Abstract
In deep-sea hydrothermal systems, super hot and reduced vent fluids from the subseafloor blend with cold and oxidized seawater. Very unique and dense ecosystems are formed within these environments. Many molecular ecological studies showed that chemoautotrophic epsilon- and gamma-Proteobacteria are predominant primary producers in both free-living and symbiotic microbial communities in global deep-sea hydrothermal fields. Inorganic sulfur compounds are important substrates for the energy conservative metabolic pathways in these microorganisms. Recent genomic and metagenomic analyses and biochemical studies have contributed to the understanding of potential sulfur metabolic pathways for these chemoautotrophs. Epsilon-Proteobacteria use sulfur compounds for both electron-donors and -acceptors. On the other hand, gamma-Proteobacteria utilize two different sulfur-oxidizing pathways. It is hypothesized that differences between the metabolic pathways used by these two predominant proteobacterial phyla are associated with different ecophysiological strategies; extending the energetically feasible habitats with versatile energy metabolisms in the epsilon-Proteobacteria and optimizing energy production rate and yield for relatively narrow habitable zones in the gamma-Proteobacteria.
Keywords: deep-sea hydrothermal vents, energy metabolism, chemoautotroph
Introduction
The deep-sea hydrothermal system is one of the most extreme environments on Earth. It is characterized by darkness, high pressures, and steep physical and chemical gradients formed in mixing zones between hot hydrothermal vent fluid and cold deep-sea water. Very unique and dense communities of invertebrates are sustained in absence of a photosynthetic energy source (Dubilier et al., 2008). Almost all vent-endemic animals are strongly associated with the primary production of the endo- and/or epi-symbiotic chemoautotrophic microorganisms (Jeanthon, 2000). With the discovery of deep-sea hydrothermal ecosystem in 1977, it had been proposed that hydrogen sulfide-oxidizing and oxygen-reducing chemoautotrophs potentially sustain the primary production in these unique ecosystems (Kvenvolden et al., 1979). However, anoxic hydrothermal fluids contain several reduced compounds such as H2, CH4, and reduced metal ions in addition to H2S (Jannasch and Mottl, 1985). Recent cultivation studies have demonstrated that these chemicals are all used as energy sources for chemoautotrophs (Jannasch and Mottl, 1985; Nakagawa et al., 2005; Campbell et al., 2006; Emerson et al., 2007), indicating the great diversity of chemoautotrophic energy metabolic processes in the ecosystems. Hydrogen gas (H2) is one of the most important energy sources, and the hydrogen-dependent ecosystems may represent analogs for the earliest biological communities on Earth (Takai et al., 2004, 2006b; Nealson et al., 2005).
Hydrogen sulfide or sulfide is primarily supplied via high temperatures of seawater–rock interactions in the subseafloor hydrothermal reaction zones (Jannasch and Mottl, 1985). Thermodynamic modeling indicated that the hydrogen sulfide or sulfide abundantly contained in the hydrothermal fluids represented the dominating energy source in the mesophilic deep-sea vent chemoautotrophic ecosystems (McCollom and Shock, 1997). In addition, partially oxidized inorganic sulfur compounds such as polysulfide, elemental sulfur, and thiosulfate are generated in the in situ mixing zones, and serve as both electron donor and acceptor in a variety of energy metabolisms. The chemical and microbial oxidation and reduction reactions of sulfur compounds probably establish the overall complex sulfur metabolism network in the ecosystem. This article reviews the representative microbial components capable of utilizing the inorganic sulfur compounds as the energy sources in the deep-sea hydrothermal environments and highlights the biochemical and genetic components of their metabolisms. In addition, the possible segregation of different sulfur metabolic pathways and their host chemoautotrophic Proteobacteria associated with the physical and chemical transition in the mixing zones of the deep-sea hydrothermal environments is discussed.
Microbial Communities in Deep-Sea Hydrothermal Fields
Recent culture-independent analyses of the both symbiotic and free-living microbial communities in the various deep-sea hydrothermal environments have revealed a great phylogenetic diversity of Archaea and Bacteria (Takai et al., 2006a); these analyses have also indicated a predominance in biomass of members within the gamma-Proteobacteria and epsilon-Proteobacteria (Stewart et al., 2005; Suzuki et al., 2005; Urakawa et al., 2005; Campbell et al., 2006; Nakagawa and Takai, 2008; Table 1). Based on the culture-dependent characterization and genetic components analyses, these Proteobacteria are found to be chemoautotrophs strongly associated with utilization of inorganic sulfur compounds (Durand et al., 1993; Inagaki et al., 2003, 2004; Takai et al., 2003b, 2006a,c). Therefore, the sulfur-related energy metabolisms in these Proteobacteria are important traits for understanding ecology and biogeochemistry in deep-sea hydrothermal vent ecosystems.
Table 1.
