Abstract
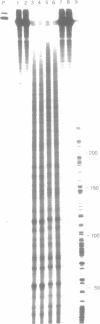
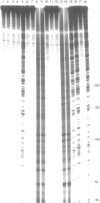
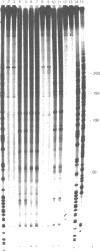
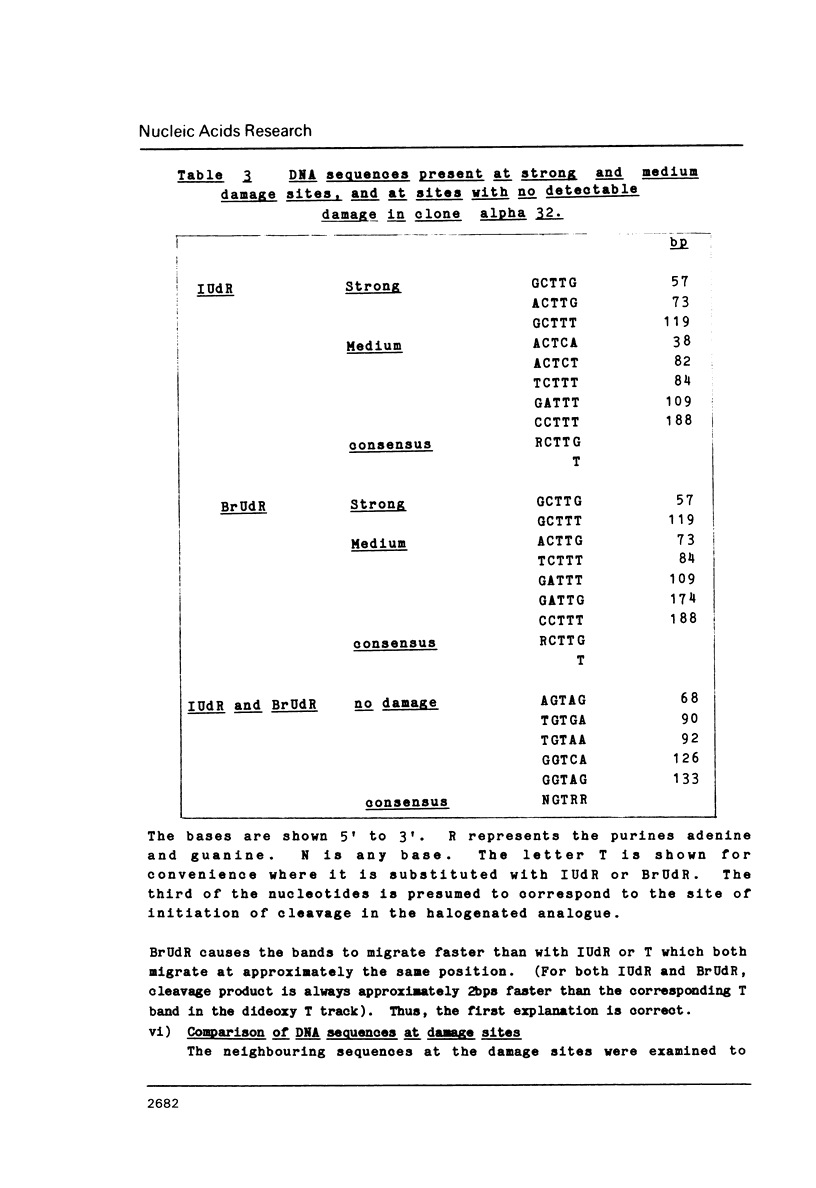
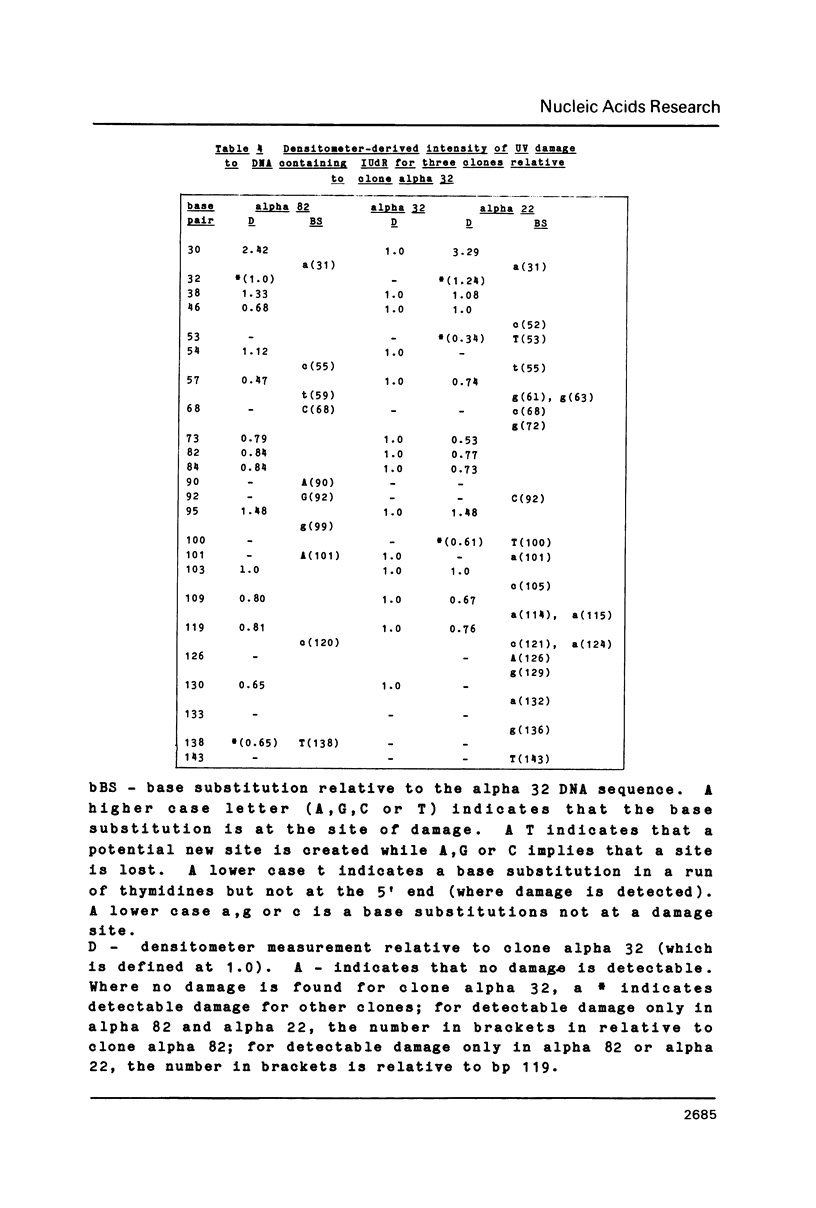
The sequence selectivity of 300 nm ultraviolet light damage to DNA containing bromodeoxyuridine or iododeoxyuridine was examined on DNA sequencing gels. This was accomplished using a system where an M13 template was employed to direct synthesis of DNA in which thymidine was fully substituted with bromodeoxyuridine or iododeoxyuridine. The sites of damage corresponded to the positions of analogue incorporation. The extent of damage varied considerably at different sites of cleavage and ranged from the undetectable to over fifteen times the limit of detection (as assessed by laser densitometer scans). Strong damage sites had the "consensus" sequence CTT while sites of no detectable damage had the "consensus" sequence GTR. Bromodeoxyuridine and iododeoxyuridine had the same sites of damage although the extent of damage varied at different sites and bromodeoxyuridine damage was slightly greater than iododeoxyuridine. DNA containing thymidine was not damaged to any detectable level in this system with 300 nm ultraviolet light. The use of three closely related DNA sequences as targets for damage confirmed that (1) the sites of analogue incorporation are the cause of ultraviolet damage; and (2) that the neighbouring DNA sequence is an important parameter in determining the extent of damage. It is proposed that the microstructure of DNA--in particular the distance between the 5-carbon of the pyrimidine base (which is attached to the halogen) and hydrogen on the 2' carbon of the 5'-deoxyribose--ultimately determines the degree of cleavage with large distances giving a small degree of damage and smaller distances a large degree of damage.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Darling S. M., Crampton J. M., Williamson R. Organization of a family of highly repetitive sequences within the human genome. J Mol Biol. 1982 Jan 5;154(1):51–63. doi: 10.1016/0022-2836(82)90416-8. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner W. D., Grunberg S. M., Haseltine W. A. Enzyme action at 3' termini of ionizing radiation-induced DNA strand breaks. J Biol Chem. 1983 Dec 25;258(24):15198–15205. [PubMed] [Google Scholar]
