Abstract
Background selection, the effects of the continual removal of deleterious mutations by natural selection on variability at linked sites, is potentially a major determinant of DNA sequence variability. However, the joint effects of background selection and genetic recombination on the shape of the neutral gene genealogy have proved hard to study analytically. The only existing formula concerns the mean coalescent time for a pair of alleles, making it difficult to assess the importance of background selection from genome-wide data on sequence polymorphism. Here we develop a structured coalescent model of background selection with recombination and implement it in a computer program that efficiently generates neutral gene genealogies for an arbitrary sample size. We check the validity of the structured coalescent model against forward-in-time simulations and show that it accurately captures the effects of background selection. The model produces more accurate predictions of the mean coalescent time than the existing formula and supports the conclusion that the effect of background selection is greater in the interior of a deleterious region than at its boundaries. The level of linkage disequilibrium between sites is elevated by background selection, to an extent that is well summarized by a change in effective population size. The structured coalescent model is readily extendable to more realistic situations and should prove useful for analyzing genome-wide polymorphism data.
GENOME-WIDE surveys of DNA sequence variability within populations in a variety of species are providing evidence that the amount and pattern of neutral or nearly neutral variability in given regions of the genome are affected by selection at sites that are linked to those under observation (Wright and Andolfatto 2008; Charlesworth et al. 2009; McVicker et al. 2009; Sella et al. 2009; Cutter and Choi 2010). While these effects are strongest in genomic regions or genomes with low frequencies of genetic recombination, where all sites are closely linked (Betancourt et al. 2009; Kaiser and Charlesworth 2009; Seger et al. 2010), they can also be detected in regions with “normal” levels of genetic recombination (Andolfatto 2007; Shapiro et al. 2007; Haddrill et al. 2011). In addition, the extent of adaptation at the sequence level, as measured by codon usage and the level of selective constraint on nonsynonymous sites, is reduced when recombination is infrequent (Betancourt and Presgraves 2002; Hey and Kliman 2002; Presgraves 2005; Haddrill et al. 2007; Larracuente et al. 2008; Betancourt et al. 2009).
These observations, which date back to the early findings in Drosophila that silent site variability at a locus is correlated with the local rate of recombination for the region in which it is situated (Aguadé et al. 1989; Begun and Aquadro 1992), suggest strongly that selection at sites genetically linked to those under observation is influencing the evolutionary process at the latter. Two main processes have been proposed as the source of this influence. The first is hitchhiking by positively selected mutations (Maynard Smith and Haigh 1974) or “selective sweeps” (Berry et al. 1991). Evidence in favor of this interpretation has been compiled by Stephan (1995) and Andolfatto (2007), among others (reviewed by Sella et al. 2009). The second involves hitchhiking effects caused by the continual introduction of new, deleterious variants by mutation at sites across the genome and their elimination by selection, as first discussed by Fisher (1930). Weakly selected or neutral variants on a haplotype that carries a closely linked mutation, which is sufficiently strongly selected against that it is virtually certain to be eliminated from the population, will also be removed from the population. This results in a reduction in the mean coalescent time for neutral variants and in the efficacy of selection on weakly selected variants; this process has become known as “background selection” (Charlesworth et al. 1993).
Several analytical and simulation studies of the effects of background selection on neutral variability (Charlesworth et al. 1993, 1995; Hudson and Kaplan 1994, 1995; Nordborg et al. 1996; Fu 1997; Neuhauser and Krone 1997; Santiago and Caballero 1998; Gordo et al. 2002; Williamson and Orive 2002; Zeng et al. 2006; Wakeley 2008b; O’Fallon et al. 2010) and on the properties of weakly selected variants (Charlesworth 1994; Stephan et al. 1999; Zeng and Charlesworth 2010) have been conducted. The effect of background selection on the mean coalescent time for a pair of alleles, caused by mutations at a large number of sites subject to sufficiently strong selection that the allele frequencies behave approximately deterministically, is especially well understood (Hudson and Kaplan 1995; Nordborg et al. 1996). The relevant formula has been used to predict patterns of variability as a function of local recombination rate in Drosophila (Hudson and Kaplan 1995; Charlesworth 1996), humans (Cai et al. 2009; McVicker et al. 2009), and Caenorhaditis elegans (Cutter and Choi 2010; Rockman et al. 2010). In addition, there is increasing evidence that background selection may affect patterns of variation and the efficacy of selection in and around a single gene, although the effects are small and detectable only with genome-wide data (Loewe and Charlesworth 2007; McVicker et al. 2009; Hammer et al. 2010).
In addition to its effect on the level of neutral variability, as determined by the mean coalescent time for a pair of alleles, background selection can also distort the shape of the neutral gene genealogy, in the direction of increasing the length of external branches relative to the rest of the tree, resulting in an excess of rare variants relative to standard neutral expectation, especially when selection against deleterious mutations is relatively weak (Charlesworth et al. 1993, 1995; Fu 1997; Santiago and Caballero 1998; Gordo et al. 2002; Williamson and Orive 2002; Zeng et al. 2006). This effect has proved hard to study analytically, and most results have been obtained either by coalescent models that assume no recombination (Charlesworth et al. 1995; Fu 1997; Neuhauser and Krone 1997; Gordo et al. 2002; Zeng et al. 2006; Wakeley 2008b; O’Fallon et al. 2010) or by forward-in-time computer simulations (Williamson and Orive 2002; Kaiser and Charlesworth 2009; Zeng and Charlesworth 2010).
Given the evidence from genomic data that background selection against nonsynonymous variants in coding sequences may have significant local effects, it is important to develop efficient methods to predict its effects on both levels of variability and patterns of departure of allele frequency spectra from neutral expectations. To initiate progress toward this end, we have developed a structured coalescent model of background selection that includes recombination. The purpose of this article is to describe this model and its implementation in a computer program that generates numerical predictions of several biologically informative genealogical statistics, as well as to check the validity of the method against the results of forward simulations. The results are at present quite limited in their range, but the approach should be readily extendable to more realistic situations.
Theory
A background selection model
In this section, we formulate a background selection model forward in time, under the assumption that the population is at mutation–selection equilibrium, so that genotype frequencies at sites under selection are constant over generations. The results are used in the next section to construct a structured coalescent model that takes into account the joint effects of background selection and recombination on local gene genealogies. Consider a haploid population of constant size N. We focus on a “deleterious region” where the total rate of mutation to deleterious alleles follows a Poisson distribution with a mean of U mutations per generation per individual. The mutation rate is uniform across the deleterious region, and back mutation does not occur. We represent the deleterious region by the interval [0, 1]. A haplotype that carries i deleterious mutations is referred to as an i-haplotype. We further assume that all mutations in the deleterious region have the same fitness effect, s (0 < s < 1). The selected sites in a haplotype interact multiplicatively, so that the fitness of an i-haplotype is (1 – s)i relative to wild type.
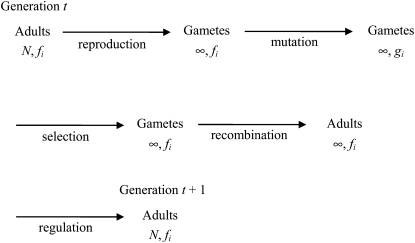
We assume a discrete-generation model as shown in Figure 1. The adults in generation t produce infinitely many gametes, without fertility differences. Mutation, selection, and recombination then operate sequentially on the resulting infinite population, causing changes in the frequency of i-haplotypes, which are denoted by fi and gi at different stages. Note that
| 1 |
Under the assumption that the system is at equilibrium, fi and gi remain constant over generations. Genetic drift operates through population size regulation, which reduces the population size to N at random with respect to genotype. We assume N to be sufficiently large that the distribution of haplotype frequencies with respect to selected sites remains approximately unchanged by the population size regulation. Consequently, drift affects only neutral sites within the region in question.
Figure 1 .
The life cycle assumed in the analysis. Shown are the life-history events between the finite adult populations of two consecutive generations. The frequencies of i-haplotypes at different stages are denoted by fi and gi.
The order of the evolutionary forces in the life cycle is to some extent arbitrary and is chosen mainly for theoretical convenience. However, if all the evolutionary forces are weak, so that second-order terms in their effects can be neglected, their effects on haplotype frequencies are approximately additive (Ewens 2004, Chaps. 4 and 5), and the dynamics of the model are approximately independent of the order of the forces.
A key feature in the life cycle is that recombination does not alter the distribution of haplotype frequencies (compare the two populations immediately before and after the recombination step in Figure 1). A derivation is given below, but heuristically this result follows from previous analyses of selection models with an infinite population size and multiplicative fitnesses (Kimura and Maruyama 1966; Charlesworth 1990; Shnol and Kondrashov 1993; Johnson 1999). These show that the genotypic composition of an infinite population at equilibrium between selection and mutation is independent of the presence or absence of recombination, which should therefore have no effect on fi.
