Abstract
Despite the significance of redox post-translational modifications (PTMs) in regulating diverse signal transduction pathways, the enzymatic systems that catalyze reversible and specific oxidative or reductive modifications have yet to be firmly established. Thioredoxin 1 (Trx1) is a conserved antioxidant protein that is well known for its disulfide reductase activity. Interestingly, Trx1 is also able to transnitrosylate or denitrosylate (defined as processes to transfer or remove a nitric oxide entity to/from substrates) specific proteins. An intricate redox regulatory mechanism has recently been uncovered that accounts for the ability of Trx1 to catalyze these different redox PTMs. In this review, we will summarize the available evidence in support of Trx1 as a specific disulfide reductase, and denitrosylation and transnitrosylation agent, as well as the biological significance of the diverse array of Trx1-regulated pathways and processes under different physiological contexts. The dramatic progress in redox proteomics techniques has enabled the identification of an increasing number of proteins, including peroxiredoxin 1, whose disulfide bond formation and nitrosylation status are regulated by Trx1. This review will also summarize the advancements of redox proteomics techniques for the identification of the protein targets of Trx1-mediated PTMs. Collectively, these studies have shed light on the mechanisms that regulate Trx1-mediated reduction, transnitrosylation, and denitrosylation of specific target proteins, solidifying the role of Trx1 as a master regulator of redox signal transduction. Antioxid. Redox Signal. 15, 2565–2604.
I. Introduction
Thioredoxin 1 (Trx1), a 12 kDa protein found primarily in the cytosol and occasionally in the nucleus (103, 224), plays critical roles in regulating protein thiol homeostasis and redox signaling both inside and outside the cellular environment (7, 107, 146). A key component of its disulfide reductase function is a highly conserved CXXC motif, located on its exterior (70), which promotes electron and disulfide exchange between Trx1 and its substrates. Trx1 is a component of the Trx protein reductive system, which also includes Trx reductase (TrxR) and nicotinamide adenine dinucleotide phosphate (NADPH) (the latter is derived mainly from cellular metabolism; Fig. 1) (216). The Trx reductive system is an essential cellular mechanism facilitating the reduction of reactive oxygen species (ROS) by supporting the peroxidase actions of peroxiredoxins (Prxs) and directly repairing oxidatively damaged proteins. Trx1 regulates a wide range of cellular functions, including cell growth (279), proliferation (77), and apoptosis (14, 191). Its dysfunction is associated with a variety of diseases in which redox imbalance has been implicated, including cancer (216), human immunodeficiency virus infection (26), neurodegenerative diseases (154), and cardiovascular diseases (4, 241). Trx1′s disulfide reduction function is widely recognized, and to a lesser degree it has been shown to modulate additional redox-dependent post-translational modifications (PTMs), including transnitrosylation (183, 184, 273) and denitrosylation (21, 23) of specific proteins. In this review, we examine the function of Trx1 in regulating protein PTMs and the proteomics approaches for the identification of Trx1 target proteins. In addition, we discuss the potential contribution of proteomics approaches for identifying novel Trx1 targets and dissecting the mechanisms of Trx1-mediated redox PTMs.
FIG. 1.
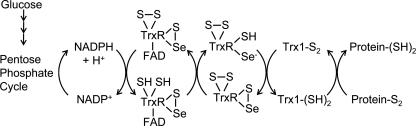
The thioredoxin (Trx) reductive system. The electron source of the Trx reductive system is nicotinamide adenine dinucleotide phosphate (NADPH), which is largely produced by the pentose phosphate pathway. Oxidized Trx1 (Trx1-S2) is directly reduced by the homodimeric selenoprotein Trx reductase (TrxR). Electrons are transferred from NADPH to TrxR via its cofactor flavin adenine dinucleotide (FAD). Reduced Trx1 (Trx1-(SH)2) catalyzes disulfide bond reduction in many proteins, including antioxidants such as the peroxiredoxins (Prxs). Adapted from Ref. (110) with permission.
II. Trx Systems
A. Trx1 and related proteins
Trx1 is conserved across kingdoms, suggesting that its redox regulatory function is essential to life (213). Mammalian Trx1 orthologs display high amino acid conservation, whereas more evolutionarily distant vertebrates maintain the catalytic CXXC reductase motif (Fig. 2A). There are five conserved cysteines in mammals, suggesting that these cysteines may have evolved to serve important cellular functions. Trx1 selectively reduces disulfide bonds within target proteins using the free thiols of Cys32 and Cys35, resulting in the formation of a disulfide bond between these cysteines. The Trx1 Cys32-Cys35 disulfide bond is resolved to free thiols using reducing equivalents derived from NADPH and TrxR (216), a homodimeric selenoprotein that uses flavin adenine dinucleotide as cofactor (Fig. 1) (253). Although the role of Cys32 and Cys35 in the reduction of protein disulfides is well understood (107, 108), there is a growing awareness of their importance in the denitrosylation of S-nitrosylated proteins (SNO-proteins) (21, 71) and in the reduction of sulfenic acids back to cysteine thiols within peptides (125).
FIG. 2.
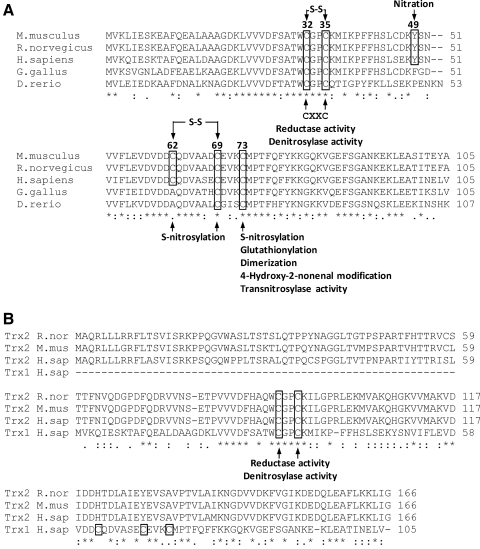
Evolutionary conservation of Trx1 and comparison with Trx2. (A) Vertebrate Trx1 sequence alignments and post-translational modifications (PTMs). Alignment (CLUSTALW 2.0.12) of mouse (Mm; P10639), rat (Rn; P11232), human (Hs; P10599), chicken (Gg; P08629), and zebra fish (Dr; Q6DGI6) protein sequences obtained from NCBI. Sequence homology (*) and conserved (:) or semi-conserved (.) substitutions are indicated. Reported PTMs at specific residues for mammalian Trx1 are denoted. Conserved vertebrate Cys are boxed. Cys32 and 35 (numbered as in the human sequence) define the CXXC motif and are essential to reductase and denitrosylating activities (108, 109, 238); Tyr49 is a reported nitration site (256); an intramolecular disulfide bond can be formed between Cys62 and 69 (96). Both Cys62 and 69 are also sites of S-nitrosylation (88, 255, 269). Cys73 is a multimodification site, reportedly undergoing nitrosylation (184, 267, 273), glutathionylation (37), dimerization (269) or 4-hydroxy-2-nonenal modification (82). (B) Alignment of human Trx1 with rat (P97615), mouse (P97493), and human (Q99757) Trx2. Of the conserved Trx1 cysteines (boxed), only the reductive site cysteines are present in Trx2.
Although Trx1 will be the focus of discussion in this review, we briefly discuss the current understanding of other proteins that contain the CXXC reductive motif (Fig. 2B), collectively known as the Trx superfamily (Table 1). This family contains a common Trx fold structure consisting of a multistranded β-sheet surrounded by three to four α-helices, with the CXXC catalytic motif projected at the end of one helix (142, 169). Although Trx superfamily members all possess disulfide reductase activity, their target specificities do not always overlap, and consequently they modulate the redox status of a diverse group of target proteins involved in various physiological processes. They have heterogeneous protein structures enabling a wide range of reduction potentials, for example, the reduction potential of Trx1 is −270 mV, whereas that of glutaredoxin 1 (Grx1) is −230 mV, and that of protein disulfide isomerase (PDI) is −175 mV (35), and perhaps different target recognition domains. Amino acids proximal to the CXXC catalytic site and the identity of the XX residues appear to be critical determinants of substrate specificity (68, 198). For example, Glu80 in thiol-disulfide oxidoreductase ResA, an extracytoplasmic Trx from Bacillus subtilis, plays a key role in controlling substrate binding and the acid-base properties of its active site Cys74 and Cys77 (142). Substitution of Pro34 in the Escherichia coli Trx catalytic site (WCGPC) with His, to mimic the active sites (WCGHC) of PDI, resulted in a mutant Trx with increased redox potential (130). Additional factors such as catalytic site cysteine pKa and nucleophilicity, cellular expression, subcellular localization, protein–protein interactions, and multiprotein complex formation may contribute to the determination of disulfide reductase specificities (36). For example, Trx2 facilitates the scavenging of oxidants in mitochondria, where it resides, and has been shown to be involved in apoptosis signaling (265), whereas PDI and other Trx-like endoplasmic reticulum residents are guardians of proper protein folding (97). Additional Trx-like molecules that are either localized specifically within the nucleus or the plasma membrane have been discovered (97). Localized expression of Trx-related molecules suggests that different redox pathways may function in a more compartmentalized fashion for modulating specific signal transduction pathways.
Table 1.
Selected Mammalian Thioredoxin Superfamily Members
| Protein | Accession (Swiss-Prot) | Mw (kDa) | Localization | Active site | Reference |
|---|---|---|---|---|---|
| Calcium-binding protein 1 (CaBP1) | Q9NZU7 | 49 | Endoplasmic reticulum | -CGHC- | (156) |
| Calcium-binding protein 2 (CaBP2) | Q9NPB3 | 72 | Endoplasmic reticulum | -CGHC- | (156) |
| Glutaredoxin 1 (Grx1) | P35754 | 12 | Cytosol | -CGYC- | (10, 109) |
| Nucleoredoxin | P97346 | 48 | Nucleus | -CGPC- | (131) |
| Protein disulfide isomerase A3 (PDIA3) | P30101 | 61 | Endoplasmic reticulum | -CGHC- | (24) |
| Protein disulfide isomerase (PDI) | P07237 | 55 | Endoplasmic reticulum | -CGHC- | (10) |
| Thioredoxin 1 (Trx1) | P10599 | 12 | Cytosol | -CGPC- | (109) |
| Thioredoxin 2 (Trx2) | Q99757 | 12 | Mitochondria | -CGPC- | (254) |
| Thioredoxin domain-containing protein 17 (TRP14) | Q9BRA2 | 14 | Cytosol | -CPDC- | (119) |
| Thioredoxin-like protein 1 (TRP32) | O43396 | 32 | Cytosol | -CGPC- | (137) |
| Thioredoxin-related transmembrane protein 1 (TMX) | Q9H3N1 | 31 | Endoplasmic reticulum | -CPAC- | (171) |
B. PTMs of Trx1
The various biological functions of Trx1 may be modulated in part by the different PTMs of Trx1 (Fig. 2A), including nitrosylation (88, 92, 184), glutathionylation (37), dimerization (269), and intramolecular disulfide formation (96). The Cys32 and Cys35-mediated reduction of protein disulfides appears to be regulated by the redox state of other cysteines of Trx1 (80). Additional regulatory functions, independent of Trx1 catalysis of disulfide reduction, for example, modulation of protein transnitrosylation (185) and denitrosylation (21), are associated with Cys62, Cys69, and Cys73, which are largely conserved in mammalian Trx1 but absent from other members of the Trx family, such as Trx2 (Fig. 2B) (88).
4-Hydroxy-2-nonenal (HNE) and other reactive aldehydes that arise from lipid peroxidation and metabolism are notoriously reactive and toxic molecules that covalently modify a vast variety of proteins and DNA and cause dysregulation of their functions (64). Two vital components of the E. coli Trx reduction system, TrxR and Trx1, are both susceptible to irreversible HNE modification, leading to inactivation of the Trx1 disulfide reductase function (64). Fang and Holmgren reported that HNE modifies E. coli Trx primarily at the reductase catalytic sites of Cys32/Cys35. By comparison, since mammalian and bacterial Trx are structurally different, Go and coworkers found that HNE and acrolein, another reactive aldehyde, modify mammalian Trx1 primarily at Cys73 (82). Modification at Cys73 inhibited Trx1 disulfide reductase activity and, further, HNE-modified Trx1 appeared to inhibit TrxR activity (82).
Nitration is an irreversible modification of proteins that occurs primarily on tyrosine residues (116). Human Trx1 contains only one tyrosine: residue 49. Nitration of Tyr49 by peroxynitrite appeared to abolish the disulfide reductive and antiapoptotic activities of Trx1 (256). Nitrated Trx1 is increased after ischemia/reperfusion damage in aging heart, but accelerating the decomposition of peroxynitrite alleviates nitrative inhibition of Trx1, and restores the cardioprotective function of Trx1 (280). Cys62 and Cys69 in Trx1 have been reported to form a disulfide bond in a highly oxidizing environment (96, 268). Whether this PTM is formed under physiological conditions in vivo and its possible function are currently unknown. However, in vitro, the formation of Cys62 and Cys69 disulfide appears to inhibit the reduction of Cys32 and Cys35 disulfide by TrxR (96, 268), suggesting a possible mechanism for the overoxidation-induced inactivation of Trx1.
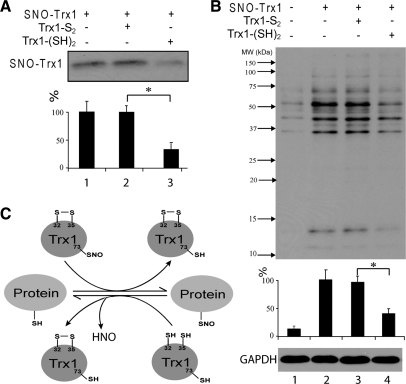
S-nitrosylation of Trx1 has been shown to occur on Cys62, 69, or 73 (88, 96, 184, 267, 269), under different experimental conditions. S-nitrosylated Trx1 (SNO-Trx1) appears to have anti-apoptotic properties (88, 185, 273) that are independent of its disulfide reductase activity, or proapoptotic activity correlated with the activation of apoptosis signal-regulating kinase 1 (ASK1) (249). SNO-Trx1 appears to have diminished disulfide reductase activity compared with unmodified Trx1 (96). Recently, we have established that only the reductase inactive Cys32-Cys35 disulfide form of Trx1 can be nitrosylated (273). We discuss Trx1 S-nitrosylation in detail in Section VI.
Glutathionylation of human Trx1 has been reported at Cys72 (equivalent to Cys73 in Fig. 2A) (37), Cys60 of chloroplast Trx f (180), and Cys68 of Trypanosoma brucei Trx (177). Although glutathionylation of the catalytic cysteines within the Trx CXXC motif has not been reported, this PTM appears to inactivate Trx1 as a disulfide reductase (37). Glutathionylation is considered to be a reversible switch for the functional regulation of Trx1 and serves as a possible nexus between the glutathione (GSH) and Trx protein reductive systems (37). Nitrosoglutathione (GSNO) derived from GSH can preferentially glutathionylate or nitrosylate target proteins (186). For example, GSNO treatment of phosphorylase b produced only S-nitrosylated forms of the protein (120), but in the same study, GSNO treatment of either crude rat liver extract (containing carbonic anhydrase III) or E. coli extract (overexpressing H-Ras) resulted in both S-nitrosylated and S-glutathionylated forms of carbonic anhydrase and H-Ras. We found that GSNO treatment of Trx1 produced mostly SNO-Trx1 (273). Mohr and colleagues hypothesize that GSNO can induce S-nitrosylation or S-glutathionylation according to the nucleophilicity of the target cysteines residues (186). Further, it is suggested that certain S-nitrosylated cysteine (SNO-Cys) sites may serve as precursors for S-glutathionylation (78, 181).
III. Trx Regulation of Target Proteins by Disulfide Bond Reduction
Trx1 is involved in many different aspects of cellular function, and can regulate at the level of transcription, translation, protein–protein interactions, and PTMs of proteins. Trx1 is best known as a hydrogen donor for ribonucleotide reductase; it therefore plays an essential role in reducing nucleotides to deoxynucleotides during the synthesis of DNA (8, 12). Further, an 80 amino acid N-terminal truncated form of Trx1 is secreted from a few cell types by unknown mechanisms (235), is present in plasma, and is involved in the growth of lymphocytes and cancer cells (216). Interestingly, Cys32 and Cy35 within the disulfide reduction site of Trx1 are required in promoting cell growth (77). However, the most widely studied biochemical role of Trx1 is the reduction of protein disulfides, cysteine sulfenic acids, and nitrosothiols.
A. Trx regulation of cellular redox balance
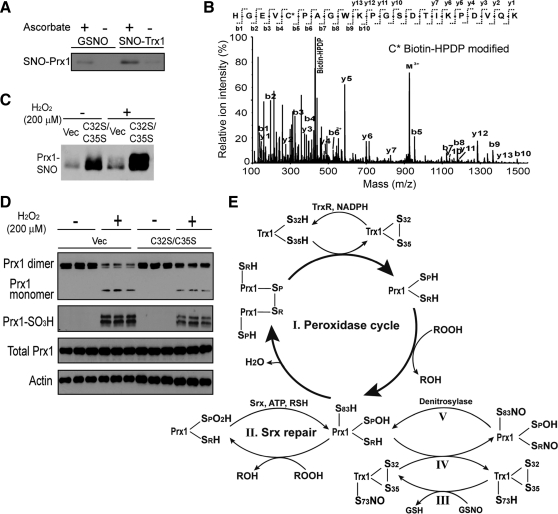
The Trx and Grx systems are the two most versatile systems for facilitating peroxide detoxification and disulfide reduction in cells [for reviews see Refs. (109, 146, 225)]. The Grx system is comprised of NADPH, glutathione reductase, Grx1, and GSH, a cysteine-containing tripeptide that supplies the reducing equivalent to Grx1 allowing it to reduce protein disulfides. Their complementary activities are important for the maintenance of thiol homeostasis and redox potentials in cells. They are also antioxidant proteins that can neutralize cellular oxidants by supplying the reducing equivalents derived from NADPH to peroxidases (79, 146, 155). Although there is a large degree of overlap among proteins whose cysteines are reduced by the Trx or Grx systems, there is evidence that some target sites are more effectively reduced by one system (225, 262), suggesting that under in vivo conditions, a selective mechanism operates for the removal of disulfides and possibly other oxidative PTMs. Trx1 is able to aid the Prx family of peroxidases in the reduction of hydrogen and lipid peroxides (76). Human Prx1 is a two-cysteine Prx that converts its peroxidatic Cys52-SH (CP) to CP-SOH during the peroxidation reaction (Fig. 3A) (91). Its cycle between CP-SOH and CP-SH involves the formation of a disulfide bond between CP and a resolving Cys173 (CR) on another Prx1 peptide chain. The disulfide-linked peptides can be reduced by Trx1. In the absence of reduction by Trx1, CP can become overoxidized to CP-SO2H, which may be repaired by sulfiredoxin (1), an adenosine triphosphate (ATP)-dependent enzyme (Fig. 3B). However, overoxidation to Prx1-CP-SO3H results in the inactivation of Prx1 as a peroxidase, which is widely believed to be irreversible (275). In addition to reducing the CP-CR disulfide linkage of the Prx1 homodimer, it has been reported that the Trx system can reduce the sulfenic acid intermediate of oxidized mammalian methionine sulfoxide reductase B, which lacks a resolving Cys, and can interact with, and be directly reduced by Trx1 (125). Therefore, the conventional notion is that Trx1 can protect its target proteins via Cys32 and Cys35-mediated reduction, as long as these catalytic site cysteines can be regenerated by TrxR, with the reducing equivalents derived from NADPH. When the Trx system is overextended during prolonged cellular exposure to oxidative stress or a lack of NADPH production from cellular metabolism, Trx1′s ability to protect target proteins is hindered. More recently, recognition of Trx1′s ability to regulate additional protein PTMs suggest alternative functions that are independent of its disulfide reductase activity; these are discussed later.
FIG. 3.
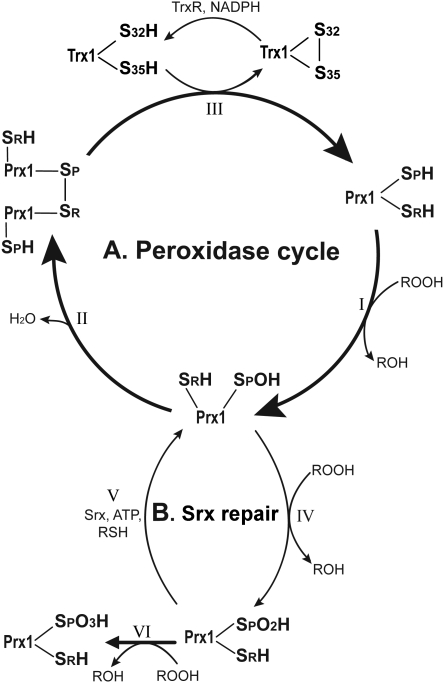
Trx1 regulation of Prx1. (A) Prx1 peroxidase cycle. (I) Prx1 reduces peroxides at the expense of converting its peroxidatic cysteine thiol (SPH) into Prx1-SpOH, which can then form an intermolecular disulfide bond with the resolving cysteine (SRH) of another Prx1 molecule to form a covalent dimer (II). This dimer can then be reduced back to its active form by the Trx1/TrxR system (III). (B) Overoxidation and sulfiredoxin (Srx) repair cycle. Prx1-SpOH can be further oxidized to Prx1-SpO2H (IV), which can either be repaired by Srx (V) or terminally oxidized to Prx1-SpO3H (VI), resulting in inactivation of its peroxidase activity. Adapted from Ref. (273) with permission.
B. Trx regulation of signal transduction and transcription
It is well established that terminal oxidative modification of amino acids is a means of tagging proteins for degradation after prolonged oxidative stress in cells and tissues (44). By comparison, reversible oxidative modification of amino acids has increasingly been recognized as an essential means for regulating protein function (54, 73, 78, 181). Such PTMs may serve as a functional switch, for example, by regulating protein structure in response to cellular stimuli. Therefore, it is important to identify both the proteins that are regulated by redox modifications and the enzymes that regulate such dynamic modifications. Cysteine is among the most studied of redox modified amino acids. The unique electrochemical properties of cysteine thiol render it susceptible to covalent modification by a diverse array of ROS and reactive nitrogen species (RNS). Oxidative covalent modification can regulate an enzyme's function by either affecting its catalytic or allosteric cysteines (185, 226). The selectivity of oxidative PTMs is largely dependent upon the local pH and pKa of a cysteine, dictated by the subdomain or microenvironment that a cysteine resides in. However, there are examples of selective oxidative cysteine modification occurring after specific protein–protein interactions, such as transnitrosylation from one protein nitrosothiol to another; this topic is discussed in depth in the latter part of this review.
Increasing numbers of signal transduction regulators are potentially regulated by Trx1-mediated disulfide reduction (Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/ars), underscoring the important role of Trx1 as a regulatory molecule. For example, protein tyrosine phosphatases contain a HC(X)5R motif in their active site, in which the pKa of Cys is uncharacteristically acidic (41). Such a property renders this family of phosphatases prone to the formation of reactive thiolate ions at the cysteine, which can be reduced by Trx1. For example, phosphatase and tensin homolog (PTEN) is a tumor suppressor that negatively regulates the survival and proliferation of cells via the phosphoinositide 3-kinase–AKT pathway (138). Lee et al. reported a H2O2 oxidation-dependent inhibition of PTEN that occurs via an intramolecular disulfide bond between Cys71 and the active site cysteine, Cys124 (138). This disulfide bond can be reduced more efficiently by the Trx system than by the Grx system. Conversely, the phosphatase activity of PTEN is greatly suppressed on formation of an intermolecular disulfide bond linking Cys212 of the PTEN C2 domain and Cys32 of Trx1 (179). Cdc25 phosphatase is another redox-sensitive phosphatase whose catalytic site Cys473 is susceptible to oxidative transformation to sulfenic acid, and disulfide bond coupling to a vicinal Cys426. Disulfide bond formation prevents Cys473 from being oxidized to more refractory forms such as sulfinic or sulfonic acids (242). It has been reported that this intramolecular disulfide bond can be more readily reduced by the Trx system than by other reductants, including dithiothreitol (DTT) or GSH (242), suggesting that the Trx system plays a very significant role in regulating the function of this phosphatase.
Trx1 modulates the nucleic acid binding and cellular localization properties of several transcription factors and regulators by maintaining the thiol form of cysteines within these oxidatively labile proteins. These include the hypoxia inducible factor (HIF)-1α (58), nuclear factor κB (NF-κB) (172), and histone deacetylase (HDAC) 4 (3). For example, Trx1 enhances the DNA binding activity of NF-κB and its consequent regulation of transcription (263). Qin et al. characterized the binding of the Trx1 catalytic domain to the NF-κB peptide fragment 56FRFRYVCEGPSHG68 by high-resolution nuclear magnetic resonance (217). They reported that the binding was stabilized by a wide range of noncovalent interactions, including hydrogen bonding, and electrostatic and hydrophobic interactions, surrounding either the Trx1 catalytic site or the target NF-κB Cys62.
The disulfide status of Trx1 and its reduction target are important for the regulation of dynamic protein–protein interactions and the functioning of the signaling molecule involved. As a classic example, the reduced form of Trx1 binds to the N-terminal of ASK1, a stress activated mitogen-activated protein kinase kinase kinase, thereby preventing its activation of apoptosis (226). However, under increasingly oxidizing conditions, Trx1 becomes oxidized and loses its ability to form a mixed disulfide and facilitate this protein–protein interaction, resulting in activation of the ASK1 apoptotic signaling pathway (189, 226, 281). In addition to directing protein–protein interactions, Trx1 can regulate signal transduction pathways as part of multiprotein complexes; an example is the HDAC protein complex described below.