- Hutchinson F. The lesions produced by ultraviolet light in DNA containing 5-bromouracil. Q Rev Biophys. 1973 May;6(2):201–246. doi: 10.1017/s0033583500001141. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma. 1978 Mar 22;66(1):23–32. doi: 10.1007/BF00285813. [DOI] [PubMed] [Google Scholar]
- Mirabelli C. K., Ting A., Huang C. H., Mong S., Crooke S. T. Bleomycin and talisomycin sequence-specific strand scission of DNA: a mechanism of double-strand cleavage. Cancer Res. 1982 Jul;42(7):2779–2785. [PubMed] [Google Scholar]
- Murray V., Martin R. F. Comparison of the sequence specificity of bleomycin cleavage in two slightly different DNA sequences. Nucleic Acids Res. 1985 Mar 11;13(5):1467–1481. doi: 10.1093/nar/13.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray V., Martin R. F. Nucleotide sequences of human alpha-DNA repeats. Gene. 1987;57(2-3):255–259. doi: 10.1016/0378-1119(87)90129-6. [DOI] [PubMed] [Google Scholar]
- Murray V., Martin R. F. Sequence specificity of 125I-labelled Hoechst 33258 damage in six closely related DNA sequences. J Mol Biol. 1988 Sep 5;203(1):63–73. doi: 10.1016/0022-2836(88)90091-5. [DOI] [PubMed] [Google Scholar]
- Murray V., Tan L., Matthews J., Martin R. F. The sequence specificity of bleomycin damage in three cloned DNA sequences that differ by a small number of base substitutions. J Biol Chem. 1988 Sep 15;263(26):12854–12859. [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D. The effect of the base sequence on the fine structure of the DNA double helix. Prog Biophys Mol Biol. 1986;47(3):159–195. doi: 10.1016/0079-6107(86)90013-1. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Manuelidis L. Sequence definition and organization of a human repeated DNA. J Mol Biol. 1980 Sep 25;142(3):363–386. doi: 10.1016/0022-2836(80)90277-6. [DOI] [PubMed] [Google Scholar]