Sulfur metabolic genes in epsilon- and gamma-Proteobacteria isolated or identified from deep-sea hydrothermal fields.
| Organism, isolation, or observation site | Genome accession number*1 | Growth tem- perature (°C) | Electron donor | Electron acceptor | Genes | Locus tag | Pathways | |
|---|---|---|---|---|---|---|---|---|
| EPSILON-PROTEOBACTERIA | ||||||||
| Sulfurovum sp. NBC37-1 Sediment, Mid-Okinawa Trough | AP009179 | 30–37 | H2, S0, | S0, , O2 | psrACB | SUN0510-0508 | Sulfur respiration | Nakagawa et al. (2005, 2007) |
| soxXYZAB | SUN0497-0501 | Sox system | Yamamoto et al. (2010) | |||||
| soxCDYZ | SUN0049-0052 | Sox system | ||||||
| soxJ | SUN0058, 1338 | Sox system (?) | ||||||
| sqr | SUN0047, 0073, 0192 | Sulfide oxidation | ||||||
| sorAB | SUN1104-1103, 1476-1477 | Sulfite oxidation | ||||||
| Nitratiruptor sp. SB155-2 Sediment, Mid-Okinawa Trough | AP009178 | 55 | H2, S0, | S0, , O2 | psrACB | NIS0573-0571 | Sulfur respiration | Nakagawa et al. (2005, 2007) |
| soxXYZAB | NIS1832-1828 | Sox system | ||||||
| soxYZ | NIS0034-0035 | Sox system | ||||||
| soxJ | NIS0032 | Sox system (?) | ||||||
| sqr | NIS0146, 0158, 0326 | Sulfide oxidation | ||||||
| Nautilia profundicola AmH Alvinella tube, East Pacific Rise | CP001279 | 45 | H2, formate | S0 | psrABC | NAMH1518-1520 | Sulfur respiration | Campbell et al. (2001, 2009) |
| Sulfurimonas autotrophica DSM16294 Sediment, Mid-Okinawa Trough | CP002205 | 25 | H2S, S0, | O2 | psrACB | Saut1622-1644 | Sulfur respiration | Inagaki et al. (2003) |
| soxXYZAB | Saut0991-0995 | Sox system | ||||||
| soxCDYZ | Saut2096-2099 | Sox system | ||||||
| soxJ | Saut1356 | Sox system (?) | ||||||
| sqr | Saut0503, 1543 | Sulfide oxidation | ||||||
| sorAB | Saut1022-1023 | Sulfite oxidation | ||||||
| Sulfurimonas denitrificans DSM1251 Tidal flat mud, Dutch Wadden Sea*2 | CP000153 | 25 | psrACB | Suden0500-0498 | Sulfur respiration | Sievert et al. (2008) | ||
| soxXYZAB | Suden0260-0264 | Sox system | ||||||
| soxCDYZ | Suden2060-2057 | Sox system | ||||||
| soxJ | Suden2049 | Sox system (?) | ||||||
| sqr | Suden0619 | Sulfide oxidation | ||||||
| Alvinella episymbiont East Pacific Rise | AAUQ | 10-65 | ND | ND | psrAB | ND | Sulfur respiration | Grzymski et al. (2008) |
| 01000000*3 | soxXYZABCD | ND | Sox system | |||||
| D | ND | Sox system (?) | ||||||
| soxJ | ||||||||
| GAMMA-PROTEOBACTERIA | ||||||||
| Thiomicrospira crunogena XCL-2 Galapagos Rift vent | CP000109 | 33 | H2, H2S, S0, | O2 | soxXYZA | Tcr0604-0601 | Sox system | Scott et al. (2006) |
| soxB | Tcr1549 | Sox system | ||||||
| soxCD | Tcr0156-0157 | Sox system | ||||||
| sqr | Tcr0619 | Sulfide oxidation | ||||||
| CoSym Vesicomyosocious symbiont | AP009247 | ND | ND | ND | soxXYZA | COSY0733-0730 | Sox system | Kuwahara et al. (2007) |
| soxB | COSY 0161 | Sox system | ||||||
| soxJ | COSY0750 | Sox system (?) | ||||||
| sqr | COSY0953 | Sulfide oxidation | ||||||
| dsrAB | COSY0795-0794 | Reverse sulfate reduction | ||||||
| aprAB | COSY0092-0091 | Reverse sulfate reduction | ||||||
| sat | COSY0089 | Reverse sulfate reduction | ||||||
| Ruthia magnifica Calyptogena symbiont | CP000488 | ND | ND | ND | soxXYZA | Rmag0808-0805 | Sox system | Newton et al. (2007) |
| soxB | Rmag0156 | Sox system | ||||||
| soxJ | Rmag0824 | Sox system (?) | ||||||
| sqr | Rmag1053 | Sulfide oxidation | ||||||
| dsrAB | Rmag0870-0869 | Reverse sulfate reduction | ||||||
| aprAB | Rmag0088-0087 | Reverse sulfate reduction | ||||||
| sat | Rmag0085 | Reverse sulfate reduction | ||||||
| Endoriftia persephone Riftia symbiont | AASF | ND | ND | ND | soxXAB*4 | ND | Sox system | Robidart et al. (2008) |
| 00000001*3 | dsrAB | ND | Reverse sulfate reduction | |||||
| aprAB | ND | Reverse sulfate reduction | ||||||
| sat | ND | Reverse sulfate reduction | ||||||
| Beggiatoa spp. Sediment, Baltic Sea | ABBY | ND | ND | ND | soxXYZAB | ND | Sox system | Mußmann et al. (2007) |
| 00000000, | fccAB | ND | Sulfide oxidation | |||||
| ABBBZ | sqr | ND | Sulfide oxidation | |||||
| 00000000*5 | dsrAB | ND | Reverse sulfate reduction | |||||
| aprAB | ND | Reverse sulfate reduction | ||||||
| sat | ND | Reverse sulfate reduction | ||||||
| dmsABC | ND | DMSO respiration | ||||||
| phsABC | ND | Thiosulfate respiration | ||||||
ND no data, psr, polysulfide reductase; sox, Sox multi enzyme system; sqr, sulfide:quinone oxidoreductase; fcc, flavocytochrome c; dsr, dissimilatory sulfite reductase; apr, adenosine 5′-phosphosulfate reductase; sat, sulfate adenylyltransferase; dms, dimethyl sulfoxide reductase; phs, thiosulfate reductase.
