mutations, 21 removed 106 bp between repeats of the CGCCG sequence (mDEL1). An additional deletion removed 260 bp between repeats of the TGGAAGAAG sequence—one in purD and one within the inserted MudJ element (mDEL2). Both deletion types are diagrammed in Figure 4 with two additional longer deletions that include the site of the UGA mutation and will be described below.
Figure 4 .
Deletion mutations that increase lacZ expression and allow tester growth on lactose. Many of the characterized revertants in the Pur-Lac system have acquired a deletion of the region between the purD coding sequence and the upstream end of the inserted MudJ element (mDEL). Of 22 sequenced mDEL deletions, 21 removed 106 bp (mDEL1) and 1 removed 260 bp (mDEL2). Both deletion types remove material downstream of the purD UGA mutation including an interrupted palindromic sequence element. The mDEL3 and 4 deletions arose secondarily in strains carrying mDEL1 and improved growth further by removing the polar UGA mutation.
The interrupted palindrome (48 bases on each side with an 8-base loop) removed by these deletions has been shown previously to serve as an imperfect transcription terminator (Zieg and Kolter 1989). Deletion of this stem structure increases the level of LacZ produced by Salmonella strains having a MudJ insertion in the eut or his operons (C. Rappleye, S. Bolen, D. Sheppard, E. Kofoid, J. R. Roth, unpublished results).
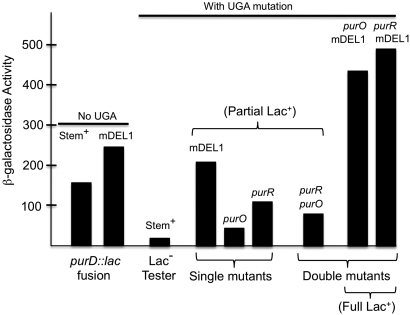
The effects of the stem deletion on the level of lac expression in the Pur-Lac tester strain are shown in Figure 5 and compared to effects of other mutations. The stem loop may serve as a weak transcription terminator despite the fact that its structure differs from typical Rho-independent transcription terminators in which the stem is followed by a run of uracil residues. The primary effect of the palindrome seems to be its enhancement of the polar effect of the purD UGA nonsense mutation. This is seen by comparing the small effect of the deletion in strains lacking the UGA mutation (first two strains in Figure 5) to its large effect in strains with the UGA mutation (compare strains 3 and 4 in Figure 5). The 10-fold increase in lac expression seen in stem deletion mutants with the UGA mutation may be because the stem structure in mRNA contributes to the polarity effect or because removal of 106 bp shortens the distance between the UGA termination codon and the next translation start (within the MudJ element). Remarkably, the effect of this deletion on lac expression is larger than that of either a purO or purR single mutation. This is because the repressed level of purHD operon expression is fairly high and the low LacZ level seen in the tester is strongly dependent on the polar effect of the UGA mutation.
Figure 5 .
β-Galactosidase activity of the Pur-Lac tester strain derivatives. The low LacZ level in the tester depends on the combined effects of operon repression and the polar effect of the purD UGA mutation. The terminator stem has a small effect in the absence of the purD UGA mutation (compare the first two strains) and a large effect in strains with the UGA mutation (compare the third and forth strains). Single mutants and purR, purO double mutants have a partially Lac+ phenotype under selective conditions on the reversion plate. Double mutants at the far left are fully Lac+. Strains assayed, in order of vertical bars, were: T12306 (No UGA, mDEL+), TT26208 (No UGA, mDEL1), TT25154 (tester strain UGA, mDEL1+), TT26174 (mDEL1 single), TT26176 (purO single), TT26169 (purR single), TT26209 (purR, purO), TT26175 (purO, mDEL1), and TT25177 (purR, mDEL1).
A surprising feature of the stem deletion mutation is its high formation rate, which is about 10−4/cell/division (data not shown). Quasipalindromes have been shown previously to be subject to frequent deletions that reduce their symmetry (Sinden et al. 1991; Leach 1994). Of the subclones isolated from 44 Lac+ revertant colonies appearing on day 6, 22 had a stem deletion mutation, but no purO or purR mutation. Below it is shown that 16 of the revertant subclones with either a purO or purR mutation also carried a stem deletion. Thus 38 of the 44 characterized subclones carry a stem deletion (all of the mDEL1 type), while only 18 carry a purR loss-of-function mutation. Later it will be shown that the subclones having only a single purO or purR mutation emerged from colonies in which other cells carry the stem deletion as well as the regulatory mutation. Thus, every day-6 revertant colony arising in this selection includes some cells with a stem deletion.
Some revertant subclones carry two mutations
Unexpectedly, 16 of the 44 subclones isolated from day-6 revertants had more than one mutation (see left side of Table 2). That is, some subclones carried a stem deletion (mDEL1) in addition to either a purR or a purO mutation. As seen in Figure 4 above, the double mutants (purO, mDEL1) or (purR, mDEL1) produced substantially higher levels of LacZ activity than any of the three single mutants (purO, purR, or mDEL1). We will suggest below that these two mutations arise sequentially in either order and thereby cause progressive improvement in growth ability. None of the characterized revertant subclones carried both a purR and a purO mutation. This is expected, since both of these mutations affect repression control and neither corrects the polar effect of the UGA mutation. The 44 initially characterized subclones are described in the left half of Table 2. The actual sequence changes are presented in Appendix 1. The composition of the original colonies from which these subclones emerged will be examined below. There it will be shown that all of the 44 colonies contained some cells with multiple mutations.