C. Trx function at the system level
Several Trx1-modulated pathways may converge and work in concert to achieve a particular biological outcome. For example, we have recently demonstrated that regulation of HDAC4 by Trx1 is important for protection against cardiac hypertrophy (3) (Fig. 4). Within the cardiovascular system, many stress-induced tissue-remodeling events are mediated via the shuttling of class II HDACs, including HDAC4, between the cytosol and the nucleus. In the nucleus, class II HDACs suppress the effects of two master positive regulators of cardiac hypertrophy, nuclear factor of activated T cell, and myocyte enhancer factor 2 (13). Phosphorylation of class II HDACs at specific serines after hypertrophic stimulation results in their export from the nucleus and an attenuation of their transcriptional suppression (176). Studies of HDAC4 mutants indicate that formation of a Cys667-Cys669 disulfide bond by hypertrophic stimuli is an alternate mode of HDAC4 nuclear export, possibly by unmasking the HDAC4 nuclear export signal to exportin 1 (3). We recently employed a transgenic mouse model overexpressing Trx1 to deduce a redox-dependent mechanism for regulating the cellular localization of HDAC4 (3) (Fig. 4). We found that Trx1 induces the expression of a protein chaperone, DnaJb5, which is able to form a complex with Trx1 via thioredoxin-interacting protein (TXNIP, also known as TBP-2). TXNIP is known to inhibit the redox activity of Trx1 (197), but we found Trx1 activity to be preserved when in complex with DnaJb5, and to reduce an intramolecular disulfide bond between DnaJb5 Cys274 and Cys276 (3). In its reduced state, DnaJb5, in complex with TXNIP and Trx1, can stably associate with cytoplasmic HDAC4, facilitating the Trx1 mediated-reduction of the HDAC4 intramolecular disulfide bond between Cys667 and Cys669. HDAC4 is then imported back into the nucleus possibly as a component of this complex, and enabled by the interaction of TXNIP with importin α1 (Fig. 4). Thus, the free thiol status of Cys32 and Cys35 in the catalytic site of Trx1 is crucial for promoting the nuclear translocation of HDAC4 during cardiac stress and for facilitation of its subsequent antihypertrophic gene expression events. Further mutational analysis of HDAC4 also revealed that its redox-mediated translocation acts independently of, and may even override, its phosphorylation status (3).
FIG. 4.
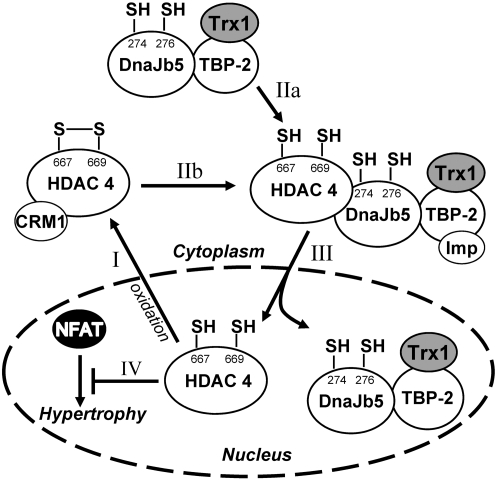
Trx1 regulation of histone deacetylase 4 (HDAC4) in cardiac myocytes. (I) With elevated oxidative stress in cardiac myocytes, nuclear HDAC4 is oxidized and exported to the cytosol by Crm1. (IIa) Trx1 upregulates and reduces DnaJb5 at Cys274 and Cys276, facilitating the formation of a multiprotein complex including DnaJb5, TBP-2 (TXNIP), and HDAC4; (IIb) which allows Trx1 to reduce HDAC4 at Cys667 and Cys669. (III) Reduction of HDAC4 restores its nuclear localization via importin α1, (IV) where it acts to suppress positive mediators of cardiac hypertrophy, such as nuclear factor of activated T cell (NFAT). Adapted from Ref. (3) with permission.
In addition to HDAC4, we have recently reported the discovery of over 50 putative Trx1 reduction targets in mouse heart (75), suggesting that Trx1 coordinates a series of protein networks, including proteins found in sugar and lipid metabolism, the mitochondrial permeability transition pore (MPTP) complex, and myofibrils that are crucial for the maintenance of cardiac function during stress (75). Specifically, we found that Trx1 reduces several key metabolic proteins involved in regulating glycolysis, β-oxidation, the tricarboxylic acid cycle, and oxidative phosphorylation, likely ensuring sufficient production of ATP for muscle contraction and other energy-consuming functions. Trx1 also regulates the MPTP, the creatine-phosphocreatine energy shuttle, and the malate-aspartate shuttle for both metabolite and ATP transport among coordinated protein networks, ensuring the uninterrupted delivery of high energy phosphates to heart muscle cells. Trx1 also appeared to reduce a large number of myofibril components and modulates redox regulator/chaperone proteins such as DnaJA2, DnaJA3, HIF-1α, heat shock 27 kDa protein 7, Prx5, DJ-1, and Grx-related protein 5, coordinating the cellular antistress response. Overall, it appears that Trx1 could regulate molecular pathways at the cellular or systems level to achieve specific biological outcome.
D. Protein oxidation and Trx reduction specificity and mechanism
Reversible oxidative modifications of protein thiols may serve as redox sensors and signal transducers for conveying cellular antistress and/or antiapoptotic responses. Redox-sensitive cysteines usually possess acidic pKa's and are likely to deprotonate under physiological pH, rendering them susceptible to oxidant modification. However, the pKa of a cysteine is influenced by the proximity and identity of surrounding amino acids and other molecules, such as metal ions; hence, not all cysteines are equally susceptible to oxidation (74). Many redox active cysteines lie within conserved motifs, for example, the CXXC oxidoreductase motif and metal-coordinating cysteines in iron-sulfur clusters, but the mechanisms that determine cysteine oxidation specificity are still not well understood. It is likely that the degree of oxidation is determined by the chemical reactivity of each ROS and their spatial and temporal distribution as governed by the metabolic dynamics within cells and tissues [for review see Ref. (51)]. Further, the regulated release of ROS by enzymatic mechanisms, including the action of NADPH oxidases on a time scale of minutes, supports the relevance of oxidative modification in signal transduction (228).
In order for reversible oxidative PTMs to truly act as a means to regulate protein function, similar to that of phosphorylation, oxidoreductase enzymes need to facilitate the specific and timely removal of the oxidative PTM. It is well known that Trx1 does not reduce all protein disulfides in vivo, indicating that a complex degree of specificity may be required before Trx1 can recognize its target protein's disulfide bonds (106). For example, only one disulfide bond between Cys392 and Cys438 of human serum albumin (HSA) is specifically reduced by the Trx system, possibly enabling HSA to act as a blood antioxidant (38). Trx1 is an efficient disulfide reductase: human Trx1 has been shown to reduce mouse ribonucleotide reductase at a rate of 2×105 M−1 s−1 (12), and Drosophila Trx can reduce a GSH peroxidase at a rate of 1.5×106 M−1 s−1 (157). Trx is at least 104 more efficient than DTT at reducing a chloroplast H+-ATPase (234), suggesting that specific three-dimensional protein–protein interaction and catalytic dithiol orientation is essential for determining Trx target specificity. Both the primary sequence and three-dimensional structures of Trx superfamily members can differ, leading to differences in the binding configuration between their catalytic domain and the disulfide domain of their target proteins. The exact mechanism that determines Trx1 recognition and reduction of specific disulfides within select target proteins is unknown. Based on single-molecule atomic force microscopy, Wiita et al. found that Trx catalyzes disulfide reduction via altering the geometry of target disulfides by either reorientation of and shortening of the disulfide, or elongating the disulfide (271). Evolutionary forces may play a role in determining the differences in Trx disulfide reduction specificity and mechanism. Perez-Jimenez et al. have shown that eukaryotic Trxs catalyze disulfide reduction through a single-electron transfer mechanism, whereas bacterial Trxs utilize both SN2 nucleophilic and single-electron transfer mechanisms (213). In addition, they suggested that the depth of the hydrophobic Trx substrate binding groove may contribute to differences in substrate binding and catalysis; a deepened binding groove in eukaryotic Trxs may contribute to its increased substrate binding specificity compared to bacterial Trxs.
Although a large number of plant Trx reduction targets have been identified using proteomic approaches (16, 90, 272, 276), a relatively small number of mammalian proteins has been identified as targets of Trx1 reduction. Among the putative Trx1 reduction targets identified (Supplementary Table S1), a consensus reduction motif has not yet been unequivocally established. Recently, we conducted a proteomics screen for Trx1 reduction targets in a mouse model overexpressing Trx1 in heart, and identified numerous candidate proteins (75). Bioinformatics analysis has enabled us to identify several CXXC motifs surrounding Trx1-sensitive cysteines that are reminiscent of the Trx catalytic site. However, only a relatively small number of target proteins contain these motifs (75). With more sensitive proteomics techniques, a larger number of Trx1 reduction targets can be discovered in different cells and tissues to facilitate the discovery of the precise molecular basis for determining Trx1 reduction specificity.
IV. Proteomics Approaches for the Identification of Trx Reduction Targets
A. Strategies
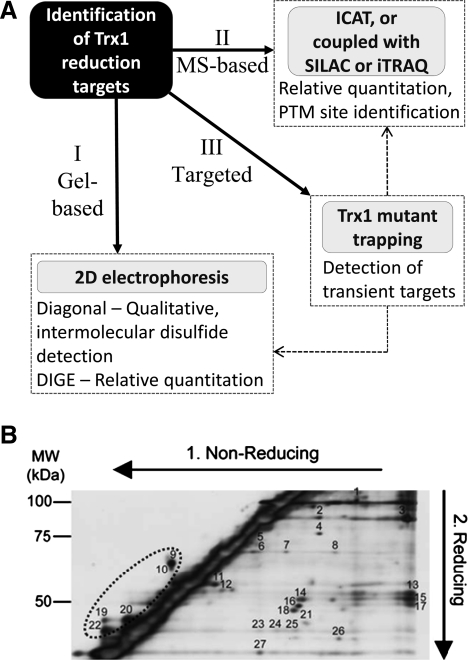
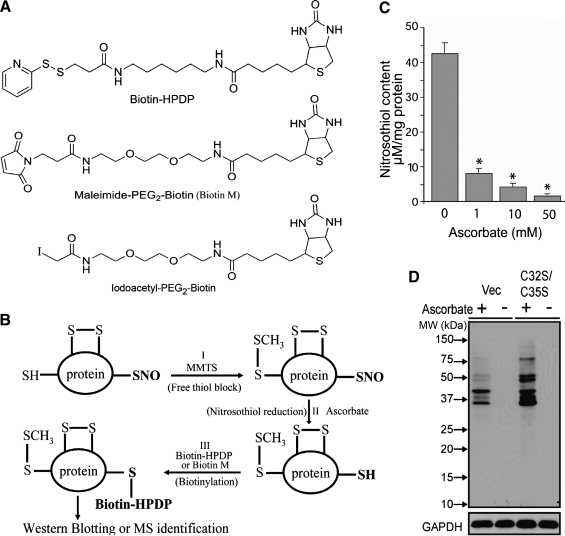
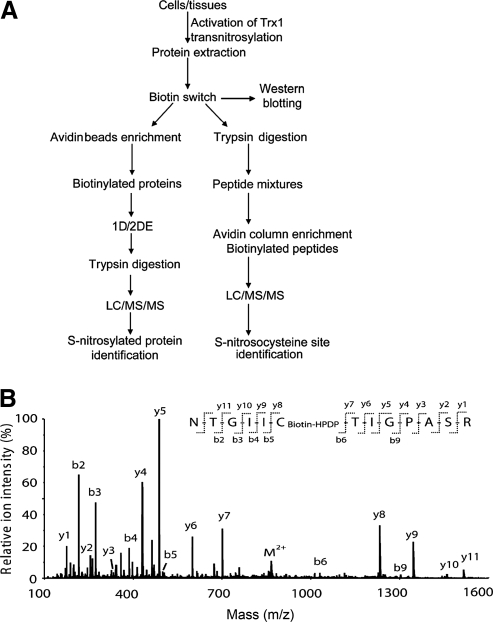
The proteomics strategies for the identification of Trx1 target proteins involve both qualitative and quantitative identification of changes among proteins whose cysteine thiol status is altered as the result of Trx1 expression or activation under different cellular contexts. In its basic form, one can obtain answers on whether a particular protein, or group of proteins isolated from a subcellular organelle contain oxidized cysteines that are amenable to Trx1-mediated reduction. More complex questions, such as to what degree is the oxidative modification reversible and the identity of the modification sites can also be addressed. Since cysteine thiols are highly reactive, they are not usually analyzed directly from total cellular protein extracts; typically, they are alkylated by covalent conjugation to iodoacetamide (IAM), N-ethylmaleimide, methylmethanethiosulfonate (MMTS), or others, to block thiol reactivity toward other oxidants during sample handling and for the purposes of tracking and quantifying redox sensitive-proteins and peptides. Protein alkylation at the earliest step of protein extraction and isolation will prevent experimental artifacts such as thiol/disulfide exchange and artificial oxidation (94). As an alternative approach to the direct analysis of protein thiols, reducing reagents such as DTT, mercaptoethanol or tris(2-carboxyethyl)phosphine, can be used to reduce oxidized cysteines, subsequent to the irreversible alkylation of total cellular free thiols. After the reduction of disulfide bonds, newly freed thiols can be labeled by a different set of alkylating reagents to quantify changes of cysteines engaged in disulfides or other oxidized forms under different cellular contexts. These strategies can be implemented to identify Trx1 reduction targets with at least two specific proteomics goals. The first goal is to identify those proteins whose cysteine free thiol levels are elevated in systems in which Trx1 is either activated or overexpressed, compared to systems in which Trx1 is inactive (e.g., due to TrxR inhibition or TXNIP binding), downregulated (e.g., siRNA knockdown) or in which there is a dominant negative Trx1 mutant (e.g., Trx1C32S/C35S) expression. The second goal is to identify those proteins whose levels of disulfide bonds are decreased within similar comparative schemes. Following the strategies above, the proteins isolated will need to be identified by different types of proteomics techniques for the identification of proteins sensitive to Trx1 reduction, mapping the responsive cysteine(s), and possibly quantifying the degree of Trx1 reduction in different biological scenarios. The proteomics techniques that are commonly used for the identification of Trx1 reduction targets include gel-based, MS-based, and targeted approaches (Fig. 5A).
FIG. 5.
Redox proteomic methodologies. (A) Flow diagram illustrating the proteomic methodologies available for the identification of Trx1 reduction targets. (I) Conventional two-dimensional gel electrophoresis (2DE) methods such as diagonal electrophoresis and DIGE are gradually being superceded by (II) mass spectrometry (MS)-based methods, although their combination often provides additional information. (III) The targeted Trx1 mutant trapping approach is effective for isolating transient reduction complexes, but needs to be coupled (dotted arrows) with gel-based or MS-based methodologies to identify and even quantify targets. (B) Diagonal gel electrophoresis. To identify proteins that form intra- or intermolecular disulfide bonds, cytosolic proteins are sequentially resolved by nonreducing followed by reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The resultant silver-stained gel reveals a prominent diagonal line, which represents the majority of proteins that do not form disulfide bonds. Proteins that form intermolecular disulfide bonds exhibit a slower electrophoretic mobility under nonreducing conditions in the first dimension and therefore appear as spots to the right of the diagonal line. Spots to the left of the diagonal line (in dotted oval) represent proteins that have a faster electrophoretic mobility under non-reducing conditions, possibly due to intramolecular disulfide bonding. Adapted from Ref. (50) with permission.
B. Gel-based proteomics approaches
A gel-based proteomics approach represents the most direct way to identify proteins whose cysteine thiol status changes as the result of Trx1 activation or downregulation. No attempts are undertaken to enrich affected proteins away from the bulk unaffected cellular proteins. Several gel-based techniques have been developed to identify redox-sensitive cysteines within proteins (19, 62, 126, 278). Among them diagonal electrophoresis (29) is designed to identify mainly disulfide-bond linked proteins based on the unique gel migration patterns of proteins with intra- and inter-molecular disulfide bonds (Fig. 5B). This approach follows sequential nonreducing and reducing gel electrophoresis separations. With this method, the proteins are first separated via non-reducing gels, in which all the proteins and disulfide-linked protein complexes are separated roughly based on size. The gel lanes are then laid onto a second dimension sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and the proteins are separated again, this time under reducing conditions. Disulfide-linked proteins will migrate to size regions that are lower than the parent covalent complex, resulting in their identification. Taking the identification of Trx1 reduction targets as an example, fewer mixed disulfides are likely to be present in samples in which Trx1 is upregulated than those in which Trx1 is downregulated. Therefore, fewer proteins will migrate in the lower mass gel regions, below the diagonal protein bands. Proteins that migrate below their mixed disulfide complex size regions in Trx downregulated cells are likely candidates for Trx1-mediated disulfide reduction. If only one protein is detected from a particular protein complex, it is likely that this protein may form oligomers via one or more disulfide bonds, which are reducible by Trx1. It is also possible that one protein can form Trx1-reducible disulfides with several proteins; when this occurs, this protein can be detected beneath different mixed disulfide complexes. Yano et al. have successfully used this method for the identification of in vitro Trx1 disulfide reduction targets in peanut seed (278). At least 20 peanut seed proteins were found to be reduction targets of Trx1, and of the 5 identified, 1 is connected to seed maturation and 3 are known allergens, raising the possibility that Trx1 may influence seed germination and allergen activity. Diagonal electrophoresis may not be as effective for detecting intra-molecular Trx reduction targets as the electrophoresis mobility differences between the disulfide and dithiol versions of the same protein are often slight (Fig. 5B). Among other limitations of the diagonal electrophoresis approach are its limited resolution and detection sensitivity. In fact, few examples of its application on actual mammalian Trx1 reduction targets are present in the literature, and traditional isoelectric focusing coupled with SDS-PAGE may be complementary.
There are several other two-dimensional gel electrophoresis (2DE)-based methods for the identification of Trx1 reduction targets based on the employment of cysteine-specific fluorochromes for tracking changes in proteins whose cysteine free thiol levels are differentially regulated within complex protein mixtures (19, 62, 278). This method can be configured to carry out multiplexed comparative experiments in a single gel to minimize the gel-to-gel variation commonly encountered during quantitative proteomics experiments. The saturation-labeling difference in gel electrophoresis (DIGE) method (GE Healthcare) employs a pair of fluorescent CyDyes to specifically label free thiols in multiple samples. For example, saturation Cy3 maleimide (Cy3m) and Cy5m fluorescent DIGE dyes can be conjugated efficiently via maleimide derivatives to free thiols from two different samples (274). Mixtures (typically equal amounts) of labeled proteins can then be analyzed in the same gel and quantitative differences can be ascertained from the fluorescent images obtained using the different fluorescent spectral properties of each fluorophore. Using one dye as the internal standard within the DIGE experimental design enables accurate quantification comparisons across different gels. The saturation DIGE technique has been used to measure the redox status of proteins, including the detection of H2O2-sensitive proteins (74, 112). With the DIGE approach, we detected over 1000 proteins and identified 26 unique proteins that were sensitive to oxidation by H2O2 (74). Among the proteins identified, several are represented by more than one 2DE gel spot. For example, malate dehydrogenase 2DE gel spots were shifted to more acidic forms as a result of H2O2 treatment, indicating the possible occurrence of acidic PTMs such as the oxidation of cysteine thiols to sulfenic, sulfinic, or sulfonic acids (74).
DIGE methods can be implemented to identify Trx1 reduction targets in two independent, yet related, fashions. For the forward labeling approach, control protein thiols can be labeled using Cy3m as the internal standard, whereas Trx1-upregulated or -downregulated cell proteins can be labeled using Cy5m. Cy5m-labeled proteins from Trx1-upregulated systems will contain more cysteine thiols than the Cy3m-labeled control proteins. As all cysteine thiol levels are compared with those of control proteins; it is possible to compare multiple samples thereby enhancing the capabilities and confidence of the statistical analysis. For example, the relative thiol levels in three protein extracts from Trx1-upregulated cells can be compared with three Trx1-downregulated cells, when calibrated against the same control cell extracts. An alternative reverse DIGE-labeling approach will first block all the free cysteine thiols using alkylating reagent such as IAM. The disulfides are then reduced with DTT, and the newly freed cysteine thiols can then be conjugated covalently to the Cy3m or Cy5m reagents. With this method one can reveal complementary changes of cysteines engaged in disulfide bonds. It is anticipated that fewer disulfide-linked cysteines will be present in Trx1-upregulated systems, whereas the opposite is true for Trx1-downregulated systems. However, it is worth being cautious when interpretating 2DE data. Proteins are typically resolved in 2DE according to their charge and size, and many proteins are present in a typical gel as several 2DE spots (in part due to the oxidation of cysteine thiols to sulfenic, sulfinic, or sulfonic acid forms). A change in the intensity of a protein spot correlates only with a particular form of the protein or with its redox modification status, and should not be generalized to apply to the overall redox level or activity of this protein. The DIGE technique has been widely applied for redox proteomics studies (74), but its relatively high cost and inefficiency at detecting low abundant proteins limits its use for the identification of Trx1 reduction targets, especially under in vivo conditions.
C. MS-based proteomics approaches
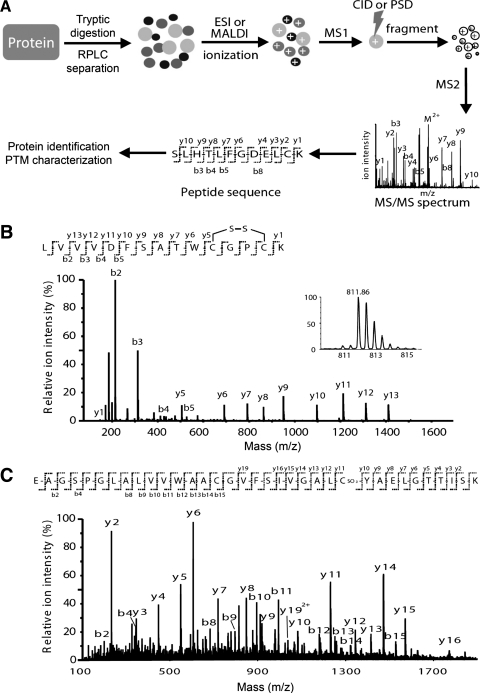
Although Trx1-sensitive proteins can be identified using the gel-based approaches described above, these approaches do not usually provide information regarding either the degree or the specific sites of Trx1 reduction. In addition, even though gel-based methods have proven to be effective in identifying redox-sensitive cysteine-containing proteins and their modified protein isoforms, detection sensitivity is typically less than that of the gel-free MS-based shotgun approaches. Therefore, the focal point of proteomics has gradually migrated toward a gel-free MS-based approach, including redox proteomics. The most common workflow for MS-based analysis of redox-sensitive or modified peptides uses stable isotope labeling of proteins, followed by protease digestion (typically trypsin) and identification and quantification of labeled peptides by liquid chromatography separation followed by tandem mass spectrometry analysis (LC/MS/MS; Fig. 6). Changes in the protein levels can then be inferred from the peptide quantification data. Stable-isotope containing reagents and methods, including isotope-coded affinity tags (ICAT) (212), stable isotope labeling by amino acids in cell culture (SILAC) (202) and isobaric tags for relative and absolute quantification (iTRAQ) (222), can be used to quantify specific cysteine modifications within protein targets of Trx1-mediated reduction (Figs. 7 and 8). We now discuss the application of these techniques.
FIG. 6.
MS/MS for peptide sequencing and identification of oxidative cysteine modifications. Proteins are typically digested with trypsin and the resulting peptides are separated by reversed phase liquid chromatography (RPLC) and detected by MS. (A) Peptides are ionized by either matrix-assisted laser desorption ionization (MALDI) or electrospray ionization techniques and analyzed by tandem mass spectrometers (MS/MS), within which a peptide ion can be fragmented by collision-induced dissociation (CID) or postsource decay (PSD) to generate fragment ion (MS2) information for peptide sequencing. Amino acid sequences and PTMs can be deduced from continuous series of either N-terminus (b-series) or C-terminus (y-series) fragment ions. (B) An MS/MS spectrum of the human Trx1 tryptic peptide 22LVVVDFSATWC32GPC35K36 containing an intramolecular Cys32 and Cys35 disulfide bond. The doubly charged peptide ion of m/z 811.86 could be detected (inset). An intramolecular disulfide bond between Cys32 and Cys35 was also detected by analyzing the MS/MS spectrum of the peptide. C-terminal fragments y5 to y13 revealed a mass reduction of 2 amu from the fully reduced sequence, suggesting a disulfide bond was formed between Cys32 and Cys35. Reprinted from Ref. (267) with permission. (C) An MS/MS spectrum of a human L-type amino acid transporter 1-like protein peptide 78EAGSPGLALVVWAACGVFSIVGALC102YAELGTTISK112 with a Cys102-SO3H modification in the peptide (C.W. and T.L., unpublished data).
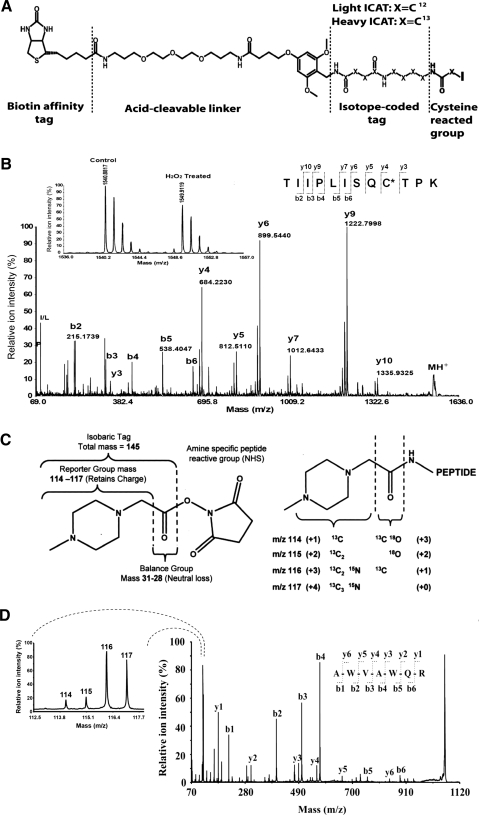
FIG. 7.