*1Deposited in DDBJ/EMBL/GenBank databases.
*2It is not a hydrothermal field.
*3For which the metagenome sequence is published.
*4SoxXAB were detected, but not SoxYZ.
*5Sequence assembles were incomplete.
Microbial Sulfur Metabolic Pathways for Energy Conservation
Inorganic sulfur compounds are used as both electron donor and acceptor by microorganisms. Sulfur-oxidation pathways have been well studied biochemically in anaerobic phototrophs (e.g., Chlorobium and Allochromatium), facultatively chemoautotrophic Proteobacteria (e.g., Acidithiobacillus and Paracoccus) and Sulfolobales (e.g., Sulfolobus and Acidianus; Friedrich, 1998; Kletzin et al., 2004; Friedrich et al., 2005; Frigaard and Dahl, 2009). Bacterial sulfur-oxidation pathways under the neutral pH are generally classified into two different types: (1) reverse sulfate reduction, and (2) Sox multienzyme system. Reverse sulfate reduction is reversal of sulfate reduction pathway, in which sulfide is oxidized to sulfate via sulfite. This pathway uses the same families of enzymes as of the sulfate reduction pathway, such as dissimilatory sulfite reductase (Dsr), adenylphosphosulfate reductase (Apr), and sulfate adenylyltransferase (Sat; Kappler and Dahl, 2001). Sox multienzyme complex catalyzes oxidation of inorganic sulfur compounds. Four protein components, SoxYZ, SoxXA, SoxB, and SoxCD are required for complete oxidation of sulfide and thiosulfate to sulfate (Complete Sox pathway; Friedrich et al., 2001). However, the sulfur oxidizers that possess the soxYZXAB genes but not soxCD have often been reported (Friedrich et al., 2005). It is considered that SoxCD acts as sulfur dehydrogenase. Sox system without SoxCD is able to oxidize sulfite and sulfone group () in thiosulfate, but not sulfide, elemental sulfur, and sulfane sulfur (-S−) in thiosulfate (Friedrich et al., 2001). In addition to these two sulfur-oxidation pathways, several enzymes have been proposed to oxidize inorganic sulfur compounds, including sulfide:quinone oxidoreductase (Sqr), flavocytochrome c (Fcc), and sulfite oxidase (SO), though the exact physiological functions are still debated. Sqr is a single-subunit flavoprotein that catalyzes oxidation of sulfide to elemental sulfur (Shahak and Hauska, 2008; Frigaard and Dahl, 2009). Recent protein structural study divided Sqr family into six groups (Marcia et al., 2010). These different types of Sqr probably have different mechanisms and physiological roles (Chan et al., 2009; Gregersen et al., 2011; Holkenbrink et al., 2011). Fcc also catalyzes oxidation of sulfide to elemental sulfur, but electrons are transferred to the level of cytochrome c (Oh-oka and Blankenship, 2004). Fcc consists of a flavoprotein subunit, FccB, and a c-type cytochrome subunit, FccA (Kusai and Yamanaka, 1973; Reinartz et al., 1998). The soxEF genes conserved in the sox gene clusters are homologous genes of the fccAB genes (Friedrich et al., 2008). The soxJ gene is a homologous gene of fccB/soxF, that does not cluster with a homolog of fccA/soxE (Gregersen et al., 2011). SO is a molybdenum enzyme and catalyzes the direct oxidation of sulfite to sulfate (Kappler and Dahl, 2001). Acidophilic sulfur-oxidizing bacteria and archaea use different sulfur-oxidation pathways that will not be further described here (see Ghosh and Dam, 2009).
Sulfur-reduction pathways can be classified into two types based on the substrates: (1) sulfur-reduction (sulfur respiration), and (2) sulfate reduction (sulfate respiration). Sulfur-reduction pathways have been intensively studied in gastrointestinal epsilon-Proteobacterium, Wolinella succinogenes (Wolin et al., 1961; Hedderich et al., 1999). Sulfur-reduction is catalyzed by polysulfide reductase (Psr), in which the actual substrate of the reaction is polysulfide but not elemental sulfur (Pfennig and Biebel, 1986). Sulfate reducing bacteria are found in several different phylogenetic lines (e.g., delta-Proteobacteria, Clostridia, Thermodesulfobacteria, and Nitrospirae; Muyzer and Stams, 2008). Sat, Apr, and Dsr catalyze the reduction of sulfate to sulfide.
Sulfur Metabolism in Epsilon-Proteobacteria in Deep-Sea Hydrothermal Fields
The most well known epsilon-Proteobacteria are gastrointestinal pathogens such as Helicobacter and Campylobacter. They are heterotrophic and do not have inorganic sulfur metabolic pathways. The genus Wolinella isolated from cattle rumen can reduce elemental sulfur to hydrogen sulfide (Wolin et al., 1961).
