Abstract
The CLAVATA1 (CLV1), CLV2, and CORYNE (CRN) receptors in Arabidopsis thaliana maintain cell proliferation in shoot apical meristems by restricting expression of the transcription factor WUSCHEL (WUS). Previously characterized receptor mutants generate extra fruit and floral organs that are proposed to arise from enlarged floral meristems (FMs). We identified new alleles in clv1, clv2, and crn and found that most mutants produce only extra fruit organs and generate FMs of similar dimensions as wild type. Characterization of gynoecium development in receptor mutants revealed increased cell proliferation and ectopic fruit organ initiation after FM termination. These regions of increased cell division also display expanded expression of the cell proliferation-promoting transcription factor SHOOTMERISTEMLESS (STM), similar to the expansion of WUS expression in the shoot apical meristems of strong clv1 mutants. We also examined genetic interactions between the ERECTA (ER) and BARELY ANY MERISTEM 1 (BAM1) receptor-like kinases and CLV pathway receptors. Our results suggest a model in which CLV1/BAM1 and CLV2/CRN complexes act in separate, parallel pathways in shoot meristems, while the CLV1, CLV2, and CRN receptors function together in a linear pathway during fruit development. These results demonstrate the importance of regulating cell proliferation in plants that undergo organogenesis throughout their life cycle.
IN the majority of animals, the formation of new organs is completed during embryogenesis or during juvenile stages. However, plants undergo organogenesis throughout their life cycle. For this indeterminate growth, plants maintain populations of stem cells in discrete structures called meristems, which retain their organization via cell–cell signaling (Satina and Blakeslee 1942; Stewart et al. 1972; Brand et al. 2000; Schoof et al. 2000). During embryogenesis in most plants, a pool of stem cells is established within the shoot apical meristem (SAM). Upon germination, the SAM produces primarily leaves, and after the vegetative-to-reproductive switch, transitions into the inflorescence meristem (IM). The IM forms floral meristems (FMs), which produce floral organs, including sepals, petals, stamens, and carpels, the structures that ultimately give rise to the fruit (reviewed in Barton 2010).
Although all cells within an adult plant can be traced back to the primary SAM or root apical meristem (RAM), additional meristems maintain populations of actively dividing or undifferentiated cells and are essential for the production of specialized tissues. Secondary meristems, such as the procambium, which generates new cells in the vasculature, maintain undifferentiated cells in an indeterminate manner, but lack the organized structure characteristic of shoot meristems (Baucher et al. 2007; reviewed in Girin et al. 2009). Other meristematic regions maintain partially differentiated cells but undergo prolonged proliferation, which terminates once the desired cells/organs are formed (Girin et al. 2009).
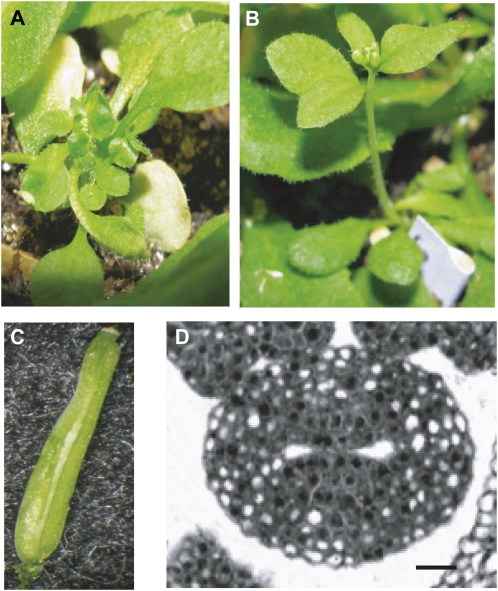
One such meristematic region that maintains prolonged proliferation is located in the medial regions of developing fruit (Pautot et al. 2001). In Arabidopsis thaliana, wild-type fruit development begins when two carpels initiated from the FM form a hollow cylinder of cells that grows apically to generate the gynoecium (Figure 1A). Simultaneously, two medial ridges grow through directed cell division toward each other, where they fuse and create the septum. The septum divides the cylinder into two chambers, reflecting the original two carpels, with two lateral domains and two medial regions located at either end of the septum. Meristematic cells at the junction between the septum and cylinder in the medial region produce placental tissue that develops into ovules (Bowman et al. 1999; Ferrandiz et al. 1999). Upon establishment of the gynoecium, cells in the cylinder differentiate into three types of mature fruit organs: valves, repla, and margins (Figure 1A; Bowman et al. 1999; Ferrandiz et al. 1999). Although a transcription factor network specifying each of these organs has been identified (Dinneny et al. 2005), the mechanisms regulating fruit organ number remain uncharacterized. The current hypothesis proposes that FM size determines carpel number, which then leads to an equivalent number of valves in the fruit (Ferrandiz et al. 1999).
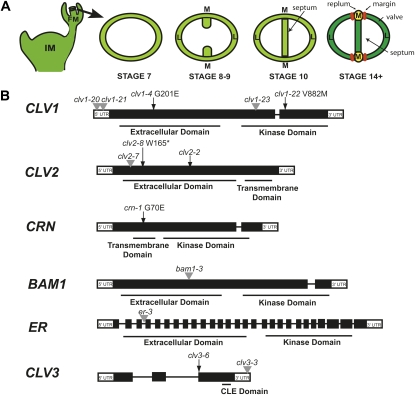
Figure 1 .
Gynoecium and fruit development in Arabidopsis thaliana. (A) The major stages of gynoecium and fruit development relevant to this study. M, medial region; L, lateral region; IM, inflorescence meristem; FM, floral meristem. (B) Gene models for CLV1, CLV2, CRN, BAM1, ER, and CLV3 and the location of alleles used in this study. Black boxes are exons and black lines are introns. Point mutations are denoted by black arrows, a 1-bp deletion is denoted by a black arrowhead, and the positions of T-DNA insertions are denoted by gray arrowheads.
Stem cell populations in shoot meristems (SAMs, IMs, and FMs) are maintained by the CLAVATA (CLV) signaling pathway, which consists of the small secreted CLV3 peptide, the receptor-like kinases (RLKs) CLV1 and CORYNE (CRN), and the receptor-like protein CLV2 (Clark et al. 1993, 1997; Kayes and Clark 1998; Jeong et al. 1999; Muller et al. 2008). The prevailing model for CLV signaling suggests that CLV1, CLV2, and CRN perceive the CLV3 peptide, and receptor activation ultimately functions to restrict the expression of the stem cell-promoting homeodomain transcription factor WUSCHEL (WUS) to a small subset of cells in the center of shoot meristems (Brand et al. 2000; Schoof et al. 2000). The WUS expression domain acts as an organizing center required for maintenance of the stem cell population (Schoof et al. 2000). Mutations in CLV1, CLV2, CLV3, or CRN result in the expansion of the WUS expression domain and subsequent increase in the stem cell population in shoot meristems (Schoof et al. 2000). Live imaging studies have shown that loss of CLV3 causes cells that had previously differentiated and decreased their proliferation to return to a stem cell state, thus creating increased cell division in the IM (Clark et al. 1995; Laux et al. 1996; Brand et al. 2000; Schoof et al. 2000; Reddy and Meyerowitz 2005).
Along with WUS, the homeodomain transcription factor SHOOTMERISTEMLESS (STM) is expressed in shoot meristems and is required to maintain cell proliferation (Long et al. 1996). Although stm and wus mutants produce similar phenotypes, there are discrete differences between these genes. For example, while WUS expression is found in a small number of cells within the SAM (Mayer et al. 1998), STM is expressed broadly throughout shoot meristems and shares overlap with the CLV1 domain (Long et al. 1996; Clark et al. 1997). Furthermore, genetic interactions between hypomorphic stm and wus mutants suggest that WUS acts to maintain stem cell function, while STM is required for maintenance of an undifferentiated state (Endrizzi et al. 1996).
Evidence from genetic studies has led to the hypothesis that the CLV1 and CLV2 receptors act in a common pathway to regulate WUS expression. However, experiments analyzing the genetic interactions between the more recently identified CRN receptor kinase and CLV1 and CLV2 suggest that a CLV2/CRN receptor complex binds CLV3 and regulates WUS in a pathway separate from CLV1 (Muller et al. 2008; Ogawa et al. 2008; Guo et al. 2010). Results from recent biochemical studies indicate that while CLV2 and CRN readily form heterodimers and CLV1 forms homodimers, CLV1 and CLV2 do not appear to interact in the absence of CRN, although there is evidence for the formation of a CLV1/CLV2/CRN receptor complex at the plasma membrane (Bleckmann et al. 2010; Zhu et al. 2010).
Most of the evidence for the structure of the CLV pathway in Arabidopsis meristems is based on genetic interactions using valve number as a readout for carpel number and meristem size. However, double mutants between clv1 and either crn or clv2 mutants have produced inconsistent phenotypes, depending on the use of strong or weak clv1 alleles. The ambiguity of these results can be partially explained by the observation that mutations in the CLV1 receptor kinase demonstrate unusual properties. Plants homozygous for missense mutations in the CLV1 extracellular domain such as clv1-4 display stronger phenotypes than null mutants (Clark et al. 1993; Dievart et al. 2003). The proteins encoded by these mutations are proposed to act in a dominant-negative manner through inhibitory interactions with closely related BARELY ANY MERISTEM (BAM) receptors (DeYoung et al. 2006; DeYoung and Clark 2008), CLV2, CRN, and/or additional unknown receptors that eliminate all repression of WUS. Genetic interactions are consistent with the model that BAM receptors can compensate for the absence of CLV1 protein and form complexes with other receptors such as CLV2 or CRN, repressing WUS (DeYoung and Clark 2008).