To establish the above result, we first note that previous work with zero recombination has shown that
| 2 |
where λf = U(1 – s)/s and λg = U/s are the mean numbers of mutations that a haplotype carries before and after the mutation step in the life cycle (Haigh 1978; Gordo et al. 2002). In other words, fi and gi follow Poisson distributions. Note that mutation increases the average number of mutations that an individual carries from λf to λg. As a result, the mean fitness of the population is lowered from exp{−U(1 – s)} before mutation to exp(−U) after mutation. On the other hand, selection changes the mean number of mutations back to λf by allowing fitter individuals to contribute more offspring to the gene pool of the next generation, thus restoring the mean fitness to the premutation level.
Recombination is formulated as follows. To produce a new haplotype in the postrecombination population, two haplotypes are sampled randomly with replacement from the postselection population. With probability 1 – R, no recombination occurs, in which case one of the two chosen haplotypes is randomly selected to be present in the postrecombination population. If recombination occurs (with probability R), it is assumed to involve a single crossover event whose breakpoint falls randomly within the interval [0, 1]. Two recombinants are produced and one of them is randomly selected to be retained in the postrecombination population. The recombination rate parameter R is therefore equivalent to the genetic map length of the focal genomic region, whose size is so small that the chance of double crossover events is negligible.
Suppose that an i-haplotype and a j-haplotype are involved in a recombination event with breakpoint x (0 < x < 1). To construct the recombinants, it is necessary to determine the numbers of mutations on both sides of the breakpoint. We assume that, for a randomly chosen i-haplotype and a given recombination breakpoint x, the number of mutations to the left of the breakpoint, denoted by Li, follows a binomial distribution with index i and parameter x,
| 3 |
where 0 ≤ l ≤ i.
Equation 3 effectively assumes that the mutations carried by an i-haplotype are uniformly distributed on the interval [0, 1]. This is reasonable, because the model is spatially homogeneous in the sense that the mutation rate to deleterious alleles is uniform and each deleterious mutation has the same fitness effect regardless of its location.
Under the above recombination model, the frequency of i-haplotypes in the postrecombination population, denoted by fi*, is
| 4 |
where
| 5 |
(see the Appendix for a derivation).
Equations 4 and 5 can be understood by noting that an i-haplotype in the postrecombination population is either a nonrecombinant or a recombinant with probabilities 1 – R and R, respectively. If it is a nonrecombinant, it must be a descendant of an i-haplotype in the postselection population. If it is a recombinant, the chance that the breakpoint falls between x and x + dx is dx. Given x, l of the i mutations, which are to the left of x, are inherited from the first parent with j (≥l) mutations, and the remaining i – l to the right of x come from the other parent with k (≥i – l) mutations.
To understand the spatial distribution of mutations in an i-haplotype taken randomly from the postrecombination population, we note that
| 6 |
(see the Appendix for a derivation).
Equation 6 shows that, among the recombinants with i mutations, the proportion that are formed by recombination events with a breakpoint between x and x + dx is dx (i.e., it is a uniform distribution). Given x, the proportion that l of the i mutations are found in the region [0, x] is B(l | i, x). These properties are the same as those leading to Equation 3. Hence the mutations carried by a recombinant are uniformly distributed on the interval [0, 1]. Because the same must be true for the mutations in a nonrecombinant, we can conclude that the uniformity assumption with respect to the spatial distribution of mutations is preserved in the postrecombination population.
Equations 4–6 show that recombination in our model does not alter the distribution of haplotype frequencies, as expected from the heuristic arguments given above. However, it should be noted that the derivation relies on the assumption of multiplicative fitness effects, in an infinite population at mutation–selection equilibrium. The result is untrue under models with synergistic interactions between mutations at different sites (Kimura and Maruyama 1966; Charlesworth 1990; Shnol and Kondrashov 1993).
A structured coalescent model
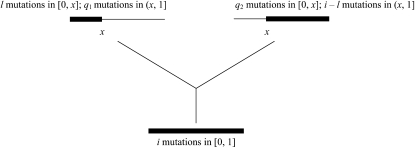
Suppose that we sample a random individual from the finite-size adult population in generation t + 1 (the bottom population in Figure 1). The probability that this haplotype carries i mutations is fi. To construct the coalescent model, we trace the ancestry of this haplotype step by step according to the life cycle in Figure 1. First, we note that the focal haplotype must have been present in the postrecombination population. A haplotype in the postrecombination population can be either a nonrecombinant or a recombinant, with probabilities 1 – R and R, respectively. If the focal haplotype is a nonrecombinant, then it must have been present in the postselection population as an i-haplotype. If the focal haplotype is a recombinant, it contains genetic material from two different individuals in the postselection population. We need to reconstruct the two parental haplotypes, so that we can trace the history of ancestry farther backward. From Equation 6, we can deduce that, for a recombinant, the breakpoint can be determined by drawing x from a uniform distribution on [0, 1] and that the number of mutations inherited from the first parental haplotype in [0, x] can be determined by drawing l from a binomial distribution with index i and parameter x and the remaining i – l mutations are inherited from (x, 1] in the other parental haplotype. These segments are marked by the thick lines in Figure 2 and are referred to as “real” segments because they are directly inherited by the recombinant under investigation. On the other hand, both parental haplotypes contain “imaginary” segments that are nonancestral to those in the sample (thin lines in Figure 2). It is, however, necessary to keep track of the numbers of mutations in both the real and the imaginary segments because, as shown below, they jointly determine the rate that a haplotype moves between different genetic backgrounds and the rate that two haplotypes coalesce.
Figure 2 .
Reconstructing the parental haplotypes in the postselection population that produce the recombinant descendant in the postrecombination population. The following steps are used in the reconstruction. First, the recombination breakpoint x is determined by sampling from a uniform distribution on [0, 1]. Second, we sample l from a binomial distribution with index i and parameter x. These l mutations are assigned to the interval [0, x] of the left-hand parent, and the remaining i – l mutations are assigned to (x, 1] of the other parent. Third, to fill the imaginary parts of the two parental haplotypes (indicated by the thin lines), q1 and q2 are drawn from Poisson distributions with means (1 – x)λf and xλf, respectively.
Consider the first parental haplotype in Figure 2, which has an imaginary part to the right of breakpoint x. Because this haplotype is in the postselection population, using the fact that the spatial distribution of mutations is uniform, it follows that the number of mutations in the imaginary segment follows a Poisson distribution with mean (1 – x)λf. Similarly, the number of mutations in the imaginary segment in the second parental haplotype follows a Poisson distribution with mean xλf. Thus, by sampling from these Poisson distributions, we can determine the total number of mutations in these haplotypes (Figure 2).
We can see from Figure 2 that, to construct the parental haplotypes of a recombinant, we need to assign mutations to different regions. In other words, a recombination event brings location information to the haplotypes in the structured coalescent model. To manage this information, we define the genetic background of a haplotype, , as
| 7 |
In Equation 7, x = (x0, x1, . . . , xB) and y = (y1, y2, . . . , yB), with 0 = x0 < x1 < . . . < xB = 1 and yb ≥ 0. The xb (0 < b < B) denote the breakpoints caused by recombination events that generated this haplotype, and the yb denote the numbers of mutations in (xb–1, xb] (1 ≤ b ≤ B). We further define the size of a genetic background as the total number of mutations carried by the focal haplotype:
| 8 |
For example, in Figure 2, the genetic background of the sampled haplotype is = {x = (0, 1), y = (i)}, whereas that of the left-hand parental haplotype is = {x = (0, x, 1), y = (l, q1)}.
The above steps enable us to reconstruct the parental haplotypes in the postselection population that gave birth to the haplotypes under investigation. To go farther backward, we note that any haplotypes that exist in the postselection population must have survived selection and therefore must have been present in the postmutation population. Suppose that we have an i-haplotype in the postmutation population. This haplotype may have received new mutations. Because the mutational process is irreversible, the parent of the i-haplotype necessarily has no more than i mutations. Specifically, the probability that the focal haplotype has a parent with j (≤i) mutations is
| 9 |
(see the Appendix for a derivation). Therefore, j can be determined by sampling from a binomial distribution with index i and parameter 1 – s. Once j is specified, the parental haplotype can be constructed by randomly purging i – j mutations from the i-haplotype.
The methods described so far can determine the haplotypes of the postreproduction population that are ancestors of one of the extant haplotypes in our sample. Repeating these procedures for other haplotypes in the sample, we can obtain a set of haplotypes in the postreproduction population that are ancestral to the individuals in our sample. The next task is to determine the probability that some of these postreproduction haplotypes were born to the same parents in the previous generation.