There are many reports that chemoautotrophic epsilon-Proteobacteria are predominant in sulfidic environments such as deep-sea hydrothermal and subsurface environments (Takai et al., 2004; Campbell et al., 2006; Nakagawa and Takai, 2008). Recently, whole genome sequences of five strains of marine epsilon-Proteobacteria, i.e., Sulfurovum sp. NBC37-1, Nitratiruptor sp. SB155-2, Nautilia profundicola AmH, Sulfurimonas autotrophica DSM16294 isolated from deep-sea hydrothermal fields, and S. denitrificans DSM1251 isolated from a coastal wetland, have been determined (Nakagawa et al., 2007; Sievert et al., 2008; Campbell et al., 2009; Table 1). Moreover, the metabolism of episymbiontic epsilon-Proteobacterium in a polychaete worm was analyzed by metagenomic analysis (Grzymski et al., 2008). In addition, several biochemical studies on sulfur metabolic pathways in deep-sea epsilon-Proteobacteria have also been reported (Takai et al., 2005; Yamamoto et al., 2010).
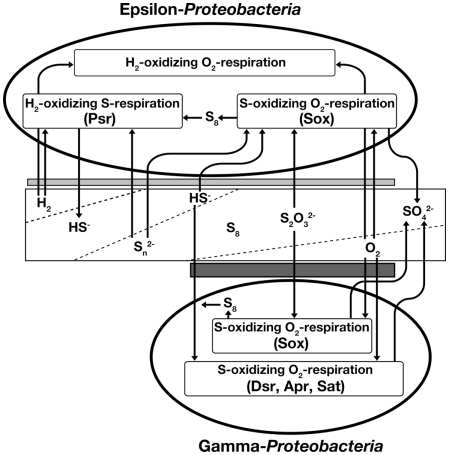
Genetic and enzymatic components characterized by the genomic and biochemical analyses of deep-sea epsilon-Proteobacteria have facilitated considerable progress in determining the possible sulfur metabolism pathways of the deep-sea chemoautotrophic epsilon-Proteobacteria (Table 1). The deep-sea epsilon-Proteobacteria characterized so far possess (1) a hydrogen-oxidizing sulfur respiration pathway using hydrogenase and polysulfide reductase (Psr). And the epsilon-Proteobacteria except for N. profundicola AmH, possess (2) a sulfur compounds-oxidizing oxygen/nitrate-respiration pathway using the Sox multienzyme system. Both of the Sox systems coupled with and without SoxCD have been found. The sox genes are not organized in a single region, but at least in two separated regions of the soxXYZAB and sox(CD)YX genes. One or more soxF gene was conserved in each sox gene cluster. Sulfurovum sp., Nitratiruptor sp., and S. autotrophica have several sqr genes. Two sets and one set of the sorAB genes coding SO were observed in Sulfurovum sp. and S. autotrophica, respectively. No gene for sulfur-oxidation pathways was found in N. profundicola. In addition to the sulfur metabolic pathways, many deep-sea epsilon-Proteobacteria are capable of chemoautotrophic growth without sulfur compounds using a third metabolic pathway: (3) a hydrogen-oxidizing oxygen/nitrate-reduction pathway (Figure 1). The bacterial members that can use sulfur compounds as both electron acceptors and donors are identified only in the phyla of epsilon-Proteobacteria and Aquificae, both of which predominantly inhabit in deep-sea hydrothermal environments. This may provide an important clue into how the versatile energy metabolic pathways are associated with the energetic advantages adapted to the dynamic and transient environmental conditions in the mixing zones of the hydrothermal vent environments.
Figure 1.
Energy metabolic pathways and habitat area of epsilon-Proteobacteria and gamma-Proteobacteria in deep-sea hydrothermal fields. From left to right on the diagram, the redox conditions shift from reductive to oxidative states. Broken lines in the rectangle indicate inclines in concentration of hydrogen gas, hydrogen sulfide, oxidized sulfur compounds, and oxygen gas, respectively. Two gray bars indicate habitat area of epsilon-Proteobacteria and gamma-Proteobacteria, respectively. Psr, polysulfide reductase; Sox, Sox multienzyme system; Dsr, dissimilatory sulfite reductase; Apr, adenosine 5′-phosphosulfate reductase; Sat, sulfate adenylyltransferase.