Strong clv1 mutants contain several phenotypes associated with enlarged shoot meristems, including fasciated or flattened stems, increased floral and fruit organ number, and the presence of an extra gynoecium inside the fruit, often called a fifth whorl, that forms when the FM fails to terminate due to prolonged WUS expression (Clark et al. 1993, 1997; Schoof et al. 2000). However, null and hypomorphic clv1 mutants lack the majority of these phenotypes, with the exception that they produce extra fruit organs, albeit fewer than dominant-negative alleles (Clark et al. 1993). clv2 and crn mutants are similar to weak clv1 alleles in that they only produce extra fruit organs (Kayes and Clark 1998; Muller et al. 2008). Mutants that generate only extra fruit organs are not consistent with the model that extra floral organs arise from enlarged meristems.
Through the characterization of mutants in known CLV pathway receptors, we have identified an alternative mechanism for the generation of fruit organs. Our results provide evidence that the CLV1, CLV2, and CRN receptors function in a single pathway in the gynoecium to repress proliferation, while CLV1/BAM1 and CLV2/CRN act in parallel pathways to restrict carpel number in shoot meristems.
Materials and Methods
Plant growth conditions and phenotypic analyses
Growth of plants:
Plants were grown as described in Belkhadir et al. (2010).
Identification of clv1, clv2, crn, and er alleles:
clv1-23, clv2-8, and crn-1:
Wild-type seeds in the Wassilewskija-2 (Ws-2) accession were mutagenized with EMS as previously described (Zhao et al. 2002). Three mutants with only extra fruit organs (14-7, 15-6, and 16-1) were identified and were crossed into the Col accession three times for mapping and for phenotypic analysis. Linkage analysis of the PCR-based markers NF5I14 and nga111 (available on www.arabidopsis.org) placed the 14-7 and 15-6 mutations near At1g65380 (CLAVATA2) and At1g75820 (CLAVATA1). In 14-7, which failed to complement clv1-20, we identified a G-to-A nucleotide substitution at position 2722, which corresponds to a V-to-M at position 882 in CLV1, and 14-7 was renamed clv1-23. In 15-6, which failed to complement clv2-2, we identified a G-to-A nucleotide substitution at position 494, which corresponds to a W-to-stop at position 165 in CLV2, and 15-6 was renamed clv2-8. For 16-1, linkage analysis of the PCR-based markers nga158 and nga139 placed 16-1 near At5g13290 (CORYNE). We sequenced CRN in 16-1 and found a G-to-A nucleotide substitution at nucleotide position 208, which corresponds to a G-to-E substitution at position 70 in CRN. This mutation was found to be identical to the crn-1 mutant identified by Muller et al. (2008); therefore, we renamed 16-1 as crn-1.
clv1-20, clv1-21, clv1-22:
Three additional alleles were found in sequenced-tagged insertion libraries (http://signal.salk.edu/cgi-bin/tdnaexpress) generated in the Columbia accession. The clv1-20 allele (SALK_008670) and the clv1-21 allele (SAIL_802C08) both contain insertions within the 5′-UTR of CLV1. We resequenced T-DNA flanking PCR products and found that the insertions in clv1-20 and clv1-21 are 217 and 146 bp, respectively, upstream of the predicted transcriptional start. The insertion in the clv1-22 allele, (WiscDsLox489-49), is located 2042 bp downstream of the predicted transcriptional start within the kinase domain of CLV1.
clv1-4:
This allele is a missense amino acid change at position 201 (G-to-E substitution) within leucine-rich-repeat (LRR) 5 of the extracellular domain (McKelvie 1962). Before conducting our analysis, the clv1-4 mutation was introgressed to plants in the Ws-2 accession for three generations, and the strain we analyzed does not contain the er-1 mutation found in the Landsberg erecta accession.
er-3:
The er-3 allele (SALK_044110) contains an insertion 906 bp downstream of the predicted transcriptional start.
RT–PCR
RNA was extracted from 100 mg of inflorescence tissue from the four new clv1 mutant alleles and Col using the RNEasy Plant Extraction kit (Qiagen). A total of 1 μg of total RNA was used to perform the first strand synthesis, primed with gene-specific primers that amplify the entire coding region (supporting information, Table S3) per manufacturer recommendations (Fermentas). Second strand synthesis was performed using the following primer pairs: CLV1 RT F and R, and ACT7 F and R (30 cycles, annealing at 55°).
Phenotypic analysis of floral organs
For all mutants and wild-type controls, the sepal, petal, anther, and valve number were counted for the buds (up to 30) on the main inflorescence. Each bud was analyzed at stages 13–14 (just prior to fertilization) to ensure that all organs would be counted before dehiscence.
Histological analysis
Floral buds and meristems were collected from the primary stem of plants 1–2 weeks after bolting. Sample preparation and imaging were performed as described by Belkhadir et al. (2010). Stage 3 floral meristem height was measured as the distance between the interior base of the medial sepal primordia and the top of the dome, and width was measured as the distance between sepal primordia (Figure S2). Inflorescence meristem height was measured as the distance between the interior base of the medial first incipient primordia to the top of the dome, and width was measured as the entire distance across the region from the first incipient primordia (Figure S2).
CLV1 promoter cloning and analysis of transcriptional fusion to GUS
Through phenotypic analysis of insertions upstream of CLV1 including clv1-20 and clv1-21, we deduced that the genomic region corresponding to 3.4 kb upstream from the CLV1 ATG was required for CLV1 expression. This region through the start codon was cloned into pENTR/D-TOPO using the TOPO-TA cloning kit (Invitrogen), subcloned into pBIB:BASTA-GUS (Gou et al. 2010), and transformed into Col plants via Agrobacterium-mediated transformation (Table S3). To visualize GUS expression, plants were incubated in X-Gluc staining solution (2 mM X-Gluc dissolved in 1 mL DMSO, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, 0.2% Triton X-100, 50 mM NaPO4, pH 7.2) for 1–16 hr, then cleared in 9:1 ethanol/acetic acid for 3–16 hr, followed by 1 hr in chloral hydrate solution (8:2:1 chloral hydrate/water/glycerol).
RNA in situ hybridization
Floral buds were fixed and sectioned as previously described (Franks et al. 2002). Riboprobes labeled with digoxigenin for STM were created per manufacturer’s recommendations (Roche). To generate riboprobes for FIL, CLV1, CLV2, and CRN, the cDNA was generated from Col RNA with similar methods used to perform RT–PCR and cloned into pCR8/GW/TOPO using the TOPO-TA cloning kit (Invitrogen; Table S3). The plasmid was used as template in a PCR reaction utilizing gene-specific primers for which either the sense or antisense primer contained the T7 recognition sequence at the 5′ end. These pools of DNA were used as template to generate riboprobes labeled with digoxigenin per manufacturer’s recommendations (Roche). Hybridization, washes, and detection were performed as previously described (Franks et al. 2002). Images were captured using QCapture Pro 5.0 software.
Results
Most clv and crn mutants produce gynoecium-specific increases in fruit organ number
To identify genes that regulate fruit organ number, we performed a screen for mutants that produce wild-type numbers of sepals, petals, and stamens, but generated more than two valves. We identified three mutants: clv1-23, a novel missense mutation in the kinase domain of CLV1; clv2-8, a novel missense mutation leading to an early stop codon in the extracellular domain of CLV2; and the identical mutation as the crn-1 mutant previously identified by Muller et al. (2008) (Figure 1B). To more completely characterize the phenotypes of loss-of-function mutations in these three genes, we also analyzed three T-DNA insertion alleles of clv1, the previously characterized clv1-4 allele (McKelvie 1962; Clark et al. 1993; 1997), and two previously characterized clv2 mutants (Figure 1B; Forstheofel et al. 1992; Kayes and Clark 1998). Our molecular analysis indicates that clv1-20 and clv1-21, two insertions located in the 5′-UTR, result in decreased levels of CLV1 mRNA; and clv1-22, an insertion in the kinase domain, is a transcriptional null (Figure 1B; Figure S1). Both clv2 alleles are predicted to be nulls.
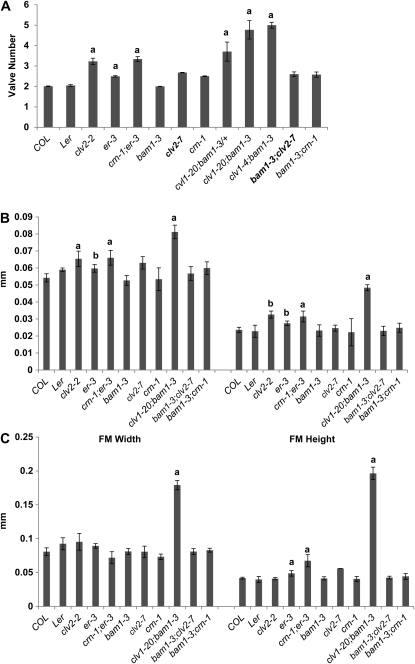
We next characterized the fruit and floral organ phenotypes of these mutants. We found that with the exception of clv1-4, all clv1, clv2, and crn-1 mutants only produced extra fruit organs and did not generate extra sepals, petals, or stamens (Figure 2A). On the basis of their molecular nature and fruit organ phenotypes, we categorized clv1-20, clv1-21, clv1-22, clv2-7, clv2-8, and crn-1 as “weak,” clv1-23 and clv2-2 as “intermediate,” and clv1-4 as “strong.” Although our quantification of fruit organs is lower on average than previously reported for each class, our groupings for the clv1 alleles are consistent with classifications of other clv1 alleles (Clark et al. 1993, 1997; Leyser and Furner 1992; Medford et al. 1992; Pogany et al. 1998; Dievart et al. 2003). The lower averages we obtained are most likely due to our counting methods. We assign a 0.5 value to partial valves, a phenotype observed in a significant percentage of fruit from all alleles (see below; Table 2; Figure S3), rather than one, as in previous studies (Kayes and Clark 1998; DeYoung and Clark 2008).