Stable isotope labeling reagents for quantitative proteomics analysis. Comparison of isotope-coded affinity tag (ICAT) redox peptide changes with iTRAQ protein expression changes allows the identification of peptides that truly are sensitive to redox modification, instead of protein expression level changes. (A) ICAT reagent structure is composed of four parts (from left to right): (1) biotin affinity tag; (2) acid-cleavable linker; (3) isotope-coded tag (C13 or C12), and (4) cysteine reactive group. This reagent can be used for cysteine thiol quantification. (B) An example spectrum of an ICAT-labeled peptide 204TIIPLISQCTPK215 from malate dehydrogenase. The inset MS spectrum demonstrates a ∼27% decrease of free Cys112 thiol in this peptide after 500 μM H2O2 treatment. Reprinted from Ref. (74). (C) Four-plex iTRAQ reagent structures. Stable isotopes (C13, N15, and O18) are differentially incorporated at the reporter region, resulting in reporter groups with masses of 114, 115, 116 or 117, which can be detected using MS/MS. The quantification of peptides is determined by the relative areas under these peaks in a MS/MS spectrum. The reporter groups are linked to a balance group with a mass of 31, 30, 29 or 28 to complete the isobaric tags, all of which have the same mass addition of 145 onto an N-termini- or Lys-labeled peptide. The isobaric tags are conjugated to an amine specific reactive group (NHS) for covalent labeling of primary amines of peptide N-termini and Lys side chains. Reprinted from Ref. (222). (D) An example spectrum of iTRAQ-labeled rat lysozyme 2 peptide 126AWVAWQR132. The spectrum on the left demonstrates expression changes of this peptide among the four experimental conditions. The fragmentation pattern on the right permits sequencing of the peptide (T.L. unpublished data).
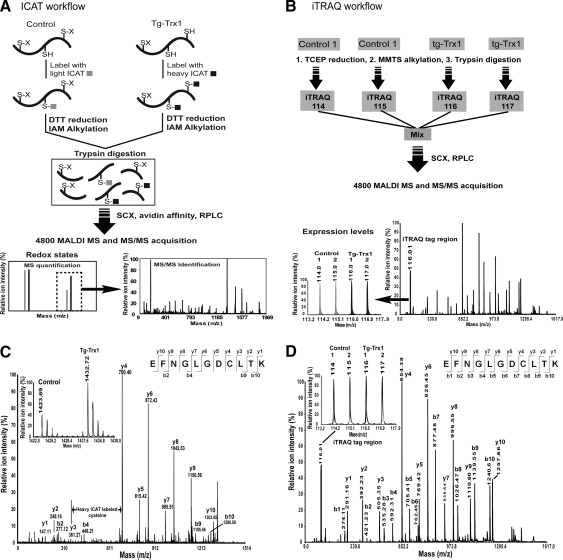
FIG. 8.
Identification of Trx1 sensitive redox active cysteines in Trx1 overexpressing transgenic mice heart tissues by comparing quantitative ICAT and iTRAQ proteomics data. (A) ICAT workflow. Protein thiols were first labeled with either the light ICAT (control) or heavy ICAT (Trx1-overexpressing tissue) reagent. Subsequently, disulfides were reduced by dithiothreitol (DTT) and alkylated with iodoacetamide (IAM). The labeled proteins were mixed together, digested with trypsin and separated sequentially using SCX chromatography, avidin affinity chromatography and RPLC. ICAT-labeled peptides were identified and quantified by MALDI-TOF MS/MS. Peptides containing Trx1-reduced cysteines had an ICAT H/L ratio larger than one, and can be identified by subsequent MS/MS analysis. (B) iTRAQ workflow. Proteins were first reduced by tris(2-carboxyethyl)phosphine, alkylated with methylmethanethiosulfonate (MMTS) and digested with trypsin. The peptide groups were labeled individually using iTRAQ reagent 114 and 115 for control samples and 116, 117 for the Trx1-overexpressing samples. The labeled samples were mixed and cleaned up using both SCX chromatography and RPLC and identified and quantified by MALDI-TOF MS and MS/MS. (C) A spectrum of ICAT-labeled 153EFNGLGDCLTK163 from ADP/ATP translocase 1 (ANT1). Trx1-reducible cysteine was identified by the mass difference of y3 and y4 fragment ions. Control samples (labeled with ICAT light chain, m/z 1423.69) had less peak area than Tg-Trx1 samples (labeled with ICAT heavy chain, m/z 1432.72), indicating that Cys160 in this peptide was 150% more reduced in Trx1 overexpressing tissues (inset). A Cys160 linked disulfide is a likely candidate of Trx1-mediated reduction. (D) A spectrum of the iTRAQ analysis of the same ANT1 peptide, showing that protein expression levels were not affected by Trx1 overexpression. Control samples (labeled by 114 and 115) had similar peak areas compared to Tg-Trx1 peptide (labeled by 116 and 117) (inset). Reprinted from Ref. (75) with permission.
For the identification of Trx1 reduction targets, cysteine-containing peptides are typically separated from other peptides using affinity techniques. For example, Aebersold and colleagues have developed the ICAT technique for the multiplexing proteomics quantification of proteins using stable isotope labeling of cysteines (Fig. 7A, B) (87). Cysteine-containing peptides are enriched, quantified, and identified on the mass spectrometer, resulting in more reliable and sensitive protein quantification than 2DE (87). Each ICAT reagent consists of a thiol-reactive group, an isotope-coded light or heavy linker, an acid linker and a biotin segment to facilitate the capture of cysteine-containing peptides by avidin affinity beads (Fig. 7A). First, the cysteine reactive head group uses the IAM moiety to react covalently with free thiols in the protein. Second, an isotope-coded segment contains nine different C12 (light) or C13 (heavy) carbons for the relative quantification of proteins using MS. Third, the acid-cleavable linker enables the shredding of the reagent after avidin affinity enrichment of ICAT-labeled peptides, resulting in more effective MS/MS fragmentation patterns for both sequence identification and the localization of the reactive cysteine thiols. The fourth component of an ICAT reagent is a biotin derivative that is used for affinity enrichment of the ICAT-labeled peptides, enhancing detection sensitivity and the specificity of the method. In contrast to the orthodox ICAT technique, designed for global protein expression analysis, for quantification of free cysteine thiol-containing peptides, no disulfide reduction step is performed before labeling with ICAT so that the native protein thiol redox states are preserved. For each redox ICAT experiment, the free cysteine thiols within the proteins are first labeled with either light or heavy versions of the ICAT reagents. Then, they are mixed together for reduction, alkylation, and proteolytic digestion. The disulfides can be reduced using DTT, and the resultant free thiols blocked using non-ICAT alkylating reagents, opening the peptide chains for proteolysis. After trypsin digestion, peptides are affinity-enriched to recover the biotinylated peptides and separated using several LC purification steps. The peptides are relatively quantified by comparing the LC elution profile of the ICAT-conjugated peptide ion signals obtained in a mass spectrometer (Fig. 7B, see example of quantification in inset). Then, differentially oxidized peptides are identified using MS/MS (Fig. 7B, see example MS/MS spectrum for peptide sequencing). Svensson and coworkers have successfully discovered over 100 in vitro Trx reduction targets in plants using the ICAT method (90), producing the largest number of plant Trx-targeted proteins to date. Advantages of the ICAT technique include the ability to obtain the extent of redox change for each peptide and the simultaneous direct identification of the redox-reactive site. It is common to observe several peptides from the same protein with a different degree of susceptibility to oxidants or reduction by Trx. For example, with ICAT, we were able to identify 78 putative Trx1 reductive sites in 55 proteins from mouse heart, with their reduced cysteine content increasing by 20%–400% (Supplementary Table S1) (75). These proteins belonged to different protein functional networks not previously implicated in Trx1 regulation, including the creatine-phosphocreatine shuttle, the MPTP complex, and the cardiac contractile apparatus. It is conceivable that by coupling the ICAT method to reduction systems (e.g., Trx vs. Grx) with different reduction potentials toward differing cysteine thiol oxidative modifications, one could identify and quantify important PTM sites and determine the extent of disulfide reduction by each reductive system.
In addition to thiol-specific MS-based methods, other nonthiol MS-based methods may be used to obtain useful information for the identification of genuine Trx1 protein reduction targets. Changes in the level of ICAT-conjugated peptides can be the result of changes in the cysteine thiol status due to redox modulation or to changes in protein expression levels. It is therefore necessary to distinguish between Trx1-induced protein expression and Trx1-mediated protein reduction. To accomplish this, we may need to compare the degree of ICAT-peptide changes to the overall changes in protein level. ICAT is a cysteine-specific, isotope-labeling method that quantifies cysteine-free thiols at the MS level, whereas the iTRAQ method quantifies global protein expression. iTRAQ reagents are used for the isobaric peptide quantification of primary amines within peptides at the MS/MS level (Fig. 7C, D). Using the iTRAQ approach, up to eight samples can be digested by trypsin and labeled with one of eight different iTRAQ reagents, depending on the desired experimental design. The iTRAQ-labeled peptides are combined and subjected to either strong cation exchange fractionation or to isoelectric focusing separation. The resulting peptide fractions can be quantified and identified using LC/MS/MS on either matrix-assisted laser desorption ionization or electrospray ionization tandem mass spectrometers. We have employed dual ICAT and iTRAQ strategies to detect an increase of specific cardiac protein thiols due to protein disulfide reduction, rather than a Trx1-mediated increase of gene/protein expression, in a transgenic mouse model overexpressing Trx1 (Fig. 8). The ICAT redox analysis results were compared with the iTRAQ expression analysis, and 55 Trx1 reductive targets were found to increase their cysteine thiol levels in Trx1-expressing mouse hearts (Supplementary Table S1) (75).
ICAT methods can be configured for different experimental strategies. In a similar manner to that described for the DIGE method, forward and reverse labeling strategies can be applied to the ICAT approach. This allows the identification of increases in disulfide-linked cysteines in Trx1-downregulated systems. To compare relative protein oxidative states in two different cells or tissues, a modified ICAT procedure, OxICAT, was developed by the Jakob group for estimation of disulfide-formed cysteines within proteins in a single cell type, using light ICAT reagent to label free thiols and subsequently heavy ICAT reagent to label DTT-reducible cysteine thiols (141).
In addition to iTRAQ, SILAC is another promising means of global protein expression analysis that can play a role in redox proteomics studies. With this method, stable heavy isotope-labeled amino acids, typically arginine and lysine, are substituted for their normal counterparts in cell culture. The heavy isotope amino acids are then incorporated into newly synthesized proteins in rapidly dividing cells. Heavy isotope-labeled proteins are typically mixed with unlabeled protein from normal cell culture, trypsin digested, and peptides containing heavy isotope at their C-terminus distinguished from unlabeled peptides by LC/MS/MS analysis. The relative MS ion abundance of SILAC-labeled and -unlabeled peptide approximates the relative abundance of corresponding protein in the two different cell cultures. This technique can easily be modified for redox proteomics. For example, a reductase can be inhibited in a SILAC-labeled culture; then, after combining with control cell extract, cysteine-containing proteins can be modified by a thiol-specific alkylating reagent. After tryptic digestion, the relative levels of thiol-containing peptides can be separated from other peptides, and hence the redox status of proteins between the two cell cultures can be measured by LC/MS/MS.
D. Targeted proteomics approaches
With global proteomics approaches, it is very difficult to identify low abundant Trx1 reduction targets, such as phosphatases and transcription factors, without their enrichment and separation from other cellular proteins. One of the most effective subproteomics approaches includes the expression of inducible Trx1C35S mutant in cells whose endogenous Trx1 levels have been knocked down, thereby maximizing the chances of capturing low abundant Trx1 targeting signaling proteins (235). The concept behind the mutant Trx1 affinity approach is based on an understanding of the reduction mechanism of Trx1. Cys32 of human Trx1 initiates attack on a disulfide-bonded cysteine to form a mixed disulfide, which transiently connects Trx1 covalently to one of the cysteines within its disulfide bond reduction target (122). To resolve this reaction intermediate, Cys35 donates its reducing equivalent to the target protein cysteine and forms a disulfide bond with Cys32. Typically, this disulfide exchange reaction is very rapid, precluding the capture of the mixed disulfide intermediates for analysis by proteomics methods. Therefore, an affinity chromatography method has been developed using Trx1C35S mutant as molecular bait to capture target proteins via the formation of an intermolecular disulfide-linked intermediate. These trapped Trx1 targets can be recovered by reductants such as DTT (15, 16, 187, 276). The resulting proteins can then be separated using either 2DE or LC/MS/MS techniques (Fig. 5A). These approaches have successfully identified Trx1 reduction targets in mammalian systems (235) and in several nonmammalian species, including plants (164) and yeast (261). However, the trapping technique has not been particularly effective for large scale identification of Trx1 targets in mammalian cells and tissues. Proteins that are trapped by this method are typically considered to contain disulfides, but Trx1 can also reduce cysteine sulfenic acids and nitrosylated residues, so the identity of the PTM should be verified (Fig. 6B, C).
E. Strengths, limitations and complementarity
In principle, the mutant Trx1 affinity approach can be used to trap and facilitate the identification of low abundant Trx1 reduction targets by 2DE or LC/MS/MS. In practice, when in the presence of Trx1 targets of several orders of magnitude higher abundance, this approach often fails to detect trace amounts of transcription factors and signaling proteins. For example, when we attempted to isolate mutant Trx1-trapped targets in HeLa cells, only highly abundant Prx1 and HSP90 were detected by 1DE or 2DE coupled with LC/MS/MS, whereas lower abundant targets could only be detected by Western blotting (C.W., unpublished data). To overcome this technical problem, specific experimental designs are needed. For example, by expressing nuclear targeted mutant Trx1, more transcription factors may be enriched. Alternatively, similar to the success of disease proteomics biomarker discovery by immunodepletion of abundant serum proteins, immunodepletion of Prx variants and other highly abundant Trx1 targets from affinity captured proteins, should increase the representation of peptides from lower abundant targets. More promisingly, developments in high sensitivity LC/MS/MS instrumentations will also likely contribute to the discovery of novel and dynamic Trx1 targets in cells and tissues.
In addition to their advantages over gel-based approaches, such as the precise mapping of modification sites, MS-based shotgun approaches (including ICAT) contain intrinsic limitations that need to be understood for correct interpretation of the experimental data. Homologous proteins often share identical sequences and domains, so when changes in protein thiol levels are quantified within a peptide, it is important to consider all proteins that contain the same sequence. At present, the precise identification of intra- and intermolecular disulfide bonds is rather labor-intensive, and is usually done in vitro using recombinant proteins. Not only does sequencing of disulfide linked peptides require MS/MS conditions that are different from those used in fragmenting nondisulfide-linked peptides (180), but also advanced software tools are required for the accurate identification of disulfide-linked peptides (84). Consequently, the global identification and quantification of intramolecular disulfide bonds is not widely reported.
Gel- and MS-based approaches each carry specific advantages and can generate complementary information in large-scale redox proteomics experiments. We have recently used both the DIGE and ICAT methods to screen for heart proteins that are sensitive to H2O2 oxidation, and we found that these two methods were complementary in terms of the number of low-abundance redox-sensitive proteins found (75). Consequently, both methods should be employed in redox proteomics studies to comprehensively identify Trx1-sensitive proteins and their reactive cysteine residues.
V. Nitrosylation
Nitrosylation is the covalent addition of a nitric oxide (NO) moiety onto a cysteine thiol. It is a dynamic PTM for the regulation of protein functions (245). NO, which is generated by nitric oxide synthases (NOSs), cannot nitrosylate cysteines directly (124, 246). Instead, biological S-nitrosylation can take place by transnitrosylation, which involves the transfer of NO from mainly low-molecular-weight S-nitrosothiols (RSNOs), such as GSNO or S-nitrosocysteine (45), onto a cysteine thiol (72). S-nitrosylation can also occur by reaction with RNS such as peroxynitrite (see below). Increasing numbers of proteins have been reported to be regulated by reversible nitrosylation, including the caspase family of cysteine proteases (144). In addition, a wide array of cell surface receptors and their downstream targets and modulators are also modulated by nitrosylation (71). It is apparent that transnitrosylation or denitrosylation (i.e., the removal of NO from SNO-proteins) are important regulatory mechanisms of normal physiology. Dysregulation of either transnitrosylation or denitrosylation leads to cellular dysfunction, especially in modulation of the immune system and inflammatory response, and in lung, cardiovascular, hormonal, and central nervous system functions (32, 33, 73, 161). Recent studies have demonstrated that the Trx system may be involved in regulating the nitrosylation status of proteins in different systems (21, 184, 276). We first provide an overview of protein nitrosylation followed by a discussion on the roles of Trx in mediating specific nitrosylation reactions as well as proteomics techniques for the identification of Trx1 transnitrosylation and denitrosylation targets.
A. S-Nitrosylation and signal transduction
One of the initial challenges in the field was to determine whether S-nitrosylation was more than a pathophysiological reflection of NO produced in excess (of nitrosative stress). Mounting evidence now supports S-nitrosylation as a PTM that has physiological relevance. The main receptor for NO is soluble guanylyl cyclase (sGC), which produces cyclic guanosine monophosphate (cGMP) on binding of NO to its heme (9), but to explain the plethora of functions exerted by NO, one has to consider mechanisms other than the production of cGMP. S-nitrosylation is now known to modulate the function of several proteins engaged in many signaling pathways. Well-described examples include the modulation of nuclear receptors, ligand- and voltage-gated ion channels, G-protein-coupled receptors (GPCR), caspases, and GTPases (such as Ras or dynamin) [for reviews see Refs. (73, 81, 149)]. Modulation of transcription by S-nitrosylation can take place at several levels and involves a diversity of targets, including NF-κB (104, 168), HIF-1α (143), HDAC2 (199), and glyceraldehyde 3′-phosphate dehydrogenase (GAPDH) (236), among many others. Several components of the NO-cGMP signaling pathway are themselves modulated by S-nitrosylation, including arginase1 (which degrades the substrate of NOS) (227), endothelial NOS (eNOS) (218), and sGC (see below). Other features that are shared between S-nitrosylation and signal transduction mechanisms include (a) the reversibility of the process, whether through changes in the redox state of the cells or through enzymatic trans- or denitrosylation by the Trx system, and (b) the specificity of cysteine S-nitrosylation in response to a stimulus, that is, an increase in NO production (54).
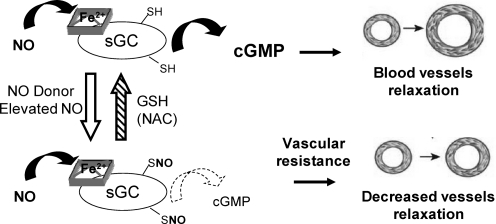
S-nitrosylation modulates protein functions by several mechanisms, including induction of conformational change, which in turn can affect the binding of a ligand. For example, the N-methyl-D-aspartic acid (NMDA) receptor affects neuronal development, synaptic plasticity, and memory and has three pairs of cysteine residues that can be modified by NO and Zn2+ (149). S-nitrosylation of Cys399 in the NMDA receptor, through Zn2+ coordination of its regulatory domain, leads to a slower off-rate of glutamate thereby enhancing receptor binding (149). S-nitrosylation can also modulate protein–protein interactions [as in the heat shock protein 90 (Hsp90)/eNOS and the glutamate receptor 2/N-ethylmaleimide-sensitive factor complexes (111, 170)], or promote cellular trafficking [as in GPCR internalization via the S-nitrosylation of β-arrestin and dynamin (203, 266)]. S-nitrosylation can also directly affect the activity of an enzyme. The activity of sGC is stimulated several hundred-fold by the binding of NO to its heme prosthetic, resulting in increased production of cGMP (Fig. 9). However, after prolonged or increased exposure to NO or RSNOs, the response of sGC to NO is decreased by more than 50% (230). One of the cysteines involved in the desensitization (βCys122) is located in the heme domain of sGC. Structural modeling of the heme domain with βCys122-SNO predicted that S-nitrosylation creates steric hindrance that could destabilize the pocket (230). The NO-cGMP pathway is involved in vasorelaxation, inhibition of platelet aggregation, and synaptic plasticity. Physiologically, S-nitrosylation of sGC could be one mechanism by which cGMP-dependent NO signaling is controlled and its specificity maintained. Pathophysiologically, an excess of NO can generate vascular resistance (constriction of blood vessels) and nitrate tolerance, that is, a loss of vascular reactivity after prolonged exposure to NO or nitroglycerin (Fig. 9) (230). Interestingly, treatment with N-acetyl-cysteine, a precursor of GSH synthesis, can restore the hemodynamic state in patients that have developed nitrate tolerance (205). And it is now thought that N-acetyl-cysteine can reverse S-nitrosylation of sGC (231).
FIG. 9.
The physiological and pathophysiological role of S-nitrosylation in nitric oxide (NO)-cGMP signaling in the vasculature. Normally, NO stimulates soluble guanylyl cyclase (sGC) to produce cGMP to signal for blood vessel relaxation. S-nitrosylation of sGC modulates vascular reactivity. Desensitization of sGC via S-nitrosylation could represent a potential mechanism of vascular NO resistance in oxidative cardiovascular disease.
Specific protein nitrosylation targets susceptible to hyper- or hypo-S-nitrosylation, such as caspases 1–9, Ras, HIF-1α, X-linked inhibitor of apoptosis, NF-κB, p65, p53, Bcl-2, Prx2, and insulin receptor substrate 1, are directly implicated in various mammalian pathophysiologies (11, 25, 73, 114, 143, 159, 162, 174, 192). For example, S-nitrosylated HIF-1α and Ras have been implicated in tumor radioresistance and maintenance, respectively (114, 143). Under normoxic conditions, HIF-1α is regulated by the oxygen-dependent hydroxylation of its proline residues 402 and 564 by prolyl-4-hydroxylase (31, 59). After exposure to radiation, S-nitrosylation of HIF-1α by inducible NOS (iNOS) at Cys533, located in its oxygen-dependent degradation domain, results in stabilization of HIF-1α in macrophages (143). Interestingly, NOS inhibitors coupled with radiotherapy result in a synergistic reduction in tumor growth rates in mouse models of breast cancer (143), and overexpression of iNOS in cancer tissues is correlated with a poor patient prognosis (57). Taken together, these results indicate that HIF-1α is regulated by S-nitrosylation, and this regulation plays an important role in tumor radiosensitivity and tumor growth.
Overactivation of the Ras family of guanine nucleotide binding proteins often leads to oncogenesis and cancer, and Ras signaling events are intricately linked to PTMs, such as S-glutathionylation (2, 215) or S-nitrosylation, at Cys118. Phosphorylated eNOS can maintain tumor growth by activating Ras through S-nitrosylation at Cys118 (135, 147). Indeed, AKT phosphorylation of eNOS at Ser1177 is required for tumor maintenance, as pharmacological inhibition of AKT signaling or the ectopic expression of eNOSS1177A mutant results in a decreased number of tumor cells (147). Overexpression of H-RasC118S mutant or knocking down eNOS all result in a loss of H-Ras nitrosylation and related tumor maintenance (147). Thus, AKT activation of eNOS promotes nitrosylation and activation of wild-type Ras proteins, thereby maintaining tumor growth.
B. Regulation of the specificity of nitrosylation
S-nitrosylation and denitrosylation can calibrate protein functions reversibly at specific cysteines. These processes are dynamic, site-specific, and spatially and temporally coupled with either extracellular or intracellular stimuli (54). Mechanisms that determine the specificity of S-nitrosylation may include the availability of the types and levels of NO donors, the microenvironment of the protein thiols, the cellular redox environment, and the presence of transnitrosylases and denitrosylases.
As mentioned earlier, cysteines can be S-nitrosylated by RNS such as peroxynitrite (OONO−) or dinitrogen trioxide (N2O3). Peroxynitrite is formed by reaction between superoxide (O2−), generated by activated macrophages, and NO, whereas reaction of peroxynitrite with NO may lead to the formation of N2O3 (20, 204). Recently, it was proposed that N2O3 could also be generated from a nitrite reductase/anhydrase redox cycle of hemoglobin (18). Additional in vivo nitrosylating agents have also been proposed; they include nitrosocomplexes of iron (259) and copper (248). N2O3 hydrolyzes rapidly to nitrite (t1/2=1 ms) (195); therefore, proteins that are nitrosylated by N2O3 or other NO-related species are likely to be present at relatively high concentrations. Certain conditions might favor these reactions in vivo, such as chronic inflammation, which increases the production of NO (220) or proximity to an NO source. Indeed, the targets of S-nitrosylation are often engaged in a complex with the various NOS molecules (100). It is likely that proximity and/or association with neuronal NOS is key to S-nitrosylation of the NMDA receptors and Ras-like G-protein Dexras1 (66). Similarly, one can propose that S-nitrosylation of Hsp90, Hsp70, and sGC is facilitated by their association with eNOS (170, 230, 260). NOSs can regulate S-nitrosylation specificity among select proteins via ligand-stimulated induction, protein complex formation, or direct protein–protein interaction [for review see Ref. (54)]. Further, both eNOS and iNOS are regulated by dynamic nitrosylation affecting either their subcellular targeting or activity (61, 183), and perhaps their ability to facilitate nitrosylation. Functionally, Erwin et al. reported that eNOS activity is normally attenuated due to S-nitrosylation in vascular endothelial cells, and that eNOS undergoes rapid denitrosylation upon agonist stimulation (60). The same group also found that eNOS is S-nitrosylated at the plasma membrane caveolae, but upon agonist stimulation, translocates to internal membrane structures, becomes denitrosylated and activated, and presumably then facilitates S-nitrosylation of neighboring proteins (61). Therefore, the compartmentalization of active NOS and specific target proteins into discrete subcellular organelles may represent a mechanism for determining S-nitrosylation specificity (134).
Within proteins prone to S-nitrosylation there are often numerous cysteine residues that are not nitrosylated. For example, among the many cysteines within hemoglobin β only Cys93 is specifically nitrosylated (223). Recent work shows that nitrosylation can be tightly regulated and plays an important role in many cellular pathways. For example, NF-κB nitrosylation at Cys62 was reported to cause inhibition in NF-κB-dependent DNA binding, promoter activity, and gene transcription (168, 172). Intricate mechanisms appear to exist that determine the selectivity of nitrosylation, as exemplified by the ryanodine receptor (RyR)/calcium release channel, which is regulated by calcium-calmodulin-linked S-nitrosylation (6). Only 1 (Cys3635) out of 50 free cysteine residues are nitrosylated per RyR subunit of the skeletal muscle (252). S-nitrosylation at Cys3635, or together with glutathionylation at different cysteines, appears to activate the RyR (102). Other proteins may contain multiple cysteines that are nitrosylation targets. It was reported that the NMDA-receptor, which affects neuronal development, synaptic plasticity, and memory, contains five cysteines that can be modified by NO, and any one nitrosylation event reduces receptor activity (47, 149).