Table 2 . Characterization of purified subclones from 44 revertants appearing on day 6.
| Genotypes of isolated subclones (total 44) | Mutation(s) in purified subclone | Composition of original revertant colonya | ||||
|---|---|---|---|---|---|---|
| purR | purO | Stem | First mutation | Second mutation | Number of revertants | |
| Single purO (1) | + | − | + | purO | mDEL1b | 1 |
| Single purR (5) | − | + | + | purR | mDEL1 | 5 |
| Single mDEL1 (22) | + | + | Δ | mDEL1 | purO | 1 |
| purR | 7 | |||||
| Extended deletions mDEL3 and mDEL4 | 2 | |||||
| Amplificationc | 12 | |||||
| purO, mDEL1 (3) | + | − | Δ | mDEL1 | purO | 3 |
| purO | mDEL1 | 0 | ||||
| purR, mDEL1 (13) | − | + | Δ | mDEL1 | purR | 4 |
| purR | mDEL1 | 9 | ||||
When single cells from a revertant colony are plated on rich medium containing X-gal, they form colonies that are either light blue (if the cell has only one mutation) or dark blue (if the cell has both a regulatory mutation and a stem deletion).
The end points of mDEL deletions (1–4) are described in Figure 4.
The amplified unit is the chromosomal region between the rrnB and rrnE loci, which includes the purHD operon.
Three mutation types contribute to appearance of Lac+ colonies
The three mutation types described above appear to be the only mutations involved in the reversion behavior of the Pur-Lac tester strain. Single and double mutants isolated from day-6 colonies show the same growth phenotypes and LacZ enzyme levels as do constructed strains that carry these mutations and have never experienced selection. Two of the subclones carrying a simple mDEL1 mutation emerged from colonies that also included cells with an enhanced Lac+ phenotype due to a deletion that removes the original mDEL1 and extends to include the polar UGA mutation (see mDEL3 and mDel4 in Figure 4). In addition, 12 subclones with a simple mDEL1 mutation came from colonies with improved Lac+ revertant strains that have no new point mutation but owe their improved growth to multiple copies of the purHD::lac(mDEL1) region. These strains will be described elsewhere (S. Quiñones-Soto and J. R. Roth, unpublished results).
A model to explain reversion behavior of the Pur-Lac system
The nature of the 44 isolated Lac+ revertant subclones described above suggested a model to explain the appearance of Lac+ colonies under selection. In this model, the plated tester cells cannot grow on solid lactose medium beyond the initial day-1 growth on contaminants in the agar (see Figure 3 above). As a consequence, lawn growth stops when residual nutrients are exhausted and tester cells make no further contribution to reversion. However, the plated population (108 cells) includes cells that carry any one of the three mutation types described above. Some single mutants may also arise in testers during day 1. Any one of these three singly mutant cell types can initiate a colony that grows slowly on selective medium above the lawn of nongrowing tester cells. These singly mutant clones expand under selection until their population reaches a size sufficient to allow some cell within the colony to acquire a second mutation. This second mutation must arise during growth under selection (after day 1) since any singly mutant cell plated would generate a clone of only ∼100 cells by the time full selection was imposed—insufficient to allow the second mutation. Thus revertant colonies were initiated by mutants that arose under nonselective conditions, either before plating or during nonselective growth on day 1. (We will provide evidence below that roughly half of the day-6 colonies are initiated before plating and half early during day 1.)
The second mutation arises during growth of a partially Lac+ singly mutant cell under selection. The second mutation greatly improves growth rate on lactose, allowing a new doubly mutant cell to expand a subclone that rapidly overgrows the initial clone and becomes a substantial proportion of the cells in the visible colony. This raises the question of whether formation of the second mutation and therefore appearance of mixed-population colonies is accelerated by any stress-induced mutagenesis or is explained by mutations arising at standard (unselected) rates. Direct measurements described above gave no support for general (genome-wide) mutagenesis during growth of revertant colonies. This leaves the possibility of some kind of directed mutation, which we will discuss later. For the moment we will assume that no stress-induced mutagenesis occurs. The possibility of directed mutagenesis will ultimately be addressed by determining whether unselected formation rates and growth phenotypes are sufficient to explain the behavior of the system.
According to the selection-only model, the time at which a revertant colony first becomes visible (about 106 cells) depends on the growth rate of the primary clone, the formation rate of the second mutation, and the growth rate of the secondary doubly mutant clone. The slow accumulation of visible colonies over 6 days is attributed to the variability of these parameters and the stochastic distribution of times at which the second mutation arises. This model makes several predictions that are tested in the sections that follow. The sequence of events proposed in the selection model is diagrammed later in Figure 9.
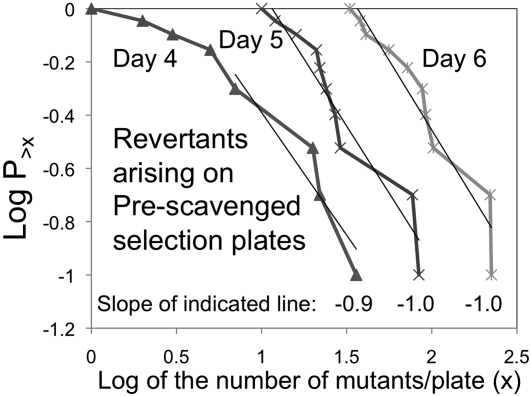
Figure 9 .
Effect of reducing plate growth on revertant distribution. Tester cells (108) were plated on selection plates that had first been seeded with 109 Lac− scavenger cells that could consume any nutrients other than lactose contaminating the agar. After 1 day of scavenging, 108 testers cells from each of 10 independent cultures were added and plates were incubated. Scavenging reduced tester growth on the selection plate 10-fold and revertant number ∼2-fold. Note that the distribution of revertant number showed a slope (−1.0)
Prediction 1: Revertant colonies contain mixtures of singly and doubly mutant cells
The 44 subclones described above were isolated from 44 day-6 revertant colonies and revealed the structure of only one cell from each such colony. To test the composition of the whole original revertant colonies, the retained samples of those colonies were suspended and distributed on rich medium with X-gal (the chromogenic substrate for β-galactosidase). Every day-6 revertant colony contained some cells that formed dark blue colonies and proved to contain cells with two mutations. Other cells from the same colony formed lighter blue colonies and carried only one mutation. Most colonies contained 50–80% double mutations but a few showed more biased ratios (0.3–99% double mutants). Of the initially characterized subclones (column one of Table 2), all of the 28 singly mutant cells formed light blue colonies on rich medium with X-gal, while the 16 doubly mutant subclones formed dark blue colonies. All of the parent colonies from which the 44 subclones were isolated showed a mixture of dark and light blue colonies when examined in this way. Each of the 44 subclones was thus derived from a colony that included another type of mutant cell.