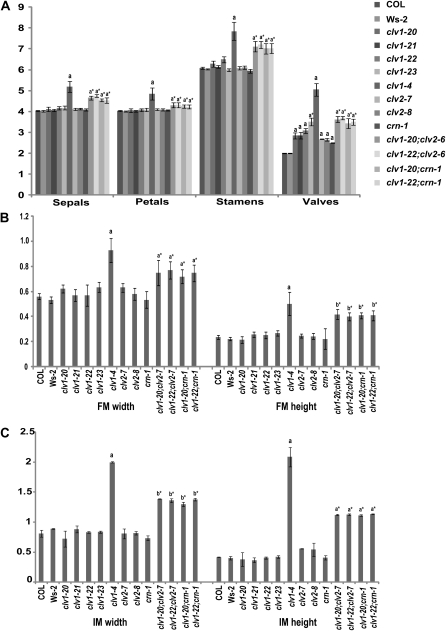
Figure 2 .
Floral and meristem phenotypes in clv1, clv2, and crn single and double mutants. (A) Floral organ number in plants homozygous for clv1, clv2, and crn single and double mutants. Values for organ numbers represent the average of 30 flowers for >20 plants. (B) Floral meristem width and height in clv1, clv2, and crn single and double mutants. (C) Inflorescence meristem width and height in clv1, clv2, and crn single and double mutants. N < 5 for each allele for both FM and IM measurements. Error bars represent the standard deviation above and below the mean. All alleles are in the Col background except for clv1-4 which was introgressed into the Ws-2 background from the Ler background. clv1-23, clv2-8, and crn-1 were initially identified in the Ws-2 accession, but were introgressed into the Col accession. a, two-tailed t test (P < 0.01), indicating that values are significantly greater than wild type. b, two-tailed t test (P < 0.05), indicating that values are significantly greater than wild type. *, denotes values that are significantly higher than wild type, but significantly lower than clv1-4.
Table 2 . Partial valves in clv1, clv2, and crn mutants.
| Allele | No. partial valves | % |
|---|---|---|
| COL | 8/600 | 1.33 |
| Ler | 12/600 | 2.0 |
| clv1-20 | 131/600 | 21.8 |
| clv1-21 | 119/600 | 19.8 |
| clv1-22 | 84/600 | 14.0 |
| clv1-23 | 86/600 | 14.3 |
| clv1-4 | 144/600 | 24.0 |
| clv2-2 | 186/600 | 31.0 |
| clv2-7 | 95/600 | 15.8 |
| clv2-8 | 129/600 | 21.5 |
| crn-1 | 77/600 | 12.8 |
Percentage of total fruit containing at least one partial valve out of 600 stage-17 fruit examined for each allele.
Our finding that clv1, clv2, and crn mutants do not generate extra sepals, petals, and stamens is not consistent with the model that larger meristems generate more organs. To directly test whether our mutants produce larger meristems, we measured FMs and IMs from weak and intermediate mutants and compared them to the FMs and IMs of clv1-4, which is known to produce larger meristems (Clark et al. 1993; Clark et al. 1995; Clark et al. 1997). FMs and IMs from all weak and intermediate mutants were similar in dimension to wild-type meristems (Figure 2, B and C; Figure S2), with the exception of clv2-2, which we believe is due to the presence of a partial loss-of-function mutation in the ERECTA (ER) receptor kinase in the Landsberg erecta (Ler) ecotype (see below).
clv1, clv2, and crn mutants generate new organs during gynoecium development
As our mutants did not produce larger meristems, we hypothesized that weak and intermediate mutants generate extra fruit organs during gynoecium development, after termination of the FM. Therefore, we generated sections from a series of 10 consecutive flower buds per plant, ranging from stages 7–14 of flower development (Smyth et al. 1990; Figure 1A). During stages 6–12 of gynoecium development, the medial regions maintain meristematic properties to allow for ovule formation (Bowman et al. 1999). Although the physical structures that will differentiate into valves and repla are present by stages 11–12, differentiation of the fruit organs occurs around stage 13, making it unlikely that a new organ would be initiated after this stage (Ferrandiz et al. 1999; Roeder et al. 2003). Since we never observed an increase in fruit organ number between stages 14 and 17 (Table S2), this indicates that extra organs are initiated between stages 6 and 13.
At stages 7–9 when septa primordia initiate and fuse, most buds homozygous for the weak clv1-20 and clv1-21 alleles produced two primordia in a fashion similar to wild type, although the primordia appeared wider than wild-type controls (Figure 3, A and B). This phenotype is significant, as the FM has terminated by these stages. However 85 and 87% of clv1-20 and clv1-21 mutants, respectively, contain at least one extra fruit organ at maturity (Table S1). Therefore, the FM is most likely not the source of the extra fruit organs found in these mutants, as we would expect to see extra primordia by this stage. Starting at stage 10, we began to see the presence of extra septa and additional chambers within the gynoecium, in clv1-20 and clv1-21 mutants (Figure 3, C and D), and by stage 12, in almost all buds. We also found extra cells in the medial regions, similar to earlier stages.
Figure 3 .
Gynoecium phenotypes in clv1, clv2, and crn single and double mutants. (A) Wild-type Columbia (Col) stages 7–9 (N = 25). (B) Representative stage-7 to -9 clv1-20 section. Totals of 28/28 clv1-20 buds and 19/21 clv1-21 buds displayed this phenotype (C) Col, stages 10–11. (D) clv1-21, stages 10–11. Totals of 5/28 clv1-21 buds and 19/30 clv1-20 buds displayed this phenotype. (E) Representative stage-7 to -9 clv1-22 section. Totals of 19/29 clv1-22 buds and 24/29 clv1-23 buds displayed this phenotype. (F) Representative stage-7 to -9 crn-1 section. A total of 12/20 crn-1 buds displayed this phenotype. Totals of 14/21 clv2-7 buds and 8/16 clv2-8 buds also displayed this phenotype. (G) clv1-4, stages 7–9 with four, evenly spaced septa primordia. (H) clv1-22, stages 10–11. (I) clv1-4, stages 10–11. (J) Representative stage-7 to -9 clv1-22; crn-1 section. Totals of 15/15 clv1-22; crn-1 buds and 18/18 clv1-20; crn-1 buds displayed this phenotype. Totals of 26/26 clv1-20; clv2-7 and 22/22 clv1-22; clv2-7 buds also displayed this phenotype. (K) Representative stage-10 to -11 clv1-22; clv2-7 section. Totals of14/14 clv1-20; clv2-7 and 17/17 clv1-22; clv2-7 buds displayed this phenotype. Totals of 12/12 clv1-20; crn-1 and 10/10 clv1-22; crn-1 buds also displayed this phenotype. (L) clv1-20; bam1-3, stages 7–9 (N = 24/24). (M) clv1-20; bam1-3, stages 10–11 (N = 15/15). (N) bam1-3; clv2-7, stages 7–9 (N = 15/15). A total of 17/17 bam1-3; crn-1 buds displayed this phenotype. (O) bam1-3; clv2-7, stages 10–11. A total of 9/9 bam1-3; crn-1 buds displayed this phenotype (N = 12/12). M, medial region; L, lateral region. Bars, 50 μm. Bar in A is the same for B. Bar in C is the same for D. Arrow indicates splitting of the septum.
In clv1-22, clv1-23, clv2-7, clv2-8, and crn-1 mutants, at stages 7–9, approximately half of the buds resembled clv1-20 and clv1-21 mutants and displayed wild-type numbers of primordia. However, in the other half, we observed either (1) multiple primordia emerging from separate medial regions that arise adjacent to one another without the presence of an extra lateral region (Figure 3E) or (2) one large primordium emerging from a single medial region that was split into two septa at the end of the primordia (Figure 3F). In the former case, the adjacent primordia were located directly across from either a single primordium (three septa) or two adjacent primordia (four septa). Although the clv1-22, clv2-7, clv2-8, and crn-1 mutants ultimately displayed only weak increases in fruit organ number, similar to clv1-20 and clv1-21 mutants, the early development phenotypes in gynoecia were more similar to intermediate clv1-23 mutants. Therefore, we will refer to clv1-20 and clv1-21 as weak and clv1-22, clv1-23, clv2-7, clv2-8, and crn-1 as developmentally intermediate on the basis of this gynoecium phenotype.
We next compared the extra primordia phenotype observed in the developmentally intermediate mutants to stages 7–9 clv1-4 mutants. In clv1-4, we find initiation of multiple septa primordia; however, the primordia were evenly spaced, with the medial regions separated by a full lateral region and never adjacent to one another (Figure 3G). This phenotype is consistent with a model in which additional septa, formed at early stages in clv1-4 mutants, are derived from extra carpels initiated in the FM.
We next asked whether clv1-4 mutants also produce extra organs during gynoecium development. We observed wide primordia with extra cells and medial regions arising adjacent to one another at later stages (Figure 3I), similar to the other clv1, clv2, and crn mutants. These results indicate that clv1-4 mutants generate extra organs both from enlarged FMs and during gynoecium development; loss of both mechanisms contribute to the strong phenotype. As the organs formed in the gynoecium have a different ontogeny from those initiated in the FM, we will refer to organs formed from the FM as carpels and organs formed during gynoecium development as ectopic valves.
We next compared septa primordia number in young gynoecia (stages 7–9) and valve number in mature fruit and found that for all mutants tested, significantly more valves were present in mature fruit vs. primordia in young gynoecia (Table 1). This was especially true for clv1-4 mutants, which produce an average of four and never more than six carpels, yet mature fruit contained an average of >5 valves, while examples of fruit containing 7–10 valves were common (Table 1). We never observed extra septa in young wild-type gynoecia (N = 25; Table 1); however, we did occasionally see extra valves in mature fruit (N = 600; Table 1).