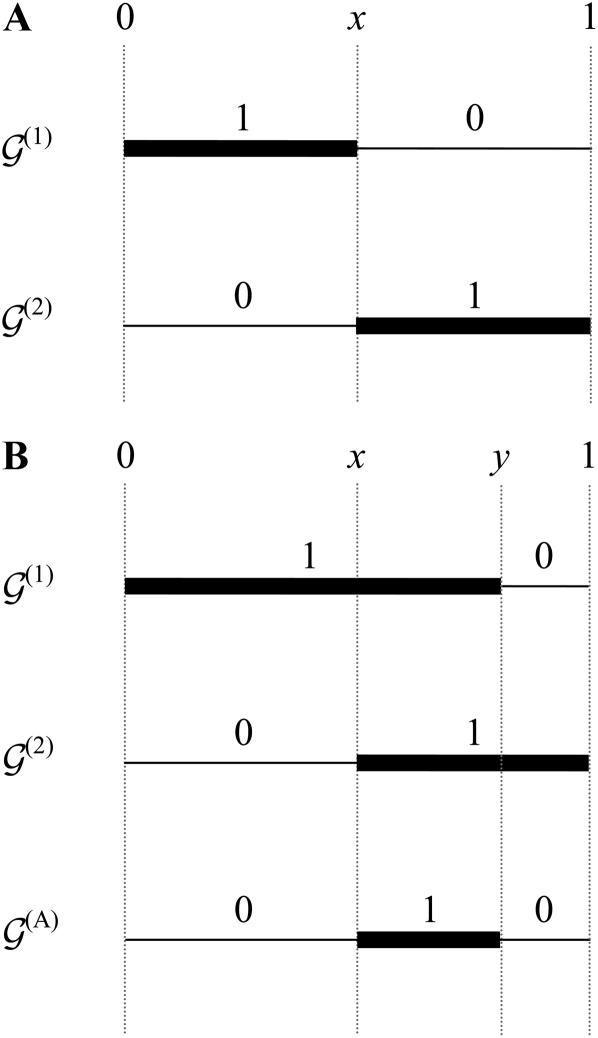
Suppose that we have two haplotypes in the postreproduction population whose genetic backgrounds are denoted by (1) and (2), respectively. Because reproduction and mutation happen in two separate steps in the life cycle, and the two focal haplotypes are in the population before mutation takes place, if |(1)| ≠ |(2)|, it is impossible for the two haplotypes to have a common parent in the previous generation. When |(1)| = |(2)|, there are two possible situations. First, the two focal haplotypes may have incompatible genetic backgrounds and therefore cannot share a common parent in the previous generation. An example is illustrated in Figure 3A. The first haplotype has one mutation in (0, x]. Consequently its parent must also have had one mutation in this region. However, this is incompatible with the fact that the second haplotype does not carry any mutation in (0, x]. In contrast, the two haplotypes with genetic backgrounds (1) and (2) depicted in Figure 3B may coalesce into the same parent with genetic background (A). This is because a mutation is located somewhere in (0, y] in the first haplotype, whereas none exists in (0, x] in the second haplotype (x < y). Therefore, if the two haplotypes share the same parent, the parental haplotype cannot have any mutation in (0, x]. This confines the mutation in the first haplotype to (x, y], because it must have inherited the region (0, x] from its parent. Applying similar arguments to (y, 1], we conclude that the parental haplotype cannot have any mutation in this region, and this requirement restricts the mutation in the second haplotype to (x, y]. Thus, we finish the construction of the potential location of the mutation in the parental haplotype, denoted by (A).
Figure 3 .
Examples of incompatible and compatible genetic backgrounds. We assume that two haplotypes have been taken from the postreproduction population; their genetic backgrounds are denoted by (1) and (2), respectively. The thick and thin lines in a haplotype indicate regions with and without deleterious mutations, respectively. The genetic backgrounds are (1) = {x = (0, x, 1), y = (1, 0)} and (2) = {x = (0, x, 1), y = (0, 1)} in A and (1) = {x = (0, y, 1), y = (1, 0)}, (2) = {x = (0, x, 1), y = (0, 1)}, and (A) = {x = (0, x, y, 1), y = (0, 1, 0)} in B. As explained in the text, (1) and (2) in A are incompatible, and consequently, the two haplotypes cannot be born to the same parent in the previous generation. In contrast, (1) and (2) in B are compatible. The probability that the two descendant haplotypes coalesce into a common parent is given by Equation 11, where (A) is the genetic background of the parent.
More generally, Figure 3 illustrates the fact that, for the postreproduction population, coalescent events can happen only among haplotypes with compatible genetic backgrounds. For two haplotypes with genetic backgrounds (1) and (2), the computer algorithm given in supporting information, File S1 can simultaneously determine whether they are compatible and, if they are, the genetic background (A) of their parental haplotype in the previous generation.
To calculate the coalescent probability, we need to know the number of individuals with a given genetic background in the finite adult population of the previous generation. Making use of the fact that the model is spatially homogeneous, we can extend the argument that leads to Equation 3 and assume that, among the i-haplotypes, the proportion with a given genetic background follows a multinomial distribution
| 10 |
where I() is a function that takes the value of one if the condition in the parentheses is true and the value of zero otherwise.
Suppose that we have two haplotypes in the postreproduction population, with genetic backgrounds (1) and (2). Equation 10 implies that, in the adult population of the previous generation, the number of individuals with genetic background (z) is NfiPi((z)), and the probability that a particular one of these is the parent of the zth descendant haplotype is 1/[NfiPi((z))] (z = 1 or 2). For a coalescent event to occur, the two sets of potential parents must have overlapped, and one of the individuals in the intersection must have given birth to the two descendant haplotypes in question. The situations where the two parental sets do or do not overlap correspond to the cases where the two descendants have compatible or incompatible genetic backgrounds (e.g., Figure 3). The overlapping region between the two parental sets, denoted by (A), can be found by the computer algorithm given in File S1. Using Equation 10, we note that the number of individuals in the intersection in the previous generation is NfiPi((A)). Hence, the probability that the two descendant haplotypes were born to the same parent in the previous generation is
| 11 |
Note that, for two incompatible genetic backgrounds, Pi((A)) = 0 and thus PCA = 0 (e.g., Figure 3A).
This calculation follows the spirit of the work of Hudson and Kaplan (1994), in the sense that the deleterious mutations are not pinpointed to specific sites in the deleterious region. Rather, the mutations are merely confined to segments in the deleterious region (i.e., genetic backgrounds), so that haplotypes with the same genetic background can be considered together, enabling the construction of coalescent processes. However, the model of Hudson and Kaplan (1994) considered only the effect of background selection on a pair of alleles at a neutral site, linked to a nonrecombining deleterious region. Here we establish the spatial distribution of deleterious mutations and introduce , so that all the neutral sites in the deleterious region can be considered simultaneously for an arbitrary sample size.
It is, however, difficult to calculate the probability of coalescent events that involve more than two individuals. Implementing a simulation algorithm that exactly follows the dynamics described above is, therefore, not straightforward. To solve the problem, we resort to the following well-established approximation techniques.
Continuous-time approximations
We approximate the processes using the standard rescaling techniques that underlie the coalescent framework (Kingman 1982; Hudson 1990). We assume that U, s, R, and 1/N are so small that their second-order terms can be neglected (i.e., the evolutionary forces are weak). Under this assumption, the possibility that two or more of the three possible events (i.e., recombination, mutation, and coalescence) occur in the same generation is in the order of 1/N2 and can be ignored. As a result, recombination, mutation, and coalescence can be regarded as three independent Poisson processes. We define three scaled parameters: θ = NU, γ = Ns, and ρ = NR. As in the neutral coalescent, time is measured in units of N generations. Going backward in time, recombination splits a haplotype into two parental haplotypes at rate ρ. For mutation, we note from Equation 9 that
| 12 |
where O(s2) represents terms of the second order in s. Therefore, the next mutation event arrives at an i-haplotype at rate iγ and changes it to an (i − 1)-haplotype by randomly removing one of the i mutations. Finally, as in the standard neutral coalescent, we assume that a coalescent event involves only two haplotypes. Under the rescaling, the rate at which two compatible haplotypes coalesce into a common parent is NPCA (see Equation 11).