For each revertant colony, the mutation found in a singly mutant cell was identical to one of the two mutations carried by a doubly mutant cell from the same colony. The mutation found in singly mutant cells is inferred to have arisen first, prior to selection. The second mutation (in doubly mutant cells) is inferred to have arisen later during growth under selection. Thus colony composition reveals the sequence of events. Table 2 describes the development of each of the 44 revertant colonies used to obtain the subclones characterized initially. The specific sequence changes are detailed in Appendix 1. As predicted by the model, no purR, purO double mutants were found, because this combination shows no increase in LacZ level over that seen for either singly mutant type (Figure 5) and is not expected to cause any growth improvement. The model predicts double mutants that arise as a single cell within the colony and become common in that colony because they improve growth and thereby contribute to the earlier appearance of the revertant colony.
Two initially tested subclones were singly mutant with the short-stem deletion mutant described above (mDEL1) and formed light blue colonies on rich medium plus X-gal. The parent revertant colonies from which these subclones emerged also included cells that formed darker blue colonies and carried a larger deletion that removed the polar purD UGA mutation (mDEL3 or mDEL4) as well as the stem. These secondary deletions include the whole region of the first deletion and improve growth by removing the polar purD UGA codon, which is the major cause of reduced lactose expression. We suggest that the shorter deletion mutants are more common because the parental palindrome enhances deletion rate. The secondary mutation arises later under selection in cells that are growing by virtue of the first deletion. These secondary deletions may be rare because they form without any stimulation by a central palindromic structure, but despite this they can occur within developing colonies when the initial clone reaches sufficient size (or some effect of selection increases their probability without causing general mutagenesis).
Twelve revertant colonies included cells with a stem deletion mDEL1, but neither a purO nor a purR secondary mutation. In these colonies, the second cell type showed only a modest growth enhancement (its colonies are slightly darker blue on X-gal) and this enhanced Lac+ phenotype was subject to rapid loss (colonies on rich medium with X-gal showed lighter blue sectors). The growth improvement of the singly mutant cells is due to amplification of the purHD::lac region with a stem deletion, rather than to any secondary point mutation. These revertant colonies are reminiscent of the mixed colonies with lac amplification mutants seen in the Cairns system (Andersson et al. 1998; Slechta et al. 2003; Kugelberg et al. 2006), which will be described elsewhere (S. Quiñones-Soto and J. R. Roth, unpublished results).
Prediction 2: Singly mutant cells grow less well under selective conditions than those carrying two mutations
The model predicts that cells with a single mutation (purR, purO, or mDEL1) should grow slowly under selective conditions and their colonies should appear quickly after some cell in the initial clone acquires the second mutation. This prediction is consistent with the LacZ levels (Figure 5) determined for cells grown in liquid minimal glycerol medium. However, the enzyme levels found in single mutants seem high enough to support better growth than is posited in the model. Apparently conditions on the selection plate are more restrictive, perhaps because the single mutants must compete with the lawn of tester cells for any nutrients that might contaminate the agar or be released during slow growth of a singly mutant clone on lactose (e.g., galactose, acetate, succinate, lactate).
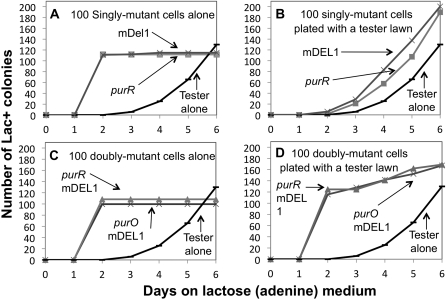
The growth ability of single mutants under selection conditions was tested by a reconstruction experiment in which 100 singly mutant cells were plated on selective medium and observed to determine their time of colony appearance (see Figure 6). Figure 6A shows the behavior of 100 singly mutant cells plated alone (no tester lawn) on selective medium. All colonies appeared on or around day 2. In a standard reversion experiment (also plotted in Figure 6A) the first Lac+ revertant colonies appeared above the tester lawn a day later (day 3) and their number accumulated slowly over several days. The model suggests that the presence of a cell lawn intensifies selection on the pre-existing singly mutant cells and the delay in colony appearance is due to the need to accumulate the second mutation, which is not required for single mutants without the competition from the tester. This interpretation was tested as described below.
Figure 6 .
Reconstruction experiments using purR, purO, and stem deletion mutants. (A) Roughly 100 singly mutant cells (mDEL1 or purR) were plated alone without testers on lactose medium. For comparison, 108 tester cells were plated alone to detect revertants in a standard reversion assay. (B) A mixture of 100 singly mutant revertant cells were genetically marked and plated together with 108 tester cells on selection medium. (C) A control experiment in which 100 doubly mutant cells were plated alone on selective medium. Behavior of tester cells alone is replotted from A. (D) A mixture of 100 doubly mutant cells and 108 tester cells was plated on lactose. Accumulation of colonies from tester cells alone is replotted for comparison. For all parts of the figure, cells were pregrown on minimal glycerol adenine, plated on minimal medium with lactose plus adenine, and incubated at 30° as described in Materials and Methods. Singly mutant cells plated alone or with the tester lawn were genetically marked with a Tn10 insertion near purR+. Strains: TT25154 (tester strain), TT26169 (purR), TT26174 (mDEL1), TT26176 (purO), TT26175 (purO mDEL1), and TT25177 (purR mDEL1).