Sulfur Metabolism in Gamma-Proteobacteria in Deep-Sea Hydrothermal Fields
Gamma-proteobacterial symbionts with invertebrates have never been isolated from deep-sea hydrothermal vents. Furthermore only a couple of free-living sulfur-oxidizing gamma-Proteobacteria have been isolated from deep-sea hydrothermal fields (Kuever et al., 2002; Takai et al., 2006c, 2009). Unculturable gamma-proteobacterial sulfur-oxidizing endosymbionts are classified into three groups based on their host invertebrates: symbionts of (1) Bathymodiolus mussels and Calyptogena clams, (2) Alviniconcha gastropods, and (3) gastropods (e.g., Ifremeria) and tubeworms (Nakagawa and Takai, 2008). In recent years, two genome sequences of sulfur-oxidizing symbionts in Calyptogena have been published (Kuwahara et al., 2007; Newton et al., 2007; Table 1). Moreover, the metabolisms of symbiont in tubeworm were analyzed by metagenomic analysis (Robidart et al., 2008). In addition, a whole genome sequence of free-living sulfur-oxidizing gamma-Proteobacterium, Thiomicrospira crunogena XCL-2 was also published (Scott et al., 2006). Putative metabolic pathways in Beggiatoa sp. from marine sediments had also analyzed by using incomplete genome sequence (Mußmann et al., 2007). From these genome-based genetic characteristics, we can develop a preliminary outline of representative sulfur-related energy metabolisms in the deep-sea hydrothermal gamma-Proteobacteria, which possess two different sulfur-oxidization pathways: (1) the reverse sulfate reduction using Dsr, Apr, and Sat, and (2) the Sox multienzyme system without SoxCD (Figure 1). T. crunogena XCL-2, however, is an exception, in which no gene for the reverse sulfate reduction pathway was found. Moreover, this organism has the soxCD genes separated from other sox genes. In contrast, functional genes amplification studies suggested that hydrothermal gamma-Proteobacteria use the reverse sulfate reduction pathway, but not the Sox pathway (Hügler et al., 2010). These results indicate that either of the two distinct sulfur-oxidization pathways is dispensable. Both of these pathways require O2 as a terminal electron acceptor in most cases. This fact indicates that the relatively reductive environment (O2-depleted condition) is inhibitory for the growth of deep-sea chemoautotrophic gamma-Proteobacteria due to the limitation of primary electron acceptor despite an increasing abundance of electron-donors (reduced sulfur compounds). Thus, it is predicted that the metabolically habitable spaces of deep-sea chemoautotrophic gamma-Proteobacteria strictly requiring co-existence of reduced sulfur compounds and O2 are much more limited than those of deep-sea epsilon-Proteobacteria in the mixing zones of hydrothermal environments. Conversely, however, it seems likely that the genomic installation and the biochemical operation of two different sulfur-oxidizing pathways in the deep-sea chemoautotrophic gamma-Proteobacteria are kinetically advantageous if both the reduced sulfur compounds and O2 are steadily supplied into the habitats. Particularly for the gamma-proteobacterial sulfur-oxidizing symbionts, the host invertebrates would prepare the metabolically suitable habitats in and around the bodies in the mixing zones of deep-sea hydrothermal environments (Arp and Childress, 1983; Zal et al., 1998; Numoto et al., 2005). This may be one of the possible rationales for the domination of deep-sea gamma-proteobacterial sulfur-oxidizing chemoautotrophs in symbiotic communities in deep-sea hydrothermal environments. All deep-sea sulfur-oxidizing gamma-Proteobacteria possess one sqr gene in the genome, except for the metagenome of Riftia symbiont. Therefore, Sqr probably has an important role for the sulfur-oxidation in the organisms. It is interesting that genes for sulfur compound (thiosulfate and dimethyl sulfoxide) respiration were found in genome of Beggiatoa sp. (Mußmann et al., 2007). This is in accordance with previous results in Beggiatoa alba that can reduce stored elemental sulfur to overcome short-term anoxic conditions (Nelson and Castenholz, 1981; Schmidt et al., 1987).
Concluding Remarks
In this review, we described the sulfur-related energy metabolisms of epsilon- and gamma-Proteobacteria dominating the microbial communities in the global deep-sea hydrothermal environments. Most of the metabolic characteristics summarized in this article are based on the reconstruction of genetic components in the genomes determined for only a few representative species. Further genetic and biochemical variability associated with the sulfur-related energy metabolisms will be clarified in many species of deep-sea hydrothermal vent epsilon- and gamma-Proteobacteria in the future. Surely, the investigation of other inorganic sulfur metabolizing groups of microbial components such as sulfate-reducers (delta-Proteobacteria, Thermodesulfobacteria, and Deferribacteres) and sulfur-reducing thermophiles (Aquificae, Thermococcaceae, and Desulfurococcaceae) will be important for understanding the biogeochemical processes and energy- and elemental fluxes of sulfur compounds in the deep-sea hydrothermal environments (Reysenbach et al., 2000, 2002; Teske et al., 2002; Takai et al., 2003a). Recently, genetic information has been rapidly accumulating and has been providing the important genetic potential for understanding the complex sulfur-related metabolic networks in deep-sea hydrothermal vent ecosystems. In contrast, enzymatic and biochemical data are highly insufficient to clarify the in vivo and in situ kinetics, operation and control of metabolic pathways. These will be an important focus for future studies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Arp A. J., Childress J. J. (1983). Sulfide binding by the blood of the hydrothermal vent tube worm Riftia pachyptila. Science 219, 295–297 10.1126/science.219.4582.295 [DOI] [PubMed] [Google Scholar]