Suppose that we have a sample of n haplotypes with genetic backgrounds (1), (2), . . . , (n). The total scaled rates for recombination and mutation events are rr = nρ and rm = , respectively. For the total coalescent rate, we need to examine all the n(n – 1)/2 pairs to identify those that involve two haplotypes with compatible genetic backgrounds, so that a NPCA value can be calculated for each of these compatible pairs. The total coalescent rate, rc, is given by the sum of the NPCA values. Then, the scaled time to the next event follows an exponential distribution with rate rt = rr + rm + rc. Given that an event has occurred, the probability that it is due to a particular process can be calculated by dividing the rate of the process of interest by rt. For instance, the chance that the first haplotype undergoes a mutational change conditional on an event occurring is γ|(1)|/rt. After randomly removing one mutation from this individual, we recalculate the event rates to determine the time of the next event. This process is repeated until the first time when only one haplotype remains. The resulting relationship between the haplotypes in the whole history of ancestry defines an ancestral recombination graph (ARG). Local gene genealogies at different neutral sites in the deleterious region can be extracted from the ARG using standard methods (Hudson 1990).
Materials and Methods
Coalescent simulations
We have written a computer program that implements the structured coalescent model with the continuous-time approximations outlined. The algorithm uses the neutral coalescent algorithm implemented in the program ms (Hudson 2002). Our program requires four parameters: n, θ, γ, and ρ. To set up the initial haplotypes, n random numbers are drawn from a Poisson distribution with mean λ = U/s (i.e., we assume λ = λg ≈ λf, which is a good approximation under the continuous-time model). Because these haplotypes are taken randomly from an equilibrium population, by using the arguments leading to Equations 3 and 10, we can assume that the mutations are uniformly distributed on these haplotypes [i.e., (z) has the form {x = (0, 1), y = (|(z)|)} for z = 1, . . . , n]. To reconstruct the coalescent history of a sample, for each haplotype in the ARG, the program keeps track of its genetic background and the ancestral information about the extant haplotypes in the sample it contains (e.g., Figure 2). For each set of parameter values presented in the Results section, 105 simulation replicates were performed.
Forward-in-time simulations
To check whether the structured coalescent model provides good approximations, we implemented a forward-in-time simulation algorithm that follows the life cycle given in Figure 1. A haploid Wright–Fisher population of constant size N is simulated. Each site in a haplotype has two states, A0 and A1, representing the wild type and the mutant type, respectively. In each generation, the number of new mutations experienced by a haplotype is determined by drawing a random number from a Poisson distribution with mean U. A haplotype with i mutations has fitness (1 – s)i. To produce a haplotype in the next generation, we first randomly sample two haplotypes from the current generation, with the probability of sampling a particular haplotype being proportional to its fitness. With probability R, the parental haplotypes undergo a recombination event. The location of the breakpoint is determined by drawing from a uniform distribution on [0, 1]. Two recombinants are constructed and one of the two is randomly chosen for retention. These steps are repeated N times to generate the N individuals in the new generation.
Each replicate of the simulation starts from a mutation-free population. This population is allowed to evolve for 20N generations so that statistical equilibrium is achieved. Random samples are then taken every 10N generations. To compare with the results obtained from the coalescent simulations, the gene genealogies at five evenly spaced sites in the simulated region are recorded (i.e., at X1 = 0, X2 = 0.25, X3 = 0.5, X4 = 0.75, and X5 = 1). The forward simulation algorithm is much more computationally demanding than the coalescent algorithm, especially when N is large. Unless stated otherwise, we used an N value of 5000 in the forward simulations and obtained 2000 random samples for each set of parameter values.
Statistics of interest
We focus on four genealogy-based statistics: T2, ηt, ζe, and rxy. T2 refers to the time to the most recent common ancestor of a sample of size 2 at a given nucleotide site. Assuming neutral evolution and measuring time in units of N generations (the same scaling is applied to the other time-related statistics of interest), the expectation of T2 is E(T2(neu)) = 1 (Wakeley 2008a, p. 76). Under background selection and recombination, previous analyses have obtained
| 13 |
where u is the mutation rate to deleterious alleles at a nucleotide site and rw is the recombination rate between the wth selected site and the focal neutral site (Hudson and Kaplan 1995; Nordborg et al. 1996). In the absence of recombination (i.e. rw ≡ 0), Equation 13 reduces to exp(−U/s) (Charlesworth et al. 1993; Hudson and Kaplan 1994). Note that E(T2(bgs))N is the expected number of generations needed for two randomly sampled alleles to coalesce into a common ancestor. Hence, an effective population size under background selection can be defined as Ne(T2) = E(T2(bgs))N.
T2 is closely related to the widely used measure of DNA sequence variability, π, the nucleotide site diversity, which is defined as the probability that two randomly sampled haplotypes have different variants at the nucleotide site under investigation (Tajima 1983). Under the infinite-sites model of mutation (Kimura 1969), π is determined by T2 through the equation π = 2Nu(neu)E(T2), where u(neu) is the neutral mutation rate per site (Tajima 1983). Thus, we can define a measure of the diversity-reducing effect of background selection as
| 14 |
Note that E[B(T2)] = E(T2(bgs)) and Ne(T2) = E[B(T2)]N.
The second statistic is ηt, the total length of all the branches in the genealogy of a sample of size n at the focal nucleotide site. Under the standard neutral coalescent model,
| 15 |
(Wakeley 2008a, p. 76). ηt is related to S, the number of segregating sites observed in the sample, whose expectation is given by E(S) = Nu(neu)E(ηt) under the infinite-sites model (Watterson 1975; Hudson 1990). Defining an = 1 + 1/2 + . . . + 1/(n – 1), ηt is also related to the alternative measure of sequence variability, θW, given by θW = S/an, whose expected value is Nu(neu)E(ηt)/an (Watterson 1975). Hence we can define a second measure of the diversity-reducing effect of background selection as
| 16 |
Third, we look at ζe, the proportion of external branches (i.e., branches leading to extant haplotypes in the sample) in the genealogy at the focal site. Specifically, ζe = ηe/ηt, where ηe is the total length of all the external branches. The neutral expectation of ηe is E(ηe(neu)) = 2 (Fu and Li 1993). However, the moments of ζe are unknown, so that we obtained the neutral expectation of ζe, denoted by E(ζe(neu)), via coalescent simulation. Because mutations on an external branch can be inherited only by the haplotype descending from this branch, these mutations must be at low frequencies in the sample. In particular, with the infinite-sites assumption, mutations on external branches lead to singletons (i.e., segregating sites where the mutant variant is present in only one individual). Let Se be the number of singletons. The proportion of segregating sites that are singletons is Se/S = (Nu(neu)ηe)/(Nu(neu)ηt) = ηe/ηt = ζe. Hence, ζe is a major determinant of the relative abundance of low-frequency variants in the sample. We therefore define a third relative diversity measure as
| 17 |
Note that, under neutrality with a stable population size, E[B(T2)] = E[B(ηt)] = E[B(ζe)] = 1. However, T2, ηt, and ζe are known to respond differently to the influence of background selection (Charlesworth et al. 1993, 1995; Fu 1997; Gordo et al. 2002; Williamson and Orive 2002; Zeng et al. 2006; Kaiser and Charlesworth 2009; Seger et al. 2010). This is the basis of using tests such as Tajima’s D (Tajima 1989) and Fu and Li’s D (Fu and Li 1993) to detect the presence of background selection. For example, the power of Tajima’s D is mainly determined by the difference between T2 and ηt, whereas the power of Fu and Li’s D is mainly determined by ζe.
The last statistic of interest, rxy, is the correlation in T2 between two different sites at positions x and y in the deleterious region (0 ≤ x < y ≤ 1). rxy can be viewed as a measure of linkage disequilibrium (LD). Under neutrality, we have
| 18 |
where ρxy = (y – x)ρ (there is a linear relationship between recombination frequency and physical distance between x and y) (Griffiths 1981; McVean 2002).
We carried out the following analysis to understand the effects of background selection on patterns of LD, which have not been examined previously. First, in the simulations (both forward and coalescent), we kept track of the local genealogies at five evenly spaced sites in the deleterious region (i.e., at X1 = 0, X2 = 0.25, X3 = 0.5, X4 = 0.75, and X5 = 1). Because of the spatial symmetry of the model, it suffices to estimate rxy for only six pairs of sites: X1X2, X1X3, X1X4, X1X5, X2X3, and X2X4. We denote these six pairwise rxy values in the order shown above by ri (i = 1, . . . , 6). Let r = (r1, . . . , r6). When we have two such r vectors, referred to as r(1) and r(2), we can define the distance between r(1) and r(2) as
| 19 |
Suppose that r(1) was obtained from the background selection model with parameters θ(1), γ(1), and ρ(1). To understand the effects of background selection, we divided the interval (0, 10ρ(1)) into an evenly spaced grid. For each point in the grid, we obtained an r vector under neutrality using Equation 18 and calculated its distance from r(1). Let ρ* be the value in the grid that produced the pattern closest to r(1), as measured by Equation 19. Then ρ* can be viewed as the effective recombination rate under background selection. Hence we can define a second effective population size as Ne(rxy) = (ρ*/ρ(1))N. In contrast to Ne(T2), which measures the reduction in nucleotide diversity brought about by background selection, Ne(rxy) measures the effects of background selection on LD. Finally, we define
| 20 |
Results
We present results obtained from both forward and coalescent simulations. The questions of interest are (i) the effects of background selection on patterns of neutral diversity and LD and (ii) whether the structured coalescent model provides a good approximation. For the latter question, instead of generating and examining sequence variability, we focus on several genealogy-based statistics, which are the ultimate determinants of sequence-based statistics (see above). The advantage of using genealogy-based statistics is that they are less variable than sequence-based statistics (the latter contains the additional variation brought about by the mutation process), allowing a more accurate assessment of the performance of the coalescent model.