Figure 6B shows the behavior of 100 singly mutant cells that were mixed with the lawn of tester cells. The added singly mutant cells were genetically marked so the colonies they formed could be distinguished from revertants arising from the tester. Note that the appearance of colonies initiated by the singly mutant cells was delayed in the presence of tester and enhanced the total number of revertant colonies only modestly. Roughly half of the observed colonies were derived from the genetically marked singly mutant cells and half from the tester. Thus in the presence of the tester, the added singly mutant cells formed colonies whose appearance is delayed 1–4 days by the presence of a tester lawn. These same cell types formed a colony within 2 days in the absence of the tester cell lawn (Figure 6A). This suggests that selection is more stringent in the presence of tester. Revertant colonies initiated by the added singly mutant cells in the presence of the lawn appear with about the same timing as colonies that arise from the plated tester strain. This is consistent with the model, which proposes that revertant colonies are initiated by singly mutant cells that arise during nonselective growth either prior to plating or during the period of tester growth during day 1 after plating.
Prediction 3: During growth under selection, colonies initiated by a singly mutant cell acquire cells with a second mutation
To test this central aspect of the model, selection plates were seeded with a lawn of tester cells plus 100 genetically marked single mutants with either a purR or a stem deletion mutation (mDEL1). In this experiment, the added cells carried a previously sequenced mutation that had been added to the genome of the tester strain with no selection for growth on lactose. That is, the 100 singly mutant cells plated had never experienced growth under selection, but carried an introduced mutation that originated under selection in another strain. On day 6, 10 revertant colonies initiated by a marked singly mutant cell plated in the presence of a tester were picked and suspended. Colony suspensions were diluted and plated on NB X-gal medium (as described above). Each colony contained two cell types. Some cells formed light blue colonies and carried the mutation present in the added parent cell. Other cells from the same colony formed dark blue clones on X-gal and carried the same mutation plus an added second mutation. Seeded purR mutants gave rise to revertant colonies that included cells with both the original purR mutation and a new stem deletion mutation. Seeded stem deletion mutants formed colonies that included cells with an added purR mutation. Doubly mutant cells (dark blue single colonies on X-gal) recovered from revertant colonies that first appeared on day 6 are fully Lac+ and form colonies by day 2 when replated alone on selective medium with or without a tester lawn (see below).
Prediction 4: Double mutants form colonies by day 2 in a reconstruction experiment
A reconstruction was done in which 100 cells of a doubly mutant revertant (either mDEL1, purR or mDEL1, purO) were plated on selective medium either alone or together with 108 tester cell (Figure 6, C and D). Figure 6C shows the behavior of 100 double-mutant cells plated without tester. All colonies appeared on day 2. The time course of revertant colony appearance with tester alone is replotted for comparison. Figure 6D shows the appearance of colonies when 100 double-mutant cells were plated with the tester lawn. The double mutants formed colonies that appeared on day 2 with or without a lawn of tester cells (compare Figure 6, C and D). This is interpreted as evidence that the double mutants form a colony on tester without delay because they already carry the second mutation. Single mutants (Figure 6B) still need the second mutation and form colonies whose number increases slowly over several days starting on day 3. The fact than no colonies appear before day 3 of a reversion experiment suggests that no double mutants are present in the plated tester population in a standard reversion experiment.
The results in Figure 6 show that selection is more stringent in the presence of tester cells. While both single and double mutants form colonies by day 2 when plated alone (Figure 6, A and C), only double mutants can do so in the presence of a tester lawn (Figure 6D). In Figure 6D, the colonies derived from added doubly mutant cells all appeared on day 2, while the revertant colonies derived from the tester cell population appeared later and presumably arose from pre-existing singly mutant cells in the tester population that required time and a second mutation before they could make a visible colony.
Prediction 5: Double mutants form prior to selection, but are too rare to contribute to the number of revertant colonies seen
In reconstruction experiments, doubly mutant cells formed colonies by day 2 when plated with or without a tester lawn. Yet in the standard reversion experiment, the earliest revertant colonies appear on day 3 and include some cells with two mutations. The absence of revertants on day 2 suggested that pre-existing double mutants in the plated population are too rare to contribute significantly to revertant colony number in a standard reversion experiment. That is, all revertant colonies seen in the standard reversion experiment are initiated by singly mutant cells that acquire the second mutation during growth under selection.
To be sure that double mutants (purR, mDEL1) could be detected in an actual reversion experiment, 10-fold more cells were plated than used in the standard experiment. Under these conditions, a few revertant colonies appeared on day 2 and consisted of homogeneous populations of cells with both a purR mutation and a stem deletion. (No purO, mDEL1 double mutants were found, as expected in light of the rarity of purO mutations). This reinforces the conclusion from reconstruction experiments that in a standard reversion experiment, pre-existing double mutants do not contribute significantly to revertant number.
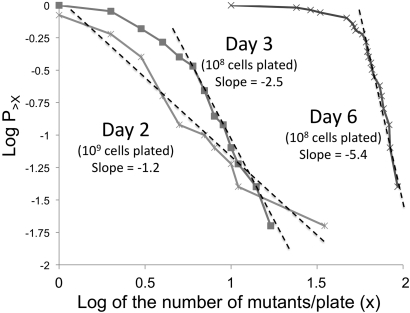
Since pre-existing double mutants could be detected when 10-fold more cells were plated, it was possible to do a fluctuation test by scoring revertant colonies on day 2. Results of this test are plotted in Figure 7. In this plot, the logarithm of the number of revertants per tube (X) is plotted against the logarithm of the fraction of tubes with more than X mutations. If the revertants arise prior to plating and show a Luria–Delbruck distribution, the line is expected to show a −1 slope (Luria and Delbruck 1943; Luria 1951; Rosche and Foster 2000). When 10-fold more cells were plated, day-2 revertants showed a slope close to the −1 value expected for a Luria–Delbruck distribution. We interpret this to mean that some double mutants formed prior to selection can be detected (when an excess of cells are plated), but are too rare to contribute to revertant number in the standard reversion experiment. On the basis of the data in Figure 7, double mutants form during nonselective pregrowth at an apparent rate of 10−9/cell/division. This approximates the product of the mutation rates for the individual mutations (data not shown).