Depending on the primary structure of the proteins, certain motifs may enhance the susceptibility of cysteines to nitrosylation. In several proteins, the proximity with basic and acidic residues was found to be associated with susceptibility to S-nitrosylation (136). A putative pattern of amino acids flanking the nitrosylated cysteine (SNO-Cys) site was proposed: XYCZ (where X=G, S, T, C, Y, or N; Y=K, R, H, D, or E; and Z=D or E). However, as the number of known SNO-Cys sites quickly grew, successive studies demonstrated that the motif cannot rigorously describe the majority of reported SNO-Cys sites (85, 165). Another attempt to generate a potential signature for S-nitrosylation, this time based on structural information, was made in the case of GSNO-mediated S-nitrosylation of hydrogen peroxide-inducible genes activator (OxyR) (100). From modeling simulations, the authors proposed another signature pattern for S-nitrosylation, (H/K/R)(C)(hydrophobic)(X)(D/E), where X represents any amino acid. Among proteins possessing one of the variants represented by the (H/K/R)(C)(hydrophobic)(X)(D/E) pattern were the C-terminal cytoplasmic domain of the β-subunit of the olfactory cyclic nucleotide-gated channel (30) and methionine adenosyl transferase I/III (214). However, a subsequent study conducted on a much larger set of proteins with SNO-Cys sites found that this pattern also did not define the majority of nitrosylation sites (165). In some cases, while no acid/base motif flanking the target Cys was present in the sequence, a similar arrangement could be found at the protein tertiary structure level. In these cases, the modifiable Cys residue was spatially oriented between acidic and basic amino acids (95), even in cases where these residues were far apart in the primary amino acid sequence.
The process of S-nitrosylation seems to exhibit a high degree of selectivity that may also be governed by protein–protein interactions, or by specific interactions with small molecules acting as a transnitrosylating agent, such as GSNO (165, 166). Experimental evidence of the influence of protein–protein interactions on transnitrosylation has been provided for nitrosylation of Band3 by SNO-hemoglobin (244). Additionally, Chu et al. reported that nitrosylation reactions in the xenon pocket can be facilitated by protein structural rearrangements that actively direct the NO molecule from the donor to a specific cysteine residue within the acceptor protein (49). In regard to the relationship between S-nitrosylation and protein–protein interactions, (a) the interaction can be a pre-requisite for specific transnitrosylation, as just discussed, but also (b) S-nitrosylation, by inducing a change in the electrostatic properties of the molecular surface, can affect the ability of the protein to interact with other protein partners. Recent theoretical studies indicate that nitrosylation induces a significant partial charge redistribution (92). Moreover, this change can propagate to adjacent regions of the protein (165). In this scenario, the recurrent presence of charged residues (acid and basic amino acids) in proximity to SNO-Cys can have an additional functional explanation, as they could transmit the electrostatic signal triggered by Cys modification, to an adjacent region of the molecular surface (165). Given the long range effect of electrostatic modifications, this could be an elegant way to modulate protein activity by using a single signal (NO) and a single switch (Cys). However, this speculation on the role of acidic and basic residues in the NO-dependent modulation of protein–protein interaction, though intriguing, needs further studies to be assessed on a larger scale.
Apart from overall protein structure changes, the process of nitrosylation is shaped by other factors, including the pH and the presence of metal ions such as Mg2+ and Ca2+ (222). These factors can facilitate nitrosylation by generating a microenvironment that could make the sulfhydryl more accessible to nitrosylating agents (63, 101). Indeed, the microenvironment surrounding a Cys can play a significant role in modulating its reactivity and polarity, by means of electrostatic perturbation and H-bond formation that can lead to significant changes in Cys pKa (167). Additionally, proteins are not static structures, and protein movements can also exert a considerable effect on Cys reactivity (69, 221) and polarity. All in all, the microenvironment is believed to play a crucial role in determining the susceptibility to modification of Cys targets. For example, hydrophobicity can exert a positive influence on the rate of S-nitrosylation. Hydrophobic environments can promote solubilization and accumulation of higher concentrations of NO and O2 that can react to generate the nitrosylating equivalents (194): this would favor the formation of the reactive species, NO2 and N2O3. Indeed, the reaction between NO and O2 is accelerated 300-fold in lipid membranes (151). Therefore, direct S-nitrosylation via NO2 and N2O3 should be favored for Cys targets characterized by a slightly hydrophobic context. Beside chemical and physical aspects, allosteric changes have also been reported to facilitate nitrosylation reactions. The hydrophobic environment or pockets that accumulate endogenous nitrosylating equivalents can be formed through allosteric transformations of the protein structure (252). One example is the hydrophobic pocket generated through redox-driven conformational changes in RyR, in which only a single cysteine target was S-nitrosylated (63, 101, 252). However, many known Cys targets for nitrosylation do not present any particular increase in hydrophobicity around the Cys (165). Most likely, Cys targets that are not nitrosylated via N2O3 may not benefit from a high hydrophobic content, but in turn profit from a more polar environment (which favors solvent exposure, thus increasing accessibility of larger nitrosylating agents such as GSNO).
Because many determinants and factors can control Cys reactivity, and also different agents (e.g., GSNO, NO2, and N2O3) can be responsible for the modification, to date, no pattern describing this modification as a whole has been developed. Therefore, the lack of effective patterns has so far hindered prediction of new nitrosylation sites in proteins. However, it has been recently proposed that by separately analyzing different nitrosylation sites, clustered on the basis of their reactivity toward a specific nitrosylating agent, it might be possible to describe common features. This idea has not been extensively tested yet, but if confirmed it could allow computational biologists to develop theoretical tools for robust description of defined subsets of SNO-Cys sites in proteins, such as those specifically reactive toward a particular modifying agent (GSNO, NO2, S-nitrosocysteine, etc.). In addition to computational approaches similar to those successfully applied to other PTMs (e.g., phosphorylation), a potential additional strategy may involve docking approaches. Recently, a quantum mechanics-based study demonstrated that NO modification can induce significant charge redistribution in the Cys side chain (with only marginal effects on backbone atoms) (92). The results from this study can provide the basis for docking calculations with SNO-containing proteins or substrates. By properly implementing the ad hoc parameters for SNO, docking approaches can model the affinity and binding modes of GSNO and a target Cys (Fig. 10), as shown in a recent study investigating a database of potential Cys targets for GSNO-mediated transnitrosylation (165). Similar approaches can be easily applied to other cases of transnitrosylation (Fig. 10), where the agent is a low-molecular-weight RSNO or an SNO-protein. For a more detailed description on this subject as well as the current state of computational analyses and tools for the investigation of reactive Cys residues and PTMs, we refer the reader to a recent review, specifically addressing these issues (165).
FIG. 10.
Prediction of transnitrosylation sites with docking approaches. Ad hoc developed force field parameters for S-nitrosylated cysteine (SNO-Cys) (92) can be transferred to a Cys containing transnitrosylating agent. For example, starting from glutathione, in silico S-nitrosylation can be simulated with a molecular builder tool (e.g., Vega ZZ, http://nova.colombo58.unimi.it/cms/), constructing nitrosoglutathione (GSNO), shown in sticks contoured by semitransparent spacefills. To deal with the effects of nitrosylation on glutathione, charge schemes and angles for SNO-Cys are transferred to GSNO (by keeping the overall charge of GSNO fixed, such that only partial charge redistribution can occur). For the application as a predictive tool, the prepared GSNO can be docked to a query protein with a docking software. For each Cys (C) of the query protein, the affinity for GSNO is calculated (in the figure, two representative Cys residues of a protein target are shown in sticks). Those Cys (if any) showing favorable energy and geometrical interactions with the substrate are predicted as potential modification sites. Only one Cys residue (labeled as C1, in the upper right of the figure) is targeted by GSNO, whereas other docking modes are not consistent with any transnitrosylation of Cys targets (e.g., Cys target labeled as C2). Therefore, the prediction would be that the query protein has a predicted nitrosylation site (C1). By analogy, other transnitrosylating agents can be tested with the docking based approach shown here (V.G. unpublished data).
Docking-based methods could prove to be particularly relevant for examining the effect of nitrosylation on protein–protein interactions. Although large-scale simulations of protein–protein interactions, in search of reactive Cys targets, are still quite (computationally) demanding tasks, these studies are becoming feasible (e.g., using the Rosetta suite of programs, RosettaDock, etc.) (188). In the future, such docking-based approaches may allow systematic investigation of partners involved in either trans- or denitrosylation.
C. Transnitrosylation and denitrosylation of proteins
As discussed above, structural features per se do not always accurately predict whether a cysteine is nitrosylated in vivo, suggesting that additional factors are at play for regulating nitrosylation specificities. Protein transnitrosylation is the transfer of an NO group from a cysteine in one protein to a specific cysteine in another protein. This mechanism may allow select SNO-proteins to serve as transnitrosylases or specific transnitrosylating agents for modulating the nitrosylation of specific target proteins (243). A classic example is the transnitrosylation from SNO-Cysβ93 of hemoglobin onto a single cysteine within the cytoplasmic domain of an anion exchanger, AE1 in the red blood cell membrane (211). Recently, GAPDH has been shown to carry out important cytosol to nucleus signal transduction activities via transnitrosylation of key proteins. GAPDH with SNO-Cys150 can bind and transnitrosylate HDAC2, sirtuin-1 at Cys387 and Cys390 and DNA-activated protein kinase (129), affecting the activities of these proteins in the nucleus. Trx1 also displays specific transnitrosylation properties, transferring NO from Cys73 onto numerous targets, including caspase 3 (Casp3) at Cys163 (273). These examples illustrate how spatial and temporally regulated S-nitrosylation events can function during signal transduction events (243), and we focus on Trx1 as a potential transnitrosylase paradigm later in this review.
Benhar et al. have suggested that denitrosylation is not simply an “off” mechanism that counters the effect of protein nitrosylation (21). In fact, denitrosylation may provide another means for the determination of nitrosylation specificity. Global analysis of the S-nitrosoproteome has revealed that a number of proteins are typically nitrosylated under steady state conditions in cells. Therefore, a protein denitrosylase may behave like a regulated phosphatase; when activated, they may denitrosylate selected SNO-proteins, thus modulating the biological activities of their targets. However, specific denitrosylation motifs have yet to be identified from the known protein denitrosylation systems, that is, the GSNO reductase (GSNOR)/GSH/nicotine adenine dinucleotide (83) or Trx systems (21). Protein denitrosylation by the Trx system is discussed in detail later in the review. GSNOR was characterized previously as GSH-dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase (67). The GSNOR system indirectly denitrosylates proteins via GSH, which extracts NO from select SNO-proteins to form GSNO, and is converted to GSNHOH by GSNOR (83). Known GSNOR/GSH denitrosylation targets include GPCR kinase 2 (270) and β-arrestin 2 (203). Both of these proteins are regulators of GPCR signaling and desensitization, especially within the β-adrenergic receptor signal transduction pathways (219). Indirect evidence from GSNOR knockout mice revealed that SNO-hemoglobin might be another target of GSNOR-mediated protein denitrosylation (255). Other studies suggested an important function for GSNOR during hypoxia and found that HIF-1α and the Von Hippel-Lindau (VHL) protein within cardiomyocytes could be denitrosylation targets of GSNOR (148). The elevated nitrosylation of HIF-1α appears to be responsible for the protection of cardiac function and the reduction in infarct size after ischemia in GSNOR knockout mice (206). The effect is augmented by nitrosylation of VHL, a negative regulator of HIF-1α, whose function is attenuated by elevated nitrosylation activity in GSNOR knockout mice (206).
In addition to protein systems, simple molecules may have specific denitrosylating ability. Bilirubin (BR) is an endogenous antioxidant molecule that acts as a scavenger of free radicals (182), but also modulates denitrosylation/nitrosylation processes (17). Barone et al. demonstrated that physiological concentrations of unconjugated BR, free or complexed with saturating concentrations of HSA, denitrosylates RSNOs as well as high-molecular-weight S-nitroso-albumin (17). The same study confirmed the formation of nitrosylated BR in a reconstituted system and in rat fibroblasts exposed to oxidative and nitrosative stress, suggesting a biological function for BR in mediating the denitrosylation of SNO-proteins.
VI. Regulation of Nitrosylation by Trx
As discussed earlier, S-nitrosylation is a dynamic, specific, and reversible PTM, important for the regulation of protein functions within diverse cellular pathways. The mechanisms that are responsible for regulating nitrosylation specificity are poorly understood. Several proteins, including superoxide dismutase 1, PDI, GAPDH, and Trx2, have been shown to possess either a transnitrosylating or a denitrosylating activity toward specific target proteins (21, 129). With regard to Trx1, it is unique in its ability to either transnitrosylate or denitrosylate a specific target, namely, Casp3 (184, 247). Here we are going to discuss the targets of Trx1 transnitrosylation and denitrosylation and how redox PTMs enable Trx1 to toggle between these two regulatory functions.
A. Transnitrosylation by Trx1
A number of studies report the transnitrosylation or denitrosylation of proteins by Trx (Supplementary Table S2). For Trx1 to serve as a transnitrosylation agent, it first needs to be nitrosylated itself, to form SNO-Trx1. The conserved Cys69 and Cys73 residues of Trx1 (Fig. 2A) have been implicated in the S-nitrosylation of both Trx1 and the possible Trx1-catalyzed transnitrosylation of other proteins in endothelial cells (88, 184, 267, 273) [it should be noted that the impact of Cys69 nitrosylation on Trx1 disulfide reductase activity is in dispute (96)]. Which of these or other conserved Trx1 cysteines is available for nitrosylation and can serve as the donor for transnitrosylation is probably cell and context dependent. One such context is pH; in vitro, the nitrosylation of Cys62 can occur at neutral pH, but more alkaline pH appears to be required for the nitrosylation of Cys69 (269). However, under physiological conditions, Cys73 may be the Trx1 cysteine most responsible for Trx1-mediated transnitrosylation (184). Using high-resolution mass spectrometry with instrumentation conditions optimized for preserving protein nitrosylation, we were able to identify the redox status of each of the five cysteines within Trx1 (267). After the treatment of recombinant Trx1 with GSNO, we only detected a nitrosylation site at Cys73 (Fig. 11A), whereas Cys32 and Cys35 were present as an intramolecular disulfide bond (Fig. 11B, C), and the other three cysteines remained as free thiols (267, 273). We also observed evidence that a minor secondary nitrosylation site was present within Trx1 (Fig. 11C); however, we were unable to determine its location due to low abundance.
FIG. 11.
S-nitrosylation of Trx1 and targets of Trx1 transnitrosylation are dependent on Trx1 cysteine redox status. (A) MS/MS spectrum of a tryptic nitrosylated Trx1 peptide 73C*MPTFQFFK81 (MH+ 1194.5; C*, nitrosylated). Inset: a spectrum of the doubly charged peptide with an m/z of 597.26 is shown. MS/MS analysis of this peptide found Cys73 to be nitrosylated. The SNO site was detected directly, no biotin switch technique (BST) was performed. Reprinted from Ref. (267). Earlier studies suggested that the redox status of Cys32 and Cys35 may affect the ability of Trx1 to transnitrosylate or denitrosylate target proteins (21). Here we present direct evidence that only after the formation of the Cys32 and Cys35 disulfide bond (forming Trx1-S2), can Trx1-S2 be S-nitrosylated in vitro (forming S-nitrosylated Trx1 [SNO-Trx1]). Mass spectra of Trx1-S2 before (B) and after (C) GSNO treatment and of Trx1-(SH)2 before (D) and after (E) GSNO treatment are shown. All spectra were deconvoluted to singly charged states by Masslynx (V4.1, Waters) for ease of comparison. Precursor and nitrosylated peaks are labeled. Under nonreducing conditions, high-resolution MS analysis of a recombinant His-tagged human Trx1 demonstrated that this protein had a mass of 13,766.75, corresponding to Trx1 with three free thiols and one disulfide bond (B). This molecule was readily S-nitrosylated by GSNO, resulting in a product with a mass of 13,795.80 (C, 13,766.75+29, i.e., Trx1-S2+NO). In addition, we also observed lower levels of dinitrosylated Trx1 with a mass of 13,824.85 (C, 13,766.75+58, i.e., Trx1-S2+2NO). For comparison, Trx1 with reduced Cys32 and Cys35 (Trx1-(SH)2) yielded a mass of 13,768.90 (D), corresponding to the addition of approximately two extra protons to Trx1-S2. Unexpectedly, we could not detect any SNO-Trx1 MS signal after the incubation of Trx1-(SH)2 with GSNO (E). To further our investigation, we examined whether the redox status of Cys32 and Cys35 affects Trx1 transnitrosylation of a Casp3 peptide containing the known nitrosylation site. (F) Spectrum of GSNO treatment of a synthetic human Casp3 peptide (Casp3p) containing the known nitrosylation site Cys163 (163CRGTELDCGIETD175, m/z 1409.58 of oxidized form oCasp3p). An m/z peak at 1440.46, corresponding to SNO-Casp3p (1411.5+29, i.e., reduced Casp3p+NO), was observed. (G) In the presence of Trx1-S2 a substantial SNO-Casp3p ion peak was observed. (H) SNO-Trx1 directly transnitrosylates Casp3p. (I) However, in the presence of Trx1-(SH)2, hardly any SNO-Casp3p was detected, as shown by the minor m/z peak at 1440.52. (J) Proposed model for Trx1 Cys32 and Cys35 redox status regulation of Casp3 nitrosylation. Adapted from Ref. (273) with permission.
The redox states of the disulfide reductive site cysteines appear to be crucial regulators that determine the nitrosylation status of the remaining Trx1 cysteines (96, 273). Hashemy and Holmgren reported that a two-disulfide form of Trx1 (Cys32-Cys35 and Cys62-Cys69) can be readily nitrosylated on Cys73 and transnitrosylate target proteins (96). The same group also reported that SNO-Trx1 has diminished disulfide reduction activity (96). Two different groups have reported increased protein nitrosylation levels after treatment of HepG2 or 10C9 cells with auranofin, a highly specific TrxR inhibitor (21, 153). These observations suggest that when the protein reduction activity of Trx1 is attenuated, it may either lose its ability to denitrosylate or increase its ability to transnitrosylate target proteins. We recently expanded on these observations, and confirmed that the redox status of the reductive site cysteines is essential to the nitrosylation of Trx1 at Cys73 and to its ability to transnitrosylate target proteins (273). We found that only the oxidized form of Trx1 (Trx1-S2), containing disulfide linked Cys32 and Cys35, could be nitrosylated on Cys73 to generate SNO-Trx1 (Fig. 11B–G). SNO-Trx1 could transnitrosylate target cysteines in a Casp3 peptide in the absence of GSNO (Fig. 11H), but transnitrosylation by either GSNO or SNO-Trx1 was diminished in the presence of reduced Trx1 (Trx1-(SH)2), with dithiol forms of Cys32 and Cys35 (Fig. 11I). Further, Trx1-(SH)2 can selectively remove the NO group from some SNO-proteins (21), including the denitrosylation of SNO-Trx1 (238, 273), suggesting that Trx1-(SH)2 and perhaps additional factors, including GSH (238) and lipoic acid (247), may play a role in regulating the stability of SNO-Trx1 and other mediators of transnitrosylation. Hence, the multiple PTM regulatory functions of Trx1 appear to be partitioned by the oxidative state of its reductive catalytic site Cys32 and Cys35 (Figs. 11J and 12).
FIG. 12.
Trx1 regulation of procaspase 3 S-nitrosylation and apoptosis. (I) Cys32 and Cys35 oxidized Trx1 can be nitrosylated by GSNO at Cys 73, to form SNO-Trx1; (II) SNO-Trx1 transnitrosylates procaspase 3 at Cys 163, which cannot be activated to induce apoptosis (184, 185). (III) S-nitrosylated procaspase 3 can be denitrosylated by the Trx system. Denitrosylated procaspase 3 can be cleaved to activated Casp3 which induces apoptosis (21). (IV) Oxidized Trx1 can be replenished by TrxR/NADPH reduction.
The most well-characterized target of Trx1 nitrosylation is Casp3. Through mutation analysis, Marletta and colleagues determined that Cys73 of Trx1 is responsible for the transnitrosylation of Casp3 (184). This group employed a Cys73Ser mutant of Trx1, which exhibited normal disulfide reduction activity, but was resistant to becoming nitrosylated, and contained no transnitrosylation activity toward Casp3 at Cys163. The specificity of Trx1-mediated transnitrosylation is likely to rely on specific protein–protein interactions between SNO-Trx1 and the target proteins. Mitchell et al. have shown that mutations of Trx1 E70A and K72A, residues that surround the Cys73 transnitrosylation site, prevents SNO-Trx1 from binding and transnitrosylating Casp3 (185), in spite of these Trx1 mutants being nitrosylated themselves. Using fluorimetry analysis of a Casp3-derived synthetic peptide, Mitchell et al. estimated that the rate of wt Trx1 transnitrosylation of Casp3 was 77.1–196.0 M−1 s−1 (184), compared to 4.6 M−1 s−1 for Trx1 E70A and K72A, and an undetectable rate for Trx1 C73D and C73S mutants (185). Further, the same group reported that GSNO was able to transnitrosylate Casp3 at a slower rate of 1.67 M−1 s−1 (184). Despite these rates being several orders less than those reported for Trx1-mediated reduction (see Section III.D), these observations suggest that Trx1 can catalyze transnitrosylation of Casp3, to regulate this critical apoptosis factor in cells. However, enzymatic turnover and transnitrosylation of a substrate with a substoichiometric level of SNO-Trx1 has not yet been reported; therefore, it is premature to name SNO-Trx1 transnitrosylase. In addition to important amino acids surrounding Cys73 of SNO-Trx1, there are likely amino acids surrounding the target transnitrosylation sites that are equally important for the determination of transnitrosylation specificity. Recently, we identified 47 putative Trx1 transnitrosylation targets, including Prx1, in HeLa cells expressing transnitrosylation-active Trx1C32S/C35S (a mutant Trx1 with both Cys32 and Cys35 replaced by Serine to mimic the function of disulfide reductase and denitrosylation-inactive Trx1) (273). We have performed a primary sequence motif analysis of the peptides whose nitrosylation sites are likely transnitrosylated by SNO-Trx1 (273). Although the 47 protein sequences were limited, we were able to identify three independent candidate motifs, AXC, CXXXXA, and AC, where X is any amino acid and C is an SNO-Cys (273). Interestingly, Cys163 of Casp3 appears to contain the AC motif (273).
B. Denitrosylation by Trx
Earlier work demonstrated that Trx1/TrxR system is capable of denitrosylating low molecular weight RSNOs, such as GSNO (196, 238, 247), 25–30 kDa liver cell proteins (238), and protein tyrosine phosphatase 1B (21) (Supplementary Table S2). Through application of advanced proteomics techniques, 11 and 46 diverse Trx1 denitrosylation targets were recently identified in RAW264.7 activated macrophages (257) and in Jurkat cells (23), respectively. Trx1 is also implicated in the denitrosylation of eNOS, allowing its homodimerization and activation (218), which is coupled to activation of the β-adrenergic receptor in endothelial cells (270).
The mechanism of denitrosylation of RSNOs by Trx was proposed by Stoyanovsky and colleagues, and involves the reductive site Cys32 and Cys35 (238, 247). They propose that the target NO is initially transferred to Trx onto the low pKa thiol of Cys32. The vicinal thiol of Cys35 then forms a disulfide bond with Cys32, concomitantly releasing HNO or possibly transferring the NO intramolecularly to Cys62, 69, or 73 (238). The same mechanism has been proposed for SNO-protein denitrosylation, as well as an alternative mechanism involving the formation of a mixed disulfide between Cys32 of Trx1 and the formerly nitrosylated target, resulting in the release of HNO (22). Another report suggested that Cys73 may contain some Casp3 denitrosylation activity (184). Although Trx1 and related molecules appear to be highly efficient at denitrosylation, the kinetics for Trx1 catalyzed protein denitrosylation has not been widely reported, other than a rate constant of 34.4 M−1 s−1 reported for the denitrosylation of Casp3 by Trx1-(SH)2 (184). Therefore, the current understanding of Trx1-mediated protein denitrosylation is largely qualitative. Further, the exact intramolecular mechanism of Trx denitrosylation remains ill-defined, as does denitrosylation target specificity. A comparison of proteins that are transnitrosylated, but not denitrosylated by Trx1, as opposed to those denitrosylated but not transnitrosylated, might provide insight into the mechanistic specificity of denitrosylation. Prx1 belongs to the former category; nitrosylated Prx1 (SNO-Prx1) cannot be denitrosylated by Trx1 (23), even though this peroxidase is a target of both Trx1 reduction (91) and transnitrosylation (273). This suggests that the Trx1-(SH)2 disulfide reductive catalytic domain, which is essential for reducing Prx1 disulfide dimers, may not be able to bind SNO-Prx1 and catalyze its denitrosylation.
Both Trx1 and Trx2 can function as denitrosylating agents (21). In contrast to its versatile relative, Trx2 has not been shown to possess transnitrosylation activity, presumably because it has not evolved to contain a Trx1 Cys73 equivalent (Fig. 2B). The data supporting the seemingly opposite functions of Trx1 transnitrosylation and denitrosylation are equally compelling. Possible cellular mechanisms exist for the regulation and segregation of the Trx1 transnitrosylation function from its denitrosylation function, which is not simply reverse-transnitrosylation. Different Trx1 cysteine residues are implicated in these two functions; Cys32 free thiol of Trx1-(SH)2 is responsible for denitrosylating activity (21), whereas Cys73 of SNO-Trx1 is responsible for transnitrosylating activity (184). Further, it is unlikely that the same Trx1 protein can catalyze both transnitrosylation and denitrosylation activities simultaneously. In support of this presumption, we recently observed that SNO-Trx1 failed to nitrosylate target proteins and was itself denitrosylated when mixed with Trx1-(SH)2 (Fig. 13) (273). Interestingly, Sengupta et al. found that in cells Trx1-(SH)2 could effectively compete (at a rate constant of 309 M−1 s−1) with Casp3 for the NO of SNO-Trx1, and when formed to any significant extent, SNO-Casp3 could be denitrosylated by the Trx1/TrxR system (238).
FIG. 13.