The effects of background selection on B(T2)
The only general analytic result regarding the joint effects of background selection and recombination on neutral diversity is the expected value of T2(bgs), given by Equation 13. We can deduce three results from this formula. First, background selection reduces neutral diversity (as measured by π) because E[B(T2)] < 1 (Equation 14). Second, the extent of reduction in diversity is spatially inhomogeneous, in that the reduction is more extreme in the interior of the deleterious region relative to the boundaries. As a result, the effects of background selection cannot be fully summarized as a simple reduction in Ne for the whole region (Nordborg et al. 1996; Loewe and Charlesworth 2007). Third, within the limit of validity of Equation 13, E(T2(bgs)) and equivalently E[B(T2)] depend exclusively on the ratios between U, s, and rw, but not the absolute values of these parameters.
The simulation results, obtained from both the forward and the coalescent approaches, confirm the first two predictions (Table 1). To evaluate the third prediction, we compare the first three pairs of results in Table 1 where the ratios of U/s and R/U are the same within each pair. With weaker selection, values calculated from Equation 13 are much lower than those obtained from simulations (e.g., the first case in Table 1). The agreement improves as γ, the scaled selection coefficient, becomes larger (e.g., the second case in Table 1). This pattern holds regardless of the presence or absence of recombination. In other words, Equation 13 tends to underestimate E[B(T2)] (i.e., overestimate the reduction in diversity) when selection is weak, as observed in previous simulations (e.g., Nordborg et al. 1996). This is probably caused by the fact that the derivation of Equation 13 replies on the assumption that, in the coalescent history of a haplotype, the time that it was loaded with more than one deleterious mutation is very short compared to the time it spent as a 0- or 1-haplotype, and consequently, coalescent events take place only among 0- or 1-haplotypes (Hudson and Kaplan 1994, 1995). Because the rate that mutations are purged is proportional to γ (Equation 12), Equation 13 is more accurate when γ is larger.
Table 1. The effects of background selection on B(T2).
| Parameters |
B(T2) at x = 0 |
B(T2) at x = 0.5 |
||||||
|---|---|---|---|---|---|---|---|---|
| θ | γ | ρ | Theory | Forward | Coalescent | Theory | Forward | Coalescent |
| 6 | 6 | 0 | 0.37 | 0.55 | 0.55 | — | — | — |
| — | [0.03, 1.72] | [0.03, 1.62] | — | — | — | |||
| 25 | 25 | 0 | 0.37 | 0.39 | 0.41 | — | — | — |
| — | [0.02, 1.28] | [0.02, 1.40] | — | — | — | |||
| 6 | 6 | 3 | 0.51 | 0.62 | 0.66 | 0.45 | 0.59 | 0.61 |
| — | [0.02, 2.12] | [0.03, 2.18] | — | [0.02, 1.82] | [0.03, 1.94] | |||
| 25 | 25 | 12.5 | 0.51 | 0.53 | 0.54 | 0.45 | 0.47 | 0.49 |
| — | [0.02, 1.93] | [0.02, 1.91] | — | [0.02, 1.65] | [0.02, 1.71] | |||
| 15 | 7.5 | 7.5 | 0.37 | 0.51 | 0.54 | 0.26 | 0.45 | 0.46 |
| — | [0.03, 1.74] | [0.03, 1.76] | — | [0.03, 1.37] | [0.03, 1.43] | |||
| 25 | 12.5 | 12.5 | 0.37 | 0.45 | 0.46 | 0.26 | 0.37 | 0.38 |
| — | [0.02, 1.52] | [0.03, 1.54] | — | [0.02, 1.17] | [0.03, 1.21] | |||
| 7.5 | 15 | 15 | 0.78 | 0.82 | 0.81 | 0.72 | 0.73 | 0.75 |
| — | [0.03, 2.98] | [0.03, 2.95] | — | [0.03, 2.68] | [0.03, 2.69] | |||
The definition of B(T2) is given by Equation 14. The scaled parameters used in the coalescent simulations were θ = NU, γ = Ns, and ρ = NR. Forward simulations were performed with a haploid population size of 5000. The results were based on 105 and 2000 replicates of the coalescent and forward simulation, respectively. The mean values of B(T2) at x = 0 (the left end of the deleterious region) and x = 0.5 (the center of the region) are presented. For each mean value, the 2.5 and 97.5 percentiles of the distribution of B(T2) are also shown. Note that B(T2) at x = 1.0 should be the same as B(T2) at x = 0 when R > 0, whereas B(T2) should be the same for every x ∈ [0, 1] when R = 0.
Encouragingly, the structured coalescent model appears to be more accurate than Equation 13, in the sense that the coalescent results agree more closely with those obtained from forward simulations, in terms of both the mean and the percentiles of B(T2) (Table 1). However, the mean values of B(T2) produced by the structured coalescent model are sometimes higher than those produced by the forward simulations. This is probably because, in the continuous-time approximation, we assume that only one event can happen at any one time and that each coalescent event involves only two haplotypes. However, in a finite population, especially one with a small population size, the chance of having more than one event may be sizeable. Furthermore the product of NfiPi() (Equation 11) can be small, which increases the chance of having coalescent events involving multiple haplotypes. These factors may affect the accuracy of coalescent models (Fu 2006). In fact, for the case with scaled parameters θ = γ = 6 and ρ = 3 (the third case in Table 1), when forward simulations were conducted using either a smaller haploid population size of 2500 or a larger one of 20,000, the mean values of B(T2) at x = 0.5 (the center of the deleterious region) were 0.587 and 0.610, respectively, showing a convergence to the value of 0.609 obtained from the coalescent model. Thus, the discrepancies shown in Table 1 may be unimportant for species with large effective population sizes such as Drosophila melanogaster whose diploid Ne is ∼106 (Kreitman 1983) and should not undermine the potential usefulness of the coalescent model.
The effects of background selection on B(ηt) and B(ζe)
In addition to reducing nucleotide site diversity (π), it is known that, in the absence of recombination, background selection also distorts neutral genealogies, such that external branches take up a larger proportion of the genealogy, and this effect tends to be greater when selection is weaker (Charlesworth et al. 1993, 1995; Fu 1997; Gordo et al. 2002; Williamson and Orive 2002; Zeng et al. 2006; Kaiser and Charlesworth 2009; Seger et al. 2010). To see whether the structured coalescent model can accurately describe these aspects of variability, we investigate the properties of two other summary statistics, ηt and ζe, which are major determinants of the number of segregating sites and the relative abundance of low-frequency variants. In Tables 2 and 3, we present values of B(ηt) and B(ζe), which are defined as the values of ηt and ζe under background selection relative to their neutral expectations (Equations 16 and 17), so that deviations from unity indicate the direction of departure from neutrality. Over the combinations of parameter values we have examined, the coalescent model offers very good approximations to both B(ηt) and B(ζe); this is true for both the mean and the percentiles. Again, as with B(T2), the properties of B(ηt) and B(ζe) depend on absolute values of the scaled parameters, not just their ratios.