Figure 7 .
Fluctuation tests for revertant number in the Pur-Lac system. Cells (109) from 50 independent cultures were plated separately and selected colonies were counted on day 2. To detect double mutants (purR, mDEL1), 10-fold more tester cells (109) were plated. This revealed double mutants that are too rare to appear in the standard experiment. The slope (−1.2) suggests that rare double mutants arise prior to plating. The data for revertants that appeared on days 3 and 6 were obtained by plating 108 cells from each of 25 tubes on selective medium.
Prediction 6: If revertant colonies are initiated by singly mutant cells that exist before plating, revertant number should show a Luria–Delbruck distribution
The model proposes that every revertant colony appearing on the selection plate is initiated by a singly mutant cell that arose during nonselective growth. If these cells arise during liquid growth prior to plating, the number of revertants seen for independent cultures is expected to show a Luria–Delbruck distribution. This distribution is not expected if reversion is initiated on the selection plate since progeny of a mutant cell remain together on the agar surface. Since some nonselective growth occurs on the plate during day 1 (see above) it was not clear whether a Luria–Delbruck distribution would be detected.
Figure 7 shows the distribution of revertant colonies that first appeared on days 3 and 6 (were not present on the previous day) of a standard reversion experiment. The day-6 revertant numbers (slope −5) show a distribution that is clearly different from that predicted by Luria–Delbruck for pre-existing mutants (slope −1), suggesting that many of these revertants were initiated on the plate, presumably during unselected growth on day 1, but possibly later during selection. The distribution of day-3 colony numbers more closely resembles that of Luria–Delbruck, but still suggests that many colonies are initiated after plating. We suggest that the earliest colonies (day 3) are more likely to be initiated by cells that arose prior to plating and can appear on day 3 if they acquire the second mutation during day 1 and form colonies that appear 2 days later (on day 3). Any single mutants that arise during day 1 are likely to form colonies that appear later in the incubation period and contribute colonies appearing later, between late day 3 and day 6. This might explain why colonies appearing on day 6 deviate more severely from a Luria–Delbruck distribution. Observing a clear Luria–Delbruck distribution would require preventing tester growth on the selection plate.
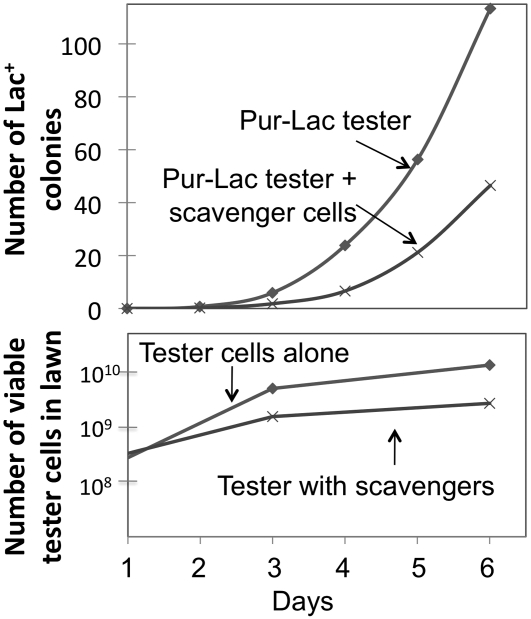
To test this prediction, day-1 growth was restricted by removing contaminating nutrients from the selection medium. Scavenger cells (109) were spread on selective plates 1 day before initiating the reversion experiment. These scavengers have no lac operon and therefore cannot give rise to revertants, but they can consume nutrients other than lactose that contaminate the medium. Scavenging reduced the residual growth of the tester ∼10-fold and revertant number ∼2-fold (Figure 8). We estimate that in the standard experiment (no scavengers) about half of late-appearing colonies were initiated after plating.
Figure 8 .
Effect of reducing plate growth on revertant number. The tester strain (108 cells) was plated on lactose adenine medium with and without 109 cells of a Lac− scavenger strain. Lawn growth was followed by testing agar plugs for their content of viable tester cells. Revertant number is recorded as in a standard reversion experiment.
Scavengers reduced lawn growth ∼10-fold and revertant number ∼2-fold. The results are the same when tester and scavenger cells are added together or when plates were prescavenged for 1 day before addition of tester cells.
This contribution of unselected day-1 growth to revertant number was tested by a fluctuation test using selection on prescavenged plates. These results (seen in Figure 9) support the idea that revertant colonies are initiated by mutants that arise during nonselective pregrowth in liquid medium. Revertant colonies first appearing on days 4, 5, and 6 (not present on the preceding day) all show a slope of −1 in the above plot, indicating a Luria–Delbruck distribution. This supports the idea that, when lawn growth is minimized, virtually all revertant colonies are initiated by mutant cells that arose prior to plating on selective medium. The time at which these colonies appear is delayed to a variable extent due to the time required for growth of the initial mutant and for the population to reach a size sufficient for acquisition of the second mutation. We estimate that in the standard experiment (no scavengers) roughly half of the colonies that appear on day 6 are initiated by singly mutant cells that arose prior to plating and the rest were initiated during nonselective growth early on day 1. While many more single mutants must arise later in day 1, few of them generate revertant colonies that appear during the 6-day period of the standard reversion experiment. The secondary mutations in these colonies arise during growth under selection. The timing of the second mutation is dictated by the formation rate of the particular mutation and the size of the colony. While we cannot exclude the possibility that growth rate limitation influences mutation rate, we think this is unlikely, since it would require directing mutation to valuable sites. This question can be answered by modeling, once rates of formation growth of all mutant types are known.