Cys32 and Cys35 reduced Trx1 (Trx1-(SH)2) denitrosylates SNO-Trx1 and attenuates SNO-Trx1 transnitrosylation of HeLa cell proteins. (A) Comparison of in vitro denitrosylation of SNO-Trx1 by Trx1-(SH)2 or Cys32 and Cys35 disulfide Trx1 (Trx1-S2). Recombinant Trx1 was either oxidized by 10 mM H2O2 for 60 min at 37°C to produce Trx1-S2, or reduced by 10 mM DTT for 60 min at 37°C to obtain Trx1-(SH)2. Ten microgram of Trx1-S2 in 100 μL of the nitrosylation buffer was first treated with 100 μM GSNO for 30 min at 37°C to obtain SNO-Trx1. Equal aliquots of SNO-Trx1 were mixed with an equimolar concentration of either Trx1-S2 (lane 2) or Trx1-(SH)2 (lane 3) for 30 min at 37°C. One hundred microgram of lysozyme was added as a carrier and the resulting protein solutions were precipitated in cold acetone. The pellets were washed with ice cold acetone and dissolved in a biotin switch blocking buffer. Trx1-(SH)2 effectively denitrosylated SNO-Trx1 (lane 3), whereas Trx1-S2 had no effect (lane 2). Values are the mean±standard error for experiments performed in triplicate (*p=0.0004, Student's t-test). (B) Trx1-(SH)2 denitrosylation of SNO-Trx1 attenuates its ability to transnitrosylate target proteins. HeLa cell proteins (200 μg) (lane 1) were mixed with 10 μg of SNO-Trx1 (lane 2), plus either 10 μg of Trx1-S2 (lane 3) or Trx1-(SH)2 (lane 4) at 37°C for 30 min. SNO-proteins were processed by BST and detected by Western blotting. Trx1-(SH)2 prevented SNO-Trx1 from transnitrosylating HeLa proteins, whereas Trx1-S2 had no effect. Values are the mean±standard error for experiments performed in triplicate (*p=0.0007, Student's t-test). (C) Proposed mechanism of Trx1-(SH)2 and SNO-Trx1 in the regulation of protein nitrosylation. It should be noted that SNO-Trx1 transnitrosylation targets and Trx1-(SH)2 denitrosylation targets may not overlap. Adapted from Ref. (273) with permission.
C. Functional significance of Trx-mediated regulation of nitrosylation
Redox imbalance associated with Trx1 disulfide reductase dysfunction has been implicated in the development of common diseases, including diabetes, inflammation, neurodegeneration, cardiovascular disease, and cancer (56, 110). However, dysregulation of NOS and protein nitrosylation are also implicated in many of the same disease tissues and models (73, 178). Therefore, the ability of Trx1 to regulate target proteins through nitrosylation has various health and disease implications. Since nitrosylation may be a common regulatory feature of apoptosis, Trx1′s role in regulating apoptosis may involve its transnitrosylation or denitrosylation of caspase family members or other apoptosis regulators within specific cellular or pathological contexts. Trx1-mediated transnitrosylation has been shown to protect endothelial cells against stress-induced apoptosis (88, 273), and nitrosylation of Trx1 is a prerequisite for the nuclear translocation of Trx1 in endothelial cells (232), suggesting the possibility of direct transcriptional modulation by SNO-Trx1. In fact, in certain model systems, SNO-Trx1-mediated transnitrosylation appears to play a more important antiapoptotic function independent of Trx1′s disulfide reductase activity. Through site-directed mutagenesis, we found that Trx1 Cys73-mediated transnitrosylation was more effective at protecting HeLa cells from TNF-α-induced apoptosis than was Cys32 and Cys35-catalyzed disulfide reduction (273). Further, S-nitrosylation of Trx1 and other proteins induced by ischemic preconditioning has been shown to be cardioprotective following ischemia/reperfusion, a condition in which cardiac cells would otherwise undergo apoptosis (250, 255). The study of the pathophysiological function of regulated nitrosylation, especially by Trx1, will likely provide insight into novel signal transduction pathways that are dysregulated under disease conditions. To serve the role as a specific regulator of cell signal pathways, as discussed previously, SNO-Trx1 stability appears to be tightly regulated by the presence of Trx1-(SH)2 and other reductants (Fig. 13), including GSH (238). Therefore, subcellular compartmentalization and distribution of SNO-Trx1 and its reductants may determine the degree of S-nitrosylation of SNO-Trx1 target proteins.
NO was found to inhibit caspases 1, 2, 3, 4, 6, 7, and 8 via nitrosylation, regulating apoptosis (144). Cytosolic Casp3 is typically not nitrosylated, but can be sequestered in the inactive form by a non-nitrosylation-based mechanism, presumably via protein–protein interactions with a cellular inhibitor (21). However, Casp3 can be reversibly inhibited in hepatocytes by either NO donor treatment or iNOS activation, thus attenuating TNFα-induced apoptosis (127). Marletta et al. have reported both in vitro and in vivo evidence suggesting that Trx1 can transnitrosylate Casp3 at Cys163, thereby preventing apoptosis in cultured T-cells (184, 185) (Fig. 12). Once nitrosylated, SNO-Casp3 cannot be proteolytically cleaved and activate apoptosis. Analogously, S-glutathionylation also inhibits cleavage of Casp3, and this is regulated by the Grx system (207). However, in a different cellular context, SNO-Casp3 have been shown to transnitrosylate an antiapoptotic E3 ligase, XIAP at Cys450, attenuating its activity, resulting in the activation of apoptosis (193). Evidently the ability to transnitrosylate/denitrosylate Cys163 of Casp3 is a major means of Trx1 to fine-tune apoptosis. Trx1 has been shown to denitrosylate Casp3 at Cys163 in different cells, resulting in Casp3 activation (21). So far, Casp3 is the only protein known to be regulated by Trx1-mediated site-specific reversible nitrosylation (Fig. 12). Both mitochondrially localized Casp3 and 9 are found nitrosylated in resting cells, inhibiting their enzymatic activities and regulatory roles in modulating apoptosis in a human B cell line (10C9) (163), and they can be activated by Trx2-mediated denitrosylation in Fas-ligand-stimulated B-cells (21). It is still unclear how FasL activates the Trx2 system for Casp3 and Casp9 denitrosylation. It appears that Trx-regulation of protein nitrosylation may play a broad role in regulating apoptosis.
Trx1 is usually characterized as an antiapoptotic molecule, primarily due to the ability of Trx1-(SH)2 to reduce and sequester ASK1 (152), preventing ASK1 from activating downstream protein kinase signaling cascades that lead to proapoptotic gene expression. Curiously, higher SNO-Trx1 levels have been correlated with an upregulation of ASK1 apoptotic activity, presumably due to dissociation of the Trx1-ASK1 complex after nitrosylation of Trx1 (249). Conversely, ASK1 has been shown to undergo nitrosylation at Cys869 in murine fibrosarcoma L929 cells after interferon-γ treatment, which attenuates ASK1′s ability to activate apoptosis in response to H2O2 (208). Therefore, it will be of interest to determine whether Trx1 can regulate the ASK1 proapoptotic pathway by altering the nitrosylation status of ASK1. The possible interplay between Trx1 and other redox regulators might determine whether apoptosis proceeds. For example, during oxidative stress both Trx1 and the heme oxygenase/biliverdin reductase system (responsible for the production of BR) are upregulated. BR can upregulate Casp3 in a dose–dependent manner in neurons (160) and can act as a denitrosylase (17). Therefore, one can speculate that under oxidative stress, SNO-Trx1 initially hinders apoptosis by nitrosylating Casp3, but ever increasing ROS leads to elevated BR, which catalyzes the denitrosylation of SNO-Casp3 and allows the resumption of apoptosis.
We recently identified Prx1, a classic 2-Cys Prx and Trx1 disulfide reduction target, to be transnitrosylated by Trx1 (Fig. 14) (273). Prx1 could be directly nitrosylated by SNO-Trx1 in vitro at Cys173, the resolving cysteine required for the formation of a disulfide bond with the peroxidatic cysteine after its reduction of peroxides (Fig. 14A, B). The same nitrosylation site was also found in vivo in HeLa cells overexpressing Trx1C32S/C35S, a denitrosylation inactive, thus transnitrosylation active Trx1 mutant (273). In addition, Cys83, a key residue for the maintenance of the human Prx1 dimer-dimer interface (139), was also nitrosylated (273). Since nitrosylation has been shown to inhibit Prx2′s peroxidase activity (65), SNO-Prx1 is likely to have diminished peroxidase activity as well. However, nitrosylation of Prx1 (into SNO-Prx1) may be a means for long-term preservation of Prx1 function, especially in highly oxidizing cellular environments. Drapier's lab has shown that the addition of NO donors to macrophages attenuated the overoxidation of Prx1 by H2O2 (55). We found that in HeLa cells, H2O2 treatment resulted in an increase in Prx1 monomer with a concomitant decrease in homodimer, and accumulation of SNO-Prx1 and Prx1-SO3H (Fig. 14C, D). Prx1-SO3H is a marker for irreversibly inactivated Prx1 (273). It was significantly attenuated in H2O2-treated cells overexpressing Trx1C32S/C35S relative to control cells (Fig. 14C), with a concomitant increase in SNO-Prx1, an effect that can be reproduced by the addition of GSNO to cells (273). Trx1-mediated transnitrosylation of Cys173 may be a means for maintaining an overoxidation-resistant Prx1 structure (264, 277), but the precise mechanism remains to be explored. We therefore speculate that during prolonged ROS exposure, when the Trx reductive system becomes overextended and SNO-Trx1 accumulates, Trx1 transnitrosylates Prx1 at Cys173, thus preventing Prx1 from becoming overoxidized (Fig. 14E). Once the cellular redox balance is restored, TrxR can facilitate the accumulation of Trx1-(SH)2 and reduce Prx1 dimers. It should be noted that since Trx1/TrxR is unable to denitrosylate SNO-Prx1 (23), it is likely to be reduced by other denitrosylation systems.
FIG. 14.
SNO-Trx1 transnitrosylation of Prx1. (A) Both SNO-Trx1 and GSNO directly transnitrosylated Prx1 in vitro. Two SNO-Trx1 transnitrosylation sites in Prx1 were identified by MS/MS analysis. (B) MS/MS spectrum of a Prx1 tryptic peptide 169HEGVC*PAGWKPGSDTIKPDVQK190 (MH+ 2349.2) was analyzed after Prx1 transnitrosylation by SNO-Trx1. Cys173 (C*: biotinylated) was found to be nitrosylated. Drapier and colleagues have shown that NO donors attenuate the overoxidation of Prx1 in macrophages (55), which the authors attributed to NO induction of Srx. We found that H2O2 treatment increased nitrosylation of proteins in both vector- and Trx1C32S/C35S-expressing cells, albeit more so in the latter cells (C). (D) Western blot of Prx1 species after separation on a nonreducing gel. Overexpression of Trx1C32S/C35S rendered Prx1 more resistant to H2O2-induced dimer to monomer conversion which correlated with the reduction of H2O2-induced formation of Prx-SO3H. Prx1 expression was not affected by either Trx1C32S/C35S expression or H2O2 treatment. (E) Proposed function for Trx1-mediated transnitrosylation of Prx1. Within physiological environments, the Trx1/Prx1 (I) and Srx (II) systems can adequately handle ROS. During periods of elevated ROS, Trx1-S2 can accumulate and be nitrosylated by NO donors into SNO-Trx1 (III), which in turn transnitrosylates Prx1 into S-nitrosylated Prx1 (SNO-Prx1) (IV). SNO-Prx1 may become reactivated by denitrosylases (V). Adapted from Ref. (273) with permission.
Based on mutational analysis and in vitro studies, we found that a redox-based mechanism can distinguish Trx1-mediated transnitrosylation from its denitrosylation function. In normal biological systems, Trx1 protects proteins via its reductase or denitrosylating activities, with sufficient reducing equivalents provided by TrxR and NADPH. When cells are undergoing oxidative stress, overpowering TrxR's ability to regenerate Trx1-(SH)2, Trx1 becomes S-nitrosylated and regulates proteins via an alternative modality of transnitrosylation. Alternatively, other regulatory systems can play a role in regulating Trx1′s ability to carry out either denitrosylation or transnitrosylation activities. TXNIP was recently reported to dynamically regulate Trx1 by repressing its denitrosylation activity (71), but this repression may be attenuated by endogenously synthesized NO (233). This dynamic regulation of denitrosylation allows cells to survive nitrosative stress, and places Trx1 in a prominent physiological role for the protection of cells against nitrosative stress (71). It is not known whether TXNIP facilitates S-nitrosylation of Trx1 or SNO-Trx1-mediated transnitrosylation of target proteins. However, in Trx1C32S/C35S-expressing HeLa cells, increased TXNIP nitrosylation was observed, suggesting the possibility of SNO-Trx1 transnitrosylation of TXNIP (273). Trx1-regulated trans- or denitrosylation of regulatory proteins may serve several purposes: as a signaling mechanism, and to temporarily prevent proteins from acquiring irreversible oxidative modifications, when the Trx1/TrxR system can no longer reduce or denitrosylate the proteins.
VII. Proteomics Strategies for the Identification of Trx Transnitrosylation/Denitrosylation Targets
A. Strategies
S-nitrosylation serves important roles in regulating protein function and cellular physiology. However, the labile and dynamic nature of reversible S-nitrosylation poses enormous technical challenges for the accurate identification and sensitive quantification of this modification. Such difficulties also reside in the fact that many proteins of biological interest are present in relatively low abundance. Given the chemically labile nature of S-nitrosylation within biological systems, it becomes more effective to replace the NO group of nitrosylated residues with a more stable alkylating group. The alkyl group can be linked to a biotin moiety (Fig. 15A), and the modified peptides affinity enriched, resulting in improved detection sensitivity and signal-to-noise ratios. Recent development of the biotin switch technique (BST) (118) allows for both qualitative and quantitative study of subtle changes in the degree of protein S-nitrosylation (21).
FIG. 15.
BST for the detection of S-nitrosylated proteins. (A) Common biotin reagents used for BST: N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP) (top), maleimide-PEG2-biotin (biotin M; middle) and iodoacetyl-PEG2-biotin (bottom). (B) BST is a three step technique for detecting nitrosylated cysteines in proteins. (I) Free thiols are alkylated with MMTS. (II) Nitrosylated residues are reduced by ascorbate. (III) Nascent free thiols are labeled by either one of the thiol reactive biotinylated reagents. Biotinylated proteins can be detected by Western blot, and nitrosylation sites determined by MS/MS (118). (C) Effective reduction of SNO by ascorbate. HeLa cell proteins were first nitrosylated in vitro with 100 μM of GSNO at 37°C for 30 min in the dark, then precipitated and washed with cold acetone to remove excess GSNO. The protein pellet was dissolved in a biotinylation buffer (118) and the resulting protein solution was treated with 1–50 mM ascorbate at room temperature for 1 h. Nitrosothiol contents were detected by Saville assay (229). Ninety percent of SNO-proteins could be reduced by just 1 mM of ascorbate in 1 h. *p<0.001 (C.W. unpublished data). (D) Expression of Trx1C32S/C35S in HeLa cells resulted in increased protein nitrosylation levels as determined by Western blotting of BST-processed proteins. A control treatment without ascorbate was used to confirm nitrosylation detection specificity. Reprinted from Ref. (273) with permission.
This section of the review compares the relative effectiveness of several thiol-specific quantitative proteomics techniques (i.e., DIGE, ICAT, and BST) for the discovery of proteins whose cysteine thiols are sensitive to nitrosylation, and in particular to Trx1-mediated transnitrosylation and denitrosylation. Each method can reveal a subset of unique Trx1-sensitive nitrosylated proteins. As with the identification of Trx1 reductive targets, one needs to either overactivate or inactivate Trx1 pathways to identify Trx1 nitrosylation targets by quantitative proteomics techniques. Overexpression or knock-down of Trx1 transcripts and Trx1 mutants is often straight forward but might not be physiologically relevant. However, a number of alternative endogenous regulators or pharmacological treatments are available. Haendeler et al. have shown that statins enhance S-nitrosylation of Trx1, and result in a significant reduction in intracellular ROS (89), suggesting that statins can be used to study Trx1-mediated regulation of transnitrosylation in certain cells. DeMorrow et al. have demonstrated that anandamide treatment can increase the expression and nuclear translocation of Trx1, but not Trx2, in cholangiocytes, inhibiting their hyperplastic proliferation (52). This endocannabinoid may be used to identify nuclear targets of Trx1. TXNIP can upregulate protein nitrosylation (71), presumably due to its ability to also inhibit Trx1′s denitrosylation activity (197). Go et al. demonstrated that acrolein and HNE covalently modifies Trx1 Cys73 and inhibited its reductase activity (82), and presumably Trx1′s transnitrosylation activity as well. Recently, specific inhibitors of Trx1 and TrxR have been developed for cancer therapy (128); such compounds are useful for identification of Trx1 targets and to increase our understanding of the role of Trx1 regulation of protein nitrosylation in cancer development and therapy.
B. Biotin switch technique
BST is widely used for the identification of SNO-proteins and in the mapping of their SNO-Cys (118). The general concept behind this method is similar to that used in the detection of redox-sensitive cysteine thiols. Proteins are first extracted in buffers containing metal chelators, especially near neutral pH, to preserve cellular protein nitrosylation status. There are then three steps involved in the successful implementation of BST (Fig. 15B). First, free cysteine thiols are alkylated with MMTS. Second, ascorbate is used to reduce the nitrosylated cysteines without also reducing disulfide bonds or other oxidative cysteine PTMs. Third, a cysteine-alkylating reagent (typically biotin-N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide [HPDP]) is used to alkylate the newly exposed cysteine thiols and serves as a sensitive marker of S-nitrosylation. In Figure 15C, we show an example of the efficient reduction of SNO-proteins by ascorbate (229). HeLa cell proteins were first nitrosylated using GSNO and then treated with increasing concentrations of ascorbate. The RSNO content was determined using the Saville assay (229), which involves the displacement of NO+ from nitrosylated cysteines with mercury. Mercury reacts with Saville reagents to generate an azo dye that can be measured spectrophotometrically. It appears that ascorbate at concentrations above 1 mM was efficient at reducing SNO-proteins in our example (Fig. 15C). To prove that biotin modifications correspond to specific nitrosylation sites, a control BST treatment is typically carried out, in which ascorbate is omitted (Fig. 15D). This control should not detect any biotinylated proteins and hence SNO-proteins, since no free thiols will be generated from the reduction of Cys-SNOs by ascorbate.
Following BST, biotinylated proteins that were formerly nitrosylated can be detected by Western blotting, or affinity-enriched using streptavidin beads and separated by either 2DE or LC/MS/MS for S-nitrosoproteome-wide identification (Fig. 16A). However, the information on specific nitrosylation sites is often lost during proteomics analysis, since the disulfide-linked biotin group can be removed by reducing agents commonly used before 2DE separations or protease digestions. Although omitting reducing agents can allow biotin-HPDP modification sites to be detected by MS/MS (Fig. 16B), this omission will likely result in reduced detection sensitivity of low abundant proteins, since disulfide-linked proteins are more resistant to trypsin digestion. Consequently, one needs to use a nonreducible substitute reagent to biotin-HPDP in BST to retain nitrosylation site information that can be determined by MS/MS analysis. A variety of biotin-HPDP alternate reagents and methods have been utilized successfully for analyzing different S-nitrosoproteomes. Similar to observations that have been made by others, we have confirmed that biotin maleimide (Biotin M; Fig. 15A) can replace biotin-HPDP for 2DE-based nitrosoproteomic analysis. As an example, HeLa cell proteins were first nitrosylated with 100 μM of GSNO and then processed with BST using either biotin-HPDP or Biotin M (Fig. 17). Biotin-HPDP-proteins could only be detected by Western blotting after nonreducing gel separations, not in reducing gels (Fig. 17A, B). By comparison, biotin-M-proteins could be detected after either non-reducing or reducing gel separations (Fig. 17C, D). Camerini et al. used a novel His-tag conjugated alkylating reagent that binds irreversibly to reduced cysteines generated by ascorbate, allowing formerly nitrosylated proteins and peptides to be purified by a nickel column (34). They identified 28 SNO-Cys within 19 proteins from rat cerebral cortex using this method. Stamler and colleagues used a modification of BST by capturing protein cysteine thiols on a thiol-reactive solid phase resin. When used in conjunction with the iTRAQ protein quantification, this method identified over 400 SNO-peptides, many of which are rapidly denitrosylated in cells (72). More recently, Chen et al., used IAM instead of MMTS for the initial free thiol block, and an irreversible thiol labeling PEO-iodoacetyl-biotin instead of biotin-HPDP for BST (Fig. 15A) (42). They reported the identification of 586 unique nitrosylation sites in 384 proteins. The continued refinement of BST techniques will likely reveal more lower abundant and transiently nitrosylated proteins in the future. It should be noted that BST is not only useful for the detection of SNO-peptides, but can be configured for the identification of other redox-sensitive cysteine-containing peptides (175).
FIG. 16.
Proteomic workflow for the identification of Trx1 transnitrosylation target proteins and their nitrosylation sites. (A) Protein extracted from cells/tissues with elevated Trx1 transnitrosylation can be processed for BST. The SNO-proteins can be detected by Western blotting or enriched by streptavidin beads and separated by 1D/2D gel and proteins bands/spots, digested by trypsin and identified by liquid chromatography/tandem mass spectrometry (LC/MS/MS). For SNO-Cys site identification, the biotinylated proteins can be digested with trypsin, and biotinylated peptides enriched by avidin bead capture and detected by LC/MS/MS. (B) Pyruvate kinase M2 is a transnitrosylation target of Trx1 in Trx1C32S/C35S-overexpressing cells. MS/MS spectrum of a pyruvate kinase M2 peptide 44NTGIIC*TIGPASR56 after biotin labeling. Biotinylated Cys (C*) was found by analyzing the MS/MS spectrum of the peptide. Reprinted from Ref. (273) with permission.
FIG. 17.
Comparison of SNO-protein detection by either reducing or non-reducing gel electrophoresis, following BST with biotin-HPDP or Biotin M. HeLa cell proteins were transnitrosylated in vitro with 100 μM of GSNO and processed by BST according to Jaffrey and Snyder (118). Western blotting analysis against biotin was performed after either 12.5% nonreducing or reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis separation. Left lanes, control; right lanes, GSNO-treated. (A) Biotin-HPDP-labeled proteins were detected only from non-reducing gel-separated proteins. (B) DTT reduction in the reducing gel removed almost all the biotin-HPDP labels before electrophoresis. Biotin M-labeled proteins were detected from both nonreducing (C) and reducing (D) gel-separated proteins (C.W. unpublished data).
C. Gel-based approaches
Following BST, formerly nitrosylated proteins can be analyzed by either gel- or MS-based methods. For example, proteins from SH-SY5Y cells treated with or without GSNO can be isolated and BST-labeled with Biotin M. Avidin-enriched proteins can be separated by 2DE and the relative protein amounts can be quantified by Western blotting (Fig. 18A, B) or by staining with Sypro Ruby (Fig. 18C, D) or other staining methods. The GSNO-induced nitrosylation of proteins can be differentiated from endogenously nitrosylated proteins by the relative intensity of their gel spots. Tandem MS can be used to identify the protein and possibly also to locate its nitrosylation site(s). Recently, an SNO-DIGE (RSNO difference in gel electrophoresis) (48) and fluorescence switch method (257) were described for the quantitative identification of SNO-proteins. In this method, Cy3m, Cy5m, or similar fluorescent dyes are used instead of biotin-HPDP or Biotin M to covalently label ascorbate-freed nitrosylation site cysteines. The labeled proteins are mixed and resolved on the same gel. The key benefit of this method is its ability to compare proteins from different sources within the same gel, thus eliminating the difficulties that arise in matching gel spots when gel-to-gel reproducibility is poor. It is also easier to obtain statistical measurements of significance when using an internal standard-based multiplexing approach. With a fluorescence switch method, Sun et al. identified over 100 SNO-proteins in endothelial cells (251), many of which have increased nitrosylation after shear stress stimulation of eNOS. Chouchani et al. successfully identified 13 mitochondrial SNO-proteins, nitrosylated by a novel mitochondria-targeted RSNO compound (48), and found that aconitase, mitochondrial aldehyde dehydrogenase, and α-ketoglutarate dehydrogenase were selectively and reversibly inhibited by nitrosylation. This technique has been used to identify proteins whose nitrosylation levels are regulated by Trx1. Tello et al. were able to identify increased nitrosylation of 11 SNO-protein species in DIGE-gels in lipopolysaccharide/interferon-treated RAW264.7 cells, cotreated with auranofin, a TrxR inhibitor (257), suggesting that they may be targets of Trx1-mediated denitrosylation. However, given that SNO-Trx1 is also activated upon TrxR inhibition (273), it is likely that some of these proteins are targets of SNO-Trx1-mediated transnitrosylation. Overall, the gel-based methods are effective at revealing multiple protein isoforms that are differentially nitrosylated (257), but are not as sensitive as the shotgun proteomics methods that use the LC/MS/MS approaches for the identification of SNO-proteins and SNO-peptides described below. Among the more sensitive gel methods for protein identification is a method called geLC-MS/MS, in which lower abundance proteins are first separated from the more abundant house-keeping proteins by SDS-PAGE. The gel-separated proteins are digested as individual gel-bands and the resulting peptides identified by LC/MS/MS methods. This method coupled with BST has been used for the identification of over 200 SNO-proteins in human spermatozoa, many of which are unique to this tissue (140). As an alternative to BST, an immuno-spin trapping method for the analysis of SNO-proteins based on the reaction of protein thiyl radicals with 5,5-dimethyl-1-pyrroline N-oxide has been demonstrated (237).
FIG. 18.
2DE analysis of SNO-proteins from SH-SY5Y cells. After SH-SY5Y cells were treated with 100 μM of GSNO, proteins were extracted and labeled with Biotin M following the BST protocol (118). After 2DE separation of the labeled proteins, Western blotting was performed to detect biotinylated proteins isolated from either (A) untreated or (B) 100 μM GSNO-treated SH-SY5Y cells. Substantially more biotinylated proteins were detected after GSNO treatment of the cells. Biotin M-labeled proteins were affinity enriched by streptavidin beads from either (C) untreated or (D) GSNO-treated samples; the gels were stained with Sypro Ruby fluorescent dye. More biotinylated proteins can be detected after GSNO treatment and subsequently can be identified by LC/MS/MS (C.W. unpublished data).