Table 2. The effects of background selection on B(ηt).
| Parameters |
B(ηt) at x = 0 |
B(ηt) at x = 0.5 |
||||
|---|---|---|---|---|---|---|
| θ | γ | ρ | Forward | Coalescent | Forward | Coalescent |
| 6 | 6 | 0 | 0.61 | 0.60 | — | — |
| [0.28, 1.10] | [0.30, 1.05] | — | — | |||
| 25 | 25 | 0 | 0.41 | 0.44 | — | — |
| [0.18, 0.78] | [0.20, 0.83] | — | — | |||
| 6 | 6 | 3 | 0.68 | 0.70 | 0.64 | 0.66 |
| [0.30, 1.28] | [0.32, 1.33] | [0.30, 1.17] | [0.31, 1.22] | |||
| 25 | 25 | 12.5 | 0.55 | 0.56 | 0.49 | 0.51 |
| [0.22, 1.10] | [0.24, 1.12] | [0.21, 0.97] | [0.22, 1.00] | |||
| 15 | 7.5 | 7.5 | 0.56 | 0.59 | 0.51 | 0.52 |
| [0.24, 1.05] | [0.27, 1.10] | [0.24, 0.93] | [0.26, 0.94] | |||
| 25 | 12.5 | 12.5 | 0.48 | 0.50 | 0.42 | 0.43 |
| [0.22, 0.92] | [0.23, 0.95] | [0.20, 0.76] | [0.21, 0.78] | |||
| 7.5 | 15 | 15 | 0.81 | 0.82 | 0.75 | 0.77 |
| [0.33, 1.67] | [0.33, 1.68] | [0.32, 1.51] | [0.31, 1.56] | |||
Table 3. The effects of background selection on B(ζe).
| Parameters |
B(ζe) at x = 0 |
B(ζe) at x = 0.5 |
||||
|---|---|---|---|---|---|---|
| θ | γ | ρ | Forward | Coalescent | Forward | Coalescent |
| 6 | 6 | 0 | 1.23 | 1.27 | — | — |
| [0.46, 1.90] | [0.53, 1.92] | — | — | |||
| 25 | 25 | 0 | 1.25 | 1.22 | — | — |
| [0.50, 1.96] | [0.47, 1.91] | — | — | |||
| 6 | 6 | 3 | 1.18 | 1.18 | 1.19 | 1.21 |
| [0.43, 1.88] | [0.43, 1.88] | [0.45, 1.89] | [0.46, 1.89] | |||
| 25 | 25 | 12.5 | 1.13 | 1.13 | 1.18 | 1.16 |
| [0.38, 1.86] | [0.39, 1.87] | [0.42, 1.87] | [0.42, 1.89] | |||
| 15 | 7.5 | 7.5 | 1.23 | 1.24 | 1.28 | 1.31 |
| [0.46, 1.94] | [0.48, 1.92] | [0.52, 1.95] | [0.55, 1.96] | |||
| 25 | 12.5 | 12.5 | 1.28 | 1.25 | 1.34 | 1.34 |
| [0.49, 1.95] | [0.49, 1.94] | [0.55, 1.99] | [0.56, 1.99] | |||
| 7.5 | 15 | 15 | 1.05 | 1.06 | 1.08 | 1.08 |
| [0.32, 1.82] | [0.33, 1.80] | [0.34, 1.82] | [0.34, 1.82] | |||
B(ζe) is defined by Equation 17. A sample size of 10 was assumed in the simulations (see Table 1 for more details of the simulation procedure). The neutral expectation of ζe for the assumed sample size is 0.378 (obtained from neutral coalescent simulations). Note that the same set of combinations of θ, γ, and ρ was also used in Tables 2 and 3.
Several patterns emerge from the comparison of Tables 1–3, where data were generated using the same set of parameter values. First, from Tables 1 and 2, it can be seen that E[B(T2)] and E[B(ηt)] are reduced to roughly the same extent by background selection, although the reduction in E[B(T2)] tends to be slightly more extreme, especially when selection is weaker. This is in agreement with the observation that Tajima’s D, which depends on the difference between T2 and ηt, tends to have a negative mean under background selection (Charlesworth et al. 1995; Fu 1997; Gordo et al. 2002; Williamson and Orive 2002; Zeng et al. 2006). As expected, with strong selection and/or a high level of recombination, the difference between E[B(T2)] and E[B(ηt)] is very small and can be difficult to detect (e.g., the last case in Tables 1 and 2).
Second, as is the case for B(T2), the mean values of B(ηt) and B(ζe) are spatially inhomogeneous when ρ > 0 (Tables 2 and 3). E[B(ηt)] is smaller in the center than at the boundaries (Table 2), suggesting that there will be on average fewer segregating neutral variants in the interior of the region. The reverse is true for E[B(ζe)], whose values tend to be larger in the center but smaller at the boundaries (Table 3). However, an examination of ηe, the total length of all the external branches, shows that E(ηe(bgs)) < E(ηe(neu)) and that E(ηe(bgs)) is smaller in the interior of the region than at the boundaries. Therefore, the increase in E[B(ζe)] in the center of the region is caused by a more rapid decline in ηt relative to ηe. With E[B(ζe)] > 1 and its spatial pattern, we expect a higher proportion of low-frequency variants in the interior of the region. Therefore, in addition to the known result that Fu and Li’s D, which is determined by ζe, should have a negative mean value under background selection, which was found previously using a model with zero recombination (Charlesworth et al. 1995; Fu 1997; Gordo et al. 2002; Williamson and Orive 2002; Zeng et al. 2006), the results in Tables 2 and 3 show that the mean value should also be more negative in the center than at the boundaries, although the difference may be small and may not be easily detectable with sequence variability.
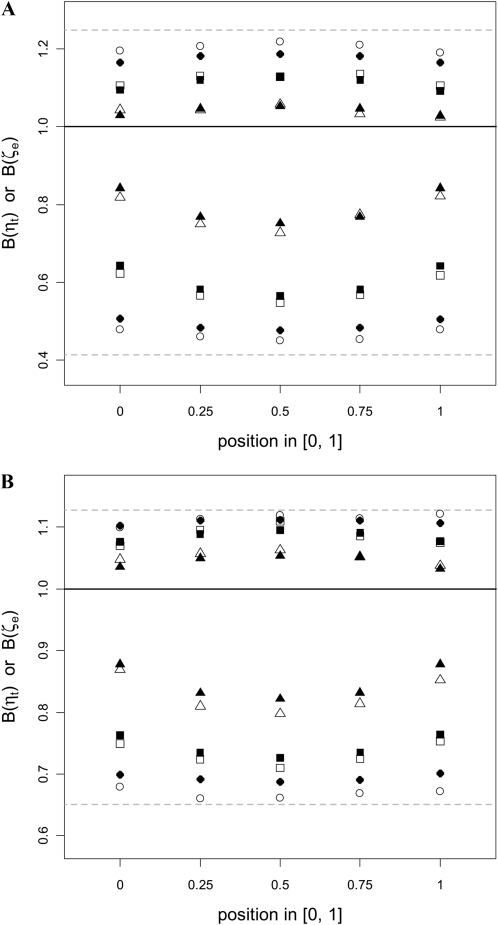
We further explored the effects of various levels of recombination on E[B(ηt)] and E[B(ζe)] (Figure 4). As expected, recombination increases neutral diversity (as measured by E[B(ηt)]), to levels well above that observed in the absence of recombination (the dashed line at the bottom). Similarly, E[B(ζe)] gets closer to the neutral expectation of unity with increasing rates of recombination. However, even with a fairly high level of recombination (θ = 25 and ρ/θ = 4; triangles in Figure 4A), the values of E[B(ηt)] and E[B(ζe)] at the boundary of the deleterious region, where the influence of background selection is minimal, are still 18% lower and 3% higher than the neutral expectation, respectively (Figure 4A), suggesting that, even in highly recombining regions of the genome, the effect of background selection should not be overlooked. The spatial pattern displayed in Figure 4 extends the results of previous studies that are based on E[B(T2)] (Nordborg et al. 1996; Loewe and Charlesworth 2007) and lends further support to the conclusion that the effect of background selection is greater in the center of the region. In particular, the parameters used to generate Figure 4B are comparable to those thought to be “typical” of a gene in the D. melanogaster genome (Loewe and Charlesworth 2007) and hence may be relevant to the intragenic spatial patterns of codon usage bias found in D. melanogaster (Comeron and Kreitman 2002; Comeron and Guthrie 2005; Loewe and Charlesworth 2007).
Figure 4 .
The joint effects of background selection and recombination on E[B(ηt)] and E[B(ζe)]. Both forward and coalescent simulations were performed and a sample size of 10 was assumed. The scaled parameters were θ = γ = 25 in A and θ = 7.5 and γ = 15 in B, with various values of ρ/θ. The solid line in the middle indicates the neutral expectation of unity. In both plots, the results were divided into three sets, and the results within a set all have symbols of the same shape (e.g., all being triangles). In a result set, the symbols beneath and above the solid line represent mean values of B(ηt) and B(ζe), respectively. The open and solid symbols in a result set represent results obtained from the forward and coalescent simulations, respectively. In both plots, the result sets indicated by the triangles, rectangles, and circles were obtained from simulations with ρ/θ values of 4, 1, 0.25, respectively. The dashed lines at the bottom and the top show the mean B(ηt) and B(ζe) values when ρ/θ = 0. Note that the scales of the y-axes are different in the two plots.