Table 1 . Average fruit organ number at stages 7-9 vs. stage 17.
| Allele | Stages 7–9 | Stage 17 |
|---|---|---|
| COLa | 2 ± 0 | 2.01 ± 0.0147 |
| clv1-20a | 2 ± 0 | 2.85 ± 0.149 |
| clv1-21a | 2.15 ± 0.366 | 2.86 ± 0.149 |
| clv1-22b | 2.86 ± 0.838 | 3.084 ± 0.12 |
| clv1-23b | 3.22 ± 0.676 | 3.5 ± 0.183 |
| clv1-4a | 4.19 ± 0.55 | 5.05 ± 0.306 |
| clv2-7b | 2.32 ± 0.466 | 2.68 ± 0.0131 |
| clv2-8b | 2.5 ± 0.474 | 2.66 ± 0.0693 |
| crn-1b | 2.31 ± 0.412 | 2.5 ± 0.0165 |
| clv1-20; clv2-7a | 3.38 ± 0.471 | 3.63 ± 0.132 |
| clv1-22; clv2-7b | 3.23 ± 0.6201 | 3.69 ± 0.0802 |
| clv1-20; crn-1a | 2.79 ± 0.838 | 3.44 ± 0.216 |
| clv1-22; crn-1a | 3.1 ± 0.541 | 3.47 ± 0.241 |
Valve primordia (stages 7–9) and valves (stage 17) were counted for all five alleles. ±, standard deviation.
P < 0.01.
P < 0.05, two-sample t test for unequal variance.
In clv1, clv2, and crn mutants, we frequently observe a phenotype in mature fruit where the apical region contains more valves than the base (Figure S3A). These additional valves vary from 10 to 90% of total valve length and were found in a significant proportion (14–31%) of fruit in all mutants analyzed (Table 2). Although previous studies of clv2 report a phenotype described as “valveless” in which part of the valve is replaced by replum tissue (Kayes and Clark 1998; DeYoung and Clark 2008; Figure S3, C and C′), the partial valve phenotype we identified did not match this description. Serial sectioning of gynoecia at various stages of development revealed that the additional organs seen in the apical region appear to form when an enlarged septa splits into two (Figure S3, D–F). Also, we never saw the valveless phenotype in our sections of clv2 mutants, indicating that the partial valve phenotype is likely separate from the valveless phenotype. Partial valves could be identified in the weak mutants starting at stage 10, and as early as stage 8 in developmentally intermediate mutants. This phenotype cannot be explained by the presence of extra carpels in the FM, but rather suggests that ectopic valves can be initiated anywhere along the medial region during stages 7–13 of gynoecium development.
Ectopic organ specification occurs during gynoecium development in clv1, -2, and crn mutants
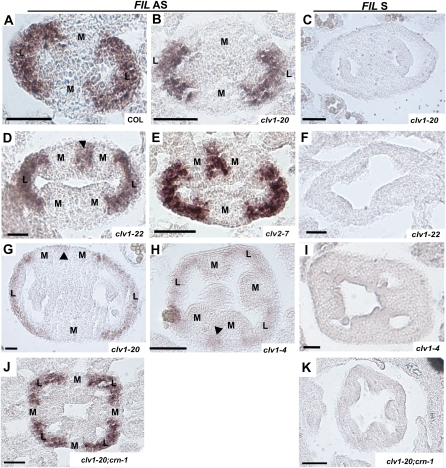
We next tested whether ectopic expression of fruit organ specification genes accompanies the initiation of ectopic valves in these mutants. We performed RNA in situ hybridization with probes for FILAMENTOUS FLOWER (FIL), a component of the fruit specification network and an early marker for lateral regions that eventually form valves (Figure 4A; Siegfried et al. 1999; Dinneny et al. 2005). In stages 7–9, wild type and weak mutants where two primordia are initiated, FIL is expressed in broad arches that correspond to the two lateral regions (Figure 4, A and B). In most stage-7 to -9-developmentally intermediate mutants, we saw FIL expressed in the two lateral regions. However, we also detected smaller patches of FIL expression in areas where two primordia abut one another or in the center of medial regions where the primordium has split at the internal end (Figure 4, D and E). We saw similar ectopic FIL expression in the weak mutants starting at stage 10 (Figure 4G). In clv1-4 mutants, we consistently found FIL expressed in broad arches in multiple lateral regions, indicative of extra carpels; however, we also found patches of FIL in adjacent medial regions (Figure 4H), similar to stage-7 to -9-developmentally intermediate and stage-10 to -12 weak mutants. These results further support the hypothesis that ectopic valves are initiated during gynoecium development in clv1-4 mutants.
Figure 4 .
Expression of FIL in clv1, clv2, and crn single and double mutants (A, B, D, E, G, H, and J) Antisense. (C, F, I, and K) Sense. (A) Col stage 8. (B) clv1-20 stages 8–9. Totals of 22/22 clv1-20 and 12/12 clv1-21 buds displayed this phenotype. (C) Col stage 9. (D) Section through the top of a clv1-22 stage-8 to -9 bud. A total of 8/23 buds displayed extra patches of FIL in the medial regions, while 13/23 buds had no ectopic expression. A total of 5/23 buds demonstrated differential expression of FIL between the apical and basal regions. A total of 11/19 crn-1 buds showed ectopic expression of FIL, and 8/17 clv1-23 buds showed ectopic FIL expression, 4 with FIL expression in apical and basal regions. (E) clv2-7 stage 8 with ectopic FIL expression. A total of 3/15 buds had no ectopic FIL expression, even when extra septa primordia were visible. (F) clv1-22 stages 7–9. (G) clv1-20 stages 10–11. (H) clv1-4 stage 9. A total of 27/27 buds had FIL expression in extra lateral regions and 2 also displayed extra patches of FIL. (I) clv1-4 stage 9. (J) clv1-20; crn-1 stages 8–9. A total of 3/13 clv1-20; crn-1 and 3/10 clv1-22; crn-1 buds displayed no ectopic regions of FIL expression. All other buds analyzed contained ectopic regions of FIL. (K) clv1-20; crn-1 stages 8–9. M, medial region; L, lateral region. Arrowheads indicate ectopic regions of FIL expression. Bars, 50 μm.
CLV1, CLV2, and CRN are expressed in developing fruit
To explain the enlarged medial regions in clv1, clv2, and crn mutants (Figure S4, B and D–H), we propose that CLV pathway receptors act to repress cell proliferation in medial regions during gynoecium development. Previous studies showed CLV1 and CLV2 transcripts in overlapping domains in the center of shoot meristems (Clark et al. 1993; Kayes and Clark 1998) and CLV1 expression in the vasculature (An et al. 2004; Zhao et al. 2005). CRN is expressed throughout shoot meristems, in young flower and carpel primordia, and in floral and pedicel vasculature (Muller et al. 2008). However, gynoecium expression of CLV1, CLV2, or CRN has not been shown. We generated a reporter gene containing the native CLV1 promoter upstream of B-galacturonidase (GUS), which was expressed in shoot meristems as previously shown with in situ hybridization (Clark et al. 1993). In addition, we found expression in medial regions of gynoecia starting at stage 10, becoming stronger at stages 13–14 (Figure S4, A and B).
To further examine gynoecium-specific expression of CLV1, CLV2, and CRN, we performed RNA in situ hybridization. We detected weak expression for all three genes in the center of the medial regions of the gynoecia starting at stages 8–9 (Figure S4, C, G, and K) that included the vasculature of the medial regions, but was not limited to this area. At stages 10–12, these genes were specifically expressed in the medial regions and in the vasculature of the lateral regions, as well as in the transmitting tract where the septa fuse (Figure S4, D, H, and L).
STM is a likely target of the CLV pathway receptors in gynoecia
In shoot meristems, CLV1, CLV2, and CRN function to maintain stem cell populations and repress proliferation (Clark et al. 1993, 1997; Schoof et al. 2000). The proliferation defects in the medial regions of clv1, clv2, and crn mutants implicate these receptors in maintenance of cell populations with higher plasticity in medial regions of gynoecia (Bowman et al. 1999). In shoot meristems, these receptors act on WUS; however, WUS is not expressed in developing gynoecia (Goss-Hardt et al. 2002). SHOOTMERISTEMLESS (STM), also involved in meristem maintenance, is expressed in meristems as well as in young gynoecia (Endrizzi et al. 1996; Long et al. 1996). Furthermore, there is evidence for a genetic interaction between the CLV1 and STM orthologs TASSEL DWARF1 and KNOTTED1, respectively, in maize reproductive meristems (Lunde and Hake 2009).
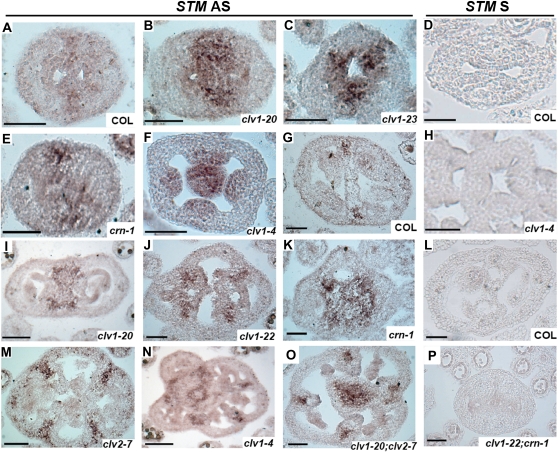
To test whether CLV1, CLV2, and CRN regulate STM in gynoecia, we examined STM expression in the clv1, clv2, and crn mutants at different stages of development with in situ hybridization. In wild type, STM is weakly expressed in medial regions at stages 7–9 (Figure 5A). In all clv1, clv2, and crn mutants, STM was expressed in medial regions, but in an expanded number of cells compared to wild type (Figure 5, B–F). In clv1-4 mutants, we also found strong expression in the center of fruit that corresponds to the fifth whorl (Clark et al. 1993; Schoof et al. 2000; Figure 5F). At stage 10 and later, STM expression is restricted to the center of the medial regions and is never observed in the septum in wild type (Figure 5G). However, in clv1, clv2, and crn mutants, STM is broadly expressed throughout the medial regions and in the septum where it is never present in wild type (Figure 5, I–N). This expansion of STM expression in the medial region/septa is very reminiscent of the expanded WUS expression domain seen in shoot meristems of strong clv1 mutants (Schoof et al. 2000).
Figure 5 .