D. MS-based approaches
BST affinity purified proteins have been used to identify hundreds of SNO-sites within SNO-proteins and peptides (42). Extending the BST paradigm, stable isotope-based biotinylating reagents or other quantitative shotgun proteomics reagents have been used in place of, or in combination with biotin-HPDP for quantitative analysis of changes in SNO-peptides. For example, the ICAT reagents discussed previously have been tailored to quantify the nitrosylation reactivity of protein tyrosine phosphatase 1B, and enabled the identification of Cys215 as the primary residue prone to nitrosylation (43). Recently, a solid-phase version of the BST method was used in conjunction with iTRAQ to quantify the rate of protein/peptide denitrosylation after the prior treatment of cells with nitrosylating agents (72). Nearly 300 peptides that underwent rapid denitrosylation within the cells were quantified by this method, suggesting that this method is effective for the identification and quantification of low-abundance peptides. Using motif search software to compare the peptide sequences reported in this study that are denitrosylated rapidly, we found that there may be several lysine-based CX5K and CX6K (X=any amino acid) motifs present among the denitrosylated peptides (Fig. 19). Obviously, it remains to be determined whether such motifs serve any purpose in directing denitrosylase(s) to their target proteins. Recently, Benhar et al. coupled SILAC with BST and quantitatively identified 46 new substrates of Trx1 (23): some of which are almost completely denitrosylated by Trx1-(SH)2 (e.g., leucyl-cystinyl aminopeptidase), whereas others are partially denitrosylated (e.g., epoxide hydrolase 2). Interestingly, they found that several Jurkat cell proteins that are pre-nitrosylated by CysNO, including Prx1, are not denitrosylated by Trx1-(SH)2, confirming that Trx1-(SH)2 is a specific denitrosylation agent of only select targets. Overall, it suggests that large-scale quantitative proteomics methods can be easily adapted for the identification of additional Trx1 transnitrosylation and denitrosylation targets, and facilitate an understanding of the underlying mechanisms of target specificity.
FIG. 19.
Identification of putative denitrosylation protein motifs. HEK293 cell proteins that have been shown to undergo rapid denitrosylation from a recent proteomics study (72) were subjected to Motif-X (http://motif-x.med.harvard.edu/motif-x.html) analysis of the amino acid residues surrounding the nitrosylated cysteines. The following parameters were used: central character=“C”; width=“17”; occurrences=“5”; significance=“0.0001”; background=“ipi.HUMAN.fasta”; foreground format=“fasta”. Two potential motifs CX6K (left panel) and CX5K (right panel) were found in 43 and 37 peptides, respectively, from a total of 398 identified denitrosylation sites. Proteins that contain these motifs are listed below their respective motif (C.W. and H.L. unpublished data).
E. Advanced proteomics approaches
Once an SNO-Cys within a specific protein has been identified by global proteomics approaches, biologists need to be able to verify that these nitrosylation events are indeed occurring within physiological or pathological contexts. Therefore, it becomes important to quantify changes in specific nitrosylation events and assess the regulation of their reversibility. One means is to develop antibodies against specific SNO-sites, but this is time consuming and may not always be possible. The MS-based selected reaction monitoring (SRM) quantitative technique, commonly used in the quantification of drug-like molecules, could be a viable alternative. This type of analysis is usually carried out on a triple-quadrupole tandem mass spectrometer. This technique relies on the fact that a peptide of a particular size (or mass/charge) can be isolated and fragmented into specific fragment ions in a tandem mass spectrometer. The selectivity of the SRM approach is based on the premise that a parent peptide-to-fragment ion transition pair may be unique and can be used to specifically quantify the peptide within biological matrices, including cell lysates or serums. Several SRM experiments can be carried out within the same LC/MS/MS analysis for the quantification of multiple peptides, and it is therefore called multiple reactions monitoring (MRM). The MRM method has been widely used in the pharmaceutical industry to quantify drugs and their metabolites during pharmaceutical development. More recently, it has been increasingly used for the quantification of peptide biomarkers. Analogous to multiplexed immunoassays for the quantification of many low abundant cellular analytes such as cytokines, the MRM approach can be used for the quantification of a collection of SNO-peptides. The application of SRM or MRM for SNO-peptide quantification has not been widely reported, presumably due to nitrosylation being very labile during the peptide ionization process. However, it is conceivable that following BST, former SNO-peptides can be identified and quantified as biotin-peptides by MRM. For example, biotinylated peptides tend to produce a characteristic collision-induced fragment ion around 428 Da (230), a characteristic that may be used for the sensitive identification of biotin-peptides that were formerly nitrosylated. Similar to detection of neutral loss of 98 Da for identifying low abundance phosphopeptides within complex mixtures, precursor peptides that produce a 428 Da diagnostic ion can be thoroughly interrogated to map their biotinylation (a surrogate for nitrosylation) sites and to quantify their charges. This method has been used effectively to identify sulfinamide modification of cysteines after reaction with nitrosyl (HNO) within human platelet proteins (105), and it is likely to also enable the identification of low abundance SNO-peptides. Further, an OxMRM method that combines differential alkylation with MRM has been effective in identifying redox-sensitive PTMs in proteins and peptides, including disulfide bond linked peptides (99).
F. Strengths, limitations and complementarity
Global proteomics methods have been proven effective at identifying novel proteins whose disulfide bond or nitrosylation status are regulated by Trx1 and related proteins. However, proteins in some functional groups are highly represented in the Trx1 target proteomes, including sugar and lipid metabolism, protein chaperons, and mitochondrial proteins (23, 75, 273). This observation is due at least in part to the fact that these proteins are typically more highly abundant and are routinely identified by global proteomics studies, regardless of whether protein PTMs are selected or enriched. Many less abundant, redox-sensitive signaling kinases, phosphatases, proteases, and transcription factors are typically identified or analyzed, not through global approaches, but by molecular biological approaches, including Western blotting and mutagenesis.
Although some of the subproteome approaches described in this review have been effective at identifying lower abundant Trx1 target proteins, the current state-of-the-art instrumentation is still not able to directly identify lower abundant signal transduction regulators. Developments in more advanced chromatographic and iso-electric focusing-based technologies for high-resolution peptide separations may allow low abundance proteins and their oxidative PTMs to be identified. Further, the recent introduction of newer MS instruments, capable of routine analysis of labile peptides by electron capture dissociation or electron impact dissociation, may enable the direct localization of SNO-Cys without the need for BST, reducing the possibility of false-positives. It is likely that with a combination of advancements in subcellular fraction and MS detection sensitivity, more and more Trx1-targeted kinases, phosphatases, and transcription factors can be identified.
To identify Trx1 transnitrosylation targets, we previously used the Trx1 dominant negative and denitrosylation-inactive mutant Trx1C32S/C35S (77). Overexpression of this mutant in HeLa cells resulted in an observable increase in protein nitrosylation, and after BST, we were able to identify 36 SNO-Cys sites by MS/MS pertaining to 28 target proteins (273). Benhar et al. used an analogous approach, in which Trx1-S2 was produced by incubating cells with TrxR inhibitors, for the identification of Trx1 denitrosylation targets (23). A potential drawback of these approaches includes the inability to distinguish genuine Trx1 transnitrosylation targets from denitrosylation targets, since both Trx1-S2 and Trx1C32S/C35S are not only inactive as a denitrosylase, but also are capable of being converted into a transnitrosylating agent (273). Therefore, the increase in SNO-proteins in these cells may be due to either increased Trx1-mediated transnitrosylation, decreased denitrosylation, or both. To overcome these technical problems, experiments should be designed such that an increase in cellular SNO-protein due to attenuation of Trx1 denitrosylation can be distinguished from SNO-Trx1-mediated transnitrosylation.
Finally, conventional BST methods are unable to accurately quantify the extent of S-nitrosylation changes at a particular SNO-Cys site; such information is crucial for distinguishing specific Trx1 catalytic targets from nonspecific ones. One means of accurately quantifying different SNO-Cys site responses to Trx1 modulation is to substitute ICAT reagents for biotin-HPDP in BST (282). Alternatively, the BST method can be coupled with SILAC or other MS-based quantification methods, to quantify the degree of nitrosylation change within specific Cys-containing peptides (23).
VIII. Conclusions
Reversible cysteine oxidative modifications, including disulfide bond formation (29) and nitrosylation (95), have been shown to regulate protein functions and to propagate redox signal transduction pathways. The Trx1 system is a key regulator of protein disulfide and nitrosylation status. Under physiological conditions, Trx1 maintains the redox status of proteins via its reductase or denitrosylating activities, sustained with the reducing equivalent from TrxR/NADPH. Within highly oxidative environments, Trx1 becomes S-nitrosylated and offers an alternative modality of protein regulation via transnitrosylation. Transnitrosylation may serve two purposes: to temporarily prevent proteins from acquiring irreversible oxidative modifications when cellular antioxidant systems are overwhelmed, and to serve as a signaling mechanism for altering protein function, contributing to the cellular stress response. It is anticipated that adaptation of the latest redox proteomics technologies to the identification of both S-nitrosoproteome and disulfide proteome-wide Trx1 target proteins will enable the discovery of novel signaling proteins. This may provide insights into the specific structural determinants and mechanisms that govern Trx1 and its interactions with target proteins, solidifying its role as a specific disulfide reductase, and denitrosylating and transnitrosylating agent. Further, the identification of specific target proteins of Trx1-mediated transnitrosylation and denitrosylation will facilitate a deeper understanding of how these two contrary activities are regulated within different cellular or physiological environments. Clinical interventions that target Trx1 for the modulation of protein disulfide reduction, nitrosylation, or denitrosylation may provide the specificities that are required for the management of diseases related to oxidative and nitrosative imbalance and the design of personalized medicines.
Supplementary Material
Abbreviations Used
- 1DE
one-dimensional gel electrophoresis
- 2DE
two-dimensional gel electrophoresis
- ASK1
apoptosis signal-regulating kinase 1
- ATP
adenosine triphosphate
- Biotin-HPDP
N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide
- Biotin M
biotin maleimide
- BR
bilirubin
- BST
biotin switch technique
- Casp3
caspase 3
- Casp3p
caspase 3 peptide
- cGMP
cyclic guanosine monophosphate
- Co-IP
coimmunoprecipitation
- CID
collision-induced dissociation
- Cy3m
Cy3 maleimide
- DIGE
difference in gel electrophoresis
- DnaJb5
DnaJ/Hsp40 homolog, subfamily B, member 5
- DTT
dithiothreitol
- EMSA
electromobility shift assay
- eNOS
endothelial nitric oxide synthase
- ESR
electron spin resonance
- FAD
flavin adenine dinucleotide
- FCA
flow cytometry assay
- FRET
fluorescence resonance energy transfer
- GAPDH
glyceraldehyde 3′-phosphate dehydrogenase
- GPCR
G protein-coupled receptor
- Grx1
glutaredoxin 1
- GSH
glutathione
- GSNO
nitrosoglutathione
- GSNOR
GSNO reductase
- HDAC
histone deacetylase
- HIF
hypoxia inducible factor
- HNE
4-hydroxy-2-nonenal
- HPLC
high performance (pressure) liquid chromatography
- HSA
human serum albumin
- Hsp90
heat shock protein 90
- IAM
iodoacetamide
- IB
immunoblot
- ICAT
isotope-coded affinity tag
- IF
immunofluorescence
- iNOS
inducible NO synthase
- IP
immunoprecipitation
- iTRAQ
isobaric tags for relative and absolute quantification
- LC
liquid chromatography
- LC/MS/MS
liquid chromatography/tandem mass spectrometry
- MALDI
matrix-assisted laser desorption ionization
- MMTS
methylmethanethiosulfonate
- MPTP
mitochondrial permeability transition pore
- MRM
multiple reactions monitoring
- MS/MS
tandem mass spectrometry
- NAC
N-acetyl-cysteine
- NADH
nicotine adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- NFAT
nuclear factor of activated T-cells
- NF-κB
nuclear factor κB
- NMDA
N-methyl-D-aspartic acid
- NO
nitric oxide
- NOS
nitric oxide synthase
- N2O3
dinitrogen trioxide
- oCasp3p
oxidized caspase 3 peptide
- PDI
protein disulfide isomerase
- Prx
peroxiredoxin
- PSD
postsource decay
- PTEN
phosphatase and tensin homolog
- PTMs
post-translational modifications
- PTP1B
protein tyrosine phosphatase 1B
- rCasp3p
reduced caspase 3 peptide
- RNAi
RNA interference assay
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RPLC
reversed phase liquid chromatography
- RSNO
S-nitrosothiol
- RyR
ryanodine receptor
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- sGC
soluble guanylyl cyclase
- SILAC
stable isotope labeling by amino acids in cell culture
- SNO-Casp3
S-nitrosylated caspase 3
- SNO-Cys
S-nitrosylated cysteine
- SNO-proteins
S-nitrosylated proteins
- SNO-Prx1
S-nitrosylated Prx1
- SNO-Trx1
S-nitrosylated Trx1
- SRM
selected reaction monitoring
- Srx
sulfiredoxin
- Tg mice
Trx1 transgenic mice
- TRP
thioredoxin related protein
- Trx1
thioredoxin 1
- Trx1-S2
disulfide Trx1
- Trx1-(SH)2
dithiol Trx1
- TrxR
thioredoxin reductase
- TXNIP or TBP-2
thioredoxin-interacting protein-2
- VHL
Von Hippel-Lindau
- Y2H
yeast two-hybrid assay
Acknowledgments
The authors are grateful for funding support from NIH grant NS046593 to H.L., for the establishment of the UMDNJ Neuroproteomics Core Facility, by a UMDNJ Foundation grant to H.L. and J.S., by NIH grants AG23039 and HL91469 to J.S. and by NIH grants GM067640 and HL089771 to A.B.
References
- 1.Abbas K. Breton J. Drapier JC. The interplay between nitric oxide and peroxiredoxins. Immunobiology. 2008;213:815–822. doi: 10.1016/j.imbio.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 2.Adachi T. Pimentel DR. Heibeck T. Hou X. Lee YJ. Jiang B. Ido Y. Cohen RA. S-glutathiolation of Ras mediates redox-sensitive signaling by angiotensin II in vascular smooth muscle cells. J Biol Chem. 2004;279:29857–29862. doi: 10.1074/jbc.M313320200. [DOI] [PubMed] [Google Scholar]
- 3.Ago T. Liu T. Zhai P. Chen W. Li H. Molkentin JD. Vatner SF. Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 4.Ago T. Sadoshima J. Thioredoxin1 as a negative regulator of cardiac hypertrophy. Antioxid Redox Signal. 2007;9:679–687. doi: 10.1089/ars.2007.1529. [DOI] [PubMed] [Google Scholar]
- 5.Akamatsu Y. Ohno T. Hirota K. Kagoshima H. Yodoi J. Shigesada K. Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. J Biol Chem. 1997;272:14497–14500. doi: 10.1074/jbc.272.23.14497. [DOI] [PubMed] [Google Scholar]
- 6.Aracena P. Tang W. Hamilton SL. Hidalgo C. Effects of S-glutathionylation and S-nitrosylation on calmodulin binding to triads and FKBP12 binding to type 1 calcium release channels. Antioxid Redox Signal. 2005;7:870–881. doi: 10.1089/ars.2005.7.870. [DOI] [PubMed] [Google Scholar]
- 7.Arner ES. Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 8.Arner ES. Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006;16:420–426. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Arnold WP. Mittal CK. Katsuki S. Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc Natl Acad Sci U S A. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aslund F. Berndt KD. Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 11.Atochina-Vasserman EN. Gow AJ. Abramova H. Guo CJ. Tomer Y. Preston AM. Beck JM. Beers MF. Immune reconstitution during Pneumocystis lung infection: disruption of surfactant component expression and function by S-nitrosylation. J Immunol. 2009;182:2277–2287. doi: 10.4049/jimmunol.0802775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avval FZ. Holmgren A. Molecular mechanisms of thioredoxin and glutaredoxin as hydrogen donors for Mammalians phase ribonucleotide reductase. J Biol Chem. 2009;284:8233–8240. doi: 10.1074/jbc.M809338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Backs J. Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 14.Baker A. Payne CM. Briehl MM. Powis G. Thioredoxin, a gene found overexpressed in human cancer, inhibits apoptosis in vitro and in vivo. Cancer Res. 1997;57:5162–5167. [PubMed] [Google Scholar]
- 15.Balmer Y. Koller A. del Val G. Manieri W. Schurmann P. Buchanan BB. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci U S A. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balmer Y. Vensel WH. Tanaka CK. Hurkman WJ. Gelhaye E. Rouhier N. Jacquot JP. Manieri W. Schurmann P. Droux M. Buchanan BB. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci U S A. 2004;101:2642–2647. doi: 10.1073/pnas.0308583101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barone E. Trombino S. Cassano R. Sgambato A. De Paola B. Di Stasio E. Picci N. Preziosi P. Mancuso C. Characterization of the S-denitrosylating activity of bilirubin. J Cell Mol Med. 2009;13:2365–2375. doi: 10.1111/j.1582-4934.2008.00680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu S. Grubina R. Huang J. Conradie J. Huang Z. Jeffers A. Jiang A. He X. Azarov I. Seibert R. Mehta A. Patel R. King SB. Hogg N. Ghosh A. Gladwin MT. Kim-Shapiro DB. Catalytic generation of N2O3 by the concerted nitrite reductase and anhydrase activity of hemoglobin. Nat Chem Biol. 2007;3:785–794. doi: 10.1038/nchembio.2007.46. [DOI] [PubMed] [Google Scholar]
- 19.Baty JW. Hampton MB. Winterbourn CC. Detection of oxidant sensitive thiol proteins by fluorescence labeling and two-dimensional electrophoresis. Proteomics. 2002;2:1261–1266. doi: 10.1002/1615-9861(200209)2:9<1261::AID-PROT1261>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 20.Beckman JS. Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 21.Benhar M. Forrester MT. Hess DT. Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benhar M. Forrester MT. Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 23.Benhar M. Thompson JW. Moseley MA. Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett CF. Balcarek JM. Varrichio A. Crooke ST. Molecular cloning and complete amino-acid sequence of form-I phosphoinositide-specific phospholipase C. Nature. 1988;334:268–270. doi: 10.1038/334268a0. [DOI] [PubMed] [Google Scholar]
- 25.Bentz BG. Simmons RL. Haines GK., 3rd Radosevich JA. The yin and yang of nitric oxide: reflections on the physiology and pathophysiology of NO. Head Neck. 2000;22:71–83. doi: 10.1002/(sici)1097-0347(200001)22:1<71::aid-hed11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 26.Bertini R. Howard OM. Dong HF. Oppenheim JJ. Bizzarri C. Sergi R. Caselli G. Pagliei S. Romines B. Wilshire JA. Mengozzi M. Nakamura H. Yodoi J. Pekkari K. Gurunath R. Holmgren A. Herzenberg LA. Herzenberg LA. Ghezzi P. Thioredoxin, a redox enzyme released in infection and inflammation, is a unique chemoattractant for neutrophils, monocytes, and T cells. J Exp Med. 1999;189:1783–1789. doi: 10.1084/jem.189.11.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloomfield KL. Osborne SA. Kennedy DD. Clarke FM. Tonissen KF. Thioredoxin-mediated redox control of the transcription factor Sp1 and regulation of the thioredoxin gene promoter. Gene. 2003;319:107–116. doi: 10.1016/s0378-1119(03)00799-6. [DOI] [PubMed] [Google Scholar]
- 28.Bogumil R. Namgaladze D. Schaarschmidt D. Schmachtel T. Hellstern S. Mutzel R. Ullrich V. Inactivation of calcineurin by hydrogen peroxide and phenylarsine oxide. Evidence for a dithiol-disulfide equilibrium and implications for redox regulation. Eur J Biochem. 2000;267:1407–1415. doi: 10.1046/j.1432-1327.2000.01133.x. [DOI] [PubMed] [Google Scholar]
- 29.Brennan JP. Wait R. Begum S. Bell JR. Dunn MJ. Eaton P. Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J Biol Chem. 2004;279:41352–41360. doi: 10.1074/jbc.M403827200. [DOI] [PubMed] [Google Scholar]
- 30.Broillet MC. Firestein S. Beta subunits of the olfactory cyclic nucleotide-gated channel form a nitric oxide activated Ca2+channel. Neuron. 1997;18:951–958. doi: 10.1016/s0896-6273(00)80334-7. [DOI] [PubMed] [Google Scholar]
- 31.Bruick RK. McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 32.Calabrese V. Cornelius C. Rizzarelli E. Owen JB. Dinkova-Kostova AT. Butterfield DA. Nitric oxide in cell survival: a janus molecule. Antioxid Redox Signal. 2009;11:2717–2739. doi: 10.1089/ars.2009.2721. [DOI] [PubMed] [Google Scholar]
- 33.Calabrese V. Mancuso C. Calvani M. Rizzarelli E. Butterfield DA. Stella AM. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 34.Camerini S. Polci ML. Restuccia U. Usuelli V. Malgaroli A. Bachi A. A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J Proteome Res. 2007;6:3224–3231. doi: 10.1021/pr0701456. [DOI] [PubMed] [Google Scholar]
- 35.Carvalho AP. Fernandes PA. Ramos MJ. Similarities and differences in the thioredoxin superfamily. Prog Biophys Mol Biol. 2006;91:229–248. doi: 10.1016/j.pbiomolbio.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Carvalho AT. Fernandes PA. Swart M. Van Stralen JN. Bickelhaupt FM. Ramos MJ. Role of the variable active site residues in the function of thioredoxin family oxidoreductases. J Comput Chem. 2009;30:710–724. doi: 10.1002/jcc.21086. [DOI] [PubMed] [Google Scholar]
- 37.Casagrande S. Bonetto V. Fratelli M. Gianazza E. Eberini I. Massignan T. Salmona M. Chang G. Holmgren A. Ghezzi P. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proc Natl Acad Sci U S A. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cha MK. Kim IH. Disulfide between Cys392 and Cys438 of human serum albumin is redox-active, which is responsible for the thioredoxin-supported lipid peroxidase activity. Arch Biochem Biophys. 2006;445:19–25. doi: 10.1016/j.abb.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 39.Chae HZ. Kim HJ. Kang SW. Rhee SG. Characterization of three isoforms of mammalian peroxiredoxin that reduce peroxides in the presence of thioredoxin. Diabetes Res Clin Pract. 1999;45:101–112. doi: 10.1016/s0168-8227(99)00037-6. [DOI] [PubMed] [Google Scholar]
- 40.Chai YC. Jung CH. Lii CK. Ashraf SS. Hendrich S. Wolf B. Sies H. Thomas JA. Identification of an abundant S-thiolated rat liver protein as carbonic anhydrase III; characterization of S-thiolation and dethiolation reactions. Arch Biochem Biophys. 1991;284:270–278. doi: 10.1016/0003-9861(91)90295-t. [DOI] [PubMed] [Google Scholar]
- 41.Chen CY. Willard D. Rudolph J. Redox regulation of SH2-domain-containing protein tyrosine phosphatases by two backdoor cysteines. Biochemistry. 2009;48:1399–1409. doi: 10.1021/bi801973z. [DOI] [PubMed] [Google Scholar]
- 42.Chen YJ. Ku WC. Lin PY. Chou HC. Khoo KH. S-alkylating labeling strategy for site-specific identification of the s-nitrosoproteome. J Proteome Res. 2010;9:6417–6439. doi: 10.1021/pr100680a. [DOI] [PubMed] [Google Scholar]
- 43.Chen YY. Chu HM. Pan KT. Teng CH. Wang DL. Wang AH. Khoo KH. Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevion M. Berenshtein E. Stadtman ER. Human studies related to protein oxidation: protein carbonyl content as a marker of damage. Free Radic Res. 2000;33(Suppl):S99–S108. [PubMed] [Google Scholar]
- 45.Chiueh CC. Rauhala P. The redox pathway of S-nitrosoglutathione, glutathione and nitric oxide in cell to neuron communications. Free Radic Res. 1999;31:641–650. doi: 10.1080/10715769900301211. [DOI] [PubMed] [Google Scholar]
- 46.Choi HI. Lee SP. Kim KS. Hwang CY. Lee YR. Chae SK. Kim YS. Chae HZ. Kwon KS. Redox-regulated cochaperone activity of the human DnaJ homolog Hdj2. Free Radic Biol Med. 2006;40:651–659. doi: 10.1016/j.freeradbiomed.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 47.Choi YB. Tenneti L. Le DA. Ortiz J. Bai G. Chen HS. Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- 48.Chouchani ET. Hurd TR. Nadtochiy SM. Brookes PS. Fearnley IM. Lilley KS. Smith RA. Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu K. Vojtchovsky J. McMahon BH. Sweet RM. Berendzen J. Schlichting I. Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature. 2000;403:921–923. doi: 10.1038/35002641. [DOI] [PubMed] [Google Scholar]