Figure 4 shows that the results obtained from the structured coalescent model agree well with those obtained from the forward simulations. The discrepancies are probably due to the artifact of using a small population size in the forward simulations, as explained above for the case of B(T2). For instance, in the case where θ = 7.5, γ = 15, and ρ/θ = 0.25 (circles in Figure 4B), the values of E[B(ηt)] at x = 0 (the bottom-left circles) obtained from the forward simulations with N = 5000 and 20,000 are 0.679 and 0.692, respectively, with the latter being very close to the value of 0.699 obtained under the coalescent model. Hence we can conclude that the structured coalescent model can accurately predict patterns of diversity for all levels of recombination.
Patterns of linkage disequilibrium under background selection
The effects of background selection on patterns of LD have not previously been examined. As an initial attempt, we used rxy, the correlation in T2 between two different sites x and y, as a measure of LD. As shown in Table 4, the structured coalescent model can accurately capture this aspect of neutral variability. Comparing these results with neutral expectations (Equation 18), we find that background selection increases the level of correlation (i.e., higher LD) between sites [compare rxy(bgs) with the neutral expectations obtained with ρ = 3 in Table 4]. This is probably caused by a reduction in the effectiveness of recombination, as a result of a shorter expected coalescent time in the presence of background selection {recall that E[B(T2)] < 1 and E[B(ηt)] < 1}. Using the method described below Equation 19, the effective recombination rate for the set of parameters in Table 4 was estimated to be ρ* = 1.98. It can be seen that the rxy(neu) values obtained using ρ* and Equation 18 agree closely with those obtained under the background selection model (Table 4). Note that, with ρ* = 1.98, B(rxy) = 0.66 (Equation 20), which is identical to the E[B(T2)] value at the boundary (see the third case in Table 1). In other words, if we estimate Ne from either nucleotide diversity at the boundary of the region (i.e., the value of π at x = 0, which is governed by T2) or patterns of LD [i.e., the six pairwise rxy(bgs) values], we should obtain a similar estimate of Ne ≈ 0.66N. This result is supported by additional simulations using a wide range of parameter combinations (Table 5). However, we have examined only one particular measure of LD, and more research is needed to examine the effects of background selection on LD.
Table 4 . Patterns of linkage disequilibrium under background selection.
|
rxy(bgs) |
rxy(neu) |
|||
|---|---|---|---|---|
| Site pairs | Forward | Coalescent | ρ = 3 | ρ* = 1.98 |
| X1X2 | 0.605 | 0.598 | 0.491 | 0.596 |
| X1X3 | 0.424 | 0.424 | 0.318 | 0.419 |
| X1X4 | 0.320 | 0.318 | 0.233 | 0.320 |
| X1X5 | 0.275 | 0.250 | 0.182 | 0.258 |
| X2X3 | 0.604 | 0.608 | 0.491 | 0.596 |
| X2X4 | 0.414 | 0.422 | 0.318 | 0.419 |
| Distance | 0 | 0.028 | 0.250 | 0.022 |
The scaled parameters were θ = γ = 6 and ρ = 3. We kept records of the local gene genealogies at five evenly spaced sites in the deleterious region (i.e., at X1 = 0, X2 = 0.25, X3 = 0.5, X4 = 0.75, and X5 = 1) and calculated the correlation coefficient between the T2 values for the six pairs of sites shown under rxy(bgs). We also obtained expected values under neutrality using Equation 18 for two different levels of recombination as shown beneath rxy(neu). Note that ρ* was the effective recombination rate found by the method described below Equation 19, and ρ*/ρ = 0.66 is the expected reduction in neutral diversity (as measured by π) caused by background selection at the boundary of the deleterious region (see the third case in Table 1). The last row gives the distance between the set of values given in a column and the set obtained from the forward simulations (Equation 19).
Table 5. Comparing effective population sizes estimated from nucleotide diversity and patterns of linkage disequilibrium.
| Parameters |
ρ/θ |
||||
|---|---|---|---|---|---|
| θ | γ | Ne/N | 0.5 | 1 | 2 |
| 6 | 6 | B(rxy) | 0.66 | 0.72 | 0.80 |
| E[B(T2)] | 0.66 | 0.73 | 0.82 | ||
| 25 | 25 | B(rxy) | 0.55 | 0.62 | 0.73 |
| E[B(T2)] | 0.54 | 0.62 | 0.73 | ||
| 15 | 7.5 | B(rxy) | 0.53 | 0.64 | 0.75 |
| E[B(T2)] | 0.54 | 0.65 | 0.78 | ||
| 25 | 12.5 | B(rxy) | 0.46 | 0.58 | 0.73 |
| E[B(T2)] | 0.46 | 0.59 | 0.73 | ||
| 7.5 | 15 | B(rxy) | 0.72 | 0.75 | 0.81 |
| E[B(T2)] | 0.71 | 0.75 | 0.81 | ||
As explained by the arguments leading to Equations 14 and 20, effective population sizes under background selection can be estimated from either rxy or T2, denoted by Ne(rxy) and Ne(T2), respectively. B(rxy) = Ne(rxy)/N and E[B(T2)] = Ne(T2)/N. Using coalescent simulations, we obtained the estimates by calculating the six pairwise rxy values (see Table 4) and the mean value of B(T2) at x = 0.
Discussion
The results that we have described above show that our structured coalescent approach provides a rapid algorithm for computing the neutral genealogies of sites situated within a group of sites subject to sufficiently strong purifying selection that they can be treated as at equilibrium under mutation and selection (the conditions for this assumption to be valid are discussed in Nordborg et al. 1996). In contrast to previous coalescent results (Charlesworth et al. 1995; Fu 1997; Neuhauser and Krone 1997; Wakeley 2008b; O’Fallon et al. 2010), we allow for the occurrence of crossing over within the region in question. Our forward simulations show that the method provides extremely accurate numerical results. While we have assumed a haploid model for convenience, the results apply equally to a diploid model when there is semidominance with respect to the effects of mutations on fitness, with the appropriate changes in parameter values, or to a diploid model with sufficiently strong selection that deleterious mutations with partially dominant effects present at low frequency in the population, so that their elimination exclusively involves heterozygotes (Kimura and Maruyama 1966; Simmons and Crow 1977; Charlesworth 1990). The assumption of haploidy is thus not especially restrictive.
Since we can compute the genealogies of neutral sites at arbitrary locations within a set of selected sites, we can investigate any desired property of a neutral site as a function of its location within the region subject to purifying selection, as well as correlations between the properties of linked neutral sites within the region. This allows predictions to be made concerning the extent of the reduction caused by background selection in the expected pairwise coalescent time, T2(bgs), as a function of location within the region (Table 1). T2(bgs) can be regarded as a measure of the local effective population size and determines the expected level of pairwise neutral diversity under the infinite sites model (Tajima 1983). In addition, statistics such as the mean total size of the genealogy at a given location, when contrasted with the mean pairwise coalescent time, and the proportion of the genealogy contributed by external branches, ζe(bgs), can be used to assess the extent of distortion of the genealogy in favor of longer external branches, which earlier studies of a nonrecombining region have shown to be produced by background selection, especially when selection is relatively weak (Charlesworth et al. 1993, 1995; Fu 1997; Gordo et al. 2002; Williamson and Orive 2002; Zeng et al. 2006; Kaiser and Charlesworth 2009; Seger et al. 2010). This distortion will be accompanied by an excess of rare, derived neutral variants compared with what is observed in the absence of background selection. Our results show that these effects are observed even in the presence of recombination (Tables 2 and 3 and Figure 4) and that they are strongest in the middle of the region subject to selection. The ζe(bgs) statistic seems to show this effect particularly clearly.
If the region subject to purifying selection is taken to represent a coding sequence, then our results suggest that not only is the local effective population size most strongly reduced in the middle of a noninterrupted stretch of coding sequence (as found previously by Loewe and Charlesworth 2007), but also this will be associated with a stronger excess of rare variants at neutral sites in the middle of genes compared with their end or with intergenic or intronic sequences. It should shortly be possible to test this prediction quantitatively against data from large-scale genome-wide resequencing projects, which are capable of detecting very small effects when data from large numbers of genes are combined. There is already evidence for reductions in noncoding nucleotide site diversity in human populations in the proximity of genes compared with more remote regions (McVicker et al. 2009; Hammer et al. 2010).
Intuitively, the reduction in effective population size for a small genomic region caused by background selection might be expected to cause a corresponding increase in the level of linkage disequilibrium among pairs of closely linked neutral sites. This has not previously been investigated theoretically. We have used our program to calculate the correlations between the genealogies at different sites within the region subject to purifying selection; the results show that these are well predicted by Equation 18 for the standard neutral case, if the population scaled recombination rate ρ is calculated using the effective size under background selection at the boundary of the region.