Expression of STM in clv1, clv2, and crn single and double mutants (A–C, E–G, I–K, M–O) Antisense. (D, H, L, and P) Sense. (A) Col stages 8–9 (N = 19). (B) clv1-20 stages 8–9 (N = 23). Similar results were observed for clv1-21 (N = 16). (C) clv1-23 stages 8–9 (N = 17). Similar results were observed for clv1-22 (N = 15). (D) Col stages 8–9 (N = 10). (E) crn-1 stages 8–9 (N = 12). A total of 14/14 clv2-7 buds displayed the same phenotype. (F) clv1-4 stages 8–9 (N = 15). (G) Col stages 11–12 (N = 15). (H) clv1-4 stages 8–9 (N = 7). (I) clv1-20 stages 10–11 (N = 18). Similar results were observed for clv1-21 (N = 15). (J) clv1-22 stages 10–11 (N = 19). Similar results were observed for clv1-23 (N = 13). (K) crn-1 stages 11–12 (N = 13). (L) Col stages 10–11 (N = 8). (M) clv2-7 stages 10–11 (N = 12). (N) clv1-4 stages 10–11 (N = 9). (O) clv1-20; clv2-7 stages 10–11. Totals of 11/11 clv1-20; clv2-7 and 10/10 clv1-22; clv2-7 buds displayed this phenotype. Totals of 12/12 clv1-20; crn-1 and 14/14 clv1-22; crn-1 buds also displayed this phenotype. (P) clv1-22; crn-1. A total of 10/10 buds displayed this phenotype. Bars, 50 μm.
The misregulation of STM in clv1, clv2, and crn mutants suggests that STM acts downstream of the CLV pathway in gynoecia. We next tested for a genetic interaction between clv1 and stm mutants using the weak clv1-20 and null clv1-22 alleles in combination with the weak stm-2 mutant (Clark et al. 1996). For most clv1-20; stm-2 and clv1-22; stm-2 mutants, plants displayed phenotypes similar to stm-2 single mutants where plants fail to maintain viable shoot, inflorescence, or floral meristems, producing bushy plants (Figure 6, A and B). Occasionally, a meristem was maintained long enough to produce a single inflorescence (Figure 6B). Most buds on the inflorescence did not produce complete sets of organs and contained sepaloid and petaloid structures. However, on occasion, a mature fruit was produced that was shorter than wild type, but always generated two valves (Figure 6C). We sectioned inflorescences from clv1-20; stm-2 and clv1-22; stm-2 mutants and found that in most cases, young buds were devoid of any identifiable floral organs, but when fruit were initiated (clv1-20; stm-2 N = 22; clv1-22; stm-2 N = 26), the number of carpels and valves were similar to wild type (Figure 6D). The observation that formation of ectopic fruit organs is strongly suppressed in clv1; stm-2 double mutants containing the weak stm-2 mutation further supports the hypothesis that STM is the downstream target of CLV1.
Figure 6 .
Mutant phenotypes for stm-2; clv1 double mutants. (A) Three-week-old stm-2 mutant plant. (B) Three-week-old stm-2; clv1-20 mutant plant. Totals of 10/20 stm-2; clv1-20 and 9/20 stm-2; clv1-22 plants produce a visible stem as in B. All other stm-2; clv1 plants resembled the image in A. (C) stm-2; clv1-22 stage 17 silique with two valves. Totals of 26/52 stm-2; clv1-20 and 22/47 stm-2; clv1-22 plants generated a mature fruit, all of which displayed two valves. Only one fruit per inflorescence was ever observed. (D) A cross-section of a stage-6 to -7 stm-2; clv1-20 fruit. Out of 16 buds sectioned, only 1 bud produced any structure recognizable as a fruit. Bar, 50 μm.
Mutations in ERECTA enhance meristem defects in CLV pathway mutants
Previous studies show that clv and crn mutants produce enlarged meristems and significant increases in valve number (Clark et al. 1993, 1997; Kayes and Clark 1998; Muller et al. 2008). However, in our studies, most clv1, clv2, and crn mutants have only modest increases in valve organ number and produce meristems that are similar in size to the wild type. One possible explanation for these differences could be ecotype-specific effects, in particular the presence of the erecta mutation in the Ler accession. Most previous studies analyzed mutants generated in or introgressed into Ler; however, in our study, all alleles are in the Col accession or were introgressed into Col from the Ws-2 accession, with two exceptions: clv1-4, which was introgressed from the Ler accession into Ws-2, and clv2-2, in the Ler accession.
Comparison of the clv2-2 and clv2-7 alleles revealed significant differences in valve number (3.23 vs. 2.68) (Figure 7A). As both alleles are predicted to be null mutants (Kayes and Clark 1998; Wang et al. 2008), the differences in their molecular nature (a single base-pair deletion vs. a coding region insertion) should not contribute to the differences seen in the fruit phenotype. We analyzed IMs and FMs from clv2-2 mutants and found that they produced IMs similar in width to Ler; however, they generated FMs that were taller than Ler, consistent with previous reports (Figure 7, B and C; Kayes and Clark 1998). As clv2-2 was the only clv2 allele to have a significant increase in meristem size (Figure 7, B and C; P < 0.01, t test), we propose that interactions between clv2 and mutations in Ler result in the larger meristems seen in previous studies (Clark et al. 1993, 1997; Kayes and Clark 1998; Muller et al. 2008).
Figure 7 .
Genetic interactions between bam1 and er and CLV pathway mutants. (A) Valve number in plants homozygous for clv1, clv2, crn, bam1-3, and er-3 single and double mutants. Values for valve number represent the average of 30 flowers for >20 plants. (B) Floral meristem width and height in clv1, clv2, crn, bam1-3, and er-3 single and double mutants. (C) Inflorescence meristem width and height in clv1, clv2, crn, bam1-3, and er-3 single and double mutants. N > 5 for each mutant for both IM and FM measurements. a, two-tailed t test (P < 0.01). b, two-tailed t test (P < 0.05). Error bars represent the standard deviation above and below the mean. a, two-tailed t test (P < 0.01). b, two-tailed t test (P < 0.05), indicating that values are significantly greater than wild type. Significance of the values for the double mutants was tested against each single mutant.
The ERECTA (ER) RLK is known to affect the shape of shoot meristems, and the Ler accession contains the er-1 partial loss-of-function allele (Torii et al. 1996; Yokoyama et al. 1998). To test whether mutations in ER are responsible for the stronger phenotype in clv and crn mutants, we generated double mutants between crn-1 and the er-3 T-DNA insertion in the Col accession. The original characterization of crn-1 (isolated in the Ler accession) described significant increases in both meristem size and valve number (Muller et al. 2008), while analysis of the crn-1 mutant identified in this study (isolated in Ws-2 and introgressed into Col), which contains the identical base-pair change, revealed meristems of wild-type dimensions and only a modest increase in valve number (Figure 2). We chose to focus on crn for this analysis because the mutations are identical and differ only in their genetic background.
er-3 single mutants generate extra valves, similar to crn-1 mutants (Figure 7A). In crn-1; er-3 double mutants, valve number was additive compared to our crn-1 and er-3 single mutants (Figure 7A), although it was lower than the values obtained for the Ler crn-1 allele (Muller et al. 2008). This difference is most likely due to our quantification methods (see above). We next compared FMs and IMs from Ler, er-3, and crn-1; er-3 mutants. We found that FM width and height in er-3 mutants were slightly, but significantly increased compared to Col and crn-1 mutants (Figure 7B; t test, P < 0.05). Although there is no significant difference in FM height between Col and Ler, there was also no significant difference in FM height between Ler and er-3 mutants (Figure 7B; t test). In IMs, there was no significant difference in width between er-3 mutants and Col, Ler, or crn-1 mutants; however, there was a significant increase in height between er-3 and Col and between Ler and crn-1 (Figure 7C; t test, P < 0.01). In crn-1; er-3 double mutants, FM width and height were significantly greater than Col, Ler, and single mutants (Figure 7B; t test, P < 0.01). While crn-1; er-3 double mutants showed no differences in IM width (Figure 7C; t test, P < 0.05), IM height was significantly increased compared to Col, Ler, and single mutants (Figure 7C; t test, P < 0.01).
Sectioning of stages 7–9 gynoecia revealed that crn-1; er-3 mutants generated some gynoecia with septa primordia arising adjacent to one another, as seen in crn-1 single mutants, while other gynoecia produced evenly spaced septa primordia similar to clv1-4 mutants (Figure S5). We also observed the formation of fifth whorls in the crn-1; er-3 double mutants, which was never seen in our crn-1 single mutants (Figure S5). These results suggest genetic interactions between mutations in clv1, clv2, or crn; and the er mutations, including the er-1 mutation in Ler, cause the formation of larger FMs, which in turn leads to the strong meristem and fruit organ phenotypes typically associated with CLV pathway receptor mutants.
CLV1, CLV2, and CRN function together in meristems and gynoecia to regulate fruit organ number
To elucidate the relationships among the CLV1, CLV2, and CRN receptors in both meristems and gynoecia, we generated clv1 and either clv2 or crn double mutants. Previous analyses looking at the interactions between clv1 and either clv2 or crn were frequently performed with dominant-negative clv1 mutants that display significantly stronger meristem and fruit organ phenotypes compared to clv2 or crn single mutants (Kayes and Clark 1998; Muller et al. 2008). Instead, we utilized the clv1-20 hypomorphic allele and the clv1-22 null allele that display similar phenotypes as clv2 and crn mutants (Figures 1B and 2). In clv1-20; clv2-7, clv1-22; clv2-7, clv1-20; crn-1, and clv1-22; crn-1 double mutants we observed significantly more sepals, petals, and stamens than in wild type or single mutants (Figure 2A; t test, P < 0.01). Furthermore, we saw an additive increase in valve number in double mutants compared to single mutants (Figure 2A).
We next tested whether the additive valve number phenotype in clv1; clv2 and clv1; crn mutants was due to extra carpel initiation or an increase in ectopic valve formation. We sectioned IMs and FMs from clv1; clv2 and clv1; crn double mutants and found significantly larger meristems compared to wild type (Figure 2, B and C), suggesting that at least some extra valves form from extra carpels. We next compared gynoecium development in clv1; clv2 and clv1; crn double mutants to single mutants. At stages 7–9, all double mutants produced extra septa primordia (Figure 3J); however, in contrast to the single clv1-22, crn-1, and clv2-7 mutants, the extra primordia were evenly spaced, with lateral regions separating each medial region (Figure 3, E, F, and J). We also observed the presence of fifth whorls in the double mutants (Figure 3J), similar to clv1-4 mutants (Clark et al. 1993, 1995).