- 50.Cumming RC. Andon NL. Haynes PA. Park M. Fischer WH. Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 51.D'Autreaux B. Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 52.DeMorrow S. Francis H. Gaudio E. Ueno Y. Venter J. Onori P. Franchitto A. Vaculin B. Vaculin S. Alpini G. Anandamide inhibits cholangiocyte hyperplastic proliferation via activation of thioredoxin 1/redox factor 1 and AP-1 activation. Am J Physiol Gastrointest Liver Physiol. 2008;294:G506–G519. doi: 10.1152/ajpgi.00304.2007. [DOI] [PubMed] [Google Scholar]
- 53.de Pina MZ. Vazquez-Meza H. Pardo JP. Rendon JL. Villalobos-Molina R. Riveros-Rosas H. Pina E. Signaling the signal, cyclic AMP-dependent protein kinase inhibition by insulin-formed H2O2 and reactivation by thioredoxin. J Biol Chem. 2008;283:12373–12386. doi: 10.1074/jbc.M706832200. [DOI] [PubMed] [Google Scholar]
- 54.Derakhshan B. Hao G. Gross SS. Balancing reactivity against selectivity: the evolution of protein S-nitrosylation as an effector of cell signaling by nitric oxide. Cardiovasc Res. 2007;75:210–219. doi: 10.1016/j.cardiores.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diet A. Abbas K. Bouton C. Guillon B. Tomasello F. Fourquet S. Toledano MB. Drapier JC. Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J Biol Chem. 2007;282:36199–36205. doi: 10.1074/jbc.M706420200. [DOI] [PubMed] [Google Scholar]
- 56.Dunn LL. Buckle AM. Cooke JP. Ng MK. The emerging role of the thioredoxin system in angiogenesis. Arterioscler Thromb Vasc Biol. 2010;30:2089–2098. doi: 10.1161/ATVBAHA.110.209643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ekmekcioglu S. Ellerhorst J. Smid CM. Prieto VG. Munsell M. Buzaid AC. Grimm EA. Inducible nitric oxide synthase and nitrotyrosine in human metastatic melanoma tumors correlate with poor survival. Clin Cancer Res. 2000;6:4768–4775. [PubMed] [Google Scholar]
- 58.Ema M. Hirota K. Mimura J. Abe H. Yodoi J. Sogawa K. Poellinger L. Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1 alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Epstein AC. Gleadle JM. McNeill LA. Hewitson KS. O'Rourke J. Mole DR. Mukherji M. Metzen E. Wilson MI. Dhanda A. Tian YM. Masson N. Hamilton DL. Jaakkola P. Barstead R. Hodgkin J. Maxwell PH. Pugh CW. Schofield CJ. Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 60.Erwin PA. Lin AJ. Golan DE. Michel T. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J Biol Chem. 2005;280:19888–19894. doi: 10.1074/jbc.M413058200. [DOI] [PubMed] [Google Scholar]
- 61.Erwin PA. Mitchell DA. Sartoretto J. Marletta MA. Michel T. Subcellular targeting and differential S-nitrosylation of endothelial nitric-oxide synthase. J Biol Chem. 2006;281:151–157. doi: 10.1074/jbc.M510421200. [DOI] [PubMed] [Google Scholar]
- 62.Essex DW. Li M. Miller A. Feinman RD. Protein disulfide isomerase and sulfhydryl-dependent pathways in platelet activation. Biochemistry. 2001;40:6070–6075. doi: 10.1021/bi002454e. [DOI] [PubMed] [Google Scholar]
- 63.Eu JP. Sun J. Xu L. Stamler JS. Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 64.Fang J. Holmgren A. Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J Am Chem Soc. 2006;128:1879–1885. doi: 10.1021/ja057358l. [DOI] [PubMed] [Google Scholar]
- 65.Fang J. Nakamura T. Cho DH. Gu Z. Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang M. Jaffrey SR. Sawa A. Ye K. Luo X. Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 67.Foglio MH. Duester G. Characterization of the functional gene encoding mouse class III alcohol dehydrogenase (glutathione-dependent formaldehyde dehydrogenase) and an unexpressed processed pseudogene with an intact open reading frame. Eur J Biochem. 1996;237:496–504. doi: 10.1111/j.1432-1033.1996.0496k.x. [DOI] [PubMed] [Google Scholar]
- 68.Foloppe N. Nilsson L. The glutaredoxin -C-P-Y-C- motif: influence of peripheral residues. Structure. 2004;12:289–300. doi: 10.1016/j.str.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 69.Foloppe N. Sagemark J. Nordstrand K. Berndt KD. Nilsson L. Structure, dynamics and electrostatics of the active site of glutaredoxin 3 from Escherichia coli: comparison with functionally related proteins. J Mol Biol. 2001;310:449–470. doi: 10.1006/jmbi.2001.4767. [DOI] [PubMed] [Google Scholar]
- 70.Fomenko DE. Gladyshev VN. Identity and functions of CxxC-derived motifs. Biochemistry. 2003;42:11214–11225. doi: 10.1021/bi034459s. [DOI] [PubMed] [Google Scholar]
- 71.Forrester MT. Seth D. Hausladen A. Eyler CE. Foster MW. Matsumoto A. Benhar M. Marshall HE. Stamler JS. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J Biol Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forrester MT. Thompson JW. Foster MW. Nogueira L. Moseley MA. Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foster MW. Hess DT. Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu C. Hu J. Liu T. Ago T. Sadoshima J. Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res. 2008;7:3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu C. Wu C. Liu T. Ago T. Zhai P. Sadoshima J. Li H. Elucidation of thioredoxin target protein networks in mouse. Mol Cell Proteomics. 2009;8:1674–1687. doi: 10.1074/mcp.M800580-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujii J. Ikeda Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002;7:123–130. doi: 10.1179/135100002125000352. [DOI] [PubMed] [Google Scholar]
- 77.Gallegos A. Gasdaska JR. Taylor CW. Paine-Murrieta GD. Goodman D. Gasdaska PY. Berggren M. Briehl MM. Powis G. Transfection with human thioredoxin increases cell proliferation and a dominant-negative mutant thioredoxin reverses the transformed phenotype of human breast cancer cells. Cancer Res. 1996;56:5765–5770. [PubMed] [Google Scholar]
- 78.Gallogly MM. Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 79.Gallogly MM. Starke DW. Mieyal JJ. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation. Antioxid Redox Signal. 2009;11:1059–1081. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gasdaska JR. Kirkpatrick DL. Montfort W. Kuperus M. Hill SR. Berggren M. Powis G. Oxidative inactivation of thioredoxin as a cellular growth factor and protection by a Cys73—>Ser mutation. Biochem Pharmacol. 1996;52:1741–1747. doi: 10.1016/s0006-2952(96)00595-3. [DOI] [PubMed] [Google Scholar]
- 81.Gaston BM. Carver J. Doctor A. Palmer LA. S-nitrosylation signaling in cell biology. Mol Interv. 2003;3:253–263. doi: 10.1124/mi.3.5.253. [DOI] [PubMed] [Google Scholar]
- 82.Go YM. Halvey PJ. Hansen JM. Reed M. Pohl J. Jones DP. Reactive aldehyde modification of thioredoxin-1 activates early steps of inflammation and cell adhesion. Am J Pathol. 2007;171:1670–1681. doi: 10.2353/ajpath.2007.070218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Godoy L. Gonzalez-Duarte R. Albalat R. S-nitrosogluthathione reductase activity of amphioxus ADH3: insights into the nitric oxide metabolism. Int J Biol Sci. 2006;2:117–124. doi: 10.7150/ijbs.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gorman JJ. Wallis TP. Pitt JJ. Protein disulfide bond determination by mass spectrometry. Mass Spectrom Rev. 2002;21:183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 85.Greco TM. Hodara R. Parastatidis I. Heijnen HF. Dennehy MK. Liebler DC. Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo Y. Einhorn L. Kelley M. Hirota K. Yodoi J. Reinbold R. Scholer H. Ramsey H. Hromas R. Redox regulation of the embryonic stem cell transcription factor oct-4 by thioredoxin. Stem Cells. 2004;22:259–264. doi: 10.1634/stemcells.22-3-259. [DOI] [PubMed] [Google Scholar]
- 87.Gygi SP. Rist B. Gerber SA. Turecek F. Gelb MH. Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 88.Haendeler J. Hoffmann J. Tischler V. Berk BC. Zeiher AM. Dimmeler S. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat Cell Biol. 2002;4:743–749. doi: 10.1038/ncb851. [DOI] [PubMed] [Google Scholar]
- 89.Haendeler J. Hoffmann J. Zeiher AM. Dimmeler S. Antioxidant effects of statins via S-nitrosylation and activation of thioredoxin in endothelial cells: a novel vasculoprotective function of statins. Circulation. 2004;110:856–861. doi: 10.1161/01.CIR.0000138743.09012.93. [DOI] [PubMed] [Google Scholar]
- 90.Hagglund P. Bunkenborg J. Maeda K. Svensson B. Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J Proteome Res. 2008;7:5270–5276. doi: 10.1021/pr800633y. [DOI] [PubMed] [Google Scholar]
- 91.Hall A. Karplus PA. Poole LB. Typical 2-Cys peroxiredoxins—structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Han S. Force field parameters for S-nitrosocysteine and molecular dynamics simulations of S-nitrosated thioredoxin. Biochem Biophys Res Commun. 2008;377:612–616. doi: 10.1016/j.bbrc.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 93.Hansen JM. Watson WH. Jones DP. Compartmentation of Nrf-2 redox control: regulation of cytoplasmic activation by glutathione and DNA binding by thioredoxin-1. Toxicol Sci. 2004;82:308–317. doi: 10.1093/toxsci/kfh231. [DOI] [PubMed] [Google Scholar]
- 94.Hansen RE. Winther JR. An introduction to methods for analyzing thiols and disulfides: reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 95.Hao G. Derakhshan B. Shi L. Campagne F. Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hashemy SI. Holmgren A. Regulation of the catalytic activity and structure of human thioredoxin 1 via oxidation and S-nitrosylation of cysteine residues. J Biol Chem. 2008;283:21890–21898. doi: 10.1074/jbc.M801047200. [DOI] [PubMed] [Google Scholar]
- 97.Haugstetter J. Blicher T. Ellgaard L. Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J Biol Chem. 2005;280:8371–8380. doi: 10.1074/jbc.M413924200. [DOI] [PubMed] [Google Scholar]
- 98.Hayashi S. Hajiro-Nakanishi K. Makino Y. Eguchi H. Yodoi J. Tanaka H. Functional modulation of estrogen receptor by redox state with reference to thioredoxin as a mediator. Nucleic Acids Res. 1997;25:4035–4040. doi: 10.1093/nar/25.20.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Held JM. Danielson SR. Behring JB. Atsriku C. Britton DJ. Puckett RL. Schilling B. Campisi J. Benz CC. Gibson BW. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol Cell Proteomics. 2010;9:1400–1410. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hess DT. Matsumoto A. Kim SO. Marshall HE. Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 101.Hess DT. Matsumoto A. Nudelman R. Stamler JS. S-nitrosylation: spectrum and specificity. Nat Cell Biol. 2001;3:E46–E49. doi: 10.1038/35055152. [DOI] [PubMed] [Google Scholar]
- 102.Hidalgo C. Donoso P. Carrasco MA. The ryanodine receptors Ca2+release channels: cellular redox sensors? IUBMB Life. 2005;57:315–322. doi: 10.1080/15216540500092328. [DOI] [PubMed] [Google Scholar]
- 103.Hirota K. Matsui M. Iwata S. Nishiyama A. Mori K. Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci U S A. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hirota K. Murata M. Sachi Y. Nakamura H. Takeuchi J. Mori K. Yodoi J. Distinct roles of thioredoxin in the cytoplasm and in the nucleus. A two-step mechanism of redox regulation of transcription factor NF-kappaB. J Biol Chem. 1999;274:27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 105.Hoffman MD. Walsh GM. Rogalski JC. Kast J. Identification of nitroxyl-induced modifications in human platelet proteins using a novel mass spectrometric detection method. Mol Cell Proteomics. 2009;8:887–903. doi: 10.1074/mcp.M800230-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Holmgren A. Enzymatic reduction-oxidation of protein disulfides by thioredoxin. Methods Enzymol. 1984;107:295–300. doi: 10.1016/0076-6879(84)07019-1. [DOI] [PubMed] [Google Scholar]
- 107.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 108.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 109.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 110.Holmgren A. Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem Biophys Res Commun. 2010;396:120–124. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- 111.Huang Y. Man HY. Sekine-Aizawa Y. Han Y. Juluri K. Luo H. Cheah J. Lowenstein C. Huganir RL. Snyder SH. S-nitrosylation of N-ethylmaleimide sensitive factor mediates surface expression of AMPA receptors. Neuron. 2005;46:533–540. doi: 10.1016/j.neuron.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 112.Hurd TR. Prime TA. Harbour ME. Lilley KS. Murphy MP. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J Biol Chem. 2007;282:22040–22051. doi: 10.1074/jbc.M703591200. [DOI] [PubMed] [Google Scholar]
- 113.Hwang CY. Ryu YS. Chung MS. Kim KD. Park SS. Chae SK. Chae HZ. Kwon KS. Thioredoxin modulates activator protein 1 (AP-1) activity and p27Kip1 degradation through direct interaction with Jab1. Oncogene. 2004;23:8868–8875. doi: 10.1038/sj.onc.1208116. [DOI] [PubMed] [Google Scholar]
- 114.Ibiza S. Perez-Rodriguez A. Ortega A. Martinez-Ruiz A. Barreiro O. Garcia-Dominguez CA. Victor VM. Esplugues JV. Rojas JM. Sanchez-Madrid F. Serrador JM. Endothelial nitric oxide synthase regulates N-Ras activation on the Golgi complex of antigen-stimulated T cells. Proc Natl Acad Sci U S A. 2008;105:10507–10512. doi: 10.1073/pnas.0711062105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Inomata Y. Tanihara H. Tanito M. Okuyama H. Hoshino Y. Kinumi T. Kawaji T. Kondo N. Yodoi J. Nakamura H. Suppression of choroidal neovascularization by thioredoxin-1 via interaction with complement factor H. Invest Ophthalmol Vis Sci. 2008;49:5118–5125. doi: 10.1167/iovs.07-1659. [DOI] [PubMed] [Google Scholar]
- 116.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 117.Jacquier-Sarlin MR. Polla BS. Dual regulation of heat-shock transcription factor (HSF) activation and DNA-binding activity by H2O2: role of thioredoxin. Biochem J. 1996;318(Pt 1):187–193. doi: 10.1042/bj3180187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jaffrey SR. Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 119.Jeong W. Yoon HW. Lee SR. Rhee SG. Identification and characterization of TRP14, a thioredoxin-related protein of 14 kDa. New insights into the specificity of thioredoxin function. J Biol Chem. 2004;279:3142–3150. doi: 10.1074/jbc.M307932200. [DOI] [PubMed] [Google Scholar]
- 120.Ji Y. Akerboom TP. Sies H. Thomas JA. S-nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch Biochem Biophys. 1999;362:67–78. doi: 10.1006/abbi.1998.1013. [DOI] [PubMed] [Google Scholar]
- 121.Kahlos K. Zhang J. Block ER. Patel JM. Thioredoxin restores nitric oxide-induced inhibition of protein kinase C activity in lung endothelial cells. Mol Cell Biochem. 2003;254:47–54. doi: 10.1023/a:1027380828645. [DOI] [PubMed] [Google Scholar]
- 122.Kallis GB. Holmgren A. Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J Biol Chem. 1980;255:10261–10265. [PubMed] [Google Scholar]
- 123.Kang SW. Chae HZ. Seo MS. Kim K. Baines IC. Rhee SG. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-alpha. J Biol Chem. 1998;273:6297–6302. doi: 10.1074/jbc.273.11.6297. [DOI] [PubMed] [Google Scholar]
- 124.Keszler A. Zhang Y. Hogg N. Reaction between nitric oxide, glutathione, and oxygen in the presence and absence of protein: how are S-nitrosothiols formed? Free Radic Biol Med. 2010;48:55–64. doi: 10.1016/j.freeradbiomed.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim HY. Kim JR. Thioredoxin as a reducing agent for mammalian methionine sulfoxide reductases B lacking resolving cysteine. Biochem Biophys Res Commun. 2008;371:490–494. doi: 10.1016/j.bbrc.2008.04.101. [DOI] [PubMed] [Google Scholar]
- 126.Kim JR. Yoon HW. Kwon KS. Lee SR. Rhee SG. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 127.Kim YM. Talanian RV. Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J Biol Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- 128.Kirkpatrick L. Dragovich T. Ramanathan R. Sharlow E. Chow S. Williams D. Himler R. Baker A. Egorin M. Results from Phase I study of PX-12, a thioredoxin inhibitor in patients with advanced solid malignancies. J Clin Oncol. 2004;22:3089. [Google Scholar]
- 129.Kornberg MD. Sen N. Hara MR. Juluri KR. Nguyen JV. Snowman AM. Law L. Hester LD. Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Krause G. Lundstrom J. Barea JL. Pueyo de la Cuesta C. Holmgren A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J Biol Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
- 131.Kurooka H. Kato K. Minoguchi S. Takahashi Y. Ikeda J. Habu S. Osawa N. Buchberg AM. Moriwaki K. Shisa H. Honjo T. Cloning and characterization of the nucleoredoxin gene that encodes a novel nuclear protein related to thioredoxin. Genomics. 1997;39:331–339. doi: 10.1006/geno.1996.4493. [DOI] [PubMed] [Google Scholar]
- 132.Kuster GM. Pimentel DR. Adachi T. Ido Y. Brenner DA. Cohen RA. Liao R. Siwik DA. Colucci WS. Alpha-adrenergic receptor-stimulated hypertrophy in adult rat ventricular myocytes is mediated via thioredoxin-1-sensitive oxidative modification of thiols on Ras. Circulation. 2005;111:1192–1198. doi: 10.1161/01.CIR.0000157148.59308.F5. [DOI] [PubMed] [Google Scholar]
- 133.Kwon M. Yoon CS. Jeong W. Rhee SG. Waisman DM. Annexin A2-S100A10 heterotetramer, a novel substrate of thioredoxin. J Biol Chem. 2005;280:23584–23592. doi: 10.1074/jbc.M504325200. [DOI] [PubMed] [Google Scholar]
- 134.Lancaster JR., Jr. Gaston B. NO and nitrosothiols: spatial confinement and free diffusion. Am J Physiol Lung Cell Mol Physiol. 2004;287:L465–L466. doi: 10.1152/ajplung.00151.2004. [DOI] [PubMed] [Google Scholar]
- 135.Lander HM. Milbank AJ. Tauras JM. Hajjar DP. Hempstead BL. Schwartz GD. Kraemer RT. Mirza UA. Chait BT. Burk SC. Quilliam LA. Redox regulation of cell signalling. Nature. 1996;381:380–381. doi: 10.1038/381380a0. [DOI] [PubMed] [Google Scholar]
- 136.Lane P. Hao G. Gross SS. S-nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci STKE. 2001;2001:re1. doi: 10.1126/stke.2001.86.re1. [DOI] [PubMed] [Google Scholar]
- 137.Lee SR. Kwon KS. Kim SR. Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 138.Lee SR. Yang KS. Kwon J. Lee C. Jeong W. Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 139.Lee W. Choi KS. Riddell J. Ip C. Ghosh D. Park JH. Park YM. Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J Biol Chem. 2007;282:22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- 140.Lefievre L. Chen Y. Conner SJ. Scott JL. Publicover SJ. Ford WC. Barratt CL. Human spermatozoa contain multiple targets for protein S-nitrosylation: an alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics. 2007;7:3066–3084. doi: 10.1002/pmic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Leichert LI. Gehrke F. Gudiseva HV. Blackwell T. Ilbert M. Walker AK. Strahler JR. Andrews PC. Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lewin A. Crow A. Oubrie A. Le Brun NE. Molecular basis for specificity of the extracytoplasmic thioredoxin ResA. J Biol Chem. 2006;281:35467–35477. doi: 10.1074/jbc.M607047200. [DOI] [PubMed] [Google Scholar]
- 143.Li F. Sonveaux P. Rabbani ZN. Liu S. Yan B. Huang Q. Vujaskovic Z. Dewhirst MW. Li CY. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li J. Billiar TR. Talanian RV. Kim YM. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- 145.Li Y. Liu W. Xing G. Tian C. Zhu Y. He F. Direct association of hepatopoietin with thioredoxin constitutes a redox signal transduction in activation of AP-1/NF-kappaB. Cell Signal. 2005;17:985–996. doi: 10.1016/j.cellsig.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 146.Lillig CH. Holmgren A. Thioredoxin and related molecules—from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 147.Lim KH. Ancrile BB. Kashatus DF. Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lima B. Lam GK. Xie L. Diesen DL. Villamizar N. Nienaber J. Messina E. Bowles D. Kontos CD. Hare JM. Stamler JS. Rockman HA. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci U S A. 2009;106:6297–6302. doi: 10.1073/pnas.0901043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lipton SA. Choi YB. Takahashi H. Zhang D. Li W. Godzik A. Bankston LA. Cysteine regulation of protein function—as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25:474–480. doi: 10.1016/s0166-2236(02)02245-2. [DOI] [PubMed] [Google Scholar]
- 150.Liu GH. Qu J. Shen X. Thioredoxin-mediated negative autoregulation of peroxisome proliferator-activated receptor alpha transcriptional activity. Mol Biol Cell. 2006;17:1822–1833. doi: 10.1091/mbc.E05-10-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu X. Miller MJ. Joshi MS. Thomas DD. Lancaster JR., Jr Accelerated reaction of nitric oxide with O2 within the hydrophobic interior of biological membranes. Proc Natl Acad Sci U S A. 1998;95:2175–2179. doi: 10.1073/pnas.95.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu Y. Min W. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ Res. 2002;90:1259–1266. doi: 10.1161/01.res.0000022160.64355.62. [DOI] [PubMed] [Google Scholar]
- 153.Lopez-Sanchez LM. Corrales FJ. Lopez-Pedrera C. Aranda E. Rodriguez-Ariza A. Pharmacological impairment of s-nitrosoglutathione or thioredoxin reductases augments protein S-Nitrosation in human hepatocarcinoma cells. Anticancer Res. 2010;30:415–421. [PubMed] [Google Scholar]
- 154.Lovell MA. Xie C. Gabbita SP. Markesbery WR. Decreased thioredoxin and increased thioredoxin reductase levels in Alzheimer's disease brain. Free Radic Biol Med. 2000;28:418–427. doi: 10.1016/s0891-5849(99)00258-0. [DOI] [PubMed] [Google Scholar]
- 155.Low FM. Hampton MB. Peskin AV. Winterbourn CC. Peroxiredoxin 2 functions as a noncatalytic scavenger of low-level hydrogen peroxide in the erythrocyte. Blood. 2007;109:2611–2617. doi: 10.1182/blood-2006-09-048728. [DOI] [PubMed] [Google Scholar]
- 156.Lundstrom-Ljung J. Birnbach U. Rupp K. Soling HD. Holmgren A. Two resident ER-proteins, CaBP1 and CaBP2, with thioredoxin domains, are substrates for thioredoxin reductase: comparison with protein disulfide isomerase. FEBS Lett. 1995;357:305–308. doi: 10.1016/0014-5793(94)01386-f. [DOI] [PubMed] [Google Scholar]
- 157.Maiorino M. Ursini F. Bosello V. Toppo S. Tosatto SC. Mauri P. Becker K. Roveri A. Bulato C. Benazzi L. De Palma A. Flohe L. The thioredoxin specificity of Drosophila GPx: a paradigm for a peroxiredoxin-like mechanism of many glutathione peroxidases. J Mol Biol. 2007;365:1033–1046. doi: 10.1016/j.jmb.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 158.Makino Y. Yoshikawa N. Okamoto K. Hirota K. Yodoi J. Makino I. Tanaka H. Direct association with thioredoxin allows redox regulation of glucocorticoid receptor function. J Biol Chem. 1999;274:3182–3188. doi: 10.1074/jbc.274.5.3182. [DOI] [PubMed] [Google Scholar]
- 159.Manabe S. Gu Z. Lipton SA. Activation of matrix metalloproteinase-9 via neuronal nitric oxide synthase contributes to NMDA-induced retinal ganglion cell death. Invest Ophthalmol Vis Sci. 2005;46:4747–4753. doi: 10.1167/iovs.05-0128. [DOI] [PubMed] [Google Scholar]
- 160.Mancuso C. Capone C. Ranieri SC. Fusco S. Calabrese V. Eboli ML. Preziosi P. Galeotti T. Pani G. Bilirubin as an endogenous modulator of neurotrophin redox signaling. J Neurosci Res. 2008;86:2235–2249. doi: 10.1002/jnr.21665. [DOI] [PubMed] [Google Scholar]
- 161.Mancuso C. Navarra P. Preziosi P. Roles of nitric oxide, carbon monoxide, and hydrogen sulfide in the regulation of the hypothalamic-pituitary-adrenal axis. J Neurochem. 2010;113:563–575. doi: 10.1111/j.1471-4159.2010.06606.x. [DOI] [PubMed] [Google Scholar]
- 162.Mannick JB. Regulation of apoptosis by protein S-nitrosylation. Amino Acids. 2007;32:523–526. doi: 10.1007/s00726-006-0427-6. [DOI] [PubMed] [Google Scholar]
- 163.Mannick JB. Schonhoff C. Papeta N. Ghafourifar P. Szibor M. Fang K. Gaston B. S-nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marchand CH. Vanacker H. Collin V. Issakidis-Bourguet E. Marechal PL. Decottignies P. Thioredoxin targets in Arabidopsis roots. Proteomics. 2010;10:2418–2428. doi: 10.1002/pmic.200900835. [DOI] [PubMed] [Google Scholar]
- 165.Marino SM. Gladyshev VN. Redox biology: computational approaches to the investigation of functional cysteine residues. Antioxid Redox Signal. 2011;15:135–146. doi: 10.1089/ars.2010.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Marino SM. Gladyshev VN. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J Mol Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Marino SM. Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marshall HE. Stamler JS. Nitrosative stress-induced apoptosis through inhibition of NF-kappa B. J Biol Chem. 2002;277:34223–34228. doi: 10.1074/jbc.M201638200. [DOI] [PubMed] [Google Scholar]
- 169.Martin JL. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 170.Martinez-Ruiz A. Villanueva L. Gonzalez de Orduna C. Lopez-Ferrer D. Higueras MA. Tarin C. Rodriguez-Crespo I. Vazquez J. Lamas S. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci U S A. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Matsuo Y. Akiyama N. Nakamura H. Yodoi J. Noda M. Kizaka-Kondoh S. Identification of a novel thioredoxin-related transmembrane protein. J Biol Chem. 2001;276:10032–10038. doi: 10.1074/jbc.M011037200. [DOI] [PubMed] [Google Scholar]