- Campbell B. J., Engel A. S., Porter M. L., Takai K. (2006). The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468 10.1038/nrmicro1414 [DOI] [PubMed] [Google Scholar]
- Campbell B. J., Jeanthon C., Kostka J. E., Luther G. W., III, Cary S. C. (2001). Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67, 4566–4572 10.1128/AEM.67.1.110-117.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. J., Smith J. L., Hanson T. E., Klotz M. G., Stein L. Y., Lee C. K., Wu D., Robinson J. M., Khouri H. M., Eisen J. A., Cary S. C. (2009). Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 5, e1000362. 10.1371/journal.pgen.1000362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L. K., Morgan-Kiss R. M., Hanson T. E. (2009). Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum. J. Bacteriol. 191, 1026–1034 10.1128/JB.01154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N., Bergin C., Lott C. (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740 10.1038/nrmicro1992 [DOI] [PubMed] [Google Scholar]
- Durand P., Reysenbach A.-L., Prieur D., Pace N. (1993). Isolation and characterization of Thiobacillus hydrothermalis sp. nov., a mesophilic obligately chemolithotrophic bacterium isolated from a deep-sea hydrothermal vent in Fiji Basin. Arch. Microbiol. 159, 39–44 10.1007/BF00244261 [DOI] [Google Scholar]
- Emerson D., Rentz J. A., Lilburn T. G., Davis R. E., Aldrich H., Chan C., Moyer C. L. (2007). A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS ONE 2, e667. 10.1371/journal.pone.0000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C. G. (1998). Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39, 235–289 10.1016/S0065-2911(08)60018-1 [DOI] [PubMed] [Google Scholar]
- Friedrich C. G., Bardischewsky F., Rother D., Quentmeier A., Fischer J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259 10.1016/j.mib.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Friedrich C. G., Quentmeier A., Bardischewsky F., Rother D., Orawski G., Hellwig P., Fischer J. (2008). “Redox control of chemotrophic sulfur oxidation of Paracoccus pantotrophus,” in Microbial Sulfur Metabolism, eds Dahl C., Friedrich C. G. (Heidelberg: Springer; ), 139–150 [Google Scholar]
- Friedrich C. G., Rother D., Bardischewsky F., Quentmeier A., Fischer J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67, 2873–2882 10.1128/AEM.67.7.3140-3148.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N. U., Dahl C. (2009). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200 10.1016/S0065-2911(08)00002-7 [DOI] [PubMed] [Google Scholar]
- Ghosh W., Dam B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33, 999–1043 10.1111/j.1574-6976.2009.00187.x [DOI] [PubMed] [Google Scholar]
- Gregersen L. H., Bryant D. A., Frigaard N. U. (2011). Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front. Microbiol. 2:116. 10.3389/fmicb.2011.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski J. J., Murray A. E., Campbell B. J., Kaplarevic M., Gao G. R., Lee C., Daniel R., Ghadiri A., Feldman R. A., Cary S. C. (2008). Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. Proc. Natl. Acad. Sci. U.S.A. 105, 17516–17521 10.1073/pnas.0802782105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedderich R., Klimmek O., Kroger A., Dirmeier R., Keller M., Stetter K. O. (1999). Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 22, 353–381 10.1111/j.1574-6976.1998.tb00376.x [DOI] [Google Scholar]
- Holkenbrink C., Ocon Barbas S., Mellerup A., Otaki H., Frigaard N. U. (2011). Sulfur globule oxidation in green sulfur bacteria is dependent on the dissimilatory sulfite reductase system. Microbiology 157, 1229–1239 10.1099/mic.0.044669-0 [DOI] [PubMed] [Google Scholar]
- Hügler M., Gartner A., Imhoff J. F. (2010). Functional genes as markers for sulfur cycling and CO2 fixation in microbial communities of hydrothermal vents of the Logatchev field. FEMS. Microbiol. Ecol. 73, 526–537 [DOI] [PubMed] [Google Scholar]
- Inagaki F., Takai K., Kobayashi H., Nealson K. H., Horikoshi K. (2003). Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing epsilon-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 53, 1801–1805 10.1099/ijs.0.02682-0 [DOI] [PubMed] [Google Scholar]
- Inagaki F., Takai K., Nealson K. H., Horikoshi K. (2004). Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the epsilon-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54, 1477–1482 10.1099/ijs.0.03042-0 [DOI] [PubMed] [Google Scholar]
- Jannasch H. W., Mottl M. J. (1985). Geomicrobiology of deep-sea hydrothermal vents. Science 229, 717–725 10.1126/science.229.4715.717 [DOI] [PubMed] [Google Scholar]
- Jeanthon C. (2000). Molecular ecology of hydrothermal vent microbial communities. Antonie Van Leeuwenhoek 77, 117–133 10.1023/A:1002463825025 [DOI] [PubMed] [Google Scholar]