Although the primordia were evenly spaced in clv1-20; crn-1, clv1-22; crn-1, clv1-20; clv2-7, and clv1-22; clv2-7 doubles, we also observed septa primordia arising adjacent to one another (Figure 3J). This is similar to clv1-4 mutants, which generate both extra carpels and ectopic valves. We measured average organ number at stages 6–9 of gynoecium development and compared this value to valve number at stage 17. There were significantly more organs at stage 17 compared to earlier stages, suggesting that ectopic valves form in clv1; clv2 and clv1; crn mutants (Table 1). To confirm this phenotype, we examined FIL expression in double mutants and observed expression in broad arches in clearly defined lateral regions (Figure 4J); however, we also occasionally saw small patches of FIL expression in medial regions, indicating that both mechanisms for generating fruit organs are aberrant in clv1; clv2 and clv1; crn mutants. As the frequency of ectopic valve initiation per bud was the same between single and double mutants, we propose that the increase in valve number in double mutants is mainly due to the initiation of extra carpels.
The CLV1/BAM1 receptors, and not CLV2/CRN, are the primary regulators of meristem size
Missense mutations in the extracellular domain of CLV1, such as clv1-4, generate the strongest phenotypes of any mutation in CLV1 (Dievart et al. 2003). Although clv1-20; crn-1, clv1-22; crn-1, clv1-20; clv2-7, and clv1-22; clv2-7 mutants displayed additive and synergistic increases in fruit and floral organ number compared to single mutants, the defects were not as severe as the clv1-4 phenotype (Figure 2B; t test, P < 0.05; Clark et al. 1993, 1995), suggesting that related receptors may be compensating for the loss of CLV1, CLV2, or CRN function.
CLV1 has three close relatives named BAM1, -2, and -3. Single bam mutants do not display a fruit organ number phenotype, although triple bam1; bam2; bam3 mutants have reduced meristem size (DeYoung et al. 2006). Genetic interactions between clv1, clv2, and bam revealed that clv1; bam1 and clv1; bam2 double mutants result in synergistic meristem and valve defects compared to weak, intermediate, and strong clv1 mutants, while a slight decrease in valve number was observed in clv2; bam1 mutants (DeYoung and Clark 2008). It was proposed that BAM1 functions to (1) restrict stem cell populations through interactions with CLV1 and (2) promote division of stem cell populations through interactions with other receptors, such as CLV2 (DeYoung and Clark 2008). However, these experiments, which used valve number as a measure of meristem function, were performed prior to the knowledge that ectopic valves form during gynoecium development.
To better understand the relationship between BAM1 and the CLV pathway receptors, we generated double mutants between bam1 and clv1, clv2, or crn mutants. Removal of a single copy of bam1-3 in a clv1-20 background significantly increased valve number to the levels observed in clv1; crn and clv1; clv2 double mutants (Figure 7A). Double mutants between bam1-3 and clv1-20 mutants produced a strongly synergistic extra valve defect, similar to clv1-4 mutants, and significantly stronger than clv1; crn-1 or clv1; clv2-7 mutant phenotypes (Figure 7A). Analysis of IMs and FMs from the clv1-20; bam1-3 mutants revealed enlarged meristems compared to wild type, clv1-20, and bam1 single mutants (Figure 7, B and C), as well as clv1; clv2 and clv1; crn double mutants (Figure 2, B and C). Analysis of gynoecium development in clv1-20; bam1-3 mutants revealed that at both early (7–9) and late (10–12) stages, development in clv1-20; bam1-3 mutants was identical to clv1-4 mutants (Figure 3, L and M). As the phenotypes in clv1-20; bam1-3 mutants mimic the phenotypes of clv1-4 single mutants, we propose that the clv1-4 dominant-negative mutation causes inhibitory interactions with bam1 protein. Although previous studies reported a synergistic phenotype in clv1-4; bam1-3 mutants compared to clv1-4 single mutants (DeYoung and Clark 2008), we suspect that the stronger phenotype is due to use of the Ler accession.
The strong phenotypes seen in clv1; bam1 mutants can be attributed to either (1) loss of CLV1/ BAM1 signaling, the primary regulators of meristem size or (2) loss of both CLV1 and CLV2/CRN/BAM1 signaling, both of which are required to regulate meristem size. To distinguish between these mechanisms, we generated double mutants between bam1-3 and either crn-1 or clv2-7. On the basis of the molecular nature of CLV2 (lacking a kinase domain) and CRN (lacking a receptor domain), we hypothesized that loss of bam1 and either clv2 or crn should result in complete loss of any CLV2/CRN/BAM1 signaling and should result in additive or synergistic defects. However, we found that there was no effect on valve number in the double mutants compared to crn-1 or clv2-7 single mutants (Figure 7A). Furthermore, analysis of meristems from double mutants revealed that there was no difference in IM or FM size between double and single mutants (Figure 7, B and C). Characterization of gynoecium development in bam1-3; crn-1 and bam1-3; clv2-7 double mutants at early and later stages showed no difference between double and single mutants (Figure 3, N and O). These results support a model in which BAM1 is partially redundant with CLV1 but not CLV2/CRN in shoot meristems. This is further supported by recent biochemical data that shows physical interactions between BAM1 and CLV1 and not BAM1 and CLV2/CRN (Guo et al. 2010). However, these experiments do not rule out a redundant role for CLV1 and BAM1 in interacting with CLV2/CRN, although this proposed function is not supported by current biochemical interactions.
CLV3 is not the primary ligand in gynoecia
In shoot meristems, the CLV1, CLV2, CRN, and BAM1 receptors bind the CLV3 signaling peptide (reviewed in Miwa et al. 2009; Guo et al. 2010). In developing gynoecia, the CLV1, CLV2, and CRN receptors appear to restrict expression of STM in a similar manner to the CLV/CRN–WUS pathway in meristems, although the ligand in this pathway remains unknown. clv3 mutants display very strong fruit organ phenotypes and generate extremely large shoot meristems (Clark et al. 1995), suggesting that clv3 mutants produce extra carpels. To determine whether clv3 mutants produce ectopic valves as well, we looked for ectopic FIL expression in medial regions at various stages of development in clv3-3 and clv3-6 mutants. We found FIL expression only in broad arches that correspond to lateral regions at stages 7–9 (Figure 8, A and C) and never in the medial regions at any stage of development (7–12). The latter was striking, as we saw significantly enlarged medial regions and septa in both clv3-3 and clv3-6 mutants. Furthermore, when we examined clv3 mutants for changes in STM regulation, we found that STM expression in the medial regions was similar to wild-type controls, and strong expression was only observed in the fifth whorl in the center of the fruit (Figure 8, B and D). These results imply that the extra cells in the medial regions of clv3 mutants are the product of misregulation of STM and are most likely derived from extra cells in the meristem.
Figure 8 .
Expression of FIL and STM in clv3 mutants. (A and C) FIL antisense); (B and D) STM antisense); (E) FIL sense); (F) STM sense); (A) clv3-3 stage 8 (N = 18); (B) clv3-3 stage 12 (N = 14); (C) clv3-6 stage 8 (N = 7); (D) clv3-6 stage 12 (N = 8); (E) clv3-6 stage 8; and (F) clv3-6 stage 12. Bar, 50 μm.
Discussion
CLV pathway receptor mutants generate extra fruit organs via two separate mechanisms
In our analysis of CLV pathway receptor mutants, we found that extra fruit organs initiate not only from extra carpels forming in FMs, but can form from the medial regions of gynoecia younger than stage 13. Our conclusions are supported by our observations that (1) most mutations in clv1, clv2, and crn affect only fruit organ number and not the numbers of other floral organs; (2) most clv1, clv2, and crn mutants produce IMs and FMs of comparable size to wild type; (3) weak and intermediate clv1 and clv2 and crn mutants do not display phenotypes during early gynoecia development that are associated with extra carpel inception, although they still produce extra valves at maturity; and (4) all receptor mutants tested, including those with larger meristems, contain significantly more organs at maturity than at the onset of gynoecium development. These results indicate that the CLV pathway receptors function in two temporally distinct mechanisms that regulate valve number—formation of extra carpels in floral meristems and generation of ectopic valves in the medial regions of developing gynoecia.
Although the medial regions of young gynoecia do not retain the conserved structure of shoot meristems, they do express the meristem identity genes STM and REPLUMLESS (RPL) (Long et al. 1996; Roeder et al. 2003). Furthermore, medial regions maintain meristematic properties, including increased proliferation (Pautot et al. 2001) and the ability to initiate organs (Bowman et al. 1999). These data suggest that the medial regions are also competent to form new fruit organs. The process by which the medial regions obtain meristematic characteristics is poorly understood. However, it is likely that this process is similar to axillary meristem formation, which occurs via one of two ways. In the “de novo” mechanism, fully differentiated cells undergo dedifferentiation to give rise to new meristematic regions (Snow and Snow 1942; McConnell and Barton 1998), while in the “detached” hypothesis, groups of cells derived from meristems maintain an undifferentiated state after they detach from the meristem (Steeves and Sussex 1989). The fact that STM is downregulated in incipient floral organ primordia (Long et al. 1996) supports the de novo hypothesis. However, it is possible that only a few cells maintain a meristematic fate, and detecting expression in these cells is difficult. Whether the medial regions become meristematic through the de novo or the detached mechanism, it is clear that CLV1, CLV2, and CRN are required in both meristem and gynoecium development.