Discussion
The Pur-Lac system described here provides an opportunity to examine in detail the process by which mutations arise under selection. This process of genetic adaptation is difficult to study in natural populations because a wide variety of mutation types may occur in a rare subset of organisms in a large growing population. In standard bacterial genetics, the complexities of adaptation are systematically avoided by use of stringent (often lethal) selection conditions that detect pre-existing large-effect mutants without contributing to their frequency. The ability of stringent selection to prevent adaptation was demonstrated by Luria and Delbruck (1943) and Lederberg and Lederberg (1952). To study mutation under more natural selection conditions, a series of nonlethal systems have been used that prevent growth of the parent cells on selective plates, but still allow mutant colonies to appear over time. This time-dependent mutant accumulation has been attributed to stress-induced mutagenesis, but has not been seen in standard lab selections. What is different about the new systems? Do they have a unique feature that allows mutagenesis? Has selection been relaxed sufficiently to allow spontaneous small-effect mutations to initiate growing clones even when parent cell growth is prevented? Are these systems affected by more types of small-effect mutations?
These questions were addressed initially in several systems, including that of Cairns and Foster (1991), which has been extensively analyzed and is the only system showing substantial evidence of genome-wide mutagenesis during selection (Torkelson et al. 1997). Modeling of the Cairns system suggested that mechanisms needed to explain these results would (in natural situations) be prohibitively costly in deleterious mutations (Roth et al. 2006a). This prompted development of models that could explain the observed associated mutations in some other way. The possibility that general mutagenesis was peripheral to revertant formation was supported by direct assessment of the mutagenesis intensity (Rosche and Foster 1999).
The selection-only model tested here gives a framework for explaining any system in which adaptation is rapid and selection conditions appear mutagenic. In the selection model, small-effect mutants (common under all conditions) arise prior to plating and initiate clones that grow under selection and acquire secondary mutations that improve growth progressively (genetic adaptation). If the needed secondary mutations are sufficiently common and have a sufficient effect on growth, colony appearance is stimulated by secondary mutations that arise with no required increase in mutation rate.
The general mutagenesis observed in the Cairns system may be an artifact due to coamplification of lac with the nearby dinB gene, which encodes an error-prone repair polymerase (Slechta et al. 2003). While others have objected to this conclusion (Hastings et al. 2004; Stumpf et al. 2007), it is striking that mutagenesis depends on the dinB copy being located immediately cis to lac on an F′ plasmid. It remains possible that dinB contributes to reversion by enhancing growth of the leaky lac mutant as shown by Cohen and Walker (2010). It is difficult to test stress-induced mutation experimentally using a system that shows some mutagenesis, even if the observed mutagenesis is neither necessary nor sufficient to explain the revertants. This prompted use of the simpler system.
The Pur-Lac system described here shows the same overall behavior as the several previous systems–mutants accumulate with time even in the absence of lawn growth. The behavior of this system was initially attributed to stress-induce mutagenesis (Yang et al. 2001, 2006). The advantage of Pur-Lac for study of selection is that all genetic changes occur in the chromosome and can be characterized (point mutations, deletions). These mutations form at rather high rates during nonselective growth. Evidence is presented that the accumulated Lac+ revertant colonies are initiated by small-effect (weakly Lac+) mutants that arise prior to selection and improve by acquiring a second mutation during growth under selection. This process does not involve genome-wide mutagenesis.
The open question is whether or not the behavior of this system can be explained by selection alone. Do the mutations form at a sufficient rate and provide a sufficient growth improvement to explain the behavior of the system without mutagenesis? Alternatively does this system involve some added feature such as a “directed” mutagenesis during growth under selection? We think that selection will prove to be enough, but we outline below the difficulties with the argument and the sorts of additional behavior that might be required.
We concluded that no genome-wide mutagenesis occurs during the course of a Pur-Lac selection experiment. This is based on a Lac+ reversion experiment done on medium containing (in addition to lactose) nutrients that cannot serve as carbon source, but would allow colony formation by Lac+ mutants that carried secondary mutations in any of ∼100 genes. The absence of such secondary mutations in revertant cells was taken as evidence that no significant mutagenesis occurred during the many generations of selective growth on the plate.
This evidence would be flawed if it turned out that genome-wide mutagenesis depends on transcription of the target genes as suggested recently for the Cairns system (Cohen and Walker 2010). That is, one could imagine that our conditions repressed these unrelated genes and made them poor targets for stress-induced mutagenesis. However, techniques very similar to those used here were applied successfully to the Cairns system, where they revealed the DinB-dependent mutagenesis associated with reversion in that system (Torkelson et al. 1997; Slechta et al. 2002, 2003).
Even without general mutagenesis, it seems clear that selection is contributing in some way to the origin of mutants in the Pur-Lac system. Over a 6-day period, ∼100 colonies appear, each of which includes cells with two mutations (purR, purO or mDEL1). The estimated frequency of the three mutant types suggest that even the commonest double mutant should occur at a discontinuous rate of ∼10−12/cell/division, yet 100 colonies containing such mutants arise from of 108 plated cells, which grew to 1010 cells during day 1. Thus, considered naively, the double mutants are ∼1000-fold more common than one would expect. The increase could reflect stress-induced mutagenesis (directed) or it might result from growth of pre-existing single mutants under selection and later addition of the second mutation. Evidence is provided that, colonies appearing on day 6 include two types of cells. One type is a single mutant (purR, purO or mDEL1) and the other is a double mutant carrying the first mutation plus an additional mutation from this list. This result reveals that the two mutations arose sequentially and the second mutation arose within a clone of cells with the first mutation growing under selection. The number of cells added to the growing singly mutant clone may enhance the frequency of double mutants simply by providing more targets for the second event. To know whether this selective growth is sufficient will require knowing the formation rate of each mutant type under nonselective conditions and the growth rate of each single and doubly mutant cell type under conditions of selection. With these parameters, it should be possible to model the behavior of the system. Such a model should reveal whether unselected mutation rates are sufficient. These measurements are under way, but we can make some preliminary estimates.