- Kappler U., Dahl C. (2001). Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203, 1–9 10.1111/j.1574-6968.2001.tb10813.x [DOI] [PubMed] [Google Scholar]
- Kletzin A., Urich T., Muller F., Bandeiras T. M., Gomes C. M. (2004). Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36, 77–91 10.1023/B:JOBB.0000019600.36757.8c [DOI] [PubMed] [Google Scholar]
- Kuever J., Sievert S. M., Stevens H., Brinkhoff T., Muyzer G. (2002). Microorganisms of the oxidative and reductive part of the sulphur cycle at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Cah. Biol. Mar. 43, 413–416 [Google Scholar]
- Kusai K., Yamanaka T. (1973). The oxidation mechanisms of thiosulphate and sulphide in Chlorobium thiosulphatophilum: roles of cytochrome c-551 and cytochrome c-553. Biochim. Biophys. Acta 325, 304–314 10.1016/0005-2728(73)90106-0 [DOI] [PubMed] [Google Scholar]
- Kuwahara H., Yoshida T., Takaki Y., Shimamura S., Nishi S., Harada M., Matsuyama K., Takishita K., Kawato M., Uematsu K., Fujiwara Y., Sato T., Kato C., Kitagawa M., Kato I., Maruyama T. (2007). Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr. Biol. 17, 881–886 10.1016/j.cub.2007.04.039 [DOI] [PubMed] [Google Scholar]
- Kvenvolden K. A., Weliky K., Nelson H., Marais D. J. (1979). Submarine seep of carbon dioxide in Norton sound, Alaska. Science 205, 1264–1266 10.1126/science.205.4412.1264 [DOI] [PubMed] [Google Scholar]
- Marcia M., Ermler U., Peng G. H., Michel H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78, 1073–1083 10.1002/prot.22665 [DOI] [PubMed] [Google Scholar]
- McCollom T. M., Shock E. L. (1997). Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochim. Cosmochim. Acta. 61, 4375–4391 10.1016/S0016-7037(97)00241-X [DOI] [PubMed] [Google Scholar]
- Muyzer G., Stams A. J. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454 [DOI] [PubMed] [Google Scholar]
- Mußmann M., Hu F. Z., Richter M., De Beer D., Preisler A., Jorgensen B. B., Huntemann M., Glöckner F. O., Amann R., Koopman W. J., Lasken R. S., Janto B., Hogg J., Stoodley P., Boissy R., Ehrlich G. D. (2007). Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 5, e230. 10.1371/journal.pbio.0050230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Takai K. (2008). Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS. Microbiol. Ecol. 65, 1–14 10.1111/j.1574-6941.2008.00502.x [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takai K., Inagaki F., Hirayama H., Nunoura T., Horikoshi K., Sako Y. (2005). Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7, 1619–1632 10.1111/j.1462-2920.2005.00856.x [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takaki Y., Shimamura S., Reysenbach A. L., Takai K., Horikoshi K. (2007). Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. U.S.A. 104, 12146–12150 10.1073/pnas.0607703104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H., Inagaki F., Takai K. (2005). Hydrogen-driven subsurface lithoautotrophic microbial ecosystems (SLiMEs): do they exist and why should we care? Trends Microbiol. 13, 405–410 10.1016/j.tim.2005.07.010 [DOI] [PubMed] [Google Scholar]
- Nelson D. C., Castenholz R. W. (1981). Use of reduced sulfur-compounds by Beggiatoa sp. J. Bacteriol. 147, 140–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton I. L., Woyke T., Auchtung T. A., Dilly G. F., Dutton R. J., Fisher M. C., Fontanez K. M., Lau E., Stewart F. J., Richardson P. M., Barry K. W., Saunders E., Detter J. C., Wu D., Eisen J. A., Cavanaugh C. M. (2007). The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315, 998–1000 10.1126/science.1138438 [DOI] [PubMed] [Google Scholar]
- Numoto N., Nakagawa T., Kita A., Sasayama Y., Fukumori Y., Miki K. (2005). Structure of an extracellular giant hemoglobin of the gutless beard worm Oligobrachia mashikoi. Proc. Natl. Acad. Sci. U.S.A. 102, 14521–14526 10.1073/pnas.0501541102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-oka H., Blankenship R. E. (2004). “Green bacteria: secondary electron donor (cytochromes),” in Encyclopedia of Biological Chemistry, eds Lennarz W. J., Lane M. D. (Boston, MA: Elsevier; ), 321–324 [Google Scholar]
- Pfennig N., Biebel H. (1986). “The dissimilatory sulfate-reducing bacteria,” in The Prokaryotes: A Handbook on Habitats, Isolation and Identification of Bacteria, ed. Starr M. P. (Heidelberg: Springer; ), 928–940 [Google Scholar]
- Reinartz M., Tschape J., Bruser T., Truper H. G., Dahl C. (1998). Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 170, 59–68 10.1007/s002030050615 [DOI] [PubMed] [Google Scholar]
- Reysenbach A. L., Gotz D., Banta A., Jeanthon C., Fouquet Y. (2002). Expanding the distribution of the Aquificales to the deep-sea vents on Mid-Atlantic Ridge and Central Indian Ridge. Cah. Biol. Mar. 43, 425–428 [Google Scholar]
- Reysenbach A. L., Longnecker K., Kirshtein J. (2000). Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66, 3798–3806 10.1128/AEM.66.9.3798-3806.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robidart J. C., Bench S. R., Feldman R. A., Novoradovsky A., Podell S. B., Gaasterland T., Allen E. E., Felbeck H. (2008). Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ. Microbiol. 10, 727–737 10.1111/j.1462-2920.2007.01496.x [DOI] [PubMed] [Google Scholar]