In IMs and FMs, the CLV pathway plays a vital role in the regulation of WUS expression and subsequently in meristem maintenance and function. However, in gynoecia, this pathway appears to be acting on a different downstream target, namely STM. The functional importance of regulating STM levels in gynoecia is supported by previous results in which the stm-2 mutant partially suppressed the strong clv1-1 fruit organ phenotype (Clark et al. 1996). We show that interactions between stm-2 and either weak or null clv1 alleles result in complete suppression of extra fruit organs (Figure 6). Although it is possible that the CLV pathway and STM perform in separate but overlapping mechanisms in gynoecia to regulate proliferation and differentiation, respectively, the changes in the STM expression pattern in clv1, clv2, and crn mutants combined with the complete suppression of ectopic valve formation in clv1-20; stm-2 and clv1-22; stm-2 mutants suggests that the CLV pathway receptors and STM act in a single pathway during fruit development.
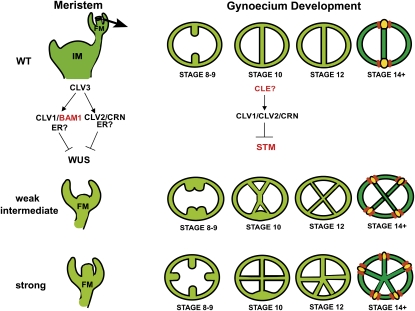
We propose that initiation of an extra valve occurs when cell proliferation in the medial regions reaches a critical threshold, regulated by STM. Thus, in weak mutants, where STM expression is minimally affected, the threshold is reached at later stages (Figure 9). Meanwhile, in mutants with a stronger effect on STM expression (Figure 5) the threshold is achieved earlier, as demonstrated by the splitting of medial regions observed in intermediate clv1, clv2, and crn mutants (Figure 3). In strong clv1 mutants that have meristem defects and misregulate STM, extra carpel formation and ectopic valve initiation occurs, producing the strongest extra organ phenotypes (Figure 9). The idea that a threshold is required to initiate ectopic organs can also explain how partial valves form. As the apical region of developing fruit is older, it has more time to undergo cell divisions than the basal region. If the threshold is not reached in the basal regions by approximately stage 13, then a partial valve forms in the apical region only.
Figure 9 .
Model for development of valves in the wild type, weak, intermediate, and strong CLV pathway mutants.
Fruit organ specification can occur independently from carpel specification
One major implication of our results is the potential direct link between CLV signaling and the transcription factor network required for fruit organ specification. STM directly regulates REPLUMLESS (RPL), a transcription factor required for specification of the replum and regulation of fruit organ patterning (Dinneny et al. 2005; Kanrar et al. 2006). Our analysis suggests that this network acts as a default patterning mechanism that is activated when undifferentiated cells are present to ensure that extra cells do not simply continue proliferating. This idea is supported by the fact that both extra carpels and ectopic valves give rise to normally patterned fruit organs.
To fully understand the mechanisms that regulate fruit organ number, it is important to identify the source of the extra cells in stages 7–9 medial regions. Although we did not observe larger FMs in clv1, clv2, and crn mutants, it is possible that just a few extra or undifferentiated cells could initiate proliferation within FMs. Although the misregulation of STM in clv1, clv2, and crn mutants suggests that additional cells are produced in the medial region through loss of CLV1 inhibition, a more definitive way to prove this would be to conduct live imaging in FMs. Specifically, monitoring the expression domains of CLV1, WUS, CLV3, and STM, as well as other markers for cell division and stem cells, would be very useful for understanding the cellular dynamics of proliferation in FMs and fruit.
Distinct roles for CLV1, CLV2, CLV3, and CRN in meristems and gynoecia
Upon the identification of CLV2 as a member of the CLV signaling pathway, the CLV1 and CLV2 receptors were proposed to function in a single complex (Kayes and Clark 1998; Jeong et al. 1999). However, the identification of CRN suggested a new hypothesis in which CLV2/CRN receptor complexes act in parallel with CLV1 to regulate WUS expression (Muller et al. 2008). Genetic evidence from interactions between crn, clv1, and clv2 mutants only supports the later hypothesis when certain clv1 alleles are utilized (Kayes and Clark 1998; Jeong et al. 1999; Muller et al. 2008). Furthermore, recent biochemical data suggest that CRN/CLV2 and CLV1/CLV1 or CLV1/BAM1 complexes can readily form, and there is also evidence that CLV1, CLV2, and CRN receptors are capable of forming a single complex (Bleckmann et al. 2010; Guo et al. 2010; Zhu et al. 2010).
Previous genetic interaction analyses interpreted carpel number (measured as valve number) as a gauge of meristem function (Kayes and Clark 1998; Jeong et al. 1999; Muller et al. 2008). As these studies only considered contributions from the meristem, we examined the effect of removing a member of each receptor pathway on extra carpel and ectopic valve formation, with the prediction that there would be a strong increase in extra carpels and ectopic valves in the case of two parallel pathways. We found that clv1; clv2 and clv1; crn double mutants containing weak or intermediate clv1 alleles resulted in additive increases in fruit and floral organ number that was due to increased meristem size (and therefore more carpels) (Figure 7) and not due to increased ectopic valve formation. These data suggest a situation in which CLV1, CLV2, and CRN utilize different dynamics in different tissues, with CLV1 and CLV2/CRN functioning in separate pathways in shoot meristems, and all three receptors working together in a linear fashion in gynoecia.
We also tested whether mutations in CLV3, the ligand of the CLV pathway, produce severe phenotypes through defects in meristems, gynoecia, or both. We found that while clv3 mutants do contain extra cells in the medial regions of developing fruit, they do not display ectopic FIL expression or expansion of STM expression, suggesting that clv3 mutants do not initiate ectopic valves. A previous study looking at the genetic interactions between stm-2 and clv3 mutants showed partial suppression of the clv3 phenotype (Clark et al. 1996). Although we cannot rule out that CLV3 functions in gynoecia at earlier stages or through an unidentified pathway, the fact that clv3 mutants generate greatly enlarged shoot meristems indicates that the extra valves in these mutants are derived from extra carpels (Clark et al. 1997). We hypothesize that another ligand(s) is acting in the CLV pathway in gynoecia, most likely a member of the CLE (CLV3/ESR, Embryonic Surrounding Region) family, of which CLV3 is a founding member (Kondo et al. 2006; Ohyama et al. 2009). Although none of the cle mutants analyzed to date have been reported to have an extra valve phenotype, several CLE genes are expressed in the medial regions in gynoecia (Jun et al. 2010). Analysis of genetic interactions and expression patterns will likely identify candidate CLE ligands in gynoecia.
BAM1 receptors are partially redundant with CLV1 in shoot meristems
Although the fruit and floral organ phenotypes in clv1; clv2 and clv1; crn double mutants containing weak or intermediate clv1 alleles were additive compared to single mutants, they were not as severe as the phenotypes of strong clv1 mutants. We predicted that this could be due to other receptors acting redundantly with CLV1, CLV2, or CRN in shoot meristems. A previous study suggests that the BAM receptors interact with CLV1 in shoot meristems (DeYoung and Clark 2008) and we tested whether this was the case in gynoecia as well. We found that the clv1-20 phenotype was extremely sensitive to the presence of BAM1, as taking away a single copy resulted in synergistic meristem and valve phenotypes, while in plants homozygous for bam1 and clv1-4 there was no significant difference in valve number between clv1-4 single and double mutants. This latter result is not consistent with previous reports, and we suspect that use of the Ler accession contributed to the additive fruit organ phenotype previously observed in clv1-4; bam1 double mutants (DeYoung and Clark 2008).
We also examined the interactions between bam1 and clv2 or crn mutants. On the basis of the predicted structures of the CLV2 and CRN receptors, we hypothesized that if BAM1 acts redundantly with either CLV2 or CRN in a complex separate from CLV1, we would see additive fruit and floral organ phenotypes. Surprisingly, we found that bam1; clv2 and bam1; crn double mutants were indistinguishable from clv2 and crn single mutants. This result would typically be interpreted as evidence that BAM1 acts in the same pathway as CLV2/CRN. However, if BAM1 were in the CLV2/CRN pathway, we would expect (1) clv1; bam1 mutants to display similar phenotypes as clv1; clv2 and clv1; crn double mutants and (2) bam1 mutants to have similar phenotypes as clv2 and crn single mutants with respect to fruit organ number. Although the interaction data between bam1 and either clv2 or crn cannot rule out the possibility that a BAM1/CLV2/CRN receptor complex exists, the fact that bam1 mutants are wild type with regard to fruit organ number and clv1; bam1 mutants display significantly stronger phenotypes compared to clv1; clv2 or clv1; crn mutants, suggests that BAM1 does not function in the CLV2/CRN pathway. This hypothesis is supported by recent biochemical evidence that BAM1 interacts readily with CLV1, but not CLV2 or CRN (Guo et al. 2010).
From these data, we propose a model in which CLV1 and/or CLV1/BAM1 complexes are the primary regulators of stem cell populations in shoot meristems, with CLV2/CRN receptor complexes playing a secondary role. Although hypomorphic, null, and kinase domain clv1 mutants do not display defects in meristem size, the interactions between bam1 and weak clv1 mutants suggest that BAM1 can perform a similar function to CLV1 in the absence of CLV1 protein. However, in strong clv1 mutants, dominant-negative interactions occur that lead to inhibition of interacting receptors (Dievart et al. 2003), including BAM1, resulting in strong phenotypes. Furthermore, recent biochemical evidence suggests that BAM1 binds CLV3 with similar affinity as CLV1 (Guo et al. 2010). This model is further supported by the fact that crn and clv2 mutant phenotypes appear to be almost entirely derived from ectopic valve formation, and that clv2 and crn mutants do not produce as strong of an induction of WUS expression as that observed for strong clv1 mutants (Schoof et al. 2000). However, we cannot rule out that other RLKs, including BAM2 or BAM3, or homologs of CRN and CLV2, may be interacting with the CRN/CLV2 pathway in the meristem, and thus masking the effects of removing CRN or CLV2.
clv1, clv2, and crn mutant phenotypes are sensitive to mutations in the ERECTA gene
Previous studies have suggested that mutations in ER enhance meristem-derived phenotypes such as fruit organ number in clv1 and clv3 mutants (Dievart et al. 2003). As the Ler accession contains a hypomorphic mutation in the ER gene (Torii et al. 1996), we suspect that this is the source of the differences in meristem size and valve number phenotypes between our Col alleles and previous results in the Ler accession. This is further supported by our evidence that an identical crn mutation in the Col accession produces significantly weaker fruit and meristem phenotypes than in the Ler background (Muller et al. 2008), and that the genetic interaction between crn-1 and er-3, both in the Col background, recapitulates the meristem and fruit organ number phenotypes seen in the Ler crn-1 mutant (Muller et al. 2008). These results are indicative of ER playing an important role in the regulation of meristem size.