- 172.Matthews JR. Wakasugi N. Virelizier JL. Yodoi J. Hay RT. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20:3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Matthias LJ. Yam PT. Jiang XM. Vandegraaff N. Li P. Poumbourios P. Donoghue N. Hogg PJ. Disulfide exchange in domain 2 of CD4 is required for entry of HIV-1. Nat Immunol. 2002;3:727–732. doi: 10.1038/ni815. [DOI] [PubMed] [Google Scholar]
- 174.McCarthy SM. Bove PF. Matthews DE. Akaike T. van der Vliet A. Nitric oxide regulation of MMP-9 activation and its relationship to modifications of the cysteine switch. Biochemistry. 2008;47:5832–5840. doi: 10.1021/bi702496v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McDonagh B. Ogueta S. Lasarte G. Padilla CA. Barcena JA. Shotgun redox proteomics identifies specifically modified cysteines in key metabolic enzymes under oxidative stress in Saccharomyces cerevisiae. J Proteomics. 2009;72:677–689. doi: 10.1016/j.jprot.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 176.McKinsey TA. Zhang CL. Lu J. Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Melchers J. Dirdjaja N. Ruppert T. Krauth-Siegel RL. Glutathionylation of trypanosomal thiol redox proteins. J Biol Chem. 2007;282:8678–8694. doi: 10.1074/jbc.M608140200. [DOI] [PubMed] [Google Scholar]
- 178.Melikian N. Seddon MD. Casadei B. Chowienczyk PJ. Shah AM. Neuronal nitric oxide synthase and human vascular regulation. Trends Cardiovasc Med. 2009;19:256–262. doi: 10.1016/j.tcm.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Meuillet EJ. Mahadevan D. Berggren M. Coon A. Powis G. Thioredoxin-1 binds to the C2 domain of PTEN inhibiting PTEN's lipid phosphatase activity and membrane binding: a mechanism for the functional loss of PTEN's tumor suppressor activity. Arch Biochem Biophys. 2004;429:123–133. doi: 10.1016/j.abb.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 180.Michelet L. Zaffagnini M. Marchand C. Collin V. Decottignies P. Tsan P. Lancelin JM. Trost P. Miginiac-Maslow M. Noctor G. Lemaire SD. Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc Natl Acad Sci U S A. 2005;102:16478–16483. doi: 10.1073/pnas.0507498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mieyal JJ. Gallogly MM. Qanungo S. Sabens EA. Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Minetti M. Mallozzi C. Di Stasi AM. Pietraforte D. Bilirubin is an effective antioxidant of peroxynitrite-mediated protein oxidation in human blood plasma. Arch Biochem Biophys. 1998;352:165–174. doi: 10.1006/abbi.1998.0584. [DOI] [PubMed] [Google Scholar]
- 183.Mitchell DA. Erwin PA. Michel T. Marletta MA. S-nitrosation and regulation of inducible nitric oxide synthase. Biochemistry. 2005;44:4636–4647. doi: 10.1021/bi0474463. [DOI] [PubMed] [Google Scholar]
- 184.Mitchell DA. Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 185.Mitchell DA. Morton SU. Fernhoff NB. Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci U S A. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mohr S. Hallak H. de Boitte A. Lapetina EG. Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 187.Motohashi K. Kondoh A. Stumpp MT. Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci U S A. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Musiani F. Ciurli S. Dikiy A. The interaction of selenoprotein W with 14-3-3 proteins: a computational approach. J Proteome Res. 2011;10:968–976. doi: 10.1021/pr101178k. [DOI] [PubMed] [Google Scholar]
- 189.Nadeau PJ. Charette SJ. Toledano MB. Landry J. Disulfide Bond-mediated multimerization of Ask1 and its reduction by thioredoxin-1 regulate H2O2-induced c-Jun NH2-terminal kinase activation and apoptosis. Mol Biol Cell. 2007;18:3903–3913. doi: 10.1091/mbc.E07-05-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nagahara N. Yoshii T. Abe Y. Matsumura T. Thioredoxin-dependent enzymatic activation of mercaptopyruvate sulfurtransferase. An intersubunit disulfide bond serves as a redox switch for activation. J Biol Chem. 2007;282:1561–1569. doi: 10.1074/jbc.M605931200. [DOI] [PubMed] [Google Scholar]
- 191.Nakamura H. Nakamura K. Yodoi J. Redox regulation of cellular activation. Annu Rev Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 192.Nakamura T. Lipton SA. Emerging roles of S-nitrosylation in protein misfolding and neurodegenerative diseases. Antioxid Redox Signal. 2008;10:87–101. doi: 10.1089/ars.2007.1858. [DOI] [PubMed] [Google Scholar]
- 193.Nakamura T. Wang L. Wong CC. Scott FL. Eckelman BP. Han X. Tzitzilonis C. Meng F. Gu Z. Holland EA. Clemente AT. Okamoto S. Salvesen GS. Riek R. Yates JR., 3rd Lipton SA. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nedospasov A. Rafikov R. Beda N. Nudler E. An autocatalytic mechanism of protein nitrosylation. Proc Natl Acad Sci U S A. 2000;97:13543–13548. doi: 10.1073/pnas.250398197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nedospasov AA. Is N2O3 the main nitrosating intermediate in aerated nitric oxide (NO) solutions in vivo? If so, where, when, and which one? J Biochem Mol Toxicol. 2002;16:109–120. doi: 10.1002/jbt.10029. [DOI] [PubMed] [Google Scholar]
- 196.Nikitovic D. Holmgren A. S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J Biol Chem. 1996;271:19180–19185. doi: 10.1074/jbc.271.32.19180. [DOI] [PubMed] [Google Scholar]
- 197.Nishiyama A. Matsui M. Iwata S. Hirota K. Masutani H. Nakamura H. Takagi Y. Sono H. Gon Y. Yodoi J. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 198.Nordstrand K. slund F. Holmgren A. Otting G. Berndt KD. NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism. J Mol Biol. 1999;286:541–552. doi: 10.1006/jmbi.1998.2444. [DOI] [PubMed] [Google Scholar]
- 199.Nott A. Watson PM. Robinson JD. Crepaldi L. Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 200.Okado-Matsumoto A. Matsumoto A. Fujii J. Taniguchi N. Peroxiredoxin IV is a secretable protein with heparin-binding properties under reduced conditions. J Biochem. 2000;127:493–501. doi: 10.1093/oxfordjournals.jbchem.a022632. [DOI] [PubMed] [Google Scholar]
- 201.Oliveira L. Bouton C. Drapier JC. Thioredoxin activation of iron regulatory proteins. Redox regulation of RNA binding after exposure to nitric oxide. J Biol Chem. 1999;274:516–521. doi: 10.1074/jbc.274.1.516. [DOI] [PubMed] [Google Scholar]
- 202.Ong SE. Blagoev B. Kratchmarova I. Kristensen DB. Steen H. Pandey A. Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 203.Ozawa K. Whalen EJ. Nelson CD. Mu Y. Hess DT. Lefkowitz RJ. Stamler JS. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol Cell. 2008;31:395–405. doi: 10.1016/j.molcel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pacher P. Beckman JS. Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Packer M. Lee WH. Kessler PD. Gottlieb SS. Medina N. Yushak M. Prevention and reversal of nitrate tolerance in patients with congestive heart failure. N Engl J Med. 1987;317:799–804. doi: 10.1056/NEJM198709243171304. [DOI] [PubMed] [Google Scholar]
- 206.Palmer LA. Doctor A. Chhabra P. Sheram ML. Laubach VE. Karlinsey MZ. Forbes MS. Macdonald T. Gaston B. S-nitrosothiols signal hypoxia-mimetic vascular pathology. J Clin Invest. 2007;117:2592–2601. doi: 10.1172/JCI29444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pan S. Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ Res. 2007;100:213–219. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 208.Park HS. Yu JW. Cho JH. Kim MS. Huh SH. Ryoo K. Choi EJ. Inhibition of apoptosis signal-regulating kinase 1 by nitric oxide through a thiol redox mechanism. J Biol Chem. 2004;279:7584–7590. doi: 10.1074/jbc.M304183200. [DOI] [PubMed] [Google Scholar]
- 209.Patel JM. Zhang J. Block ER. Nitric oxide-induced inhibition of lung endothelial cell nitric oxide synthase via interaction with allosteric thiols: role of thioredoxin in regulation of catalytic activity. Am J Respir Cell Mol Biol. 1996;15:410–419. doi: 10.1165/ajrcmb.15.3.8810647. [DOI] [PubMed] [Google Scholar]
- 210.Patwari P. Higgins LJ. Chutkow WA. Yoshioka J. Lee RT. The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pawloski JR. Hess DT. Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 212.Peng J. Elias JE. Thoreen CC. Licklider LJ. Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 213.Perez-Jimenez R. Li J. Kosuri P. Sanchez-Romero I. Wiita AP. Rodriguez-Larrea D. Chueca A. Holmgren A. Miranda-Vizuete A. Becker K. Cho SH. Beckwith J. Gelhaye E. Jacquot JP. Gaucher EA. Sanchez-Ruiz JM. Berne BJ. Fernandez JM. Diversity of chemical mechanisms in thioredoxin catalysis revealed by single-molecule force spectroscopy. Nat Struct Mol Biol. 2009;16:890–896. doi: 10.1038/nsmb.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Perez-Mato I. Castro C. Ruiz FA. Corrales FJ. Mato JM. Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J Biol Chem. 1999;274:17075–17079. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 215.Pimentel DR. Adachi T. Ido Y. Heibeck T. Jiang B. Lee Y. Melendez JA. Cohen RA. Colucci WS. Strain-stimulated hypertrophy in cardiac myocytes is mediated by reactive oxygen species-dependent Ras S-glutathiolation. J Mol Cell Cardiol. 2006;41:613–622. doi: 10.1016/j.yjmcc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 216.Powis G. Mustacich D. Coon A. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic Biol Med. 2000;29:312–322. doi: 10.1016/s0891-5849(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 217.Qin J. Clore GM. Kennedy WM. Huth JR. Gronenborn AM. Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B. Structure. 1995;3:289–297. doi: 10.1016/s0969-2126(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 218.Ravi K. Brennan LA. Levic S. Ross PA. Black SM. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc Natl Acad Sci U S A. 2004;101:2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Reiter E. Lefkowitz RJ. GRKs and beta-arrestins: roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab. 2006;17:159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 220.Ridnour LA. Thomas DD. Mancardi D. Espey MG. Miranda KM. Paolocci N. Feelisch M. Fukuto J. Wink DA. The chemistry of nitrosative stress induced by nitric oxide and reactive nitrogen oxide species. Putting perspective on stressful biological situations. Biol Chem. 2004;385:1–10. doi: 10.1515/BC.2004.001. [DOI] [PubMed] [Google Scholar]
- 221.Roos G. Foloppe N. Van Laer K. Wyns L. Nilsson L. Geerlings P. Messens J. How thioredoxin dissociates its mixed disulfide. PLoS Comput Biol. 2009;5:e1000461. doi: 10.1371/journal.pcbi.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ross PL. Huang YN. Marchese JN. Williamson B. Parker K. Hattan S. Khainovski N. Pillai S. Dey S. Daniels S. Purkayastha S. Juhasz P. Martin S. Bartlet-Jones M. He F. Jacobson A. Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 223.Rossi R. Lusini L. Giannerini F. Giustarini D. Lungarella G. Di Simplicio P. A method to study kinetics of transnitrosation with nitrosoglutathione: reactions with hemoglobin and other thiols. Anal Biochem. 1997;254:215–220. doi: 10.1006/abio.1997.2424. [DOI] [PubMed] [Google Scholar]
- 224.Rozell B. Hansson HA. Luthman M. Holmgren A. Immunohistochemical localization of thioredoxin and thioredoxin reductase in adult rats. Eur J Cell Biol. 1985;38:79–86. [PubMed] [Google Scholar]
- 225.Sabens EA. Mieyal JJ. Glutaredoxin and thioredoxin enzyme systems: catalytic mechanisms and physiological functions. In: Masella R, editor; Massa G, editor. Glutathione and Sulfur Amino Acids in Human Health and Disease. Hoboken, NJ: John Wiley & Sons, Inc.; 2009. pp. 121–156. [Google Scholar]
- 226.Saitoh M. Nishitoh H. Fujii M. Takeda K. Tobiume K. Sawada Y. Kawabata M. Miyazono K. Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Santhanam L. Lim HK. Miriel V. Brown T. Patel M. Balanson S. Ryoo S. Anderson M. Irani K. Khanday F. Di Costanzo L. Nyhan D. Hare JM. Christianson DW. Rivers R. Shoukas A. Berkowitz DE. Inducible NO synthase dependent S-nitrosylation and activation of arginase1 contribute to age-related endothelial dysfunction. Circ Res. 2007;101:692–702. doi: 10.1161/CIRCRESAHA.107.157727. [DOI] [PubMed] [Google Scholar]
- 228.Sauer H. Wartenberg M. Hescheler J. Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell Physiol Biochem. 2001;11:173–186. doi: 10.1159/000047804. [DOI] [PubMed] [Google Scholar]
- 229.Saville B. A scheme for the colorimetric determination of microgram amounts of thiols. Analyst. 1958;83:670–675. [Google Scholar]
- 230.Sayed N. Baskaran P. Ma X. van den Akker F. Beuve A. Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc Natl Acad Sci U S A. 2007;104:12312–12317. doi: 10.1073/pnas.0703944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sayed N. Kim DD. Fioramonti X. Iwahashi T. Duran WN. Beuve A. Nitroglycerin-induced S-nitrosylation and desensitization of soluble guanylyl cyclase contribute to nitrate tolerance. Circ Res. 2008;103:606–614. doi: 10.1161/CIRCRESAHA.108.175133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schroeder P. Popp R. Wiegand B. Altschmied J. Haendeler J. Nuclear redox-signaling is essential for apoptosis inhibition in endothelial cells—important role for nuclear thioredoxin-1. Arterioscler Thromb Vasc Biol. 2007;27:2325–2331. doi: 10.1161/ATVBAHA.107.149419. [DOI] [PubMed] [Google Scholar]
- 233.Schulze PC. Liu H. Choe E. Yoshioka J. Shalev A. Bloch KD. Lee RT. Nitric oxide-dependent suppression of thioredoxin-interacting protein expression enhances thioredoxin activity. Arterioscler Thromb Vasc Biol. 2006;26:2666–2672. doi: 10.1161/01.ATV.0000248914.21018.f1. [DOI] [PubMed] [Google Scholar]
- 234.Schwarz O. Schurmann P. Strotmann H. Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J Biol Chem. 1997;272:16924–16927. doi: 10.1074/jbc.272.27.16924. [DOI] [PubMed] [Google Scholar]
- 235.Schwertassek U. Balmer Y. Gutscher M. Weingarten L. Preuss M. Engelhard J. Winkler M. Dick TP. Selective redox regulation of cytokine receptor signaling by extracellular thioredoxin-1. EMBO J. 2007;26:3086–3097. doi: 10.1038/sj.emboj.7601746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sen N. Hara MR. Kornberg MD. Cascio MB. Bae BI. Shahani N. Thomas B. Dawson TM. Dawson VL. Snyder SH. Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Sengupta R. Billiar TR. Stoyanovsky DA. Studies toward the analysis of S-nitrosoproteins. Org Biomol Chem. 2009;7:232–234. doi: 10.1039/b817981f. [DOI] [PubMed] [Google Scholar]
- 238.Sengupta R. Ryter SW. Zuckerbraun BS. Tzeng E. Billiar TR. Stoyanovsky DA. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry. 2007;46:8472–8483. doi: 10.1021/bi700449x. [DOI] [PubMed] [Google Scholar]
- 239.Seo MS. Kang SW. Kim K. Baines IC. Lee TH. Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J Biol Chem. 2000;275:20346–20354. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 240.Seth D. Rudolph J. Redox regulation of MAP kinase phosphatase 3. Biochemistry. 2006;45:8476–8487. doi: 10.1021/bi060157p. [DOI] [PubMed] [Google Scholar]
- 241.Shioji K. Nakamura H. Masutani H. Yodoi J. Redox regulation by thioredoxin in cardiovascular diseases. Antioxid Redox Signal. 2003;5:795–802. doi: 10.1089/152308603770380106. [DOI] [PubMed] [Google Scholar]
- 242.Sohn J. Rudolph J. Catalytic and chemical competence of regulation of cdc25 phosphatase by oxidation/reduction. Biochemistry. 2003;42:10060–10070. doi: 10.1021/bi0345081. [DOI] [PubMed] [Google Scholar]
- 243.Stamler JS. Hess DT. Nascent nitrosylases. Nat Cell Biol. 2010;12:1024–6. doi: 10.1038/ncb1110-1024. [DOI] [PubMed] [Google Scholar]
- 244.Stamler JS. Jia L. Eu JP. McMahon TJ. Demchenko IT. Bonaventura J. Gernert K. Piantadosi CA. Blood flow regulation by S-nitrosohemoglobin in the physiological oxygen gradient. Science. 1997;276:2034–2037. doi: 10.1126/science.276.5321.2034. [DOI] [PubMed] [Google Scholar]
- 245.Stamler JS. Lamas S. Fang FC. Nitrosylation: the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 246.Stamler JS. Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- 247.Stoyanovsky DA. Tyurina YY. Tyurin VA. Anand D. Mandavia DN. Gius D. Ivanova J. Pitt B. Billiar TR. Kagan VE. Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J Am Chem Soc. 2005;127:15815–15823. doi: 10.1021/ja0529135. [DOI] [PubMed] [Google Scholar]
- 248.Stubauer G. Giuffre A. Sarti P. Mechanism of S-nitrosothiol formation and degradation mediated by copper ions. J Biol Chem. 1999;274:28128–28133. doi: 10.1074/jbc.274.40.28128. [DOI] [PubMed] [Google Scholar]
- 249.Sumbayev VV. S-nitrosylation of thioredoxin mediates activation of apoptosis signal-regulating kinase 1. Arch Biochem Biophys. 2003;415:133–136. doi: 10.1016/s0003-9861(03)00199-1. [DOI] [PubMed] [Google Scholar]
- 250.Sun J. Protein S-nitrosylation: a role of nitric oxide signaling in cardiac ischemic preconditioning. Sheng Li Xue Bao. 2007;59:544–552. [PubMed] [Google Scholar]
- 251.Sun J. Morgan M. Shen RF. Steenbergen C. Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 252.Sun J. Xin C. Eu JP. Stamler JS. Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci U S A. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tamura T. Stadtman TC. A new selenoprotein from human lung adenocarcinoma cells: purification, properties, and thioredoxin reductase activity. Proc Natl Acad Sci U S A. 1996;93:1006–1011. doi: 10.1073/pnas.93.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tanaka T. Hosoi F. Yamaguchi-Iwai Y. Nakamura H. Masutani H. Ueda S. Nishiyama A. Takeda S. Wada H. Spyrou G. Yodoi J. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J. 2002;21:1695–1703. doi: 10.1093/emboj/21.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tao L. Gao E. Bryan NS. Qu Y. Liu HR. Hu A. Christopher TA. Lopez BL. Yodoi J. Koch WJ. Feelisch M. Ma XL. Cardioprotective effects of thioredoxin in myocardial ischemia and reperfusion: role of S-nitrosation. Proc Natl Acad Sci U S A. 2004;101:11471–11476. doi: 10.1073/pnas.0402941101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Tao L. Jiao X. Gao E. Lau WB. Yuan Y. Lopez B. Christopher T. RamachandraRao SP. Williams W. Southan G. Sharma K. Koch W. Ma XL. Nitrative inactivation of thioredoxin-1 and its role in postischemic myocardial apoptosis. Circulation. 2006;114:1395–1402. doi: 10.1161/CIRCULATIONAHA.106.625061. [DOI] [PubMed] [Google Scholar]
- 257.Tello D. Tarin C. Ahicart P. Breton-Romero R. Lamas S. Martinez-Ruiz A. A “fluorescence switch” technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins. Proteomics. 2009;9:5359–5370. doi: 10.1002/pmic.200900070. [DOI] [PubMed] [Google Scholar]
- 258.Ueda S. Nakamura H. Masutani H. Sasada T. Yonehara S. Takabayashi A. Yamaoka Y. Yodoi J. Redox regulation of caspase-3(-like) protease activity: regulatory roles of thioredoxin and cytochrome c. J Immunol. 1998;161:6689–6695. [PubMed] [Google Scholar]
- 259.Vanin AF. Malenkova IV. Serezhenkov VA. Iron catalyzes both decomposition and synthesis of S-nitrosothiols: optical and electron paramagnetic resonance studies. Nitric Oxide. 1997;1:191–203. doi: 10.1006/niox.1997.0122. [DOI] [PubMed] [Google Scholar]
- 260.Venema RC. Venema VJ. Ju H. Harris MB. Snead C. Jilling T. Dimitropoulou C. Maragoudakis ME. Catravas JD. Novel complexes of guanylate cyclase with heat shock protein 90 and nitric oxide synthase. Am J Physiol Heart Circ Physiol. 2003;285:H669–H678. doi: 10.1152/ajpheart.01025.2002. [DOI] [PubMed] [Google Scholar]
- 261.Verdoucq L. Vignols F. Jacquot JP. Chartier Y. Meyer Y. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem. 1999;274:19714–19722. doi: 10.1074/jbc.274.28.19714. [DOI] [PubMed] [Google Scholar]
- 262.Vieira Dos Santos C. Laugier E. Tarrago L. Massot V. Issakidis-Bourguet E. Rouhier N. Rey P. Specificity of thioredoxins and glutaredoxins as electron donors to two distinct classes of Arabidopsis plastidial methionine sulfoxide reductases B. FEBS Lett. 2007;581:4371–4376. doi: 10.1016/j.febslet.2007.07.081. [DOI] [PubMed] [Google Scholar]
- 263.von Knethen A. Callsen D. Brune B. NF-kappaB and AP-1 activation by nitric oxide attenuated apoptotic cell death in RAW 264.7 macrophages. Mol Biol Cell. 1999;10:361–372. doi: 10.1091/mbc.10.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wagner E. Luche S. Penna L. Chevallet M. Van Dorsselaer A. Leize-Wagner E. Rabilloud T. A method for detection of overoxidation of cysteines: peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem J. 2002;366:777–785. doi: 10.1042/BJ20020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang D. Masutani H. Oka S. Tanaka T. Yamaguchi-Iwai Y. Nakamura H. Yodoi J. Control of mitochondrial outer membrane permeabilization and Bcl-xL levels by thioredoxin 2 in DT40 cells. J Biol Chem. 2006;281:7384–7391. doi: 10.1074/jbc.M509876200. [DOI] [PubMed] [Google Scholar]
- 266.Wang G. Moniri NH. Ozawa K. Stamler JS. Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci U S A. 2006;103:1295–1300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wang Y. Liu T. Wu C. Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1353–1360. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Watson WH. Pohl J. Montfort WR. Stuchlik O. Reed MS. Powis G. Jones DP. Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J Biol Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- 269.Weichsel A. Brailey JL. Montfort WR. Buried S-nitrosocysteine revealed in crystal structures of human thioredoxin. Biochemistry. 2007;46:1219–1227. doi: 10.1021/bi061878r. [DOI] [PubMed] [Google Scholar]
- 270.Whalen EJ. Foster MW. Matsumoto A. Ozawa K. Violin JD. Que LG. Nelson CD. Benhar M. Keys JR. Rockman HA. Koch WJ. Daaka Y. Lefkowitz RJ. Stamler JS. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 271.Wiita AP. Perez-Jimenez R. Walther KA. Grater F. Berne BJ. Holmgren A. Sanchez-Ruiz JM. Fernandez JM. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wong JH. Balmer Y. Cai N. Tanaka CK. Vensel WH. Hurkman WJ. Buchanan BB. Unraveling thioredoxin-linked metabolic processes of cereal starchy endosperm using proteomics. FEBS Lett. 2003;547:151–156. doi: 10.1016/s0014-5793(03)00696-3. [DOI] [PubMed] [Google Scholar]
- 273.Wu C. Liu T. Chen W. Oka S. Fu C. Jain MR. Parrott AM. Baykal AT. Sadoshima J. Li H. Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol Cell Proteomics. 2010;9:2262–2275. doi: 10.1074/mcp.M110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Xu SZ. Sukumar P. Zeng F. Li J. Jairaman A. English A. Naylor J. Ciurtin C. Majeed Y. Milligan CJ. Bahnasi YM. Al-Shawaf E. Porter KE. Jiang LH. Emery P. Sivaprasadarao A. Beech DJ. TRPC channel activation by extracellular thioredoxin. Nature. 2008;451:69–72. doi: 10.1038/nature06414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Yamamoto M. Yang GP. Hong C. Liu J. Holle E. Yu XZ. Wagner T. Vatner SF. Sadoshima J. Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J Clin Invest. 2003;112:1395–1406. doi: 10.1172/JCI17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Yamazaki D. Motohashi K. Kasama T. Hara Y. Hisabori T. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:18–27. doi: 10.1093/pcp/pch019. [DOI] [PubMed] [Google Scholar]
- 277.Yang KS. Kang SW. Woo HA. Hwang SC. Chae HZ. Kim K. Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 278.Yano H. Wong JH. Lee YM. Cho MJ. Buchanan BB. A strategy for the identification of proteins targeted by thioredoxin. Proc Natl Acad Sci U S A. 2001;98:4794–4799. doi: 10.1073/pnas.071041998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yoshioka J. Schreiter ER. Lee RT. Role of thioredoxin in cell growth through interactions with signaling molecules. Antioxid Redox Signal. 2006;8:2143–2151. doi: 10.1089/ars.2006.8.2143. [DOI] [PubMed] [Google Scholar]
- 280.Zhang H. Tao L. Jiao X. Gao E. Lopez BL. Christopher TA. Koch W. Ma XL. Nitrative thioredoxin inactivation as a cause of enhanced myocardial ischemia/reperfusion injury in the aging heart. Free Radic Biol Med. 2007;43:39–47. doi: 10.1016/j.freeradbiomed.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Zhang R. Al-Lamki R. Bai L. Streb JW. Miano JM. Bradley J. Min W. Thioredoxin-2 inhibits mitochondria-located ASK1-mediated apoptosis in a JNK-independent manner. Circ Res. 2004;94:1483–1491. doi: 10.1161/01.RES.0000130525.37646.a7. [DOI] [PubMed] [Google Scholar]
- 282.Zhang X. Huang B. Zhou X. Chen C. Quantitative proteomic analysis of S-nitrosated proteins in diabetic mouse liver with ICAT switch method. Protein Cell. 2010;1:675–687. doi: 10.1007/s13238-010-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zhou J. Damdimopoulos AE. Spyrou G. Brune B. Thioredoxin 1 and thioredoxin 2 have opposed regulatory functions on hypoxia-inducible factor-1alpha. J Biol Chem. 2007;282:7482–7490. doi: 10.1074/jbc.M608289200. [DOI] [PubMed] [Google Scholar]
- 284.Zhou Y. Kok KH. Chun AC. Wong CM. Wu HW. Lin MC. Fung PC. Kung H. Jin DY. Mouse peroxiredoxin V is a thioredoxin peroxidase that inhibits p53-induced apoptosis. Biochem Biophys Res Commun. 2000;268:921–927. doi: 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.