- Schmidt T. M., Arieli B., Cohen Y., Padan E., Strohl W. R. (1987). Sulfur metabolism in Beggiatoa alba. J. Bacteriol. 169, 5466–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. M., Sievert S. M., Abril F. N., Ball L. A., Barrett C. J., Blake R. A., Boller A. J., Chain P. S., Clark J. A., Davis C. R., Detter C., Do K. F., Dobrinski K. P., Faza B. I., Fitzpatrick K. A., Freyermuth S. K., Harmer T. L., Hauser L. J., Hügler M., Kerfeld C. A., Klotz M. G., Kong W. W., Land M., Lapidus A., Larimer F. W., Longo D. L., Lucas S., Malfatti S. A., Massey S. E., Martin D. D., McCuddin Z., Meyer F., Moore J. L., Ocampo L. H., Jr., Paul J. H., Paulsen I. T., Reep D. K., Ren Q., Ross R. L., Sato P. Y., Thomas P., Tinkham L. E., Zeruth G. T. (2006). The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 4, e383. 10.1371/journal.pbio.0040383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahak Y., Hauska G. (2008). “Sulfide oxidation from cyanobacteria to humans: sulfide – quinone oxidoreductase (SQR),” in Sulfur Metabolism in Phototrophic Organisms, eds Hell R., Dahl C., Knaff D. B., Leustek T. (Dordrecht: Springer; ), 319–335 [Google Scholar]
- Sievert S. M., Scott K. M., Klotz M. G., Chain P. S. G., Hauser L. J., Hemp J., Hügler M., Land M., Lapidus A., Larimer F. W., Lucas S., Malfatti S. A., Meyer F., Paulsen I. T., Ren Q., Simon J., USF Genomics Class (2008). Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74, 1145–1156 10.1128/AEM.01844-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart F. J., Newton I. L., Cavanaugh C. M. (2005). Chemosynthetic endosymbioses: adaptations to oxic-anoxic interfaces. Trends Microbiol. 13, 439–448 10.1016/j.tim.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Sasaki T., Suzuki M., Nogi Y., Miwa T., Takai K., Nealson K. H., Horikoshi K. (2005). Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl. Environ. Microbiol. 71, 5440–5450 10.1128/AEM.71.4.1790-1797.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K., Campbell B. J., Cary S. C., Suzuki M., Oida H., Nunoura T., Hirayama H., Nakagawa S., Suzuki Y., Inagaki F., Horikoshi K. (2005). Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl. Environ. Microbiol. 71, 7310–7320 10.1128/AEM.71.11.7310-7320.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K., Gamo T., Tsunogai U., Nakayama N., Hirayama H., Nealson K. H., Horikoshi K. (2004). Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8, 269–282 10.1007/s00792-004-0386-3 [DOI] [PubMed] [Google Scholar]
- Takai K., Kobayashi H., Nealson K. H., Horikoshi K. (2003a). Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate- and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 53, 839–846 10.1099/ijs.0.02773-0 [DOI] [PubMed] [Google Scholar]
- Takai K., Inagaki F., Nakagawa S., Hirayama H., Nunoura T., Sako Y., Nealson K. H., Horikoshi K. (2003b). Isolation and phylogenetic diversity of members of previously uncultivated epsilon-proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218, 167–174 10.1016/S0378-1097(02)01147-3 [DOI] [PubMed] [Google Scholar]
- Takai K., Miyazaki M., Hirayama H., Nakagawa S., Querellou J., Godfroy A. (2009). Isolation and physiological characterization of two novel, piezophilic, thermophilic chemolithoautotrophs from a deep-sea hydrothermal vent chimney. Environ. Microbiol. 11, 1983–1997 10.1111/j.1462-2920.2009.01921.x [DOI] [PubMed] [Google Scholar]
- Takai K., Nakagawa S., Reysenbach A.-L., Hoek J. (2006a). Microbial ecology of mid-ocean ridges and back-arc basins. Geophys. Monogr. Ser. 166, 185–213 [Google Scholar]
- Takai K., Nakamura K., Suzuki K., Inagaki F., Nealson K. H., Kumagai H. (2006b). Ultramafics-Hydrothermalism-Hydrogenesis-HyperSLiME (UltraH3) linkage: a key insight into early microbial ecosystem in the Archean deep-sea hydrothermal systems. Paleontol. Res. 10, 269–282 10.2517/prpsj.10.269 [DOI] [Google Scholar]
- Takai K., Miyazaki M., Nunoura T., Hirayama H., Oida H., Furushima Y., Yamamoto H., Horikoshi K. (2006c). Sulfurivirga caldicuralii gen. nov., sp. nov., a novel microaerobic, thermophilic, thiosulfate-oxidizing chemolithoautotroph, isolated from a shallow marine hydrothermal system occurring in a coral reef, Japan. Int. J. Syst. Evol. Microbiol. 56, 1921–1929 10.1099/ijs.0.64255-0 [DOI] [PubMed] [Google Scholar]
- Teske A., Hinrichs K. U., Edgcomb V., de Vera Gomez A., Kysela D., Sylva S. P., Sogin M. L., Jannasch H. W. (2002). Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68, 1994–2007 10.1128/AEM.68.4.1994-2007.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakawa H., Dubilier N., Fujiwara Y., Cunningham D. E., Kojima S., Stahl D. A. (2005). Hydrothermal vent gastropods from the same family (Provannidae) harbour epsilon- and gamma-proteobacterial endosymbionts. Environ. Microbiol. 7, 750–754 10.1111/j.1462-2920.2005.00753.x [DOI] [PubMed] [Google Scholar]
- Wolin M. J., Wolin E. A., Jacobs N. J. (1961). Cytochrome-producing anaerobic Vibrio succinogenes, sp. n. J. Bacteriol. 81, 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Nakagawa S., Shimamura S., Takai K., Horikoshi K. (2010). Molecular characterization of inorganic sulfur-compound metabolism in the deep-sea epsilonproteobacterium Sulfurovum sp. NBC37-1. Environ. Microbiol. 12, 1144–1153 10.1111/j.1462-2920.2010.02155.x [DOI] [PubMed] [Google Scholar]
- Zal F., Leize E., Lallier F. H., Toulmond A., Van Dorsselaer A., Childress J. J. (1998). S-Sulfohemoglobin and disulfide exchange: the mechanisms of sulfide binding by Riftia pachyptila hemoglobins. Proc. Natl. Acad. Sci. U.S.A. 95, 8997–9002 10.1073/pnas.95.15.8997 [DOI] [PMC free article] [PubMed] [Google Scholar]