Although it has been previously hypothesized that ER may also be part of the CLV receptor complex (Dievart et al. 2003), the exact nature of this role remains unclear. The fact that mutations in ER enhance phenotypes of all CLV pathway receptor mutants suggests that ER may play a role in meristem maintenance separate from the CLV pathway (Figure 9). However, differences between Ler and Col alleles appear to be more dramatic in clv2 and crn mutants than clv1 mutants, suggesting that ER acts in the CLV1/BAM1 pathway and not the CLV2/CRN pathway. Biochemical and genetic experiments looking at the physical interactions between CLV1, BAM1, CLV2, and CRN and ER will help to clarify the nature of receptor dynamics in the meristem and in fruit.
Implications for fruit development in Arabidopsis and other species
Our results, that increased proliferation in young gynoecia generates extra organs, implicate the regulation of cell division in meristems and/or gynoecia as critical for regulation of fruit organ number. We show that the CLV pathway receptors CLV1, CLV2, and CRN, previously identified because of their involvement in meristem maintenance, perform a similar function in developing gynoecia, likely through the action of an unidentified CLE ligand or ligands. Whether other components of the CLV pathway also perform fruit-specific roles remains to be addressed. Our discovery of a new role for CLV1, CLV2, and CRN in gynoecia suggests that there may be other as yet undiscovered regulators of fruit organ number. In particular, the links between the CLV pathway, STM, and the fruit organ specification pathway suggests that genes downstream of the CLV pathway in gynoecia may act in fruit organ number regulation in other species. This hypothesis is supported by evidence that a YABBY-like gene, most closely related to AtYABBY2, was identified as the fasciated quantitative trait locus in tomato, a known regulator of fruit organ number (Cong et al. 2008). AtYABBY2 is closely related to FIL and YABBY3, which are part of the Arabidopsis transcription factor network that specifies fruit organs (Dinneny et al. 2005). Identification of more components of the proposed CLV–STM network will provide links to key players in the regulation of fruit size in tomatoes and other crop plants.
Acknowledgments
We thank the Tax lab for critical discussions and Andrea Mitchell, Erica Hsiao, and Erika Starks for technical assistance. We thank Ravi Palanivelu, Karen Schumaker, Ramin Yadegari, and Daniela Zarnescu for critical discussion and comments on the manuscript. Cristina Ferrandiz and Danielle Nevarez contributed to early studies that led to these results. This work was supported by National Science Foundation (NSF) IBN-0347675. A.D. was supported by the NSF Integrative Graduate Education and Research Traineeship (DGE-0114420), NSF IOS-0922678, and by funds from the University of Arizona Department of Plant Sciences.
Literature Cited
- An H., Roussot C., Suarez-Lopez P., Corbesier L., Vincent C., et al. , 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Barton M. K., 2010. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Baucher M., El Jaziri M., Vandeputte O., 2007. From primary to secondary growth: origin and development of the vascular system. J. Exp. Bot. 58: 3485–3501 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Durbak A., Wierzba M., Schmitz R. J., Aguirre A., et al. , 2010. Intragenic suppression of a trafficking-defective brassinosteroid receptor mutant in Arabidopsis. Genetics 185: 1283–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C. A. M., Simon R., 2010. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to plasma membrane. Plant Physiol. 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Baum S. F., Eshed Y., Putterill J., Alvarez J., 1999. Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45: 155–205 [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M., Simon R., 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Clark S. E., Running M. P., Meyerowitz E. M., 1993. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Clark S. E., Running M. P., Meyerowitz E. M., 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067 [Google Scholar]
- Clark S. E., Jacobsen S. E., Levin J. Z., Meyerowitz E. M., 1996. The CLAVATA and SHOOTMERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Clark S. E., Williams R. W., Meyerowitz E. M., 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cong B., Barrero L. S., Tanksley S. D., 2008. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40: 800–804 [DOI] [PubMed] [Google Scholar]
- DeYoung B. J., Bickle K. L., Schrage K. J., Muskett P., Patel K., et al. , 2006. The CLAVATA1-related BAM1, BAM2, and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45: 1–16 [DOI] [PubMed] [Google Scholar]
- DeYoung B., Clark S., 2008. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dievart A., Dalal M., Tax F. E., Lacey A. D., Huttly A., et al. , 2003. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J. R., Weigel D., Yanofsky M. F., 2005. A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132: 4687–4696 [DOI] [PubMed] [Google Scholar]
- Endrizzi K., Moussian B., Haecker A., Levin J. Z., Laux T., 1996. The SHOOTMERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristem and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10: 967–979 [DOI] [PubMed] [Google Scholar]
- Ferrandiz C., Pelaz S., Yanofsky M. F., 1999. Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Forstheofel N. R., Wu Y., Schulz B., Bennett M. J., Feldmann K., 1992. T-DNA insertion mutagenesis in Arabidopsis: prospects and perspectives. Aust. J. Plant Physiol. 10: 353–366 [Google Scholar]
- Franks R. G., Wang C., Levin J. Z., Liu Z., 2002. SUESS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129: 253–263 [DOI] [PubMed] [Google Scholar]
- Girin T., Sorefan K., Ostergaard L., 2009. Meristematic sculpting in fruit development. J. Exp. Bot. 60: 1493–1502 [DOI] [PubMed] [Google Scholar]
- Gou X., He K., Yang H., Yuan T., Lin H., et al. , 2010. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss-Hardt R., Lenhard M., Laux T., 2002. WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Han L, Hymes M, Denver R, Clark S. E, 2010. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63: 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A. E., Clark S. E., 1999. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A. H., Meng L., Sharma V. K., et al. , 2010. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar S., Onguka O., Smith H. M. S., 2006. Arabidopsis inflorescence architecture requires the activities of KNOX_BELL homeodomain heterodimers. Planta 224: 1163–1173 [DOI] [PubMed] [Google Scholar]
- Kayes J. M., Clark S. E., 1998. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., et al. , 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K. F., Berger J., Jurgens G., 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Leyser H. M. O., Furner I. J., 1992. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116: 397–403 [Google Scholar]
- Long J. A., Moan E. I., Medford J. I., Barton M. K., 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lunde C., Hake S., 2009. The interaction of knotted1 and thick tassel dwarf1 in vegetative and reproductive meristems of maize. Genetics 181: 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F. X., Schoof H., Haecker A., Lenhard M., Jurgens G., et al. , 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- McConnell J. R., Barton M. K., 1998. Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McKelvie A. D., 1962. Studies on induction of mutation in Arabidopsis thaliana (L.) Heynh. Radiat. Bot. 1: 233–241 [Google Scholar]
- Medford J. I., Behringer F. J., Callos J. D., Feldmann K. A., 1992. Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell 4: 631–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Tamaki T., Fukuda H., Sawa S., 2009. Evolution of CLE signaling. Plant Signal. Behav. 4: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R., Bleckmann A., Simon R., 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y., 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayahsi Y., 2009. A glycopeptides regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Pautot V., Dockx J., Harnant O., Kronenberger J., Grandjean O., et al. , 2001. KNAT2: evidence for a link between Knotted-like genes and carpel development. Plant Cell 13: 1719–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J. A., Simon E. J., Katzman R. B., deGuzman B. M., Yu L. P., et al. , 1998. Identifying novel regulators of shoot meristem development. J. Plant Res. 111: 307–313 [Google Scholar]
- Reddy G. V., Meyerowitz E. M., 2005. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310: 663–667 [DOI] [PubMed] [Google Scholar]
- Roeder A. H. K., Ferrandiz C., Yanofsky M. F., 2003. The role of the REPLUMLESS homeodomain protein in patterning of the Arabidopsis fruit. Curr. Biol. 13: 1630–1636 [DOI] [PubMed] [Google Scholar]
- Satina S., Blakeslee A. F., 1942. Periclinal chimeras in Datura stramonium in relation to the development of the leaf and flower. Am. J. Bot. 28: 862–871 [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Klaus F., Mayer X., et al. , 2000. The stem cell population of Arabidopsis shoot meristems are maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Siegfried K. R., Eshed Y., Baum S. F., Ostuga D., Drews G. N., et al. , 1999. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M., 1990. Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow M., Snow R., 1942. The determination of axillary buds. New Phytol. 41: 13–22 [Google Scholar]
- Steeves T. A., Sussex I. M., 1989. Patterns in Plant Development. Cambridge University Press, Cambridge [Google Scholar]
- Stewart R. N., Meyer F. G., Dermen H., 1972. Camelia + ‘Daisy Eagleson’, a graft chimera of Camellia sasaqua and C. japonica. Am. J. Bot. 59: 515–524 [Google Scholar]
- Torii K. U., Mitsukawa N., Oosumi T., Matsuura Y., Yokoyama R., et al. , 1996. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Ellendorff U., Kemp B., Mansfield J. W., Forsyth A., et al. , 2008. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., Takahashi T., Kato A., Torri K. U., Komeda Y., 1998. The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J. 15: 301–310 [DOI] [PubMed] [Google Scholar]
- Zhao C., Craig J. C., Petzold H. E., Dickerman A. W., Beers E. P., 2005. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Peng P., Schmitz R. J., Decker A. D., Tax F. E., et al. , 2002. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 130: 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang Y., Li R., Song X., Wang Q., et al. , 2010. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 61: 223–233 [DOI] [PubMed] [Google Scholar]