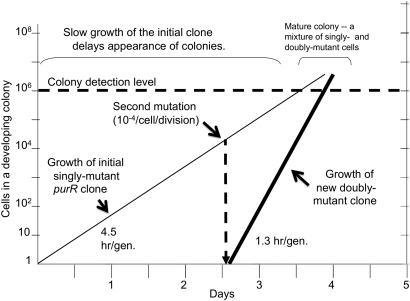
The selection model (Figure 10) describes a pre-existing purR mutant grows on the selection plate with an estimated doubling time of 4.5 hr and reaches a population size of 104 sometime on day 2. This population size might allow spontaneous appearance of an mDEL1 mutation, which forms at a unselected rate of ∼10−4/cell/division). The resulting mixed clone would be expected to become visible sometime during days 3 and 4. The problem is more difficult when one tries to explain the more common situation in which the first mutation is an mDEL1 deletion. Current estimates suggest a 17-hr generation time under selection and clones have to reach a much more substantial colony size before the rarer secondary purR or purO mutation could be realized.
Figure 10 .
Model for development of a single colony. A purR mutant has a 4.5 hr/generation under plate conditions and the second mutation (mDEL1) arises at a rate of 10−4/cell per generation to produce a doubly mutant cell with a 1.3 hr/generation. Colonies become visible at a population size above 106.
A solution to this problem would be provided by stress-induced mutagenesis, but that would require direction of mutability to particular sites, since no evidence for genome-wide mutagenesis was seen. Alternatively, growth of mDEL1 mutants might be faster under selection than our estimates indicate. Both possibilities could be realized if the lac region amplified under selection. This would accelerate growth by providing more copies of a partially active operon and increase mutation rate by providing more targets for mutagenesis. There is some evidence for this possibility in that some of the mDEL1 single mutations showed improvement by amplification under selection (Table 2). In addition, the chromosomal purD::lac region is closely flanked by 5-kb direct sequence repeats [rRNA (rrn) cistrons]. Duplications of this region form at a high rate (10−3/cell/division) and are carried at a high steady state frequency (1%) in unselected cultures (Reams et al. 2010).
While the Pur-Lac system is simpler in some ways than the systems used previously to study reversion under selection, it does have several ideosyncracies of its own. The lac genes under selection are part of a Mud-lac element integrated into the chromosome. This element lacks replication and transposition functions, leading us to believe that no encoded features contribute to reversion or rearrangements. The major idiosyncrasy is the stem-loop structure that reduces lac expression and is prone to deletions that increase lac function (mDEL1). This deletion is not a typical mutation type due to the palindromic structure and it arises at a very high frequency under nonselective conditions (about 10−4/cell/division). This deletion type contributes to every revertant colony. The purR loss-of-function mutation also arises at a high rate. The several genetic systems in which revertants accumulate over time may be ones in which the new phenotype can be generated by a series of mutations that arise frequently.
It may seem strange that all of the systems used to study the origin of mutation under selection become complex upon close examination. This is true for both the system discussed here and for the previously described systems (Shapiro and Brinkley 1984; Hall 1990; Foster and Cairns 1994; Taddei et al. 1997; Wrande et al. 2008). We suggest that this complexity is neither happenstance nor a contrivance of the experimenters. Whenever cells grow under some limiting conditions, a vast array of possible genetic changes can contribute to small increases in growth rate. Some changes may be indirect and provide a benefit that is offset by some unrelated cost (e.g., extended protein half-life, changes in level of inhibitory metabolites, increases in translational error rate). These many complicating possibilities are avoided in standard laboratory selection by use of stringent selection (often lethal) conditions that eliminate growth of small-effect mutations and thereby simplify genetic analysis. Natural selection is under no such constraint and can detect mutants with a variety of small-effect changes, many of which are extremely common. Once growth is allowed, adaptation is rapid because the reservoir of small-effect variants is large. These small-effect mutations make the nature of the response more difficult to analyze. Adaptation may often involve copy number increases (which are frequent), but can also exploit other sorts of common mutations (such as the null purR mutations and the stem-loop deletions seen in the Pur-Lac system). Understanding the processes by which adaptation occurs in a variety of systems may give us better insight into general rules that govern natural selection.
Acknowledgments
We thank Zhiwei Yang, Zhong Lu, and Aoquan Wang for generously providing the tester strain they developed. We thank lab members Drew Reams, Sophie Maisnier-Patin, Eric Kofoid, Emiko Sano, Doug Huseby, Cristina Tun-Garrido, Sue Slechta, Shawn Gerum, and Natalie Duleba for advice and helpful comments on the preparation of this manuscript. This work was supported in part by National Institutes of Health grant GM27068.