The results presented here are obviously very limited in scope and need to be extended in several ways. First, our results for genealogies need to be translated into predictions concerning observable DNA sequence diversity statistics; this is a relatively straightforward matter of adding neutral mutations onto the genealogies, as is standard practice in coalescent theory (reviewed by Wakeley 2008a). Second, a distribution of mutational effects on fitness needs to be included in the model, in view of the evidence that the selection coefficients against deleterious nonsynonymous mutations follow a wide distribution (Loewe and Charlesworth 2006; Keightley and Eyre-Walker 2007; Boyko et al. 2008). Third, instead of considering a single genomic region, the division of the genome into different regions with differing strengths of purifying selection, such as noncoding and coding sequences, needs to be modeled. Fourth, the effects of nonreciprocal recombination need to be included, since this is of major importance over short genetic distances in eukaryotes and across the whole genome in bacteria. Fifth, the effects of population size change and population structure need to be modeled. Some of these extensions, notably the inclusion of a distribution of fitness effects, present more of a challenge than others. The ultimate goal is to have a flexible and rapid set of programs that allows predictions to be made about the properties of neutral genetic diversity across the genome.
Acknowledgments
We thank Nick Barton and Matt Hartfield for valuable discussions. This study made use of the resources provided by the Edinburgh Compute and Data Facility (ECDF) (http://www.ecdf.ed.ac.uk/). The ECDF is partially supported by the e-Science Data, Information and Knowledge Transformation (eDIKT) initiative (http://www.edikt.org.uk). K.Z. acknowledges support from a Biomedical Personal Research Fellowship awarded by the Royal Society of Edinburgh and the Caledonian Research Foundation.
Appendix
Derivation of Equation 5
First, we note that
| (A1) |
Similarly, we have
| (A2) |
Therefore Equation 5 can be rewritten as
| (A3) |
Derivation of Equation 6
Using the second equation in Equation A3, we have
| (A4) |
Derivation of Equation 9
Because mutation is unidirectional, the i-haplotypes in the postmutation population must have been produced by premutation haplotypes with j (≤i) mutations. Noting that the proportion of j-haplotypes in the premutation population is given by fj and that the number of new mutations (i.e., i – j) follows a Poisson distribution with mean U, we have
| (A5) |
Literature Cited
- Aguadé M., Miyashita N., Langley C. H., 1989. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 17: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520 [DOI] [PubMed] [Google Scholar]
- Berry A. J., Ajioka J. W., Kreitman M., 1991. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics 129: 1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Presgraves D. C., 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99: 13616–13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Welch J. J., Charlesworth B., 2009. Reduced effectiveness of selection caused by a lack of recombination. Curr. Biol. 19: 655–660 [DOI] [PubMed] [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J. J., Macpherson J. M., Sella G., Petrov D. A., 2009. Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet. 5: e1000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1990. Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55: 199–221 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63: 213–227 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1996. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 68: 131–149 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Betancourt A. J., Kaiser V. B., Gordo I., 2009. Genetic recombination and molecular evolution. Cold Spring Harbor Symp. Quant. Biol. 74: 177–186 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Morgan M. T., 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141: 1619–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Guthrie T. B., 2005. Intragenic Hill-Robertson interference influences selection intensity on synonymous mutations in Drosophila. Mol. Biol. Evol. 22: 2519–2530 [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Kreitman M., 2002. Population, evolutionary and genomic consequences of interference selection. Genetics 161: 389–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Choi J. Y., 2010. Natural selection shapes nucleotide polymorphism across the genome of the nematode Caenorhabditis briggsae. Genome Res. 20: 1103–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. Springer-Verlag, Berlin [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford [Google Scholar]
- Fu Y. X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X., 2006. Exact coalescent for the Wright-Fisher model. Theor. Popul. Biol. 69: 385–394 [DOI] [PubMed] [Google Scholar]
- Fu Y. X., Li W. H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I., Navarro A., Charlesworth B., 2002. Muller’s ratchet and the pattern of variation at a neutral locus. Genetics 161: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. C., 1981. Neutral two-locus multiple allele models with recombination. Theor. Popul. Biol. 19: 169–186 [Google Scholar]
- Haddrill P. R., Halligan D. L., Tomaras D., Charlesworth B., 2007. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 8: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P. R., Zeng K., Charlesworth B., 2011. Determinants of synonymous and nonsynonymous variability in three species of Drosophila. Mol. Biol. Evol. 28: 1731–1743 [DOI] [PubMed] [Google Scholar]
- Haigh J., 1978. The accumulation of deleterious genes in a population—Muller’s Ratchet. Theor. Popul. Biol. 14: 251–267 [DOI] [PubMed] [Google Scholar]
- Hammer M. F., Woerner A. E., Mendez F. L., Watkins J. C., Cox M. P., et al. , 2010. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat. Genet. 42: 830–831 [DOI] [PubMed] [Google Scholar]
- Hey J., Kliman R. M., 2002. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics 160: 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 1990. Gene genealogies and the coalescent process. Oxford Surv. Evol. Biol. 7: 1–44 [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1994. Gene trees with background selection, pp. 140–153 Non-Neutral Evolution: Theories and Molecular Data, edited by Golding B. Chapman & Hall, London [Google Scholar]
- Hudson R. R., Kaplan N. L., 1995. Deleterious background selection with recombination. Genetics 141: 1605–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., 1999. The approach to mutation-selection balance in an infinite asexual population, and the evolution of mutation rates. Proc. Biol. Sci. B 266: 2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser V. B., Charlesworth B., 2009. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 25: 9–12 [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1969. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics 61: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Maruyama T., 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. On the genealogy of large populations. J. Appl. Probab. 19A: 27–43 [Google Scholar]
- Kreitman M., 1983. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304: 412–417 [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Sackton T. B., Greenberg A. J., Wong A., Singh N. D., et al. , 2008. Evolution of protein-coding genes in Drosophila. Trends Genet. 24: 114–123 [DOI] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2006. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2007. Background selection in single genes may explain patterns of codon bias. Genetics 175: 1381–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- McVean G. A., 2002. A genealogical interpretation of linkage disequilibrium. Genetics 162: 987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G., Gordon D., Davis C., Green P., 2009. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5: e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser C., Krone S. M., 1997. The genealogy of samples in models with selection. Genetics 145: 519–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996. The effect of recombination on background selection. Genet. Res. 67: 159–174 [DOI] [PubMed] [Google Scholar]
- O’Fallon B. D., Seger J., Adler F. R., 2010. A continuous-state coalescent and the impact of weak selection on the structure of gene genealogies. Mol. Biol. Evol. 27: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 15: 1651–1656 [DOI] [PubMed] [Google Scholar]
- Rockman M. V., Skrovanek S. S., Kruglyak L., 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330: 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago E., Caballero A., 1998. Effective size and polymorphism of linked neutral loci in populations under directional selection. Genetics 149: 2105–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger J., Smith W. A., Perry J. J., Hunn J., Kaliszewska Z. A., et al. , 2010. Gene genealogies strongly distorted by weakly interfering mutations in constant environments. Genetics 184: 529–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., Petrov D. A., Przeworski M., Andolfatto P., 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5: e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C., Hubisz M. J., Lu J., et al. , 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnol E. E., Kondrashov A. S., 1993. The effect of selection on the phenotypic variance. Genetics 134: 995–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M. J., Crow J. F., 1977. Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 11: 49–78 [DOI] [PubMed] [Google Scholar]
- Stephan W., 1995. An improved method for estimating the rate of fixation of favorable mutations based on DNA polymorphism data. Mol. Biol. Evol. 12: 959–962 [DOI] [PubMed] [Google Scholar]
- Stephan W., Charlesworth B., McVean G., 1999. The effect of background selection at a single locus on weakly selected, partially linked variants. Genet. Res. 73: 133–146 [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2008a. Coalescent Theroy: An Introduction. Roberts & Company, Greenwood Village [Google Scholar]
- Wakeley J., 2008b. Conditional gene genealogies under strong purifying selection. Mol. Biol. Evol. 25: 2615–2626 [DOI] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Williamson S., Orive M. E., 2002. The genealogy of a sequence subject to purifying selection at multiple sites. Mol. Biol. Evol. 19: 1376–1384 [DOI] [PubMed] [Google Scholar]
- Wright S. I., Andolfatto P., 2008. The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annu. Rev. Ecol. Evol. Syst. 39: 193–213 [Google Scholar]
- Zeng K., Charlesworth B., 2010. The effects of demography and linkage on the estimation of selection and mutation parameters. Genetics 186: 1411–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Fu Y. X., Shi S., Wu C. I., 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]