Appendix 1
Literature Cited
- Andersson D. I., Hughes D., 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43: 167–195 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Slechta E. S., Roth J. R., 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282: 1133–1135 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D., Roth J. R. (Editors), 2011. The Origin of Mutants under Selection: Interactions of Mutation, Growth, and Selection. ASM, Washington, DC. [DOI] [PubMed] [Google Scholar]
- Cairns J., 1998. Mutation and cancer: the antecedents to our studies of adaptive mutation. Genetics 148: 1433–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Foster P. L., 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Overbaugh J., Miller S., 1988. The origin of mutants. Nature 335: 142–145 [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N., 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76: 4530–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. E., Walker G. C., 2010. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr. Biol. 20: 80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Botstein D., Roth J. R., 1980. Advanced Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Foster P. L., 1993. Adaptive mutation: the uses of adversity. Annu. Rev. Microbiol. 47: 467–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 1998. Adaptive mutation: Has the unicorn landed? Genetics 148: 1453–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2004. Adaptive mutation in Escherichia coli. J. Bacteriol. 186: 4846–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2006. Methods for determining spontaneous mutation rates. Methods Enzymol. 409: 195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42: 373–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Cairns J., 1992. Mechanisms of directed mutation. Genetics 131: 783–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Cairns J., 1994. The occurrence of heritable Mu excisions in starving cells of Escherichia coli. EMBO J. 13: 5240–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo R. S., Hastings P. J., Rosenberg S. M., 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42: 399–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo R. S., Do R., Yamada M., Friedberg E. C., Hastings P. J., et al. , 2009. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182: 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., 1982. Evolution of a regulated operon in the laboratory. Genetics 101: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., 1990. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics 126: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J., Slack A., Petrosino J. F., Rosenberg S. M., 2004. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2: e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Shiau A., Choi K. Y., Zalkin H., Smith J. M., 1990. Genes of the Escherichia coli pur regulon are negatively controlled by a repressor-operator interaction. J. Bacteriol. 172: 4555–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R., Maenhaut-Michel G., Yamada M., Yamamoto Y., Matsui K., et al. , 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94: 13792–13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid E., Bergthorsson U., Slechta E. S., Roth J. R., 2003. Formation of an F' plasmid by recombination between imperfectly repeated chromosomal Rep sequences: a closer look at an old friend (F'(128) pro lac). J. Bacteriol. 185: 660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E., Kofoid E., Reams A. B., Andersson D. I., Roth J. R., 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. USA 103: 17319–17324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamrani S., Ranquet C., Gama M. J., Nakai H., Shapiro J. A., et al. , 1999. Starvation-induced Mucts62-mediated coding sequence fusion: a role for ClpXP, Lon, RpoS and Crp. Mol. Microbiol. 32: 327–343 [DOI] [PubMed] [Google Scholar]
- Leach D. R., 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16: 893–900 [DOI] [PubMed] [Google Scholar]
- Lederberg J., Lederberg E. M., 1952. Replica plating and indirect selection of bacterial mutants. J. Bacteriol. 63: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., 1951. The frequency distribution of spontaneous bacteriophage mutants as evidence for the exponential rate of phage reproduction. Cold Spring Harb. Symp. Quant. Biol. 16: 463–470 [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbruck M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I., Radman M., Taddei F., Picard B., Doit C., et al. , 1997. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277: 1833–1834 [DOI] [PubMed] [Google Scholar]
- Miller J. H., 1972. Assay of β-galactosidase, pp. 352–355 Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Reams A. B., Kofoid E. C., Savageau M., Roth J. R., 2010. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184: 1077–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L., 1999. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96: 6862–6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L., 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., 2010. Genetic adaptation: a new piece for a very old puzzle. Curr. Biol. 20: R15–R17 [DOI] [PubMed] [Google Scholar]
- Roth J. R., Kofoid E., Roth F. P., Berg O. G., Seger J., et al. , 2006a Regulating general mutation rates: examination of the hypermutable state model for Cairnisian adaptive mutation. Genetics 163: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Kugelberg E., Reams A. B., Kofoid E., Andersson D. I., 2006b Origin of mutations under selection: the adaptive mutation controversy. Annu. Rev. Microbiol. 60: 477–501 [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., 1984. Observations on the formation of clones containing araB-lacZ cistron fusions. Mol. Gen. Genet. 194: 79–90 [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., Brinkley P. M., 1984. Programming of DNA rearrangements involving mu prophages. Cold Spring Harb. Symp. Quant. Biol. 49: 313–320 [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Zheng G. X., Brankamp R. G., Allen K. N., 1991. On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics 129: 991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta E. S., Bunny K. L., Kugelberg E., Kofoid E., Andersson D. I., et al. , 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 100: 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta E. S., Liu J., Andersson D. I., Roth J. R., 2002. Evidence that selected amplification of a bacterial lac frameshift allele stimulates Lac(+) reversion (adaptive mutation) with or without general hypermutability. Genetics 161: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele D. F., Jinks-Robertson S., 1992. An examination of adaptive reversion in Saccharomyces cerevisiae. Genetics 132: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf J. D., Poteete A. R., Foster P. L., 2007. Amplification of lac cannot account for adaptive mutation to Lac+ in Escherichia coli. J. Bacteriol. 189: 2291–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Berg O. G., Roth J. R., Andersson D. I., 2009. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182: 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F., Halliday J. A., Matic I., Radman M., 1997. Genetic analysis of mutagenesis in aging Escherichia coli colonies. Mol. Gen. Genet. 256: 277–281 [DOI] [PubMed] [Google Scholar]
- Taddei F., Matic I., Radman M., 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92: 11736–11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkelson J., Harris R. S., Lombardo M.-J., Nagendran J., Thulin C., et al. , 1997. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 16: 3303–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J., 1994. The Beak of the Finch: A Story of Evolution in Our Time. Alfred A. Knopf, New York [Google Scholar]
- Wrande M., Roth J. R., Hughes D., 2008. Accumulation of mutants in “aging” bacterial colonies is due to growth under selection, not stress-induced mutagenesis. Proc. Natl. Acad. Sci. USA 105: 11863–11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Lu Z., Wang A., 2001. Study of adaptive mutations in Salmonella typhimurium by using a super-repressing mutant of a trans regulatory gene purR. Mutat. Res. 484: 95–102 [DOI] [PubMed] [Google Scholar]
- Yang Z., Lu Z., Wang A., 2006. Adaptive mutations in Salmonella typhimurium phenotypic of purR super-repression. Mutat Res. 595: 107–116 [DOI] [PubMed] [Google Scholar]
- Zieg J., Kolter R., 1989. The right end of MudI(Ap,lac). Arch. Microbiol. 153: 1–6 [DOI] [PubMed] [Google Scholar]